Abstract
Gamma-tocotrienol (GT3), a naturally occurring vitamin E isomer, a promising radioprotector, has been shown to protect mice against radiation-induced hematopoietic and gastrointestinal injuries. We analyzed changes in protein expression profiles of spleen tissue after GT3 treatment in mice exposed to gamma radiation to gain insights into the molecular mechanism of radioprotective efficacy. Male CD2F1 mice, 12-to-14 weeks old, were treated with either vehicle or GT3 at 24 h prior to 7 Gy total-body irradiation. Nonirradiated vehicle, nonirradiated GT3 and age-matched naïve animals were used as controls. Blood and tissues were harvested on days 0, 1, 2, 4, 7, 10 and 14 postirradiation. High-resolution mass-spectrometry-based radioproteomics was used to identify differentially expressed proteins in spleen tissue with or without drug treatment. Subsequent bioinformatic analyses helped delineate molecular markers of biological pathways and networks regulating the cellular radiation responses in spleen. Our results show a robust alteration in spleen proteomic profiles including upregulation of the Wnt signaling pathway and actin-cytoskeleton linked proteins in mediating the radiation injury response in spleen. Furthermore, we show that 24 h pretreatment with GT3 attenuates radiation-induced hematopoietic injury in the spleen by modulating various cell signaling proteins. Taken together, our results show that the radioprotective effects of GT3 are mediated, via alleviation of radiation-induced alterations in biochemical pathways, with wide implications on overall hematopoietic injury.
INTRODUCTION
Exposure to ionizing radiation from radiotherapy or a radiological/nuclear accident is a major concern because of its resultant adverse effects on tissue structure and function (1, 2). Exposure to radiation induces a complex network of cellular and molecular responses resulting in normal tissue damage (3, 4). Damage due to radiation may lead to multiorgan failure that includes hematopoietic, gastrointestinal, cardiovascular and neurological damage collectively known as acute radiation syndrome (ARS). Hematopoietic tissues are the most sensitive to radiation; suppression can lead to deficiency in innate and adaptive immunity and mortality with depletion of bone marrow hematopoietic stem and progenitor cells and damage to hematopoietic organs, including spleen (5, 6). In several published studies, reduced spleen colony forming units (CFUs) have been reported in mice receiving total-body irradiation (TBI) (7, 8). Therefore, most radiation countermeasures are designed to protect bone marrow stem and progenitor cells, which restore peripheral blood cells, as well as providing protection to the spleen. Although there are various countermeasures for ARS, which are in different stages of development (9–13), there are only three medical countermeasures approved by the U.S. Food and Drug Administration (FDA), Neupogen®, Neulasta® and Leukine®, which can be administered after acute radiation exposure to restore peripheral blood cells.
Previously, we reported that treatment with a natural isomer of vitamin E, gamma-tocotrienol (GT3), protects mice from lethal doses of radiation by preventing radiation-induced peripheral blood cytopenia (10), restoring bone marrow progenitor cells (14), inducing granulocyte colony stimulating factor (G-CSF) (15) and causing a recovery of spleen CFUs (7). More recently, we have shown that administration of GT3 24 h prior to exposure can improve radiation-induced neutropenia and thrombocytopenia in irradiated nonhuman primates (16). However, signaling pathways that trigger pathophysiological events after TBI and the molecular mechanisms by which GT3 prevents damage to the spleen to protect the hematopoietic system and mouse survival are not fully understood. To fully develop GT3 as a prophylactic radiation countermeasure under the FDA, the animal rule (17) mandates the elucidation of the mechanism of action of the drug being evaluated. Since different tissues/organs respond to radiation damage by altering their protein expression level and posttranslational modification status, proteomic profiling can be used to delineate radiation-induced protein biomarkers and biological pathways in serum and tissues (18). Therefore, the goal this study was to interrogate global changes in the protein expression profiles in irradiated mice with or without prior administration of the novel radio protectant GT3, then correlate the radioprotective efficacy with pathway perturbations.
There is an increasing interest within the radiobiology community in the use of a high-throughput proteomics approach as a research tool (19). Comprehensive proteomic analysis enables the identification of previously unanticipated biological pathways involved in the radiation injury response (20–26). In previously published studies, we have used human cell lines as model systems to study the effects of ionizing radiation on differential protein expression (19, 27, 28). Since radiation-induced cellular responses are complex, we have more recently used complementary genomics and proteomic approaches to understand the signaling targets of radiation damage through biochemical pathway network analysis. Furthermore, we recently reported that GT3 treatment reverses the expression of several miRNAs, which are involved in postirradiation hematopoiesis in irradiated mouse spleen (29). Network pathway analysis implicated ERK/P38MAPK as the target signaling pathway for the recovery of radiation-induced damage in spleen by GT3 (29). Since in vitro cellular studies alone may not be sufficient to explain the proteomic alterations in complex tissues, using a global high-throughput proteomics approach offers several advantages for advancing the field of radiation biology (20, 22, 24). Radioproteomics is a powerful tool for comparing irradiated and normal tissues to identify organ-specific protein markers of radiation-induced tissue injury (20, 22, 24, 30, 31). There are previously published studies on radioproteomics in various mouse tissues/organs, including brain, lung, spleen, kidney and intestine (20, 32, 33). The authors identified two candidate biomarkers of radiation exposure, transaldolase 1 (TA1) and phosphoglycerate kinase 1(PGK1), in the mouse brain and intestine, respectively, by comparing proteomic profiling and validation by immunoblotting (32). In their recently published work, Srivastava et al. demonstrated that radioproteomics can be used to develop drugs for radiation injury using protein signatures from primary CD34+ progenitor cells derived from healthy donors that were subsequently irradiated (34). The authors also showed that the survival of mice receiving TBI after pretreatment with alpha-tocopherol succinate, a synthetic derivative of alpha-tocopherol, could be linked to activation of epigenetic mechanisms relevant to radioprotection or activation of DNA repair and inflammation leading to cell survival (34).
We employed radioproteomics as a discovery tool to identify differentially expressed proteins in spleen tissue with or without drug treatment. We used nano-reverse phase liquid chromatography (RPLC) for resolving peptides in conjunction with high-resolution mass spectrometry for peptide detection and protein identification based on MS/MS spectra. Subsequent bioinformatics analyses allowed us to identify differentially expressed proteins, and thus, pathways that mediate radiation-induced injury response in spleen. Since our overall goal is to understand the radioprotective mechanism of GT3, we mined our data to look for pathways dysregulated by radiation that were attenuated in mice who received GT3 treatment prior to exposure. We found that the Wnt signaling pathway is a major player of radiation injury response in the spleen and that this pathway response is alleviated by GT3. To our knowledge, this is the first reported study to elucidate the molecular pathways involved in the radiation-induced damage in spleen and its recovery by GT3 using a global proteomics approach.
MATERIALS AND METHODS
Animals
CD2F1 male mice (10, 29), 8–10 weeks old (Envigo RMS Inc., Indianapolis, IN) were housed in an air-conditioned facility at the Armed Forces Radiobiology Research Institute (AFRRI, Bethesda, MD), which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). All mice were kept in rooms with a 12 h light-dark schedule at 21 ± 2°C with 10–15 hourly cycles of fresh air and a relative humidity of 50 ± 10%. Mice were held in quarantine for two weeks and were used after microbiology, serology and histopathology examination of representative samples ensured the absence of Pseudomonas aeruginosa and common murine diseases. Mice were given certified rodent rations of Harlan Teklad Rodent Diet no. 8604 (Madison, WI) and acidified water (with HCl, pH 2.5–3.0) ad libitum. Animal procedures were performed following a protocol approved by the Armed Forces Radiobiology Research Institute Institutional Animal Care and Use Committee (IACUC). Research was conducted according to the Guide for the Care and Use of Laboratory Animals, prepared by the Institute of Laboratory Animal Resources, the National Research Council, U.S. National Academy of Sciences (29).
Drug Preparation and Administration
GT3 was formulated in 5% Tween™ 80 (Yasoo Health Inc., Johnson City, TN) and a single subcutaneous (SC) injection was administered 24 h prior to irradiation at the nape of the neck at a dose of 100 mg/kg (for 25 g average weight of a mouse) in a volume of 0.1 ml. The vehicle was administered SC as an equivalent dose volume of 5% Tween 80 (0.1 ml) in saline. No inflammation or local reactions were observed at the injection site (35).
Irradiation
Mice received TBI bilaterally in the AFRRI 60Co irradiation facility in well-ventilated Plexiglas® boxes (8 mice per box) at a dose rate of ~0.6 Gy/min to total midline doses of 7 Gy, as described below. An alanine/ESR (electron spin resonance) dosimetry system (ASTM Standard E 1607; American Society for Testing and Material, West Conshohocken, PA) was used to measure dose rates (to water) in the cores of acrylic mouse phantoms. Phantoms were 3 inches long and 1 inch in diameter and were located in 50% of the compartments of the exposure rack. The ESR signals were measured with a calibration curve based on standard calibration dosimeters provided by the National Institute of Standard and Technology (NIST, Gaithersburg, MD). The accuracy of the calibration curve was verified by intercomparison with the National Physical Laboratory (NPL, London, UK). The only corrections applied to the dose rates in phantoms were for the decay of the 60Co source and for a small difference in mass energy-absorption coefficients for water and soft tissue. The radiation field was uniform within ±2%. After irradiation, mice were returned to their original cages with access to food and water ad libitum (10, 14). All exposures were performed in the morning to minimize the diurnal effect.
Blood and Tissue Harvest after Irradiation
Six mice per group were used: nonirradiated vehicle control (NRV); nonirradiated GT3 control (NRG); vehicle with 7 Gy irradiation; and GT3 treatment with 7 Gy irradiation. Nonirradiated controls (NRV and NRG) were also placed in the Plexiglas boxes for the same amount of time as the irradiated mice, but were not exposed to radiation. This is to mimic the stress similar to that which the irradiated animals would experience during irradiation. Age-matched healthy animals used as naïve controls were not placed in the box nor receive any kind of treatment or irradiation. Blood and tissues were harvested on days 0, 1, 2, 4, 7, 10 and 14 after 7 Gy gamma irradiation. Blood and tissue from nonirradiated controls (NRV and NRG) were collected at each time point. Whole blood was collected through the inferior vena cava (IVC) in accordance with the approved IACUC protocol. The animals were deeply anesthetized with 5% isoflurane (Abbott Laboratories, Chicago, IL) in a rodent anesthesia machine; the tail was pinched to check for reflexive movement, indicative of insufficient anesthesia. If there was no response, the animal was moved to a station with an individual nose cone for continued anesthesia. An incision was made on the right side of the animal, closest to the IVC, the vein exposed, and blood was drawn with a 23g needle. Blood was collected in serum separator tubes and allowed to clot for 30 min at room temperature. After centrifugation at 1,500g for 10 min, serum was transferred to a microcentrifuge tube and stored at −80°C until use. After blood collection under isoflurane anesthesia, mice were euthanized by cervical dislocation and spleens were harvested at each time point and immediately snap frozen in liquid nitrogen and stored at −80°C until use.
Protein Denaturation and Digestion
Spleens (n = 3 per group), which were harvested upon euthanasia on days 1, 7 and 14 post-TBI from vehicle- and GT3-treated mice (six groups in total), were used in this study. Spleens from naive animals (healthy age-matched controls receiving no treatment), NRV and NRG (three groups) were collected only on day 1 postirradiation. A total of 27 samples (9 groups × 3 biological replicates) were used in the experiment. Samples were prepared for digestion using the filter-assisted sample preparation (FASP) method (36). Briefly, the samples were suspended in 8 M urea, 50 mM Tris-HCl, pH 7.6, 3 mM DTT, sonicated briefly and incubated in a ThermoMixer® at 40°C, 1,000 rpm for 20 min. Samples were centrifuged to clarify and the supernatant was transferred to a 30 k Amicon® MWCO device (Millipore, Billerica, MA) and centrifuged at 13,000 rpm for 30 min. The remaining sample was buffer exchanged with 8 M urea, 100 mM Tris-HCl, pH 7.6, then alkylated with 15 mM iodoacetamide. The urea concentration was reduced to 2 M. A part of the samples, (approximately 200 μg), was digested using 5 μg of trypsin, overnight, at 37°C on the ThermoMixer at 1,000 rpm. Digested peptides were collected by centrifugation.
Peptide Desalting
Approximately 20 μg of the digested peptides were desalted using C18 stop-and-go extraction (STAGE) tips. Briefly, for each sample a C18 STAGE tip was activated with methanol, then conditioned with 60% acetonitrile, 0.5% acetic acid followed by 5% acetonitrile and 0.5% acetic acid. Samples were loaded onto the tips and desalted with 0.5% acetic acid. Peptides were eluted with 60% acetonitrile, 0.5% acetic acid and lyophilized in a Savant Speed Vac® (Thermo Fisher Scientific™ Inc., Waltham, MA) to dryness, approximately 2 h.
Liquid Chromatography-Tandem Mass Spectrometry
Each digestion mixture was analyzed by UPLC-MS/MS. Liquid chromatography (LC) was performed on an Easy-nLC 1000 UPLC system (Thermo Fisher Scientific). Mobile phase A was 97.5% MilliQ® water (Millipore), 2% acetonitrile, 0.5% acetic acid. Mobile phase B was 99.5% acetonitrile, 0.5% acetic acid. The 240-min LC gradient ran from 0% B to 35% B over 200 min, then to 80% B for the remaining time. Samples were loaded directly to the column. The column was 20 cm × 75 μm I.D. and packed with 3-μm C18 media (Magic; Michrom Bioresources/Bruker, Auburn, CA). The LC was interfaced to a Quadrupole-Orbitrap™ Mass Spectrometer (Q-Exactive™; Thermo Fisher Scientific) via nano-electrospray ionization. An electrospray voltage of 2.2 kV was applied. The mass spectrometer was programmed to acquire by data-dependent acquisition and tandem mass spectra from the top 20 ions in the full scan from 350–1,600 m/z. Dynamic exclusion was set to 60 s and singly-charged ions were excluded. Isolation width was set to 1.6 Da and full MS resolution to 70,000 and MS/MS resolution to 17,500. Normalized collision energy was set to 25, automatic gain control to 1e6, maximum fill MS to 20 ms, maximum fill MS/MS to 60 ms and the underfill ratio to 0.1%.
Data Processing and Library Searching
Mass spectrometer RAW data files were converted to Mascot generic format (MGF) format using msConvert. Detailed search parameters are printed in the search output XML files at Proteome Cluster [https://www.proteomecluster.com/projects/8#searches (account required)]. Briefly, all searches required 20 ppm precursor mass tolerance, 0.05 Da fragment mass tolerance, strict tryptic cleavage, 0 or 1 missed cleavages, fixed modification of cysteine alkylation, variable modification of methionine oxidation and expectation value scores of 0.01 or lower. MGF files were searched using the mouse sequence library from Ensembl (https://bit.ly/2u9hMaa). MGF files were searched using X!Tandem (37) employing both the native (38) and k-score(39) scoring algorithms and by open mass spectrometry search algorithm (40). MGF files were searched using X!Hunter (41) against the GPM (42) mouse spectral library (https://bit.ly/2L0YRrC). All searches were performed on Amazon Web Services-based cluster compute instances using the Proteome Cluster interface. XML output files were parsed and nonredundant protein sets determined using MassSieve (43). Proteins were required to have one or more unique peptides across the analyzed samples with E-value scores of 0.001 or less.
Statistical and Bioinformatics Analyses of Proteomics Data
For the proteomic data, the protein intensity values were log2 transformed, centered and scaled prior to analysis by Student’s t test, fold change and partial least squares discriminant analysis (Unscrambler® X; CAMO Software, Oslo, Norway). Prior to calculating the significance and fold expression, the log2 intensity values were centered (mean equals 0) and scaled (standard deviation equals 1). The P values were corrected for multiple testing by the Benjamini-Hochberg method. Proteins were visualized using hierarchical clustering explorer version 3.0. The volcano plots were created in R version 3.3.0, and indicated in red are the proteins with a P value <0.05 and a fold change >2. All other data sets are expressed as mean ± standard error of mean (SEM) (44). All experiments were performed independently at least twice and biological replicates (n = 3) were used for each data set. GraphPad Prism version 6.07 (LaJolla, CA) for windows was used for data analysis. Statistical significance was considered at P < 0.05. The differences among days, regimens (vehicle and GT3) were analyzed by performing a multiple comparisons test using Tukey’s honest significant difference test to find means that are significantly different from each other.
Ingenuity® Pathway Analysis (IPA)
We used IPA (Ingenuity Systems/QIAGEN, Redwood City, CA) to determine the biological and functional relevance of significantly dysregulated proteins by integrating them into pathways and networks that can be correlated for radioprotective efficacy of GT3. Statistically significant proteins that are altered by GT3 treatment compared to vehicle were uploaded in the IPA software that evaluates the biological functions pathways and significance value.
Splenic Total RNA Extraction
Spleens (n = 3 per group) harvested from euthanizing mice on days 1, 4, 7 and 14 after 7 Gy TBI from vehicle- and GT3-treated mice (eight groups) were used in this study. Spleens from NRV and NRG (two groups) were collected only on day 1 postirradiation. Approximately 50 mg of the frozen tissue was homogenized by brief sonication (5 pulses/s) in ice and total RNA was extracted using mirVana™ total RNA isolation kits (Life Technologies, Frederick, MD) following the manufacturer’s protocol. RNA concentrations were determined by measuring absorption at 260 and 280 nm on a NanoDrop spectrophotometer ND-1000 (Thermo Fisher Scientific).
Real-Time Quantitative PCR
RNA was reverse transcribed into cDNA (RT2 First Strand Kit, SA Biosciences, Frederick, MD) and the cDNA used for quantitative PCR analysis. Wnt-signaling-regulated genes were analyzed by quantitative real-time RT-PCR using the SA Biosciences RT2 Profiler™ PCR Array and RT2 SYBR® Green Master Mixes. Analyses were performed on an iCycler iQ™ Real-Time PCR Detection System (Bio-Rad® Laboratories Inc., Hercules, CA). Delta cycle threshold (DCt) values were calculated by subtracting average Ct values of housekeeping genes [glucuronidase, beta (Gusb); heat shock protein 90 alpha (cytosolic), class B member 1 (Hsp90ab1); glyceraldehyde-3-phosphate dehydrogenase (Gapdh); actin, beta (Actb); and hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1)]. Data were evaluated by the ΔΔCt method against nonirradiated vehicle control (n = 3) and fold changes in gene expression were calculated using PCR Array Pathways (SABiosciences; http://gncpro.sabiosciences.com/gncpro/gncpro.php?toggler=startpathwaysabio) after normalizing data to the array’s housekeeping gene pool.
Mouse Serum and Erythropoietin (EPO) Enzyme-Linked Immunosorbent Assay (ELISA)
Sandwich ELISAs for mouse EPO were performed using commercially available kits (R&D Systems™, Minneapolis, MN) following assay instructions provided by the manufacturer. For EPO ELISA, data sets are expressed as mean ± SEM (44). All experiments were performed independently at least twice and biological replicates (n = 6) were used for each data set. GraphPad Prism version 6.07 for windows was used for data analysis. Significant difference in protein expression in the treatment groups were determined by analysis of variant (ANOVA) with a threshold P value <0.05.
Western Blot Analysis
Proteins were extracted from snap-frozen spleens (n = 3) in RIPA buffer (Santa Cruz Biotechnology, Santa Cruz, CA) in the presence of phosphatase and protease inhibitors. To adjust the final concentration of protein, BCA method was used to calculate the total protein. Samples with 1× final Laemmli buffer were subjected to SDS-PAGE electrophoresis and transferred to PVDF membranes by semi-dry transfer. For immunodetection, membranes were blocked for 2 h at room temperature in 4% milk and incubated overnight at 4°C with primary antibodies Adducin-α, Frg1 (Santa Cruz Biotechnology) and β-Catenin (Abcam, Cambridge, UK). The membranes were then washed with 1× TBST four times and incubated with horseradish peroxidase-conjugated secondary antibodies for 2 h. After washing four times with 1× Tris-buffered saline and Tween® 20, enhanced chemiluminescence Western blotting kit (Thermo Fisher Scientific) was used to detect the specific protein according to the instructions from the manufacturer. Beta-actin (Santa Cruz Biotechnology) was used as an internal control. ImageJ software (National Institutes of Health, Bethesda, MD) was used to measure the intensities of specific bands corresponding to the proteins of interest.
RESULTS
Radiation Exposure Induces Robust Alterations in Protein Expression Response
Initially, to study radiation-induced changes in protein expression, groups of male CD2F1 mice were exposed to 7 Gy TBI. Spleen tissue collected on days 1, 7 and 14 were subjected to mass-spectrometry-based proteomics analyses (see Materials and Methods). The resultant data matrix yielded relative abundances for all identified proteins across experimental groups. There were a total of 27 spleen samples divided into nine groups (three biological replicates per group). A total of 15,268 proteins were identified from all 27 samples. The abundance values for each protein resulting from matching fragment ion intensity values for all peptide identification scans mapping to that protein were log2 transformed prior to performing statistical comparisons across multiple groups. We used a multivariate analysis approach to identify discriminating dysregulated proteins for each comparative group comparison using partial least squares discriminant analysis (Unscrambler X). Initially, we compared NRV to those that were vehicle-treated with tissue collected on days 1, 7 or 14 post-TBI (Rd1V, Rd7V and Rd14V, respectively).
As shown in Fig. 1, a partial least squares discriminant analysis resulted in clear separation of NRV and irradiated groups based on time after irradiation. In Fig. 1, factor 1 (intergroup separation) is plotted against factor 2 (intragroup variability) and together both factors account for 99% of the variability in the data set. Employing Martens’ uncertainty test, we were able to identify proteins that were differentially expressed in irradiated mice on days 1, 7 and 14 post-TBI compared to NRV (day 1) (Table 1). These data show that radiation induced robust changes in proteomic profiles resulting in clear group separation not only between NRV and irradiated groups but also among the irradiated groups based on time after exposure (days 1, 7 and 14). To visualize candidate protein markers of radiation response, we performed hierarchical clustering (Explorer version 3.0; University of Maryland, College Park, MD) to generate a heat map of the significantly differentiating proteins among NRV, irradiated mice at days 1 (Rd1V), 7 (Rd7V) and 14 (Rd14V) samples (Fig. 2).
FIG. 1.
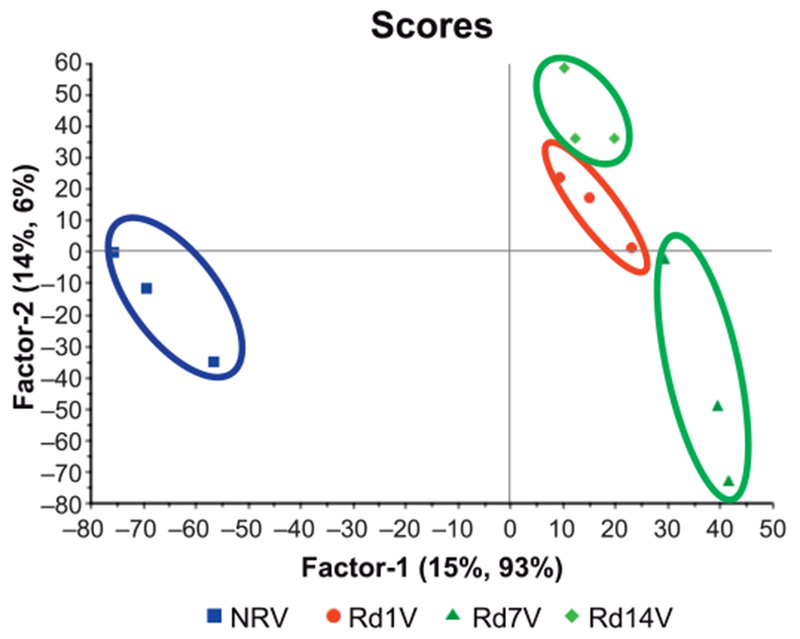
Multivariate analysis results in unambiguous separation of irradiated groups of mice (n = 3) from controls: The nonlinear iterative partial least squares (NIPALS) plot shows separation of the spleen proteome of nonirradiated vehicle control mice (NRV) from that of irradiated mice on days 1, 7, and 14 (Rd1V, Rd7V and Rd14V).
TABLE 1.
Comparative Proteomic Analysis of Control Mice (NRV) and Irradiated Mice (RDV) Reveals Differentially Expressed Proteins
| Gene symbol | Description | Relative fold change (RDV/NRV) | P value | FDR adjusted P value |
|---|---|---|---|---|
| Upregulated | ||||
| Gucy1a2 | Guanylate cyclase 1, soluble, alpha 2 | 68.71 | 2.34E–08 | 3.58E–05 |
| Tmem205 | Transmembrane protein 205 | 54.70 | 3.94E–08 | 4.38E–05 |
| Gatm | Glycine amidinotransferase (l-arginine:glycine amidinotransferase) | 117.8 | 9.43E–07 | 9.62E–04 |
| Col5a2 | Collagen, type V, alpha 2 | 8.28 | 6.08E–06 | 3.10E–03 |
| Vwa5a | von Willebrand factor A domain containing 5A | 5.40 | 1.46E–05 | 7.15E–03 |
| Aldh3b2 | Aldehyde dehydrogenase 3 family, member B2 | 82.73 | 2.04E–05 | 9.61E–03 |
| Aldh3b1 | Aldehyde dehydrogenase 3 family, member B1 | 83.07 | 2.19E–05 | 8.13E–03 |
| Vcam1 | Vascular cell adhesion molecule 1 | 3.66 | 3.88E–05 | 1.16E–02 |
| Ctnna1 | Catenin (cadherin associated protein), alpha 1 | 5.01 | 7.09E–05 | 1.97E–02 |
| Hmgcs2 | 3-Hydroxy-3-methylglutaryl-coenzyme A synthase 2 | 98.45 | 7.55E–05 | 2.05E–02 |
| Pcca | Propionyl-coenzyme A carboxylase, alpha polypeptide | 94.82 | 9.75E–05 | 2.54E–02 |
| Ctnna2 | Catenin (cadherin associated protein), alpha2 | 86.78 | 1.33E–04 | 3.26E–02 |
| Anpep | Alanyl (membrane) aminopeptidase | 2.06 | 1.47E–04 | 3.28E–02 |
| Ctnnb1 | Catenin (cadherin associated protein), beta 1 | 4.41 | 1.55E–04 | 3.39E–02 |
| Lama5 | Laminin, alpha 5 | 2.17 | 2.15E–04 | 4.31E–02 |
| Ctnnd1 | Catenin (cadherin associated protein), delta 1 | 4.36 | 3.33E–04 | 4.38E–02 |
| Downregulated | ||||
| Gimap7 | GTPase, IMAP family member 7 | 0.016 | 1.29E–25 | 1.58E–21 |
| Ppp6r3 | Protein phosphatase 6, regulatory subunit 3 | 0.016 | 9.29E–25 | 5.68E–21 |
| Mcpt8 | Mast cell protease 8 | 0.015 | 1.20E–23 | 3.67E–20 |
| Grap2 | GRB2-related adaptor protein 2 | 0.016 | 2.49E–23 | 6.10E–20 |
| Pcsk4 | Proprotein convertase subtilisin/kexin type 4 | 0.034 | 7.31E–15 | 1.49E–11 |
| Coro1a | Coronin, actin-binding protein 1A | 0.011 | 1.67E–12 | 2.92E–09 |
| Sh3kbp1 | SH3-domain kinase binding protein 1 | 0.019 | 2.37E–06 | 1.52E–03 |
| Fam107b | Family with sequence similarity 107, member B | 0.023 | 2.15E–05 | 8.24E–03 |
| Add1 | Adducin 1 (alpha) | 0.114 | 3.28E–05 | 1.06E–02 |
| Nmral1 | NmrA-like family domain containing 1 | 0.083 | 3.75E–05 | 1.18E–02 |
| Frg1 | FSHD region gene 1 | 0.013 | 1.18E–04 | 2.95E–02 |
| Samp | SAP domain containing ribonucleoprotein | 0.017 | 1.47E–04 | 3.45E–02 |
| Polr2d | Polymerase (RNA) II (DNA directed) polypeptide D | 0.021 | 1.91E–04 | 4.02E–02 |
| Pml | Promyelocytic leukemia | 0.101 | 2.13E–04 | 4.34E–02 |
| Ctcf | CCCTC-binding factor | 0.100 | 2.72E–04 | 4.75E–02 |
| Lpxn | Leupaxin | 0.058 | 3.34E–04 | 4.34E–02 |
| Hmha1 | Histocompatibility (minor) HA-1 | 0.126 | 3.75E–04 | 4.83E–02 |
| Septin 1 | Septin 1 | 0.106 | 3.95E–04 | 4.98E–02 |
Notes. The protein intensity values were log2 transformed, centered by the median and scaled based on the standard deviation so that the data distribution had a median of 0 and a standard deviation of 1. The average intensity for each protein was used to calculate the relative fold change. Student’s t test was used to calculate the P values, and they were false discovery rate (FDR)-adjusted to account for multiple tests.
FIG. 2.
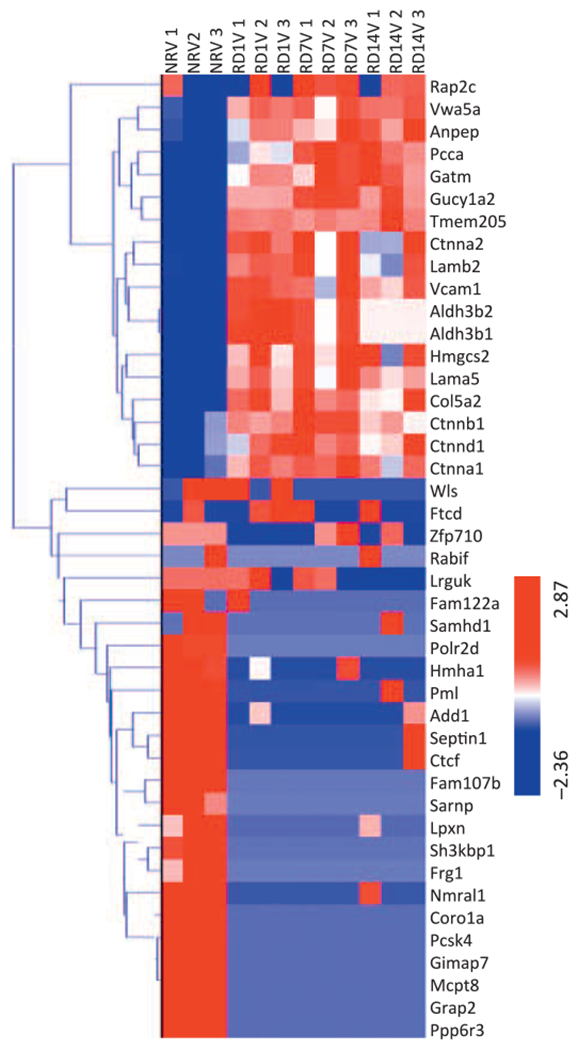
Ionizing radiation induces time-dependent changes in protein expression profiles. Hierarchical clustering of proteins using the average linkage method shows that radiation exposure (Rd1V, Rd7V and Rd14V) generated a robust protein expression response compared to the nonirradiated vehicle control (NRV). n = 3 biological replicates.
The clustering of proteins was performed using the average linkage method with the Euclidean distance metric. Exposure to radiation generated a robust protein expression response compared to the nonirradiated controls.
We found an increase in the catenin family of proteins as well as 3-hydroxy-3-methylglutaryl-coenzyme A synthase, in the irradiated animals. On the other hand, several proteins including ribonucleoproteins, mast cell protease, GTPase, protein phosphatase and pro-protein convertase were significantly downregulated. Given the important biological functions of these enzymes, it is reasonable to hypothesize that radiation exposure is likely to interfere with an array of cellular biochemical pathways ultimately affecting the integrity of tissue structure and function. We used the IPA tool to discern pathway perturbations in irradiated spleen samples compared to nonirradiated. Network analysis (Fig. 3A) suggested upregulation of organ injury response elements including the catenin family of glycoproteins, vascular cell adhesion molecule-1 (VCAM-1), that have been implicated in radiation-induced stress injury (45).
FIG. 3.
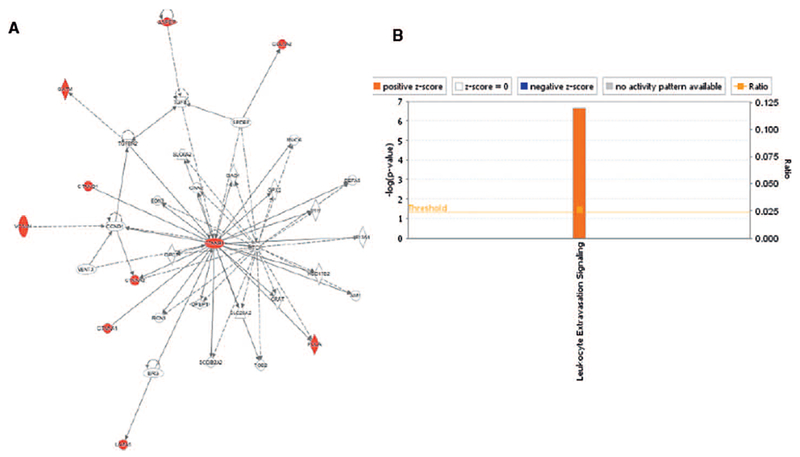
Pathway analysis shows an upregulation of leukocyte extravasation signaling. Differentially expressed proteins in the spleen samples (day 14 postirradiation) were queried for pathway enrichment using the IPA tool. Network analysis showed upregulation of proteins associated with organ injury (panel A), while leukocyte extravasation signaling was found to be significantly upregulated (panel B).
There was a significant upregulation of leucocyte extravasation signaling (movement of leukocytes out of the circulatory system and towards the site of tissue damage or infection) observed in irradiated spleen samples. These results implicate the role of Wnt signaling in exacerbating oxidative stress and inflammation mediated stress signaling in this tissue (Fig. 3B).
Differential Expression of Spleen Proteome after GT3 Treatment
We have reported extensively on the radioprotective effects of GT3 (10, 14, 16, 17, 29). Subsequently, using the same experimental design (29), we sought to determine if administration of GT3 prior to irradiation would alleviate some of the pathway alterations indicative of radiation injury. Therefore, groups of mice from NRG were compared to irradiated GT3-treated mice for a given time point (days 1, 7 and 14) post-TBI. Figure 4 shows a representative partial least squares regression model with the Martens’ uncertainty test showing group separation based on inherent differences in proteomic profiles on day 14 post-TBI. Table 2 indicates up- and downregulated proteins from GT3 treatment in mice receiving TBI compared to NRG.
FIG. 4.
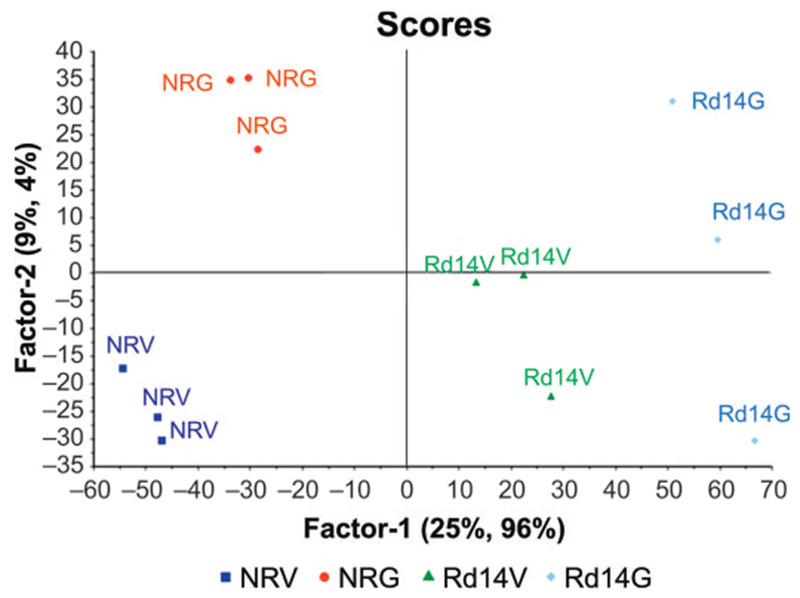
GT3 treatment modulates changes in protein expression profiles after irradiation. The 2D scores plot shows separation of the spleen proteome of nonirradiated vehicle control mice (NRV) from that of nonirradiated GT3 control (NRG), irradiated vehicle (Rd14V ) and GT3-treated mice on day 14 (Rd14G).
TABLE 2.
Differential Proteomic Profiles upon GT3 Treatment Prior to Radiation Exposure
| Associated gene name | Description | Fold change RG/NRG | t test | FDR adjusted P value |
|---|---|---|---|---|
| Upregulated | ||||
| Rap2c | RAP2C, member of RAS oncogene family | 7.48 | 1.24236E–05 | 0.011672 |
| Lamb2 | Laminin, beta 2 | 12.9 | 3.16587E–05 | 0.02776 |
| Downregulated | ||||
| Wls | Wntless homolog (Drosophila) | 0.022 | 6.45426E–29 | 8.49E–25 |
| A630010A05Rik | RIKEN cDNA A630010A05 gene | 0.037 | 8.59694E–29 | 3.77E–25 |
| Lrguk | Leucine-rich repeats and guanylate kinase domain containing | 0.037 | 8.59694E–29 | 2.26E–25 |
| Fam122a | Family with sequence similarity 122, member A | 0.035 | 6.20682E–26 | 1.17E–22 |
| Samhd1 | SAM domain and HD domain, 1 | 0.015 | 4.79923E–24 | 7.89E–21 |
| Rabif | RAB interacting factor | 0.015 | 6.51711E–23 | 8.57E–20 |
| 4930563D23Rik | RIKEN cDNA 4930563D23 gene | 0.039 | 3.489E–22 | 4.17E–19 |
| Zfp710 | Zinc finger protein 710 | 0.034 | 1.03769E–19 | 1.14E–16 |
| Ftcd | Formiminotransferase cyclodeaminase | 0.034 | 9.31634E–13 | 9.43E–10 |
Notes. RG = GT3-treated, irradiated; NRG = nonirradiated GT3 control; FDR = false discovery rate.
To visualize and delineate proteins that contributed maximally to the group differences, we used volcano plots (Fig. 5) to show differentiating proteins between the NRV and irradiated, day 14 post-TBI (Rd14V), groups (Fig. 5A) and between the vehicle- (Rd14V) and GT3-treated irradiated groups on day 14 post-TBI (Rd14G) (Fig. 5B). The volcano plot highlights proteins in red having a relative fold change of ≥0.5 or ≤2.0 and P value ≤0.05 in the two comparative groups (Student’s t test).
FIG. 5.
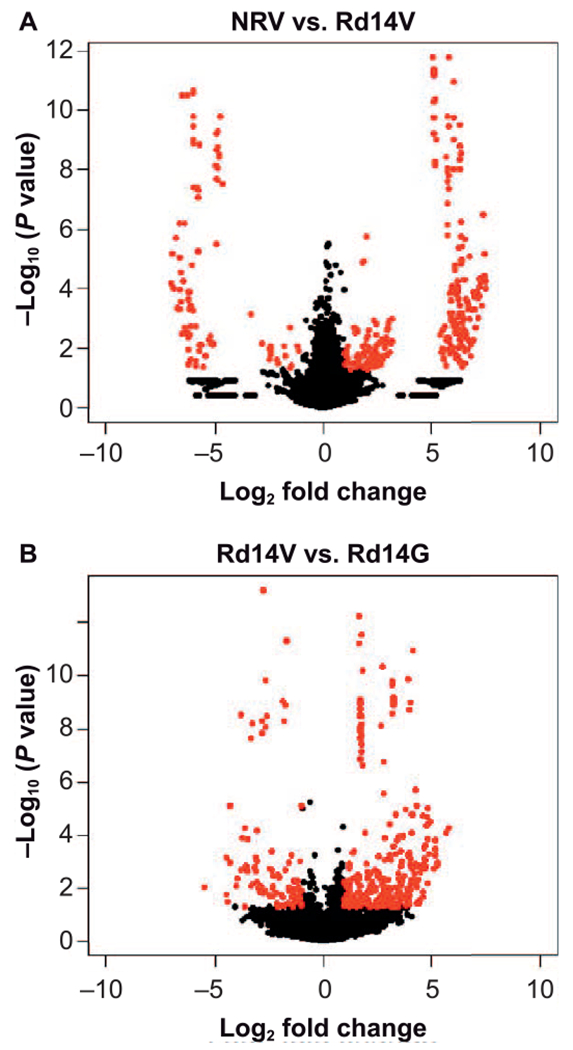
Volcano plots facilitating visualization of significantly differentiating proteins. The plot indicates both fold change expression (X-axis, log2) as well as the significance level [Y-axis, −log10 (P value)] for nonirradiated vehicle control (NRV) and irradiated mice at day 14 (Rd14V) (panel A) and irradiated vehicle (Rd14V) and irradiated mice receiving GT3 (Rd14G) (Panel B), respectively. The proteins shown in red have a relative fold change of ≥0.5 or ≤2.0 and P value ≤ 0.05 in the two comparative groups. n = 3 biological replicates.
Hierarchical clustering based heat map was used to visualize the changes in protein expression after TBI as well as the effect of GT3 treatment (Fig. 6A). Interestingly, protein expression was predominantly downregulated, especially for proteins involved in cell proliferation, while proteins mediating DNA repair (RAD21) or inhibiting cell proliferation (retinoblastoma binding protein 7) were upregulated on days 7 and 14.
FIG. 6.
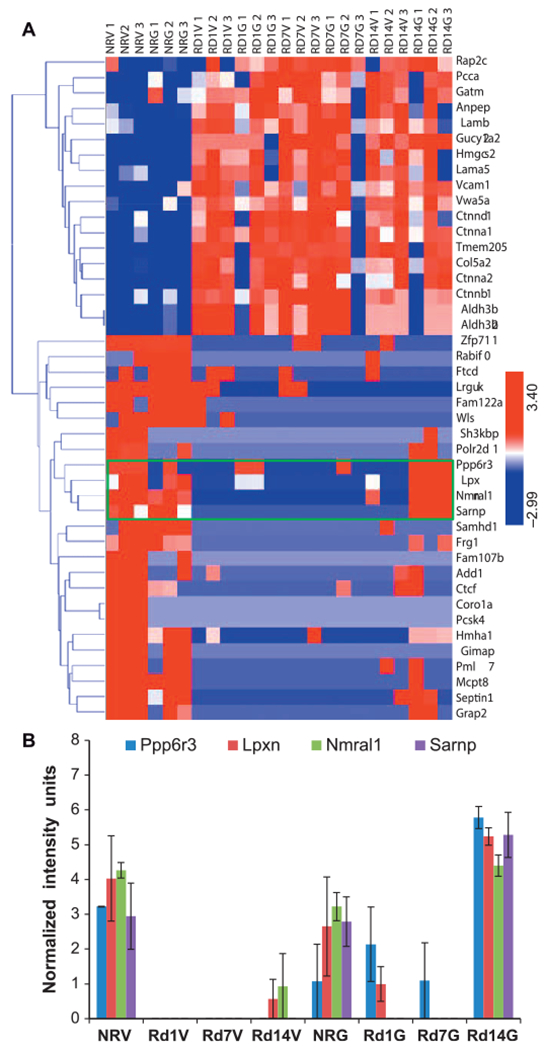
GT3 treatment modulates protein expression changes at day 14. Exposure to ionizing radiation generated a robust protein expression response compared to the nonirradiated controls (panel A), while GT3 treatment induced partial attenuation of response notable at day 14 postirradiation (panel B). NRV = nonirradiated vehicle control; NRG = nonirradiated GT3 control; Rd1V, Rd7V and Rd14V = irradiated mice on days 1, 7 and 14. n = 3/group.
These results suggest that GT3 may exert radioprotective effects by inducing cell cycle arrest and promoting DNA damage repair in response to radiation. Additionally, the endogenous levels of a number of proteins, including Frizzled family of proteins, Wnt signaling proteins and catenin family of proteins, which were significantly elevated after irradiation, appeared to be approach near normal levels if the animals received GT3 prior to irradiation. These results suggest that the observed radioprotective effects of GT3 are mediated at least in part via suppressing signaling pathways that would trigger oxidative stress, DNA doublestrand breaks, impaired bone marrow recovery and hematopoietic stem cell (HSC) regeneration (46). We also observed that GT3 treatment mediated attenuation of expression for proteins such as protein phosphatase 6, regulatory subunit 3 (Ppp6r3), leupaxin (Lpxn), Nmral1 (NmrA-like family domain containing 1) and SAP domain-containing ribonucleoprotein (Sarnp) on day 14 post-TBI (Fig. 6B). Leupaxin is preferentially expressed in hematopoietic cells and has been shown to be upregulated in prostate and breast tumors (47). Four actin-cytoskeleton linked proteins, adducin-α, Ftcd, β-catenin and Frg1, were differentially modulated by radiation, as shown in Fig. 6 (48). Three of these, adducin-α, β -catenin and Frg1, were modulated by treatment with GT3, however, Ftcd expression remain unchanged.
RT-PCR-Based Validation of Attenuation of Wnt Signaling Pathway by GT3
One striking finding of our proteomics analyses was upregulation of the Wnt signaling pathway in spleen after irradiation and its alleviation with prophylactic administration of GT3. Since Wnt-regulated proteins were increased in spleen after TBI in mice, we proceeded to validate our finding using targeted gene expression array analysis of Wnt signaling-regulated genes (see Materials and Methods). Consistent with our findings from the proteomic screen, we observed upregulation of Wnt-regulated gene expression on days 1, 4, 7 and 14 post-TBI (Fig. 7, Supplementary Fig. S1 and Table S1; http://dx.doi.org/10.1667/RR15008.1.S1). GT3 administration prior to irradiation resulted in decreases in expression of these genes, particularly on day 14 (P < 0.05), which is consistent with our previous phenotypic observation (10, 14, 29) (Table 3).
FIG. 7.
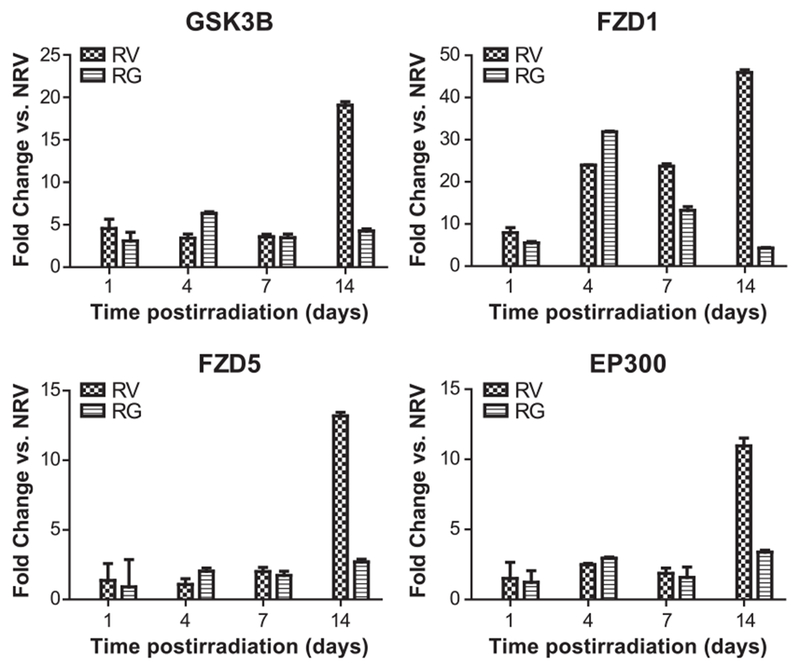
GT3 treatment induces attenuation of Wnt signaling pathway at day 14 postirradiation. RT-PCR-based gene expression profiling was performed with tissue extracts obtained from irradiated and nonirradiated mice (see Materials and Methods) to assess the expression of genes involved in cellular processes that facilitate or regulate Wnt signaling. RV = irradiated mice with vehicle treatment; RG = irradiated mice with GT3 treatment.
TABLE 3.
Attenuation of Gene Expression by GT3 Pretreatment at Day 14 Postirradiation
| Gene symbol | Gene name | RV | RG |
|---|---|---|---|
| CCND1 | Cyclin D1 | 13.8 | 4.4 |
| CCND3 | Cyclin D3 | 2.5 | 1 |
| CSNK1A1 | Casein kinase 1, alpha 1 | 2.3 | 0.6 |
| CSNK1D | Casein kinase 1, delta 1 | 4.8 | 1.2 |
| CTBP1 | C-terminal binding protein 1 | 20.9 | 5.0 |
| CTNNB1 | Catenin (cadherin associated protein), beta 1 | 11.4 | 1.7 |
| DAAM1 | Disheveled associated activator of morphogenesis 1 | 72.8 | 23.5 |
| EP300 | E1A binding protein p300 | 10.9 | 3.3 |
| FBXW11 | F-box and WD-40 domain protein 11 | 17.3 | 3.8 |
| FBXW2 | F-box and WD-40 domain protein 2 | 24.0 | 10.7 |
| FRAT1 | Frequently rearranged in advanced T-cell lymphomas | 3.8 | 0.7 |
| FRZB | Frizzled-related protein | 45.8 | 5.2 |
| FZD1 | Fizzled class receptor 1 | 45.8 | 4.2 |
| FZD6 | Fizzled class receptor 6 | 3.7 | 0.5 |
| GSK3B | Glycogen synthase kinase 3 beta | 19.0 | 4.2 |
| LRP6 | Low-density lipoprotein receptor-related protein 6 | 3.6 | 0.7 |
| PORCN | Porcupine homolog | 5.7 | 0.7 |
| PPP2R5D | Protein phosphatase 2 regulatory subunit B (B56), delta isoform | 2.6 | 0.5 |
| SFRP1 | Secreted frizzled-related protein 1 | 4.6 | 0.8 |
| SLC9A3R1 | Solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | 8.5 | 0.3 |
| TCF7L1 | Transcription factor 7-like 1 (T-cell specific, HMG box) | 4.0 | 0.4 |
| WISP1 | Wnt1 inducible signaling pathway protein 1 | 9.1 | 1.9 |
| WNT11 | Wingless-related MMTV integration site 11 | 2.8 | 0.3 |
| WNT6 | Wingless-related MMTV integration site 6 | 21.9 | 4.0 |
| WNT7A | Wingless-related MMTV integration site 7A | 3.1 | 0.4 |
Notes. Significant fold changes of irradiated vehicle and irradiated GT3 groups compared to nonirradiated vehicle (NRV) control are shown. All genes listed here have a P value ≤ 0.00001.
Wnt proteins are a large family of secreted signaling molecules that are expressed in diverse tissues. These proteins regulate the development of various processes in vertebrate and invertebrate growth (48). Modulation of Wnt signaling can affect HSC function as shown in a mice model (48). To determine if GT3 modulates Wnt pathway gene expression in the spleen after radiation injury, we used Wnt signaling quantitative real-time PCR (RT-PCR) array to query expression levels of 84 Wnt-related genes in four groups (NRV, NRG, RV and RG) at four time points (days 1, 4, 7 and 14 post-TBI). Expression of a subset of these genes was altered after irradiation compared to NRV. The genes showing significant fold changes of irradiated vehicle and irradiated GT3 groups compared to NRV for all days are listed in Supplementary Table S1 (http://dx.doi.org/10.1667/RR15008.1.S1). There was a >10-fold increase in glycogen synthase kinase 3 beta (GSK3β), frizzled class receptor 1 (FZD1), frizzled class receptor 5 (FZD5), E1A binding protein p300 (EP300), cyclin-D1 (CCND1) and C-terminal binding protein 1 (CTBP1) in irradiated vehicle compared to NRV on day 14 post-TBI (P ≤ 0.05). A dramatic increase (>60-fold) was observed in the expression of disheveled associated activator of morphogenesis 1 (DAAM1), 14 days post-TBI. The expression of these genes was significantly inhibited by GT3 treatment on day 14 post-TBI (Table 3). Other Wnt ligands (Wnt1, Wnt2, Wnt5a, Wnt6 and Wnt7a) were also upregulated after TBI, along with Wnt-binding proteins disheveled segment polarity protein 1 (Dvl1), F-box and WD repeat domain containing 11 (Fbxw11), F-box and WD repeat domain containing 2 (Fbxw2) and frizzled-related protein (Frzb). Expression of the downstream Wnt signaling targets Myc, Jun, T-cell transcription factor 7 (Tcf7), and Wnt1 inducible signaling pathway protein 1 (Wisp1) were also increased after TBI, as were other genes such as secreted frizzled related protein 1 (Sfrp1), porcupine (Porcn), and serine/threonine-protein phosphatase 2A regulatory subunit A alpha (PPP2r1a). There was significant inhibition on day 14 by GT3 treatment in the expression of all Wnt-regulated genes that were upregulated by irradiation in animals (data not shown for all). Thus, using an orthogonal approach, we were able to validate our discovery mode results from the proteomic screen.
GT3 Represses Radiation-Induced EPO in Mouse Serum
We evaluated EPO levels in mouse serum up to day 14 post-TBI. Radiation induced an increase in EPO levels in a time-dependent manner (Fig. 8). Baseline level was 0.289 (±0.009) ng/ml−1 in nonirradiated vehicle. EPO levels gradually increased after irradiation from day 0 (0.38 ± 0.02 ng/ml−1) to day 14 postirradiation (11.57 ± 1.03 ng/ml−1), the maximum was reached on day 14 for animals after 7 Gy TBI.
FIG. 8.
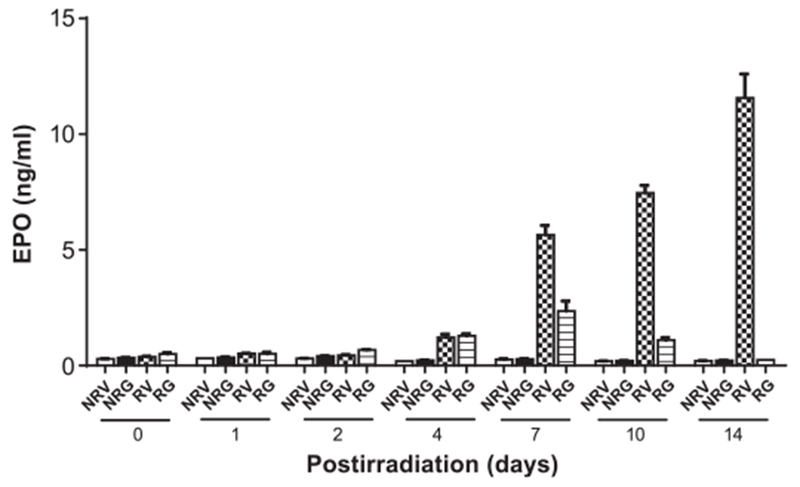
EPO induction after TBI and modulation by GT3. A time-dependent increase in EPO levels was observed up to day14. EPO levels were significantly inhibited by GT3 treatment on days 7, 10 and 14 (P < 0.05). The data reflect the average of three or more independent experiments and are presented as mean ± SEM (n = 6). NRV = nonirradiated vehicle control; NRG = nonirradiated GT3 control; RV = irradiated mice treated with vehicle; RG = irradiated mice with GT3 treatment.
There was a significant time-dependent increase in EPO levels in irradiated mice from day 4 to 7 (1.21 vs. 5.65 ng/ml−1, P < 0.0001) and day 7 to 14 (5.65 vs. 11.57 ng/ml−1, P < 0.0001). Increase in EPO from day 2 to 4 (0.43 vs. 1.21 ng/ml−1, P = 0.65) was not significant. On days 7, 10 and 14 post-TBI, EPO levels were significantly (P < 0.01) inhibited by GT3 treatment (Fig. 8). At day 14, EPO levels were 11.57 (±1.03) ng/ml−1 and 0.24 (±0.003) ng/ml−1 for vehicle- and GT3-treated animals irradiated at 7 Gy, respectively. The data show that EPO levels in GT3-treated animals on day 14 postirradiation returned to basal level (0.217 ±0.003 ng/ml−1) as was found in serum from nonirradiated GT3-treated animals. Based on previously published data, our results indicate that EPO induction in peripheral blood after irradiation might be correlated with Wnt1 upregulation in spleen (49). Both EPO and Wnt1 were found to be inhibited by GT3 treatment at day 14 post-TBI.
GT3 Modulate the Expression of Actin-Cytoskeleton Linked Proteins
Cell migration is powered by a machinery which is built from the actin-skeleton. Actin-cytoskeleton can change cell morphology by disassembling or assembling itself (50, 51). It is important to investigate the role of proteins related to actin-skeleton after radiation injury. To validate the proteomics results from liquid chromatography-tandem mass spectrometry using Western blot, we randomly chose three radiation-induced actin-cytoskeleton proteins shown in Fig. 6, two of which were upregulated (adducin-α and Frg1) (50, 52) and one of which was downregulated (β-catenin) (53) after GT3 treatment. The levels of adducin-α and Frg1 decreased significantly in irradiated vehicle-treated mice after day 14, while the levels in drug-treated mice were significantly higher. At day 7, there was no change in the expression of adducin-α and Frg1 in vehicle- and GT3-treated mice; both groups had lower levels compared to nonirradiated vehicle-treated animals (Fig. 9). Expression of β-catenin increased in vehicle-treated mice after day 14, which was inhibited by GT3 treatment. There was also a trend toward decrease in the levels of β-catenin in drug-treated groups on day 7 post-TBI but it was not significant (Fig. 9).
FIG. 9.
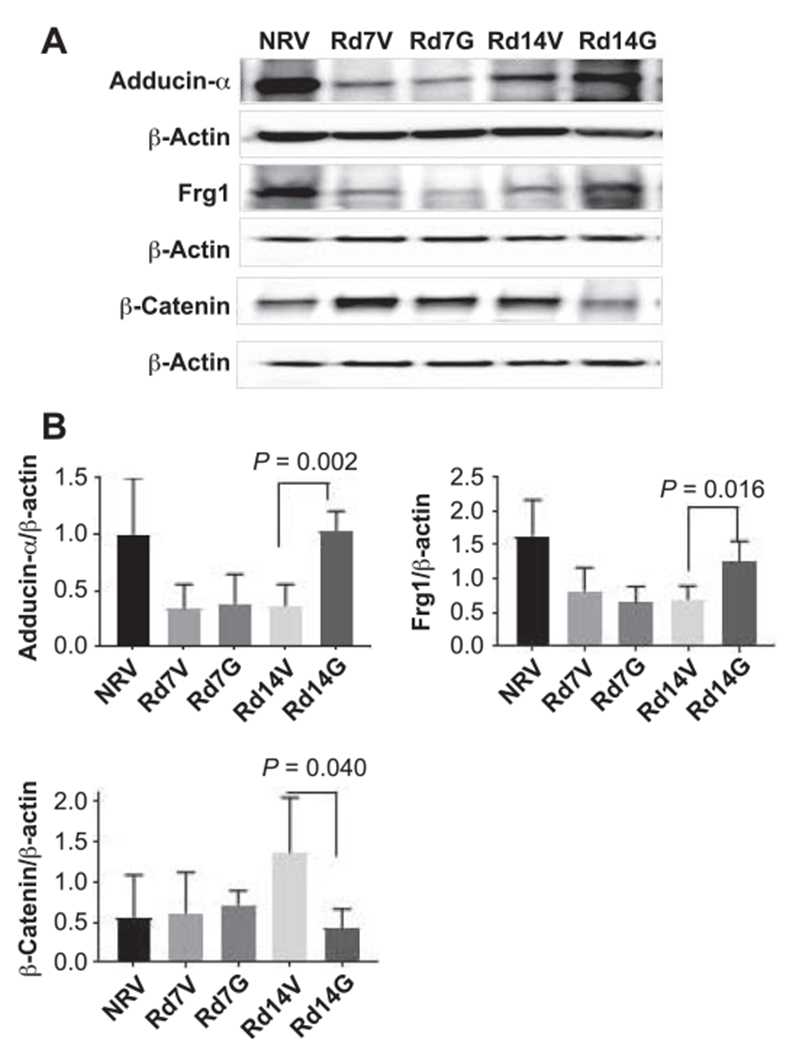
GT3 treatment modulates protein expression in actin-cytoskeleton related proteins. Panel A: Western blot analysis of adducin-α, Frg1 and β-catenin in spleens. Panel B: Quantification of samples was done after analyzing four independent experiments from different samples (n = 3) and the bar graph shows the quantification of the ratio of interest protein and respective β-actin. NRV = nonirradiated vehicle; Rd7V = irradiated day 7 vehicle; Rd7G = irradiated day 7 GT3-treated; Rd14V = irradiated day 14 vehicle and Rd14G = irradiated day 14 GT3-treated.
DISCUSSION
Exposure to ionizing radiation is known to trigger a complex set of signaling events resulting in organ injury. A global molecular profiling approach can help provide insights into radiation-induced molecular events associated with hematopoietic ARS (H-ARS). Bone marrow is the most sensitive hematopoietic organ, and myelosuppression is caused by depletion of progenitor cells in bone marrow (54). Earlier we reported that GT3 protects bone marrow hematopoietic stem and progenitor cells, which results in the amelioration of radiation-induced peripheral blood cytopenia (14). In this study, we analyzed spleen as one of the hematopoietic organs to study differential expression of proteins and genes in response to H-ARS. Arguably, molecular profiling of bone marrow can be potentially more informative regarding mechanisms of hematopoietic recovery; however, the number of depleted bone marrow cells harvested from mice after TBI did not meet the requirement needed for the molecular assays reported in this study. Herein, we report findings from high-resolution mass spectrometry-based untargeted proteomic profiling of spleen tissue performed using a mouse model. Since the hematopoietic system is susceptible to radiation injury, our initial goal was to identify proteins that underscore a molecular signature of radiation exposure in spleen tissue. We observed significant changes in the endogenous abundance of several metabolic enzymes, including guanylate cyclase, glycine amidinotransferase, aldehyde dehydrogenase and propionyl coenzyme A, suggestive of perturbations in carbon and amino acid metabolism. We also observed a significant increase in the levels of hydroxy-3-methylglutaryl-CoA synthase in gamma irradiated vs. nonirradiated animals. Hydroxy-3-methylglutaryl-CoA is an important intermediate in the isoprenoid synthesis that is also implicated in mediating nitrosative stress by modulating the eNOS activity. One of the striking findings of our study was the significant increase in catenin and Wnt family of proteins after irradiation. This increase was consistently observed at all time points measured after TBI (days 1, 4, 7 and 14). We have previously reported that GT3 treatment significantly enhanced survival in mice that had received a lethal dose of TBI (10). GT3 accelerates blood cell recovery in mice exposed to a sublethal dose of TBI by day 15, protects hematopoietic stem and progenitor cells and helps in regeneration of sternal bone marrow cellularity by day 14 after 7 Gy TBI (10, 14). Moreover, GT3 enhances the number of spleen colony-forming units as well as spleen size in CD2F1 mice receiving 8.5 Gy TBI by day 12 compared to irradiated control groups (7). Although peripheral blood cells and bone marrow progenitor cells decreased after 7 Gy TBI in CD2F1 mice, an accelerated recovery by GT3 was observed by day 14 after TBI compared to irradiated vehicle control (10). Micro-RNA (miRNA) profiles in spleen RNA also revealed a dramatic differential expression of miRNAs in GT3-treated mice (29) on day 14 post-TBI.
It is well known that EPO production is induced, leading to the expansion of extramedullary erythropoiesis in the spleen of mice after depletion of red blood cells (RBCs) following stress, including radiation (37–39). These data suggest that it would be important to know the EPO regulation for a period of time during bone marrow progenitor cell depletion and its recovery after a sublethal dose of radiation. It is known that EPO maintains β-catenin activity through inhibition of GSK-3β, and promotes translocation of β-catenin from endothelial cell cytoplasm to the nucleus through a Wnt1 pathway in rat brain endothelial cells after d-glucose treatment (49). Since we have shown that radiation causes maximal suppression of peripheral blood cell count by day 4 and there is significant recovery by day 14 after sublethal (7 Gy) dose of radiation, our study focuses on irradiated spleen tissue with or without GT3 treatment harvested 14 days after TBI (10, 14). Significantly less bone marrow colony forming units were observed on day 7 and 14 in 7 Gy irradiated CD2F1 mice (10, 14). A lethal dose of radiation would provide valuable insight into the recovery from H-ARS, however, this was beyond the scope of this study. During recovery from radiation-induced aplastic anemia, an increase in erythropoietic stimulatory activity leads to an expansion of EPO-responsive cells to manufacture new red blood cells, as reported in the literature (55–57). EPO levels were shown to increase significantly from day 2 to 5 in serum from mice after 8 Gy TBI (48). In addition, EPO administered after a lethal dose of radiation stimulates reticulocyte recovery and improves survival (58). These data suggest that it would be important to understand the EPO regulation during bone marrow progenitor cell depletion and its recovery after a sublethal dose of radiation. In this study, we have shown that at day 14, EPO serum levels were significantly higher in irradiated vehicle-treated animals and attenuated in irradiated GT3-treated animals.
Activation of the Wnt pathway directly stimulates the proliferation of hematopoietic stem and progenitor cell populations and helps reconstitute the hematopoietic system (59). It was also reported that the Wnt signaling pathway is reactivated after injury in the hematopoietic system in a mouse model. Hematopoietic injury was modeled using the chemotherapeutic drug cyclophosphamide (60) in conjunction with the cytokine G-CSF in mice. Using this model, the authors demonstrated that after injury, soluble fraction of the bone marrow microenvironment develops an enhanced ability to support HSCs. Using TCF-GFP reporters, Reya et al. demonstrated that Wnt/β-catenin/TCF/LEF signaling was active in HSCs (61). Wnt signaling was also documented in HSCs in vitro after treatment with purified Wnt3a (61). Based on these studies, it has been suggested that the Wnt/β-catenin/TCF/LEF signaling pathway is active during proliferation of HSCs in a mouse model (62). To validate our proteomic findings, we used the RT-PCR-based targeted gene expression profiling approach to confirm upregulation of the Wnt signaling pathway (Fig. 7) and Western blot analysis to confirm the effect of GT3 on radiation-induced actin-cytoskeleton linked proteins adducin-α, Frg1 and β-catenin. Interestingly, we observed restoration of catenin expression to near-normal levels if the animals received GT3 treatment, both at the protein and mRNA level. This observation was confirmed by Western blot analysis of spleen lysates. To our knowledge, this has not been reported elsewhere to date, and suggests that the strong radioprotective effects of GT3 reported by us and others (7, 10, 14) are mediated in part by alleviation of the Wnt signaling pathway. This is also consistent with the phenotypic rescue, which we previously reported, at day 14 post-TBI in mice pretreated with GT3 (14). Modulation of adducin-α and Frg1 protein expression by GT3 treatment has been confirmed by Western blot analysis in spleen. EPO induction in peripheral blood after irradiation might be correlated with Wnt1 upregulation in spleen (49). Both EPO and Wnt1 were found to be inhibited by GT3 treatment at day 14 post-TBI. Taken together, these data suggest that Wnt signaling can be activated after radiation injury in mouse hematopoietic tissues to reconstitute the hematopoietic system, and administration of GT3 will promote Wnt signaling and, therefore, hematopoietic regeneration. Future studies are planned to elucidate the relationship between EPO and Wnt gene activation after radiation injury and delineate the downstream targets.
CONCLUSION
Our findings indicate that radiation-induced pathway perturbations are in part rescued by GT3 treatment resulting in alleviation of radiation-mediated injury. We used an orthogonal “omics” approach to confirm perturbation and alleviation of the Wnt pathway, which provides an in silico validation of our initial findings. Our results illustrate the utility of high-throughput “omics” approaches to identify and characterize unanticipated pathway perturbations that underscore the observed phenotype. Taken together, the proteome analysis of spleens from irradiated mice provides a vital understanding about the radioprotective activity of GT3, revealing candidate proteins, signaling pathways and networks involved in conventional and nonconventional functions. Thus the results from the current study show pleiotropic effects of GT3 that operate through the restoration or modulation of key proteins that regulate the DNA replication, recombination and repair, B-cell development, metabolic activity and immunological responses induced by ionizing radiation. Further studies validating these observations are warranted to implicate the role of identified candidate proteins involved in determining the radioprotective efficacy of GT3. Since GT3 is a promising prophylactic countermeasure for ARS, which protects animals from hematopoietic as well as gastrointestinal injury (7, 10), our future goal is to uncover other signaling pathways that might be associated with gastrointestinal damage and recovery using a proteomic approach.
Supplementary Material
Table S1. RT PCR results showing attenuation of Wnt signaling pathway after GT3 treatement.
Fig. S1. RT PCR results showing attenuation of Wnt signaling pathway after GT3 on day 14 post-TBI.
ACKNOWLEDGMENTS
We acknowledge Gregory Holmes-Hampton for editing the manuscript. This study was supported by the U.S. Department of Defense Threat Reduction Agency (H.10016_09_AR_R, administered by the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. and the AFRRI Intramural funding). Additional funding was provided by: the National Institutes of Health (grant nos. R01GM106024, R21ES025268, UL1TR000039, P20GM103625, S10OD018445 and P20GM103429 to AJT); the National Space Biomedical Research (grant no. RE03701) and the NIAID/NIH (grant no. 1U01AI133561-01), to AKC; and by an Institutional Development Award from the National Institute of General Medical Sciences of the National Institutes of Health (grant no. P20 GM109005 to MHJ). Finally, we thank the anonymous reviewers who helped us polish the manuscript. The opinions contained herein are the private views of the authors, and are not necessarily those of the Armed Forces Radiobiology Research Institute, the Uniformed Services of the University of the Health Sciences, or the Department of Defense. The authors have no conflict of interest, financial, or otherwise, to declare.
Footnotes
Editor’s note. The online version of this article (DOI: 10.1667/RR15008.1) contains supplementary information that is available to all authorized users.
REFERENCES
- 1.Baskar R, Lee KA, Yeo R, Yeoh KW. Cancer and radiation therapy: current advances and future directions. Int J Med Sci 2012; 9:193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentzen SM. Preventing or reducing late side effects of radiation therapy: radiobiology meets molecular pathology. Nat Rev Cancer 2006; 6:702–13. [DOI] [PubMed] [Google Scholar]
- 3. Hauer-Jensen M, Kumar KS, Wang J, Berbee M, Fu Q, Boerma M. Intestinal toxicity in radiation and combined injury: Significance, mechanisms, and countermeasures. Larche R, editor. Hauppauge, NY: NOVA Science Publishers Inc; 2008. [Google Scholar]
- 4.Hendry JH, Feng-Tong Y. Response of bone marrow to low LET irradiation In: Hendry JH, Lord BI, editors. Radiation Toxicology: bone marrow and leukomia. London: Taylor & Francis; 1995. p. 91–116. [Google Scholar]
- 5. Fliedner TM, Nothdurft W, Heit H. Biological factors affecting the occurrence of radiation syndromes In: Broerse JJ, MacVittie TJ, editors. Response of different species to total body irradiation. Leiden, Netherlands: Martinus Nijhoff Publishers; 1984. p. 209–19. [Google Scholar]
- 6.Mauch P, Constine L, Greenberger J, Knospe W, Sullivan J, Liesveld JL, et al. Hematopoietic stem cell compartment: acute and late effects of radiation therapy and chemotherapy. Int J Radiat Oncol Biol Phys 1995; 31:1319–39. [DOI] [PubMed] [Google Scholar]
- 7.Berbee M, Fu Q, Boerma M, Wang J, Kumar KS, Hauer-Jensen M. gamma-Tocotrienol ameliorates intestinal radiation injury and reduces vascular oxidative stress after total-body irradiation by an HMG-CoA reductase-dependent mechanism. Radiat Res 2009; 171:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bykov VN, Drachev IS, Kraev SY, Maydin MA, Gubareva EA, Pigarev SE, et al. Radioprotective and radiomitigative effects of BP-C2, a novel lignin-derived polyphenolic composition with ammonium molybdate, in two mouse strains exposed to total body irradiation. Int J Radiat Biol 2018; 94:114–23. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh SP, Perkins MW, Hieber K, Kulkarni S, Kao TC, Reddy EP, et al. Radiation protection by a new chemical entity, Ex-Rad: efficacy and mechanisms. Radiat Res 2009; 171:173–9. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh SP, Kulkarni S, Hieber K, Toles R, Romanyukha L, Kao TC, et al. Gamma-tocotrienol, a tocol antioxidant as a potent radioprotector. Int J Radiat Biol 2009; 85:598–606. [DOI] [PubMed] [Google Scholar]
- 11.Satyamitra MM, Kulkarni S, Ghosh SP, Mullaney CP, Condliffe D, Srinivasan V. Hematopoietic recovery and amelioration of radiation-induced lethality by the vitamin e isoform delta-tocotrienol. Radiat Res 2011; 175:736–45. [DOI] [PubMed] [Google Scholar]
- 12. Singh VK, Brown DS, Kao TC. Tocopherol succinate: a promising radiation countermeasure. Int Immunopharmacol 2009; 9:1423–30. [DOI] [PubMed] [Google Scholar]
- 13.Singh VK, Ducey EJ, Fatanmi OO, Singh PK, Brown DS, Purmal A, et al. CBLB613: a TLR 2/6 agonist, natural lipopeptide of Mycoplasma arginini, as a novel radiation countermeasure. Radiat Res 2012; 177:628–42. [DOI] [PubMed] [Google Scholar]
- 14.Kulkarni S, Ghosh SP, Satyamitra M, Mog S, Hieber K, Romanyukha L, et al. Gamma-tocotrienol protects hematopoietic stem and progenitor cells in mice after total-body irradiation. Radiat Res 2010; 173:738–47. [DOI] [PubMed] [Google Scholar]
- 15.Kulkarni SS, Cary LH, Gambles K, Hauer-Jensen M, Kumar KS, Ghosh SP. Gamma-tocotrienol, a radiation prophylaxis agent, induces high levels of granulocyte colony-stimulating factor. Int Immunopharmacol 2012; 14:495–503. [DOI] [PubMed] [Google Scholar]
- 16.Singh VK, Kulkarni S, Fatanmi OO, Wise SY, Newman VL, Romaine PL, et al. Radioprotective efficacy of gamma-tocotrienol in nonhuman primates. Radiat Res 2016; 185:285–98. [DOI] [PubMed] [Google Scholar]
- 17.Singh VK, Hauer-Jensen M. Gamma-tocotrienol as a promising countermeasure for acute radiation syndrome: current status. Int J Mol Sci 2016; 17(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pernot E, Hall J, Baatout S, Benotmane MA, Blanchardon E, Bouffler S, et al. Ionizing radiation biomarkers for potential use in epidemiological studies. Mutat Res 2012; 751:258–86. [DOI] [PubMed] [Google Scholar]
- 19.Azimzadeh O, Atkinson MJ, Tapio S. Proteomics in radiation research: present status and future perspectives. Radiat Environ Biophys 2014; 53:31–8. [DOI] [PubMed] [Google Scholar]
- 20.Azimzadeh O, Scherthan H, Sarioglu H, Barjaktarovic Z, Conrad M, Vogt A, et al. Rapid proteomic remodeling of cardiac tissue caused by total body ionizing radiation. Proteomics 2011; 11:3299–311. [DOI] [PubMed] [Google Scholar]
- 21.Azimzadeh O, Scherthan H, Yentrapalli R, Barjaktarovic Z, Ueffing M, Conrad M, et al. Label-free protein profiling of formalin-fixed paraffin-embedded (FFPE) heart tissue reveals immediate mitochondrial impairment after ionising radiation. J Proteomics 2012; 75:2384–95. [DOI] [PubMed] [Google Scholar]
- 22.Azimzadeh O, Sievert W, Sarioglu H, Yentrapalli R, Barjaktarovic Z, Sriharshan A, et al. PPAR alpha: a novel radiation target in locally exposed Mus musculus heart revealed by quantitative proteomics. J Proteome Res 2013; 12:2700–14. [DOI] [PubMed] [Google Scholar]
- 23.Barjaktarovic Z, Anastasov N, Azimzadeh O, Sriharshan A, Sarioglu H, Ueffing M, et al. Integrative proteomic and microRNA analysis of primary human coronary artery endothelial cells exposed to low-dose gamma radiation. Radiat Environ Biophys 2013; 52:87–98. [DOI] [PubMed] [Google Scholar]
- 24.Barjaktarovic Z, Schmaltz D, Shyla A, Azimzadeh O, Schulz S, Haagen J, et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PloS One 2011; 6:e27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barjaktarovic Z, Shyla A, Azimzadeh O, Schulz S, Haagen J, Dorr W, et al. Ionising radiation induces persistent alterations in the cardiac mitochondrial function of C57BL/6 mice 40 weeks after local heart exposure. Radiother Oncol 2013; 106:404–10. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian V, Borchard S, Azimzadeh O, Sievert W, Merl-Pham J, Mancuso M, et al. PPARalpha is necessary for radiation-induced activation of noncanonical TGFbeta signaling in the heart. J Proteome Res 2018; 17:1677–89. [DOI] [PubMed] [Google Scholar]
- 27.Pluder F, Barjaktarovic Z, Azimzadeh O, Mortl S, Kramer A, Steininger S, et al. Low-dose irradiation causes rapid alterations to the proteome of the human endothelial cell line EA.hy926. Radiat Environ Biophys 2011; 50:155–66. [DOI] [PubMed] [Google Scholar]
- 28.Sriharshan A, Boldt K, Sarioglu H, Barjaktarovic Z, Azimzadeh O, Hieber L, et al. Proteomic analysis by SILAC and 2D-DIGE reveals radiation-induced endothelial response: four key pathways. J Proteomics 2012; 75:2319–30. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh SP, Pathak R, Kumar P, Biswas S, Bhattacharyya S, Kumar VK, et al. Gamma tocotrienol modulates radiation-induced micro-RNA expression in mouse spleen Radiat Res 2016; 185:485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hengel SM, Aldrich JT, Waters KM, Pasa-Tolic L, Stenoien DL. Quantitative proteomic profiling of low-dose ionizing radiation effects in a human skin model. Proteomes 2014; 2:382–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins LM, Mazur SJ, Rossi M, Gaidarenko O, Xu Y, Appella E. Quantitative proteomics analysis of the effects of ionizing radiation in wild type and p53 K317R knock-in mouse thymocytes. Mol Cell Proteomics 2008; 7:716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim YB, Pyun BJ, Lee HJ, Jeon SR, Jin YB, Lee YS. Proteomic identification of radiation response markers in mouse intestine and brain. Proteomics 2011; 11:1254–63. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Lorimore SA, Evans CA, Whetton AD, Wright EG. A proteomic analysis of murine bone marrow and its response to ionizing radiation. Proteomics 2005; 5:4254–63. [DOI] [PubMed] [Google Scholar]
- 34.Srivastava A, Leighton X, Eidelman O, Starr J, Jozwik C, Srivastava M, et al. Personalized radioproteomics: identification of a protein biomarker signature for preemptive rescue by tocopherol succinate in CD34+ irradiated progenitor cells isolated from a healthy control donor. J Proteomics Bioinform 2015; 8:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swift SN, Pessu RL, Chakraborty K, Villa V, Lombardini E, Ghosh SP. Acute toxicity of subcutaneously administered vitamin e isomers delta- and gamma-tocotrienol in mice. Int J Toxicol 2014; 33:450–8. [DOI] [PubMed] [Google Scholar]
- 36.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods 2009; 6:359–62. [DOI] [PubMed] [Google Scholar]
- 37.Bjornson RD, Carriero NJ, Colangelo C, Shifman M, Cheung K- H, Miller PL, et al. X!!Tandem, an improved method for running X!Tandem in parallel on collections of commodity computers. J Proteome Res 2008; 7:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 2004; 20:1466–7. [DOI] [PubMed] [Google Scholar]
- 39.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 2006; 22:2830–2. [DOI] [PubMed] [Google Scholar]
- 40.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, et al. Open mass spectrometry search algorithm. J Proteome Res 2004; 3:958–64. [DOI] [PubMed] [Google Scholar]
- 41.Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J Proteome Res 2006; 5:1843–9. [DOI] [PubMed] [Google Scholar]
- 42.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res 2004; 3:1234–42. [DOI] [PubMed] [Google Scholar]
- 43.Slotta DJ, McFarland MA, Markey SP. MassSieve: Panning MS/MS peptide data for proteins. Proteomics 2010; 10:3035–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol 2001; 13:167–71. [DOI] [PubMed] [Google Scholar]
- 45.Yuan Y, Lee SH, Wu S. The role of ROS in ionizing radiation-induced VLA-4 mediated adhesion of RAW264.7 cells to VCAM-1 under flow conditions. Radiat Res 2013; 179:62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lento W, Ito T, Zhao C, Harris JR, Huang W, Jiang C, et al. Loss of beta-catenin triggers oxidative stress and impairs hematopoietic regeneration. Genes Dev 2014; 28:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dierks S, von Hardenberg S, Schmidt T, Bremmer F, Burfeind P, Kaulfuss S. Leupaxin stimulates adhesion and migration of prostate cancer cells through modulation of the phosphorylation status of the actin-binding protein caldesmon. Oncotarget 2015; 6:13591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ossetrova NI, Condliffe DP, Ney PH, Krasnopolsky K, Hieber KP, Rahman A, et al. Early-response biomarkers for assessment of radiation exposure in a mouse total-body irradiation model. Health Phys 2014; 106:772–86. [DOI] [PubMed] [Google Scholar]
- 49.Chong ZZ, Hou J, Shang YC, Wang S, Maiese K. EPO relies upon novel signaling of Wnt1 that requires Akt1, FoxO3a, GSK-3beta, and beta-catenin to foster vascular integrity during experimental diabetes. Curr Neurovasc Res 2011; 8:103–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staessen JA, Bianchi G. Adducin and hypertension. Pharmacogenomics 2005; 6:665–9. [DOI] [PubMed] [Google Scholar]
- 51.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature 2010; 463:485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu Q, Jones TI, Tang VW, Brieher WM, Jones PL. Facioscapulohumeral muscular dystrophy region gene-1 (FRG-1) is an actin-bundling protein associated with muscle-attachment sites. J Cell Sci 2010; 123(Pt 7):1116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A 1999; 96:5522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Schulte BA, Zhou D. Hematopoietic stem cell senescence and long-term bone marrow injury. Cell Cycle 2006; 5:35–8. [DOI] [PubMed] [Google Scholar]
- 55.DeGowin RL, Johnson S. Development of response to erythropoietin and repletion of the stem cell compartment after irradiation. J Lab Clin Med 1968; 72:893–904. [PubMed] [Google Scholar]
- 56.Schooley JC, Hayes JM, Cantor LN, Havens VW. Studies on the behavior of erythropoietin-sensitive cells in the mouse during recovery from 200 roentgens of whole-body irradiation. Radiat Res 1967; 32:875–83. [PubMed] [Google Scholar]
- 57.Weisman M, Martinson D, Fried W, Gurney CW. A comparison of the rates at which colony forming units and erythropoietin responsive cells recover after 200 r total body x-irradiation. J Lab Clin Med 1967; 69:438–46. [PubMed] [Google Scholar]
- 58.Naidu NV, Reddi OS. Effect of post-treatment with erythropoietin(s) on survival and erythropoietic recovery in irradiated mice. Nature 1967; 214:1223–4. [DOI] [PubMed] [Google Scholar]
- 59.Congdon KL, Voermans C, Ferguson EC, DiMascio LN, Uqoezwa M, Zhao C, et al. Activation of Wnt signaling in hematopoietic regeneration. Stem Cells 2008; 26:1202–10. [DOI] [PubMed] [Google Scholar]
- 60.Korcyl M, Waligorski MP. Track structure effects in a study of cell killing in normal human skin fibroblasts. Int J Radiat Biol 2009; 85:1101–13. [DOI] [PubMed] [Google Scholar]
- 61.Reya T Regulation of hematopoietic stem cell self-renewal. Recent Prog Horm Res 2003; 58:283–95. [DOI] [PubMed] [Google Scholar]
- 62.Staal FJ, Sen JM. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol 2008; 38:1788–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. RT PCR results showing attenuation of Wnt signaling pathway after GT3 treatement.
Fig. S1. RT PCR results showing attenuation of Wnt signaling pathway after GT3 on day 14 post-TBI.


