Abstract
This short review traces how our knowledge of the molecular mechanisms of cellular movements originated and developed over the past 50 years. Work on actin-based and microtubule-based movements developed in different ways, but in both fields, the discovery of the key proteins drove progress. Starting from an inventory of zero molecules in 1960, both fields matured spectacularly, so we now know the atomic structures of the important proteins, understand the kinetics and thermodynamics of their interactions, have documented how the molecules behave in cells, and can test theories with molecularly explicit computer simulations of cellular processes.
Keywords: Actin, Tubulin, Myosin, Kinesin, Dynein
Introduction: birth of the field in the 1960s
Scientists who have started work on the mechanisms of cellular movements since the turn of the twenty-first century would not recognize the field I entered as a college student. I spend a summer of 1963 at the Pasadena Foundation in the laboratory of Charles Pomerat, one of the pioneers of tissue culture. He had about a half dozen phase contrast microscopes with 16-mm movie cameras recording time-lapse movies of cultured cells day and night, and I had the privilege of reviewing these films with him twice a week. I was fascinated by the wide variety of movements including cellular locomotion, organelle movements, nuclear rotation, mitosis, and cytokinesis and the explorations of axonal growth cones. I made quantitative measurements of growth cone dynamics and calculated means and standard deviations using a mechanical Marchant calculator (Pomerat et al. 1964). A trip to the library revealed that nothing was known about the molecular mechanisms of any of the movements. Several papers reported the isolation of crude protein fractions from nonmuscle cells that resembled mixtures of actin and myosin from skeletal muscle (Lowey 1952; Bettex-Galland and Luscher 1959), but none of the motility proteins had been purified. Discussions at a conference on “Primitive Motile Systems in Biology” at Princeton University in 1963 revealed that leading figures in the field questioned whether biochemical approaches would reveal anything useful about cellular movements (Allen and Kamiya 1964).
Everything changed when Ian Gibbons isolated dynein from axonemes (Gibbons and Rowe 1965); Gary Borisy, Richard Weisenberg, and Ed Taylor (Borisy and Taylor 1967; Weisenberg et al. 1968) purified tubulin; and Sadashi Hatano and Fumio Oosawa (Hatano and Oosawa 1966; Hatano and Tazawa 1968) followed by Mark Adelman and Ed Taylor (Adelman and Taylor 1969a, b) purified actin and myosin from the acellular slime mould Physarum.
Unaware of the work on Physarum, I investigated amoeboid movements with professor Susumo Ito from 1965 to 1968 as a medical student at Harvard Medical School. Our attempts to image the motility machinery by transmission electron microscopy of thin sections of fixed cells were unrevealing, so I attempted to repeat experiments by Thompson and Wolpert (Thompson and Wolpert 1963). They made extracts of Amoeba proteus in the cold and reported that after adding ATP and warming to room temperature, the crude cytoplasm would undergo streaming in a sealed chamber on a microscope slide. Fortunately, I did not know that the experts in the field were skeptical about these claims, and, after ten failures in the spring of 1968, I learned how to reactivate the extracts. We observed spectacular streaming of organelles followed by the formation of linear bundles that were pulled toward organizing centers. Electron microscopy revealed that thin filaments assembled in the extract and formed bundles that converged on clusters of thick filaments (Pollard and Ito 1970). Experiments with a homemade viscometer showed that the extracts gelled when the thin filaments formed. We imagined that the filaments were actin and myosin, although some were skeptical. When I presented our observations at the annual meeting of the American Society for Cell Biology in December 1968, I spoke in a session on microtubules, because no one else at the meeting was working on cellular actin filaments. At the same time, Hal Ishikawa developed a method to decorate actin filaments in glycerol-extracted cells with muscle heavy meromyosin, and his electron micrographs of thin sections convinced cell biologists that actin filaments filled the cytoplasm of animal cells (Ishikawa et al. 1969).
At that point, in the late 1960s, we dreamed someday to know the amino acid sequences and structures of actin and myosin, but that seemed far off, given the limited technology available. At that time, there were no purified DNAs, no DNA sequences, no SDS gel electrophoresis of proteins, no useful antibodies, no applications of fluorescence microscopy in cell biology, no electronic databases of publications or published papers, no electronic cameras, and no personal computers or kits for routine biochemical procedures. Light and electron microscopy images were recorded on film. Biochemists used enzyme assays or viscosity measurements to detect cytoskeletal proteins of interest and had only low-pressure gel filtration and ion exchange chromatography to purify proteins and analytical ultracentrifugation to assess purity.
Success of the reductionist strategy
Fifty years later, the field has advanced far beyond our wildest dreams in 1968. The field has collected an extensive inventory of the molecules comprising the motile machinery, atomic structures of key proteins, quantitative measurements of proteins in live cells on a second time scale, and enough measurements of concentrations, rate constants, and equilibrium constants to formulate mathematical models of complex cellular systems for simulations to test their ability to account for the microscopic measurements.
Three factors drove this progress. First, the pioneers in the motility field were inspired by the high standards set by the biophysicists, physiologists, and biochemists working on the mechanism of muscle contraction (CSHSQB 1972). Many of these leaders came from physics and we aspired to emulate them. The following generations of investigators accepted these standards, which made the field much more quantitative and mechanistic than many other areas of cell biology research.
Second, the field adopted every new technology to study mechanisms including presteady state kinetics (Finlayson et al. 1969), three-dimensional reconstructions of polymers from electron micrographs (Moore et al. 1970), fluorescent antibody staining of fixed cells (Lazarides and Weber 1974), molecular cloning and expression of recombinant proteins (Cleveland et al. 1978), fluorescence spectroscopy (Kouyama and Mihashi 1981), video microscopy (Inoué 1981; Allen et al. 1985), microinjecting and imaging fluorescently labeled proteins in live cells (Wang et al. 1982), confocal microscopy (White et al. 1987), molecular genetics (DeLozanne and Spudich 1987), mathematical modeling (Bray et al. 1993), phylogenetic analysis (Goodson and Spudich 1993), GFP-fusion proteins (Ding et al. 1998), quantitative fluorescence microscopy (Wu and Pollard 2005), super resolution microscopy (Bates et al. 2007), and electron cryomicroscopy (Fujii et al. 2010).
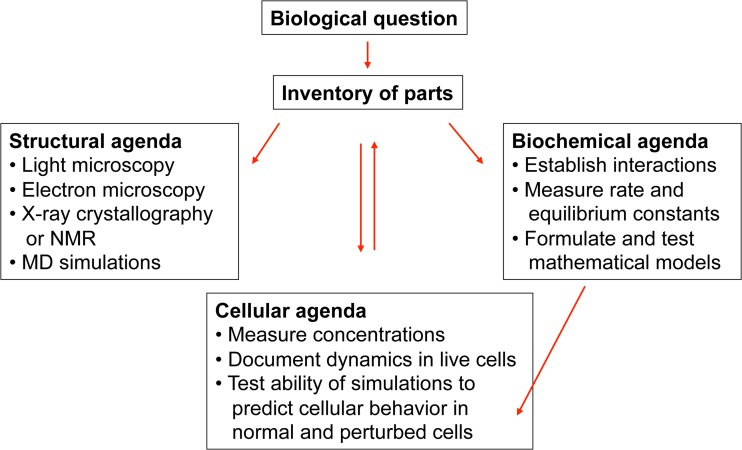
Third, the nascent motility field pursued the full reductionist strategy (Fig. 1) to characterize the mechanisms of cellular movements. The fundamental biological questions were already posed in the nineteenth century: how do cells move, change their shapes, transport intracellular components, separate their chromosomes, and divide in two during cytokinesis? Studies of muscle contraction led the way in the period from the 1930s to the 1960s using elegant physiological experiments on live cells, biochemistry, and structural studies. Biochemists isolated muscle myosin and actin and recombined them in threads that contracted in the presence of ATP (Szent-Gyorgyi 1945). Electron microscopy and x-ray fiber diffraction revealed how filaments of actin and myosin interact as they slide during contraction (Huxley 1969). Biochemical measurements showed how ATP hydrolysis by myosin released energy that is converted into force during interactions with actin filaments (Lymm and Taylor 1971). Physiologists proposed and tested mathematical models that accounted for the force produced in living muscles (Huxley and Simmons 1971). A detailed understanding of the molecular mechanisms was already achieved by 1972 by the time of a meeting at the Cold Spring Harbor Laboratory (CSHSQB 1972).
Fig. 1.
Reductionist agenda (reprinted from PLoS Biology (Pollard 2013))
Studies of the molecular basis of cellular movements lagged behind muscle, since the reductionist agenda was stuck before the inventory stage (Fig. 1) until the 1960s. Of course, some progress was made without knowing the molecules. For example, light microscopy revealed important features of mitosis (Inoué and Sato 1967) and radioactive tracers established the existence of fast and slow axonal transport (Ochs 1972; Weiss 1967). However, it was the discovery of motility proteins that opened the field for rapid progress. In less than a decade, the field grew explosively culminating in an intense meeting on “Cell Motility” in 1975 (Goldman et al. 1976).
Our knowledge of the mechanisms of cellular motility, intracellular movements, mitosis, and cytokinesis each developed in different ways. Biochemists took the lead to identify the major protein components of the motility system, in part because no good methods were available to image actin filaments in live cells. They used simple assays to purify myosin motors (starting with myosin-II (Hatano and Tazawa 1968; Adelman and Taylor 1969a, b; Adelstein et al. 1971) followed by the first unconventional myosin-I (Pollard and Korn 1973) and many other unconventional myosins (Goodson and Spudich 1993)), actin monomer-binding proteins (profilin (Carlsson et al. 1977) and thymosin-ß4 (Safer et al. 1991)), severing proteins (cofilin (Bamburg et al. 1980; Nishida et al. 1984) and gelsolin (Yin and Stossel 1979)), capping proteins (capping protein (Isenberg et al. 1980) and gelsolin (Yin and Stossel 1979)), and cross-linking proteins (spectrin (Marchesi and Steers Jr. 1968), alpha-actinin (Lazarides and Burridge 1975), filamin (originally actin-binding protein) (Hartwig and Stossel 1975), fascin (Otto et al. 1979), fimbrin (Bretscher and Weber 1980), dystrophin (Koenig and Kunkel 1990)). Once libraries of gene and cDNA sequences became available in the 1980s, the inventories of actin-binding proteins grew rapidly. Later, biochemists discovered the Arp2/3 complex (Machesky et al. 1994), which nucleates actin filament branches (Mullins et al. 1998), and geneticists discovered formins (Castrillon and Wasserman 1994), which nucleate and processively elongate actin filament barbed ends (Pruyne et al. 2002; Sagot et al. 2002; Paul and Pollard 2009).
In contrast to this biochemical attack on the mechanism of cellular motility, geneticists discovered most of the proteins that participate in cytokinesis. Actin and myosin-II were implicated in cytokinesis by microscopy (Schroeder 1973; Fujiwara and Pollard 1976) and antibody injections (Mabuchi and Okuno 1977), but Paul Nurse followed by his students used genetic screens for conditional mutations of fission yeast to discover more than 150 genes required for cytokinesis (Guertin et al. 2002; Pollard and Wu 2010). RNAi experiments in Drosophila and C. elegans linked a subset of these proteins to cytokinesis in animals (Pollard 2003). Our understanding of cytokinesis developed rapidly over the past twenty years, so we now know enough the biochemistry and cellular events to formulate and simulate mathematical models that reproduce the assembly (Vavylonis et al. 2008) and constriction (Stachowiak et al. 2014) of the cytokinetic contractile ring. Nevertheless, many questions remain about all aspects of cytokinesis, especially the regulatory mechanism (Pollard 2017; Basant and Glotzer 2018).
Progress in understanding of microtubule-based movements during mitosis and intracellular transport of organelles benefitted from being able to image microtubules first by electron microscopy and fluorescence microscopy of fixed cells and subsequently by DIC and fluorescence microscopy of live cells. Microscopy revealed the structure of the mitotic spindle including the polarity of the microtubules (Euteneuer and McIntosh 1981), assembly properties of microtubules (Walker et al. 1988) including their dynamic instability (Mitchison and Kirschner 1984), and the great variety of cargo transported along microtubules (Barlan and Gelfand 2017). The discoveries of kinesin (Vale et al. 1985) and cytoplasmic dynein (Paschal and Vallee 1987) initiated the characterization of many varieties of kinesin and a smaller family of cytoplasmic dyneins. (A small historical note is that in the 1973 Physiology Course at the Marine Biology Laboratory, Roy Burns and I isolated a dynein-like 380-kDa protein from brain that bound to microtubules but had only low ATPase activity (Burns and Pollard 1974). No motility assays were available to test if our protein moved on microtubules.) Yeast genetics decisively implicated kinesins in the assembly of the mitotic spindle and the movements of chromosomes (Hoyt et al. 1992; Saunders and Hoyt 1992). We now appreciate that cells orchestrate dynamic microtubules, attachment sites at kinetochores, and motor proteins to assemble the mitotic apparatus and separate chromosomes reliably into the daughter cells (McIntosh 2016), which are divided by cytokinesis.
The past three decades led to the discovery and characterization of many novel proteins that modify all aspects of microtubule assembly. These proteins have functions comparable to the proteins that regulate actin assembly, but the two systems evolved separately. The only homologous proteins are myosins and kinesins (Kull et al. 1996) and actin filament cross-linking proteins and microtubule end-binding proteins, which both use calponin homology domains to interact with the polymers (Hayashi and Ikura 2003). Other microtubule-binding proteins sequester tubulin dimers (stathmin (Belmont and Mitchison 1996)), nucleate assembly by forming a template for the minus end (γ-tubulin ring complex (Zheng et al. 1995)), stabilize minus ends (CAMSAPs (Akhmanova and Steinmetz 2015)), stabilize the wall of the microtubule (MAP2/tau family (Cleveland et al. 1977)), kinesins that promote dissociation of subunits from the ends (kinesin 8, kinesin 13, and kinesin 14 (Howard and Hyman 2007)), and AAA-ATPases that sever microtubules by extracting subunits from the wall (katanin and related proteins (McNally and Vale 1993)).
Talented people matter
The reductionist strategy would not have been so successful without dozens of investigators who entered the field over the past five decades. They brought biophysical and biochemical methods, molecular biology and genetics, numerous advances in light and electron microscopy and mathematical modeling. Even more important, they brought curiosity, analytical acumen, and creative ideas to explain complicated processes.
I have been particularly fortunate to have mentored 27 graduate students and 50 postdocs as well as eager undergraduate and medical students and skilled research assistants, who worked and learned in my laboratory. These individuals have achieved great success in academic science and education, publishing, the biopharmaceutical industry, and leadership positions in the biomedical research community. A long list of collaborators, including the authors of some of the papers in this volume, has helped with their expertise and ideas. And, of course, all of us are grateful to the National Institutes of Health, which has funded my laboratory continuously since 1972.
Acknowledgements
The author thanks Laurent Blanchoin, Enrique De La Cruz, and Mike Ostap for organizing this volume.
Funding
Research in the author’s laboratory has been supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01GM026132 and R01GM026338. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest
Thomas D. Pollard declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
References
- Adelman MR, Taylor EW. Further purification and characterization of slime mold myosin and slime mold actin. Biochemistry. 1969;8:4976–4988. doi: 10.1021/bi00840a047. [DOI] [PubMed] [Google Scholar]
- Adelman MR, Taylor EW. Isolation of an actomyosin-like protein complex from slime mold plasmodium and the separation of the complex into actin- and myosin-like fractions. Biochemistry. 1969;8(12):4964–4975. doi: 10.1021/bi00840a046. [DOI] [PubMed] [Google Scholar]
- Adelstein RS, Pollard TD, Kuehl WM. Isolation and characterization of myosin and two myosin fragments from human blood platelets. Proc Natl Acad Sci U S A. 1971;68(11):2703–2707. doi: 10.1073/pnas.68.11.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol. 2015;16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Allen RD, Kamiya N. Primitive motile systems in cell biology. New York: Academic Press; 1964. pp. 1–642. [Google Scholar]
- Allen RD, Weiss DG, Hayden JH, Brown DT, Fujiwake H, Simpson M. Gliding movement of and bidirectional transport along single native microtubules from squid axoplasm: evidence for an active role of microtubules in cytoplasmic transport. J Cell Biol. 1985;100(5):1736–1752. doi: 10.1083/jcb.100.5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR, Harris HE, Weeds AG. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 1980;121(1):178–182. doi: 10.1016/0014-5793(80)81292-0. [DOI] [PubMed] [Google Scholar]
- Barlan K, Gelfand VI (2017) Microtubule-based transport and the distribution, tethering, and organization of organelles. Cold Spring Harb Perspect Biol 9(5). 10.1101/cshperspect.a025817 [DOI] [PMC free article] [PubMed]
- Basant A, Glotzer M. Spatiotemporal regulation of RhoA during cytokinesis. Curr Biol. 2018;28:R570–R580. doi: 10.1016/j.cub.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor superresolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Bettex-Galland M, Luscher EF. Extraction of an actomyosin-like protein from human platelets. Nature. 1959;184:276–277. doi: 10.1038/184276b0. [DOI] [PubMed] [Google Scholar]
- Borisy GG, Taylor EW. The mechanism of action of colchicine: binding of colchicine-3H to cellular protein. J Cell Biol. 1967;34:525–534. doi: 10.1083/jcb.34.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D, Bourret RB, Simon MI. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol Biol Cell. 1993;4:469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Weber K. Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J Cell Biol. 1980;86:335–340. doi: 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns RG, Pollard TD. A dynein-like protein from brain. FEBS Lett. 1974;40:274–280. doi: 10.1016/0014-5793(74)80243-7. [DOI] [PubMed] [Google Scholar]
- Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Castrillon D, Wasserman S. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Cleveland D, Hwo S, Kirschner M. Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. J Mol Biol. 1977;116:207–225. doi: 10.1016/0022-2836(77)90213-3. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Kirschner MW, Cowan NJ. Isolation of separate mRNAs for alpha- and beta-tubulin and characterization of the corresponding in vitro translation products. Cell. 1978;15:1021–1031. doi: 10.1016/0092-8674(78)90286-6. [DOI] [PubMed] [Google Scholar]
- CSHSQB (1972) “The mechanism of muscle contraction.” Cold Spring Harbor Symp. Quant. Biol, vol 37. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- DeLozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111:701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Euteneuer U, McIntosh JR. Structural polarity of kinetochore microtubules in PTK1-cells. J Cell Biol. 1981;89:338–345. doi: 10.1083/jcb.89.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson B, Lymn RW, Taylor EW. Studies on the kinetics of formation and dissociation of the actomyosin complex. Biochemistry. 1969;8(March 1969):811–819. doi: 10.1021/bi00831a008. [DOI] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Pollard TD. Fluorescent antibody localization of myosin in the cytoplasm, cleavage furrow, and mitotic spindle of human cells. J Cell Biol. 1976;71(3):848–875. doi: 10.1083/jcb.71.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Rowe AJ. Dynein: a protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- Goldman R, Pollard TD, Rosenbaum J. Cell motility. Cold Spring Harbor: Cold Spring Harb Press; 1976. [Google Scholar]
- Goodson HV, Spudich JA. Molecular evolution of the myosin family: relationships derived from comparisons of amino acid sequences. Proc Natl Acad Sci U S A. 1993;90:659–663. doi: 10.1073/pnas.90.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev. 2002;66(2):155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig JH, Stossel TP. Isolation and properties of actin, myosin and a new actin-binding protein in rabbit alveolar macrophages. J Biol Chem. 1975;250:5695–5705. [PubMed] [Google Scholar]
- Hatano S, Oosawa F. Isolation and characterization of plasmodium actin. Biochim Biophys Acta. 1966;127:488–498. doi: 10.1016/0304-4165(66)90402-8. [DOI] [PubMed] [Google Scholar]
- Hatano S, Tazawa M. Isolation, purification and characterization of myosin B from myxomycete plasmodium. Biochim Biophys Acta. 1968;154:507–519. doi: 10.1016/0005-2795(68)90011-1. [DOI] [PubMed] [Google Scholar]
- Hayashi I, Ikura M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1) J Biol Chem. 2003;278:36430–36434. doi: 10.1074/jbc.M305773200. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. Mechanism of muscular contraction. Science. 1969;164:1356–1366. [PubMed] [Google Scholar]
- Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- Inoué S. Video image processing greatly enhances contrast, quality, and speed in polarization-based microscopy. J Cell Biol. 1981;89:346–356. doi: 10.1083/jcb.89.2.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoué S, Sato H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J Gen Physiol. 1967;50(Suppl):259–292. [PMC free article] [PubMed] [Google Scholar]
- Isenberg GH, Aebi U, Pollard TD. An actin binding protein from Acanthamoeba regulates actin filament polymerization and interactions. Nature. 1980;288(5790):455–459. doi: 10.1038/288455a0. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Bischoff R, Holtzer H. Formation of arrowhead complexes with heavy meromyosin in a variety of cell types. J Cell Biol. 1969;43:312–328. [PMC free article] [PubMed] [Google Scholar]
- Koenig M, Kunkel LM. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem. 1990;265(8):4560–4566. [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114(1):33–38. [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E, Burridge K. Alpha-actinin: immunofluorescent localization of a muscle structural protein in nonmuscle cells. Cell. 1975;6:289–298. doi: 10.1016/0092-8674(75)90180-4. [DOI] [PubMed] [Google Scholar]
- Lazarides E, Weber K. Actin antibody: the specific visualization of actin filaments in non-muscle cells. Proc Natl Acad Sci U S A. 1974;71:2268–2272. doi: 10.1073/pnas.71.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey AG. An actomyosin-like substance from the plasmodium of a myxomycete. J Cell Comp Physiol. 1952;40:127–156. doi: 10.1002/jcp.1030400109. [DOI] [PubMed] [Google Scholar]
- Lymm RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by acto-myosin. Biochemist. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 1977;74(1):251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin agarose. J Cell Biol. 1994;127(1):107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi VT, Steers E., Jr Selective solubilization of a protein component of the red cell membrane. Science. 1968;159(811):203–204. doi: 10.1126/science.159.3811.203. [DOI] [PubMed] [Google Scholar]
- McIntosh JR (2016) Mitosis. Cold Spring Harb Perspect Biol 8(9). 10.1101/cshperspect.a023218 [DOI] [PMC free article] [PubMed]
- McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Moore PB, Huxley HE, DeRosier DJ. Three-dimensional reconstruction of F-actin, thin filaments and decorated thin filaments. J Mol Biol. 1970;50:279–295. doi: 10.1016/0022-2836(70)90192-0. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95(11):6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E, Maekawa S, Muneyuki E, Sakai H. Action of a 19K protein from porcine brain on actin polymerization - a new functional class of actin-binding proteins. J Biochem. 1984;95:387–398. doi: 10.1093/oxfordjournals.jbchem.a134619. [DOI] [PubMed] [Google Scholar]
- Ochs S. Fast axoplasmic transport of materials in mammalian nerve and its integrative role. Ann N Y Acad Sci. 1972;193:43–58. doi: 10.1111/j.1749-6632.1972.tb27822.x. [DOI] [PubMed] [Google Scholar]
- Otto JJ, Kane RE, Bryan J. Formation of filopodia in coelomocytes: localization of fascin, a 58,000 Dalton actin cross-linking protein. Cell. 1979;17:285–293. doi: 10.1016/0092-8674(79)90154-5. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Vallee RB. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 1987;330(6144):181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- Paul A, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskel. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Functional genomics of cell morphology using RNA interference: pick your style, broad or deep. J Biol. 2003;2(4):25. doi: 10.1186/1475-4924-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. No question about exciting questions in cell biology. PLoS Biol. 2013;11:e1001734. doi: 10.1371/journal.pbio.1001734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Nine unanswered questions about cytokinesis. J Cell Biol. 2017;216:3007–3016. doi: 10.1083/jcb.201612068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Ito S. Cytoplasmic filaments of Amoeba proteus. I. The role of filaments in consistency changes and movement. J Cell Biol. 1970;46(2):267–289. doi: 10.1083/jcb.46.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Korn ED. Acanthamoeba myosin. I. Isolation from Acanthamoeba castellanii of an enzyme similar to muscle myosin. J Biol Chem. 1973;248(13):4682–4690. [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerat CM, Rounds DE, Raiborn CW, Pollard TD. Observations on newborn rat dorsal root ganglia in vitro following gamma irradiation. In: Haley, Snider, editors. Response of the Nervous System to Ionizing Radiation. Boston: Little, Brown and Company; 1964. p. 175. [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297(5581):612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Safer D, Elzinga M, Nachmias VT. Thymosin beta 4 and Fx, an actin-sequestering peptide, are indistinguishable. J Biol Chem. 1991;266(7):4029–4032. [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4(8):626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Schroeder TE. Actin in dividing cells: contractile ring filaments bind heavy meromyosin. Proc Natl Acad Sci U S A. 1973;70(6):1688–1692. doi: 10.1073/pnas.70.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MR, Laplante C, Chin HF, Guirao B, Karatekin E, Pollard TD, O’Shaughnessy B. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev Cell. 2014;29:547–561. doi: 10.1016/j.devcel.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi A. Studies on muscle. Acta Physiol Scand. 1945;9(Suppl. 25):1–115. [Google Scholar]
- Thompson CM, Wolpert L. The isolation of motile cytoplasm from Amoeba proteus. Exp Cell Res. 1963;32:156–160. doi: 10.1016/0014-4827(63)90078-8. [DOI] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42(1):39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu J-Q, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107(4):1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Lanni F, McNeil PL, Ware BR, Taylor DL. Mobility of cytoplasmic and membrane-associated actin in living cells. Proc Natl Acad Sci U S A. 1982;79:4660–4664. doi: 10.1073/pnas.79.15.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberg R, Borisy GG, Taylor EW. The colchicine-binding protein of mammalian brain and its relation to microtubules. Biochemistry. 1968;7:4466–4479. doi: 10.1021/bi00852a043. [DOI] [PubMed] [Google Scholar]
- Weiss P. Neuronal dynamics and axonal flow. 3. Cellulifugal transport of labeled neuroplasm in isolated nerve preparations. Proc Natl Acad Sci U S A. 1967;57:1239–1245. doi: 10.1073/pnas.57.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Amos WB, Fordham M. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J Cell Biol. 1987;105:41–48. doi: 10.1083/jcb.105.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310(5746):310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Yin HL, Stossel TP. Control of cytoplasmic actin gel-sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature. 1979;281(5732):583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]