Abstract
The human genome codes for 21 S100 protein family members, which exhibit cell- and tissue-specific expression patterns. Despite sharing a high degree of sequence and structural similarity, the S100 proteins bind a diverse range of protein targets and contribute to a broad array of intracellular and extracellular functions. Consequently, the S100 proteins regulate multiple cellular processes such as proliferation, migration and/or invasion, and differentiation, and play important roles in a variety of cancers, autoimmune diseases, and chronic inflammatory disorders. This review focuses on the development of S100 neutralizing antibodies and small molecule inhibitors and their potential therapeutic use in controlling disease progression and severity.
Keywords: S100 protein, Calcium binding, Small molecule inhibitor, Neutralizing antibody
In humans, there are 21 S100 protein family members (Zimmer et al. 2012). The nomenclature for this family of small calcium-binding proteins derives from the observation that the two founding family members, S100A1 and S100B, are soluble in 100% saturated ammonium sulfate (Moore 1965). The majority of S100 genes cluster on the long arm of human chromosome 1 (S100A1–S100A14, S100A7s, and S100A16), with the remaining family members distributed on chromosomes 4 (S100P), 5 (S100Z), 21 (S100B), and the X chromosome (S100G) (Henry et al. 2012; Ravasi et al. 2004). Although the genomic loci encoding the S100 proteins are highly conserved in mammals, there are species differences (e.g., between humans and mice) that complicate the biological evaluation of several family members and their contribution to human disease. For example, the human S100A7 locus encodes three proteins (S100A7, S100A7A, and S100A7L2), whereas the mouse locus encodes a single protein (S100A7A) (Zimmer et al. 2012). Additionally, S100A12 and S100P are not expressed in mice.
S100 family members share a high degree of sequence and structural similarity, and typically form homodimers, with the exception of the S100A8/S100A9 heterodimer (Donato et al. 2013; Zimmer et al. 2012). Each S100 subunit is composed of four α-helices and contains two EF-hands (helix-loop-helix motifs that are Ca2+-binding domains): a C-terminal canonical EF-hand composed of 12 amino acids and an N-terminal S100 EF-hand composed of 14 amino acids that is unique to the S100 family (Kawasaki et al. 1998). The two EF-hands are connected by a loop or hinge region consisting of 12–14 amino acids, which exhibits the most sequence divergence within the family and is critical for interactions with target proteins (Marenholz et al. 2004). In the absence of a protein target, S100 proteins exhibit modest Ca2+-binding affinities that are well below intracellular calcium concentrations. However, Ca2+-binding affinities increase by 5–300-fold in the presence of peptide and protein targets (Malashkevich et al. 2008; Markowitz et al. 2005; Wright et al. 2009). This increase in affinity can be understood in terms of structural rearrangements, as Ca2+ binding induces a significant conformational reorganization that reorients helix 3 to expose a hydrophobic cleft required for target recognition (Fig. 1). Several studies suggest that in the absence of a protein target, Ca2+-bound S100 proteins sample a number of conformational states with predominantly weak Ca2+-binding affinities; target binding reduces dynamics throughout the protein and shifts the ensemble towards conformations with high Ca2+-binding affinities (Liriano et al. 2012; Palfy et al. 2016). As a consequence of this coupling, target binding is typically Ca2+-dependent. Despite the fact that Ca2+ binding induces a similar conformational reorganization in all S100 family members examined to date, structural studies of S100-target complexes have shown that S100 family members utilize distinct mechanisms for target recognition (Bhattacharya et al. 2003; Dempsey et al. 2012; Kiss et al. 2012; Lee et al. 2008; Oh et al. 2013; Ozorowski et al. 2013; Rety et al. 2000; Rety et al. 1999; Rustandi et al. 2000; Wright et al. 2009). The distribution of hydrophobic and charged residues, as well as differences in surface geometries, all contribute to the range of target binding modes observed amongst S100 family members (Ozorowski et al. 2013; Ramagopal et al. 2013; Wafer et al. 2013). The growing number of S100-target structures has provided important insights into the chemical and physical determinants controlling target selectivity, which can be exploited for the development of selective S100 therapeutics. This review focuses on the development of S100 protein small molecule inhibitors, as well as more recent efforts on biologics that specifically target S100 proteins in the extracellular milieu.
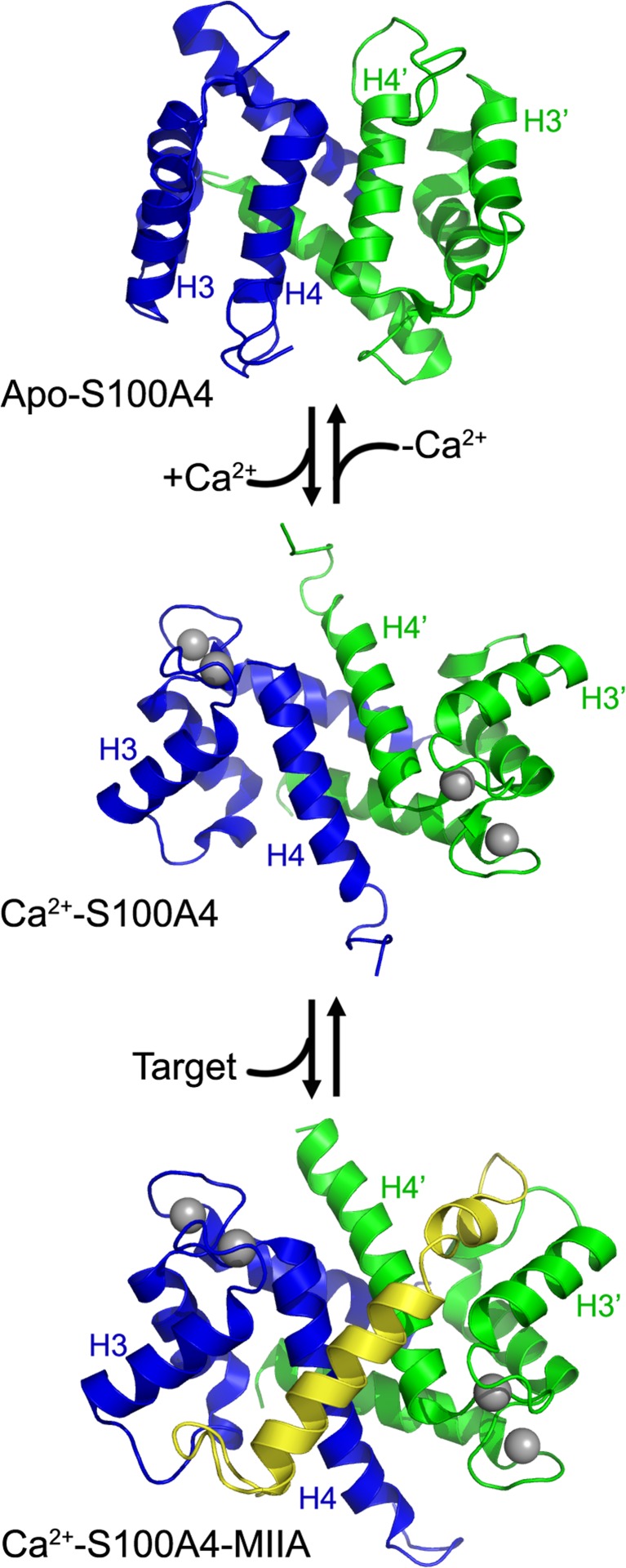
Fig. 1.
S100 protein organization. Ribbon diagrams of apo-S100A4 (PDB 1M31), Ca2+-S100A4 (PDB 2Q91), and the Ca2+S100A4myosin-IIA (MIIA) peptide complex (PDB 3ZHW). The individual S100A4 subunits are shown in blue and green, the Ca2+ ions are shown as gray spheres, and the myosin-IIA peptide is in yellow. Ca2+ binding induces a significant conformational reorganization that reorients helix 3 to expose a hydrophobic cleft that is required for target binding
Intracellular and extracellular functions
The diversity of the S100 proteins enables cells to selectively respond to changes in intracellular Ca2+ levels. The S100 proteins are expressed in a cell- and tissue-specific manner in vertebrates (Donato 2003) and have non-redundant roles in a wide range of biological processes such as proliferation, migration and/or invasion, and differentiation. S100 proteins regulate the activity of numerous intracellular protein targets, and some targets are regulated by multiple S100 family members (Donato et al. 2013; Hermann et al. 2012; Liu et al. 2015). The Ca2+-dependent regulation of these interactions enables S100 proteins to function as calcium sensors that transduce changes in intracellular calcium concentrations into biochemical and biological responses. There are a number of well-characterized S100-target protein interactions, including S100B and p53 (Bresnick et al. 2015), S100A4 and nonmuscle myosin-IIA (Dulyaninova and Bresnick 2013), and S100A10 and annexin A2 (Liu et al. 2015). However, the complete repertoire of intracellular protein targets and corresponding cellular functions are not well described for the majority of S100 proteins.
S100s proteins lack signal sequences and are typically considered cytoplasmic proteins. Nonetheless, several family members are secreted via nonclassical pathways and/or are released by cells to function as extracellular factors (Donato et al. 2013; Marenholz et al. 2004; Ryckman et al. 2003; Yan et al. 2008). Extracellular S100 proteins have been reported to bind several cell surface receptors, including the advanced glycosylation end product-specific receptor (also known as RAGE) (Koch et al. 2010; Park et al. 2010; Penumutchu et al. 2014; Xie et al. 2007; Yatime et al. 2016), TLR4 (Ehrchen et al. 2009), CD36 (Tondera et al. 2017), FGFR1 (Riuzzi et al. 2011), CD166 antigen (also known as ALCAM) (von Bauer et al. 2013), the interleukin-10 receptor (Dmytriyeva et al. 2012), EMMPRIN (also known as cell surface glycoprotein extracellular matrix metalloproteinase inducer) (Hibino et al. 2013), neuroplastin-β (Sakaguchi et al. 2016), CD68 (Okada et al. 2016) and ErbB4 (Pankratova et al. 2018). Some S100 proteins, such as S100B and S100A12, are reported to bind multiple cell surface receptors (Koch et al. 2010; Riuzzi et al. 2011; Tondera et al. 2017; von Bauer et al. 2013; Xie et al. 2007). Despite the identification of potential cell surface receptors for several family members, for most S100 proteins, the biochemical mechanisms mediating these S100-receptor interactions and the downstream consequences of S100 signaling are not known.
Specificity and regulation of S100-receptor interactions
Multiple S100 proteins bind TLR4 (Cerezo et al. 2014; Foell et al. 2013; Vogl et al. 2007) and RAGE (Leclerc et al. 2009). The ectodomain of RAGE is composed of three immunoglobulin domains (V, C1, and C2), and several S100 proteins have been reported to bind each domain, suggesting that these S100 proteins may have overlapping binding sites (Leclerc and Heizmann 2011). For example, S100B, S100A1, S100A2, S100A5, and S100A6 all bind the V domain (Leclerc et al. 2009; Ostendorp et al. 2007; Yatime et al. 2016). Given that multiple extracellular S100 proteins are typically associated with specific pathologies (e.g., elevated S100A8/S100A9, S100A4, and S100B in the serum of rheumatoid arthritis patients) (Austermann et al. 2018; Bresnick et al. 2015), this raises the question as to whether distinct S100 proteins can elicit differential signaling responses via interactions with the same cell surface receptors.
Recent studies with S100A8/S100A9 suggest that oligomerization can locally restrict S100 protein activity. Biochemical and cellular studies indicate that extracellular S100A8/S1009 elicits many of its effects via interactions with TLR4 (Cheng et al. 2008; Vogl et al. 2007) and RAGE (Bjork et al. 2009; Ghavami et al. 2008; Turovskaya et al. 2008). While the S100A8/S100A9 heterodimer can bind TLR4, the higher calcium ion concentrations found in the extracellular milieu (in the range of 2–3 mM (Brini et al. 2013; Goldstein 1990)) induces the formation of S100A8/S100A9 tetramers. This masks the TLR4 binding interface on the S100A8/S100A9 heterodimer, providing a mechanism for modulating S100 biological activity (Vogl et al. 2018). In contrast, S100A8 or S100A9 homodimers, which also bind TLR4, do not form tetramers (Vogl et al. 2006). Thus, this autoinhibitory mechanism allows for selective regulation of S100A8/S100A9 heterodimer activity. Oligomerization may also contribute to the regulation of other S100 protein-receptor interactions. For example, in the presence of calcium, S100B forms stable tetramers that bind RAGE with higher affinity than the S100B dimer (Ostendorp et al. 2007). Similarly, calcium and zinc induce the formation of S100A12 hexamers, which are required for RAGE and TLR4 binding (Kessel et al. 2018; Moroz et al. 2009). In addition, S100 oligomers are reported to bind different multimeric states of RAGE (e.g., S100A12 hexamers bind RAGE tetramers and S100B dimers bind RAGE dimers) (Xie et al. 2007; Xue et al. 2016). Together, these data suggest that S100 protein oligomerization is an important mechanism for regulating the functional diversity of this family of proteins.
In addition to oligomerization, covalent modification may also regulate the extracellular functions of S100 proteins. An intramolecular disulfide bond modulates the antimicrobial activity of S100A7 (Cunden et al. 2017) and transglutaminase 2-mediated crosslinking of S100A11 dimers is required for signaling via the p38 MAPK pathway in chondrocytes (Cecil and Terkeltaub 2008). Other types of post-translation modifications may also regulate oligomerization and/or activity. For example, citrullination promotes the formation of a S100A3 homotetramer (Kizawa et al. 2008), and a number of S100 proteins are S-nitrosylated, including S100B (Bajor et al. 2016), S100A1 (Lenarcic Zivkovic et al. 2012) and S100A8/A9 (Lim et al. 2011). Other types of oxidative modification such as S-glutathionylation, cysteinylation and the formation of intra- and intersulfinamide bonds have also been observed in S100 proteins (Lim et al. 2011; Orre et al. 2007). Additionally, sumoylation and phosphorylation of S100 proteins have been reported (Miranda et al. 2010; Sakaguchi et al. 2004; Schenten et al. 2018). Whether S100 proteins with post-translational modifications are released into the extracellular environment and how these modifications modulate S100 structure or function has largely not been determined.
S100 neutralizing antibodies
During both local and systemic inflammation, tissue and serum levels of several S100 proteins correlate with disease severity (Donato et al. 2013; Kessel et al. 2013; Nefla et al. 2016). In addition, extracellular S100 proteins can function as damage-associated molecular pattern (DAMP) proteins, thereby triggering proinflammatory responses via binding to pattern recognition receptors expressed on epithelial cells and innate and adaptive immune cells. This can induce autoimmune conditions and inflammatory disorders (Donato et al. 2013; Foell et al. 2007; Nefla et al. 2016; Xia et al. 2017; Zackular et al. 2015). Function-blocking antibodies targeting cell surface receptors and ligands are major classes of protein therapeutics for the treatment of cancers and immune disorders (Brufsky 2010; Mansh 2011; Saif 2013; Scott et al. 2012). Given the substantial literature showing that extracellular S100 proteins mediate inflammatory responses in cancer and autoimmune and chronic inflammatory diseases (Austermann et al. 2018; Bresnick et al. 2015; Grigorian et al. 2008), S100 neutralizing antibodies may provide a novel therapeutic strategy. To date, antibodies targeting S100A8/A9, S100A4, S100A7 (Padilla et al. 2017), and S100P (Dakhel et al. 2014) have demonstrated efficacy for a number of pathological conditions. Since antibodies targeting S100A8/A9 and S100A4 have been examined in the most detail, our discussion will focus on studies examining the biological activity of these antibodies.
S100A8/A9
The S100A8/A9 heterodimers are the best characterized S100 family members with respect to extracellular functions. Extracellular S100A8/S100A9 is strongly associated with inflammatory and autoimmune diseases, including rheumatoid arthritis, spondyloarthritis, systemic sclerosis, and systemic lupus erythematosus (Austermann et al. 2018). Function-blocking S100A9 antibodies inhibit dextran sulfate sodium (DSS)-induced acute colitis and attenuate azoxymethane/DSS-induced colitis-associated cancer (Zhang et al. 2017b), reduce neutrophilic inflammation and airway reactivity in a murine asthma model (Lee et al. 2017), and diminish immune cell infiltration and preserve bone/collagen in a model of rheumatoid arthritis (Cesaro et al. 2012).
In solid cancers, elevated S100A8/A9 expression within the tumor microenvironment or in plasma correlates with aggressive disease (Cheng et al. 2008; Hauschild et al. 1999; Laouedj et al. 2017; Miller et al. 2017; Tidehag et al. 2014). In particular, extracellular S100A8/A9 plays an important role in the recruitment of myeloid cells and myeloid-derived suppressor cells (MDSCs), which promote tumor growth and the establishment of the pre-metastatic niche (Acharyya et al. 2012; Cheng et al. 2008; Hiratsuka et al. 2006; Ichikawa et al. 2011). Tumor-derived TGFβ and VEGFA upregulate the expression and secretion of S100A8/A9 in lung-associated myeloid and endothelial cells (Hiratsuka et al. 2006). S100A8/A9 induces the expression of serum amyloid 3, which in turn recruits CD11b+ myeloid cells to pre-metastatic sites (Hiratsuka et al. 2008). This process produces a proinflammatory environment that recruits circulating tumor cells (CTCs) to the lung; S100A8 and S100A9 neutralizing antibodies block the recruitment of both myeloid cells and CTCs (Hiratsuka et al. 2006; Hiratsuka et al. 2008). In acute myeloid leukemia (AML), S100A8 antibodies, but not S100A9 antibodies, induce AML cell differentiation, reduce leukemic burden and increase survival (Laouedj et al. 2017). In addition, peptibodies, peptide-Fc fusion proteins that target S100A8 and S100A9, reduce tumor burden in multiple cancer models (Qin et al. 2014). Lastly, in murine models of breast cancer, S100A9 antibodies have been used in conjunction with single-photon emission computed tomography (SPECT) for the in vivo detection of S100A8/A9 as a marker for the establishment of the pre-metastatic niche (Becker et al. 2015; Eisenblaetter et al. 2017). Together, these studies highlight the potential use of S100A8 and S100A9 antibodies as both therapeutic and diagnostic reagents.
S100A4
S100A4 has a direct and causative role in tumor metastasis (Bresnick et al. 2015). In animal models of breast and other cancers, S100A4 overexpression in tumor cells promotes an aggressive metastatic phenotype, while inhibition of S100A4 expression significantly reduces metastatic burden (Ambartsumian et al. 1996; Davies et al. 1993; Davies et al. 1996; Grigorian et al. 1996; Maelandsmo et al. 1996; Takenaga et al. 1997; Xue et al. 2003). S100A4 is also expressed in normal cells and tissues, including fibroblasts, lymphocytes, macrophages, osteoclasts, and other bone marrow-derived cells (Bruhn et al. 2014; Erlandsson et al. 2013; Hashimoto et al. 2013; Li et al. 2010; Takenaga et al. 1994). Stromal S100A4, and in particular extracellular S100A4, is thought to promote tumor metastasis by stimulating an inflammatory, pro-tumorigenic environment (Bettum et al. 2014; Grum-Schwensen et al. 2005; Hansen et al. 2015; O'Connell et al., 2011). Consistent with a role in mediating inflammatory responses, extracellular S100A4 is associated with the pathogenesis of several autoimmune and chronic inflammatory diseases such as osteoarthritis (Amin and Islam 2014; Yammani et al. 2009), rheumatoid arthritis (Klingelhofer et al. 2007; Oslejskova et al. 2009), psoriasis (Zibert et al. 2010), Crohn’s disease (Cunningham et al. 2010), bacterial colitis (Zhang et al. 2017a), and fibrosis (Chen et al. 2015). S100A4 blocking monoclonal antibodies have been shown to limit tumor metastasis and T cell recruitment in syngeneic mouse models of breast cancer (Grum-Schwensen et al. 2015; Klingelhofer et al. 2012), to inhibit the growth of pancreatic tumors in immunocompromised mice (Hernandez et al. 2013), to decrease azoxymethane/DSS-induced colon inflammation and tumorigenesis (Zhang et al. 2018), and to reduce epidermal thickness in a mouse model of human psoriasis (Zibert et al. 2010). While these studies support a role for extracellular S100A4 in promoting an inflammatory phenotype, the receptors responsible for S100A4-mediated inflammatory responses are not well characterized.
In vitro, biochemical studies support the interaction of S100A4 with RAGE and TLR4 (Bjork et al. 2013; Leclerc et al. 2009). While extracellular S100A4 is reported to stimulate downstream signaling events in a number of systems, the role of RAGE in mediating cellular responses to S100A4 is controversial (Grotterod et al. 2010). Most notably, a number of studies on extracellular S100A4 and associated downstream signaling events have reported that the bioactive form of S100A4 is an oligomer of a higher order than the canonical dimer (Cerezo et al. 2014; Forst et al. 2010; Novitskaya et al. 2000). However, these studies used a His-tagged S100A4, which forms large multimers (~ 200 kDa) (Novitskaya et al. 2000). It is well established that histidine tags can affect the oligomeric states and functions of proteins (Amor-Mahjoub et al. 2006; Majorek et al. 2014; Sprules et al. 1998). Indeed, biophysical studies with untagged S100A4 in the presence of calcium have reported the formation of only S100A4 dimers and tetramers, but not higher order oligomers (House et al. 2011; Malashkevich et al. 2008; Streicher et al. 2010). The high molecular weight S100A4 species detected in the plasma of cancer patients or synovial fluid from osteoarthritis and rheumatoid arthritis patients may represent higher order S100A4 oligomers, but could also represent S100A4 dimers or tetramers bound to target proteins present in the extracellular milieu (Ambartsumian et al. 1996; Klingelhofer et al. 2007). These observations underscore the need to rigorously validate biochemical reagents and highlight the need to re-evaluate the biological functions of extracellular S100A4 using untagged S100A4.
Small molecule inhibition of S100 proteins
Given the roles of S100 proteins in proinflammatory processes in human disease, strategies for the pharmacological modulation of S100 protein function have received considerable attention. One approach is the inhibition of S100 gene transcription (Gao et al. 2018; Sack et al. 2011; Stein et al. 2011). Transcription of S100A4 is directly mediated by the β-catenin/TCF complex (Stein et al. 2006), and compounds that induce β-catenin degradation and/or block the formation of the β-catenin/TCF complex (e.g., calcimycin—a calcium ionophore; niclosamide—an antihelminth drug; and sulindac—a nonsteroidal anti-inflammatory drug) inhibit S100A4 transcription (Dahlmann et al. 2016; Sack et al. 2011; Stein et al. 2011). In addition, duloxetine, a serotonin-norepinephrine reuptake inhibitor, was identified recently as a S100B transcriptional inhibitor (Gao et al. 2018). However, the effectiveness of this general strategy may be limited by the long half-life of S100 proteins, as a study in NIH3T3 cells showed that several S100 proteins have half-lives on the order of 90–140 h (Schwanhausser et al. 2011). Such long half-lives can make it difficult to sufficiently reduce protein levels to achieve a therapeutic response. In addition, these transcriptional inhibitors are known to affect the expression of multiple gene targets, which could cause significant toxicities (Dahlmann et al. 2016). Despite these potential limitations, both S100A4 and S100B transcriptional regulators have exhibited efficacy in a number of cancer models (Dahlmann et al. 2016; Gao et al. 2018; Stewart et al. 2016). Moreover, niclosamide, an FDA-approved drug, is currently under evaluation for safety and efficacy in a phase II clinical trial for patients with metastatic colorectal cancer whose disease has progressed under previous therapy (Burock et al. 2018).
Most protein-protein interactions are typified by large interfaces composed of relatively “flat” featureless surfaces that are difficult to disrupt with small molecules. However, the target binding clefts of S100 proteins, which are exposed upon Ca2+ binding, can readily bind small molecules. As a consequence, there has been significant success in the identification of small molecules that block S100-target protein interactions. Several anti-allergy drugs such as cromolyn, amlexanox, tranilast, and olopatadine are reported to bind multiple S100 proteins (Fig. 2) (Mack and Marshall 2010; Okada et al. 2002; Rani et al. 2010; Shishibori et al. 1999). Cromolyn, which blocks the coimmunoprecipitation of S100P with RAGE, also attenuates the growth of pancreatic tumors and sensitizes tumors cells to gemcitabine, a chemotherapeutic agent (Arumugam et al. 2013; Kim et al. 2012). Amlexanox is a S100A13 antagonist that blocks interactions with fibroblast growth factor 1 (FGF1), and inhibits the release of the S100A13-FGF1 complex in vivo (Mouta Carreira et al. 1998). In addition, amlexanox sensitizes MLL/AF4-positive acute lymphoblastic leukemia to TNFα treatment via the downregulation of S100A6 expression through an unknown mechanism (Tamai et al. 2017). While these anti-allergic compounds exhibit promising effects on S100-mediated pathologies they are not selective S100 inhibitors. For example, amlexanox also inhibits IκB kinase ε and TANK-binding kinase 1, proteins that promote a proinflammatory response associated with the development of obesity (Beyett et al. 2018; Reilly et al. 2013). These observations suggest that the anti-inflammatory responses observed with amlexanox and other anti-allergics are likely due to the modulation of multiple cellular pathways.
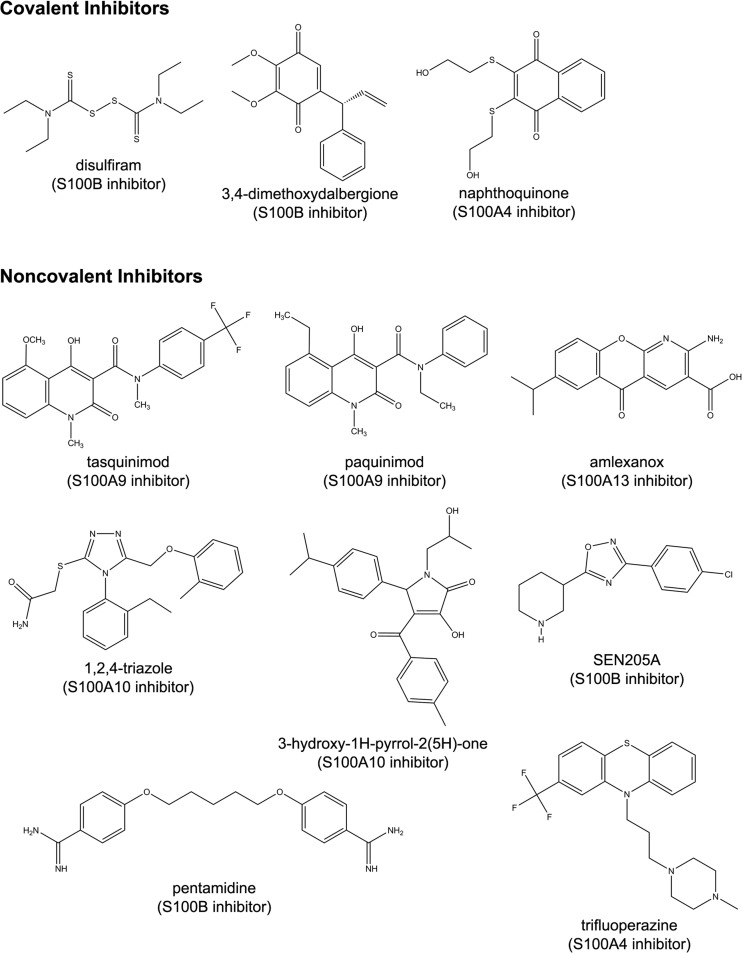
Fig. 2.
Chemical structures of S100 inhibitors
Phenothiazines, a class of anti-psychotic compounds, also interact with multiple S100 family members (Garrett et al. 2008; Marshak et al. 1985; Pingerelli et al. 1990; Wilder et al. 2010) as well as other EF-hand-containing proteins such as troponin C and calmodulin (Fig. 2) (Cook et al. 1994; Feldkamp et al. 2015; Vandonselaar et al. 1994; Vertessy et al. 1998). Structural studies of the S100A4-trifluoperazine (TFP) complex demonstrate that two TFP molecules reside in the target binding cleft of each S100A4 subunit, and that TFP binding induces the assembly of Ca2+-S100A4-TFP dimers into a five-fold symmetric pentameric ring (Malashkevich et al. 2010). Prochlorperazine (PCP) also induces the formation of a pentameric ring of Ca2+-S100A4 dimers. Phenothiazine-mediated oligomerization may be unique to S100A4, as chlorpromazine does not induce the formation of higher order S100B oligomers (Wilder et al. 2010). Notably, the architectures of the TFP-binding pockets, the number of bound TFP molecules, and the orientation of the TFP molecules are quite different between S100A4, troponin C, and calmodulin (Cook et al. 1994; Feldkamp et al. 2015; Malashkevich et al. 2010; Vandonselaar et al. 1994; Vertessy et al. 1998). Given the differences in TFP-binding modes and the large number of phenothiazine derivatives that are available, it may be possible to selectively target these proteins with appropriate phenothiazine analogs (Brem et al. 2017; Montoya et al. 2018; Pluta et al. 2017).
Other examples of small molecule S100 inhibitors include covalent inhibitors that modify cysteine residues in helix 4 of S100B and S100A4. Despite the proximity of these cysteines to the C-terminal EF-hand, their modification does not affect Ca2+ binding, but does disrupt Zn2+-mediated conformational rearrangements in S100B, and target binding to both S100A4 and S100B (Cavalier et al. 2014; Dulyaninova et al. 2011). While these compounds exhibit efficacy in disrupting S100-target interactions in vitro, selectivity is an issue. The covalent S100A4 and S100B inhibitor, 2,3-bis[2-hydroxyethylsulfanyl]-1,4-naphthoquinone, also inhibits the activities of multiple protein tyrosine phosphatases through the modification of an active site cysteine (Brisson et al. 2005; Vogt et al. 2008). Similarly, other covalent S100B inhibitors are reported to have a number of targets, including transglutaminase 2 (Palanski and Khosla 2018), alcohol dehydrogenase (Koppaka et al. 2012), and protein kinase C (Herbert et al. 1990). Nonetheless, these compounds represent new chemical scaffolds for the development of S100 inhibitors with improved affinity and specificity.
In addition to covalent S100 inhibitors, non-covalent inhibitors include paquinimod (ABR-215757) and tasquinimod (ABR-215050), quinoline-3-carboxamide derivatives that disrupt the interaction of S100A8/S100A9 with TLR4 and RAGE (Bjork et al. 2009; Kallberg et al. 2012); 4-aroyl-3-hydroxy-5-phenyl-1H-pyrrol-2(5H)-one analogs and substituted 1,2,4-triazoles that inhibit the interaction between S100A10 and annexin A2 (Reddy et al. 2012; Reddy et al. 2011); pentamidine, which blocks the interaction of S100B with p53 (Charpentier et al. 2008; Markowitz et al. 2004), and SEN205A and distamycin A, which target the S100B-p53 interaction site (Agamennone et al. 2010; Cerofolini et al. 2015) (Fig. 2). While these compounds have been evaluated for their ability to disrupt specific S100 protein-target interactions, it is unknown if they can inhibit the binding of all ligands for a particular S100 protein.
Although current efforts are focused on improving the affinity, selectivity, and biological half-life of these S100 inhibitors, a number of these compounds have been evaluated in murine models of disease and some have advanced to human clinical trials. Paquinimod reduces inflammation and disease progression and/or severity in a number of inflammatory models (Fransen Pettersson et al. 2018; Tahvili et al. 2018; Wache et al. 2015). Tasquinimod inhibits tumor growth and metastasis in several models of prostate cancer, possibly by limiting the recruitment of MDSCs and tumor-associated macrophages to the tumor microenvironment (Raymond et al. 2014). However, tasquinimod has also been reported to be a potent negative allosteric regulator of HDAC4; inhibition of HDAC4-mediated deacetylation of HIF-1α and other factors compromises cancer cell survival and tumor angiogenesis (Isaacs et al. 2013). Regardless of whether tasquinimod acts through S100A9, HDAC4, or both, tasquinimod improves progression-free survival in patients with metastatic castration-resistant prostate cancer (Fizazi et al. 2017; Pili et al. 2011), but does not exhibit clinical efficacy in heavily pre-treated patients with advanced hepatocellular, ovarian, renal cell, and gastric cancers (Escudier et al. 2017). S100B is overexpressed in cultured melanoma cells and is a strong biomarker for melanoma (Gaynor et al. 1980; Hauschild et al. 1999). Pentamidine, an FDA-approved anti-parasitic that targets S100B and disrupts its interaction with p53 (Markowitz et al. 2004), exhibits efficacy against ex vivo melanoma samples (Smith et al. 2010). Pentamidine is under evaluation in patients with relapsed or refractory melanoma and in patients with solid tumors, including pancreatic, colon, and hepatocellular cancers (www.clinicaltrials.gov, NCT00810953, NCT00809796, NCT02210182).
Within the S100 family, S100B has been most thoroughly studied with respect to small molecule inhibitors. An examination of S100B-inhibitor complexes has revealed three discrete pockets that accommodate small molecules (Fig. 3) (Cavalier et al. 2014; Hartman et al. 2013). Site 1 is the target binding site (e.g., TRTK peptide or p53 C-terminal peptide) and involves interactions with residues from the hinge region and helices 3 and 4. SEN205A is an example of a site 1 inhibitor (Agamennone et al. 2010). Site 2 interactions involve residues from the hinge and helix 4, and site 3 interactions utilize residues from the C-terminal loop and helix 1. Examples of inhibitors that occupy these sites include compounds that covalently modify Cys84 of S100B (site 2) (Cavalier et al. 2014), and amlexanox and chlorpromazine, which bind site 3 in S100A13 and S100B, respectively (Rani et al. 2010; Wilder et al. 2010). For the S100B-pentamidine and S100A4-TFP complexes, in which two inhibitor molecules are bound per S100 subunit, both sites 2 and 3 are occupied (Charpentier et al. 2008; Malashkevich et al. 2010). Despite minimal overlap with the S100B target–binding cleft, pentamidine disrupts p53 binding, suggesting that inhibition occurs via allosteric effects (Markowitz et al. 2004). Altogether, these data demonstrate that the binding of small molecules at different sites within a given S100 subunit allows for multiple mechanisms for inhibition, including small molecule–mediated S100 oligomerization (S100A4-TFP and S100A4-PCP), competitive inhibition with protein targets (S100A13-amlexanox; S100B-SEN205A) and allosteric regulation (S100B-pentamidine) (Charpentier et al. 2008; Malashkevich et al. 2010) (Agamennone et al. 2010; Rani et al. 2010). Moreover, the unique surface geometries and chemical features of each S100 family member should readily allow selective targeting of these proteins.
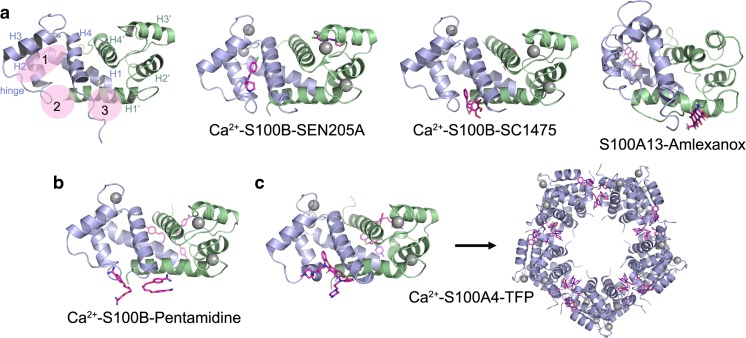
Fig. 3.
S100 protein-inhibitor complexes. a Ribbon diagram of Ca2+-S100B showing the general locations of the three binding sites that can accommodate small molecules and representative structures showing small molecules bound to each site. The individual S100 subunits are shown in light blue and green, the Ca2+ ions are shown as gray spheres and the inhibitors as pink sticks. Site 1: S100B-SEN205A (PDB 3HCM)—involves residues from the hinge and helices 2 and 3. Site 2: S100B-SC1475 (3,4-dimethoxydalbergione) (PDB 4PE4)—involves residues from the hinge and helix 4. Site 3: S100A13-amlexanox (PDB 2KOT)—involves residues from the C-terminal loop and helix 1. b Structures of Ca2+–S100B-pentamidine (PDB 3CR4) and c Ca2+–S100A4-trifluoperazine (TFP) (PDB 3KO0) showing two inhibitor molecules bound per S100 subunit and occupation of both sites 2 and 3. TFP binding induces the assembly of five Ca2+–S100A4-TFP dimers into a pentameric ring
Conclusion
Significant advances have been made in understanding the intracellular and extracellular functions of S100 proteins and their roles in modulating proinflammatory and other responses that contribute to the development and progression of cancer and autoimmune and chronic inflammatory diseases. Despite this progress, a detailed understanding of the cell surface receptors that mediate extracellular S100 signaling is lacking. Furthermore, we do not fully understand the dynamics and regulation of S100 protein secretion, or the role of oligomerization and post-translational modifications in the regulation of intracellular/extracellular S100 activity. The continued development of antibodies and small molecule inhibitors will be important for attributing specific biological activities to particular S100 proteins and for defining the contribution of intracellular and extracellular S100 activities in biological processes. Furthermore, these reagents may have potential therapeutic applications for a number of cancers and immune disorders. S100 protein biology continues to provide a rich area of investigation and the evaluation of the cell biological and biochemical functions of these proteins will provide new insights into human disease.
Acknowledgements
We thank Drs. SC Almo and JM Backer (Albert Einstein College of Medicine) for helpful discussions and for reading the manuscript. This work was supported by National Institutes of Health grants P01 CA100324 and R01 GM119279.
Conflict of interest
Anne R. Bresnick declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
References
- Acharyya S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agamennone M, et al. Fragmenting the S100B-p53 interaction: combined virtual/biophysical screening approaches to identify ligands. ChemMedChem. 2010;5:428–435. doi: 10.1002/cmdc.200900393. [DOI] [PubMed] [Google Scholar]
- Ambartsumian NS, et al. Metastasis of mammary carcinomas in GRS/A hybrid mice transgenic for the mts1 gene. Oncogene. 1996;13:1621–1630. [PubMed] [Google Scholar]
- Amin AR, Islam AB. Genomic analysis and differential expression of HMG and S100A family in human arthritis: upregulated expression of chemokines, IL-8 and nitric oxide by HMGB1 DNA. Cell Biol. 2014;33:550–565. doi: 10.1089/dna.2013.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor-Mahjoub M, Suppini JP, Gomez-Vrielyunck N, Ladjimi M. The effect of the hexahistidine-tag in the oligomerization of HSC70 constructs. J Chromatogr B Anal Technol Biomed Life Sci. 2006;844:328–334. doi: 10.1016/j.jchromb.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Arumugam T, Ramachandran V, Maxwell D, Bornmann WG, Logsdon CD. Designing and developing S100P inhibitor 5-methyl cromolyn (C5OH) for pancreatic cancer therapy. Mol Cancer Ther. 2013;12:654–662. doi: 10.1158/1535-7163.MCT-12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austermann J, Spiekermann C, Roth J. S100 proteins in rheumatic diseases. Nat Rev Rheumatol. 2018;14:528–541. doi: 10.1038/s41584-018-0058-9. [DOI] [PubMed] [Google Scholar]
- Bajor M, Zareba-Koziol M, Zhukova L, Goryca K, Poznanski J, Wyslouch-Cieszynska A. An interplay of S-nitrosylation and metal ion binding for astrocytic S100B. Protein PloS one. 2016;11:e0154822. doi: 10.1371/journal.pone.0154822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, et al. Optical in vivo imaging of the alarmin S100A9 in tumor lesions allows for estimation of the individual malignant potential by evaluation of tumor-host cell interaction. J Nucl Med. 2015;56:450–456. doi: 10.2967/jnumed.114.146688. [DOI] [PubMed] [Google Scholar]
- Bettum IJ, et al. Metastasis-associated protein S100A4 induces a network of inflammatory cytokines that activate stromal cells to acquire pro-tumorigenic properties. Cancer Lett. 2014;344:28–39. doi: 10.1016/j.canlet.2013.10.036. [DOI] [PubMed] [Google Scholar]
- Beyett TS, et al. Carboxylic acid derivatives of amlexanox display enhanced potency toward TBK1 and IKKepsilon and reveal mechanisms for selective inhibition. Mol Pharmacol. 2018;94:1210–1219. doi: 10.1124/mol.118.112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- Bjork P, et al. Identification of human S100A9 as a novel target for treatment of autoimmune disease via binding to quinoline-3-carboxamides. PLoS Biol. 2009;7:e97. doi: 10.1371/journal.pbio.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork P, et al. Common interactions between S100A4 and S100A9 defined by a novel chemical probe. PLoS One. 2013;8:e63012. doi: 10.1371/journal.pone.0063012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem B et al. (2017) Novel thiazolo[5,4-b]phenothiazine derivatives: synthesis, structural characterization, and in vitro evaluation of antiproliferative activity against human leukaemia. Int J Mol Sci 18 doi:10.3390/ijms18071365 [DOI] [PMC free article] [PubMed]
- Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer nature reviews. Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini M, Ottolini D, Cali T, Carafoli E. Calcium in health and disease. Met Ions Life Sci. 2013;13:81–137. doi: 10.1007/978-94-007-7500-8_4. [DOI] [PubMed] [Google Scholar]
- Brisson M, et al. Redox regulation of Cdc25B by cell-active quinolinediones. Mol Pharmacol. 2005;68:1810–1820. doi: 10.1124/mol.105.016360. [DOI] [PubMed] [Google Scholar]
- Brufsky A. Trastuzumab-based therapy for patients with HER2-positive breast cancer: from early scientific development to foundation of care. Am J Clin Oncol. 2010;33:186–195. doi: 10.1097/COC.0b013e318191bfb0. [DOI] [PubMed] [Google Scholar]
- Bruhn S, et al. A generally applicable translational strategy identifies S100A4 as a candidate gene in allergy. Sci Transl Med. 2014;6:218ra214. doi: 10.1126/scitranslmed.3007410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burock S, Daum S, Keilholz U, Neumann K, Walther W, Stein U. Phase II trial to investigate the safety and efficacy of orally applied niclosamide in patients with metachronous or sychronous metastases of a colorectal cancer progressing after therapy: the NIKOLO tria. BMC Cancer. 2018;18:297. doi: 10.1186/s12885-018-4197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier MC, et al. Covalent small molecule inhibitors of ca(2+)-bound S100B. Biochemistry. 2014;53:6628–6640. doi: 10.1021/bi5005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil DL, Terkeltaub R. Transamidation by transglutaminase 2 transforms S100A11 calgranulin into a procatabolic cytokine for chondrocytes. J Immunol. 2008;180:8378–8385. doi: 10.4049/jimmunol.180.12.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo LA, et al. The metastasis-associated protein S100A4 promotes the inflammatory response of mononuclear cells via the TLR4 signalling pathway in rheumatoid arthritis. Rheumatology (Oxford) 2014;53:1520–1526. doi: 10.1093/rheumatology/keu031. [DOI] [PubMed] [Google Scholar]
- Cerofolini L, Amato J, Borsi V, Pagano B, Randazzo A, Fragai M. Probing the interaction of distamycin A with S100beta: the “unexpected” ability of S100beta to bind to DNA-binding ligands. J Mol Recognit. 2015;28:376–384. doi: 10.1002/jmr.2452. [DOI] [PubMed] [Google Scholar]
- Cesaro A, Anceriz N, Plante A, Page N, Tardif MR, Tessier PA. An inflammation loop orchestrated by S100A9 and calprotectin is critical for development of arthritis. PLoS One. 2012;7:e45478. doi: 10.1371/journal.pone.0045478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier TH, et al. Divalent metal ion complexes of S100B in the absence and presence of pentamidine. J Mol Biol. 2008;382:56–73. doi: 10.1016/j.jmb.2008.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, et al. S100A4 promotes liver fibrosis via activation of hepatic stellate cells. J Hepatol. 2015;62:156–164. doi: 10.1016/j.jhep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook WJ, Walter LJ, Walter MR. Drug binding by calmodulin: crystal structure of a calmodulin-trifluoperazine complex. Biochemistry. 1994;33:15259–15265. doi: 10.1021/bi00255a006. [DOI] [PubMed] [Google Scholar]
- Cunden LS, Brophy MB, Rodriguez GE, Flaxman HA, Nolan EM. Biochemical and functional evaluation of the intramolecular disulfide bonds in the zinc-chelating antimicrobial protein human S100A7 (Psoriasin) Biochemistry. 2017;56:5726–5738. doi: 10.1021/acs.biochem.7b00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MF, Docherty NG, Burke JP, O'Connell PR. S100A4 expression is increased in stricture fibroblasts from patients with fibrostenosing Crohn’s disease and promotes intestinal fibroblast migration. Am J Physiol Gastrointest Liver Physiol. 2010;299:G457–G466. doi: 10.1152/ajpgi.00351.2009. [DOI] [PubMed] [Google Scholar]
- Dahlmann M, Kobelt D, Walther W, Mudduluru G, Stein U (2016) S100A4 in cancer metastasis: Wnt signaling-driven interventions for metastasis restriction cancers (Basel) 8 doi:10.3390/cancers8060059 [DOI] [PMC free article] [PubMed]
- Dakhel S, et al. S100P antibody-mediated therapy as a new promising strategy for the treatment of pancreatic cancer. Oncogenesis. 2014;3:e92. doi: 10.1038/oncsis.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BR, Davies MP, Gibbs FE, Barraclough R, Rudland PS. Induction of the metastatic phenotype by transfection of a benign rat mammary epithelial cell line with the gene for p9Ka, a rat calcium-binding protein, but not with the oncogene EJ-ras-1. Oncogene. 1993;8:999–1008. [PubMed] [Google Scholar]
- Davies MP, Rudland PS, Robertson L, Parry EW, Jolicoeur P, Barraclough R. Expression of the calcium-binding protein S100A4 (p9Ka) in MMTV-neu transgenic mice induces metastasis of mammary tumours. Oncogene. 1996;13:1631–1637. [PubMed] [Google Scholar]
- Dempsey BR, Rezvanpour A, Lee TW, Barber KR, Junop MS, Shaw GS. Structure of an asymmetric ternary protein complex provides insight for membrane interaction. Structure. 2012;20:1737–1745. doi: 10.1016/j.str.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Dmytriyeva O, et al. The metastasis-promoting S100A4 protein confers neuroprotection in brain injury. Nat Commun. 2012;3:1197. doi: 10.1038/ncomms2202. [DOI] [PubMed] [Google Scholar]
- Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60:540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of S100 proteins. Curr Mol Med. 2013;13:24–57. doi: 10.2174/156652413804486214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulyaninova NG, Bresnick AR. The heavy chain has its day: regulation of myosin-II assembly. Bioarchitecture. 2013;3:77–85. doi: 10.4161/bioa.26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulyaninova NG, Hite KM, Zencheck WD, Scudiero DA, Almo SC, Shoemaker RH, Bresnick AR. Cysteine 81 is critical for the interaction of S100A4 and myosin-IIA. Biochemistry. 2011;50:7218–7227. doi: 10.1021/bi200853y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrchen JM, Sunderkotter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- Eisenblaetter M, et al. Visualization of tumor-immune interaction - target-specific imaging of S100A8/A9 reveals pre-metastatic niche establishment. Theranostics. 2017;7:2392–2401. doi: 10.7150/thno.17138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsson MC, et al. Expression of metastasin S100A4 is essential for bone resorption and regulates osteoclast function. Biochim Biophys Acta. 2013;1833:2653–2663. doi: 10.1016/j.bbamcr.2013.06.020. [DOI] [PubMed] [Google Scholar]
- Escudier B, et al. A phase II multicentre, open-label, proof-of-concept study of tasquinimod in hepatocellular, ovarian, renal cell, and gastric cancers. Target Oncol. 2017;12:655–661. doi: 10.1007/s11523-017-0525-2. [DOI] [PubMed] [Google Scholar]
- Feldkamp MD, Gakhar L, Pandey N, Shea MA. Opposing orientations of the anti-psychotic drug trifluoperazine selected by alternate conformations of M144 in calmodulin. Proteins. 2015;83:989–996. doi: 10.1002/prot.24781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fizazi K, et al. A randomized, double-blind, placebo-controlled phase II study of maintenance therapy with tasquinimod in patients with metastatic castration-resistant prostate cancer responsive to or stabilized during first-line docetaxel chemotherapy. Ann Oncol. 2017;28:2741–2746. doi: 10.1093/annonc/mdx487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foell D, et al. Proinflammatory S100A12 can activate human monocytes via Toll-like receptor 4. Am J Respir Crit Care Med. 2013;187:1324–1334. doi: 10.1164/rccm.201209-1602OC. [DOI] [PubMed] [Google Scholar]
- Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- Forst B, et al. Metastasis-inducing S100A4 and RANTES cooperate in promoting tumor progression in mice. PLoS One. 2010;5:e10374. doi: 10.1371/journal.pone.0010374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen Pettersson N, et al. The immunomodulatory quinoline-3-carboxamide paquinimod reverses established fibrosis in a novel mouse model for liver fibrosis. PLoS One. 2018;13:e0203228. doi: 10.1371/journal.pone.0203228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H et al. (2018) S100B suppression alters polarization of infiltrating myeloid-derived cells in gliomas and inhibits tumor growth Cancer Lett doi:10.1016/j.canlet.2018.07.034 [DOI] [PMC free article] [PubMed]
- Garrett SC, Hodgson L, Rybin A, Toutchkine A, Hahn KM, Lawrence DS, Bresnick AR. A biosensor of S100A4 metastasis factor activation: inhibitor screening and cellular activation dynamics. Biochemistry. 2008;47:986–996. doi: 10.1021/bi7021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor R, Irie R, Morton D, Herschman HR. S100 protein is present in cultured human malignant melanomas. Nature. 1980;286:400–401. doi: 10.1038/286400a0. [DOI] [PubMed] [Google Scholar]
- Ghavami S, et al. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol. 2008;83:1484–1492. doi: 10.1189/jlb.0607397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DA (1990) Serum calcium. In: rd, Walker HK, Hall WD, Hurst JW (eds) Clinical methods: the history, physical, and laboratory examinations. Boston, [PubMed]
- Grigorian M, Ambartsumian N, Lukanidin E. Metastasis-inducing S100A4 protein: implication in non-malignant human pathologies. Curr Mol Med. 2008;8:492–496. doi: 10.2174/156652408785747942. [DOI] [PubMed] [Google Scholar]
- Grigorian M, Ambartsumian N, Lykkesfeldt AE, Bastholm L, Elling F, Georgiev G, Lukanidin E. Effect of mts1 (S100A4) expression on the progression of human breast cancer cells. Int J Cancer. 1996;67:831–841. doi: 10.1002/(SICI)1097-0215(19960917)67:6<831::AID-IJC13>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Grotterod I, Maelandsmo GM, Boye K. Signal transduction mechanisms involved in S100A4-induced activation of the transcription factor NF-kappaB. BMC Cancer. 2010;10:241. doi: 10.1186/1471-2407-10-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grum-Schwensen B, et al. S100A4-neutralizing antibody suppresses spontaneous tumor progression, pre-metastatic niche formation and alters T-cell polarization balance. BMC Cancer. 2015;15:44. doi: 10.1186/s12885-015-1034-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grum-Schwensen B, Klingelhofer J, Berg CH, El-Naaman C, Grigorian M, Lukanidin E, Ambartsumian N. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res. 2005;65:3772–3780. doi: 10.1158/0008-5472.CAN-04-4510. [DOI] [PubMed] [Google Scholar]
- Hansen MT, et al. A link between inflammation and metastasis: serum amyloid A1 and A3 induce metastasis, and are targets of metastasis-inducing S100A4. Oncogene. 2015;34:424–435. doi: 10.1038/onc.2013.568. [DOI] [PubMed] [Google Scholar]
- Hartman KG, McKnight LE, Liriano MA, Weber DJ. The evolution of S100B inhibitors for the treatment of malignant melanoma. Future Med Chem. 2013;5:97–109. doi: 10.4155/fmc.12.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild A, Engel G, Brenner W, Glaser R, Monig H, Henze E, Christophers E. S100B protein detection in serum is a significant prognostic factor in metastatic melanoma. Oncology. 1999;56:338–344. doi: 10.1159/000011989. [DOI] [PubMed] [Google Scholar]
- Henry J, et al. Update on the epidermal differentiation complex. Frontiers in bioscience : a journal and virtual library. 2012;17:1517–1532. doi: 10.2741/4001. [DOI] [PubMed] [Google Scholar]
- Herbert JM, Augereau JM, Gleye J, Maffrand JP. Chelerythrine is a potent and specific inhibitor of protein kinase. C Biochem Biophys Res Commun. 1990;172:993–999. doi: 10.1016/0006-291X(90)91544-3. [DOI] [PubMed] [Google Scholar]
- Hermann A, Donato R, Weiger TM, Chazin WJ. S100 calcium binding proteins and ion channels. Front Pharmacol. 2012;3:67. doi: 10.3389/fphar.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JL, et al. Therapeutic targeting of tumor growth and angiogenesis with a novel anti-S100A4 monoclonal antibody. PLoS One. 2013;8:e72480. doi: 10.1371/journal.pone.0072480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, et al. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res. 2013;73:172–183. doi: 10.1158/0008-5472.CAN-11-3843. [DOI] [PubMed] [Google Scholar]
- Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol. 2006;8:1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- Hiratsuka S, et al. The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat Cell Biol. 2008;10:1349–1355. doi: 10.1038/ncb1794. [DOI] [PubMed] [Google Scholar]
- House RP, et al. Two functional S100A4 monomers are necessary for regulating nonmuscle myosin-IIA and HCT116 cell invasion. Biochemistry. 2011;50:6920–6932. doi: 10.1021/bi200498q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res : MCR. 2011;9:133–148. doi: 10.1158/1541-7786.MCR-10-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JT, et al. Tasquinimod is an allosteric modulator of HDAC4 survival signaling within the compromised cancer microenvironment. Cancer Res. 2013;73:1386–1399. doi: 10.1158/0008-5472.CAN-12-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg E, et al. S100A9 interaction with TLR4 promotes tumor growth. PLoS One. 2012;7:e34207. doi: 10.1371/journal.pone.0034207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Nakayama S, Kretsinger RH. Classification and evolution of EF-hand proteins. Biometals. 1998;11:277–295. doi: 10.1023/A:1009282307967. [DOI] [PubMed] [Google Scholar]
- Kessel C et al. (2018) Calcium and zinc tune autoinflammatory Toll-like receptor 4 signaling by S100A12. J Allergy Clin Immunol doi:10.1016/j.jaci.2018.06.027 [DOI] [PubMed]
- Kessel C, Holzinger D, Foell D. Phagocyte-derived S100 proteins in autoinflammation: putative role in pathogenesis and usefulness as biomarkers. Clin Immunol. 2013;147:229–241. doi: 10.1016/j.clim.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Kim CE, Lim SK, Kim JS. In vivo antitumor effect of cromolyn in PEGylated liposomes for pancreatic cancer. Journal of controlled release : official journal of the Controlled Release Society. 2012;157:190–195. doi: 10.1016/j.jconrel.2011.09.066. [DOI] [PubMed] [Google Scholar]
- Kiss B, Duelli A, Radnai L, Kekesi KA, Katona G, Nyitray L. Crystal structure of the S100A4-nonmuscle myosin IIA tail fragment complex reveals an asymmetric target binding mechanism. Proc Natl Acad Sci U S A. 2012;109:6048–6053. doi: 10.1073/pnas.1114732109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizawa K, Takahara H, Troxler H, Kleinert P, Mochida U, Heizmann CW. Specific citrullination causes assembly of a globular S100A3 homotetramer: a putative Ca2+ modulator matures human hair cuticle. J Biol Chem. 2008;283:5004–5013. doi: 10.1074/jbc.M709357200. [DOI] [PubMed] [Google Scholar]
- Klingelhofer J, Grum-Schwensen B, Beck MK, Knudsen RS, Grigorian M, Lukanidin E, Ambartsumian N. Anti-S100A4 antibody suppresses metastasis formation by blocking stroma cell invasion. Neoplasia. 2012;14:1260–1268. doi: 10.1593/neo.121554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingelhofer J, et al. Up-regulation of metastasis-promoting S100A4 (Mts-1) in rheumatoid arthritis: putative involvement in the pathogenesis of rheumatoid arthritis. Arthritis Rheum. 2007;56:779–789. doi: 10.1002/art.22398. [DOI] [PubMed] [Google Scholar]
- Koch M, Chitayat S, Dattilo BM, Schiefner A, Diez J, Chazin WJ, Fritz G. Structural basis for ligand recognition and activation of RAGE. Structure. 2010;18:1342–1352. doi: 10.1016/j.str.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppaka V, et al. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64:520–539. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laouedj M, et al. S100A9 induces differentiation of acute myeloid leukemia cells through TLR4. Blood. 2017;129:1980–1990. doi: 10.1182/blood-2016-09-738005. [DOI] [PubMed] [Google Scholar]
- Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta. 2009;1793:993–1007. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Leclerc E, Heizmann CW. The importance of Ca2+/Zn2+ signaling S100 proteins and RAGE in translational medicine. Front Biosci (Schol Ed) 2011;3:1232–1262. doi: 10.2741/223. [DOI] [PubMed] [Google Scholar]
- Lee TH, et al. Role of S100A9 in the development of neutrophilic inflammation in asthmatics and in a murine model. Clin Immunol. 2017;183:158–166. doi: 10.1016/j.clim.2017.08.013. [DOI] [PubMed] [Google Scholar]
- Lee YT, et al. Structure of the S100A6 complex with a fragment from the C-terminal domain of Siah-1 interacting protein: a novel mode for S100 protein target recognition. Biochemistry. 2008;47:10921–10932. doi: 10.1021/bi801233z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic Zivkovic M, Zareba-Koziol M, Zhukova L, Poznanski J, Zhukov I, Wyslouch-Cieszynska A. Post-translational S-nitrosylation is an endogenous factor fine tuning the properties of human S100A1 protein. J Biol Chem. 2012;287:40457–40470. doi: 10.1074/jbc.M112.418392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZH, Dulyaninova NG, House RP, Almo SC, Bresnick AR. S100A4 regulates macrophage chemotaxis. Mol Biol Cell. 2010;21:2598–2610. doi: 10.1091/mbc.E09-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SY, Raftery MJ, Geczy CL. Oxidative modifications of DAMPs suppress inflammation: the case for S100A8 and S100A9. Antioxid Redox Signal. 2011;15:2235–2248. doi: 10.1089/ars.2010.3641. [DOI] [PubMed] [Google Scholar]
- Liriano MA, Varney KM, Wright NT, Hoffman CL, Toth EA, Ishima R, Weber DJ. Target binding to S100B reduces dynamic properties and increases Ca(2+)-binding affinity for wild type and EF-hand mutant proteins. J Mol Biol. 2012;423:365–385. doi: 10.1016/j.jmb.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Myrvang HK, Dekker LV. Annexin A2 complexes with S100 proteins: structure, function and pharmacological manipulation. Br J Pharmacol. 2015;172:1664–1676. doi: 10.1111/bph.12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack GS, Marshall A. Lost in migration. Nat Biotechnol. 2010;28:214–229. doi: 10.1038/nbt0310-214. [DOI] [PubMed] [Google Scholar]
- Maelandsmo GM, et al. Reversal of the in vivo metastatic phenotype of human tumor cells by an anti-CAPL (mts1) ribozyme. Cancer Res. 1996;56:5490–5498. [PubMed] [Google Scholar]
- Majorek KA, Kuhn ML, Chruszcz M, Anderson WF, Minor W. Double trouble-Buffer selection and His-tag presence may be responsible for nonreproducibility of biomedical experiments. Protein Sci. 2014;23:1359–1368. doi: 10.1002/pro.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malashkevich VN, et al. Phenothiazines inhibit S100A4 function by inducing protein oligomerization. Proc Natl Acad Sci U S A. 2010;107:8605–8610. doi: 10.1073/PNAS0913660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malashkevich VN, et al. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansh M. Ipilimumab and cancer immunotherapy: a new hope for advanced stage melanoma. Yale J Biol Med. 2011;84:381–389. [PMC free article] [PubMed] [Google Scholar]
- Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- Markowitz J, et al. Identification and characterization of small molecule inhibitors of the calcium-dependent S100B-p53 tumor suppressor interaction. J Med Chem. 2004;47:5085–5093. doi: 10.1021/jm0497038. [DOI] [PubMed] [Google Scholar]
- Markowitz J, et al. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry. 2005;44:7305–7314. doi: 10.1021/bi050321t. [DOI] [PubMed] [Google Scholar]
- Marshak DR, Lukas TJ, Watterson DM. Drug-protein interactions: binding of chlorpromazine to calmodulin, calmodulin fragments, and related calcium binding proteins. Biochemistry. 1985;24:144–150. doi: 10.1021/bi00322a020. [DOI] [PubMed] [Google Scholar]
- Miller P, et al. Elevated S100A8 protein expression in breast cancer cells and breast tumor stroma is prognostic of poor disease outcome. Breast Cancer Res Treat. 2017;166:85–94. doi: 10.1007/s10549-017-4366-6. [DOI] [PubMed] [Google Scholar]
- Miranda KJ, Loeser RF, Yammani RR. Sumoylation and nuclear translocation of S100A4 regulate IL-1beta-mediated production of matrix metalloproteinase-13. J Biol Chem. 2010;285:31517–31524. doi: 10.1074/jbc.M110.125898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya MC, DiDone L, Heier RF, Meyers MJ, Krysan DJ. Antifungal phenothiazines: optimization, characterization of mechanism, and modulation of neuroreceptor activity. ACS Infect Dis. 2018;4:499–507. doi: 10.1021/acsinfecdis.7b00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BW. A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun. 1965;19:739–744. doi: 10.1016/0006-291X(65)90320-7. [DOI] [PubMed] [Google Scholar]
- Moroz OV, et al. Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Biochem. 2009;10:11. doi: 10.1186/1471-2091-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouta Carreira C, et al. S100A13 is involved in the regulation of fibroblast growth factor-1 and p40 synaptotagmin-1 release in vitro. J Biol Chem. 1998;273:22224–22231. doi: 10.1074/jbc.273.35.22224. [DOI] [PubMed] [Google Scholar]
- Nefla M, Holzinger D, Berenbaum F, Jacques C. The danger from within: alarmins in arthritis. Nat Rev Rheumatol. 2016;12:669–683. doi: 10.1038/nrrheum.2016.162. [DOI] [PubMed] [Google Scholar]
- Novitskaya V, et al. Oligomeric forms of the metastasis-related Mts1 (S100A4) protein stimulate neuronal differentiation in cultures of rat hippocampal neurons. J Biol Chem. 2000;275:41278–41286. doi: 10.1074/jbc.M007058200. [DOI] [PubMed] [Google Scholar]
- O'Connell JT, et al. VEGF-A and Tenascin-C produced by S100A4+ stromal cells are important for metastatic colonization. Proc Natl Acad Sci U S A. 2011;108:16002–16007. doi: 10.1073/pnas.1109493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YS, et al. SMARCA3, a chromatin-remodeling factor, is required for p11-dependent antidepressant action. Cell. 2013;152:831–843. doi: 10.1016/j.cell.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Arai S, Itoh H, Adachi S, Hayashida M, Nakase H, Ikemoto M. CD68 on rat macrophages binds tightly to S100A8 and S100A9 and helps to regulate the cells' immune functions. J Leukoc Biol. 2016;100:1093–1104. doi: 10.1189/jlb.2A0415-170RRR. [DOI] [PubMed] [Google Scholar]
- Okada M, Tokumitsu H, Kubota Y, Kobayashi R. Interaction of S100 proteins with the antiallergic drugs, olopatadine, amlexanox, and cromolyn: identification of putative drug binding sites on S100A1 protein. Biochem Biophys Res Commun. 2002;292:1023–1030. doi: 10.1006/bbrc.2002.6761. [DOI] [PubMed] [Google Scholar]
- Orre LM, Pernemalm M, Lengqvist J, Lewensohn R, Lehtio J. Up-regulation, modification, and translocation of S100A6 induced by exposure to ionizing radiation revealed by proteomics profiling. Mol Cell Proteomics. 2007;6:2122–2131. doi: 10.1074/mcp.M700202-MCP200. [DOI] [PubMed] [Google Scholar]
- Oslejskova L, et al. Metastasis-inducing S100A4 protein is associated with the disease activity of rheumatoid arthritis. Rheumatology (Oxford) 2009;48:1590–1594. doi: 10.1093/rheumatology/kep316. [DOI] [PubMed] [Google Scholar]
- Ostendorp T, et al. Structural and functional insights into RAGE activation by multimeric S100B. EMBO J. 2007;26:3868–3878. doi: 10.1038/sj.emboj.7601805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozorowski G, Milton S, Luecke H. Structure of a C-terminal AHNAK peptide in a 1:2:2 complex with S100A10 and an acetylated N-terminal peptide of annexin. A2. Acta Crystallogr D Biol Crystallogr. 2013;69:92–104. doi: 10.1107/S0907444912043429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla L, et al. S100A7: from mechanism to cancer therapy. Oncogene. 2017;36:6749–6761. doi: 10.1038/onc.2017.283. [DOI] [PubMed] [Google Scholar]
- Palanski BA, Khosla C. Cystamine and disulfiram inhibit human transglutaminase 2 via an oxidative mechanism. Biochemistry. 2018;57:3359–3363. doi: 10.1021/acs.biochem.8b00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfy G, Kiss B, Nyitray L, Bodor A. Multilevel changes in protein dynamics upon complex formation of the calcium-loaded S100A4 with a nonmuscle myosin IIA tail fragment. Chembiochem. 2016;17:1829–1838. doi: 10.1002/cbic.201600280. [DOI] [PubMed] [Google Scholar]
- Pankratova S, et al. The S100A4 protein signals through the ErbB4 receptor to promote neuronal survival. Theranostics. 2018;8:3977–3990. doi: 10.7150/thno.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Adsit FG, Boyington JC. The 1.5 A crystal structure of human receptor for advanced glycation endproducts (RAGE) ectodomains reveals unique features determining ligand binding. J Biol Chem. 2010;285:40762–40770. doi: 10.1074/jbc.M110.169276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penumutchu SR, Chou RH, Yu C. Structural insights into calcium-bound S100P and the V domain of the RAGE complex. PLoS One. 2014;9:e103947. doi: 10.1371/journal.pone.0103947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pili R, et al. Phase II randomized, double-blind, placebo-controlled study of tasquinimod in men with minimally symptomatic metastatic castrate-resistant prostate cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:4022–4028. doi: 10.1200/JCO.2011.35.6295. [DOI] [PubMed] [Google Scholar]
- Pingerelli PL, Mizukami H, Wagner AS, Bartnicki DE, Oliver JP. Investigation of the Ca2(+)-dependent interaction of trifluoperazine with S100a: a 19F NMR and circular dichroism study. J Protein Chem. 1990;9:169–175. doi: 10.1007/BF01025308. [DOI] [PubMed] [Google Scholar]
- Pluta K, Jelen M, Morak-Mlodawska B, Zimecki M, Artym J, Kocieba M, Zaczynska E. Azaphenothiazines - promising phenothiazine derivatives. An insight into nomenclature, synthesis, structure elucidation and biological properties. Eur J Med Chem. 2017;138:774–806. doi: 10.1016/j.ejmech.2017.07.009. [DOI] [PubMed] [Google Scholar]
- Qin H, et al. Generation of a new therapeutic peptide that depletes myeloid-derived suppressor cells in tumor-bearing mice. Nat Med. 2014;20:676–681. doi: 10.1038/nm.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopal UA, et al. Structure of the S100A4/myosin-IIA complex. BMC Struct Biol. 2013;13:31. doi: 10.1186/1472-6807-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani SG, Mohan SK, Yu C. Molecular level interactions of S100A13 with amlexanox: inhibitor for formation of the multiprotein complex in the nonclassical pathway of acidic fibroblast growth factor. Biochemistry. 2010;49:2585–2592. doi: 10.1021/bi9019077. [DOI] [PubMed] [Google Scholar]
- Ravasi T, et al. Probing the S100 protein family through genomic and functional analysis. Genomics. 2004;84:10–22. doi: 10.1016/j.ygeno.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Raymond E, Dalgleish A, Damber JE, Smith M, Pili R. Mechanisms of action of tasquinimod on the tumour microenvironment. Cancer Chemother Pharmacol. 2014;73:1–8. doi: 10.1007/s00280-013-2321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TR, Li C, Fischer PM, Dekker LV. Three-dimensional pharmacophore design and biochemical screening identifies substituted 1,2,4-triazoles as inhibitors of the annexin A2-S100A10 protein interaction. ChemMedChem. 2012;7:1435–1446. doi: 10.1002/cmdc.201200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TR, Li C, Guo X, Myrvang HK, Fischer PM, Dekker LV. Design, synthesis, and structure-activity relationship exploration of 1-substituted 4-aroyl-3-hydroxy-5-phenyl-1H-pyrrol-2(5H)-one analogues as inhibitors of the annexin A2-S100A10 protein interaction. J Med Chem. 2011;54:2080–2094. doi: 10.1021/jm101212e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SM, et al. An inhibitor of the protein kinases TBK1 and IKK-varepsilon improves obesity-related metabolic dysfunctions in mice. Nat Med. 2013;19:313–321. doi: 10.1038/nm.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rety S, et al. Structural basis of the Ca(2+)-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure Fold Des. 2000;8:175–184. doi: 10.1016/S0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- Rety S, et al. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat Struct Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- Riuzzi F, Sorci G, Donato R. S100B protein regulates myoblast proliferation and differentiation by activating FGFR1 in a bFGF-dependent manner. J Cell Sci. 2011;124:2389–2400. doi: 10.1242/jcs.084491. [DOI] [PubMed] [Google Scholar]
- Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta) Nat Struct Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- Sack U, et al. S100A4-induced cell motility and metastasis is restricted by the Wnt/beta-catenin pathway inhibitor calcimycin in colon cancer cells. Mol Biol Cell. 2011;22:3344–3354. doi: 10.1091/mbc.E10-09-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif MW. Anti-VEGF agents in metastatic colorectal cancer (mCRC): are they all alike? Cancer Manag Res. 2013;5:103–115. doi: 10.2147/CMAR.S45193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M, et al. PKCalpha mediates TGFbeta-induced growth inhibition of human keratinocytes via phosphorylation of S100C/A11. J Cell Biol. 2004;164:979–984. doi: 10.1083/jcb.200312041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M, et al. Identification of an S100A8 receptor Neuroplastin-beta and its heterodimer formation with EMMPRIN. J Invest Dermatol. 2016;136:2240–2250. doi: 10.1016/j.jid.2016.06.617. [DOI] [PubMed] [Google Scholar]
- Schenten V, et al. Secretion of the phosphorylated form of S100A9 from neutrophils is essential for the proinflammatory functions of extracellular S100A8/A9. Front Immunol. 2018;9:447. doi: 10.3389/fimmu.2018.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- Shishibori T, et al. Three distinct anti-allergic drugs, amlexanox, cromolyn and tranilast, bind to S100A12 and S100A13 of the S100 protein family. Biochem J. 1999;338(Pt 3):583–589. doi: 10.1042/bj3380583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Stewart BJ, Glaysher S, Peregrin K, Knight LA, Weber DJ, Cree IA. The effect of pentamidine on melanoma ex vivo. Anti-Cancer Drugs. 2010;21:181–185. doi: 10.1097/CAD.0b013e3283340cee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprules T, Green N, Featherstone M, Gehring K. Nickel-induced oligomerization of proteins containing 10-histidine tags. Biotechniques. 1998;25:20–22. doi: 10.2144/98251bm02. [DOI] [PubMed] [Google Scholar]
- Stein U, et al. Intervening in beta-catenin signaling by sulindac inhibits S100A4-dependent colon cancer metastasis. Neoplasia. 2011;13:131–144. doi: 10.1593/neo.101172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein U, et al. The metastasis-associated gene S100A4 is a novel target of beta-catenin/T-cell factor signaling in colon cancer. Gastroenterology. 2006;131:1486–1500. doi: 10.1053/j.gastro.2006.08.041. [DOI] [PubMed] [Google Scholar]
- Stewart RL, et al. S100A4 drives non-small cell lung cancer invasion, associates with poor prognosis, and is effectively targeted by the FDA-approved anti-helminthic agent niclosamide. Oncotarget. 2016;7:34630–34642. doi: 10.18632/oncotarget.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher WW, Lopez MM, Makhatadze GI. Modulation of quaternary structure of S100 proteins by calcium ions. Biophys Chem. 2010;151:181–186. doi: 10.1016/j.bpc.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahvili S, Torngren M, Holmberg D, Leanderson T, Ivars F. Paquinimod prevents development of diabetes in the non-obese diabetic (NOD) mouse. PLoS One. 2018;13:e0196598. doi: 10.1371/journal.pone.0196598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaga K, Nakamura Y, Sakiyama S. Cellular localization of pEL98 protein, an S100-related calcium binding protein, in fibroblasts and its tissue distribution analyzed by monoclonal antibodies. Cell Struct Funct. 1994;19:133–141. doi: 10.1247/csf.19.133. [DOI] [PubMed] [Google Scholar]
- Takenaga K, Nakamura Y, Sakiyama S. Expression of antisense RNA to S100A4 gene encoding an S100-related calcium-binding protein suppresses metastatic potential of high-metastatic Lewis lung carcinoma cells. Oncogene. 1997;14:331–337. doi: 10.1038/sj.onc.1200820. [DOI] [PubMed] [Google Scholar]
- Tamai H, et al. Amlexanox downregulates S100A6 to sensitize KMT2A/AFF1-positive acute lymphoblastic leukemia to TNFalpha treatment. Cancer Res. 2017;77:4426–4433. doi: 10.1158/0008-5472.CAN-16-2974. [DOI] [PubMed] [Google Scholar]
- Tidehag V, et al. High density of S100A9 positive inflammatory cells in prostate cancer stroma is associated with poor outcome. Eur J Cancer. 2014;50:1829–1835. doi: 10.1016/j.ejca.2014.03.278. [DOI] [PubMed] [Google Scholar]
- Tondera C, Laube M, Pietzsch J. Insights into binding of S100 proteins to scavenger receptors: class B scavenger receptor CD36 binds S100A12 with high affinity. Amino Acids. 2017;49:183–191. doi: 10.1007/s00726-016-2349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turovskaya O, et al. RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis. 2008;29:2035–2043. doi: 10.1093/carcin/bgn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandonselaar M, Hickie RA, Quail JW, Delbaere LT. Trifluoperazine-induced conformational change in Ca(2+)-calmodulin. Nat Struct Biol. 1994;1:795–801. doi: 10.1038/nsb1194-795. [DOI] [PubMed] [Google Scholar]
- Vertessy BG, Harmat V, Bocskei Z, Naray-Szabo G, Orosz F, Ovadi J. Simultaneous binding of drugs with different chemical structures to Ca2+-calmodulin: crystallographic and spectroscopic studies. Biochemistry. 1998;37:15300–15310. doi: 10.1021/bi980795a. [DOI] [PubMed] [Google Scholar]
- Vogl T, Leukert N, Barczyk K, Strupat K, Roth J. Biophysical characterization of S100A8 and S100A9 in the absence and presence of bivalent cations. Biochim Biophys Acta. 2006;1763:1298–1306. doi: 10.1016/j.bbamcr.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Vogl T, et al. Autoinhibitory regulation of S100A8/S100A9 alarmin activity locally restricts sterile inflammation. J Clin Invest. 2018;128:1852–1866. doi: 10.1172/JCI89867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- Vogt A, McDonald PR, Tamewitz A, Sikorski RP, Wipf P, Skoko JJ, 3rd, Lazo JS. A cell-active inhibitor of mitogen-activated protein kinase phosphatases restores paclitaxel-induced apoptosis in dexamethasone-protected cancer cells. Mol Cancer Ther. 2008;7:330–340. doi: 10.1158/1535-7163.MCT-07-2165. [DOI] [PubMed] [Google Scholar]
- von Bauer R, et al. CD166/ALCAM mediates proinflammatory effects of S100B in delayed type hypersensitivity. J Immunol. 2013;191:369–377. doi: 10.4049/jimmunol.1201864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wache C, et al. Myeloid-related protein 14 promotes inflammation and injury in meningitis. J Infect Dis. 2015;212:247–257. doi: 10.1093/infdis/jiv028. [DOI] [PubMed] [Google Scholar]
- Wafer LN, Tzul FO, Pandharipande PP, Makhatadze GI. Novel interactions of the TRTK12 peptide with S100 protein family members: specificity and thermodynamic characterization. Biochemistry. 2013;52:5844–5856. doi: 10.1021/bi400788s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder PT, et al. In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction. Int J High Throughput Screen. 2010;2010:109–126. doi: 10.2147/IJHTS.S8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright NT, Cannon BR, Wilder PT, Morgan MT, Varney KM, Zimmer DB, Weber DJ. Solution structure of S100A1 bound to the CapZ peptide (TRTK12) J Mol Biol. 2009;386:1265–1277. doi: 10.1016/j.jmb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Braunstein Z, Toomey AC, Zhong J, Rao X. S100 proteins as an important regulator of macrophage inflammation. Front Immunol. 2017;8:1908. doi: 10.3389/fimmu.2017.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Burz DS, He W, Bronstein IB, Lednev I, Shekhtman A. Hexameric calgranulin C (S100A12) binds to the receptor for advanced glycated end products (RAGE) using symmetric hydrophobic target-binding patches. J Biol Chem. 2007;282:4218–4231. doi: 10.1074/jbc.M608888200. [DOI] [PubMed] [Google Scholar]
- Xue C, Plieth D, Venkov C, Xu C, Neilson EG. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 2003;63:3386–3394. [PubMed] [Google Scholar]
- Xue J, et al. Change in the molecular dimension of a RAGE-ligand complex triggers RAGE signaling. Structure. 2016;24:1509–1522. doi: 10.1016/j.str.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yammani RR, Long D, Loeser RF. Interleukin-7 stimulates secretion of S100A4 by activating the JAK/STAT signaling pathway in human articular chondrocytes. Arthritis Rheum. 2009;60:792–800. doi: 10.1002/art.24295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Armishaw C, Goyette J, Yang Z, Cai H, Alewood P, Geczy CL. Mast cell and monocyte recruitment by S100A12 and its hinge domain. J Biol Chem. 2008;283:13035–13043. doi: 10.1074/jbc.M710388200. [DOI] [PubMed] [Google Scholar]
- Yatime L, Betzer C, Jensen RK, Mortensen S, Jensen PH, Andersen GR. The structure of the RAGE:S100A6 complex reveals a unique mode of Homodimerization for S100. Proteins Structure. 2016;24:2043–2052. doi: 10.1016/j.str.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Zackular JP, Chazin WJ, Skaar EP. Nutritional immunity: S100 proteins at the host-pathogen interface. J Biol Chem. 2015;290:18991–18998. doi: 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. S100A4 promotes colon inflammation and colitis-associated colon tumorigenesis. Oncoimmunology. 2018;7:e1461301. doi: 10.1080/2162402X.2018.1461301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. S100A4 contributes to colitis development by increasing the adherence of Citrobacter rodentium in intestinal epithelial cells. Sci Rep. 2017;7:12099. doi: 10.1038/s41598-017-12256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Suppression colitis and colitis-associated Colon Cancer by anti-S100a9 antibody in mice. Front Immunol. 2017;8:1774. doi: 10.3389/fimmu.2017.01774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibert JR, Skov L, Thyssen JP, Jacobsen GK, Grigorian M. Significance of the S100A4 protein in psoriasis. J Invest Dermatol. 2010;130:150–160. doi: 10.1038/jid.2009.206. [DOI] [PubMed] [Google Scholar]
- Zimmer DB, Eubanks JO, Ramakrishnan D, Criscitiello MF. Evolution of the S100 family of calcium sensor proteins. Cell Calcium. 2012;53:170–179. doi: 10.1016/j.ceca.2012.11.006. [DOI] [PubMed] [Google Scholar]