Abstract
Background
Circulating microRNA (miRNA) biomarkers have been extensively reported in cardiovascular diseases (CVDs). However, serum exosomal miRNA (exo-miRNA) as biomarker in patients with heart failure (HF) with reduced ejection fraction (HFrEF) remain largely unexplored. We sought to investigate the potential of three types of serum exo-miRNAs as biomarkers for diagnosis in HFrEF patients who were admitted in hospital because of acute heart failure (AHF).
Methods
A total of 28 HFrEF patients hospitalized for AHF, including de novo AHF and acute decompensated HF, and 30 volunteers as control group (CG) from 2015 to 2017 were enrolled in this study. Serum exo-miRNAs were extracted and analyzed by NaNOZS-90, electron microscopy, and western blotting. Three types of serum exo-miRNAs (exo-miR-92b-5p, -192-5p, and -320a) were assessed by quantitative real time polymerase chain reaction (qRT-PCR).
Results
The particle size was confirmed as 40–150 nm using NaNOZS-90 and transmission electron microscopy. Exosomal biomarkers CD63 and Hsp70 were readily detected. The expression level of serum exo-miRNAs were transformed into log2−delta CT in the qPCR assay. The data showed that exo-miR-92b-5p was elevated in HFrEF patients compared with controls. Moreover, exo-miR-92b-5p was inversely correlated with the left ventricular fraction shortening (LVFS) and left ventricular ejection fraction (LVEF), whereas it was positively correlated with left atrial diameter (LAD), left ventricular diastolic diameters (LVDD) and systolic diameters (LVSD). A receiver operating characteristic (ROC) curve was generated for discrimination between HFrEF patients and controls based on exo-miR-92b-5p (P<0.001, sensitivity =71.4%, specificity =83.3%).
Conclusions
Exo-miR-92b-5p levels in the serum may serve as a marker for HFrEF diagnosis.
Keywords: Acute heart failure (AHF), circulating, diagnostic biomarker, exosome, miR-92b-5p, heart failure with reduced ejection fraction (HFrEF)
Introduction
Heart failure (HF) is a multifaceted and complicated condition representing a common endpoint of many cardiovascular diseases (CVDs) (1). Given its high morbidity and mortality rate, HF is a major health care burden (2,3). Left ventricular ejection fraction (LVEF) ≤40% is defined as HF with reduced ejection fraction (HFrEF), showing higher mortality compared with other types of HF (4,5). Furthermore, acute heart failure (AHF) is diagnosed based on acute onset or HF symptom recurrence, which requires immediate evaluation and urgent treatment (6,7). Therefore, patients with HFrEF hospitalized for AHF need immediate attention.
Clinical biomarkers are of great importance in diagnosing and treating HF. Currently, B-type natriuretic peptide (BNP) and N-terminal pro B-type natriuretic peptide (NT-proBNP) are the most classical biomarkers for HF diagnosis with high sensitivity (4). Nevertheless, due to the low specificity, other diseases such as pulmonary embolism, right ventricular strain, and myocardial infarction must be excluded, and patients’ age, sex, body mass index (BMI), and renal function can interfere with HF diagnosis (4). Other types of biomarkers have also been explored for HF diagnosis, such as ST2, Galectin-3, high-sensitivity troponin, growth differentiation factor 15, procalcitonin, and NGAL; however, they were not specific for HF and were often affected by other multiple comorbidities (8).
Meanwhile, many circulating microRNAs (miRNAs) have been investigated as biomarkers for HF. Upregulation of miR-23a, miR-23b, miR-24, miR-195, and miR-214 were observed in HF (9), and upregulation of miR-423-5p, miR-320, miR-22, and miR-92b were found in chronic heart failure (CHF) (10). In addition, the levels of miR-34a, miR-192, and miR-194 were elevated in HF due to acute myocardial infarction (11). Moreover, the plasma miR-210 expression was also elevated in HF patients with NYHA III and IV relative to those with NYHA II and controls (12). Several studies have been done on patients with CHF undergoing acute exhaustive exercise. For example, serum miR-21, miR-378, and miR-940 expression have been shown to rise promptly following exercise (13). However, all these studies took the serum or plasma as a single sample, without considering the various components inside, for example, the exosome fraction and non-exosome supernatant fraction (11).
Exosomes (40–150 nm) were found during mammalian reticulocyte maturation, which were believed to detach from the plasma membrane (14). They might interact with recipient cells through surface proteins such as CD63, and release their inner contents, including DNA, RNA, and proteins, to regulate the function of target cells (15). Exosomes have been shown to facilitate miRNAs transfer between cells (16). Turchinovich et al. verified the existence of non-exosome supernatant fraction using 0.22-µm filters and ultracentrifugation at 110,000 g (17). Beg et al., developed different protocols to measure both free and exosome-encapsulated circulating miRNAs (18). Their data showed that free circulating miRNA levels might change over time depending on rates of tissue repair, disease status and age (18). On the contrary, exosomal miRNAs (exo-miRNAs), including the exosomes from circulation hematopoietic cells, reflect general health status, which not rely on the damaged tissue (18). This study further showed that miR-146a levels were higher in exosomes than in plasma patients with HF compared to controls (18).
In summary, several studies demonstrated the values of circulating miRNAs as biomarkers in patients with HF. However, the studies about circulating biomarkers of HFrEF rarely focused on exo-miRNAs. Therefore, we firstly tested many exo-miRs based on the previous reports: miR-1-1 (19), miR-1-2 (19), miR-210 (12), miR-499a (20), miR-499b (20), miR-208b (20-22), miR-133a (21,23), miR-133b (21), miR-423 (24). miR-92b-5p (25), miR-320a (26), miR-192-5p (27). Unfortunately, we could not get reliable results for most of them due to the small sample size and the undetectable level by Q-PCR (raw data showed “undetermined”). However, three serum exo-miRNAs (exo-miR-92b-5p, exo-miR-320a, and exo-miR-192-5p) were found to be consistently detectable. Therefore, we analyzed their potential utility as diagnostic biomarkers for HFrEF in this study.
Methods
Patient’s involvement and sample collection
A total of 28 patients hospitalized for AHF in the Affiliated Hospital of Medical School of Ningbo University from March 2015 through March 2017 were recruited. New AHF and acute decompensated CHF patients were eligible for the present study. The American Heart Association (AHA) (1) and the European Society of Cardiology (ESC) (6) guidelines were used for AHF diagnosis. All patients underwent clinical examination, laboratory examination, and Doppler echocardiography assessment for the diagnosis of clinical HF. LVEF was assessed using two-dimensional echo with the Simpson’s method. According to the HF guidelines of ESC (2016) (6), patients with LVEF ≤40% were classified as HFrEF. Exclusion criteria: congenital or primary valvular heart diseases, rheumatic diseases, severe pulmonary disease, and cancer.
A total of 30 healthy controls were also recruited that showed the absence of coronary disease, valvular disease, or no echocardiography evidence of cardiomyopathy. Healthy controls were also age and gender matched with the patients included.
The Institutional Review Board of the Affiliated Hospital of Medical School of Ningbo University approved this study (No.KY20180601), with all participants providing informed written consent. This study was compliant with the Declaration of Helsinki.
Peripheral blood was collected from HFrEF patients immediately upon enrollment in the hospital and from control group (CG) before breakfast.
Pro-coagulation tubes were used to collect whole blood samples. The data of BNP from patients were immediately tested after sample collection. About 800–1,000 µL serum was separated after 2 h standing followed by spinning at 1,660 g for 10 minutes at room temperature (Sorvall ST16R, Thermo Scientific). The serum was stored at −80 °C.
Exosome extraction and nanoparticle tracking analysis
The serum was first centrifuged at 2,000 g at room temperature for 20 min to remove debris. Then, serum was incubated with the one-third volume of exosome isolation reagent (R11064.5, RiboBio, Guangzhou, China) at 4 °C for 30 minutes. Exosomes were then collected after a 2-min 15,000 g spin at 4 °C for (Sorvall ST16R, Thermo Scientific). A Zetasizer NaNOZS-90 (Malvern Instrucments Ltd., Malvern, UK) was used to identify exosome particle sizes.
Transmission electron microscopy
In total, 30 µL PBS was used for resuspension of exosomes. And 10 µL sample was dropwise added to a copper grid and subsided for 1 min, followed by sucking out of the floating fluid by a filter paper. Then, 10 µL uranyl acetate was then added and subsided for another 1 min, and removed as above. Finally, samples were dried at room temperature for 10 min and imaged with a Hitachi H-7650 at 80 KV.
Western blotting
Radioimmunoprecipitation assay (RIPA) buffer was used to lyse exosomes, and the protein concentration was assessed using a NanoDrop 2000. About 20 µg proteins were electrophoresed on an SDS-PAGE gel and transferred onto PVDF membranes, followed by a 2-h blocking step at room temperature using 5% bovine serum albumin. Primary antibodies were then added at 4 °C overnight [1:1,000, Anti-CD63 antibody (TS63), ab59479, Abcam, UK; 1:450 anti-Hsp70 antibody4876, #74220, CST, USA]. After washing, secondary antibodies were then added [1:2,000, goat anti-mouse Immunoglobulin G (IgG), horseradish peroxidase (HRP)-conjugated, CWBIO, China; 1:4,000 anti-rabbit IgG, HRP-linked antibody 7074, #74220, CST] for another 1 h at room temperature. A LI-COR model 3600 (LI-COR, USA) was used to measure protein levels.
miRNA extraction and quantitative real time polymerase chain reaction (qRT-PCR)
The miRNA was isolated from exosomes using blood (serum) miRNA quick extraction kit (Bioteke, Beijing, China) according to the manufacturer’s instructions. Exo-miRNA was eluted in 30 µL Nuclease-free H2O. A NanoDrop 2000 (Thermo Fisher Scientific Inc) was used to assess RNA concentration and purity.
The expression level was evaluated using the standard polyA qRT-PCR assay. The procedure for adding polyA was as follows: 37 °C for 1 h followed by 95 °C for 5 min → chilling on ice for 5 min, following with the RT process (42 °C for 5 min → 85 °C for 5 min). The Q-PCR procedure was as follows: 95 °C, 10 min, then 95 °C, 20 s, repeat 40 times, then 61 °C, 1 min, then perform a melt curve (ABI Q5, USA). Exo-miRNAs expression were normalized to exo-miR-451b (28). Primer sequences were shown in Table 1.
Table 1. Primer sequences.
| Primers | Sequence |
|---|---|
| RT-adaptor primer | 5'-gcgag cacag aatta atacg actca ctata ggttt ttttt ttttv n-3' |
| Universal reverse primer | 5'-gcgag cacag aatta atacg ac-3' |
| miR-92b-5p primer | 5'-aggga cggga cgcgg tgcag tg-3' |
| miR-192-5p primer | 5'-ctgac ctatg aattg acagc c-3' |
| miR-320a primer | 5'-aaagc tgggt tgaga gggcg a-3' |
| Internal reference miR-451b | 5'-tagca agaga accat tacca tt-3' |
Statistical analysis
Data was analyzed by SPSS 18.0 (SPSS Co., Chicago, USA). Enumeration data were indicated as percentage, whereas measurement data were expressed as mean ± standard error of mean (SEM) or median with quartile range. All data were analyzed using the normal distribution test, and different methods were chosen for further analysis. The expression data of exo-miRNAs were calculated by log2−delta CT. Serum exo-miRNAs expression were compared between HFrEF and CG by Mann-Whitney U test. The characteristic features of HFrEF and CG were analyzed by Pearson’s chi-squared test, Fisher’s exact test, Student’s t-test, and Mann-Whitney U test. Associations between exo-miR-92b-5p and echocardiographic indices were analyzed by Spearman correlation test. A receiver operating characteristic (ROC) curve was allowed for sensitivity and specificity analysis of exo-miR-92b-5p expression level for distinguishing patients with HFrEF from control. P value <0.05 was the threshold for statistical significance.
Results
Clinical characteristics of HFrEF and CG
The characteristics of HFrEF patients were: average age of 59.7 (range, 28–88) years; 18 males and 10 females. The characteristics of the CG were: average age of 56.5 (range, 21–79) years; 22 males and 8 females.
The clinical characteristics of HFrEF and CG were analyzed (Table 2). The gender, age, BMI, smoking, alcohol consumption, renal failure, diabetes mellitus, and hypertension rates were similar between two groups. Significant changes, reflecting the difference between HFrEF versus CG, included echocardiographic indices (Table 2), such as left ventricular diastolic diameter (LVDD, P<0.001), left ventricular systolic diameter (LVSD, P<0.001), left atrial diameter (LAD, P<0.001), left ventricular eject fraction (LVEF, P<0.001), and left ventricular fraction shortening (LVFS, P<0.001).
Table 2. Characteristics of HFrEF and CG.
| Variable | CG (n=30) | HFrEF (n=28) | P value |
|---|---|---|---|
| Age (year) | 56.5±2.1 | 59.7±3.3 | 0.416a |
| Gender (male) | 22 (73.3%) | 18 (64.3%) | 0.457b |
| Smoking | 11 (36.7%) | 11 (39.3%) | 0.837b |
| Alcohol consumption | 8 (26.7%) | 11 (39.3%) | 0.306b |
| Diabetes mellitus | 4 (13.3%) | 10 (35.7%) | 0.067c |
| Hypertension | 18 (60.0%) | 10 (35.7%) | 0.064b |
| Renal failure | 1 (3.3%) | 3 (10.7%) | 0.344c |
| BMI (kg/m2) | 23.6±0.6 | 24.2±0.7 | 0.373a |
| BNP (ng/L) | NA | 1,018.5 (702.0–1,236.7) | NA |
| LAD (mm) | 33.5 (31.0–37.0) | 46.0 (42.0–49.8) | <0.001d |
| LVDD (mm) | 48.3±0.7 | 65.9±1.4 | <0.001a |
| LVSD (mm) | 30.0 (28.0–32.0) | 52.5 (50.0–58.0) | <0.001d |
| LVEF (%) | 68.2±0.8 | 33.4±0.8 | <0.001a |
| LVFS (%) | 38.5 (35.0–41.0) | 17.0 (15.0–19.0) | <0.001d |
Enumeration data were displayed as percentage, whereas measurement data were expressed as mean ± SEM or median with quartile range. All measurement data were analyzed using the normal distribution test, and different methods were chosen for further analysis. Data with normal distribution were expressed as mean ± SEM, whereas data with abnormal distribution were displayed as median with quartile range. a, Student t test; b, Pearson’s chi-square test; c, Fisher’s exact test; d, Mann-Whiney U test. CG, control group; HFrEF, heart failure with reduced ejection fraction; BMI, body mass index; BNP, B-type natriuretic peptide; LAD, left atrial diameter; LVDD, left ventricular diastolic diameter; LVEF, left ventricular ejection fraction; LVFS, left ventricular fraction shortening; LVSD, left ventricular systolic diameter; SEM, standard error of mean.
Identification of serum exosomes
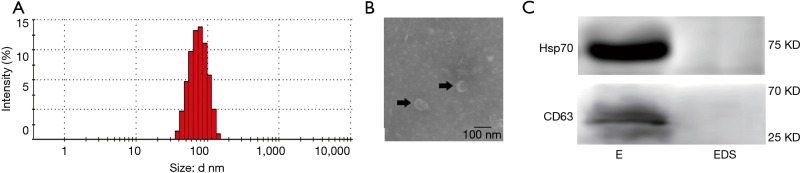
Serum exosome samples were collected from three HFrEF patients and were pooled together, and the exosomes were validated using NaNOZS-90 and transmission electron microscopy. The data showed that the size of the extracted double-layer membrane particles ranged from 40 to 150 nm (average 80 nm; Figure 1A,B).
Figure 1.
Characterization of serum exosomes. (A) The particle sizes of extracted exosomes were evaluated using NaNOZS-90; (B) serum exosomes were analyzed using microscopy and highlighted using black arrows. Scale bar =100 nm; (C) the biomarkers of exosomes (Hsp70 and CD63) were confirmed using Western blotting assay. E, exosomes; EDS, exosome-depleted supernatant.
Furthermore, the protein markers of exosomes (CD63 and Hsp70) were confirmed by Western blot (Figure 1C).
Exo-miR-92b-5p levels were elevated in HFrEF patients
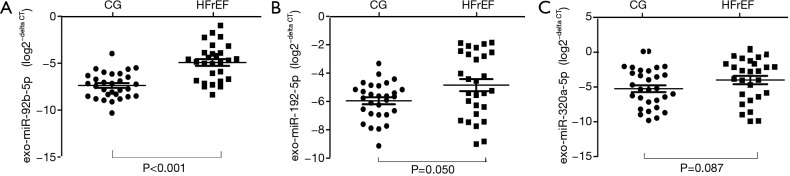
Exo-miR-92b-5p, exo-miR-192-5p, and exo-miR-320a levels were tested by qRT-PCR (Figure 2). The transformed data of exo-miRNAs expression level were showed as log2−delta CT. Although the levels of exo-miR-192-5p and exo-miR-320a were similar between HFrEF and CG (Figure 2B, exo-miR-192, P=0.050; Figure 2C, exo-miR-320a, P=0.087), exo-miR-92b-5p expression was enriched in HFrEF patients compared with CG (Figure 2A, P<0.001).
Figure 2.
Comparison the log2−delta CT of exo-miRNAs between HFrEF and CG. (A) exo-miR-92b-5p (Mann-Whitney U test; HFrEF vs. CG, P<0.001); (B) exo-miR-192-5p (Mann-Whitney U test; HFrEF vs. CG, P=0.050); (C) exo-miR-320a (Mann-Whitney U test; HFrEF vs. CG, P=0.087). HFrEF, heart failure with reduced ejection fraction; CG, control group.
Expression of exo-miR-92b-5p correlated with echocardiographic indices
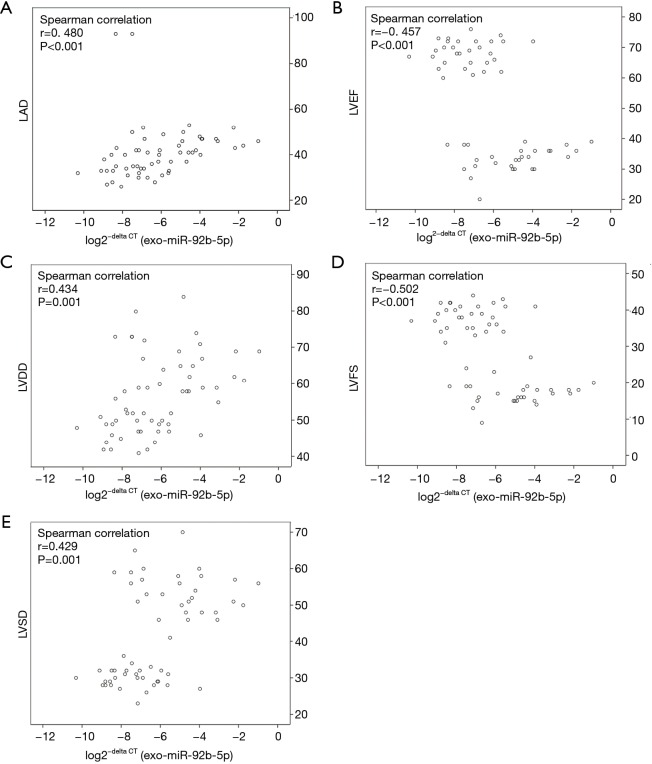
The association of exo-miR-92b-5p and echocardiographic indexes was also identified using Spearman correlation analysis (Figure 3). The log2−delta CT of exo-miR-92b-5p was positively correlated with LAD (Figure 3A, r=0.480, P<0.001), LVDD (Figure 3C, r=0.434, P=0.001), and LVSD (Figure 3E, r=0.429, P=0.001), while it was inversely correlated with LVEF (Figure 3B, r=−0.457, P<0.001) and LVFS (Figure 3D, r=−0.502, P<0.001).
Figure 3.
Relationship analysis between log2−delta CT of exo-miR-92b-5p and echocardiographic indices. (A) log2−delta CT of exo-miR-92b-5p vs. LAD (Spearman correlation: r=0.480, P<0.001); (B) log2−delta CT of exo-miR-92b-5p vs. LVEF (Spearman correlation: r=–0.457, P<0.001); (C) log2−delta CT of exo-miR-192-5p vs. LVDD (Spearman correlation: r=0.434, P=0.001); (D) log2−delta CT of exo-miR-192-5p vs. LVFS (Spearman correlation: r=–0.502, P<0.001); (E) log2−delta CT of exo-miR-192-5p vs. LVSD (Spearman correlation: r=0.429, P=0.001). LAD, left atrial diameter; LVDD, left ventricular diastolic diameter; LVSD, left ventricular systolic diameter; LVEF, left ventricular ejection fraction; LVFS, left ventricular fraction shortening.
Exo-miR-92b-5p may be an HFrEF biomarker
The sensitivity and specificity of exo-miR-92b-5p as a biomarker were analyzed using ROC curve to confirm its ability to discriminate HFrEF from CG (Figure 4). The ROC curve demonstrated by exo-miR-92b-5p obtained an area under the curve of 0.844, with a cutoff value of −6.09 to discriminate HFrEF from CG (Figure 4). A sensitivity of 71.4% and a specificity of 83.3% were achieved for identifying HFrEF.
Figure 4.
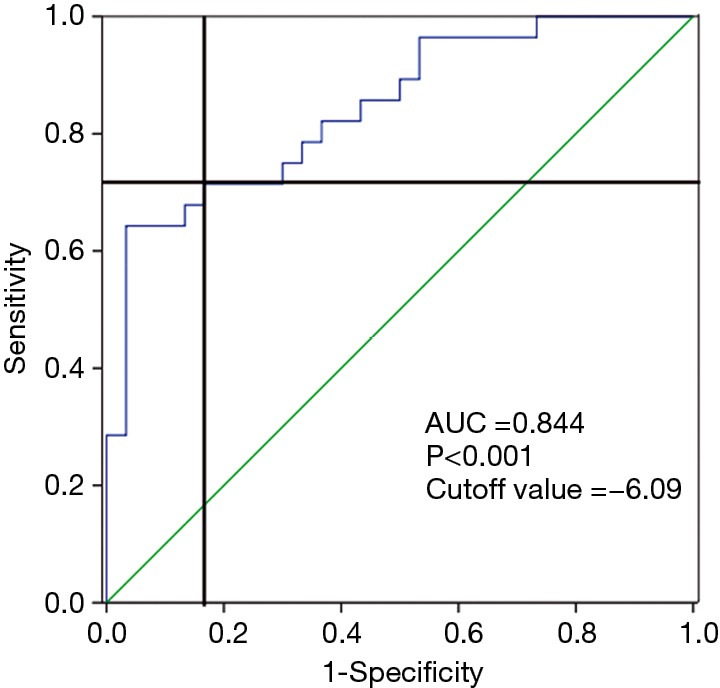
ROC curve for discriminating HFrEF from CG using log2−delta CT of exo-miR-92b-5p (AUC of 0.844, cutoff value =−6.09, sensitivity =71.4%, specificity =83.3%, P<0.001). ROC, receiver operating characteristic; HFrEF, heart failure with reduced ejection fraction; CG, control group; AUC, area under the curve.
Discussion
Circulating miRNAs have been widely studied in CVDs. Some studies also explored the exo-miRNAs in HF. However, studies on exo-miRNAs as biomarkers for HFrEF are limited. Three exo-miRNAs, based on the previous reports on circulating miRNAs in CVDs, were compared between HFrEF and CG. The exo-miR-92b-5p level was elevated in HFrEF patients compared with CG.
Both sensitivity and specificity were important indexes for biomarkers. Although miRNAs could exist as free miRNAs and exo-miRNAs, their sensitivity and specificity might be different. According to the study of Gall et al., most miRNAs were found in exosomes in circulation, allowing for more sensitive tests than those in serum (29). Besides, Jansen et al. found that circulating microvesicular miR-126 and miR-199a were related to CVDs, whereas circulating free miRNAs were not (30).
Cel-miR-39 was used as a reference initially. However, the expression of cel-miR-39 was easily affected by many factors, leading to unstable results. Then, attempts were made to find a reliable serum exo-miRNAs reference that was internal. According to the review of Occhipinti et al., various endogenous controls could be used (31), such as U6 (32,33), miR-16 (34,35), miR-484 (35), miR-642-3p (36), and miR-451 (28). Three exo-miRNAs (exo-miR-484, exo-miR-451b, and exo-miR-642-3p) were selected as candidate internal references based on available reports and tested in six samples (two healthy and four with HFrEF). The data showed that exo-miR-451b-5p was relatively stable with the range of 20–25 cycles in exosomes among the aforementioned samples, compared with the other two candidate exo-miRNAs that were lowly expressed (27–32 cycles) in exosomes (Figure S1). Therefore, exo-miR-451b was selected as an internal reference to adjust the final results.
Many circulating miRNAs have been reported as biomarkers for HF, such as miR-210 (12), miR-19 (37,38), miR-145 (39), miR-148b-3p, and miR-409-3p (40), without considering their exact origin. A systematic study was performed by the team of Wong Lee. They enrolled 28 controls, 39 HFrEF patients, and 19 patients for miRNA array, followed by a validating training cohort containing 30 controls, 30 HFrEFs patients, and 30 heart failure with preserved ejection fraction (HFpEF) patients for further qRT-PCR analysis. They concluded that some miRNAs could distinguish HF from CG, while others could distinguish HFrEF or HFpEF from CG (41). Although their study suggested many miRNAs as biomarkers for HFrEF, miRNAs studied in the present study were not involved. The present study focused on patients with HFrEF, who were serious and in the acute stage of HFrEF. In addition, the reports from Lee et al. did not investigate exo-miRNA biomarkers, but instead focused on the total plasma or serum miRNA.
Unlike miR-92a, which was located in a miR-17-92 cluster and was more frequently reported in CVDs (42,43), miR-92b-5p was rarely reported in CVDs. Hoffmann et al. demonstrated that miR-92b-5p could target the genetic variant 3’UTR of SHOX2 gene and reduce its expression, leading to atrial fibrillation (AF) (44). AF and HFrEF frequently coexist (45). This study first tested the expression levels of exo-miR-92b-5p in the serum and found an increase in exo-miR-92b-5p level in HFrEF. The study suggested that exo-miR-92b-5p in the serum can be used as biomarker to distinguish HFrEF from CG.
Exo-miR-92b-5p could distinguish HFrEF from CG with high sensitivity and specificity. In addition, exo-miR-92b-5p was positively correlated with echocardiographic indices, such as LAD, LVDD, and LVSD, indicating that its expression was positively associated with dilation of the left ventricle. However, it was negatively correlated with other echocardiographic indices, such as LVEF and LVFS, implying that exo-miR-92b-5p level also increased along with the declining function of the left ventricle.
The BNP or NT-proBNP was usually used as biomarker for diagnosing with AHF. However, the specificity of these markers was always affected by other factors, such as renal failure, age, and other comorbidities (4). In this study, we showed that exo-miR-92b-5p was a candidate biomarker to distinguish HFrEF from CG. However, we could not compare the sensitivity and specificity of BNP and exo-miRNAs. Indeed, we have tested the BNP both in HFrEF patient group and healthy CG. Regrettably, the indexes of BNP in healthy control did not show their exact data in our laboratory examination. If possible, we will further explore the sensitivity and specificity of NT-proBNP/BNP and exo-miRNAs in future.
There are some limitations in the present study. First, only three exo-miRNAs were tested as candidate biomarkers for HFrEF based on previous reports, ignoring other candidate biomarkers for HFrEF. If feasible, representative samples should be selected and tested using microarray followed by training cohort to confirm their significance. Second, if feasible, more patients should be involved and they should be classified by LVEF into subgroups (HFrEF and HFpEF).
Conclusions
Collectively, serum exo-miR-92b-5p level was elevated in HFrEF patients. Therefore, it could be a candidate HFrEF diagnostic biomarker.
Figure S1.
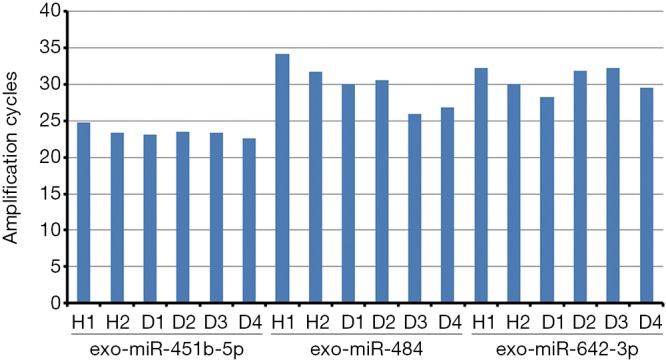
Choice of internal reference. Six samples from healthy controls and patients with HFrEF were separately tested. H1 and H2 were from healthy controls. D1–D4 were from patients with HFrEF. HFrEF, heart failure with reduced ejection fraction.
Acknowledgments
Funding: This work was supported by the grants from the Ningbo University Science Foundation (grant numbers XYY15016, XYY16008).
Ethical Statement: The Institutional Review Board of the Affiliated Hospital of Medical School of Ningbo University approved this study (No.KY20180601), with all participants providing informed written consent. This study was compliant with the Declaration of Helsinki.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Yancy CW, Jessup M, Bozkurt B, et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guidelines. Circulation 2013;128:e240-327. [DOI] [PubMed] [Google Scholar]
- 2.Nichols M, Townsend N, Scarborough P, et al. Cardiovascular disease in Europe 2014: epidemiological update. Eur Heart J 2014;35:2929. 10.1093/eurheartj/ehu299 [DOI] [PubMed] [Google Scholar]
- 3.Rohde LE, Bertoldi EG, Goldraich L, et al. Cost-effectiveness of heart failure therapies. Nat Rev Cardiol 2013;10:338-54. 10.1038/nrcardio.2013.60 [DOI] [PubMed] [Google Scholar]
- 4.Metra M, Teerlink JR. Heart failure. Lancet 2017;390:1981-95. 10.1016/S0140-6736(17)31071-1 [DOI] [PubMed] [Google Scholar]
- 5.Basuray A, French B, Ky B, et al. Heart failure with recovered ejection fraction: clinical description, biomarkers, and outcomes. Circulation 2014;129:2380-7. 10.1161/CIRCULATIONAHA.113.006855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail 2016;18:891-975. 10.1002/ejhf.592 [DOI] [PubMed] [Google Scholar]
- 7.Ural D, Cavusoglu Y, Eren M, et al. Diagnosis and management of acute heart failure. Anatol J Cardiol 2015;15:860-89. 10.5152/AnatolJCardiol.2015.6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer RA, Daniels LB, Maisel AS, et al. State of the Art: Newer biomarkers in heart failure. Eur J Heart Fail 2015;17:559-69. 10.1002/ejhf.273 [DOI] [PubMed] [Google Scholar]
- 9.Pfeifer P, Werner N, Jansen F. Role and Function of MicroRNAs in Extracellular Vesicles in Cardiovascular Biology. Biomed Res Int 2015;2015:161393. 10.1155/2015/161393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goren Y, Kushnir M, Zafrir B, et al. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail 2012;14:147-54. 10.1093/eurjhf/hfr155 [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto S, Sakata Y, Suna S, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res 2013;113:322-6. 10.1161/CIRCRESAHA.113.301209 [DOI] [PubMed] [Google Scholar]
- 12.Endo K, Naito Y, Ji X, et al. MicroRNA 210 as a biomarker for congestive heart failure. Biol Pharm Bull 2013;36:48-54. 10.1248/bpb.b12-00578 [DOI] [PubMed] [Google Scholar]
- 13.Xu T, Zhou Q, Che L, et al. Circulating miR-21, miR-378, and miR-940 increase in response to an acute exhaustive exercise in chronic heart failure patients. Oncotarget 2016;7:12414-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987;262:9412-20. [PubMed] [Google Scholar]
- 15.Mathivanan S, Lim JW, Tauro BJ, et al. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics 2010;9:197-208. 10.1074/mcp.M900152-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol 2012;23:91-7. 10.1097/MOL.0b013e328350a425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223-33. 10.1093/nar/gkr254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beg F, Wang R, Saeed Z, et al. Inflammation-associated microRNA changes in circulating exosomes of heart failure patients. BMC Res Notes 2017;10:751. 10.1186/s13104-017-3090-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai J, Zhang R, Li Y, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun 2010;391:73-7. 10.1016/j.bbrc.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 20.Corsten MF, Dennert R, Jochems S, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet 2010;3:499-506. 10.1161/CIRCGENETICS.110.957415 [DOI] [PubMed] [Google Scholar]
- 21.Ferreira LR, Frade AF, Santos RH, et al. MicroRNAs miR-1, miR-133a, miR-133b, miR-208a and miR-208b are dysregulated in Chronic Chagas disease Cardiomyopathy. Int J Cardiol 2014;175:409-17. 10.1016/j.ijcard.2014.05.019 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Q, Schotterl S, Backes D, et al. Inhibition of miR-208b improves cardiac function in titin-based dilated cardiomyopathy. Int J Cardiol 2017;230:634-41. 10.1016/j.ijcard.2016.12.171 [DOI] [PubMed] [Google Scholar]
- 23.Kuwabara Y, Ono K, Horie T, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet 2011;4:446-54. 10.1161/CIRCGENETICS.110.958975 [DOI] [PubMed] [Google Scholar]
- 24.Tijsen AJ, Creemers EE, Moerland PD, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res 2010;106:1035-9. 10.1161/CIRCRESAHA.110.218297 [DOI] [PubMed] [Google Scholar]
- 25.Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol 2008;45:185-92. 10.1016/j.yjmcc.2008.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song CL, Liu B, Diao HY, et al. The protective effect of microRNA-320 on left ventricular remodeling after myocardial ischemia-reperfusion injury in the rat model. Int J Mol Sci 2014;15:17442-56. 10.3390/ijms151017442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pichiorri F, Suh SS, Rocci A, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010;18:367-81. 10.1016/j.ccr.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 2014;9:e92921. 10.1371/journal.pone.0092921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012;7:e30679. 10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen F, Yang X, Proebsting S, et al. MicroRNA expression in circulating microvesicles predicts cardiovascular events in patients with coronary artery disease. J Am Heart Assoc 2014;3:e001249. 10.1161/JAHA.114.001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Occhipinti G, Giulietti M, Principato G, et al. The choice of endogenous controls in exosomal microRNA assessments from biofluids. Tumour Biol 2016;37:11657-65. 10.1007/s13277-016-5164-1 [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Zhou Y, Lu J, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol 2014;31:148. 10.1007/s12032-014-0148-8 [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Hou L, Li A, et al. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int 2014;2014:864894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka Y, Kamohara H, Kinoshita K, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013;119:1159-67. 10.1002/cncr.27895 [DOI] [PubMed] [Google Scholar]
- 35.Eichelser C, Stuckrath I, Muller V, et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014;5:9650-63. 10.18632/oncotarget.2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia HL, He CH, Wang ZY, et al. MicroRNA expression profile in exosome discriminates extremely severe infections from mild infections for hand, foot and mouth disease. BMC Infect Dis 2014;14:506. 10.1186/1471-2334-14-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao Y, Chen H, Li M. MiR-19a overexpression contributes to heart failure through targeting ADRB1. Int J Clin Exp Med 2015;8:642-9. [PMC free article] [PubMed] [Google Scholar]
- 38.Beaumont J, Lopez B, Ravassa S, et al. MicroRNA-19b is a potential biomarker of increased myocardial collagen cross-linking in patients with aortic stenosis and heart failure. Sci Rep 2017;7:40696. 10.1038/srep40696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M, Cheng YJ, Sara JD, et al. Circulating MicroRNA-145 is Associated with Acute Myocardial Infarction and Heart Failure. Chin Med J (Engl) 2017;130:51-6. 10.4103/0366-6999.196573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen MC, Chang TH, Chang JP, et al. Circulating miR-148b-3p and miR-409-3p as biomarkers for heart failure in patients with mitral regurgitation. Int J Cardiol 2016;222:148-54. 10.1016/j.ijcard.2016.07.179 [DOI] [PubMed] [Google Scholar]
- 41.Wong LL, Armugam A, Sepramaniam S, et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail 2015;17:393-404. 10.1002/ejhf.223 [DOI] [PubMed] [Google Scholar]
- 42.Niculescu LS, Simionescu N, Sanda GM, et al. MiR-486 and miR-92a Identified in Circulating HDL Discriminate between Stable and Vulnerable Coronary Artery Disease Patients. PLoS One 2015;10:e0140958. 10.1371/journal.pone.0140958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Y, Wang HY, Li Y, et al. Peripheral blood miRNAs as a biomarker for chronic cardiovascular diseases. Sci Rep 2014;4:5026. 10.1038/srep05026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann S, Clauss S, Berger IM, et al. Coding and non-coding variants in the SHOX2 gene in patients with early-onset atrial fibrillation. Basic Res Cardiol 2016;111:36. 10.1007/s00395-016-0557-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma A, Kalman JM, Callans DJ. Treatment of Patients With Atrial Fibrillation and Heart Failure With Reduced Ejection Fraction. Circulation 2017;135:1547-63. 10.1161/CIRCULATIONAHA.116.026054 [DOI] [PubMed] [Google Scholar]