Abstract
Plant receptor-like proteins (RLPs) are a family of transmembrane receptors which are distinguished from receptor-like kinases (RLKs) by their lack of a cytoplasmic kinase domain. RLPs continue to be implicated in a broad range of plant immunological and developmental processes as critical sensors or participants in receptor complexes on the plasma membrane. RLPs often associate with RLKs to activate or attenuate signal perception and relay. Some RLPs also physically cluster with RLKs and bear similar expression patterns. Here, we discuss the characteristics, function, and expression of characterized RLPs in the context of their associated RLKs in plant immunity and development.
Keywords: Receptor-like protein, Leucine-rich repeat, Receptor-like kinase, Pattern recognition receptor, Plant Immunity, Plant development
1. Introduction
Perception of extracellular signals by transmembrane receptors is a nearly ubiquitous feature of cellular life. The term phytosemiotics was coined to describe the study of the signal processes used by plants to sense and respond to stimuli at the cellular level [1]. In plants, a continuous and coordinated communication with the environment must be uniquely mandated by their general sessility. Further, their lack of a circulatory system with specialized immune cells likely compounds this challenge, and may account for some of the differences in immune signaling mechanisms between plants and animals [2].
Plant immunity and development often rely on similar or overlapping cellular mechanisms for signal recognition and transduction. Frequently, transmembrane receptors are the first actors in play. While receptor tyrosine kinases (RTK) are the most common class of transmembrane receptor protein kinases (RPKs) in animals, they are comparatively rare in plants. Instead, plant RPKs most often use serine/threonine kinases (STKs) for immediate downstream signal transduction, although several plant RPKs have tyrosine kinase activity [3–5]. The term receptor-like kinase (RLK) refers to plant receptor and nonreceptor protein kinases [6]. Phylogenetic analysis suggests that an ancient gene duplication event prior to the divergence of plants and animals led to the independent evolution of plant RLKs and animal RTKs [4]. RLKs comprise more than 60% of all kinases in Arabidopsis thaliana [4, 7]. The proportion of homologous RLKs found in tandem clusters and segmentally duplicated regions in the A. thaliana genome suggests a selective pressure towards the expansion of this gene family [8]. RLKs with various extracellular ligand-recognition domains (e.g. leucine-rich repeats, lectin/lectin-like, epidermal growth factor-like, etc.) have been identified. However, the leucine-rich repeat containing RLKs (LRR-RLKs) are not only the most common, but also the most studied RLKs in plants [6, 9].
Receptor-like proteins (RLPs) share structural similarity with RLKs but lack a cytoplasmic kinase domain [10, 11]. This likens them to the Toll-like receptors (TLRs) involved in mammalian immunity, which also contain an extracellular LRR domain and a short cytoplasmic tail [12]. It should come as no surprise, then, that several RLPs and RLKs have been identified as pattern recognition receptors (PRRs) critical in plant innate immunity [13, 14]. Certain RLPs and RLKs also have crucial functions in plant development [15, 16]. This is consistent with the originally observed dorsal-ventral patterning function of the Drosophila Toll gene for which human TLRs were named [17]. The combined immune and developmental functions of RLPs and RLKs suggest they may play a broad and critical role in the detection of both self and non-self.
Far more is currently known about RLKs relative to RLPs, but a growing body of evidence suggests that the role of RLPs is as important as RLKs. By examining the character, function, and expression of RLPs in the context of their associated RLKs, we aim to summarize that body of evidence, and to discuss the state of knowledge on RLPs in plant immunity and development.
2. RLP Characteristics and Expression
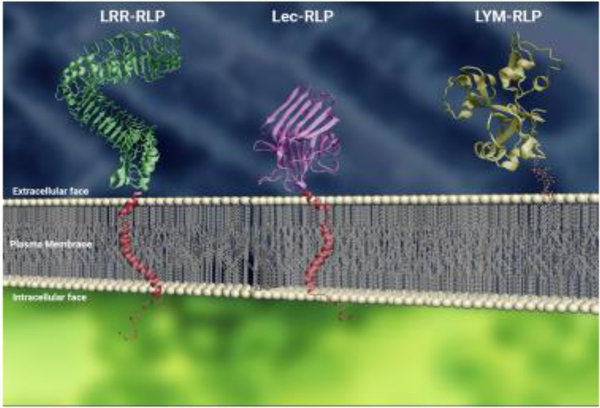
In simplest terms, RLPs can be thought of as transmembrane receptors with an extracellular ligand binding domain and a short cytoplasmic tail (Figure 1). It can be generally assumed, then, that RLPs are single-pass (type I) transmembrane proteins [4, 10, 11]. However, some proteins without transmembrane domains might be considered as RLPs by virtue of their attachment to the extracellular face of the plasma membrane using a glycosylphosphatidylinositol (GPI) anchor [18, 19]. In addition, RLPs traditionally contain an N-terminal signal peptide.
Figure 1: Conceptual representations of RLPs with different ectodomains on the plasma membrane.
From left: LRR-RLP (green), Lec-RLP (purple) and LYM-RLP (gold); transmembrane helices are depicted in red. The transmembrane helices were added to depict a representation of the structure, but do not represent the actual structure of the transmembrane domains of these proteins. The protein structure of OsCEBiP (PDB: 5JCD) represents the LYM in this figure, anchored to the membrane with a GPI anchor. The intracellular area is further indicated by the depiction of short cytoplasmic tails (red). Molecular structures were visualized using Visual Molecular Dynamics (VMD) software [101]. PM and GPI anchor were modeled using Autodesk 3DS Max (2017). Proteins containing domains with similar structure were used to represent RLPs: LRR-RLP (BRI1; PDB: 3RGX4), LecRLP (PHA-E; PDB: 3WCR5) [102, 103].
The most common extracellular ligand-binding domain found in RLPs is LRRs (Figure 1) [8]. There are about 223 LRR-RLKs and 57 LRR-RLPs in Arabidopsis [4, 7, 10, 11]. In LRR-RLKs, the intracellular kinase domain exhibits more conservation than the extracellular LRR domain [20]. A quantification of the number of LRRs in Arabidopsis LRR-RLKs reveals a bimodal distribution where RLKs containing three to six and 21 to 24 repeats are frequent, but proteins with three to nine LRRs are the most common. By contrast, most Arabidopsis LRR-RLPs have a high number LRRs ranging from 16 to 28 repeats [15]. Plant-derived extracellular LRRs are 22–23 amino acids long, and each repeat forms a LxxLxxLxLxxNxLSGxIPxxLGx consensus [21]. In contrast to bacterial and animal LRR proteins which form a horseshoe-shaped structure, plant LRRs usually form a twisted or superhelical assembly because of an additional β-sheet forming the inner surface of the solenoid [21]. Likely due to their variability, LRRs have been observed to recognize a range of ligands, including sterols, lipids, sugars, peptides, lipopeptides, nucleic acids, and others.
RLP and/or RLK ectodomains have also been found to contain legume-like (L-type) lectins, G-type lectins, calcium-dependent (C-type) lectins, and the lectin-like Lysin-motifs (LysM). Plant lectin or lectin-like domains are usually involved in the perception of carbohydrates and glycans [22]. Among them, LysM RLKs or RLPs are well studied, while C-type lectins are rare in plants. There are 42–45 L-type lectin RLKs and six L-type lectin RLPs in Arabidopsis [22–24]. Compared to 39 G-type lectin-containing RLKs, only three G-type lectin-containing RLPs are predicted in Arabidopsis [23]. Further, of these predicted G-type lectin-containing RLPs in A. thaliana, one (AT4G21370) is an ortholog of the S-locus receptor kinase (SRK) in A. lyrata. In A. lyrata and other crucifers, this gene is a critical regulator of self-incompatibility which is expressed in the stigma. In A. thaliana, this gene is spliced differently, introducing an early stop codon before the cytoplasmic kinase domain [25]. The role of this gene in the evolution of self-fertility in A. thaliana remains to be determined. By contrast, only one C-type lectin-containing RLK (AT1G52310) has been predicted, and no C-type lectin-containing RLPs were found in Arabidopsis [23].
LysMs, named for their similarity to bacterial autolysins, are considered a distinct plant lectin subfamily and found in both plant RLKs and RLPs [8, 26] (Figure 1). In bacteria, autolysins catalyze the hydrolysis of the glycosidic bonds in peptidoglycan (PGN) to facilitate cell wall restructuring processes in growth and cell division [27]. LysMs in eukaryotes are likely the result of a horizontal gene transfer event, and in plants they are often adapted to recognize polysaccharide ligands such as PGN and fungal chitin [28, 29]. There are 5 LysM RLKs and 3 LysM RLPs that are characterized in Arabidopsis [28].
Other motifs, such as malectin-like domains, cysteine rich domains of unknown function (DUF26), and thaumatin domains have been found as ectodomains of RLKs and RLPs [8]. Except for malectin-like domain containing RLKs, their functions have not been well characterized. Malectin-like ectodomains were originally found in the Catharanthus roseus RLK1-like (CrRLK1L) subfamily of RLKs [30–32]. Malectin domains bear similarity to the animal carbohydrate-binding malectin proteins involved in the endoplasmic reticulum quality control [33]. Several Arabidopsis malectin-like domain RLKs, such as FERONIA and ANXURs, have a broad role in plant development, hormone signaling and immunity [30–32, 34]. There is one malectin-like domain RLP (At4g00300) in Arabidopsis with uncharacterized function [8].
Interestingly, some RLKs such as IMPAIRED OOMYCETE SUSCEPTIBILITY 1 (IOS1) contain both LRR and malectin-like domains and are named malectin-like/LRR-RLKs [4, 35]. The FLG22-INDUCED RECEPTOR-LIKE KINASE (FRK1), whose expression level is often used as an indicator of early defense [36], also belongs to the malectin-like/LRR-RLK subfamily. We have identified putative malectin-like/LRR-RLPs in Physcomitrella patens (XP_001756784.1) and Oryza sativa (XP_015645303.1), suggesting that malectin-like/LRR ectodomains might be a common feature in RLKs and RLPs in different plant species.
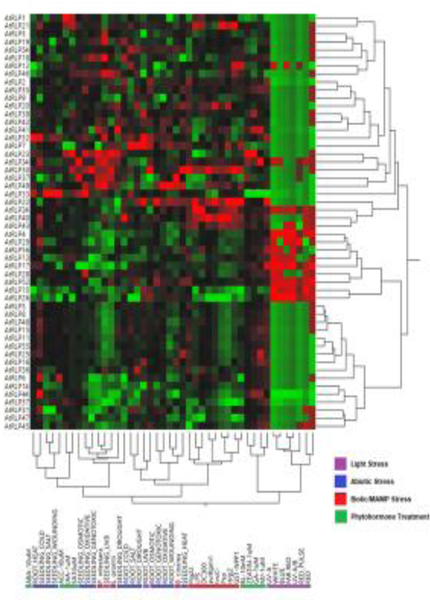
An often-overlooked dimension of RLP functions is their tissue-specific expression. When and where each RLP is expressed may modify its function specificity, e. g. by interacting with specific RLKs. In this way, characterization of tissue-specific expression pattern of individual RLKs and RLPs is a critical step in understanding their functions. The expression pattern of all 223 LRR-RLKs at various developmental stages have been systematically characterized with promoter::GUS transgenic plants [37]. The tissue-specific expression patterns of LRR-RLKs may provide insights into their functions in certain developmental stages. Analysis of available microarray datasets indicates that Arabidopsis LRR-RLPs are differentially expressed under a panoply of biotic, abiotic, light, and hormone treatments (Figure 2) [38]. In general, the expression pattern of LRR-RLPs during abiotic and biotic stress is similar, but distinct from the pattern observed during light stress (Figure 2). Biotic stresses, including treatments with Pseudomonas syringae, Phytopthora infestans, Botrytis cineria, Erysiphe orontii, or pathogen elicitors, caused an expression change in more Arabidopsis LRR-RLPs (~90% of all LRR-RLPs tested) than abiotic stresses (~80% of all LRR-RLPs tested) or hormone treatments (~40% of all LRR-RLPs tested) (Figure 2) [32]. The virulent P. syringae pv. tomato DC3000 suppressed the expression of more LRR-RLPs than it induced (15 vs. 9), whereas its type III secretion mutant DC3000 hrcC, or a non-adapted bacterium P. syringae pv. phaseolicola, induced far more LRR-RLPs than it suppressed LRR-RLPs (~20 vs. ~2) [32], suggesting the importance of LRR-RLPs in the arms race of plant immunity and bacterial pathogenicity (see below details for plant immune system).
Figure 2: Expression heatmap of LRR-RLPs in A. thaliana.
RLPs (Y-axis), and treatments (X-axis) are depicted after hierarchical two-way clustering. Relative transcriptional induction (red) and suppression (green) represent visualization of fold change in expression from the global median using values acquired from the AtGenExpress Visualization Tool [104]. This heatmap was generated using the JMP statistical software.
3. RLPs in Plant Immunity
3.1. The plant immune system
The first line of plant immunity is triggered by the recognition of conserved microbe-associated molecular patterns (MAMPs) [39]. Canonically, MAMPs are small, highly conserved molecules that are important for microbial survival or growth. MAMPs are perceived by PRRs and activate pattern-triggered immunity (PTI). PTI is characterized by the rapid deployment of a series of cellular responses including the activation of mitogen-activated protein kinase (MAPK) cascades, calcium-dependent protein kinases (CDPKs), the generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS), ion flux, callose deposition, phytohormone production, transcriptional reprogramming, production of phytoalexins, and likely other as-yet undiscovered responses [14, 40, 41].
Accordingly, pathogens have developed various virulence strategies to subvert PTI and promote pathogenicity. For example, pathogenic bacteria inject type III effectors (T3Es) and fungal pathogens haustorially deliver effectors to interfere with PTI signaling and responses [42]. These effectors are sometimes recognized, directly or indirectly, by host intracellular proteins. These proteins are predominantly encoded by nucleotide-binding site leucine-rich repeat (NBS-LRR or NLR) proteins, also called resistance (R) proteins, and trigger the second tier of defense known as effector-triggered immunity (ETI) [39]. ETI is characterized by a more drastic and robust activation of cellular defenses relative to PTI. Often, this activation is accompanied by a cell death process local to the infection site known as the “hypersensitive response” (HR) [39].
3.2. RLPs involved in responses triggered by apoplastic fungal effectors
The first characterized RLP Cf-9, encodes a LRR-RLP in tomato [43]. Cf-9 confers resistance specific to races of Cladosporium fulvum carrying the Avr9 effector gene, which was characterized in a gene-for-gene resistance now referred as ETI. Later, a second tomato LRR-RLP designated Cf-4 was shown to be required for resistance to C. fulvum expressing Avr4 [44]. While MAMP perception by RLKs or RLPs canonically leads only to the activation of PTI, both Avr4 and Avr9 recognition results in the activation of HR, a response more commonly associated with ETI [43, 44]. In addition, Cf-9 does not seem to directly perceive Avr9, as there is no direct binding between them [45]. Thus, as their designation (Avr for “avirulence”) would suggest, this classifies Avr4/Avr9 as apoplastic effectors, rather than MAMPs [46]. This example confounds the traditional R-gene/Avr-gene interaction scheme, as Cf-4 and Cf-9 are structurally similar to PRRs, but functionally defined as R genes. Consequently, apoplastic effector recognition by RLPs is distinctly considered to activate “effector-triggered defense” (ETD). ETD is distinguished from ETI not only by the location of effector recognition, but also by its longer response time, propensity for heterodimer-mediated activation, and rate of cell death [46].
The tomato resistance gene Ve, which confers race-specific resistance to Verticillium dahliae or V. albo-atrum carrying Ave1, was identified as two closely linked LRR-RLPs, Ve1 and Ve2 [47]. Although Ve1 and Ve2 share 84% amino acid identity, subsequent analysis indicated that Ve1, not Ve2, confers Ave1-mediated resistance [48]. Ave1 is an apoplastic effector, thus, Ve1 also induces ETD. The Brassica napus blackleg resistance gene LepR3 is an LRR-RLP, which confers resistance to Leptosphaeria maculans carrying the effector AvrLm1 in a gene-for-gene manner [49]. AvrLm1 is also important in resistance triggered by an independent R gene Rlm1, whose identity is still unknown [49].
3.3. RLPs involved in responses triggered by fungal and oomycete MAMPs
Chitin is a well-studied fungal MAMP with distinct perception and signaling systems in Arabidopsis and rice [14, 28, 40]. In Arabidopsis, chitin is perceived by two LYM-RLKs, AtLYK5 and AtLYK1, also called CHITIN ELICITOR RECEPTOR KINASE 1 (AtCERK1) [50, 51]. In rice, OsCERK1 does not have apparent chitin binding activity, instead, the LysM-RLP OsCEBiP directly binds to chitin and homodimerizes upon chitin binding [52] [53]. OsCERK1 cooperates with OsCEBiP to mediate chitin-triggered responses [54]. Two additional rice LysM-RLPs designated OsLYP4 and OsLYP6 have also been shown to bind to chitin and are required for chitin-triggered responses in rice [55]. Unlike classical LRR-RLPs which are assumed to be inserted into the plasma membrane through the transmembrane domain, OsCEBiP likely associates with the membrane through a GPI anchor [19]. Arabidopsis LysM-RLPs, by contrast, do not appear to be required for AtCERK1 function in chitin signaling [56].
Tomato LRR-RLPs ETHYLENE INDUCING XYLANASE 1 (SlEIX1) and SlEIX2 both bind to fungal xylanases, although only SlEIX2 is important in xylanase-mediated responses [57]. Interestingly, SlEIX1 could suppress SlEIX2-activated defense, suggesting that SlEIX1 may act as a functional decoy receptor [58]. Arabidopsis LRR-RLP AtRLP23 specifically binds to the 20 amino acid peptide “NECROSIS AND ETHYLENE-INDUCING PEPTIDE1 (NEP1)-LIKE PROTEINS” (NLPs), designated nlp20, and is required for nlp20-mediated responses [59]. Nep1, was originally identified in Fusarium oxysporum f. sp erythroxyli. This family of proteins was subsequently identified in several microbial clades, from bacteria to oomycetes [60]. Despite that NLPs are highly conserved in several plant pathogenic microbes, the corresponding receptor RLP23 does not have an obvious ortholog in other plant species (Figure 3). However, heterologous expression of AtRLP23 in potato conferred resistance to the oomycete Phytophthora infestans and the fungus Sclerotinia sclerotiorum [59]. The LRR-RLP ELICITIN RESPONSE (SmELR) from wild potato Solanum microdontum recognizes elicitin INF1, a conserved MAMP in several species of Phytophthora, and transfer of SmELR into cultivated potato enhanced its resistance to late blight [61]. The function of both AtRLP23 and SmELR outside of their original genetic context points to the possibility that interspecies transfer of RLP genes may sometimes represent a means to improve plant resistance to fungal and oomycete pathogens. Arabidopsis RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES 1 (AtRBPG1), which encodes LRR-RLP AtRLP42, forms a complex with polygalacturonases (PGs) derived from Botrytis cinerea, and is important for plant necrotic response to PGs [62]. Thus, AtRBPG1 is a putative receptor of PGs.
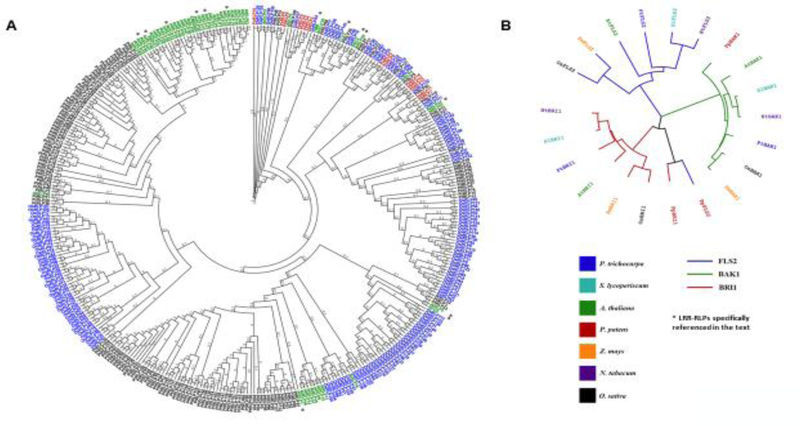
Figure 3. Phylogenetic analysis of RLPs and several well-studied RLKs.
(A) Phylogenetic tree of LRR-RLPs from A. thaliana (AtRLPs, green), P. patens (PpRLPs, red), P. trichocarpa (PtRLPs, black), and O. sativa (OsRLPs, blue). LRR-RLPs were identified for each species using the following bioinformatic pipeline: 1.) All Ref-seq proteins were scanned for matches to known protein domains using a client-side instance of EMBL-EBI InterPro Scan [105]. 2.) The dataset was filtered for proteins with predicted LRR domains, but without predicted kinase domains. 3.) The resulting proteins were manually screened for the expected C-terminal TM domain using TMHMM [106]. 4.) Datasets were further screened for the presence of a predicted signal peptide using SignalP 4.1 and PrediSi [107, 108].
(B) Phylogenetic tree containing LRR-RLK FLS2, BRI1 and BAK1 homologs in multiple plant species. The relatedness of RLKs FLS2, BAK1, and BRI1 are indicated with blue, green, and red leaves respectively. The species were chosen to represent the breadth of plant evolution, including bryophytes (Physcomitrella patens; red), monocots (Oryza sativa; black, Zea mays; orange), dicots (Arabidopsis thaliana; green, Nicotiana tabacum; purple, Solanum lyoperiscum; teal), and woody dicots (Populus trichocarpa; blue). Tree files were generated using the MEGA software [109], and visualized with iTOL [110]. Protein sequences were aligned using ClustalW [111]. Phylogenetic relationships were inferred with a maximum-likelihood method based on the JTT matrix-based model.
Several other LRR-RLPs have been shown to be involved in fungal resistance, but their corresponding MAMPs or apoplastic effectors have not been identified. Arabidopsis LRR-RLP AtRLP30 is involved in the response triggered by an unknown proteinaceous elicitor from Sclerotinia sclerotium designated SCLEROTINIA CULTURE FILTRATE ELICITOR 1 (SCFE1). AtRLP30 was further demonstrated to be important for Arabidopsis resistance to S. sclerotium and B. cinerea. [63]. Arabidopsis RESISTANCE TO FUSARIUM OXYSPORUM 2 (RFO2) is an LRR-RLP encoded by AtRLP3 and confers resistance to the fungal pathogen Fusarium oxysporum. Although AtRLP3/RFO2 and AtRLP2 are physically adjacent and highly similar, AtRLP2 does not contribute to resistance against F. oxysporum. Despite this, the extracellular LRR domains of AtRLP2 and AtRLP3/RFO2 are exchangeable for resistance [64]. Interestingly, the extracellular LRR domain of AtRLP3/RFO2 is also highly similar to that of the LRR-RLK PSY1R, which recognizes tyrosine-sulfated peptide PSY1, whose signaling negatively regulates plant immunity. Thus, the authors propose that AtRLP3/RFO2 acts as a decoy receptor for a F. oxysporum effector, which is otherwise recognized by PSY1R to promote pathogenicity [64]. The importance of Arabidopsis LRR-RLP AtRLP52 in resistance against the fungal powdery mildew pathogen Erysiphe cichoracearum was demonstrated after it was first characterized for its responsiveness upon treatment with chito-octamers [65]. It remains unknown whether AtRLP52 participates in chitin perception through interaction with AtLYK5/AtCERK1 complex. In wheat, RLP1.1 was shown to play a role in defense against the rust pathogen Puccinia striiformis f. sp. tritici [66]. Additionally, apple LRR-RLP HcrVf2, located in a cluster of LRR-RLP genes, mediates resistance to apple scab caused by the ascomycete Venturia inaequalis (Cke.) Wint. [67].
3.4. RLPs involved in resistance to bacteria
Although several RLPs have been implicated in plant bacterial resistance, their exact role in PTI or ETI is not entirely clear. In one case, it has been shown that Arabidopsis LysM-RLPs LYM1 and LYM3 are required for bacterial PGN-mediated responses through direct binding to PGNs [68]. The lym1, lym3, and lym1 lym3 mutants were all compromised in defense against P. syringae pv. tomato DC3000, and its type III secretion mutant strain hrcC- [68]. Similarly, rice LysM-RLPs, OsLYP4 and OsLYP6, also bind to PGN, and mediate PGN-triggered responses [55]. Thus, OsLYP4 and OsLYP6 could bind to both chitin and PGN and mediate resistance to fungal and bacterial pathogens.
Arabidopsis RECEPTOR OF ENIGMATIC MAMP OF XANTHOMONAS (ReMAX) is an LRR-RLP (AtRLP1) implicated in the perception of eMAX, a MAMP found in several Xanthomonas species [69]. Further, AtRLP30 was found to contribute to resistance against the non-adapted bacterial pathogen P. syringae pv. phaseolicola [10]. Arabidopsis SUPPRESSOR OF NPR1, CONSTITUTIVE 2 (SNC2) is an LRR-RLP (AtRLP51) [70]. The snc2–1D mutant, which bears a glycine to arginine mutation in the second glycine of the conserved GXXG motif of the transmembrane domain, exhibits a constitutive defense response, and increased pathogen resistance. Similarly, the corresponding mutation in AtRLP55/SNC3, a close homolog of AtRLP51/SNC2, also caused a constitutive activation of defense [70]. The GXXG motif is known to be important for the interaction of transmembrane helices [71], suggesting that AtRLP51/SNC2 and AtRLP55/SNC3 might interact with each other or another membrane proteins. Using a suppressor screen, it was shown that AtRLP51/SNC2 signal transduction is likely mediated by BIAN DA 1 (BDA1) (Chinese for “becoming big”), an ankyrin-repeat-containing protein with four transmembrane domains, to trigger defense through the activation of the transcription factor WRKY70 [72]. Both AtRLP51/SNC2 and BDA1 are important for plant defense to P. syringae pv. tomato DC3000. It has been proposed that AtRLP51/SNC2 may perceive an unknown MAMP from bacterium to activate immunity through BDA1 [72]. Interestingly, a mutation in one transmembrane domain of BDA1 also caused constitutive activation of defense [72], raising the possibility that transmembrane domains of AtRLP51/SNC2 and BDA1 interact with each other and suppress the defense response in the absence of pathogen infection.
3.5. RLPs involved in resistance to parasitic weeds
Parasitic plants also produce MAMP-like elicitors that induce classical defense responses. Tomato LRR-RLP Cuscuta Receptor 1 (SlCuRe1) is involved in the response triggered by a small, potentially O-glycosylated peptide from the parasitic plant Cuscuta reflexa [73]. It remains unknown whether SlCuRe1 is a genuine receptor of this peptide, and whether it shares the same signaling pathway as PRRs perceiving classical MAMPs. Nevertheless, ectopic expression of SlCuRe1 in susceptible hosts increased resistance to C. reflexa [73], further substantiating the potential of RLPs in applied crop biotechnology.
4. RLPs in Plant Development
Several LRR-RLPs have been shown to function as key regulators in different plant developmental processes including stomatal patterning, meristematic tissue regulation, and hormone signaling. Arabidopsis TOO MANY MOUTHS (TMM) encoded by LRR-RLP AtRLP17 expresses most highly in dividing postprotodermal cells and differentially modulates stomatal development in different organs with a negative role in cotyledons and positive role in hypocotyls and stems [74]. Arabidopsis CLAVATA 2 (CLV2) encoded by the LRR-RLP AtRLP10 was implicated in maintaining shoot apical meristem (SAM), root apical meristem (RAM), and floral organ size—a function consistent with its broader expression pattern [75, 76]. Interestingly, expression of AtRLP2 or AtRLP12 under the AtRLP10/CLV2 promoter in clv2 mutant plants compensated for the mutant phenotype, emphasizing the importance of expression specificity in determining RLP functions [77]. In addition, overexpression of AtRLP11 or AtRLP3 rescued the clv2–1 mutant phenotype [38], suggesting that CLV2 may regulate the protein stability of other RLPs. This is consistent with the observation that CLV2 regulates the stability of the associated LRR-RLK CLV1 [76]. The maize CLV2 ortholog FASCIATED EAR 2 (FEA2) was also found to maintain normal shoot and floral meristematic tissue proliferation [78].
Arabidopsis AtRLP41 is likely involved in abscisic acid (ABA) signaling or perception, as the T-DNA insertion lines exhibited a hyper-sensitivity to exogenous ABA treatment [10]. Since AtRLP41 is highly induced during senescence, it may regulate ABA-mediated leaf senescence. Arabidopsis LRR-RLP AtRLP44 is important for normal growth, and mediates the activation of brassinosteroid (BR) signaling [79]. BR regulates the expression of many cell wall-related genes, which likely modulate the output of the BR signaling through a feedback regulation. AtRLP44 is involved in this feedback regulation by sensing cell wall perturbation, specifically pectin modification [79]. This is conceptually similar with plant immune responses triggered by damage-associated molecular patterns (DAMPs), some of which are also cell wall-related components [80]. Some DAMPs have been shown to be transcriptionally induced during plant PTI and are thought to amplify plant PTI signaling [81].
5. RLPs and RLKs come in pairs
5.1. RLPs and RLKs pair in functions
Without a kinase domain, RLPs likely act as receptors or regulators, and transduce signals by complexing with RLKs and other transmembrane/membrane-associated proteins [82, 83]. Arabidopsis LRR-RLK SUPPRESSOR OF BIR1–1 (SOBIR1) and its orthologs have been found in complex with multiple LRR-RLPs, and the function of several of these RLPs requires the formation of this complex, including tomato Ve1 and Cf proteins, and Arabidopsis AtRLP23 [59, 84]. In addition, these LRR-RLPs associate with and functionally require Arabidopsis LRR-RLK, BRI1-ASSOCIATED RECEPTOR KINASE 1 (AtBAK1), also called SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (AtSERK3), or its orthologs. SERK family LRR-RLKs are shared co-receptors of multiple LRR-RLKs and usually dimerize with LRR-RLKs upon corresponding ligand perception [85]. Similarly, LRR-RLPs associate with BAK1/SERK complexes upon ligand perception, whereas they constitutively associate with SOBIR1 [59, 61, 86]. It has been shown that SOBIR1 can stabilize LRR-RLPs, which may lead to enhanced kinase activity of SOBIR1 and/or BAK1/SERK3 for signal transduction [84, 86]. BAK1 also associates with AtRLP44, which is involved in the feedback regulation of cell wall integrity through BR signaling [79]. Similarly, rice LysM-RLP OsCEBiP interacts with LysM-RLK OsCERK1 to activate chitin signaling [54]. Arabidopsis AtCERK1 is required for LYM1/LYM3-mediated PGN signaling although it remains unknown whether AtCERK1 associates with LYM1/LYM3 [68].
Development-related LRR-RLPs also functionally associate with LRR-RLKs. Stomatal development regulator TMM associates with ERECTA (ER) family LRR-RLKs and their coreceptors in the SERK family to perceive EPIDERMAL PATTERNING FACTOR (EPF) peptide signals to regulate stomatal patterning [87, 88]. There are 11 EPF family members in Arabidopsis [89]. Although many EPFs could be perceived by ERs, they exhibit different or even opposite functions in stomatal patterning, likely due to the competitive binding between ligands and receptors [90, 91]. The crystal structure of ER and TMM ectodomains with different EPF peptides indicates that TMM determines the specificity of ER in the recognition of different EPFs [92]. To recognize certain EPFs, such as EPF1 and EPF2, TMM must interact with ER. Conversely, the recognition to other EPFs, such as EPFL4 and EPFL6, is inhibited by the interaction of TMM with ER. This represents an example of the role of an RLP in determining ligand-receptor recognition specificity. The function of RLPs in plant immunity might be different from TMM since PRR RLKs usually recognize specific MAMPs. In addition, the SAM regulator CLV2 forms a functional receptor complex with LRR-RLK CLV1 or a transmembrane kinase CORYNE (CRN) in perceiving secreted peptide ligand CLV3 [93, 94].
5.2. RLPs and RLKs pair on chromosome and in expression
It has been hypothesized that the expansion of the LRR-RLK gene family was mediated by a repetitive duplication through some mechanism like tandem or whole genome duplication [8, 11]. Like RLKs, many LRR-RLPs are also physically clustered on chromosomes. Among the 57 LRR-RLPs in Arabidopsis, RLP11-RLP16, RLP22-RLP28, RLP30-RLP33, RLP36-RLP43, and RLP47-RLP50 appear to form distinct clusters of more than three genes adjacent to each other (Figure 4).
Figure 4. Physical map of A. thaliana LRR-RLPs and LRR-RLKs on chromosomes.
RLPs and RLKs (depicted on the left and right of each chromosome, respectively) were positioned using the TAIR chromosome map tool and further arranged for ease visualization. RLPs physically close to RLKs were indicated using boxes. Asterisks indicate proximal RLPs or RLKs with similar expression pattern. Physically clustered RLPs and RLKs were tested for correlations in expression using absolute expression values acquired from the Arabidopsis eFP browser (datasets “Developmental Map” and “Biotic Stress” http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). A Spearman’s rho value with magnitude greater than |ρ| = 0.5 with a p-value of <0.05 was used to determine positive and negative correlations.
Many RLPs and RLKs are found in large homogeneous gene clusters (i.e. clusters containing either RLK or RLP genes), and generally these clusters physically alternate between RLKs and RLPs on the chromosomes. Heterogeneous clusters (i.e. clusters containing both RLK and RLP genes) are rarer, and often have a single gene of one type among several of the other type in the cluster. For example, two RLPs, AtRLP2 (AT1G17240) and AtRLP3/RFO2 (AT1G17250), cluster with RLK AT1G17230 on chromosome 1. AtRLP55 (AT5G45770) clusters with RLKs AT5G45780, AT5G45800 and AT5G45840 on chromosome 5. Strikingly, in some cases, physically clustering LRR-RLPs and LRR-RLKs contain highly homologous ectodomains. For instance, the ectodomains of AtRLP52 (At5g25910) and the RLK AT5G25930, which are separated by only 10 kb on chromosome 5 (Figure 4), share 86% identity in amino acid sequence. Based on these observations, it seems likely that some RLKs and RLPs may share a common ancestor. One or more RLKs may have originated from a fusion event between an RLP and a kinase; similarly, one or more RLPs may be a truncation product of a former RLK.
Several other physically clustering LRR-RLPs and LRR-RLKs are correlated in expression (Figure 4). For example, the RLP SNC2 (At4g18760) and the RLK MORPHOGENESIS OF ROOT HAIR 1 (MRH1) (At4g18640) are correlated in mean-normalized expression during development [95]. Despite their physical proximity and co-expression, SNC2 and MRH1 share little sequence identity. These observations of physical distance and expression beg exciting new questions about the functional roles of these genes with each other and in the broader cellular context.
5.3. RLPs are more divergent than RLKs in different plant species
The phylogenetic analyses indicate that there are subsets of RLPs and RLKs which are conserved across species (Figure 3). This conservation is apparent in several well-studied LRR-RLKs, such as FLS2, BRI1, and BAK1 in Arabidopsis, rice, moss, and poplar, suggesting their structural and functional conservation across those species (Figure 3B). However, most RLPs are clustered by plant species, not individual genes (Figure 3A). It is possible that their lack of an intracellular kinase makes RLPs relatively freer of selective pressure, and therefore more predisposed to neofunctionalization. This is consistent with the hypothesis that poplar and Arabidopsis LRR-RLPs were evolved independently [96]. One explanation is that the potential fitness cost of an RLP duplication event may be less than that of an RLK duplication event, as RLK redundancy could be more likely to produce maladapted functional changes in cellular perception. It appears that LRR-RLPs involved in plant development, such as TMM and CLV2, are more conserved than LRR-RLPs implicated in plant immunity, such as RFO2 and RBPG1, across plant species (Figure 3A). The phylogenetic analyses also suggest that each lineage likely expanded and diversified its own subset of RLPs. Interestingly, the moss Physcomitrella patens, a bryophyte, contains fourteen LRR-RLPs, all of which are in the subset that interleave among species (Figure 3A). This might suggest that a small subset of LRR-RLPs are basally conserved, and that the conditions which favored a major expansion of this core group occurred in a slightly different way for each subsequent lineage after the divergence of polysporangiophytes.
6. Concluding remarks and future perspectives
While RLPs remain the less-examined counterpart to RLKs, their roles in plant immunity and development are increasingly appreciated. The functions of several RLPs with LRR ectodomain have been well characterized. In particular, LRR-RLPs often complex with LRR-RLKs for ligand perception and/or signaling relay. LRR-RLPs may regulate LRR-RLK protein stability or function as a specificity switch for ligand-receptor recognition. Similarly, LYM-RLPs can complex with LYM-RLKs in signal sensing and transduction. However, the functions of RLPs with other ectodomains remain to be elucidated. Interestingly, several LRR-RLPs and LRR-RLKs are physically clustered on chromosomes, and sometimes they correlate in expression pattern. It remains to be determined whether these LRR-RLPs and LRR-RLKs share functional similarity.
Crystal structures of RLK receptor-coreceptor complexes have revealed insights into the potential activation mechanisms of these complexes [21, 97]. The structure of a recent plant RLK-RLP complex revealed a surprising role of the RLP TMM in regulating the ligand recognition specificity by its associated RLK [92]. It will be interesting to determine the structure of other RLPs, in particular immunity-related RLPs, in complex with RLKs upon ligand perception. In several cases, the shared RLK co-receptor BAK1 family of RLKs also participate in RLP-RLK receptor complexing [85]. The structure of these complexes will vastly improve attempts to elucidate the complexities of receptorsome activation and regulation. A recent investigation of 40,000 interactions between 200 Arabidopsis LRR-RLK ectodomains was used to construct an “extracellular LRR network”. This identified some previously uncharacterized LRR-RLKs that function in plant growth and immunity [98]. Further analysis revealed that some LRR-RLKs function as articulation points in this network to stabilize the LRR-RLK signaling pathways. A similar approach could be used to study the contribution of LRR-RLPs to these networks. The aforementioned crystal structure and interaction networks of RLKs were based only on their LRR ectodomains. If future studies are able to observe full length protein while retaining the integrity of the receptor complex in its biochemical context at the plasma membrane, this would help avoid perturbations from the native conformation, steric effects, and orientation relative to the bilayer normal of these complexes.
An improvement of the available tools for the continued examination of RLPs will likely be required to reveal the functional roles of RLPs in an expeditious fashion. It remains to be technically changeling to examine the function and interaction of transmembrane receptors. Some techniques, such as split-ubiquitin membrane yeast-two hybrid assays, or membrane anchored ligand and receptor (MALAR) yeast-two hybrid approaches show some promise in identifying interactions between membrane proteins—and even ligands and receptors [99, 100]. However, these techniques are still laborious and tend to produce false positives. Other assays, such as co-immunoprecipitation, gel filtration, and quartz crystal microbalance (QCM) biosensor assays would be required to verify interactions of candidates.
Despite that efforts to improve the experimental design underlying our investigation of RLPs are critical, that which has already been elucidated can be applied to crop improvement efforts. As previously mentioned, simple ectopic expression of functionally important RLPs has proven somewhat effective in conferring favorable resistance traits. It remains to be determined whether it could provide a durable resistance. Elegant solutions to plant-disease can only be obtained through a more complete understanding of the governing biomolecular dynamics.
Highlights:
Receptor-like proteins (RLPs) are transmembrane proteins with an extracellular domain and a short cytoplasmic tail functioning in diverse biological processes, including plant development and immunity.
While different RLP subfamilies contain distinct extracellular domains, the leucine-rich repeat domain RLP subfamily is the most common and well-studied.
Due to their lack of an intracellular kinase domain, RLPs often complex with receptor-like kinases (RLKs) for ligand perception and/or signaling relay.
RLPs may regulate RLK protein stability or function as a specificity switch for ligand-receptor recognition.
Some RLPs and RLKs are physically clustered, and sometimes they correlate in expression and physical proximity—although the biological function is not clear.
The relatedness of LRR-RLPs across plant species reveals that the apparent diversity of RLPs is likely the result of an arbitrary expansion and neofunctionalization from one or more subsets of these RLPs by each plant species.
Acknowledgments
We apologize to colleagues whose work was not discussed here because of space limitations. The work was supported by National Science Foundation (IOS-1252539) to P.H., and National Institutes of Health (R01GM097247) and the Robert A. Welch foundation (A-1795) to L.S. The authors have declared no conflict of interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Krampen M, Phytosemiotics, Semiotica, 36 (1981). [Google Scholar]
- [2].Spoel SH, Dong X, How do plants achieve immunity? Defence without specialized immune cells, Nature Reviews Immunology, 12 (2012) 89. [DOI] [PubMed] [Google Scholar]
- [3].Oh M-H, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC, Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis, Proceedings of the National Academy of Sciences, 106 (2009) 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shiu SH, Bleecker AB, Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases, Proc Natl Acad Sci U S A, 98 (2001) 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L, Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity, Proc Natl Acad Sci U S A, 111 (2014) 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shiu S-H, Bleecker AB, Plant Receptor-Like Kinase Gene Family: Diversity, Function, and Signaling, Science’s STKE, 2001 (2001) re22–re22. [DOI] [PubMed] [Google Scholar]
- [7].Lehti-Shiu MD, Zou C, Hanada K, Shiu SH, Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes, Plant Physiol, 150 (2009) 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shiu S-H, Bleecker AB, Expansion of the Receptor-Like Kinase/Pelle Gene Family and Receptor-Like Proteins in Arabidopsis, Plant Physiology, 132 (2003) 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fischer I, Diévart A, Droc G, Dufayard J-F, Chantret N, Evolutionary Dynamics of the Leucine-Rich Repeat Receptor-Like Kinase (LRR-RLK) Subfamily in Angiosperms, Plant Physiology, 170 (2016) 1595–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu C-M, Woods-Tör A, Zipfel C, de Wit PJGM, Jones JDG, Tör M, Thomma BPHJ, A Genome-Wide Functional Investigation into the Roles of Receptor-Like Proteins in Arabidopsis, Plant Physiology, 147 (2008) 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fritz-Laylin LK, Krishnamurthy N, Tor M, Sjolander KV, Jones JD, Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis, Plant Physiol, 138 (2005) 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Medzhitov R, Preston-Hurlburt P, Janeway CA Jr, A human homologue of the Drosophila Toll protein signals activation of adaptive immunity, Nature, 388 (1997) 394. [DOI] [PubMed] [Google Scholar]
- [13].Bohm H, Albert I, Fan L, Reinhard A, Nurnberger T, Immune receptor complexes at the plant cell surface, Current Opinion in Plant Biology, 20 (2014) 47–54. [DOI] [PubMed] [Google Scholar]
- [14].Yu X, Feng B, He P, Shan L, From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition, Annu Rev Phytopathol, 55 (2017) 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J, The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors, Trends Biochem Sci, 39 (2014) 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang GD, Fiers M, Ellendorff U, Wang ZZ, de Wit PJGM, Angenent GC, Thomma BPHJ, The Diverse Roles of Extracellular Leucine-rich Repeat-containing Receptor-like Proteins in Plants, Crit Rev Plant Sci, 29 (2010) 285–299. [Google Scholar]
- [17].Anderson KV, Jurgens G, Nusslein-Volhard C, Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product, Cell, 42 (1985) 779–789. [DOI] [PubMed] [Google Scholar]
- [18].Borner GHH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P, Prediction of Glycosylphosphatidylinositol-Anchored Proteins in Arabidopsis. A Genomic Analysis, Plant Physiology, 129 (2002) 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gong B-Q, Xue J, Zhang N, Xu L, Yao X, Yang Q-J, Yu Y, Wang H-B, Zhang D, Li J-F, Rice Chitin Receptor OsCEBiP Is Not a Transmembrane Protein but Targets the Plasma Membrane via a GPI Anchor, Molecular Plant, 10 (2017) 767–770. [DOI] [PubMed] [Google Scholar]
- [20].Shiu S-H, Karlowski WM, Pan R, Tzeng Y-H, Mayer KFX, Li W-H, Comparative Analysis of the Receptor-Like Kinase Family in Arabidopsis and Rice, The Plant Cell, 16 (2004) 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hohmann U, Lau K, Hothorn M, The Structural Basis of Ligand Perception and Signal Activation by Receptor Kinases, Annual Review of Plant Biology, Vol 68, 68 (2017) 109–137. [DOI] [PubMed] [Google Scholar]
- [22].Lannoo N, Van Damme EJM, Lectin domains at the frontiers of plant defense, Frontiers in Plant Science, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bellande K, Bono J-J, Savelli B, Jamet E, Canut H, Plant Lectins and Lectin Receptor-Like Kinases: How Do They Sense the Outside?, International Journal of Molecular Sciences, 18 (2017) 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vaid N, Pandey PK, Tuteja N, Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice, Plant Mol Biol, 80 (2012) 365–388. [DOI] [PubMed] [Google Scholar]
- [25].Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, Nasrallah ME, Self-Incompatibility in the Genus Arabidopsis: Characterization of the S Locus in the Outcrossing A. lyrata and Its Autogamous Relative A. thaliana, The Plant Cell, 13 (2001) 627–644. [PMC free article] [PubMed] [Google Scholar]
- [26].Desaki Y, Miyata K, Suzuki M, Shibuya N, Kaku H, Plant immunity and symbiosis signaling mediated by LysM receptors, Innate Immun, 24 (2018) 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shockman GD, Höltje JV, Chapter 7 Microbial peptidoglycan (murein) hydrolases, in: Ghuysen JM, Hakenbeck R (Eds.) New Comprehensive Biochemistry, Elsevier, 1994, pp. 131–166. [Google Scholar]
- [28].Shinya T, Nakagawa T, Kaku H, Shibuya N, Chitin-mediated plant-fungal interactions: catching, hiding and handshaking, Curr Opin Plant Biol, 26 (2015) 64–71. [DOI] [PubMed] [Google Scholar]
- [29].Gust AA, Peptidoglycan Perception in Plants, PLoS Pathog, 11 (2015) e1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lindner H, Müller LM, Boisson-Dernier A, Grossniklaus U, CrRLK1L receptor-like kinases: not just another brick in the wall, Curr Opin Plant Biol, 15 (2012) 659–669. [DOI] [PubMed] [Google Scholar]
- [31].Nissen KS, Willats WG, Malinovsky FG, Understanding CrRLK1L Function: Cell Walls and Growth Control, Trends Plant Sci, 21 (2016) 516–527. [DOI] [PubMed] [Google Scholar]
- [32].Li C, Wu HM, Cheung AY, FERONIA and Her Pals: Functions and Mechanisms, Plant Physiol, 171 (2016) 2379–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schallus T, Jaeckh C, Fehér K, Palma AS, Liu Y, Simpson JC, Mackeen M, Stier G, Gibson TJ, Feizi T, Pieler T, Muhle-Goll C, Malectin: A Novel Carbohydrate-binding Protein of the Endoplasmic Reticulum and a Candidate Player in the Early Steps of Protein N-Glycosylation, Molecular Biology of the Cell, 19 (2008) 3404–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mang H, Feng B, Hu Z, Boisson-Dernier A, Franck CM, Meng X, Huang Y, Zhou J, Xu G, Wang T, Shan L, He P, Differential Regulation of Two-Tiered Plant Immunity and Sexual Reproduction by ANXUR Receptor-Like Kinases, Plant Cell, 29 (2017) 3140–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hok S, Danchin EG, Allasia V, Panabieres F, Attard A, Keller H, An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease, Plant Cell Environ, 34 (2011) 1944–1957. [DOI] [PubMed] [Google Scholar]
- [36].Li F, Cheng C, Cui F, de Oliveira MV, Yu X, Meng X, Intorne AC, Babilonia K, Li M, Li B, Chen S, Ma X, Xiao S, Zheng Y, Fei Z, Metz RP, Johnson CD, Koiwa H, Sun W, Li Z, de Souza Filho GA, Shan L, He P, Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity, Cell Host Microbe, 16 (2014) 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wu Y, Xun Q, Guo Y, Zhang J, Cheng K, Shi T, He K, Hou S, Gou X, Li J, Genome-Wide Expression Pattern Analyses of the Arabidopsis Leucine-Rich Repeat Receptor-Like Kinases, Mol Plant, 9 (2016) 289–300. [DOI] [PubMed] [Google Scholar]
- [38].Wu J, Liu Z, Zhang Z, Lv Y, Yang N, Zhang G, Wu M, Lv S, Pan L, Joosten MH, Wang G, Transcriptional regulation of receptor-like protein genes by environmental stresses and hormones and their overexpression activities in Arabidopsis thaliana, Journal of experimental botany, 67 (2016) 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jones JD, Dangl JL, The plant immune system, Nature, 444 (2006) 323–329. [DOI] [PubMed] [Google Scholar]
- [40].Couto D, Zipfel C, Regulation of pattern recognition receptor signalling in plants, Nat Rev Immunol, 16 (2016) 537–552. [DOI] [PubMed] [Google Scholar]
- [41].Wu S, Shan L, He P, Microbial signature-triggered plant defense responses and early signaling mechanisms, Plant Sci, 228C (2014) 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dou D, Zhou JM, Phytopathogen effectors subverting host immunity: different foes, similar battleground, Cell Host Microbe, 12 (2012) 484–495. [DOI] [PubMed] [Google Scholar]
- [43].Jones D, Thomas C, Hammond-Kosack K, Balint-Kurti P, Jones J, Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging, Science, 266 (1994) 789–793. [DOI] [PubMed] [Google Scholar]
- [44].Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JD, Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9, Plant Cell, 9 (1997) 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Luderer R, Rivas S, Nurnberger T, Mattei B, Van den Hooven HW, Van der Hoorn RAL, Romeis T, Wehrfritz JM, Blume B, Nennstiel D, Zuidema D, Vervoort J, De Lorenzo G, Jones JDG, De Wit PJGM, Joosten MHAJ, No evidence for binding between resistance gene product Cf-9 of tomato and avirulence gene product AVR9 of Cladosporium fulvum, Mol Plant Microbe In, 14 (2001) 867–876. [DOI] [PubMed] [Google Scholar]
- [46].Stotz HU, Mitrousia GK, de Wit P, Fitt BDL, Effector-triggered defence against apoplastic fungal pathogens, Trends Plant Sci, 19 (2014) 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, Fischer R, Prüfer D, Tomato Ve disease resistance genes encode cell surface-like receptors, Proceedings of the National Academy of Sciences, 98 (2001) 6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CD, Nazar RN, Robb J, Liu CM, Thomma BP, Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1, Plant Physiol, 150 (2009) 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Larkan NJ, Lydiate DJ, Parkin IA, Nelson MN, Epp DJ, Cowling WA, Rimmer SR, Borhan MH, The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1, The New phytologist, 197 (2013) 595–605. [DOI] [PubMed] [Google Scholar]
- [50].Cao YR, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G, The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1, eLife, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, Zhou JM, Chai J, Chitin-induced dimerization activates a plant immune receptor, Science, 336 (2012) 1160–1164. [DOI] [PubMed] [Google Scholar]
- [52].Hayafune M, Berisio R, Marchetti R, Silipo A, Kayama M, Desaki Y, Arima S, Squeglia F, Ruggiero A, Tokuyasu K, Molinaro A, Kaku H, Shibuya N, Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization, P Natl Acad Sci USA, 111 (2014) E404–E413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liu S, Wang J, Han Z, Gong X, Zhang H, Chai J, Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP, Structure, 24 (2016) 1192–1200. [DOI] [PubMed] [Google Scholar]
- [54].Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, Shibuya N, Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice, The Plant Journal, 64 (2010) 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu B, Li J-F, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J, Feng D, Qi K, He Y, Wang J, Wang H-B, Lysin Motif–Containing Proteins LYP4 and LYP6 Play Dual Roles in Peptidoglycan and Chitin Perception in Rice Innate Immunity, The Plant Cell, 24 (2012) 3406–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ, LYM2-dependent chitin perception limits molecular flux via plasmodesmata, Proc Natl Acad Sci U S A, 110 (2013) 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ron M, Avni A, The Receptor for the Fungal Elicitor Ethylene-Inducing Xylanase Is a Member of a Resistance-Like Gene Family in Tomato, Plant Cell, 16 (2004) 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bar M, Sharfman M, Ron M, Avni A, BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1, The Plant journal : for cell and molecular biology, 63 (2010) 791–800. [DOI] [PubMed] [Google Scholar]
- [59].Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang H, Krol E, Grefen C, Gust AA, Chai J, Hedrich R, Van den Ackerveken G, Nürnberger T, An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity, Nature Plants, 1 (2015) 15140. [DOI] [PubMed] [Google Scholar]
- [60].Gijzen M, Nürnberger T, Nep1-like proteins from plant pathogens: Recruitment and diversification of the NPP1 domain across taxa, Phytochemistry, 67 (2006) 1800–1807. [DOI] [PubMed] [Google Scholar]
- [61].Du J, Verzaux E, Chaparro-Garcia A, Bijsterbosch G, Keizer LC, Zhou J, Liebrand TW, Xie C, Govers F, Robatzek S, van der Vossen EA, Jacobsen E, Visser RG, Kamoun S, Vleeshouwers VG, Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato, Nat Plants, 1 (2015) 15034. [DOI] [PubMed] [Google Scholar]
- [62].Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, Chen W, Liang X, Shang H, Liu W, Shi C, Xiao G, Gou C, Ye W, Xu X, Zhang X, Wei H, Li Z, Zhang G, Wang J, Liu K, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S, Genome sequence of the cultivated cotton Gossypium arboreum, Nat Genet, 46 (2014) 567–572. [DOI] [PubMed] [Google Scholar]
- [63].Zhang W, Fraiture M, Kolb D, Loffelhardt B, Desaki Y, Boutrot FF, Tor M, Zipfel C, Gust AA, Brunner F, Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1–1/EVERSHED mediate innate immunity to necrotrophic fungi, Plant Cell, 25 (2013) 4227–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shen Y, Diener AC, Arabidopsis thaliana RESISTANCE TO FUSARIUM OXYSPORUM 2 Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection, PLOS Genetics, 9 (2013) e1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S, Loss-of-Function Mutations in Chitin Responsive Genes Show Increased Susceptibility to the Powdery Mildew Pathogen Erysiphe cichoracearum, Plant Physiology, 138 (2005) 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jiang ZN, Ge S, Xing LP, Han DJ, Kang ZS, Zhang GQ, Wang XJ, Wang XU, Chen PD, Cao AZ, RLP1.1, a novel wheat receptor-like protein gene, is involved in the defence response against Puccinia striiformis f. sp tritici, Journal of Experimental Botany, 64 (2013) 3735–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Belfanti E, Silfverberg-Dilworth E, Tartarini S, Patocchi A, Barbieri M, Zhu J, Vinatzer BA, Gianfranceschi L, Gessler C, Sansavini S, The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety, Proc Natl Acad Sci U S A, 101 (2004) 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Willmann R, Lajunen HM, Erbs G, Newman M-A, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono J-J, Cullimore JV, Jehle AK, Götz F, Kulik A, Molinaro A, Lipka V, Gust AA, Nürnberger T, Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection, Proceedings of the National Academy of Sciences, 108 (2011) 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Jehle AK, Lipschis M, Albert M, Fallahzadeh-Mamaghani V, Furst U, Mueller K, Felix G, The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas, Plant Cell, 25 (2013) 2330–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhang Y, Yang Y, Fang B, Gannon P, Ding P, Li X, Zhang Y, Arabidopsis snc2–1D Activates Receptor-Like Protein-Mediated Immunity Transduced through WRKY70, The Plant Cell, 22 (2010) 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Teese MG, Langosch D, Role of GxxxG Motifs in Transmembrane Domain Interactions, Biochemistry, 54 (2015) 5125–5135. [DOI] [PubMed] [Google Scholar]
- [72].Yang Y, Zhang Y, Ding P, Johnson K, Li X, Zhang Y, The Ankyrin-Repeat Transmembrane Protein BDA1 Functions Downstream of the Receptor-Like Protein SNC2 to Regulate Plant Immunity, Plant Physiology, 159 (2012) 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hegenauer V, Furst U, Kaiser B, Smoker M, Zipfel C, Felix G, Stahl M, Albert M, Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor, Science, 353 (2016) 478–481. [DOI] [PubMed] [Google Scholar]
- [74].Nadeau JA, Sack FD, Control of stomatal distribution on the Arabidopsis leaf surface, Science, 296 (2002) 1697–1700. [DOI] [PubMed] [Google Scholar]
- [75].Kayes JM, Clark SE, CLAVATA2, a regulator of meristem and organ development in Arabidopsis, Development, 125 (1998) 3843–3851. [DOI] [PubMed] [Google Scholar]
- [76].Jeong S, Trotochaud AE, Clark SE, The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase, Plant Cell, 11 (1999) 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wang G, Long Y, Thomma BP, de Wit PJ, Angenent GC, Fiers M, Functional analyses of the CLAVATA2-like proteins and their domains that contribute to CLAVATA2 specificity, Plant Physiol, 152 (2010) 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D, The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize, Genes & Development, 15 (2001) 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wolf S, van der Does D, Ladwig F, Sticht C, Kolbeck A, Schürholz A-K, Augustin S, Keinath N, Rausch T, Greiner S, Schumacher K, Harter K, Zipfel C, Höfte H, A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling, Proceedings of the National Academy of Sciences, 111 (2014) 15261–15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yamaguchi Y, Huffaker A, Endogenous peptide elicitors in higher plants, Curr Opin Plant Biol, 14 (2011) 351–357. [DOI] [PubMed] [Google Scholar]
- [81].Li B, Meng X, Shan L, He P, Transcriptional Regulation of Pattern-Triggered Immunity in Plants, Cell Host Microbe, 19 (2016) 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Liebrand TWH, van den Burg HA, Joosten MHAJ, Two for all: receptor-associated kinases SOBIR1 and BAK1, Trends in Plant Science, 19 (2014) 123–132. [DOI] [PubMed] [Google Scholar]
- [83].Gust AA, Felix G, Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases, Curr Opin Plant Biol, 21 (2014) 104–111. [DOI] [PubMed] [Google Scholar]
- [84].Liebrand TW, van den Berg GC, Zhang Z, Smit P, Cordewener JH, America AH, Sklenar J, Jones AM, Tameling WI, Robatzek S, Thomma BP, Joosten MH, Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection, Proc Natl Acad Sci U S A, 110 (2013) 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ma X, Xu G, He P, Shan L, SERKing Coreceptors for Receptors, Trends Plant Sci, 21 (2016) 1017–1033. [DOI] [PubMed] [Google Scholar]
- [86].Postma J, Liebrand TW, Bi G, Evrard A, Bye RR, Mbengue M, Kuhn H, Joosten MH, Robatzek S, Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity, New Phytol, 210 (2016) 627–642. [DOI] [PubMed] [Google Scholar]
- [87].Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU, Direct interaction of ligand-receptor pairs specifying stomatal patterning, Genes & Development, 26 (2012) 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L, Differential Function of Arabidopsis SERK Family Receptor-like Kinases in Stomatal Patterning, Curr Biol, 25 (2015) 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T, The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule, Genes Dev, 21 (2007) 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Lee JS, Hnilova M, Maes M, Lin YC, Putarjunan A, Han SK, Avila J, Torii KU, Competitive binding of antagonistic peptides fine-tunes stomatal patterning, Nature, 522 (2015) 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Pillitteri LJ, Dong J, Stomatal development in Arabidopsis, Arabidopsis Book, 11 (2013) e0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lin GZ, Zhang L, Han ZF, Yang XR, Liu WJ, Li ET, Chang JB, Qi YJ, Shpak ED, Chai JJ, A receptor-like protein acts as a specificity switch for the regulation of stomatal development, Genes & Development, 31 (2017) 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lee CH, Clark SE, Core pathways controlling shoot meristem maintenance, Wires Dev Biol, 2 (2013) 671–684. [DOI] [PubMed] [Google Scholar]
- [94].Guo Y, Han L, Hymes M, Denver R, Clark SE, CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification, The Plant journal : for cell and molecular biology, 63 (2010) 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Laubinger S, Zeller G, Henz SR, Sachsenberg T, Widmer CK, Naouar N, Vuylsteke M, Schölkopf B, Rätsch G, Weigel D, At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana, Genome Biology, 9 (2008) R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Petre B, Hacquard S, Duplessis S, Rouhier N, Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor, Frontiers in plant science, 5 (2014) 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Han ZF, Sun YD, Chai JJ, Structural insight into the activation of plant receptor kinases, Curr Opin Plant Biol, 20 (2014) 55–63. [DOI] [PubMed] [Google Scholar]
- [98].Smakowska-Luzan E, Mott GA, Parys K, Stegmann M, Howton TC, Layeghifard M, Neuhold J, Lehner A, Kong J, Grunwald K, Weinberger N, Satbhai SB, Mayer D, Busch W, Madalinski M, Stolt-Bergner P, Provart NJ, Mukhtar MS, Zipfel C, Desveaux D, Guttman DS, Belkhadir Y, An extracellular network of Arabidopsis leucine-rich repeat receptor kinases, Nature, 553 (2018) 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Li J, Gao J, Han L, Zhang Y, Guan W, Zhou L, Yu Y, Han W, Development of a membrane-anchored ligand and receptor yeast two-hybrid system for ligand-receptor interaction identification, Sci Rep, 6 (2016) 35631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, Sanders D, Revuelta JL, Boles E, André B, Frommer WB, K+channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions, P Natl Acad Sci USA, 101 (2004) 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Humphrey W, Dalke A and Schulten K, VMD - Visual Molecular Dynamics, Journal of Molecular Graphics, 14 (1996) 33–38. [DOI] [PubMed] [Google Scholar]
- [102].Nagae M, Soga K, Morita-Matsumoto K, Hanashima S, Ikeda A, Yamamoto K, & Yamaguchi Y, Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold, Glycobiology, 24 (2014) 368–378. [DOI] [PubMed] [Google Scholar]
- [103].She J, Han ZF, Kim TW, Wang JJ, Cheng W, Chang JB, Shi SA, Wang JW, Yang MJ, Wang ZY, Chai JJ, Structural insight into brassinosteroid perception by BRI1, Nature, 474 (2011) 472–U496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, Harter K, The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses, The Plant Journal, 50 (2007) 347–363. [DOI] [PubMed] [Google Scholar]
- [105].Mulder NJ, & Apweiler R, The InterPro Database and Tools for Protein Domain Analysis, Current Protocols in Bioinformatics, (2008) 2.7.1–2.7.18. [DOI] [PubMed] [Google Scholar]
- [106].Krogh BLA, von Heijne G, and Sonnhammer ELL, Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes, Journal of Molecular Biology, 305 (2001) 567–580. [DOI] [PubMed] [Google Scholar]
- [107].Hiller K, Grote A, Scheer M, Munch R, & Jahn D, PrediSi: prediction of signal peptides and their cleavage positions, Nucleic Acids Research, 32 (2004) W375–W379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Henrik Nielsen IK, Protein Function Prediction Methods in Molecular Biology, 1611 (2017) 59–73. [DOI] [PubMed] [Google Scholar]
- [109].Kumar S, Stecher G, Tamura K, MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets, Molecular biology and evolution, 33 (2016) 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Letunic I, & Bork P, Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees, Nucleic Acids Research, 44 (2016) W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Thompson JD, Higgins DG, Gibson TJ, CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Research, 22 (1994) 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]