Abstract
Global developmental delay and intellectual disability are phenotypically and genetically heterogeneous and a specific diagnosis is not reached in many cases. This paper outlines a systematic approach to global developmental delay and intellectual disability.
Keywords: Chromosomal micro-array, clinical exome, global developmental delay, intellectual diasbility, learning disability, WES, WGS
Key points
Intellectual disability (ID) has a prevalence of 2.17% in the adult population in England
A systematic clinical approach can help to identify a genetic cause for global developmental delay (GDD) and ID
Chromosomal microarray (CMA) is the first line diagnostic genetic test for individuals with GDD/ID
Second-line genetic tests include next-generation sequencing of GDD/ID gene panels or trio clinical exome or whole exome sequencing (CES/WES)
Use of genomic tests such as CMA, CES and WES can reveal incidental findings unrelated to the diagnosis of GDD/ID
Introduction
Global developmental delay (GDD) is defined as the failure to achieve developmental milestones within the expected age range. Objectively, this refers to significant delay in two or more developmental domains in children aged 5 years or younger. Developmental domains include gross or fine motor skills, speech and language, cognition, personal-social and activities of daily living. Intellectual disability (ID) involves problems with general mental abilities that affect both intellectual functioning (such as learning and reasoning) and adaptive functioning (activities of daily living such as communication and independent living). According to the American Association on Intellectual and Developmental Disabilities, ID is characterised by significant limitations in intellectual functioning (reasoning, learning, problem solving) and adaptive behaviour (conceptual, social and practical skills) that originates before the age of 18 years.1 The UK Department of Health’s strategy defines learning disability rather than intellectual disability as the presence of impaired intelligence with impaired social functioning, which started before adulthood and has a lasting effect on development.2
The patients with GDD/ID are likely to be seen in general and community paediatric clinics, and learning disabilities psychiatry, neurology/epilepsy and rehabilitation medicine clinics for adults. The estimated incidence of GDD is 1–3% of children aged 5 years or younger. The prevalence of ID is 2.7% of school-aged children and 2.17% of the adult population in England.3 A recent study of whole exome sequencing (WES) in children with undiagnosed developmental disorders identified rare heterozygous de novo mutations in developmental genes as an important cause. Developmental disorders caused by de novo mutations have an average prevalence of 1 in 213 to 1 in 448 births depending on parental age and globally account for about 400,000 affected children born every year.4 A specific diagnosis would be helpful for management of the condition, prediction of possible problems, prognosis and prenatal diagnosis or pre-implantation diagnosis for the parents and extended family depending on the condition.
Clinical approach
GDD and ID can occur in isolation or in combination with other congenital malformations, neurological features such as epilepsy, and behavioural problems such as autism spectrum disorder and attention-deficit hyperactivity disorder. GDD and ID are phenotypically and genetically heterogeneous and a specific diagnosis is not reached in many cases.
There are many reasons for making a specific diagnosis in a child who presents with GDD. These include potential treatment options, to provide parents/carers and the paediatric team with information about likely clinical problems and complications as well as long-term prognosis, enabling families to access special education and social care, and allowing accurate counselling of parents about recurrence risks and options for prenatal diagnosis and pre-implantation genetic diagnosis. A diagnosis could also help parents to access research trials and to get in touch with other similarly affected families.
A systematic diagnostic approach is needed in patients with GDD/ID to identify a specific underlying genetic cause.5 A three-generation family history is the most important preliminary step in the diagnostic pathway for a child who presents to the clinical geneticist with GDD and/or ID (Box 1). This family tree may have to be extended if there are other known affected members. Photographs of other affected family members may also be helpful. If the proband is an adult, serial photographs in a chronological order may assist in identifying the diagnosis as some dysmorphic conditions are evident within a critical age range (ie a diagnostic window).
Box 1.
Key points in history and physical examination
| History |
| Family history |
|
|
| Antenatal history |
|
|
|
|
|
| Birth history |
|
|
|
|
| Neonatal history |
|
|
|
|
|
| Postnatal history |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Physical examination |
| Growth parameters |
|
|
|
|
| Head-to-toe examination |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
BMI = body mass index; GDD = global development delay; ID = intellectual disability; IVF = in vitro fertilisation
Obtaining a detailed pregnancy and neonatal history is the next step in the diagnostic process. Specific details to elicit include maternal use of any prescription drugs (such as anticonvulsants), maternal alcohol intake, antenatal scan abnormalities, birth history and a chronological postnatal history with special emphasis on growth, developmental progress, eyesight, hearing, behaviour and progress at nursery or school (Box 1). This is followed by a detailed physical, dysmorphological and neurologic examination using a ‘head-to-toe’ approach (Box 1). All findings should be documented and photographs obtained with informed consent for further discussion with colleagues and specialists. The parents/carers should be reassured that confidentiality and anonymity will be maintained at all times when discussing their children’s clinical findings and photographs with other geneticists. Baseline investigations are then arranged, including baseline biochemistry (blood and urine), followed by additional investigations in specific cases depending on the differential diagnosis (Box 2 and 3).
Box 2.
Baseline investigations
| Biochemistry |
|
|
| Genetic |
| Chromosomal microarray |
| Molecular diagnostic test for Fragile X |
| Radiological |
| MRI brain (microcephaly, pigmentary skin anomalies, abnormal neurological examination) |
CK = creatine kinase; MRI = magnetic resonance imaging
Box 3.
Additional biochemical investigations in specific cases
|
|
|
|
|
Chromosomal microarray is now considered the first-line diagnostic genetic test in all individuals with GDD/ID.6 It is estimated that the diagnostic yield of a microarray test is about 12% and identification of significant copy number variants could be as high as 15–20%. Several studies have highlighted that microarray may identify ‘variants of unknown or uncertain significance’, which may be difficult to interpret in some cases. It can reveal ‘incidental findings’ like deletion of a cancer predisposition gene (eg BRCA1 or MLH1), which could have potentially important ethical implications when investigating a child for GDD/ID. The routine use of chromosomal microarrays for the investigation of GDD/ID in the last two decades has resulted in the identification of many new recurring micro-deletions and micro-duplications that can show intra-familial variability of expression and reduced penetrance. Examples include the 1q21 microdeletion, 15q11.2 microdeletion and the 16p13.11 microduplication.
Routine first-line testing for Fragile X syndrome is debated and although the cost-benefit ratio of routine Fragile X syndrome testing in patients with GDD/ID has been questioned by some experts, we believe that this may be a useful diagnostic test.7 Most patients with Fragile X syndrome do not have a pedigree showing an X-linked pattern of ID. Triplet repeats are also missed by next-generation sequencing testing. The test is not expensive (the cost of the Fragile X polymerase chain reaction test is around £100) and still has a pick up rate of 2%.
Box 4 lists the additional genetic tests that may help to make a genetic diagnosis in patients with GDD/ID. Many studies show an excess of males presenting with GDD/ID. In part, this may be due to the genetic variants on the X-chromosome. When a specific clinical diagnosis is suspected for individuals with a ‘syndromic’ X-linked disorder, a single gene test may be considered (for eg ATR-X or Coffin-Lowry syndrome in a male or Rett syndrome in a female). In other patients, where a ‘syndromic’ cause is suspected (X-linked or autosomal), next-generation sequencing of a panel of genes known to cause GDD/ID or clinical exome sequencing may be the preferred option to establish a genetic diagnosis. There are many gene panel tests available through the UK genetic testing network. Clinical exome sequencing is rapidly becoming a key diagnostic tool for establishing a genetic diagnosis in children with GDD/ID, which is reducing the mean cost per diagnosis. This enables testing of patients for rare pathogenic variants in all known developmental disorder genes, including those listed in the Developmental Disorder Genotype-Phenotype database (https://decipher.sanger.ac.uk/ddd#ddgenes). WES and whole genome sequencing (WGS) are revolutionising the diagnostic process in the investigation of GDD/ID. The former approach was taken by the very successful Deciphering Developmental Disorders (DDD) Study, which has been able to identify one or more specific genetic diagnoses in about 35% of cases of GDD/ID.8 The latter approach is being taken by the ongoing 100,000 Genomes Project.9
Box 4.
Genetic investigations in specific cases
|
|
|
|
|
|
|
MLPA = multiplex ligation-dependent probe amplification; PCR = polymerase chain reaction
Brain magnetic resonance imaging (MRI), including proton magnetic resonance spectroscopy (MRS) where available, should be undertaken in patients with GDD/ID who have microcephaly or macrocephaly or abnormal neurological findings, such as hypotonia, spasticity, ataxia or optic atrophy. However, some authors recommend brain imaging in all cases of GDD/ID. If all the tests undertaken fail to identify a specific diagnosis the situation should be reviewed with the family and a plan made to either review the child when older or consider making referrals to other specialists, such as a paediatric neurologist. The parents should also be provided with information about potential research projects in which their child could participate to help make a diagnosis.
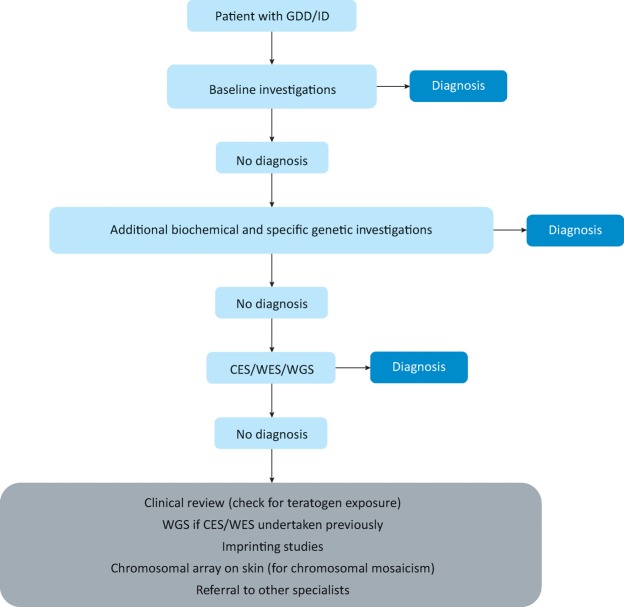
There is no single approach to diagnostic evaluation for GDD/ID. Fig 1 shows one possible diagnostic pathway. Clinicians who follow a systematic approach and develop familiarity with specific clinical features and diagnostics will become more efficient in the process. A chromosomal microarray is the first-tier genomic test for GDD/IDD. A reduction in the cost of next-generation sequencing will likely lead to widespread use of family-based trio evaluation (proband and parents) by WES/WGS as the second-tier test for GDD/ID in the near future, leading to a higher diagnostic yield.10 However, many challenges (including ethical, clinical utility and cost-benefit analysis) need to be addressed before the routine implementation of WES/WGS in clinical practice.
Fig 1.
A suggested diagnostic pathway for global developmental delay/intellectual disability. CES = clinical exome sequencing; GDD = global developmental delay; ID = intellectual disability; WES = whole exome sequencing; WGS = whole genome sequencing
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- 1.American Association on Intellectual and Developmental Disabilities Frequently asked questions on intellectual disability. http://aaidd.org/intellectual-disability/definition/faqs-on-intellectual-disability#.WPyVn9Lyu70. [Accessed 20 September 2017] [Google Scholar]
- 2.Department of Health Valuing people: a new strategy for learning disabilities for the 21st century. London:: Department of Health; 2001. [Google Scholar]
- 3.Hatton C. Emerson E. Glover G, et al. People with learning disabilities in England 2013. London:: Public Health England; 2014. [Google Scholar]
- 4.Deciphering Developmental Disorders study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srour M. Shevell M. Genetics and the investigation of developmental delay/intellectual disability. Arch Dis Child. Vol. 99. 2014. pp. 386–9. [DOI] [PubMed] [Google Scholar]
- 6.Moeschler JB. Shevell M Committee on Genetics. Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics. 2014. pp. e903–18. [DOI] [PMC free article] [PubMed]
- 7.UK Genetic Testing Network. www.ukgtn.nhs.uk. [Accessed 20 September 2017]. [Google Scholar]
- 8.Deciphering Developmental Disorders www.ddduk.org. [Accessed 20 September 2017]. [Google Scholar]
- 9.Genomics England. www.genomicsengland.co.uk. [Accessed 20 September 2017]. [Google Scholar]
- 10.Vissers LELM. Gilissen C. Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]