Abstract
Cellular viability requires tight regulation of actin cytoskeletal dynamics. Distinct families of nucleation-promoting factors enable the rapid assembly of filament nuclei that elongate and are incorporated into diverse and specialized actin-based structures. In addition to promoting filament nucleation, the formin family of proteins directs the elongation of unbranched actin filaments. Processive association of formins with growing filament ends is achieved through continuous barbed end binding of the highly conserved, dimeric formin homology (FH) 2 domain. In cooperation with the FH1 domain and C-terminal tail region, FH2 dimers mediate actin subunit addition at speeds that can dramatically exceed the rate of spontaneous assembly. Here, I review recent biophysical, structural, and computational studies that have provided insight into the mechanisms of formin-mediated actin assembly and dynamics.
Keywords: Formin, Actin, Profilin, Polymerization
Introduction
Dynamic remodeling of the actin cytoskeleton enables eukaryotic cells to alter their shapes, withstand pressure, establish polarity, move, form contacts with neighboring cells, and divide. De novo actin filament assembly is slow because the spontaneous formation of a filament nucleus is energetically unfavorable (Cooper et al. 1983; Frieden 1983; Oda et al. 2016; Sept and McCammon 2001). Nucleation-promoting factors including Arp2/3 complex (Pollard 2007), Ena/Vasp (Krause et al. 2003), Spire (Quinlan et al. 2005), Cordon-Bleu (Cobl) (Ahuja et al. 2007), Junction-mediating and regulatory protein (JMY) (Zuchero et al. 2009), leiomodin (Chereau et al. 2008), adenomatous polyposis coli (APC) (Okada et al. 2010), and formins (Goode and Eck 2007) help overcome this kinetic barrier by initiating actin polymerization at specific times and locations in cells. The mechanisms of action of these proteins are diverse; Arp2/3 complex initiates branched actin formation by binding to the sides of pre-existing filaments and serving as a template for elongation, whereas Ena/Vasp, Spire, Cobl, Junction-mediating and regulatory protein (JMY), and leiomodin use tandem actin monomer-binding domains (typically Wiskott-Aldrich syndrome protein homology 2 (WH2) domains) to bring multiple actin monomers together to nucleate unbranched filaments. Similarly, APC promotes nucleation by binding actin monomers via a basic domain. In contrast, the formin family of proteins stabilizes energetically unstable actin dimers and trimers by encircling them with their dimeric formin homology 2 (FH2) domains, thus establishing stable filament nuclei.
In addition to their role in enhancing nucleation, formins processively bind filament barbed ends (Breitsprecher et al. 2012; Mizuno et al. 2011; Paul and Pollard 2009a; Pruyne et al. 2002; Zigmond et al. 2003), inhibiting both capping (Zigmond et al. 2003) and annealing (Kovar et al. 2003), and influencing the rate of elongation via a unique mechanism that involves the coordinated actions of multiple domains (Goode and Eck 2007; Kovar et al. 2006; Paul and Pollard 2009b). Some formins also possess the ability to bind along the lengths of actin filaments or microtubules (Bartolini and Gundersen 2010; Bartolini et al. 2008; Gaillard et al. 2011; Harris et al. 2006; Wen et al. 2004), promoting cytoskeletal network bundling, coordination, and, in some cases, disassembly (Chhabra and Higgs 2006; Gurel et al. 2014).
Formins utilize their diverse and specialized activities to assemble a variety of actin-based cellular structures, including but not limited to cytokinetic contractile rings, filopodia, polarized actin cables, and stress fibers that promote adhesion junction maturation and nuclear positioning (Campellone and Welch 2010; Faix and Grosse 2006; Goode and Eck 2007). Proper timing and regulation of the assembly of these structures is essential.
Despite their unifying mechanistic similarities, formin isoforms do not possess redundant biological functions. For example, the three formins in fission yeast have distinct, non-overlapping roles (cytokinesis, actin cable formation, and mating) (Chang et al. 1997; Feierbach and Chang 2001; Petersen et al. 1998). Although the roles of the 15 human formin genes are still being elucidated, their importance is underscored by their connections to a number of diseases, including preleukemic disorders (Peng et al. 2007), microcephaly (Ercan-Sencicek et al. 2015), nonsyndromic deafness (Lynch et al. 1997), dilated and hypertrophic cardiomyopathies (Arimura et al. 2013; Wooten et al. 2013), the blood disorder macrothrombocytopenia, the neurological disorder Charcot-Marie-Tooth disease (Boyer et al. 2011), the kidney disease focal segmental glomerulosclerosis (Brown et al. 2010), and nonsyndromic intellectual disability (Law et al. 2014).
To understand the diverse roles played by formins and their contributions to cellular and organismal viability, it is essential to elucidate the mechanisms by which these proteins influence actin assembly and dynamics. Here, I discuss recent mechanistic studies that have advanced our understanding of the interactions between formins and the actin cytoskeleton.
Formin domain architecture
Formins are large (generally > 1000 amino acids (Higgs 2005)), multi-domain proteins whose defining feature is their conserved FH2 domain (Castrillon and Wasserman 1994; Higgs 2005; Higgs and Peterson 2005; Pruyne 2016), a ~ 350 amino acid sequence that is essential for formin-mediated effects on actin assembly. Most eukaryotes possess multiple FH2 domain-containing proteins; budding yeast and fission yeast express 2 and 3 formins, respectively, whereas Drosophila and placental mammals express 6 and 15 (Higgs and Peterson 2005). All known animal formins can be categorized into one of nine formin subtypes (Pruyne 2016), and the 15 formin genes in placental mammals fall into seven of these subtypes (Higgs and Peterson 2005; Pruyne 2016).
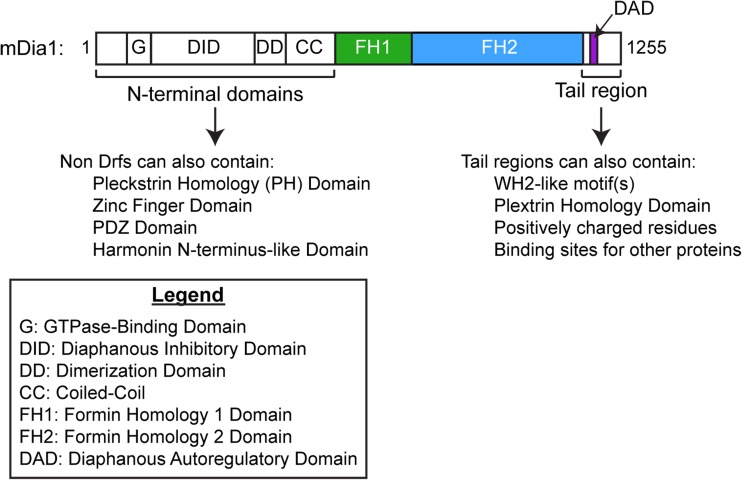
A formin’s FH2 domain is typically located in the C-terminal half of its amino acid sequence and is often immediately preceded by a proline-rich formin homology 1 (FH1) domain, a sequence that is highly variable in length (15–229 amino acids) and proline content (35–100%) (Higgs 2005) (Fig. 1). Many formins also encode additional actin-binding sites in their C-terminal tail regions, which extend from the final residue of the FH2 domain to the protein’s carboxyl-terminus. Whereas some C-terminal tail regions include one or two actin monomer-biding, WH2-like motifs, other tail sequences promote non-specific, electrostatic-based interactions along the lengths of actin filaments (Chhabra and Higgs 2006; Heimsath and Higgs 2012; Vizcarra et al. 2014). Other tail regions bind proteins that can regulate the formin’s actin assembly properties, such as Bud6p, which increases actin filament nucleation by Bni1p in budding yeast (Graziano et al. 2011) (Fig. 1).
Fig. 1.
Domain architecture of mDia1, a diaphanous-related formin (Drf). Domain boundaries are shown to scale. The three domains that influence actin dynamics are the FH1 domain, FH2 domain, and tail region
The majority of metazoan formins are categorized as “Diaphanous-related formins” (Drfs) owing to their N-terminal domain architecture, which includes a RhoGTPase-binding domain (GBD) followed by a diaphanous inhibitory domain (DID) and a dimerization domain (Fig. 1). Association of the DID with a diaphanous autoregulatory domain (DAD) sequence present in Drf C-terminal tail regions gives rise to an autoinhibited conformation (Chesarone et al. 2010; Higgs 2005). Binding of a RhoGTPase to the GBD and DID can disrupt the DID/DAD association and partially relieve autoinhibition (Higgs 2005; Li and Higgs 2003, 2005), but full activation of Drfs requires additional binding partners (Maiti et al. 2012).
In non-Drfs, one or more conserved N-terminal domain is often replaced by another domain, such as a different GTPase binding domain, a postsynaptic density protein 95/Drosophila disc large tumor suppressor 1/zonula occludens-1 protein (PDZ) domain, a pleckstrin homology (PH) domain, or a phosphatase and tensin (PTEN) domain. These alternative domain architectures likely function in regulating the localization and/or activity of these formins (Pruyne 2016).
A number of excellent reviews have described the physiological roles played by formins and the regulation of their sub-cellular localization and actin assembly activities (Bogdan et al., 2013; Faix and Grosse 2006; Schonichen and Geyer 2010). In this review, I focus on the mechanistic functions of the three domains that directly influence actin assembly dynamics: the FH1 and FH2 domains, and the C-terminal tail.
Actin filament assembly mediated by FH2 domains
Formins influence actin polymerization by binding to actin nuclei or filament barbed ends with their FH2 domains. A pioneering study first demonstrated this interaction via electron microscopy, which showed gold-labeled formins localized near barbed ends (Pruyne et al. 2002). Once bound, formins prevent filament annealing (Kovar et al. 2003) and can inhibit blocking of filament elongation by capping protein (Bombardier et al. 2015; Harris et al. 2004; Kovar et al. 2005; Moseley et al. 2004; Shekhar et al. 2015; Zigmond et al. 2003).
Formin FH2 domains are the minimal unit necessary to influence actin assembly (Moseley et al. 2004; Shimada et al. 2004; Xu et al. 2004). Purified FH2 domains dimerize (Moseley et al. 2004), and this self-association is required for formin activity (Shimada et al. 2004; Xu et al. 2004). For formins including the Saccharomyces cerevisiae formin Bni1p and the mammalian formin mDia1, the FH2 dimer is stable and exhibits a very slow dissociation rate (Moseley et al. 2004; Xu et al. 2004). Other formin FH2 dimers such as FMNL1, INF2, and mDia2 have the ability to dissociate in solution, a property that likely facilitates binding along filament sides (see section “Actin filament side-binding and bundling”) (Gurel et al. 2014; Harris et al. 2006; Sharma et al. 2014).
In kinetic assays of bulk actin assembly, a purified FH1FH2 construct of Bni1p was found to slow, but not inhibit, filament elongation both in the absence and presence of capping protein (Pruyne et al. 2002; Zigmond et al. 2003). These results suggested that the association of formins with barbed ends is processive, thus permitting formins to influence elongation while remaining bound to growing filaments. This model for processive binding was supported by live-cell imaging of active mDia1 constructs fused to GFP and expressed in Xenopus fibroblasts, which showed translocation of fluorescent spots at rates of 2.0 μm/s through the cytoplasm (Higashida et al. 2004). These spots were interpreted to be individual formin dimers bound at the barbed ends of elongating actin filaments. Further evidence for formin processivity was generated by the observation that barbed ends of actin filaments visualized by total internal reflection fluorescence (TIRF) microscopy elongated from fixed attachment points on glass surfaces coated with purified FH1FH2 constructs of four formin isoforms (Kovar et al. 2006). In more recent studies, purified formins have been labeled with quantum dots or fluorescent labels and directly visualized at the barbed ends of filaments during elongation (Bombardier et al. 2015; Breitsprecher et al. 2012; Paul and Pollard 2009a; Shekhar et al. 2015).
Filament elongation mediated by most FH2 dimers is slower than that observed for filaments with free barbed ends (Gurel et al. 2015; Kovar et al. 2003, 2006; Silkworth et al. 2018; Thompson et al. 2013). A “gating” model for formin-mediated subunit addition explains this phenomenon by postulating that FH2 dimers fluctuate among polymerization-competent and -incompetent, “open” and “closed”, conformations. The rate of subunit addition mediated by a formin is thus dictated by the probability that an incoming actin monomer will find the FH2-barbed end complex in an open conformation, a property known as the “gating factor” (Vavylonis et al. 2006). The extent to which an FH2 dimer gates elongation varies widely among formin isoforms: FH2 dimers of the Schizosaccharomyces pombe formin Cdc12p and the mammalian formin Delphilin inhibit elongation by approximately 99% (Dutta et al. 2017; Kovar et al. 2003, 2006; Silkworth et al. 2018), suggesting that these formins strongly favor a closed conformation, whereas mouse mDia1 slows elongation only modestly (i.e., by about 5–10%), indicating that its FH2 domain frequently populates an open conformation (Kovar et al. 2006). The equilibrium between open and closed conformations is formin-specific, and FH2 dimers from other formins produce intermediate effects on elongation (Gurel et al. 2015; Kovar et al. 2006; Thompson et al. 2013).
Structure of the FH2 domain and its interactions with the barbed end
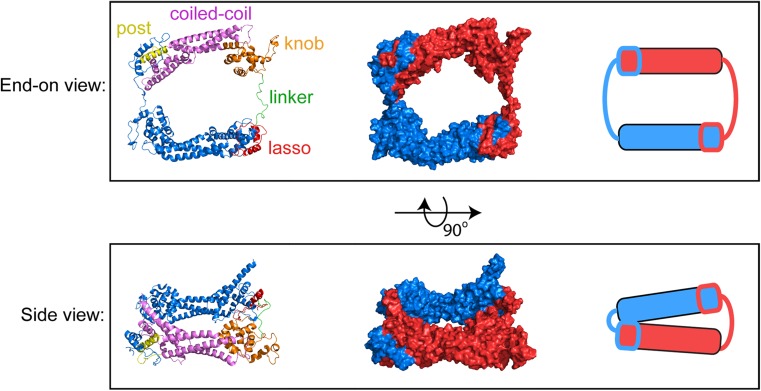
Consistent with the dimerization requirement for FH2-mediated filament assembly (Moseley et al. 2004), crystal structures confirmed that FH2 domains exist as tethered dimers (Lu et al. 2007; Thompson et al. 2013; Xu et al. 2004; Yamashita et al. 2007). Each FH2 monomer consists of an elongated, mainly alpha-helical structure that self-associates in a head-to-tail manner to form a closed, ring-like dimer (Fig. 2). Dimerization is mediated via packing of two conserved tryptophan or other aromatic residues in the “lasso” region of one monomer into hydrophobic pockets defined by glycine residues in the “post” region of the second monomer (Pruyne 2016; Xu et al. 2004). A flexible “linker” connects the lasso to the “knob” region located in the core of the domain. The length of the linker is specific to each formin isoform, and it is characterized as a disordered region based on the absence of electron density in structures of the FH2 domains of the mammalian formins mDia1 and FMNL3, as well as its ability to adopt an extended conformation in the actin-bound structure of Bni1p (Nezami et al. 2010; Otomo et al. 2005; Thompson et al. 2013). FH2 linkers can also adopt secondary structural features including an alpha-helix in the structure of Bni1p solved in the absence of actin (Xu et al. 2004), and a short, two-stranded beta-sheet that forms between the ends of an otherwise unstructured linker in the mammalian formin Daam1 (Lu et al. 2007; Yamashita et al. 2007). In the latter case, the beta-sheet confers an unusual orientation to the Daam1 FH2 dimer that occludes the actin-binding sites. Disruption of the beta-sheet by mutagenesis increases the actin assembly properties of Daam1, suggesting that the linker region might be a regulatory site for this formin (Lu et al. 2007).
Fig. 2.
Atomic structure of the dimeric FH2 domain of Bni1p. End-on and side views of the dimeric FH2 domain of the S. cerevisiae formin Bni1p. The structure depicts the actin-bound FH2 dimer, based on pdb ID 1Y64 (Otomo et al. 2005), following 160 ns of all-atom molecular dynamics simulations (Baker et al. 2015). Left = Ribbon representations of the FH2 dimer. One monomer is color-coded to highlight the lasso, linker, knob, coiled-coil and post subdomains. The second monomer is depicted in blue. Center = surface representations of the FH2 dimer; the monomers are depicted in red and blue. Right = cartoon representations of the FH2 dimer
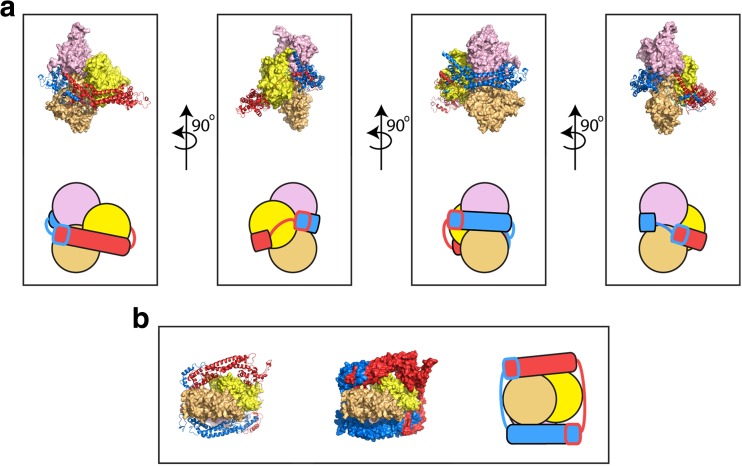
A co-crystal of the FH2 domain of Bni1p in complex with actin provided the first structural insights into the interactions of formins with the barbed ends of actin filaments (Otomo et al. 2005) (Fig. 3). In this structure, rather than associating as a discrete dimer, neighboring FH2 domains associated through lasso-post interactions to form a continuous chain of FH2 domains encircling an actin filament. FH2 domains of the mammalian formin INF2 have been observed to adopt a similar mode of continuous filament encirclement, confirming that at high formin concentrations, exchange of lasso/post interaction sites enables higher-level oligomerization of FH2 domains (Gurel et al. 2014; Sharma et al. 2014). Manual rearrangement of the flexible linker region yielded a model of a discrete dimer of Bni1p FH2 domains bound to three successive actin subunits, which adopted a 180° rotation relative to their nearest neighbors in the opposite strand rather than the traditional 167° rotation that is normally populated in filaments (Oda et al. 2009; Otomo et al. 2005). In this model, each FH2 domain interacts with two actin subunits via distinct binding sites, one in the knob and one in the post. The FH2 subunits are staggered, such that the “leading” FH2 domain interacts with the terminal and second-to-last actin subunits, and the “trailing” FH2 domain interacts with the second- and third-to-last subunits (Fig. 3).
Fig. 3.
Structure of the dimeric FH2 domain of Bni1p bound to actin. Structural representations of the FH2 dimer of the S. cerevisiae formin Bni1p bound to the three terminal subunits of an actin filament following 160 ns of all-atom molecular dynamics simulations (Baker et al. 2015). The initial structural model was constructed based on the crystal structure of the actin-bound Bni1p FH2 domain (pdb ID 1Y64) (Otomo et al. 2005). The structures of the FH2 domains are represented as ribbon diagrams and the individual domains are colored red and blue. The structures of the three actin subunits are depicted as surface representations. The terminal, barbed end subunit is depicted in light orange. The other two actins are shown in yellow and pink. a Multiple orientations of the side view are depicted. The FH2 dimer forms contacts with all three actin subunits. Cartoon representations are also shown to orient the reader. b End-on view of the FH2-actin complex. The FH2 dimer is shown both with ribbon diagrams (right) and as a surface representation (center). A cartoon representation is also shown (left)
Molecular dynamics simulations of a Bni1p FH2 dimer bound at the barbed end of an actin filament allowed for refinement of the model for the interactions between formin and actin (Baker et al. 2015). An initial structure was built through a series of alignments of the structures of the Bni1p FH2 domain in complex with actin (Otomo et al. 2005) and an actin filament (solved using fiber diffraction; (Oda et al. 2009)), which imposed a 167° angle of rotation between successive actin subunits. Over the course of 160 ns of simulations, the angles of rotation between the neighboring terminal subunits became heterogeneous and shifted toward larger values, reminiscent of the original crystal structure, although a full transition to the 180° conformation did not occur (Baker et al. 2015). A number of salt-bridges also formed during the simulations and appeared to stabilize the FH2-actin interactions. Consistent with the contribution of electrostatic interactions to FH2-barbed end association, binding of an FH2 dimer of mDia1 has been shown to increase actin filament flexibility and weaken interactions between neighboring actin subunits in an ionic-strength-dependent manner (Bugyi et al. 2006; Papp et al. 2006).
Structural models for elongation and gating
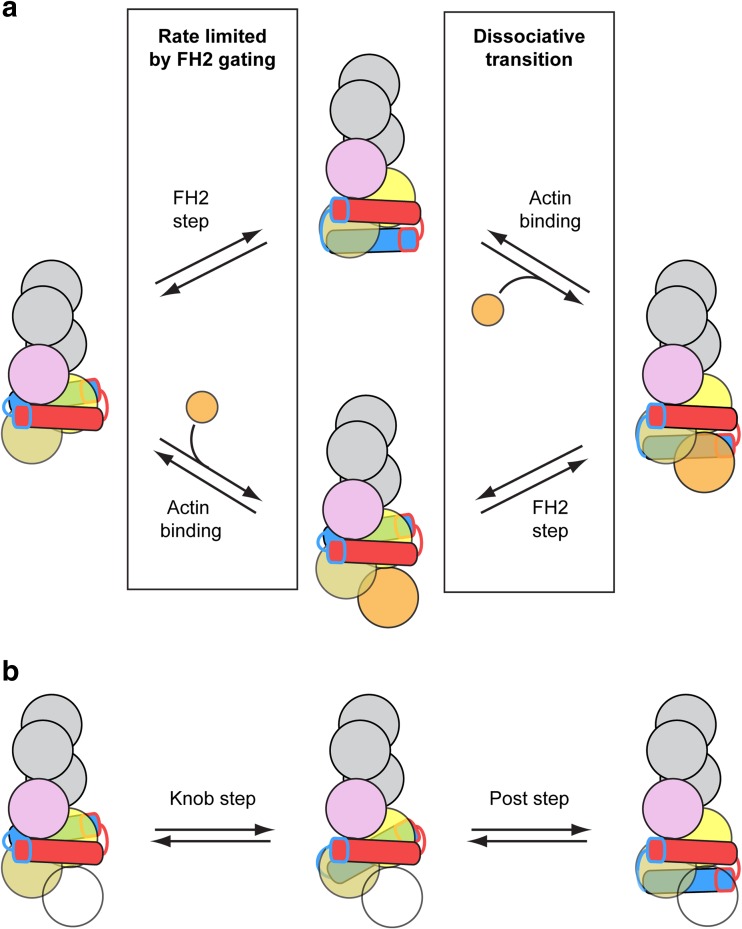
Based on the structure of the Bni1p FH2 domain, a “stair-stepping” model for FH2-mediated filament elongation postulates that binding of both subunits of the FH2 dimer sterically blocks the barbed end, corresponding to a closed conformation (Goode and Eck 2007; Xu et al. 2004). According to this model, dissociation of the intermolecular contacts between one FH2 monomer and actin allows the trailing FH2 domain to step forward in the barbed end direction to alleviate the steric blockage and populate an open conformation. Stepping forward requires the dislocated FH2 domain to bind the terminal actin subunit with its knob subdomain while its post subdomain presents an available binding site for a new actin subunit (Fig. 4a, upper reaction scheme). An incoming actin subunit can then bind both the newly accessible barbed end and the dissociated post subdomain, restoring all four FH2-actin contacts and repopulating a closed conformation. The stair-stepping model assumes that the partially dissociated FH2 domain does not interfere with subunit addition, and that incoming actin subunits can bind the non-canonical 180° rotational orientation of the terminal actin subunits.
Fig. 4.
Two models for FH2-mediated actin polymerization. a Cartoon representation of the addition of an actin subunit (orange circle) to the barbed end of an actin filament bound by an FH2 dimer. Upper reaction scheme = the “stair-stepping” model in which the trailing FH2 domain (blue domain) steps forward before an incoming actin subunit (orange circle) binds to the barbed end. Lower reaction scheme = the “stepping second” model, in which actin binds to the barbed end prior to stepping of the trailing FH2 domain. In both models, the first reaction is limited by FH2 gating. Dissociation of the FH2 dimer from the barbed end is most likely to occur during the second reaction, which involves the population of a dissociative transition state (Cao et al. 2018; Paul and Pollard 2008, 2009b). b Cartoon representation of a two-step model for the translocation of the trailing FH2 domain, in which an intermediate state is populated (Thompson et al. 2013). The intermediate state is formed by dissociation of the knob subdomain from its trailing binding site (pink circle), followed by translocation and association with a binding site at the barbed end (light orange circle). In this state, the FH2 dimer is bound to two actin subunits. Complete stepping of the trailing FH2 domain is achieved via subsequent translocation of the post subdomain, as depicted in the second reaction. This two-step model for stepping can occur as part of either the stair-stepping or stepping second model for actin subunit addition. To account for this, the incoming actin subunit is depicted as a transparent circle
An alternative, “stepping second,” model proposes that the transition from a closed to an open FH2-barbed end conformation and subsequent subunit addition occur while both FH2 domains remain bound to the terminal actin subunits (Paul and Pollard 2008, 2009b) (Fig. 4a, lower reaction scheme). This model suggests that association of a formin FH2 dimer traps the barbed end in a high-energy, closed conformation characterized by a large (i.e., > 167°) actin subunit rotational orientation, which is unfavorable for subunit addition. Transition from this conformation to an open state involves concerted conformational changes within the FH2 dimer and the barbed end, resulting in a canonical 167° subunit orientation and alleviation of steric blocking by the FH2 domain. In contrast to the stair-stepping model, the stepping second model assumes that FH2 domain dissociation and stepping follow, rather than precede, subunit addition (Fig. 4a).
Recent all-atom molecular dynamics simulations of Bni1p, mDia1 and the fission yeast formin Cdc12p bound to actin filaments provided further evidence that FH2 domains increase the angle of helical rotation of the terminal actin subunits, making the end less favorable for subunit addition (Aydin 2018). The terminal subunits of the filament bound by Cdc12p, which strongly gates elongation (Kovar et al. 2003, 2006), populated rotational angles that likely compromise subunit addition (> 170°). In contrast, the barbed end of the filament bound by mDia1, which weakly gates subunit addition (Kovar et al. 2006), populated rotational angles that were favorable for binding of an incoming actin subunit (~ 168°). The helical distortion was largest at the terminal subunits and dissipated gradually away from the end. The correlation between the angles of terminal subunit rotation and the extent of filament gating by the bound formins supports the hypothesis that formins inhibit elongation by imposing a strain that flattens the barbed end subunits (Paul and Pollard 2008, 2009b). Over the course of the simulations, the FH2 domains of all three formins also interfered sterically with an incoming actin subunit, with the largest clashes produced by Cdc12p. Thermal motions enabled movement of the FH2 domains out of sterically blocked sites, suggesting that dissociation or stepping of an FH2 domain is not required to open the barbed end for elongation. These results support the hypothesis that both steric blocking by the FH2 dimer and a distorted helical orientation of the terminal subunits contribute to the closed conformation, as postulated by the stepping second model (Paul and Pollard 2009b).
A crystal structure of actin in complex with an FH2 dimer of FMNL3 provided insight into a potential intermediate state along the pathway for elongation mediated by formins (Thompson et al. 2013). In this structure, the FMNL3 FH2 dimer is bound to two actin monomers, which are spatially separated and do not interact with one another. Consistent with the Bni1p structure, each FMNL3 FH2 domain interacts with both actin monomers via binding sites in the knob and post subdomains. However, the structure of FMNL3 FH2-actin is suggestive of a distinct intermediate step in the processive model for elongation in which the FH2 dimer interacts with only two actin subunits via all four knob and post sites. This intermediate state precedes both subunit addition and final displacement of the trailing FH2 domain (Fig. 4b). The order in which these final two steps occurs, which remains the major distinguishing feature of the stair-stepping and stepping second models, is yet to be determined.
Mechanism of FH2 processivity
Processive association with the barbed end during elongation causes formin FH2 domains to rotate around the axis of the helical actin filament as new subunits are incorporated (Mizuno et al. 2011, 2018). However, when formin FH2 domains are surface-immobilized, they can assemble filaments that elongate without rotating (Kovar and Pollard 2004). A model described this “rotation paradox” as slipping of the FH2 dimer around the filament to relieve strain that accumulates during subunit addition (Shemesh et al. 2005). Restriction of rotational freedom in a magnetic tweezers assay slows actin elongation mediated by mDia1 considerably (Yu et al., 2018; Yu et al. 2017), indicating that this mechanism might not be utilized efficiently by all formins. An alternative explanation postulates that slippage of the FH2 dimer relative to the glass surface can alleviate torsional stress (Mizuno et al. 2011).
Direct visualization of elongating actin filaments by TIRF microscopy revealed that the rate of dissociation of an FH2 dimer from a barbed end is proportional to the rate of formin-mediated subunit addition (Cao et al. 2018; Paul and Pollard 2008). Consistent with the stabilizing effects of salt-bridge formation on FH2-actin binding (Baker et al. 2015), processive association with barbed ends is dependent on ionic strength (Cao et al. 2018). In the presence of profilin, formin processivity increases in a manner that requires both the formation of profilin-actin complexes and the association of profilin with polyproline tracts in the formin’s FH1 domain (Cao et al. 2018).
The flexible FH2 linker region also strongly influences processivity, as revealed in polymerization assays with purified variants of the Bni1p FH2 domain in which the wild-type linker was replaced by linkers from other formins. These experiments demonstrated that the rate of dissociation is roughly proportional to the length of the linker (Paul and Pollard 2009a).
Kinetic modeling of formin-mediated elongation suggested that formin dissociation most likely occurs via a dissociative transition state that follows binding of a new actin subunit to the barbed end (Cao et al. 2018; Paul and Pollard 2008). This transition state is populated during each cycle of actin subunit addition, so the probability of dissociation increases as formins mediate more cycles of actin addition. In the “stepping second” model for filament elongation, this transition state corresponds to stepping of the trailing FH2 domain, at which point the formin is bound to the barbed end via only the leading FH2 domain, which provides a minimal number of interactions (Fig. 4a, lower reaction scheme) (Paul and Pollard 2009a). In the “stair-stepping” model, the transition state corresponds to binding of the new actin subunit by the pre-stepped, formerly-trailing FH2 domain (Fig. 4a, upper reaction scheme) (Goode and Eck 2007; Xu et al. 2004).
FH1-mediated elongation in the presence of profilin
The majority of unpolymerized actin in cells is bound to profilin (Carlsson et al. 1977; Kaiser et al. 1999; Suarez et al. 2015), a protein that also binds polyproline (Perelroizen et al. 1994; Petrella et al. 1996; Tanaka and Shibata 1985). Profilin binds in the barbed end groove of actin monomers and strongly inhibits both actin filament nucleation and elongation of the pointed end (Pollard and Cooper 1984). Profilin has a much weaker affinity for the barbed end of polymerized actin, consistent with structural differences between monomeric and filamentous actin (Courtemanche and Pollard 2013; Jegou et al. 2011; Oda et al. 2009). Thus, profilin dissociates rapidly from the barbed end after profilin-actin complex addition. Although the rate of filament elongation is largely unaffected by the association of ATP-actin monomers with profilin, high concentrations of excess profilin slow and can even reverse elongation by binding to, and increasing the dissociation rate of, the terminal subunit (Courtemanche and Pollard 2013; Jegou et al. 2011).
Owing to its affinity for polyproline, profilin also interacts with formin FH1 domains (Chang et al. 1997; Watanabe et al. 1997). These domains are typically located directly N-terminal to the FH2 domain, and contain discrete polyproline tracts that form rigid type-II polyproline helices and serve as sites for profilin binding (Horan et al. 2018). The polyproline tracts are separated by regions of low sequence conservation, which lend an overall flexible structure to the domain. The numbers of polyproline tracts in mammalian formin FH1 domains range from 2, as seen in mDia2 and Delphilin, to 19, as seen in FMN2 (Horan et al. 2018). Polyproline tract lengths are also variable, ranging from as few as 3 to as many as 13 successive prolines (Horan et al. 2018). Non-proline residues often interrupt otherwise contiguous sequences of prolines. The significance of these non-proline residues is not well understood, but structural studies have demonstrated that the number of intermolecular contacts that form upon profilin binding to a polyproline tract, as well as the binding polarity and registry, are strongly influenced by the non-proline residues (Kursula et al. 2008; Mahoney et al. 1999). Consistent with this, a leucine residue in one of the two polyproline tracts of the FH1 domain of Cdc12p is essential for profilin binding (Yonetani et al. 2008).
Polyproline tracts bind profilin in a groove of highly conserved aromatic residues on the face opposite the actin-binding site (Ferron et al. 2007; Kursula et al. 2008; Schutt et al. 1993), allowing profilin to bind both actin and a polyproline tract in the FH1 domain simultaneously (Ferron et al. 2007; Perelroizen et al. 1994; Tanaka and Shibata 1985). Neighboring polyproline tracts in FH1 domains can also bind profilin simultaneously (Horan et al. 2018; Kursula et al. 2008).
The affinity of profilin for polyproline increases with polyproline tract length, so longer tracts bind profilin more tightly than do shorter tracts (Perelroizen et al. 1994; Petrella et al. 1996). Most FH1 domains contain a combination of tracts, which together establish a gradient of affinities for profilin, such that tracts located closer to the FH2 domain bind profilin more weakly than do tracts located farther away from the FH2 domain (Courtemanche and Pollard 2012). Profilin-actin binds polyproline tracts up to 11-fold more tightly than does profilin alone, and the affinity of monomeric actin for profilin increases 2-fold when the profilin is polyproline-bound (Ferron et al. 2007).
Despite profilin’s inhibitory effects on actin nucleation, low concentrations of profilin stimulate polymerization of actin in bulk samples that contain formin FH1FH2 constructs (Sagot et al. 2002). The FH1 domain is essential for this enhancement, which results from an increase in the elongation rate of individual filaments associated with formin FH1FH2 constructs (Kovar et al. 2006; Paul and Pollard 2008; Romero et al. 2004). Whereas elongation rates of filaments bound by formin FH2 constructs are limited by gating, profilin-stimulated elongation rates mediated by FH1FH2 constructs can significantly exceed the diffusion-limited elongation rates of filaments not bound by formins in the same conditions (Drenckhahn and Pollard 1986).
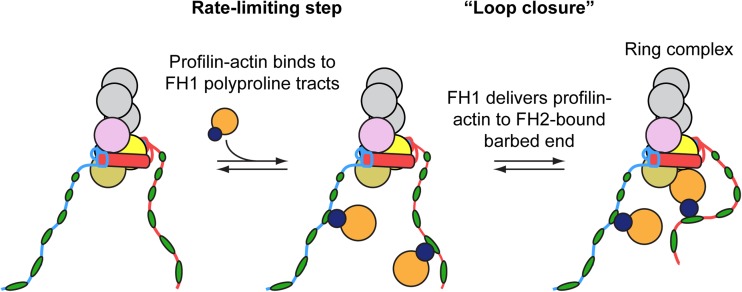
A series of experimental and computational studies provided insight into the mechanism by which profilin stimulates elongation of filaments bound by formin FH1FH2 dimers. Binding polyproline tracts in the FH1 domain effectively increases the local profilin-actin concentration at the barbed end (Vavylonis et al. 2006; Yu et al., 2018) (Fig. 5). The inherent flexibility of the FH1 domain promotes rapid collisions (predicted to occur on the order of 104 s−1) between the bound profilin-actin complexes and the barbed end, enabling direct delivery of profilin-actin to the FH2-associated barbed end when an open FH2 conformation is populated (Vavylonis et al. 2006). Delivery proceeds via the formation of a “ring complex” in which the formin interacts with the barbed end both directly via its FH2 domain and indirectly via binding of an FH1-associated profilin-actin complex (Fig. 5). Formation of this transient ring complex has been proposed to stabilize the formin at the barbed end, preventing dissociation and promoting processivity (Cao et al. 2018). Subsequent dissociation of profilin, which interacts only weakly with barbed ends, from the newly bound actin subunit allows FH2-mediated incorporation of the subunit into the filament. Dissociation of profilin from the barbed end can either precede or follow FH1-profilin dissociation (Vavylonis et al. 2006).
Fig. 5.
Model for FH1-mediated delivery of profilin actin to the FH2-bound barbed end. Cartoon representation of the addition of a profilin-bound actin subunit (dark blue and orange circles) to the barbed end of an actin filament bound by a representative formin FH1FH2 construct. The unstructured FH1 domains extend outwards from the FH2 dimer and contain multiple polyproline tracts (in this case, six green ovals per FH1 domain). The number of prolines in each polyproline tract tends to increase with the distance from the FH2 domain. Profilin-actin complexes bind the polyproline tracts and are delivered directly to the barbed end in a “loop closure” reaction via diffusion of the FH1 domain and formation of a ring complex (Vavylonis et al. 2006). Subsequent profilin and polyproline tract dissociation events occur rapidly and are not depicted
A thermodynamic analysis of the interactions between formin domains, profilin, actin monomers, and the barbed end determined that the energy associated with formin-mediated subunit addition is sufficient to drive processive elongation (Paul and Pollard 2009a). This is consistent with studies demonstrating that the rate of phosphate release from polymerized actin is unaffected by formin-mediated subunit addition to the barbed end (Paul and Pollard 2009a), and that formins can polymerize ADP-actin monomers (Kovar et al. 2006; Kubota et al. 2017), both of which indicate that processive elongation is not coupled to ATP hydrolysis.
The rate of elongation mediated by formin FH1FH2 constructs depends on the concentration of profilin (Kovar et al. 2006; Paul and Pollard 2008). Under most conditions, the rate-limiting step in elongation is binding of profilin-actin to the FH1 domain (Vavylonis et al. 2006). Thus, maximal elongation rates are observed in conditions where the majority of actin monomers are profilin-bound. In conditions in which the concentration of profilin exceeds the concentration of actin monomers, free profilin competes with profilin-actin complexes for binding polyproline tracts, dramatically reducing the efficiency of FH1-mediated profilin-actin transfer to the barbed end and slowing elongation (Kovar et al. 2006; Paul and Pollard 2008).
Consistent with the laws of thermodynamic reversibility and conservation of energy, formin-mediated actin subunit addition is a reversible process. As a result, depolymerization of filaments with mDia1(FH1FH2)-bound barbed ends has been observed in the presence of excess profilin (Jegou et al. 2011, 2013; Kubota et al. 2017; Pernier et al. 2016). Formation of the ring complex via the association of FH1-profilin with the terminal actin subunit of a FH2-bound barbed end likely promotes subunit dissociation and depolymerization. As is the case for filaments with free barbed ends (Courtemanche and Pollard 2013; Jegou et al. 2011; Pernier et al. 2016), formin-bound actin filaments depolymerize most rapidly when subunits are ADP-bound (Kubota et al. 2017).
The two FH1 domains in each formin dimer deliver actin independently of one another, with the leading FH1 domain exhibiting a higher probability of transfer at any given time (Courtemanche and Pollard 2012; Horan et al. 2018). Each polyproline tract binds and delivers actin in a diffusion-limited manner (Courtemanche and Pollard 2012; Paul and Pollard 2008). The gradient of affinities for profilin exhibited by the polyproline tracts within the FH1 domain of Bni1p promotes weak binding of profilin-actin near the FH2 domain, where collisions with the barbed end are likely to be frequent, and tighter binding of profilin-actin farther away from the FH2 domain, where collisions are relatively infrequent (Courtemanche and Pollard 2012). The FH1 domains of most formins are similarly organized, suggesting that optimization of position-specific profilin binding affinities might be a general mechanism for tuning FH1-mediated elongation activity (Courtemanche and Pollard 2012). A second mechanism for tuning the rate of elongation is isoform-selective profilin binding, which supports optimal formin-mediated elongation only in the context of a specific profilin isoform (Ezezika et al. 2009; Neidt et al. 2009). This selectivity appears not to be confined to interactions of profilin with the FH1 domain, but might also be a requirement for efficient FH2-mediated subunit incorporation (Neidt et al. 2009).
Effects of force on formin-mediated filament elongation
Owing to their participation in dynamic cellular processes including cytokinesis, focal adhesion assembly and filopodial extension, formins are exposed to forces as they polymerize actin filaments (Bidone et al. 2014; Mellor 2010; Tang et al. 2014; 2015). An initial model for processive, FH2-mediated filament elongation proposed that the formin mechanism is mechano-sensitive (Kozlov and Bershadsky 2004), suggesting that compressive or tensile forces could modulate polymerization by influencing the rate at which the trailing FH2 domain steps forward. Consistent with the hypothesis that formins can generate and withstand picoNewton-level compressive forces, actin filaments were observed to buckle as they elongated while tethered to a glass surface by a formin attachment point at their barbed end and an inactive myosin attachment point at the pointed end (Kovar and Pollard 2004). In a separate study, mechanical compression of Xenopus XTC cells induced a release of monomeric actin, which in turn increased mDia1-mediated polymerization activity, confirming that formins can function while under compressive force (Higashida et al. 2013).
A series of in vitro experiments have shown that application of force influences the rate of formin-mediated elongation in isoform-specific ways. Tensile forces ranging from 0.1 to 5.5 pN, exerted along formin-anchored filaments either via hydrodynamic flow or by an optical trap, increased elongation rates mediated by both Bni1p and mDia1 up to threefold in the presence of ATP-actin monomers and profilin (Courtemanche et al. 2013; Jegou et al. 2013; Kubota et al. 2017; Yu et al., 2018). In the absence of profilin, force sped elongation by mDia1 2-fold (Cao et al. 2018; Kubota et al. 2017; Yu et al. 2017), but slowed and ultimately halted elongation by Bni1p (Courtemanche et al. 2013). Much faster mDia1-mediated elongation rates (up to 800 subunits/s, constituting an increase in the elongation rate of up to 10-fold) were observed in assays utilizing magnetic tweezers (Yu 2018; Yu et al. 2017). Such efficient acceleration of mDia1-mediated elongation under tension required filaments that were free of torsion and super-coiling, ensuring formin rotational freedom about the axis of the filament as the FH2 dimer stepped onto new subunits (Yu 2018; Yu et al. 2017).
Formins remain processively associated with barbed ends of both polymerizing and depolymerizing filaments even under tension (Courtemanche et al. 2013; Jegou et al. 2013; Kubota et al. 2017; Yu et al., 2018; Yu et al. 2017; Zimmermann et al. 2017). Tensile force has also been shown to promote efficient elongation of mDia1-bound, ADP-actin filaments in otherwise depolymerizing conditions, consistent with a force-induced decrease in critical concentration (Kubota et al. 2017).
Kinetic modeling of experimental results suggested that FH2 gating by both mDia1 and Bni1p is force-sensitive (Courtemanche et al. 2013; Jegou et al. 2013). In the case of mDia1, force biases the FH2 dimer toward populating an open conformation, thus speeding elongation by favoring a polymerization-competent barbed end state. In contrast, force shifts the conformational equilibrium of Bni1p-associated barbed ends toward a closed state that precludes polymerization, but profilin-actin association with the FH1 domain allosterically reverses this effect.
Although formins can mediate both filament polymerization and depolymerization under force (Jegou et al. 2013; Kubota et al. 2017), tension was shown to decrease the processivity of mDia1 and mDia2 by several orders of magnitude, overcoming the stabilizing effects of profilin (Cao et al. 2018). Fitting of experimentally determined, tension-dependent dissociation rates with a model that considered two possible pathways for dissociation of an FH2 dimer from the barbed end indicated that tension promotes dissociation from an open gating conformation that precedes actin binding rather than from the dissociative transition state that follows actin subunit addition (Cao et al. 2018). Consistent with a model in which force alters the conformational equilibrium of the FH2 dimer (Courtemanche et al. 2013; Jegou et al. 2013; Kozlov and Bershadsky 2004), this finding provides additional evidence for a tension-sensitive gating mechanism. It remains to be seen whether the processivity of Bni1p is altered in the same way despite its favoring of a closed conformation under force.
In vitro reconstitution assays of the effects of myosin-induced tension along filaments anchored and polymerized by two cytokinesis-specific formins revealed that force dramatically slows polymerization mediated by the fission yeast formin Cdc12p, but that the mammalian formin mDia2 is relatively mechano-insensitive (Zimmermann et al. 2017). Chimeric constructs consisting of the FH1 domain of one formin fused to the FH2 domain of the other revealed that the force-sensitivity of Cdc12p is conferred via its FH1 domain. Quantitative modeling suggested that the tension generated by myosin’s ATPase activity and power stroke cycle propagates along the actin filament and ultimately stretches the anchored, flexible FH1 domain of the barbed end-associated formin (Zimmermann et al. 2017). Stretching is predicted to increase the relative accessibility of the profilin-binding polyproline tracts that might otherwise be occluded within the disordered FH1 domain (Bryant et al. 2017). Concomitantly, an increase in the end-to-end distance of the FH1 domain likely decreases the probability of collisions between FH1-bound profilin-actin and the barbed end (i.e., formation of the ring complex), slowing delivery and elongation. Thus, force can influence different aspects of the formin mechanism in isoform-specific ways, which might confer mechano-sensitive properties appropriate for each formin’s cellular role.
Mechanism of actin filament nucleation by formins
Purified formin FH2 domains robustly stimulate actin nucleation rates in bulk kinetic assays (Li and Higgs 2003; Lu et al. 2007; Maiti et al. 2012; Moseley et al. 2004; Patel et al. 2018; Pring et al. 2003; Pruyne et al. 2002; Ramabhadran et al. 2012; Sagot et al. 2002; Silkworth et al. 2018; Yamashita et al. 2007). Profilin inhibits nucleation both in the absence and presence of formin FH2 domains (Pollard and Cooper 1984; Pruyne et al. 2002; Sagot et al. 2002; Zigmond et al. 2003), suggesting that formins nucleate filaments from free actin monomers rather than from profilin-actin (Paul and Pollard 2008, 2009b). In contrast, FH1FH2 constructs can nucleate filaments in the presence of profilin (Li and Higgs 2003; Paul and Pollard 2008; Pring et al. 2003). Once formed, these new filaments elongate rapidly via FH1-mediated delivery of profilin-actin to the barbed end, thus stimulating overall polymer assembly.
Based on time courses of bulk actin assembly in the presence of Bni1p, Pring et al. proposed a kinetic model for formin-mediated filament nucleation in which FH2 dimers bind and stabilize actin dimers (Pring et al. 2003). Consistent with this model, the crystal structures of Bni1p and FMNL3 FH2-actin complexes revealed that FH2 dimers could encircle and bind actin dimers or trimers, stabilizing these otherwise short-lived intermediates of the polymerization pathway (Otomo et al. 2005; Thompson et al. 2013). Disruption of two intermolecular salt bridges that form between residues in the FH2 dimer and actin dramatically decreased filament nucleation by Bni1p in bulk assembly assays, highlighting the importance of electrostatic interactions for the formation of stable, initial contacts between formin FH2 domains and filament nuclei (Baker et al. 2015). These mutations did not impact FH2 gating or the rate of Bni1p-mediated filament elongation, providing evidence that nucleation is a biochemically separable function of formins that involves the formation of a unique subset of contacts with actin (Baker et al. 2015).
Nucleation activity varies among formins, and some formins use alternative mechanisms to overcome relatively weak intrinsic nucleation activities. These additional mechanisms are likely to be especially important in the cellular context, where the vast majority of monomeric actin is profilin-bound. For example, the assembly of a discrete FMNL3 FH2-actin complex with spatially separated actin monomers could inhibit the formation of a productive filament nucleus, thus explaining this formin’s weak nucleation activity (Thompson et al. 2013). On the other hand, the C-terminal WH2 motif of FMNL3, which tightly binds actin monomers, dramatically enhances nucleation by this formin (Thompson et al. 2013), consistent with a model in which the FH2 dimer binds two actin subunits while the WH2-like motifs interact independently with additional actin monomers, thus bringing four subunits together. WH2-like motifs have been identified in the C-terminal regions of other formins, suggesting that this mechanism for enhancing weak FH2-mediated nucleation may be conserved (Breitsprecher and Goode 2013; Pruyne 2016).
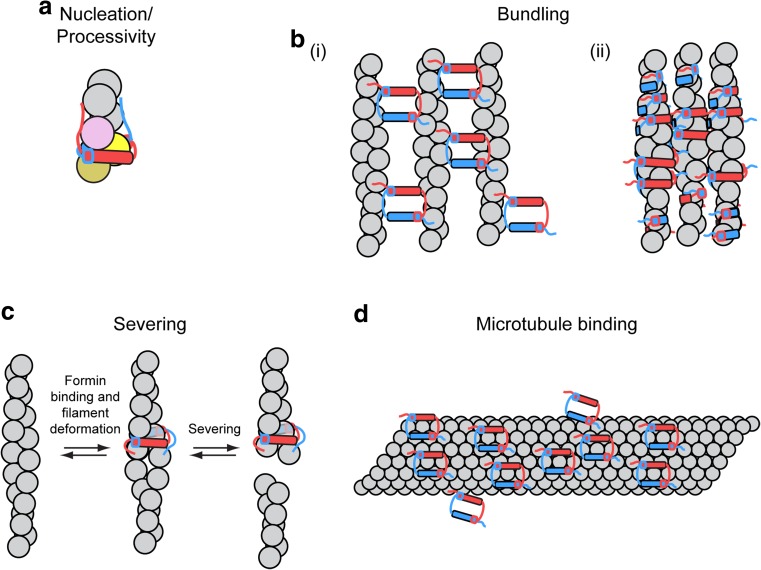
The C-terminal DAD motifs of the mammalian formins mDia1 and Daam1, as well as the budding yeast formins Bni1p and Bnr1p also bind actin monomers and increase rates of nucleation mediated by FH1FH2 constructs in bulk assembly assays performed in the absence and presence of profilin (Gould et al. 2011). The C-terminal region of the Drosophila formin Capuccino (Capu), a homolog of the FMN family of formins, lacks both DAD and WH2-like motifs, but promotes nucleation likely through non-specific electrostatic interactions with actin (Vizcarra et al. 2014) (Fig. 6a).
Fig. 6.
Formin tail-mediated interactions with actin and microtubules. Cartoon representations of interactions between formin FH2-tail constructs with actin filaments and microtubules. Tail regions are depicted as unstructured regions and are color-coded to match their corresponding FH2 domain. a Electrostatic interactions between tail regions and actin promote formin-mediated filament nucleation and FH2 processivity. b Formins can associate with the sides of actin filaments (i) via binding sites on the exterior surfaces of their FH2 domains or (ii) by encircling the filaments with their FH2 domains. In several cases, actin filament decoration by formins promotes filament bundling. Binding is often strengthened via interactions between formin tail regions and actin. c A model for INF2-mediated severing in which an FH2-tail construct of the formin INF2 encircles an actin filament and creates a localized deformation, which promotes severing (Gurel et al. 2014). d FH2-tail constructs of a representative formin decorate a microtubule (gray tube) via interactions mediated by the post subdomains and strengthened by the tail regions
Secondary binding proteins can also enhance or inhibit formin nucleation activities. In budding yeast, Bud6p stimulates filament nucleation through simultaneous associations with actin monomers and the C-terminus of Bni1p in a mechanism that complements the activity of the FH1 domain (Evangelista et al. 1997; Graziano et al. 2011; Moseley et al. 2004). In so doing, Bud6p enables Bni1p to overcome the inhibitory effect of profilin-actin complex formation on spontaneous actin dimer and trimer assembly in cells. Other ligands of formin C-terminal tail regions that regulate nucleation include Disheveled, which binds and relieves autoinhibition of the formin Daam1, thus promoting its nucleation and elongation activities (Liu et al. 2008); adenomatous polyposis coli (APC), which simultaneously binds mDia1 and actin monomers, and remains bound to the pointed end of actin filaments as they are elongated by mDia1 following nucleation (Breitsprecher et al. 2012; Okada et al. 2010; Wen et al. 2004); Spire, a WH2-motif-containing protein that binds and inhibits actin filament nucleation and processive barbed end association by Fmn-family formins, while enhancing nucleation by Spire (Quinlan et al. 2007; Rosales-Nieves et al. 2006; Vizcarra et al. 2011); Ena/Vasp, which nucleates and bundles actin filaments on its own, but also binds and suppresses the nucleation activities of Dia formins (Bilancia et al. 2014); and the F-BAR family protein Hof1p and kinesin-like myosin passenger-protein Smy1p, which both inhibit nucleation and slow filament elongation mediated by Bnr1p via interactions with its FH1 and FH2 domains, respectively (Chesarone-Cataldo et al. 2011; Graziano et al. 2014).
Additional formin activities
Actin filament side-binding and bundling
In addition to their barbed end-binding activity, several formins bind along the lengths of actin filaments (Gurel et al. 2014; Harris et al. 2006; Silkworth et al. 2018; Vizcarra et al. 2014). This binding pattern is thought to arise from non-specific, electrostatic interactions between positively charged formin FH2 domains and negatively charged, solvent-accessible patches on filamentous actin subunits (Harris et al. 2006). Formins whose FH2 domains have high isoelectric points therefore may be predisposed to bind along the lengths of actin filaments in this manner (Harris et al. 2006). For some formins, binding along the filament length is strengthened via necessary, but insufficient, supplementary interactions between the C-terminal formin tail domain and actin (Vizcarra et al. 2014).
Side-binding of some, but not all, formins leads to actin filament bundling (Harris et al. 2006). Analysis of bundles induced by binding of FRL1 or mDia2 by electron microscopy revealed mixtures of parallel and anti-parallel filament orientations (Harris et al. 2006), suggesting that filament decoration and bundling by these formins is insensitive to filament polarity. Bundles induced by binding of Capu also lack uniform polarity (Vizcarra et al. 2014).
Formins bind along the lengths of actin filaments in two general orientations. Following filament nucleation, the Arabidopsis formin AFH1 rapidly transfers from the barbed end to the side of the filament, where it can continue to nucleate filaments, suggesting that regions on the external surface of the FH2 domain are involved in binding to filament sides (Michelot et al. 2005, 2006). In contrast, FH2 dimers of mDia2, FRL1, and INF2 physically dissociate and encircle actin filaments either as discrete dimers or as higher order oligomers in which neighboring FH2 domains interact in an end-to-end manner (Gurel et al. 2014; Harris et al. 2006; Otomo et al. 2005; Sharma et al. 2014). Following filament decoration in either orientation, additional interactions with actin filaments involving residues located on the outside surface of the FH2 dimers can promote actin bundling (Fig. 6b). Consistent with the role of electrostatic interactions in driving filament binding and bundling, formins with low isoelectric points and FH2 domains that do not readily dissociate do not exhibit significant side-binding or bundling activities (Harris et al. 2006).
Actin filament severing
Upon encircling and binding along the lengths of actin filaments, INF2 and FRL1 accelerate actin disassembly by promoting filament severing and depolymerization (Chhabra and Higgs 2006; Gurel et al. 2014, 2015; Harris et al. 2004). INF2-mediated severing occurs at sites where INF2 has bound and requires phosphate release following ATP hydrolysis by the filamentous actin (Gurel et al. 2014). Binding of INF2 to actin introduces a 1.5% change in the helical rise of the filament (Gurel et al. 2014). In a current model for INF2-mediated severing, the change in the filament structure induced by INF2 binding is proposed to expose and promote binding of the C-terminal INF2 DAD sequence to the barbed end groove of a proximal filamentous actin subunit (Fig. 6c) (Gurel et al. 2014). This association further disrupts the filament structure and promotes severing. Thermal motions likely also play a role in INF2-mediated severing, as evidenced by a delay that occurs between INF2 binding and subsequent severing (Gurel et al. 2014). Release of actin monomers from the WH2 motifs results in a net depolymerization and promotes nucleotide exchange and rapid FH1-mediated reincorporation into filaments upon binding to profilin (Gurel et al. 2015). Changes in the concentration of INF2 can thus promote either rapid filament depolymerization or maintenance of short filaments.
Microtubule binding
In addition to their roles as regulators of actin filament dynamics, formins have emerged as coordinators of actin and microtubule networks during essential cellular processes including motility, morphogenesis, transport, and nuclear migration (Goode et al., 2000; Rodriguez et al. 2003; Wallar and Alberts 2003). Initial evidence that formins interact with, and alter the dynamics of, microtubules emerged from a study showing that a constitutively active construct of mDia2 co-localized with a subset of microtubules in vivo and bound to microtubules in vitro (Palazzo et al. 2001). Since then, all formins tested have been demonstrated to bind microtubules directly in vitro, although their interaction mechanisms differ (Bartolini et al. 2008; Gaillard et al. 2011; Roth-Johnson et al. 2014; Young et al. 2008; Zhou et al. 2006). Formin constructs encompassing the FH1 and FH2 domains and C-terminal tail regions of mDia1, mDia2, and INF2 and Capu bind the microtubule lattice with similar high affinities (< 150 nM) and stoichiometries ranging from 1:1 (tubulin dimer/formin dimer) for mDia2 to 3:1 for INF2 and mDia1 (Gaillard et al. 2011; Roth-Johnson et al. 2014) (Fig. 6d). The C-terminal tail regions of mDia2 and Capu are required for high-affinity binding through nonspecific charge-based interactions, whereas those of mDia1 and INF2 are not. FH1FH2 constructs of mDia2, INF2, and Capu also bind microtubules via electrostatic interactions mediated by several conserved charged residues located in the post region of the FH2 domain, which might serve to orient the formins on the microtubule lattice (Roth-Johnson et al. 2014). Binding of microtubules by some formins, including INF2 and Capu, induces microtubule bundling (Gaillard et al. 2011; Rosales-Nieves et al. 2006).
Formin-microtubule association can influence both formin activity and microtubule dynamics (Bartolini and Gundersen 2010; Chesarone et al. 2010). Microtubule binding inhibits actin filament nucleation fully by mDia2 and Capu, partially by mDia1 and not at all by INF2, but has no effect on the rate of actin elongation mediated by Capu (Gaillard et al. 2011; Rosales-Nieves et al. 2006). On the other hand, mDia2 binding stabilizes microtubules by slowing their depolymerization (Bartolini et al. 2008), whereas binding and bundling of microtubules by INF2 significantly decreases the rate of catastrophe (Gaillard et al. 2011). In cultured fibroblasts and primary neurons, mDia1 binding also decreases the frequency of microtubule catastrophes, thus promoting microtubule network stability (Pianu et al. 2014; Qu et al. 2017).
In addition to binding the microtubule lattice directly, mDia1 associates indirectly with microtubule plus ends via an interaction with a “formin elongation effector domain” (FEED) motif found in the amino acid sequence of CLIP-170, a protein that also interacts with the microtubule end-binding protein EB1. Binding of CLIP-170 dramatically increases the rate of mDia1-mediated actin filament elongation, an effect that requires co-localization of both proteins at filament barbed ends (Henty-Ridilla et al. 2016). This enhancement is not specific to mDia1, and is also observed upon binding of CLIP-170 to other formins including mDia2, Daam1, INF1, and INF2.
Conclusions and open questions
In recent years, as the important biological roles played by formins have come into focus, numerous advances have provided details of the mechanisms by which formins promote actin filament nucleation and elongation through processive association with barbed ends. A number of unique and surprising features of individual formin isoforms also have come to light. Building on this progress, the field is now poised to answer a number of remaining mechanistic questions. Examples of unanswered questions include: what conformational changes are associated with the FH2 gating equilibrium and what are the kinetics of these changes? How are actin binding and formin stepping coordinated in time? Does the formin step first or step second? Is there a relationship between gating and the rate of transfer of profilin-actin from the FH1 domain to the barbed end? How is delivery of profilin-actin from the multiple polyproline tracts in the FH1 domain coordinated? What is the molecular basis for the effects of force on formin FH2 gating? How do binding partners enhance or disrupt actin assembly by formins? And perhaps most challenging of all: how does possessing particular actin polymerization activities render a formin isoform most suitable to fulfill its cellular role?
Acknowledgements
The author thanks Antoine Jégou for helpful discussions and Jessica Henty-Ridilla, David Pruyne, and Mark Zweifel for critical reading of the manuscript.
Funding
This work was supported by National Institutes of Health research grant GM-122787.
Conflict of interest
Naomi Courtemanche declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the authors.
References
- Ahuja R, et al. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131(2):337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura T, et al. Dilated cardiomyopathy-associated FHOD3 variant impairs the ability to induce activation of transcription factor serum response factor. Circ J. 2013;77(12):2990–2996. doi: 10.1253/circj.CJ-13-0255. [DOI] [PubMed] [Google Scholar]
- Aydin F, Courtemanche N, Pollard TD, Voth GA. Gating mechanisms during actin filament elongation by formins. Elife. 2018;7:e37342. doi: 10.7554/eLife.37342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Courtemanche N, Parton DL, McCullagh M, Pollard TD, Voth GA. Electrostatic interactions between the Bni1p formin FH2 domain and actin influence actin filament nucleation. Structure. 2015;23(1):68–79. doi: 10.1016/j.str.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Formins and microtubules. Bba-Mol Cell Res. 2010;1803(2):164–173. doi: 10.1016/j.bbamcr.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, Gundersen GG. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181(3):523–536. doi: 10.1083/Jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidone TC, Tang HS, Vavylonis D. Dynamic network morphology and tension buildup in a 3D model of cytokinetic ring assembly. Biophys J. 2014;107(11):2618–2628. doi: 10.1016/j.bpj.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilancia CG, et al. Enabled negatively regulates diaphanous-driven actin dynamics in vitro and in vivo. Dev Cell. 2014;28(4):394–408. doi: 10.1016/j.devcel.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan S, Schultz J, Grosshans J. Formin' cellular structures: physiological roles of diaphanous (Dia) in actin dynamics. Commun Integr Biol. 2013;6(6):e27634. doi: 10.4161/cib.27634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombardier JP, et al. Single-molecule visualization of a formin-capping protein ‘decision complex’ at the actin filament barbed end. Nat Commun. 2015;6:8707. doi: 10.1038/Ncomms9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer O, et al. INF2 mutations in Charcot-Marie-tooth disease with glomerulopathy. New Engl J Med. 2011;365(25):2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126(1):1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336(6085):1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42(1):72–U91. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Clemens L, Allard J. Computational simulation of formin-mediated actin polymerization predicts homologue-dependent mechanosensitivity. Cytoskeleton. 2017;74(1):29–39. doi: 10.1002/cm.21344. [DOI] [PubMed] [Google Scholar]
- Bugyi B, et al. Formins regulate actin filament flexibility through long range allosteric interactions. J Biol Chem. 2006;281(16):10727–10736. doi: 10.1074/jbc.M510252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao LY, et al. Modulation of formin processivity by profilin and mechanical tension. Elife. 2018;7:e34176. doi: 10.7554/eLife.34176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low-molecular weight protein in non-muscle cells. J Mol Biol. 1977;115(3):465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120(12):3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137(1):169–182. doi: 10.1083/Jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, et al. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320(5873):239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11(1):62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chesarone-Cataldo M, Guerin C, Yu JH, Wedlich-Soldner R, Blanchoin L, Goode BL. The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a Formin damper. Dev Cell. 2011;21(2):217–230. doi: 10.1016/j.devcel.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281(36):26754–26767. doi: 10.1074/jbc.M60466200. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Buhle EL, Walker SB, Tsong TY, Pollard TD. Kinetic evidence for a monomer activation step in actin polymerization. Biochemistry. 1983;22(9):2193–2202. doi: 10.1021/Bi00278a021. [DOI] [PubMed] [Google Scholar]
- Courtemanche N, Lee JY, Pollard TD, Greene EC. Tension modulates actin filament polymerization mediated by formin and profilin. P Natl Acad Sci USA. 2013;110(24):9752–9757. doi: 10.1073/pnas.1308257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD. Determinants of formin homology 1 (FH1) domain function in actin filament elongation by formins. J Biol Chem. 2012;287(10):7812–7820. doi: 10.1074/jbc.M111.322958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD. Interaction of profilin with the barbed end of actin filaments. Biochemistry-Us. 2013;52(37):6456–6466. doi: 10.1021/bi400682n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn D, Pollard TD. Elongation of actin-filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J Biol Chem. 1986;261(27):2754–2758. [PubMed] [Google Scholar]
- Dutta P, Das S, Maiti S. Non diaphanous formin delphilin acts as a barbed end capping protein. Exp Cell Res. 2017;357(2):163–169. doi: 10.1016/j.yexcr.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Ercan-Sencicek AG, et al. Homozygous loss of DIAPH1 is a novel cause of microcephaly in humans. Eur J Hum Genet. 2015;23(2):165–172. doi: 10.1038/ejhg.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, et al. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276(5309):118–122. doi: 10.1126/Science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Ezezika OC, et al. Incompatibility with formin Cdc12p prevents human profilin from substituting for fission yeast profilin insights from crystal structures of fission yeast profilin. J Biol Chem. 2009;284(4):2088–2097. doi: 10.1074/jbc.M807073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10(6):693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11(21):1656–1665. doi: 10.1016/S0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26(21):4597–4606. doi: 10.1038/Sj.Emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C. Polymerization of actin - mechanism of the mg-2+-induced process at Ph-8 and 20-degrees-C. P Natl Acad Sci-biol. 1983;80(21):6513–6517. doi: 10.1073/Pnas.80.21.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J, et al. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell. 2011;22(23):4575–4587. doi: 10.1091/mbc.E11-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Drubin DG, Barnes G. Functional cooperation between the microtubule and actin cytoskeletons. Curr Opin Cell Biol. 2000;12(1):63–71. doi: 10.1016/S0955-0674(99)00058-7. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21(5):384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, et al. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22(21):4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, et al. The F-BAR protein Hof1 tunes formin activity to sculpt actin cables during polarized growth. Mol Biol Cell. 2014;25(11):1730–1743. doi: 10.1091/mbc.E14-03-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel PS, et al. INF2-mediated severing through actin filament encirclement and disruption. Curr Biol. 2014;24(2):156–164. doi: 10.1016/j.cub.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel PS, Mu A, Guo BQ, Shu R, Mierke DF, Higgs HN. Assembly and turnover of short actin filaments by the formin INF2 and profilin. J Biol Chem. 2015;290(37):22494–22506. doi: 10.1074/jbc.M115.670166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. The mouse formin, FRL alpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem. 2004;279(19):20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281(20):14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- Heimsath EG, Higgs HN. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J Biol Chem. 2012;287(5):3087–3098. doi: 10.1074/jbc.M111.312207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Rankova A, Eskin JA, Kenny K, Goode BL. Accelerated actin filament polymerization from microtubule plus ends. Science. 2016;352(6288):1004–1009. doi: 10.1126/science.aaf1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashida C, et al. F- and G-actin homeostasis regulates mechanosensitive actin nucleation by formins. Nat Cell Biol. 2013;15(4):395. doi: 10.1038/ncb2693. [DOI] [PubMed] [Google Scholar]
- Higashida C, et al. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303(5666):2007–2010. doi: 10.1126/Science.1093923. [DOI] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30(6):342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16(1):1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan BG, Zerze GH, Kim YC, Vavylonis D, Mittal J. Computational modeling highlights the role of the disordered formin homology 1 domain in profilin-actin transfer. FEBS Lett. 2018;592(11):1804–1816. doi: 10.1002/1873-3468.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegou A, Carlier MF, Romet-Lemonne G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat Commun. 2013;4:1883. doi: 10.1038/Ncomms2888. [DOI] [PubMed] [Google Scholar]
- Jegou A, et al. Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 2011;9(9):e1001161. doi: 10.1371/journal.pbio.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser DA, Vinson VK, Murphy DB, Pollard TD. Profilin is predominantly associated with monomeric actin in Acanthamoeba. J Cell Sci. 1999;112(21):3779–3790. doi: 10.1242/jcs.112.21.3779. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124(2):423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161(5):875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. P Natl Acad Sci USA. 2004;101(41):14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Wu JQ, Pollard TD. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell. 2005;16(5):2313–2324. doi: 10.1091/Mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM, Bershadsky AD. Processive capping by formin suggests a force-driven mechanism of actin polymerization. J Cell Biol. 2004;167(6):1011–1017. doi: 10.1083/jcb.200410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. ENA/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Bi. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- Kubota H, Miyazaki M, Ogawa T, Shimozawa T, Kinosita K, Ishiwata S. Biphasic effect of profilin impacts the formin mDia1 force-sensing mechanism in actin polymerization. Biophys J. 2017;113(2):461–471. doi: 10.1016/j.bpj.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursula P, et al. High-resolution structural analysis of mammalian profilin 2a complex formation with two physiological ligands: the formin homology 1 domain of mDial and the proline-rich domain of VASP. J Mol Biol. 2008;375(1):270–290. doi: 10.1016/j.jmb.2007.10.050. [DOI] [PubMed] [Google Scholar]
- Law R, et al. Biallelic truncating mutations in FMN2, encoding the actin-regulatory protein Formin 2, cause nonsyndromic autosomal-recessive intellectual disability. Am J Hum Genet. 2014;95(6):721–728. doi: 10.1016/j.ajhg.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13(15):1335–1340. doi: 10.1016/S0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280(8):6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- Liu W, et al. Mechanism of activation of the formin protein Daam1. P Natl Acad Sci USA. 2008;105(1):210–215. doi: 10.1073/pnas.0707277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Meng WY, Poy F, Maiti S, Goode BL, Eck MJ. Structure of the FH2 domain of Daam1: implications for formin regulation of actin assembly. J Mol Biol. 2007;369(5):1258–1269. doi: 10.1016/j.jmb.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch ED, Lee MK, Morrow JE, Welcsh PL, Leon PE, King MC. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278(5341):1315–1318. doi: 10.1126/Science.278.5341.1315. [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Rozwarski DA, Fedorov E, Fedorov AA, Almo SC. Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat Struct Biol. 1999;6(7):666–671. doi: 10.1038/10722. [DOI] [PubMed] [Google Scholar]
- Maiti S, Michelot A, Gould C, Blanchoin L, Sokolova O, Goode BL. Structure and activity of full-length formin mDia1. Cytoskeleton. 2012;69(6):393–405. doi: 10.1002/cm.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor H. The role of formins in filopodia formation. Bba-Mol Cell Res. 2010;1803(2):191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Michelot A, et al. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol. 2006;16(19):1924–1930. doi: 10.1016/j.cub.2006.07.054. [DOI] [PubMed] [Google Scholar]
- Michelot A, et al. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell. 2005;17(8):2296–2313. doi: 10.1105/tpc.105.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Higashida C, Yuan YF, Ishizaki T, Narumiya S, Watanabe N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331(6013):80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Tanaka K, Yamashiro S, Narita A, Watanabe N. Helical rotation of the diaphanous-related formin mDia1 generates actin filaments resistant to cofilin. P Natl Acad Sci USA. 2018;115(22):E5000–E5007. doi: 10.1073/pnas.1803415115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, et al. A conserved mechanism for Bni1-and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15(2):896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidt EM, Scott BJ, Kovar DR. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J Biol Chem. 2009;284(1):673–684. doi: 10.1074/jbc.M804201200. [DOI] [PubMed] [Google Scholar]
- Nezami A, Poy F, Toms A, Zheng W, Eck MJ. Crystal structure of a complex between amino and Carboxy terminal fragments of mDia1: insights into autoinhibition of diaphanous-related formins. PLoS One. 2010;5(9):e12992. doi: 10.1371/journal.pone.0012992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Aihara T, Wakabayashi K. Early nucleation events in the polymerization of actin, probed by time-resolved small-angle x-ray scattering. Sci rep-Uk. 2016;4:34539. doi: 10.1038/Srep34539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular-to fibrous-actin transition. Nature. 2009;457(7228):441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Okada K, et al. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189(7):1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433(7025):488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001;3(8):723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- Papp G, et al. Conformational changes in actin filaments induced by formin binding to the barbed end. Biophys J. 2006;91(7):2564–2572. doi: 10.1529/biophysj.106.087775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, Durer ZAO, van Loon AP, Bremer KV, Quinlan ME. Drosophila and human FHOD family formin proteins nucleate actin filaments. J Biol Chem. 2018;293(2):532–540. doi: 10.1074/jbc.M117.800888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation (vol 18, pg 9, 2008) Curr Biol. 2008;18(3):233–233. doi: 10.1016/j.cub.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J Biol Chem. 2009;284(18):12533–12540. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by Formins. Cell Motil Cytoskeleton. 2009;66(8):606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Kitchen SM, West RA, Sigler R, Eisenmann KM, Alberts AS. Myeloproliferative defects following targeting of the Drf1 gene encoding the mammalian diaphanous-related formin mDia1. Cancer Res. 2007;67(16):7565–7571. doi: 10.1158/0008-5472.CAN-07-1467. [DOI] [PubMed] [Google Scholar]
- Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33(28):8472–8478. doi: 10.1021/Bi00194a011. [DOI] [PubMed] [Google Scholar]
- Pernier J, Shekhar S, Jegou A, Guichard B, Carlier MF. Profilin interaction with actin filament barbed end controls dynamic instability, capping, branching, and motility. Dev Cell. 2016;36(2):201–214. doi: 10.1016/j.devcel.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141(5):1217–1228. doi: 10.1083/Jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella EC, Machesky LM, Kaiser DA, Pollard TD. Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 1996;35(51):16535–16543. doi: 10.1021/Bi961498d. [DOI] [PubMed] [Google Scholar]
- Pianu B, Lefort R, Thuiliere L, Tabourier E, Bartolini F. The a beta(1-42) peptide regulates microtubule stability independently of tau. J Cell Sci. 2014;127(5):1117–1127. doi: 10.1242/jcs.143750. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Bioph Biom. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative-analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23(26):6631–6641. doi: 10.1021/Bi00321a054. [DOI] [PubMed] [Google Scholar]
- Pring M, Evangelista M, Boone C, Yang CS, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry-Us. 2003;42(2):486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- Pruyne D. Revisiting the phylogeny of the animal formins: two new subtypes, relationships with multiple wing hairs proteins, and a lost human formin. PLoS One. 2016;11(10):e0164067. doi: 10.1371/journal.pone.0164067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, et al. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297(5581):612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Qu XY, et al. Stabilization of dynamic microtubules by mDia1 drives tau-dependent a beta(1-42) synaptotoxicity. J Cell Biol. 2017;216(10):3161–3178. doi: 10.1083/jcb.201701045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila spire is an actin nucleation factor. Nature. 2005;433(7024):382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, spire and cappuccino. J Cell Biol. 2007;179(1):117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramabhadran V, Gurel PS, Higgs HN. Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. J Biol Chem. 2012;287(41):34234–34245. doi: 10.1074/jbc.M112.365122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5(7):599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119(3):419–429. doi: 10.1016/J.Cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves AE, Johndrow JE, Keller LC, Magie CR, Pinto-Santini DM, Parkhurst SM. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, spire and cappuccino. Nat Cell Biol. 2006;8(4):367–U47. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Johnson EA, Vizcarra CL, Bois JS, Quinlan ME. Interaction between microtubules and the Drosophila Formin cappuccino and its effect on actin assembly. J Biol Chem. 2014;289(7):4395–4404. doi: 10.1074/jbc.M113.499921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4(8):626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Schonichen A, Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Bba-Mol Cell Res. 2010;1803(2):152–163. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NCW, Lindberg U. The structure of crystalline profilin beta-actin. Nature. 1993;365(6449):810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81(2):667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, et al. Nanostructured self-assembly of inverted Formin 2 (INF2) and F-actin-INF2 complexes revealed by atomic force microscopy. Langmuir. 2014;30(25):7533–7539. doi: 10.1021/la501748x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar S, et al. Formin and capping protein together embrace the actin filament in a menage a trois. Nat Commun. 2015;6:8730. doi: 10.1038/Ncomms9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh T, Otomo T, Rosen MK, Bershadsky AD, Kozlov MM. A novel mechanism of actin filament processive capping by formin: solution of the rotation paradox. J Cell Biol. 2005;170(6):889–893. doi: 10.1083/jcb.200504156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, et al. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol Cell. 2004;13(4):511–522. doi: 10.1016/S1097-2765(04)00059-0. [DOI] [PubMed] [Google Scholar]
- Silkworth WT, Kunes KL, Nickel GC, Phillips ML, Quinlan ME, Vizcarra CL. The neuron-specific formin Delphilin nucleates nonmuscle actin but does not enhance elongation. Mol Biol Cell. 2018;29(5):610–621. doi: 10.1091/mbc.E17-06-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, et al. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell. 2015;32(1):43–53. doi: 10.1016/j.devcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Shibata H. Poly(L-proline)-binding proteins from chick-embryos are a profilin and a profilactin. Eur J Biochem. 1985;151(2):291–297. doi: 10.1111/J.1432-1033.1985.Tb09099.X. [DOI] [PubMed] [Google Scholar]
- Tang HS, Bidone TC, Vavylonis D. Computational model of polarized actin cables and cytokinetic actin ring formation in budding yeast. Cytoskeleton. 2015;72(10):517–533. doi: 10.1002/cm.21258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HS, Laporte D, Vavylonis D. Actin cable distribution and dynamics arising from cross-linking, motor pulling, and filament turnover. Mol Biol Cell. 2014;25(19):3006–3016. doi: 10.1091/mbc.E14-05-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ME, Heimsath EG, Gauvin TJ, Higgs HN, Kull FJ. FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation. Nat Struct Mol Biol. 2013;20(1):111–U143. doi: 10.1038/nsmb.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Kovar DR, O'Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21(4):455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Bor B, Quinlan ME. The role of formin tails in actin nucleation, processive elongation, and filament bundling. J Biol Chem. 2014;289(44):30602–30613. doi: 10.1074/jbc.M114.588368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, et al. Structure and function of the interacting domains of spire and Fmn-family formins. P Natl Acad Sci USA. 2011;108(29):11884–11889. doi: 10.1073/pnas.1105703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallar BJ, Alberts AS. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13(8):435–446. doi: 10.1016/S0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Watanabe N, et al. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for rho small GTPase and is a ligand for profilin. EMBO J. 1997;16(11):3044–3056. doi: 10.1093/Emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of rho and promote cell migration. Nat Cell Biol. 2004;6(9):820–8U1. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Wooten EC, et al. Formin homology 2 domain containing 3 variants associated with hypertrophic cardiomyopathy. Circ-Cardiovasc Gene. 2013;6(1):10–18. doi: 10.1161/CIRCGENETICS.112.965277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YW, et al. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116(5):711–723. doi: 10.1016/S0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yamashita M, et al. Crystal structure of human DAAM1 formin homology 2 domain. Genes Cells. 2007;12(11):1255–1265. doi: 10.1111/j.1365-2443.2007.01132.x. [DOI] [PubMed] [Google Scholar]
- Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol Biol Cell. 2008;19(5):2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KG, Thurston SF, Copeland S, Smallwood C, Copeland JW. INF1 is a novel microtubule-associated formin. Mol Biol Cell. 2008;19(12):5168–5180. doi: 10.1091/mbc.E08-05-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Le S, Efremov AK, Zeng X, Bershadsky A, Yan J (2018) Effects of mechanical stimuli on profilin- and formin-mediated actin polymerization. Nano Lett. 10.1021/acs.nanolett.8b02211 [DOI] [PubMed]
- Yu M, et al. mDia1 senses both force and torque during F-actin filament polymerization. Nat Commun. 2017;8:1650. doi: 10.1038/S41467-017-01745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Leder P, Martin SS. Formin-1 protein associates with microtubules through a peptide domain encoded by exon-2. Exp Cell Res. 2006;312(7):1119–1126. doi: 10.1016/j.yexcr.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Zigmond SH, et al. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13(20):1820–1823. doi: 10.1016/j.cub.2003.90.057. [DOI] [PubMed] [Google Scholar]
- Zimmermann D, et al. Mechanoregulated inhibition of formin facilitates contractile actomyosin ring assembly. Nat Commun. 2017;8:703. doi: 10.1038/S41467-017-00445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, La Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11(4):451–U198. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]