Abstract
Multidrug-resistant Klebsiella pneumoniae is a nosocomial pathogen, produces septicemia, pneumonia and UTI. Excessive use of antibiotics contributes towards emergence of multidrug-resistance. Bacteriophage-therapy is a potential substitute of antibiotics with many advantages. In this investigation, microbiological and genome characterization of TSK1 bacteriophage and its biofilm elimination capability are presented. TSK1 showed narrow host range and highest stability at pH 7 and 37 °C. TSK1 reduced the growth of K. pneumoniae during the initial 14 hours of infection. Post-treatment with TSK1 against different age K. pneumoniae biofilms reduced 85–100% biomass. Pre-treatment of TSK1 bacteriophage against the biofilm of Klebsiella pneumoniae reduced > 99% biomass in initial 24 hr of incubation. The genome of TSK1 phage comprised 49,836 base pairs with GC composition of 50.44%. Total seventy-five open reading frames (ORFs) were predicted, 25 showed homology with known functional proteins, while 50 were called hypothetical, as no homologs with proved function exists in the genome databases. Blast and phylogenetic analysis put it in the Kp36 virus genus of family Siphoviridae. Proposed packaging strategy of TSK1 bacteriophage genome is headful packaging using the pac sites. The potential of TSK1 bacteriophage could be used to reduce the bacterial load and biofilm in clinical and non-clinical settings.
Introduction
Klebsiella pneumoniae is an important biofilm-forming bacteria responsible for a broad range of infections, placing it among the eight most significant nosocomial pathogens1. It is ubiquitous and causes septicemia, pneumonia and urinary tract infections. It is one of the most important nosocomial pathogen causing 5–7% hospital-acquired infections and resists a wide range of antibiotics2. K. pneumoniae have two main resistance mechanisms: production of enzymes and biofilm formation. Since 1980, the third-generation cephalosporins (cefotaxime, ceftazidime, and ceftriaxone) have been introduced as useful and effective drugs against most nosocomial infections. The excessive use of cephalosporins in clinical settings and production of a β-lactamase enzyme is the leading cause of antibiotic resistance in K. pneumoniae3.
Due to the emergence of cephalosporin-resistant K. pneumoniae strains, carbapenems were broadly used for the treatment of serious infections caused by multidrug-resistant K. pneumoniae4. However, in recent years, widespread use of carbapenems and fluoroquinolones have accelerated the global emergence of its resistant strains. Carbapenem-resistant K. pneumoniae (CRKP) is an emerging antibiotic-resistant nosocomial pathogen5. The acquisition of genes encoding enzymes capable of breaking down most β-lactams including carbapenems (K. pneumoniae carbapenemases (KPCs), carbapenemases of the oxacillinase-48 (OXA-48) and New Delhi Metallo-β-lactamase (NDM) carbapenemases) leads to develop antibiotic resistance against carbapenems6. Fluoroquinolones having broader antimicrobial spectra and improved pharmacokinetic properties are extensively used in the treatment of the most nosocomial infections. K. pneumoniae develops resistance against quinolones by incorporating mutations in enzymes (DNA gyrase and topoisomerase) which are targets of these antibiotics7. The extensive use of fluoroquinolones in treating bacteremia and UTI infections are responsible for decreased susceptibility of K. pneumoniae against them8.
Biofilm is the heterogeneous, organized microbial communities that are entrenched in a hydrated self- produced matrix9. It provides a shield from desiccation and allows bacteria to achieve 1000 times higher densities than the freely existing forms of bacteria. Due to the presence of capsular polysaccharide (CPS) and lipopolysaccharide (LPS), K. pneumoniae biofilm, safeguard it against the host immune system10. These diverse microenvironments confer properties of multidrug resistance, protection against the opsonization and other immune reactions11. The threat of antibiotic resistance and inability of breaking the biofilm structure necessitates to explore novel strategies for controlling biofilm producing pathogens1. Bacteriophage therapy is a viable option of controlling bacterial entities involved in biofilm formation12. It is preferable over antibiotic treatment, due to its high specificity and self-replication at the site of infection along with its narrow host-range that leaves normal flora unharmed13. Through their interaction with host cells, phage-borne exopolysaccharide depolymerases degrade biofilm matrix that acts as a barrier for antimicrobial agents, infects the bacterial cells and may cause extensive biofilm disruption9.
The global emergence of extended-spectrum beta-lactamases (ESBLs), carbapenem and fluoroquinolones resistance in K. pneumoniae is a serious problem and only a few remaining last-resort antibiotics can be used to treat fluoroquinolones and carbapenem-resistant K. pneumoniae (CRKP) infections14,15. The worldwide spread of Klebsiella pneumoniae spp., resistant to a variety of antibiotics, threatens to revert modern medicine to a pre-antibiotic era. This research was conducted to isolate a novel virulent bacteriophage against multidrug-resistant K. pneumoniae. The isolated bacteriophage (TSK1) was characterized based on morphology, physiological parameters, host range and genome characteristics. Furthermore, the tendency of bacteriophage to disintegrate the biofilm was also assessed.
Results
TSK1 bacteriophage forms opaque halo zone around clear plaques
The bacteriophage TSK1 exhibiting potent lytic activity against K. pneumoniae was isolated from sewage water sample of TECH Housing Society Lahore. TSK1 bacteriophage formed bull’s eye shaped clear plaques (3 mm in diameter) that were surrounded by large opaque halo zone (Fig. 1). Plaque, characterized by morphology, was purified and amplified for further analysis.
Figure 1.
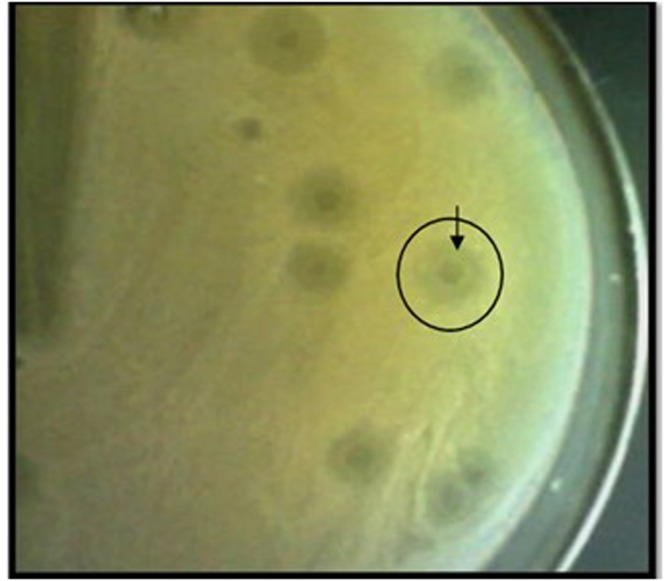
Plaque morphology. Plaques (clear surrounded by a large halo, 3 mm diameter) formed by TSK1 bacteriophage with k. pneumoniae lawn on double layer agar plate.
TSK1 showed highest stability at pH 7 and 37 °C temperature
Effect of different pH and temperatures on the stability of TSK1 was observed by incubating at different pH and temperatures for 1 hour followed by determination of bacteriophage titer after neutralizing the pH. Bacteriophage TSK1 showed the highest titer at pH 7 (2.7 × 108 pfu/ml), while the reduction of titer was observed at pH 6, 8 and 9, whereas no stable virus was observed at pH 5 and 10 (Supplementary Fig. 1). However, the highest viral titer was noted after incubation at 37 °C (3.2 × 108), than the other tested temperatures (25, 45, 55 & 60) (Supplementary Fig. 2). TSK1 bacteriophage showed narrow host range against different clinical isolates of K. pneumoniae, determined by spot test (Supplementary Table 2). One- step growth curve was performed to determine the burst size and latent period of TSK1 bacteriophage. The latent period of TSK1 was 30 minutes while the burst size was 113 pfu/infected cell (Supplementary Fig. 4). Bacterial growth reduction assay holds a tremendous relation towards phage therapy as it shows the reduction of bacterial growth through incubation time with bacteriophages. The Klebsiella pneumoniae infected with TSK1 bacteriophage had slower growth from the start to 14 hours of incubation. Then the growth increased gradually but remained lower than the control values (Supplementary Fig. 5).
Sequence and phylogenetic analysis of TSK1 genome
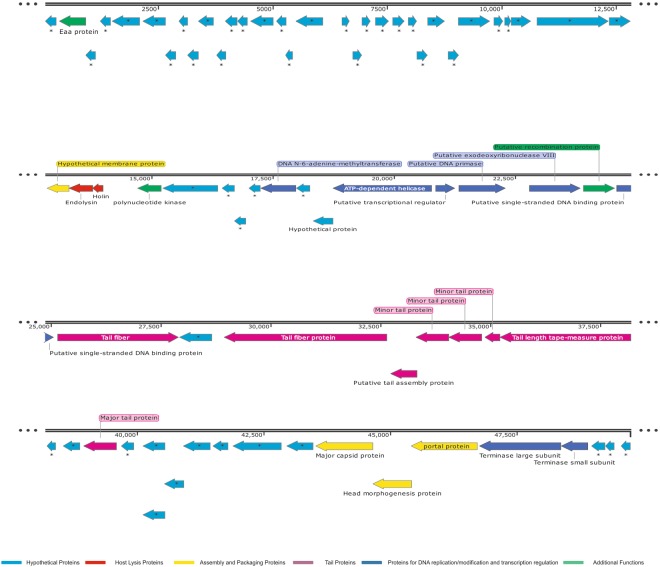
DNA genome of TSK1 is 49836 bp long with GC content of 50.44%, similar to previously sequenced K. pneumoniae phages KP1513 (97% identity) and KP36 (96% identity)16. The genome comprising of 75 predicted open reading frames (ORFs), while no tRNA gene was predicted. Each ORF has a start codon, stop codon, ribosomal binding sites with higher Shine Dalgarno (SD) score and the minimum length of the protein-coding gene is about 120 bp (40 codons). Among 75 predicted ORFs, only 25 showed homology with known functional proteins, while remaining 50 were termed hypothetical due to the absence of any homologous gene in the genome databases. The genome annotation information of functional ORFs is presented in (Supplementary Material Table 3). The functional ORFs were further divided into five functional groups: DNA replication/modification/transcriptional regulations (DNA primase/helicase, exodeoxyribonuclease, DNA terminase, DNA N-6 methyltransferase, ssDNA binding protein, DNA adenine methyltransferase), structure and packaging (portal protein, major capsid protein, membrane protein and head morphogenesis protein), host lysis (holin, endolysin) tail structure (major and minor tail fiber protein, tail assembly and tail length tap measure proteins) and some additional proteins (recombination protein, polynucleotide kinase and Eaa like protein). These results show that the bacteriophage genome contains all the major structural and functional proteins. The orientation of genome annotation showed that most genes (53) are on the plus strand, while 22 on the reverse strand. The linear genome map with different families of proteins with known and unknown functions is presented in Fig. 2. The PhageTerm analysis showed that the genome packaging of TSK1 bacteriophage takes place through headful packaging by using the pac-site. According to the results obtained from PhageTerm, the TSK1 phage genome packaging starts at a specific packaging initiation sequences (5′ TGATCA 3′) at pac-site on concatameric precursors, like the reported P1 bacteriophage17. Headful packaging in TSK1 genome occurred in the forward direction. The BlastN analysis revealed that the most related genome sequences to TSK1 genome in the database were Klebsiella phage KP1513 (accession KP658157.1, 85% query coverage and 97% identity) and Klebsiella phage KP36 (accession JF501022.1, 86% query coverage and 96% identity). The results of sequence similarity suggested that TSK1 phage is a member of Siphoviridae family18.
Figure 2.
Linear genomic map of TSK1 bacteriophage obtained by Snapgene viewer. Arrow represent predicted ORFs, the direction of arrow represent direction of transcription. Different functional groups of TSK1 bacteriophage are denoted by different colors, according to their function.
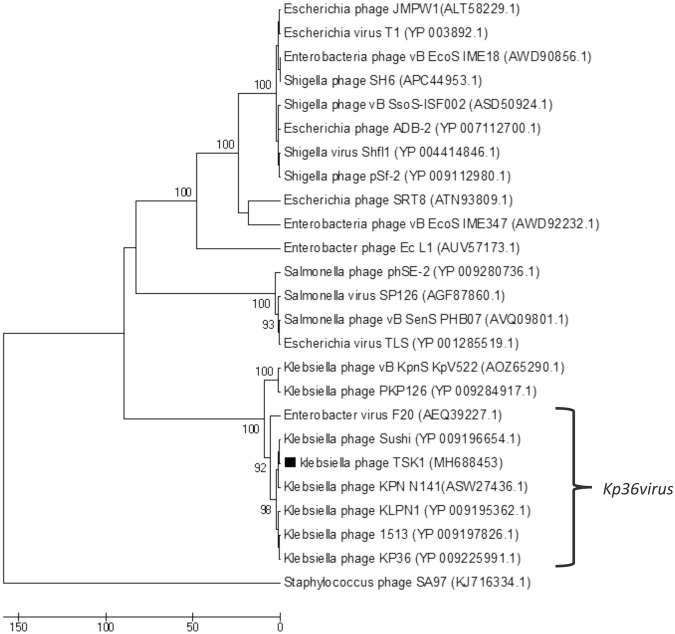
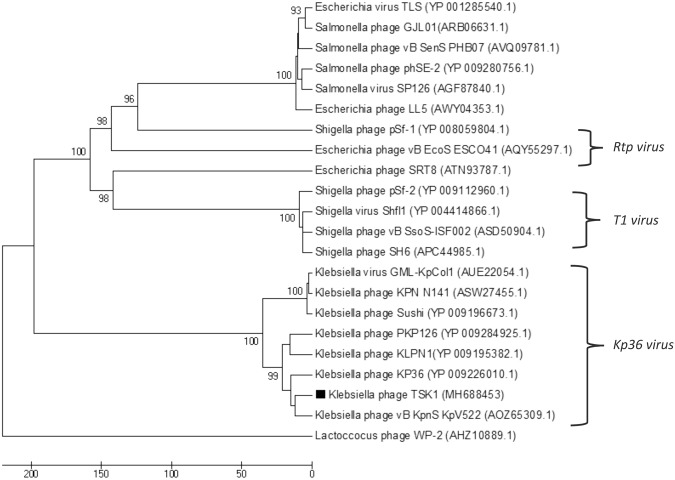
Evolutionary relationship of TSK1 bacteriophage terminase large subunit and tail fiber protein with related phages were analyzed by using MEGA v 7.0. The amino acid sequence of TSK1 terminase and tail fiber were analyzed using BLASTp to identify putative homologs in the NCBI protein database. The phylogenetic tree showed that this phage had homology to subfamily Tunavirinae phages (Klebsiella phage 1513, Klebsiella phage KLPN1, Klebsiella phage KP36, Klebsiella phage PKP126 and Klebsiella phage Sushi). According to UPGMA dendrograms, Klebsiella phage TSK1 was classified as a Kp36virus genus in the Tunavirinae subfamily (Figs 3 and 4).
Figure 3.
The Phylogenetic tree of TSK1 bacteriophage terminase large subunit. The tree was constructed by using the UPGMA method with 2000 bootstrap replications. The terminase large subunit of Staphylococcus phage SA97 was used as an out-group. The GenBank accession numbers are also provided after phage names, in parentheses. This tree showed the relationship of TSK1 terminase large subunit with other closely and distant bacteriophages.
Figure 4.
The Phylogenetic tree of TSK1 bacteriophage tail fiber protein. The tree was constructed by using the UPGMA method with 2000 bootstrap replications. The tail fiber protein of Lactococcus garvieae phage WP-2 was used as an out-group. The GenBank accession numbers are also provided after phage names, in parentheses. This tree showed the relationship of TSK1 tail fiber protein with other closely and distant bacteriophages.
K. pneumoniae biofilm formation was maximum at 72 hr post- inoculation
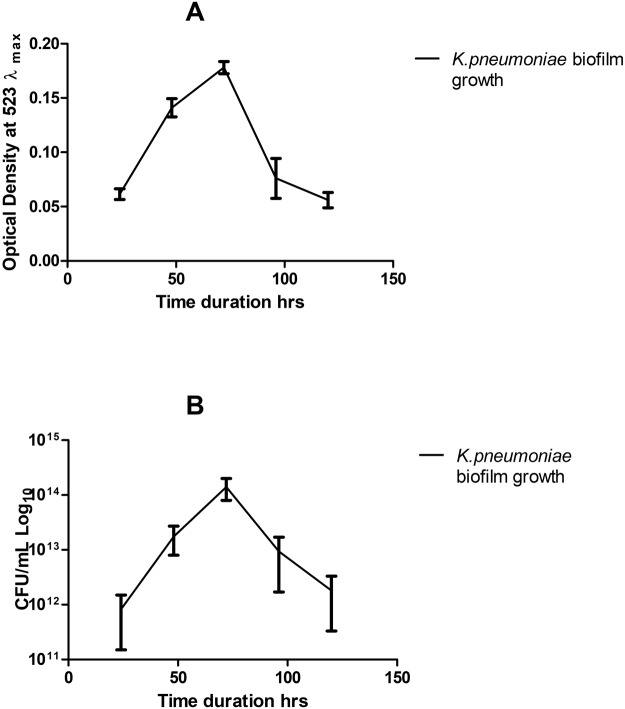
Biomass production of K. pneumoniae was analyzed by CV assay at various time points. Maximum biofilm formation was observed after 72 hr, while a decline of biomass started on further incubation (Fig. 6A). Similar observations were got in viable cell count assay. The bacterial count for young immature 24 hr old biofilm was 7.5 × 108 cfu ml−1 followed by a maximum biomass production on the 3rd day (1.4 × 1014 cfu ml−1) and a further decline resulted in a bacterial count of 1.32 × 1012 on the 5th day of mature biofilm (Fig. 5). Based on these findings, all further experiments of biofilm eradication through TSK1 were performed on up to 72 hr old biofilms.
Figure 6.
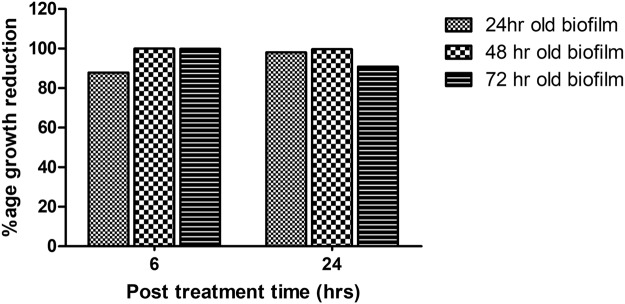
Comparison of percentage reduction in K. pneumoniae biofilms (1- to 3-day old) subjected to TSK1 challenge for 6 and 24 hours.
Figure 5.
Evaluation of biofilm establishment in polystyrene microtitre plate through CV assay and Viable cell count. Different days old biofilm analyzed through CV assay (A) and viable cell counting (B).
Effect of exposure time on biomass reduction of K. pneumoniae biofilm by TSK1 bacteriophage
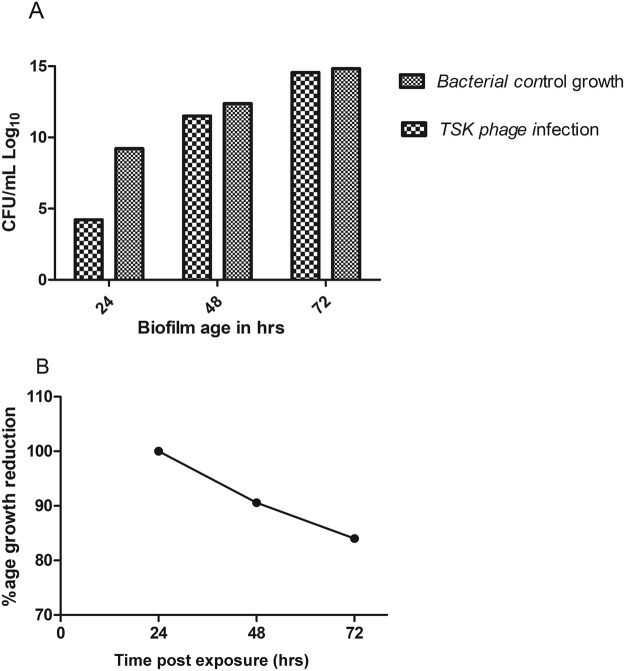
The efficient reduction of biofilm depends on the efficiency of bacteriophage invasion. The biofilm of different ages might influence the infection efficiency of bacteriophage. Significant biofilm reduction was observed upon challenging pre-formed biofilm of different ages with phage TSK1 (MOI: 0.6), at 6 and 24 hr post challenge. More than 95% biomass reduction was observed in 24 hr, 48 hr and 72 hr old biofilm treated with TSK1. There was no major difference in the biofilm removal capacity of TSK1 when applied to the biofilm of different ages (P = 0.534). The ability of the TSK1 phage to remove a bacterial load from the biofilm of different ages was 91% to 100%. The bacterial load from 48 hr old biofilm was consistently removed at 6 hr and 24 hr post inoculation, while the reduction was variable on 24 hr and 72 hr old biofilm, however still the percentage reduction was higher than 85% (Fig. 6). However, the difference between the bacterial load reductions was statistically non-significant when calculated at 6 and 24 hr post exposure (P = 0.95).
Pre-treatment of TSK1 showed inhibition of biofilm formation
Pretreatment of surface with TSK1 before biofilm development showed promising results. Average percentage reduction of the bacterial load at 24, 48 and 72 hr post inoculation was 99.9%, 90% and 84% respectively (Fig. 7). Percentage reduction in the biofilm at 1–3 days post inoculation was statistically significant (P => 0.0001) at different time intervals (24 hr, 48 hr, and 72 hr).
Figure 7.
Study of Phage treatment on the pre-biofilm formation. (A) Bacteriophage TSK1 challenge was given at 0 times of biofilm formation and studied for 72 hr. (B) The percentage reduction of bacterial load compared with untreated control at different time intervals. Bacterial count was performed at 24, 48, and 72 hr post inoculation.
Discussion
Due to excessive use of antibiotics in hospitalized patients, K. pneumoniae exhibits resistance against many antimicrobial drugs. Previous studies explained that multiple drug resistant (MDR) K. pneumoniae exhibits several mechanisms such as the production of extended spectrum β-lactamases and carbapenemase19. The MDR strains of K. pneumoniae cause different infections in humans, which are difficult to control by using antibiotics. So, bacteriophage therapy is an effective alternative treatment option against multiple drug-resistant pathogens. In the present study, a lytic bacteriophage (TSK1) against K. pneumoniae was isolated from a sewage sample and then characterized for different physiological parameters and the whole genome. The K. pneumoniae specific bacteriophage TSK1 has the distinctive character to form opaque halo zone surrounding the clear zone of plaque. The hazy halo zones suggest the ability of TSK1 phage to produce soluble, polysaccharide-degrading enzymes20. Previous research showed that the formation of halo zones is an indicator for the presence of phage tail-associated exopolysaccharide (EPS) depolymerases9. When the bacterial growth cycle enters the stationary phase, phage replication often stops or slows down substantially but their tail spikes still can depolymerize bacterial exopolysaccharide, resulting in the appearance of an increased transparent zone known as the halo zone9. It was deduced from previous research that phage polysaccharide depolymerase can degrade bacterial capsules21. Verma and colleagues reported that bacteriophage ФNDP with no depolymerase was unable to significantly decrease K. pneumoniae biofilm mass, however, the bacteriophage KPO1K2 with depolymerase activity was able to significantly decrease biofilm mass22. Additionally, the halo zones could also be due to diffusion of virus-encoded non-virion associated lytic enzymes such as endolysins, which might have tried to degrade the cell wall of neighboring cells23.
Both temperature and pH are crucial factors that influence the bacteriophage stability. Temperature affects the whole phage replication process and also regulate phage viability, occurrence, and storage24. TSK1 showed the highest activity at 37 °C while a reduction in phage titer was observed at all other temperatures (Supplementary Fig. 2). The phage was completely inactivated at 65 °C temperature, similar to Kp34virus genus phages as described earlier25. It showed more activity at alkaline pH than acidic pH (Supplementary Fig. 1), however, 7.0 pH found optimal for this phage. TSK1 is not suitable for direct oral delivery of the phage for therapeutic purpose without some kind of protective layers such as alginate and chitosan26 since the baseline pH of 1.5 in the human stomach27 will deactivate the phage. Due to its stability at alkaline pH, it can be used as a therapeutic agent for K. pneumoniae mediated urinary tract infection (UTI) and infected wounds. It can be used in the impregnation of urinary catheters to inhibit bacteriological biofilm as previously suggested28. TSK1 phage was examined for its lytic ability against 16 strains belonging to K. pneumoniae and other related and unrelated genera. TSK1 showed activity against few tested strains of K. pneumoniae, while not showed lytic activity against tested members of other genera; depicting its narrow host range (Supplementary Table 2). These results demonstrate that TSK1 phage showed host specificity within the genus Klebsiella. Previously, the bacteriophages KP1513 and KP-34 against K. pneumoniae also showed narrow host range25,29. TSK1 can be applied in phage therapy in combinations with other bacteriophages as phage cocktails and with antibiotics for the eradication of K. pneumoniae infections. The one-step growth curve of bacteriophage reveals that the latent period for TSK1 phage is short with moderate burst size. These results clearly indicate that TSK1 possesses replication properties similar to phage B505528. The application of phages in phage therapy and their feasibility can be predicted by the results of bacterial growth reduction assay. TSK1 phage reduced the bacterial growth till 14 hours, while the appearance of resistance in bacteria started at 14 hr, post-treatment (Supplementary Material Fig. 4), which indicates the development of bacteriophage resistant mutants. The exact mechanism of bacteriophage resistance is not clear, however different mechanisms like downregulation of bacteriophage receptors and CRISPR/Cas mediated digestion of bacteriophage genome, production of proteins to abort bacteriophage replication and others are proposed which need further exploration30.
The whole genome sequencing and genome annotation of TSK1 bacteriophage revealed that it has a 49,836 bp long genome, having 75 ORFs and no tRNA gene. The predicted homology of protein-coding sequences (CDSs) with genes from reported whole genomes showed maximum resemblance with Siphoviridae bacteriophages KP36 and KP151316. Unsurprisingly, it did not show any evolutionary relationship with the members of the family Podoviridae and Myoviridae. The genome annotation of TSK1 bacteriophage showed that it has all basic structural and functional genes that encode host lysis proteins, DNA replication/modification and packaging proteins, tail structure proteins and some additional functional proteins. The gene products with potential antibacterial properties could be used in phage therapy in the future31. A large portion of the genome (67%) represents hypothetical proteins (HPs). There are four ORFs, which not showed any homology to sequences in databases while remaining forty-nine ORFs showed homologies to the HPs of Siphoviridae phages. More than half of a K. Pneumoniae phage JD001 genome also comprised of functionally uncharacterized hypothetical proteins32. Furthermore, novel HPs may also serve as markers and pharmacological targets for drug design, discovery, and screening33. Phylogenetic trees of terminase large subunit and tail fiber protein of TSK1 bacteriophage and other similar K. pneumoniae phages showed that the TSK1 proteins showed homology with other Klebsiella phages of Kp36 virus genus (Figs 3 and 4). This indicates that TSK1 is a novel virulent bacteriophage of a proposed new genus called Kp36 like virus. The bioinformatics approach of utilizing PhageTerm for identifying termini of the bacteriophage genome has identified its genome as a linear molecule with “pac” site and “headful packaging” strategy, also reported for P1and T1 type viruses34. The proposed packaging of TSK1 occurs in the forward direction in a headful of concatameric genomic DNA and only in the direction of concatamers35. Terminases are involved in the cutting of concatameric DNA at specific locations initiated in a processive series from a specific packaging sequence also described earlier in P1 and T1 phages. This packaging series initiation site is called as “pac-site”, which the terminase recognize and starts a headful packaging series. The Terminase large (TerL) protein of TSK1 phage showed homology to the TerL of phages that use a pac-headful DNA packaging mechanism36.
Biofilm formation and development is a significant virulence factor of numerous microbial species. Biofilm-forming bacteria were found to inhibit the effectiveness of antibiotic treatment by reducing its penetration to bacteria and exhibiting the ability to evade from host responses. In this scenario, bacteriophages can be an effective alternative as they not only replicate at the site of infection but also produce special enzymes/proteins which disrupt the biofilm structure. In the present investigation, K. pneumoniae biofilm kinetics study was carried out through viable count assay. A viable count assay provides the direct estimate of live cells. However, the method is laborious and needs a lot of medium & glassware. In the current study, the viable count assay was carried out through the micro-drop method37. The TSK1 bacteriophage showed degradation of different age biofilms (24, 48 and 72 hr old), and showed >95% biomass reduction during 24 hr post-treatment. The biofilm removal tendency was not different for biofilm of different ages as the difference in biomass reduction was not statistically significant (P = 0.534). Similarly, the biomass reduction capability also not came out different when tested for different incubation times (6 and 24 hr post exposure, P = 0.95). The pre-treated surfaces with bacteriophage TSK1 also showed decreased development of biofilm by K.pneumoniae for three different incubation durations (24, 48 and 72 hr) post inoculation. Amazingly the biofilm inhibition tendency was decreased with the passage of time (99.9% to 84%) and the difference in biofilm inhibition tendency at different incubation times was statistically significant (P = 0.0001). The decrease in biofilm inhibition tendency might be due to the production of some compounds after a certain time of bacterial growth, which decreased the bacteriophage entry and needs further research. It could also be due to the emergence of bacteriophage resistant cells, which also requires further exploration. A similar pattern of decrease in biofilm inhibition capacity of a bacteriophage JHP against Pseudomonas aeruginosa biofilm was reported earlier38. A K.pneumoniae bacteriophage B5055 showed enhanced biofilm removal capacity when used in combination with Co(II) as compared with it alone1.
Conclusion
The emergence of multiple antibiotic-resistant K. pneumoniae strains has limited the use of antibiotics to control this pathogen. In this study, a novel lytic bacteriophage TSK1, against multidrug-resistant K. pneumoniae was isolated from sewage water. Its narrow host range makes it capable to control specific pathogen, while its stability at various pH and temperatures, burst size and latent period makes it suitable for bacteriophage therapy. The significant novelty of this study is that TSK1 bacteriophage produces depolymerase enzyme which plays a role in the disruption of K. pneumoniae capsular polysaccharide and biofilm. The TSK1 with reported physiological and genetic characters could be a new candidate for bacteriophage therapy either singly or in the form of cocktails with other bacteriophages.
Materials and Methods
Bacterial strain and culture conditions
An environmental isolate of K. pneumoniae ShA2 strain (accession number KF975429) provided by Dr. Yasir Rehman (Assistant Professor at Department of Microbiology and Molecular Genetics, University of the Punjab, Lahore) was used as a host for isolation and characterization of bacteriophage. The strain was 16 S rRNA sequenced from Macrogen, Korea. Its sequence was found 99% identical to K. pneumoniae. The selected bacterial strain was cultured in Luria Bertani (LB) medium and incubated at 37 °C. Antibiogram of K. pneumoniae ShA2 strain is available in Supplementary Table 1.
Bacteriophage isolation and Purification
The bacteriophage was isolated from wastewater collected from TECH Housing Society Lahore by a previously reported method39 with slight modification. Briefly, after centrifugation of the liquid sample, 10 ml of the supernatant was mixed with 10 ml of 2X L-broth. Then 100 µl of 6 hours fresh culture of the host bacteria was added, and the flask was incubated in a shaking incubator at 37 °C for 24 hours followed by centrifugation. The supernatant was filtered using a 0.45-μm filter and spotted onto LB plates overlaid with the respective strain to detect phage plaques. An agar overlay method was used for the isolation of a pure phage preparation and to determine phage titer. A tenfold dilution series (1–10–9) of lysate was prepared in Luria Bertani broth, these dilutions were mixed with an exponentially grown culture of the host bacteria and incubated at 37 °C for 15 minutes. The incubated mixture was then mixed in soft agar medium and plated over the solid agar plate. Single plaque isolation, elution, and re-plating were performed repeatedly40. The filtrate containing bacteriophages was subjected to the bacteriophage titer determination (pfu ml−1) through a double layer agar method followed by recording the plaque morphology.
Determination of Phage host range
Host range of the isolated phage was assessed on 16 clinical isolates from a medical Microbiology lab in Lahore, through standard spot test40. Lytic activity was determined against K. pneumoniae and other strains listed in Supplementary Material Table 2. Briefly, a 100 ul of test strain(s) was mixed with 3 ml of semi-solid medium and poured on LB agar medium. After solidification, 10ul of isolated and purified phage lysate was spotted onto the marked area against every bacterial strain. Following overnight incubation at 37 °C, plates were observed for a clear spot in the bacterial lawn.
Assessment of Bacteriophage Thermal and pH stability
The effect of different temperatures and pH on the stability of bacteriophage was determined as reported previously41. The effect of temperature on bacteriophage was studied at different temperatures (28, 37, 40, 45, 55, 60 and 65 °C). A known titer of purified bacteriophage was incubated at different temperatures for one hour followed by bacteriophage titer determination. Similarly, known phage titer was incubated at different pH values (5, 6, 7, 8, 9 and 10) for one hour at 37 °C followed by titer count after neutralizing the pH to 7.0.
Assessment of Bacteriophage Bacterial growth reduction Tendency
The bacterial growth reduction assay was determined by an already reported method41. Overnight bacterial culture (1 × 108 cfu) was added into two L-broth flasks (50 ml). One flask was inoculated with bacteriophage (1 × 108 pfu), while the other was taken as a control and incubated at 37 °C with shaking. The optical density (OD600) was noted for 24 hr at an interval of 2 hours.
One-step growth curve
One-step growth curve of isolated bacteriophage against K. pneumoniae was performed in duplicates according to the previously reported method41. Fresh host strain (1 × 106 cfu) was mixed with bacteriophage (1 × 108 pfu) at an MOI of 100 in 500 μL of L-broth and incubated at 37 °C for one minute. The mixture was centrifuged at 13,000 rpm for 30 seconds to eradicate free bacteriophages and the pellet was incubated at 37 °C after re-suspension in 100 mL of fresh broth. Samples were collected for one hour with a 5-minute interval, centrifuged and the phage titer was determined. Burst size was determined from the ratio of the mean yield of phage used for bacterial infection to the mean of phage particles liberated after infection.
Genome characterization and analysis
The genome of TSK1 bacteriophage was extracted utilizing Phage DNA Extraction Kit (Norgen Biotek, Canada, Cat No; 46850) according to manufacturer’s protocol. Supernatant with high concentration of viruses was used to isolate the genomic DNA. Whole genome sequencing of TSK1 bacteriophage was done by next-generation sequencing from the University of Minnesota, Genomic Centre (UMGC). The qualified sequence reads were assembled using CLC genomic workbench 10. The gene annotation and phylogenetic analysis of the complete genome were performed utilizing different bioinformatics tools. PHASTER, Gene Marks and Gene Glimmer were used for identification of open reading frames (ORFs) within the genome42,43. The confirmation of the predicted ORF was done by finding ribosomal binding sites (RBS) using the PECAAN program [https://discover.kbrinsgd.org/autoannotate/]. The predicted ORFs were annotated for specific functions using the BLASTp [https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE = Proteins] and InterProScan programs with various protein domain databases [http://www.ebi.ac.uk/interpro/search/sequence-search]. InterProScan is a tool that allows protein sequences to be scanned against several different databases i.e HAMAP Prosite-Profiles, PANTHER, PfamA, PIRSF, ProDOM, SMART, TIGRFAM, Prosite-Patterns and structural domains i.e Gene3d, SUPERFAMILY44. The proteins were characterized by their molecular weight and the isoelectric point at online Sequence Manipulation Suite [http://www.bioinformatics.org/sms2/dna_mw.html] and [http://www.bioinformatics.org/sms2/protein_iep.html] respectively. Genome map was drawn with the help of Snapgene [http://www.snapgene.com/]. DNA termini and phage packaging mechanism were determined by PhageTerm through its online application incorporated in galaxy (https://galaxy.pasteur.fr/). PhageTerm software uses raw sequence reads of a phage and its genomic reference sequence to determine the termini position45. The complete genome sequence of the K. pneumoniae phage TSK1 was deposited in GenBank under accession no. MH688453. The amino acid sequences of two predicted ORFs (terminase large subunit (ORF 19) and tail fiber protein (ORF 38) and other bacteriophages sequences showing maximum similarity at NCBI Blast were aligned in MEGA version 7.1. The Phylogenetic tree of the aligned sequences was constructed using the UPGMA (unweighted pair group method with arithmetic mean)46 method with 2000 bootstrap value. The Staphylococcus phage SA97 terminase large subunit and Lactococcus garvieae phage WP-2 tail fiber were used as an out-groups47 for both phylogenetic trees.
Assessment of biofilm removal capacity of TSK1 bacteriophage
Biofilm development is an important characteristic of K. pneumoniae responsible for a wide range of nosocomial infections. The activity of TSK1 to eradicate the K. pneumoniae biofilm was assessed. To determine the effectiveness of bacteriophage TSK1 treatment for eradication of K. pneumoniae biofilms, the biofilms were grown in microtiter plates. The biofilm was developed by adding LB broth (196 µl) and 4 μl of bacterial culture (1012 cfu ml−1) in the wells of a pre-sterilized microtiter plate, followed by incubation at 37 ± 1 °C for up to five days.
Kinetics of biofilm formation
To study the kinetics of biofilm formation, viable cell count was performed according to the method described earlier38. In this method, after every 24 hours, planktonic bacteria were removed and a set of two control and two wells of a microtiter plate (corresponding to each day) were washed thoroughly three times with 0.85% NaCl. The adherent biofilm was incubated with sterile 0.85% NaCl for 10 min on the rocker. The suspension was vortexed for 3 min using Cyclomixer (Remi Instruments & Appliances, Germany). For the quantitative measurement of biofilm through the viable count, the homogeneous biofilm suspension was serially diluted and micro drop method37 was carried out on L. agar plates followed by incubation at 37 ± 1 °C for 24 hr. This procedure was repeated until the 5th day of the experiment.
Bacteriophage treatment of biofilm grown on microtiter plates
Two types of phage treatments were carried out to check the susceptibility of biofilm from bacteriophage TSK1: Post-treatment of phage infection and pre-treatment of phage infection. In Post-treatment phage infection experiments, K. pneumoniae biofilm of different ages (1–3 days old) grown in a microtiter plate was exposed to bacteriophage at 0.6 MOI. The viable count was determined after 6 and 24 hr of incubation in the treated and untreated biofilm. The reduction in log values of the bacterial count was reported compared with untreated control. In the pre-treatment experiments, LB broth containing phage TSK1 was added in the wells of the microtiter plate along with the bacterial culture at an MOI of 0.6 and incubated the plate at 37 ± 1 °C for 72 hr. A Viable count was determined after every 6 and 24 hr time interval, followed by the comparison of reduction in log values of the bacterial count with untreated control. The percentage reduction in a particular treatment was calculated by the following formula.
Where T = bacterial load in the treated sample and U = Bacterial load in the untreated sample
Statistical analysis
All experiments were performed in duplicates. On different days of biofilm formation, all the data from a particular treatment and time points were grouped and log reductions compared to untreated biofilm at the respective time points were calculated. The effect of different treatments on biofilm eradication was tested by GraphPad Prism 6 using an appropriate test (unpaired t-test and one way ANOVA) at a confidence interval of 95%.
Electronic supplementary material
Acknowledgements
We acknowledge Mr. Muhammad Sarfraz, Microbiologist at Al-Razi Diagnostic for providing the clinical isolates for checking host specificity in this study. We also acknowledge the higher education commission of Pakistan for providing (research project number NRPU- 4501) to complete this study.
Author Contributions
Dr S.R. supervised all experiments and manuscript write up. R.T., M.S., K.A. and I.A.A. conducted the experiments as described, analyzed and interpreted the results. Y.R. and C.S.S. help in genome analysis and sequencing. Z.A. help out in some experiments. R.T. and M.S. wrote the manuscript. All authors have reviewed the final version of the manuscript.
Data Availability
The data is submitted with the Accession number no. MH688453, however it will be public on publication of the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36229-y.
References
- 1.Chhibber S, Nag D, Bansal S. Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and a bacteriophage. BMC Microbiol. 2013;13:174. doi: 10.1186/1471-2180-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang T-W, et al. Capsule deletion via a λ-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578. BMC Res Notes. 2014;7:13. doi: 10.1186/1756-0500-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mshana SE, et al. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis. 2013;13:466. doi: 10.1186/1471-2334-13-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saleem AF, Qamar FN, Shahzad H, Qadir M, Zaidi AK. Trends in antibiotic susceptibility and incidence of late-onset Klebsiella pneumoniae neonatal sepsis over a six-year period in a neonatal intensive care unit in Karachi, Pakistan. Int J Infect Dis. 2013;17:e961–e965. doi: 10.1016/j.ijid.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Band VI, et al. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. MBio. 2018;9:e02448–02417. doi: 10.1128/mBio.02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C-R, et al. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, Y. et al. Comparative analysis of quinolone resistance in clinical isolates of Klebsiella pneumoniae and Escherichia coli from Chinese children and adults. BioMed Res Int. 2015 (2015). [DOI] [PMC free article] [PubMed]
- 8.Chen F-J, Lauderdale T-L, Ho M, Lo H-J. The roles of mutations in gyrA, parC, and ompK35 in fluoroquinolone resistance in Klebsiella pneumoniae. Microb Drug Resist. 2003;9:265–271. doi: 10.1089/107662903322286472. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen A, et al. The T7-related Pseudomonas putida phage ϕ15 displays virion-associated biofilm degradation properties. PLoS One. 2011;6:e18597. doi: 10.1371/journal.pone.0018597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt KE, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weitao T. Multicellularity of a unicellular organism in response to DNA replication stress. Res Microbiol. 2009;160:87. doi: 10.1016/j.resmic.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Kudva IT, Jelacic S, Tarr PI, Youderian P, Hovde CJ. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl Environ Microbiol. 1999;65:3767–3773. doi: 10.1128/aem.65.9.3767-3773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark JR, March JB. Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol. 2006;24:212–218. doi: 10.1016/j.tibtech.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Doi Y, et al. Gram-Negative Bacterial Infections: Research Priorities, Accomplishments, and Future Directions of the Antibacterial Resistance Leadership Group. Clin Infect Dis. 2017;64:S30–S35. doi: 10.1093/cid/ciw829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piperaki E-T, Syrogiannopoulos GA, Tzouvelekis LS, Daikos GL. Klebsiella pneumoniae: Virulence, Biofilm and Antimicrobial Resistance. Pediatr Infect Dis J. 2017;36:1002–1005. doi: 10.1097/INF.0000000000001675. [DOI] [PubMed] [Google Scholar]
- 16.Hoyles L, et al. Klebsiella pneumoniae subsp. pneumoniae–bacteriophage combination from the caecal effluent of a healthy woman. PeerJ. 2015;3:e1061. doi: 10.7717/peerj.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobocka MB, et al. Genome of bacteriophage P1. J. Bacteriol. 2004;186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh P-F, Lin H-H, Lin T-L, Chen Y-Y, Wang J-T. Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Sci Rep. 2017;7:4624. doi: 10.1038/s41598-017-04644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karumidze N, et al. Isolation and characterisation of lytic bacteriophages of Klebsiella pneumoniae and Klebsiella oxytoca. Curr Microbiol. 2013;66:251–258. doi: 10.1007/s00284-012-0264-7. [DOI] [PubMed] [Google Scholar]
- 20.D’andrea MM, et al. ϕBO1E, a newly discovered lytic bacteriophage targeting carbapenemase-producing Klebsiella pneumoniae of the pandemic Clonal Group 258 clade II lineage. Sci Rep. 2017;7:2614. doi: 10.1038/s41598-017-02788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes K, Sutherland I, Clark J, Jones M. Bacteriophage and associated polysaccharide depolymerases–novel tools for study of bacterial biofilms. J Appl Microbiol. 1998;85:583–590. doi: 10.1046/j.1365-2672.1998.853541.x. [DOI] [PubMed] [Google Scholar]
- 22.Verma V, Harjai K, Chhibber S. Structural changes induced by a lytic bacteriophage make ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling. 2010;26:729–737. doi: 10.1080/08927014.2010.511196. [DOI] [PubMed] [Google Scholar]
- 23.Jurczak-Kurek A, et al. Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016;6:34338. doi: 10.1038/srep34338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jończyk E, Kłak M, Międzybrodzki R, Górski A. The influence of external factors on bacteriophages. Folia microbiologica. 2011;56:191–200. doi: 10.1007/s12223-011-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drulis-Kawa Z, et al. Isolation and characterisation of KP34–a novel phiKMV-like bacteriophage for Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2011;90:1333–1345. doi: 10.1007/s00253-011-3149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik DJ, et al. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv Colloid Interface Sci. 2017;249:100–133. doi: 10.1016/j.cis.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Beasley DE, Koltz AM, Lambert JE, Fierer N, Dunn RR. The evolution of stomach acidity and its relevance to the human microbiome. PLoS One. 2015;10:e0134116. doi: 10.1371/journal.pone.0134116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma V, Harjai K, Chhibber S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr Microbiol. 2009;59:274–281. doi: 10.1007/s00284-009-9430-y. [DOI] [PubMed] [Google Scholar]
- 29.Cao, F. et al. Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. BioMed Res Int. 2015 (2015). [DOI] [PMC free article] [PubMed]
- 30.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 31.Viertel TM, Ritter K, Horz H-P. Viruses versus bacteria—novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemother. 2014;69:2326–2336. doi: 10.1093/jac/dku173. [DOI] [PubMed] [Google Scholar]
- 32.Cui Z, et al. Complete genome sequence of Klebsiella pneumoniae phage JD001. J Virol. 2012;86:13843–13843. doi: 10.1128/JVI.02435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahbaaz M, ImtaiyazHassan M, Ahmad F. Functional annotation of conserved hypothetical proteins from Haemophilus influenzae Rd KW20. PLoS One. 2013;8:e84263. doi: 10.1371/journal.pone.0084263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casjens, S. R. & Gilcrease, E. B. In Bacteriophages 91–111 (Springer, 2009). [DOI] [PMC free article] [PubMed]
- 35.Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen DT, Lessor LE, Cahill JL, Rasche ES, Everett GFK. Complete genome sequence of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae siphophage Sushi. Genome Announc. 2015;3:e00994–00915. doi: 10.1128/genomeA.00994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qazi JI, Asif H, Shahid R. Economical Method for Estimation of Bacterial Viable Count. Pak. J. Zool. 2008;40:6. [Google Scholar]
- 38.Shafique, M., Alvi, I. A., Abbas, Z. & Rehman, S. Assessment of biofilm removal capacity of a broad host range bacteriophage JHP against Pseudomonas aeruginosa. Apmis (2017). [DOI] [PubMed]
- 39.Bibi Z, Abbas Z. & Rehman, S. u. The phage P. E1 isolated from hospital sewage reduces the growth of Escherichia coli. Biocontrol Sci Technol. 2016;26:181–188. doi: 10.1080/09583157.2015.1086311. [DOI] [Google Scholar]
- 40.Capra M, Quiberoni A, Reinheimer J. Thermal and chemical resistance of Lactobacillus casei and Lactobacillus paracasei bacteriophages. Lett Appl Microbiol. 2004;38:499–504. doi: 10.1111/j.1472-765X.2004.01525.x. [DOI] [PubMed] [Google Scholar]
- 41.Khawaja KA, Rauf M, Abbas Z, Rehman S. A virulent phage JHP against Pseudomonas aeruginosa showed infectivity against multiple genera. J. Basic Microbiol. 2016;56:1090–1097. doi: 10.1002/jobm.201500764. [DOI] [PubMed] [Google Scholar]
- 42.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altermann E, Klaenhammer TR. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. Omics A Journal of Integrative Biology. 2003;7:161–169. doi: 10.1089/153623103322246557. [DOI] [PubMed] [Google Scholar]
- 44.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garneau JR, Depardieu F, Fortier L-C, Bikard D, Monot M. PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep. 2017;7:8292. doi: 10.1038/s41598-017-07910-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evo. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang Y, et al. Isolation and genome characterization of the virulent Staphylococcus aureus bacteriophage SA97. Viruses. 2015;7:5225–5242. doi: 10.3390/v7102870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is submitted with the Accession number no. MH688453, however it will be public on publication of the paper.