Abstract
Epidermal growth factor receptor (EGFR) blockade resistance is common in the treatment of RAS wide type colorectal cancer (CRC). During the treatment of cetuximab, acquired resistant genomic alterations always occurs earlier than disease progression observed by medical images. Identification of genomic alterations dynamically might have certain clinical significance. Because of the limitation of repeated tissue biopsy, liquid biopsy is increasingly recognized. Droplet digital polymerase chain reaction (ddPCR) is the main detection methods for circulating tumor DNA (ctDNA), however, the application of next-generation sequencing (NGS) for ctDNA detection becomes more and more popular. Here we develop a NGS-based ctDNA assay and evaluated its sensitivity and specificity while using ddPCR as control. These two technologies were both used for genomic alteration detection for the peripheral blood samples from cetuximab-treated colorectal cancer patients dynamically. Fifteen patients were enrolled in this study, including eight males and seven females. The sensitivity and specificity of our NGS assay were 87.5% and 100% respectively, and liner regression analysis comparing variant allele frequency (VAF) revealed high concordance between NGS and ddPCR (R2 = 0.98). NGS actually found more mutation information than ddPCR such as the additional dynamic changes of TP53 which were observed in the disease progression patients. Moreover, the variant allele fraction of TP53 was also found by NGS to be changed along with the clinical efficacy evaluation dynamically during the whole treatment process. In conclusion, our newly developed NGS-based ctDNA assay shows similar performance with ddPCR but have more advantages of its high throughput of multigenetic detection for the dynamic monitoring during the treatment of cetuximab in metastasis CRC patients.
Introduction
Colorectal cancer (CRC) is the third-most common cancer and the second leading cause of cancer-related death worldwide [1]. Several clinical trials have proved that cetuximab combines with standard chemotherapy achieved important improvement in overall survival for RAS wild type metastatic colorectal cancer (mCRC) patients, while RAS mutant type tumors derive no benefit [2], [3], [4]. However, acquired resistance to treatment always occurs after several months of initiating cetuximab therapy. Many biomarkers, including KRAS and NRAS mutations, BRAF mutation, PTEN loss and HER-2 amplification have already been identified to be associated with cetuximab resistance [5], [6]. It is important to identify patients who have developed resistance as soon as possible to switch to subsequent available treatment early. Therefore, detection of a biomarker involved in early diagnosis, minimal residual disease, early recurrence and emerging drug resistance is advisable choice.
It is reported that circulating tumor DNA (ctDNA) is a promising tumor biomarker for monitoring tumor burden and response to anticancer therapy [7], [8]. Studies showed that RAS pathway mutations related to acquired resistance to anti-EGFR therapy can be detected in the blood of patients with colorectal cancer prior to disease progression [4], [9]. Retrospective analysis further revealed the correlation of ctDNA levels with anti-EGFR response [4], [10]. Therefore, ctDNA based RAS detection may help identify patients who have no response to anti-EGFR therapy or monitor patients who develop acquired resistance to this targeted therapy [11]. Not only for the detection of newly appeared resistant genomic alterations, it is also reported that variant allele frequency (VAF) of the genomic alterations identified in ctDNA would change during anti-cancer therapies. Mostly, the drop of VAF was associated with clinical response to certain treatment, and the increasing of VAF often indicated the recurrence or resistance of treatment [12], [13], [14].
In addition, Liquid biopsy for ctDNA has its advantages in tissue sampling and sampling bias, especially for those post-treatment patients. And the concordance rate of 80% between liquid biopsy and tissue biopsy has been observed in mCRC patients [15], [16]. Therefore, liquid biopsy is an innovative method to study both primary and acquired resistance to anti-EGFR monoclonal antibodies [17]. ctDNA can be detected in almost all available biologic fluids, the most commonly samples used for liquid biopsy were blood and urine. ctDNA may account for less than 0.1% of the total cell-free DNA (cfDNA), which makes detection of the tumor-specific mutations in cfDNA challenging, therefore it requires a highly sensitive technique [18]. Currently, several methods such as allele-specific amplification refractory mutation system PCR (ARMS), bead emulsification amplification and magnetics (BEAMing) technology, allele-specific PCR (AS-PCR), and droplet digital PCR (ddPCR) have been applied to detect the mutation abundance in ctDNA with high sensitivity and specificity. Despite the technologies presented above are useful for longitudinal monitoring, they only allow to identify certain hotspot mutations in the primary tumor or occurring during the acquired resistance mechanism. However, For those well-known tumor suppressor genes in CRC such as TP53, PTEN, etc., there were no hotspots within them while loss-of-function mutations could be found in almost every region. Meanwhile, the mutations leading to truncation or possible mRNA decay in any region of the tumor suppressor genes could be considered as clinical significance. From this perspective, it's impossible to detect the whole regions of tumor suppressor genes for the technologies mentioned above. Next generation sequencing (NGS) technology is a reliable and powerful high throughput tool that can identify genomic alterations occurring in any region of the target genes at a frequency as low as one mutant copy in several thousand wild-type copies and have illuminated the mutational landscape of many types of tumor [19], [20]. The aim of this study was to develop a high-performance NGS-based ctDNA assay and evaluate the clinical application for measuring dynamic changes of ctDNA in cetuximab-treated mCRC patients.
Materials and Methods
Patients and Sample Collection
A total of 15 patients were included in this study, which was approved by Ethics Committee of Fudan University Shanghai Cancer Center and written informed consent was obtained from each patient before samples collection. All enrolled patients had wild-type RAS and RAF gene status when tested by tissue at the baseline. Plasma samples were collected at the baseline, every 8 weeks during treatment and at the time of disease progression. Plasma samples from all these patients were collected and stored at −80 °C until tested by ddPCR and NGS. We analyzed a total of 98 plasma samples throughout this study. All the plasma samples were tested by NGS panel. For ddPCR detection, only positive sites detected at the time of disease progression were retrospectively tested for all the plasma samples during treatment.
Droplet Digital PCR
The cfDNA was stored in DNA Elution of 20ul. The concentrations of all the circulating DNA samples were assessed using qubit 3.0 (Thermo Scientific)being in the range of 0.4 to 12 ng/μl. Next, ddPCR assays were performed track the mutation on ctDNA. ddPCR was performed using the conventional method on the Bio-Rad QX200TM (Bio-Rad). Primers and Taqman probe pairs were custom-designed. Briefly, 900 nM probes and 250 nM primers were mixed with 2× Droplet PCR Supermix (Bio-Rad Laboratories, Hercules, CA, USA), 5 μl of template DNA, and H2O to become 20 μl for each reaction. The reaction mixture was placed into the sample well of DG8 cartridge (Bio-Rad). A volume of 70 μl of droplet generation oil was loaded into the oil well, and droplets were formed in the droplet generator (BioRad). After processing, the droplets were transferred to a 96-well PCR plate (Eppendorf). The PCR amplification was carried out on C1000 TouchTM Thermal Cycler (Bio-Rad) with the following thermal profile: hold at 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 58 °C for 1 min (ramp 2 °C/s), 1 cycle at 98 °C for 10 min, and ending at 4 °C. After amplification, the plate was loaded on the droplet reader (Bio-Rad) and the droplets from each well of the plate were read automatically. QuantaSoft software was used to count the PCR-positive and PCR-negative droplets to provide absolute quantification of target DNA. The quantification measurements of each target were expressed as the copies number per 1 microliter of reaction. A mutation was only considered to be present according to the sensitivity of the assay.
Next-generation Sequencing
Plasma samples were performed for NGS based ultra-deep panel sequencing in a College of American Pathologists (CAP) certified laboratory of OrigiMed. Diluted libraries were sequenced to a mean coverage of 3000X on an Illumina NextSeq-500 Platform (Illumina Incorporated, San Diego, CA). Resultant sequences were further analyzed for genomic alterations, including single nucleotide variants (SNVs), short and long insertions/deletions (indels), copy number variations (CNVs), and structural variants of gene rearrangement/fusion and long indels. Firstly, Burrows-Wheeler Aligner (BWA) was applied to align the reads to human genome reference sequence (hg19), and Picard was employed to remove PCR duplicates. SNVs and short indels were identified by MUTECT and Pindel. The log-ratio per region of each gene was calculated, and copy number changes were detected using customized algorithms. Customized algorithm was developed to detect gene rearrangement/fusion and long indels. Clinically relevant genomic alterations were served as druggable genomic alterations in current anticancer therapies or clinical trials.
Results
Characteristics of Patients
Between April 2016 to September 2017, there were 15 mCRC patients who treated with cetuximab and standard chemotherapy enrolled in this study. The characteristics of patients (Table 1) showed that there were 8 males and 7 females. The median age of all the patients was 50 (range 22–68). Most patients (14/15, 93.3%) had their primary tumor site in left while only one patient had its primary tumor site in right. There were 8 patients treated with cetuximab in combination with first-line chemotherapy and 3 patients treated with cetuximab in combination with second–line chemotherapy, and the last four patients were given cetuximab cross lines. The median progression free survival (PFS) was 9.2 months of all enrolled patients. Most patients (13/15, 86.7%) had a PFS longer than 6 months, only 2 patients had poor clinical response for cetuximab with PFS shorter than 6 months.
Table 1.
Patient Characteristics
| Characteristic | Patients |
|---|---|
| Age, year, median (range) | 50 (22–68) |
| Sex, n (%) | |
| Male | 8 (53.3%) |
| Female | 7 (46.7%) |
| ECOG performance status, n (%) | |
| 0 | 1 (6.7%) |
| 1 | 14 (93.3%) |
| Anatomical position of primary lesion, n (%) | |
| Right | 1 (6.7%) |
| Left | 14 (93.3%) |
| Number of metastasis, n (%) | |
| ≤1 | 4 (26.7%) |
| >1 | 11 (73.3%) |
| Prior adjuvant chemotherapy, n (%) | 12 (80%) |
| Combined chemotherapy, n (%) | |
| FOLFOX | 5 (33.3%) |
| FOLFIRI | 6 (40%) |
| Irinotecan | 4 (26.7%) |
| Cetuximab use | |
| First line | 8 (53.3%) |
| Second line or more | 3 (20%) |
| Cross line | 4 (26.7%) |
| Median PFS (months, range) | 9.2 (3.0–15.3) |
| <6 months, n (%) | 2 (13.3%) |
| ≥6 months, n (%) | 13 (86.7%) |
Evaluation the Technical Performance of NGS-based ctDNA Assay
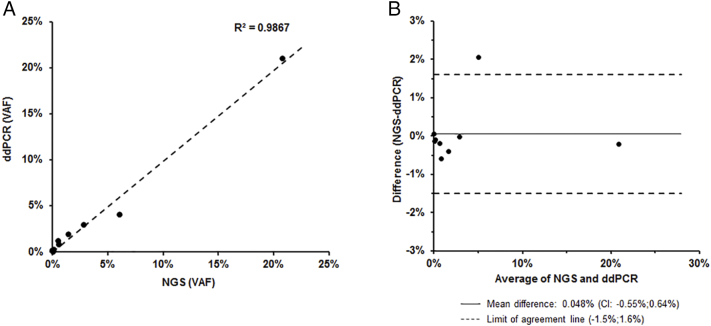
The detection range of ddPCR and NGS is different. Only hotspots of KRAS (G12A/C/D/R/S/V, G13D, Q61H/K/L/R, A146T, K117 N), EGFR (R451C, S492R, S464 L, K467 T, I491M), NRAS (G12A/C/D/R/S/V, G13D, Q61R), MEK1 (K57 N) and BRAF (V600E) are available for ddPCR detection. While the newly devolved NGS assay includes all exons and selected introns of 18 genes (ALK, BRAF, DDR2, EGFR, HER2, KRAS, MEK1, MET, NRAS, NTRK1, RET, ROS1, AKT1, CDKN2A, PIK3CA, PTEN, RB1, TP53). Using the ddPCR detection range as a comparison standard, the variant allele frequency (VAF) detected by both NGS and ddPCR methods was compared in all samples of 15 patients. Qualitative results show a high concordance between NGS and ddPCR (R2 = 0.9867, Figure 1A). The result indicated that the abundance of positive mutation obtained by NGS was highly consistent with that obtained by ddPCR. According to the Bland–Altman method, the average value of VAF difference detected by these two methods is 0.048% [95% CI 0.55%-0.64%], and the limits of agreement is −1.5% to 1.6% (Figure 1B).
Figure 1.
Comparison of the NGS and ddPCR ctDNA analysis. (A) Linear regression from the comparison of NGS and ddPCR VAFs (R2 = 0.9867). (B) Bland–Altman plot of the differences (NGS (VAF) - ddPCR (VAF)).
Due to the high effective sequencing depth of NGS, the detection limit has reached 0.05%. Thus, when comparing with ddPCR, different VAF cutoff values (0.05%, 0.1% and 0.2%) were selected for evaluation the sensitivity and specificity of our NGS assay. We found that 0.1% cutoff values of VAF can achieve a sensitivity of 87.5% and specificity of 100% (Table 2). Lower VAF cutoff value did not increase the number of positive test sites in this cohort, but increased false-positive sites which decreased specificity. While high cutoff value may prevent false-positive sites, but increased the proportion of false negatives which decreased sensitivity. Therefore, NGS panel detection needs to select the appropriate cutoff value of VAF, and 0.1% was considered acceptable in our NGS-based ctDNA detecting system.
Table 2.
Sensitivity and Specificity of NGS Assay at Different VAF Cutoff
| Cutoff | True-Positive | False-Negative | True-Negative | False-Positive | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| 0.05% | 7 | 1 | 328 | 1 | 87.5% | 99.7% |
| 0.10% | 7 | 1 | 329 | 0 | 87.5% | 100.0% |
| 0.20% | 6 | 2 | 329 | 0 | 75.0% | 100.0% |
Comparison of Mutation Profiles Between Baseline and Progression
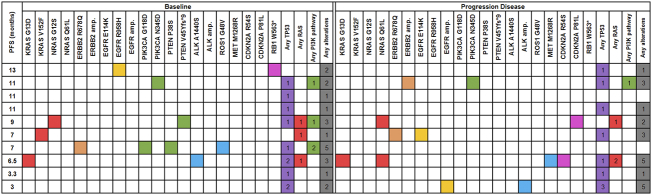
We monitored the mechanisms of resistance to therapy in baseline and subsequent treatments in all patients. However, ctDNA samples from the baseline and progression of cetuximab-based combination therapy of 10 patients were available for NGS-based assay to compare the mutation distribution at baseline and progression. As shown in Figure 2, the genomic scope was different between baseline ctDNA and progression ctDNA samples. TP53 was the most commonly mutated gene detected by NGS, and 80% (8/10) of the patients harbored mutations in TP53, which indicated that the dynamic monitoring VAF changes of TP53 mutations would work in most cetuximab-treated mCRC patients. Comparing with baseline, amplifications of ERBB2 (HER2), EGFR and ALK, newly mutations in NRAS, EGFR, HER2, MET, RB1 and CDKN2A/B were observed in ctDNA samples at disease progression. Most of these genes were known mechanism of acquired resistance to cetuximab treatment.
Figure 2.
Comparison of mutation profiles between baseline and progression. Columns with red (RAS), orange (ERBB2), golden (EGFR), green (PI3K pathway), blue (ALK, ROS1 and MET), pink (CDKN2A and RB1), purple (TP53) and gray (any genes) indicate the presence of gene alterations. Columns with white indicate wild type. PFS, progression-free survival (months). The number in each box indicates the number of mutations of each gene or pathways.
Interestingly, we found KRAS G13D at baseline sample of one patient who was considered as RAS negative by tissue based RAS detecting, which indicated the possibility of the existence of a KRAS sub clone in the tumor, and additional NRAS Q61L was identified in the progression sample of this patient. RAS pathway mutations related to acquired resistance to anti-EGFR therapy can be detected in the blood of patients with colorectal cancer prior to disease progression [4], [9]. The PFS of this patient was only 6.5 months, the short PFS might be related to KRAS sub clone and the newly discovered acquired NRAS Q61L mutation. Therefore, detection of ctDNA may help identify patients who have no response to anti-EGFR therapy or monitor patients who develop acquired resistance to this targeted therapy.
Dynamic Changes of Mutations during Treatment
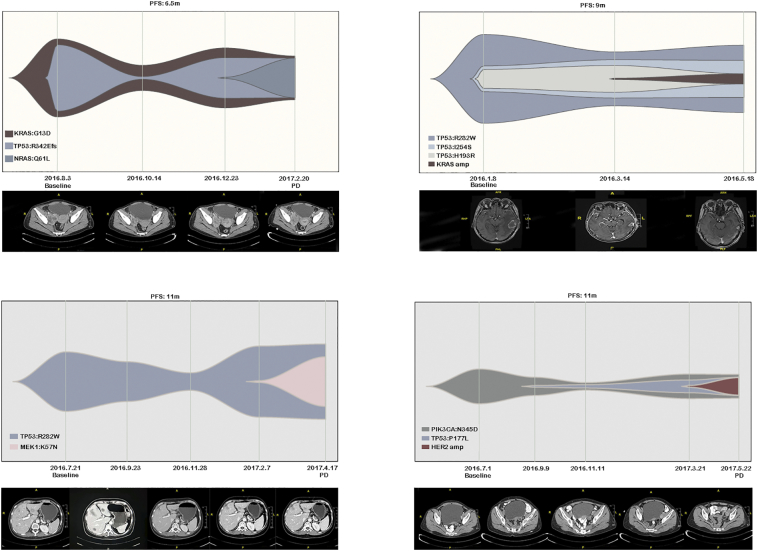
We performed serial monitoring of mutations in ctDNA as much as possible in our patients. Four cases were shown in Figure 3, patient 1 was detected KRAS G13D and TP53 R342Efs mutations in baseline with a PFS of 6.5 months. The dynamic changes of KRAS G13D and TP53 R342Efs were monitored during treatment while NGS additionally observed NRAS Q16L mutation at the time of disease progression. While ddPCR only detected consistent change of KRAS G13D. Unexpectedly, there was a KRAS mutation at baseline, but there was also a decrease in the abundance of KRAS with the treatment of cetuximab suggesting that KRAS mutant clones may also respond to cetuximab. NRAS is a secondary mutation that occurs later in the patient's treatment, suggesting that the patient's resistance to cetuximab treatment is mainly due to NRAS mutations rather than KRAS mutations. Patient 2 was detected TP53 R282W, I254S and H193R mutations at baseline and additional KRAS amplification at the time of disease progression. Patient 3 was detected TP53 R282W at baseline and additional MEK1 K57 N mutation during disease progression in both NGS and ddPCR. Patient 4 was detected original PIK3CA N345D and TP53 P177L mutations and HER-2 amplification when disease progressed in both methods. Our NGS assay not only identified the potential mechanisms of cetuximab treatment for these patients as ddPCR did, but also found the dynamic VAF changes of baseline genomic alterations, especially those mutations in tumor suppressor genes such as TP53, which could not be identified by ddPCR. Thus, monitoring the dynamic VAF changes of TP53 mutations by NGS assay would be a promising marker for evaluate the clinical response of cetuximab treatment.
Figure 3.
Dynamic changes of mutations and mutation abundance tested by NGS during treatment in four patients. During the treatment of cetuximab, the abundance of mutations detected in the ctDNA at the baseline showed a downward trend with the treatment process. The corresponding imaging to the blood collection time showed that new drug-resistant mutations were detected in blood in advance of imaging development.
Discussion
Anti-EGFR antibodies improve the treatment efficiency and survival of mCRC patients. However, acquired resistance to cetuximab emerges eventually after a period of treatment, even in patients who response at the beginning. RAF/FAS/ERK/MEK and mTOR/PI3K/AKT pathways play important roles in response to anti-EGFR antibodies and have been well known. And recent studies showed that other genomic alternations, such as MET, ERBB2, FLT3 and MAP2K1 in ctDNA of patients are also associated with resistance to EGFR blockade [21], [22], [23].
ctDNA has potential advantages over traditional solid tumor biopsies. These include less invasive means of obtaining diagnostic information by blood testing, which can be repeated more frequently and easily with minimal risk to the patient [24]. In addition, the use of ctDNA may lead to a more comprehensive manifestation of tumor heterogeneity within the tumor itself [24]. Thierry et al. showed that BRAF V600E mutation and multiple KRAS mutations could be reliably identified from ctDNA in patients with metastatic colorectal cancer [15]. Mohan et al. detected KRAS and MET mutations from ctDNA in a small cohort of patients who acquired resistance to anti-EGFR therapy [10]. Misale et al. showed the emergence of KRAS amplification or other KRAS mutations in 60% of patients with metastases colorectal cancer who developed resistance to anti-EGFR therapy [4]. Interestingly, the KRAS mutation from ctDNA was detectable in the blood of anti-EGFR treated patients before radiographic disease progression [4]. Therefore, ctDNA is a valid surrogate tumor biomarker for monitoring tumor burden and responses to anticancer therapies.
Different methods have been developed to characterize ctDNA, including ddPCR and NGS. ddPCR is an accurate and sensitive approach, and able to overcome some qPCR drawbacks in sensitivity, reproducibility and feasibility. However, there are some disadvantages including incapable of discovery of unknown mutations and limitation of allele-specific design [25]. In addition, cetuximab resistance mechanism has its diversity, but ddPCR can only monitor limited sites. Unlike ddPCR, NGS-based ctDNA assay has the potential to more broadly assess the molecular profile of the tumor. As a new and powerful tool, NGS has already improved the knowledge of the clonal heterogeneity and of the kinetics of many tumors [25]. NGS enables detection of low-frequency somatic mutations in heterogeneous tumor populations and in ctDNA with the potential for guidance of treatment [26]. Both methods make minimally invasive, dynamic genotyping and consequently prognostic predictions possible, and NGS enables rapid and highly sensitive identification of somatic genomic alterations in a broader genome region in individuals.
In this clinical validation, we demonstrate the ability of NGS to sensitively detect a wide range of molecular alterations in colorectal cancer. We also confirm in colorectal cancer patients the high sensitivity of the NGS assay, matching the sensitivity of ddPCR. This high sensitivity and an ability to detect a full spectrum of genomic variants make the NGS testing a compelling alternative to ddPCR for detection of ctDNA.
Baseline and progression ctDNA highlighted significant tumor heterogeneity and widespread potential of therapeutic resistance mechanisms in these patients. In this study, TP53 was the most commonly mutated gene to be identified in ctDNA. We observed the TP53 mutation abundance in a partial of patients varied with the course of treatment. Our data supported the use of NGS not only to find mechanisms of acquired resistance but, more importantly, to monitor disease progression which will be helpful for prevention or reversal of resistance.
In conclusion, we demonstrated the ability of NGS to detect with a wide range of mutations in colorectal cancer. We also found that high concordance exists between NGS and ddPCR methods. Moreover, NGS could detect more mutation information than ddPCR, which make it possible to dynamic monitoring early genetic change and then make the decent therapeutic adjustments in advance.
Acknowledgement
The authors thank all the patients who participated in this study.
The authors have no conflicts of interest to declare.
This study was supported by the National Natural Science Foundation of China (Grant No. 81472210); the Key Subject of Clinical Engineering of Zhejiang Province, No. G3221; CSCO Oncology Research Fund.
Footnotes
Funding: This study was supported by the National Natural Science Foundation of China (Grant no. National Natural Science Foundation of China No. 81472210;the Key Subject of Clinical Medical Engineering of Zhejiang Province No. G3221);CSCO Oncology Research Fund.
Contributor Information
Weijia Fang, Email: weijiafang@zju.edu.cn.
Zhiyu Chen, Email: chanhj75@aliyun.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sobrero AF, Maurel J, Fehrenbacher L, Scheithauer W, Abubakr YA, Lutz MP, Vega-Villegas ME, Eng C, Steinhauer EU, Prausova J. EPIC: phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(14):2311–2319. doi: 10.1200/JCO.2007.13.1193. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pinter T, Lim R, Bodoky G. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 4.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizzo S, Bronte G, Fanale D, Corsini L, Silvestris N, Santini D, Gulotta G, Bazan V, Gebbia N, Fulfaro F. Prognostic vs predictive molecular biomarkers in colorectal cancer: is KRAS and BRAF wild type status required for anti-EGFR therapy? Cancer Treat Rev. 2010;36(Suppl. 3):S56–S61. doi: 10.1016/S0305-7372(10)70021-9. [DOI] [PubMed] [Google Scholar]
- 6.Russo A, Rizzo S, Bronte G, Silvestris N, Colucci G, Gebbia N, Bazan V, Fulfaro F. The long and winding road to useful predictive factors for anti-EGFR therapy in metastatic colorectal carcinoma: the KRAS/BRAF pathway. Oncology. 2009;77(Suppl. 1):57–68. doi: 10.1159/000258497. [DOI] [PubMed] [Google Scholar]
- 7.Ziegler A, Zangemeister-Wittke U, Stahel RA. Circulating DNA: a new diagnostic gold mine? Cancer Treat Rev. 2002;28(5):255–271. doi: 10.1016/s0305-7372(02)00077-4. [DOI] [PubMed] [Google Scholar]
- 8.Khakoo S, Georgiou A, Gerlinger M, Cunningham D, Starling N. Circulating tumour DNA, a promising biomarker for the management of colorectal cancer. Crit Rev Oncol Hematol. 2018;122:72–82. doi: 10.1016/j.critrevonc.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Diaz LA, Jr., Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohan S, Heitzer E, Ulz P, Lafer I, Lax S, Auer M, Pichler M, Gerger A, Eisner F, Hoefler G. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet. 2014;10(3) doi: 10.1371/journal.pgen.1004271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Emburgh BO, Sartore-Bianchi A, Di Nicolantonio F, Siena S, Bardelli A. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol Oncol. 2014;8(6):1084–1094. doi: 10.1016/j.molonc.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Zhuo M, Ye X, Bai H, Wang Z, Sun Y, Zhao J, An T, Duan J, Wu M. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget. 2016;7(15):20810–20824. doi: 10.18632/oncotarget.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan W, Zhang A, Powell MJ. Genetic alteration and mutation profiling of circulating cell-free tumor DNA (cfDNA) for diagnosis and targeted therapy of gastrointestinal stromal tumors. Chin J Cancer. 2016;35(1):68. doi: 10.1186/s40880-016-0131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan K, Rata M, Cunningham D, Koh DM, Tunariu N, Hahne JC, Vlachogiannis G, Hedayat S, Marchetti S, Lampis A. Functional imaging and circulating biomarkers of response to regorafenib in treatment-refractory metastatic colorectal cancer patients in a prospective phase II study. Gut. 2018;67(8):1484–1492. doi: 10.1136/gutjnl-2017-314178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte P, Robert B. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20(4):430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 16.Thierry AR, El Messaoudi S, Mollevi C, Raoul JL, Guimbaud R, Pezet D, Artru P, Assenat E, Borg C, Mathonnet M. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann Oncol. 2017;28(9):2149–2159. doi: 10.1093/annonc/mdx330. [DOI] [PubMed] [Google Scholar]
- 17.Klein-Scory S, Maslova M, Pohl M, Eilert-Micus C, Schroers R, Schmiegel W, Baraniskin A. Significance of liquid biopsy for monitoring and therapy decision of colorectal cancer. Transl Oncol. 2018;11(2):213–220. doi: 10.1016/j.tranon.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. Liquid Biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem. 2017;41(2):755–768. doi: 10.1159/000458736. [DOI] [PubMed] [Google Scholar]
- 19.Busser B, Lupo J, Sancey L, Mouret S, Faure P, Plumas J, Chaperot L, Leccia MT, Coll JL, Hurbin A. Plasma circulating tumor DNA levels for the monitoring of melanoma patients: landscape of available technologies and clinical applications. Biomed Res Int. 2017;2017 doi: 10.1155/2017/5986129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demuth C, Spindler KG, Johansen JS, Pallisgaard N, Nielsen D, Hogdall E, Vittrup B, Sorensen BS. Measuring KRAS mutations in circulating tumor DNA by droplet digital PCR and next-generation sequencing. Transl Oncol. 2018;11(5):1220–1224. doi: 10.1016/j.tranon.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3(6):658–673. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Cora D, Di Nicolantonio F, Buscarino M, Petti C. A molecularly annotated platform of patient-derived xenografts ("xenopatients") identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1(6):508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 23.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, Ercan D, Rogers A, Roncalli M, Takeda M. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3(99):99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabriel E, Bagaria SP. Assessing the impact of circulating tumor DNA (ctDNA) in patients with colorectal cancer: separating fact from fiction. Front Oncol. 2018;8:297. doi: 10.3389/fonc.2018.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dogliotti I, Drandi D, Genuardi E, Ferrero S. New molecular technologies for minimal residual disease evaluation in B-Cell lymphoid malignancies. J Clin Med. 2018;7(9) doi: 10.3390/jcm7090288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page K, Guttery DS, Fernandez-Garcia D, Hills A, Hastings RK, Luo J, Goddard K, Shahin V, Woodley-Barker L, Rosales BM. Next generation sequencing of circulating cell-free DNA for evaluating mutations and gene amplification in metastatic breast cancer. Clin Chem. 2017;63(2):532–541. doi: 10.1373/clinchem.2016.261834. [DOI] [PMC free article] [PubMed] [Google Scholar]