Abstract
In order to identify potential diagnostic and prognostic biomarkers, and treatment targets for head and neck squamous cell carcinoma (HNSCC), the present study obtained the gene expression profiles in HNSCC through public data mining, and core genes were identified using a series of bioinformatics analysis methods and databases. A total of nine hub genes (SPP1, ITGA6, TMPRSS11D, MMP1, LAMC2, FAT1, ACTA1, SERPINE1 and CEACAM1) were identified to be significantly correlated with HNSCC. Furthermore, overall survival analysis demonstrated that the expression values of hub genes were associated with overall survival in HNSCC. Furthermore, certain of the identified genes, including, TMPRSS11D, ACTA1 and CEACAM1, have not been thoroughly investigated in HNSCC previously. Taken together, the nine hub genes obtained by screening in the present study may serve as potential tumor markers and important prognostic indicators for HNSCC.
Keywords: head and neck cancer, data mining, biomarkers, gene expression profiling, bioinformatics
Introduction
Head and neck squamous cell carcinoma (HNSCC), a common malignant tumor of the head and neck region, is primarily comprised of lip, oral cavity, larynx, nasopharynx and other pharynx carcinomas. In 2012, the number of new HNSCC cases reported worldwide was ~686,000, while the HNSCC-associated mortality cases were 375,000 (1). Currently, smoking and alcohol consumption are deemed to be risk factors for HNSCC development; furthermore, human papilloma virus (HPV) infection is considered to have an important role in the occurrence and prognosis of HNSCC (2). In spite of the application of surgery, chemoradiation and multimodal treatment approaches, the prognosis of HNSCC remains poor due to local recurrence and metastasis, and the 5-year overall survival is ~50% (1). Furthermore, poor prognosis is partially attributed to the lack of understanding of the molecular mechanism underlying the development of this cancer. The oncogenesis and progression of HNSCC is a complicated process involving multiple molecules, including microRNA-98 (3), Twist family BHLH transcription factor 1 (4) and Mastermind-like 1 (4). With the application of immunotherapy, multimodal therapeutic approaches may be improved in the future (2). Thus, identifying specific tumor markers and novel molecular targets for the treatment of HNSCC is important.
Huang et al (5) has recently proposed that lysine (K)-specific demethylase 5B (KDM5B) is overexpressed in HNSCC tissues as compared with its levels in adjacent noncancerous tissues, and may be a significant prognostic biomarker of HNSCC on account of the association between KDM5B and overall survival times. A previous study by Trivedi et al (6) suggested an association between the expression of several tumor markers (including pemphigus vulgaris antigen, parathyroid hormone-related peptide and tumor-associated calcium signal transducer 1) and the metastasis of HNSCC to the lymph nodes. Furthermore, numerous established or emerging biomarkers associated with HNSCC have previously been explored, including hypoxia-inducible factor 1, carbonic anhydrase IX, programmed death ligand-1 and cytotoxic T-lymphocyte antigen 4, which are involved in immune checkpoints, hypoxia and radiation sensitivity (7). Tumor markers vary widely due to the multiple anatomical sites and histological types of HNSCC; thus, the identification of more valuable tumor markers of HNSCC is required.
The approach for screening tumor markers and molecular targets has improved with the application of microarray and transcriptional sequencing technologies (8,9). Microarray analysis and high-throughput sequencing technology provide valuable information and have been successfully used in marker screening for numerous tumors (10,11). The transcriptome data containing large amounts of information is stored in a public database for sharing. Data mining and bioinformatics analysis allow for transcriptome data to be fully utilized by researchers and enable more reliable biological information to be obtained (12–14). Biological and medical research has improved through the generation and development of bioinformatics (15). Numerous biological websites and software are available for use in bioinformatics analysis, including The Cancer Genome Atlas (TCGA) (16), Gene Expression Omnibus (GEO) 2R (17), The Database for Annotation Visualization and Integrated Discovery (DAVID) (18), Gene Expression Profiling Interactive Analysis (GEPIA) (19). These websites and software have been successfully applied in numerous biological studies (13,14).
Despite the extensive application of bioinformatics analysis in biomarker screening, investigations regarding HNSCC are limited. Demokan et al (20) have demonstrated that guanine nucleotide-binding protein γ-7 (GNG7) is downregulated in HNSCC through expression profile screening, and the expression level of GNG7 was then validated by quantitative methylation-specific polymerase chain reaction. A recent study also identified differentially expressed miRNAs and mRNAs in laryngeal squamous cell carcinoma through data mining and bioinformatics analysis (21). Therefore, the present study aimed to identify novel diagnostic, prognostic or predictive biomarkers for HNSCC, and highlight the core genes associated with HNSCC through bioinformatics analysis.
In the present study, three mRNA expression profiles associated with HNSCC were retrieved and filtered from the National Center for Biotechnology Information (NCBI)/GEO datasets, which are public and freely available databases (17,22). The differentially expressed genes (DEGs) in each data series were analyzed using GEO2R. The intersection of the three sets of DEGs extracted from the three data series included 19 mRNAs, which were defined as the hub genes and deemed to be highly associated with HNSCC. Through bioinformatics analysis, validation and survival analysis of a large sample size based on TCGA, a total of nine hub genes were identified as potential tumor markers and important prognostic indicators for HNSCC.
Materials and methods
Data mining
Transcriptome expression profiles of HNSCC patients were retrieved from the NCBI-GEO datasets (https://www.ncbi.nlm.nih.gov/gds/) and used for further screening. Three data series were selected for investigation in the present study, including GSE6631, GSE58911 and GSE83519. The screening criteria were set as follows: The mRNA expression profiles selected were derived from matched-pairs sample, while data derived from non-paired sample or cell lines were excluded. The series GSE6631 and GSE83519 included 22 paired samples each, and GSE58911 contained 15 paired samples. These data series were downloaded (retrieval date, January 7, 2018), and the DEGs between the HNSCC and matched control samples included in the three data series were filtered using GEO2R (www.ncbi.nlm.nih.gov/geo/info/geo2r.html), an NCBI-GEO integration tool. The default parameters used for DEG identification was a false discovery rate (Benjamini-Hochberg procedure) and P<0.05 (17,22). Then, MultiExperiment Viewer (23) and R package ‘ggplot2’ were applied for cluster analysis and visualization of DEGs. With these software, the overall distribution of DEGs in cancer tissues and paired normal tissues may be presented using a hierarchical clustering graph. The distribution of each DEG in different samples and the distribution of all DEGs in a given sample may also be distinguished using a clustering graph. The distribution tendency of DEGs may be presented directly using a volcano diagram.
Screening of hub genes
Three sets of DEGs derived from the three data series were further filtered according to rigorous screening criteria, including a fold change (FC) of ≥2 and P≤0.05. The newly acquired sets of DEGs were used in subsequent analyses. The intersections of the three sets of DEGs originating from the data series (GSE6631, GSE58911 and GSE83519) were presented using a Venn diagram, which was implemented with the online software VENN DIAGRAMS (http://bioinformatics.psb.ugent.be/beg/tools/venn-diagrams). Subsequently, the common genes of the three DEG sets were extracted and defined as hub genes if they were highly correlated with HNSCC.
Validation of relative expression levels of hub genes
In order to confirm and increase the reliability of the data analysis, the relative expression levels of hub genes in HNSCC were validated using GEPIA (http://gepia.cancer-pku.cn/index.html; retrieval date, January 9, 2018), an online analysis software based on the TCGA and Genotype-Tissue Expression (GTEx) databases, using |logFC|≥1 and P≤0.05 as the cut-off criteria (19). The results are presented as box plots.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses
The GO project describes genes and gene products on the basis of three different aspects, namely biological processes, molecular functions and cellular components, in a species-independent manner. GO has been widely used for genomics and proteomics analyses in biomedical research (24,25). KEGG, which primarily consists of various functions, including KEGG PATHWAY and KEGG GENES, is used for understanding high-level functions from molecular-level data (26). The KEGG PATHWAY, a manually reference database for pathway mapping, represents the current knowledge on molecular interactions, reactions and association networks (26). To gain an improved understanding of the hub genes, GO enrichment and KEGG pathway analyses were performed using DAVID, an online integration tool (retrieval date, January 9, 2018) (18,27).
Construction and analysis of protein-protein interaction (PPI) network
For a more in-depth understanding of the associations among hub genes, the PPI network of hub genes was structured via online database STRING, according to the provided manual (retrieval date, January 9, 2018) (28). PPIs may be evaluated and integrated via the PPI networks (28). Furthermore, the expression correlation of key nodes was verified using GEPIA (retrieval date, January 9, 2018).
Extraction and analysis of potential tumor markers
In order to identify tumor markers associated with prognosis, the hub genes were further filtered according to the score of nodes and the strength of interaction in the PPI network. Next, a log-rank test of overall survival (200 months was the longest follow-up period) associated with these selected hub genes was performed using GEPIA (retrieval date, January 9, 2018).
Results
DEGs derived from three data series associated with HNSCC
Initially, eight data series (including GSE83519, GSE58911, GSE6631, GSE23036, GSE13397, GSE40185, GSE10774 and GSE7073), which were able to be analyzed with GEO2R, were obtained by the retrieval of mRNA expression profiles associated with HNSCC in NCBI-GEO datasets. Among them, five data series were excluded out according to the screening criteria, including the cell line-based series GSE40185, GSE10774 and GSE7073, as well as the unpaired tissue sample-based series GSE23036 and GSE13397. The remaining three data series (GSE83519, GSE58911 and GSE6631) that met the screening criteria were used in the subsequent investigation.
GEO2R is a NCBI integration analysis tool for expression profile data and may be used to screen DEGs of two or more groups of sample sources (17,22). Three sets of DEGs derived from the three included data series were acquired by GEO2R analysis, and these are listed in Table I.
Table I.
Differentially expressed genes in the three data series.
In order to present the distribution of DEGs in each sample, the microarray data associated with the three NCBI-GEO datasets were downloaded, and the relative gene expression values of DEGs were extracted from the microarray data. Based on this, clustering analysis was performed using MultiExperiment Viewer, an open-source genomic analysis software (23). Subsequently, a volcano diagram was developed using the R package ‘ggplot2’. The hierarchical clustering graphs and volcano diagrams are presented in Fig. 1. Hierarchical clustering graphs and volcano diagrams of DEGs of the three data series presented that the DEGs are both upregulated and downregulated in cancerous tissues, but mainly downregulated in cancerous tissues, which suggested that the inactivation of tumor suppressor genes and the activation of oncogenes played important roles in the development of HNSCC, especially the inactivation of tumor suppressors.
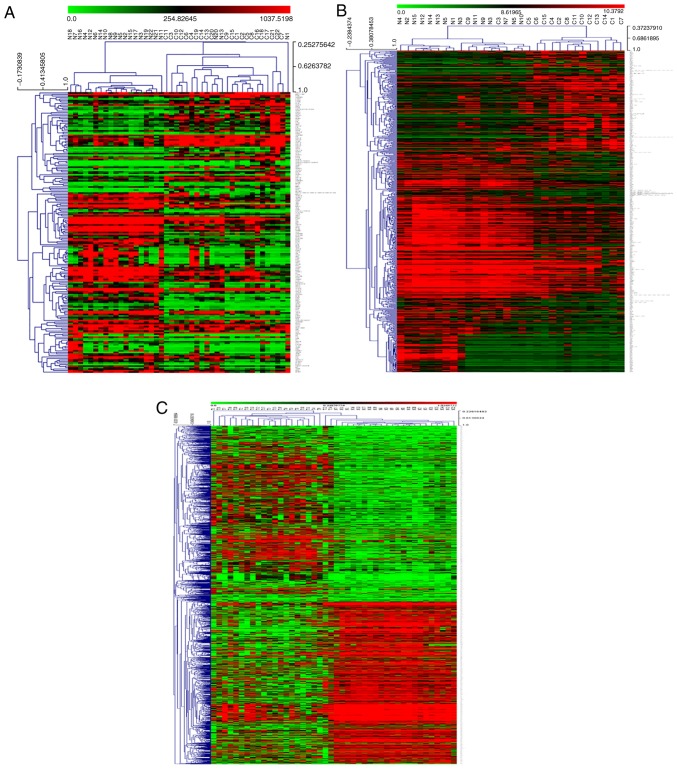
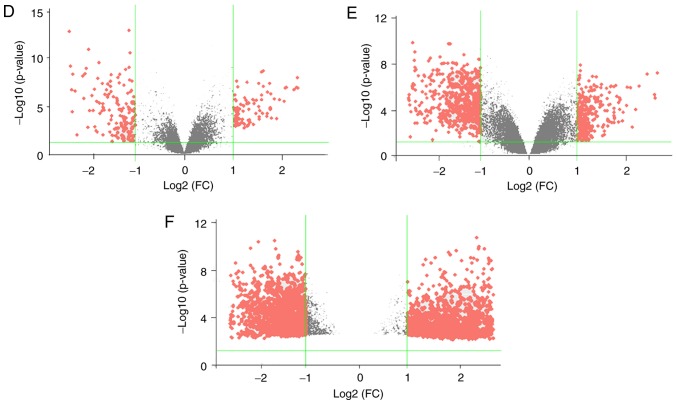
Figure 1.
Visual presentation of the differentially expressed genes (|logFC|≥1; P≤0.05) included in the three data series examined in the present study. Hierarchical clustering graphs of genes in the (A) GSE6631, (B) GSE58911 and (C) GSE83519 data series are shown. Rows represent the mRNAs and columns represent the samples. Red and green represent upregulated and downregulated genes in cancerous tissues compared with paired adjacent normal samples, respectively. Volcano plots for (D) GSE6631, (E) GSE58911 and (F) GSE83519 data series are displayed. The x-axis represents the log2 of FC and the y-axis represents the negative log10 of the P-value. Red and grey foci on the graphs represent differently and non-differentially expressed mRNAs, respectively, in head and neck squamous cell carcinoma. FC, fold change.
Nineteen hub genes are screened from the DEGs of the three data series
The common DEGs from the three data series may be associated with HNSCC and were defined as the hub genes. A total of 19 hub genes were extracted using the VENN DIAGRAMS drawing tool (Fig. 2A), and were then visualized as hierarchical clustering graphs for each data series (Fig. 2B-D). Among the 19 hub genes, 11 hub genes (including SPP1, COL4A1, COL1A1, FN1, ITGA6, MMP1, MMP11, LAMC2, FAT1, SERPINE1 and MMP9) were overexpressed in the HNSCC samples compared with the paired corresponding control samples. Conversely, 8 hub genes (including GPD1 L, MYH7, ATP2A1, HOPX, CEACAM1, TMPRSS11D, ABLIM1 and ACTA1) were downregulated in the cancerous samples compared with the controls.
Figure 2.
Hub genes derived from the three data series. (A) Venn diagram demonstrating the intersections of data series GSE6631, GSE58911 and GSE83519. Intersections represent the differentially expressed genes in two or three data series. Hierarchical clustering graph represents the hub genes derived from the data series (B) GSE6631. Hierarchical clustering graphs represent the hub genes derived from the data series (C) GSE58911 and (D) GSE83519.
Relative expression levels of hub genes in HNSCC samples against paired corresponding normal samples are validated using GEPIA
Although the screening of hub genes performed in the current study was vigorous and highly reliable, the relative expression levels of 19 hub genes in tissues were further validated through the online analysis tool GEPIA, which is based on TCGA and GTEx databases (tumor, 519 cases; normal, 44 cases in GEPIA). The results demonstrated that the relative expression trend of the 19 hub genes in GEPIA were consistent with the expression profiles (Fig. 3), supporting the reliability of the data analysis.
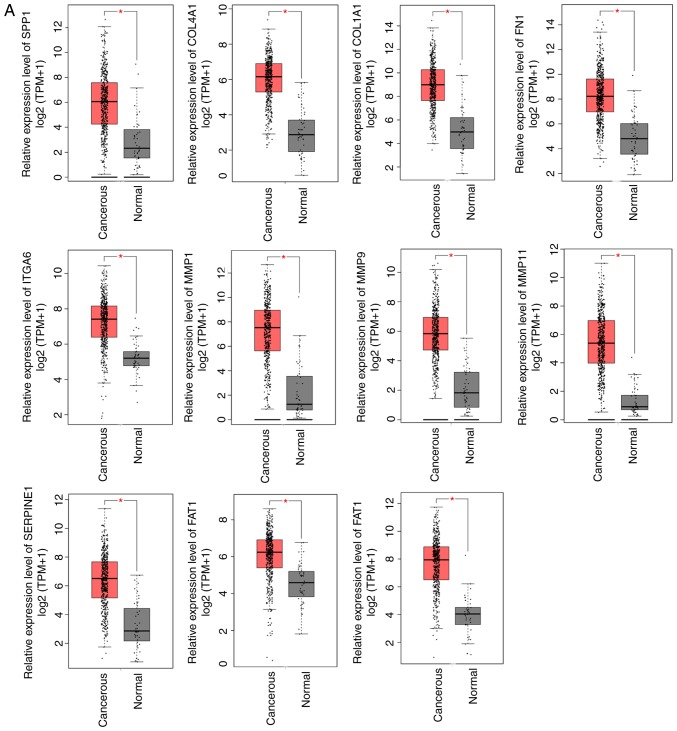
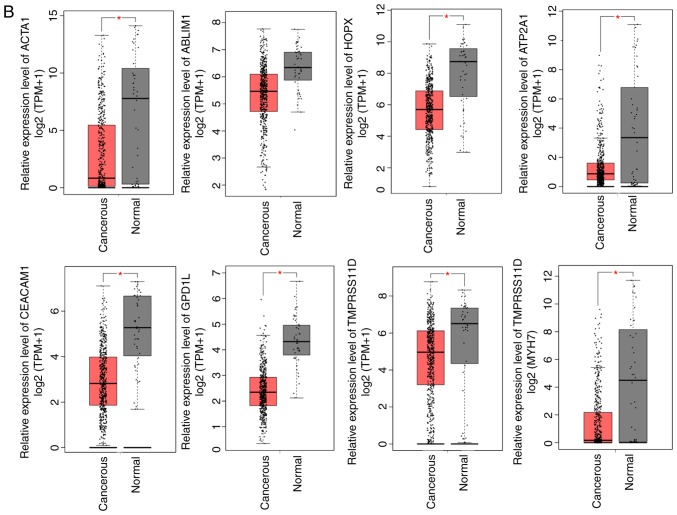
Figure 3.
Relative expression levels of hub genes. (A) Upregulated and (B) downregulated genes were identified using Gene Expression Profiling Interactive Analysis (GEPIA) based on The Cancer Genome Atlas database. Red and black boxes represent the relative expression levels of genes in the tumor and normal samples, respectively. The y-axis represents the relative expression levels of genes in terms of log2 (TPM+1) (tumor samples, 519; normal samples, 44 from GEPIA; P≤0.05). TPM, transcripts per million.
Nineteen hub genes are involved in multiple GO terms and KEGG pathways
GO and KEGG pathway analyses of 19 hub genes were further performed using the online analytical tool DAVID (threshold, count ≥3; EASE score, ≤0.05), in order to achieve a preliminary understanding of the biological functions in which these hub genes may participate (Table II). The results of GO analysis indicated that the main biological processes of the selected hub genes consisted of extracellular matrix disassembly or organization, leukocyte migration, collagen catabolic process, cell adhesion, proteolysis, positive regulation of cell migration and angiogenesis. The primary cellular component terms included the extracellular region, extracellular matrix, stress fiber, extracellular space, proteinaceous extracellular matrix, filopodium, extracellular exosome, lamellipodium and perinuclear region of the cytoplasm. Finally, the primary molecular function terms included serine-type endopeptidase activity, metalloendopeptidase activity, actin binding and calcium ion binding. According to the GO terms and the distribution of hub genes, the role of a protein in HNSCC may be preliminarily hypothesized. For instance, MMP9 may participate in extracellular matrix disassembly (GO:0022617) via exertion of metalloendopeptidase activity (GO:0004222) in the extracellular space (GO:0005615). Furthermore, KEGG pathway analysis indicated that the screened hub genes were primarily involved in extracellular matrix-receptor interaction (hsa04512), focal adhesion (hsa04510), the PI3K-Akt signaling pathway (hsa04151), pathways in cancer (hsa05200), small cell lung cancer (hsa05222) and amoebiasis (hsa05146).
Table II.
GO and KEGG pathway analysis of hub genes.
| Term | Count | P-value | Genes | Fold enrichment | FDR |
|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | |||||
| GO:0022617-extracellular matrix disassembly | 6 | 1.36×10−8 | MMP9, MMP11, FN1, MMP1, LAMC2, SPP1 | 69.77285319 | 1.78×10−5 |
| GO:0030198-extracellular matrix organization | 7 | 3.87×10−8 | FN1, SERPINE1, LAMC2, SPP1, COL4A1, ITGA6, COL1A1 | 31.56390977 | 5.06×10−5 |
| GO:0050900-leukocyte migration | 6 | 1.48×10−7 | MMP9, FN1, MMP1, CEACAM1, ITGA6, COL1A1 | 43.46505608 | 1.94×10−4 |
| GO:0030574-collagen catabolic process | 5 | 5.64×10−7 | MMP9, MMP11, MMP1, COL4A1, COL1A1 | 69.04605263 | 7.37×10−4 |
| GO:0007155-cell adhesion | 7 | 5.67×10−6 | FN1, LAMC2, CEACAM1, SPP1, FAT1, ITGA6, COL1A1 | 13.47827084 | 0.007408 |
| GO:0006508-proteolysis | 4 | 0.015348 | MMP9, MMP11, TMPRSS11D, MMP1 | 7.070315789 | 18.30441 |
| GO:0030335-positive regulation of cell migration | 3 | 0.016282 | LAMC2, ITGA6, COL1A1 | 14.40961098 | 19.31187 |
| GO:0001525-angiogenesis | 3 | 0.023357 | FN1, SERPINE1, CEACAM1 | 11.88954449 | 26.57648 |
| GOTERM_CC_DIRECT | |||||
| GO:0005576-extracellular region | 10 | 7.46×10−6 | MMP9, MMP11, FN1, TMPRSS11D, SERPINE1, MMP1, LAMC2, SPP1, COL4A1, COL1A1 | 5.957502452 | 0.007625 |
| GO:0031012-extracellular matrix | 6 | 7.87×10−6 | MMP11, FN1, SERPINE1, MMP1, COL4A1, COL1A1 | 19.44238976 | 0.008049 |
| GO:0001725-stress fiber | 3 | 0.001279 | MYH7, ABLIM1, ACTA1 | 53.28654971 | 1.300056 |
| GO:0005615-extracellular space | 7 | 0.001379 | MMP9, FN1, SERPINE1, LAMC2, SPP1, COL1A1, ACTA1 | 4.984487946 | 1.401185 |
| GO:0005578-proteinaceous extracellular matrix | 4 | 0.002179 | MMP9, MMP11, FN1, MMP1 | 14.31578947 | 2.205664 |
| GO:0030175-filopodium | 3 | 0.002199 | FAT1, ITGA6, ACTA1 | 40.52779837 | 2.225839 |
| GO:0070062-extracellular exosome | 9 | 0.003242 | MMP9, FN1, TMPRSS11D, SERPINE1, CEACAM1, SPP1, FAT1, GPD1L, ACTA1 | 3.070943099 | 3.265211 |
| GO:0030027-lamellipodium | 3 | 0.010687 | ABLIM1, FAT1, ACTA1 | 17.98421053 | 10.40403 |
| GO:0048471-perinuclear region of cytoplasm | 4 | 0.021939 | ATP2A1, LAMC2, SPP1, FAT1 | 6.178150691 | 20.29296 |
| GOTERM_MF_DIRECT | |||||
| GO:0004252-serine-type endopeptidase activity | 4 | 0.00235 | MMP9, MMP11, TMPRSS11D, MMP1 | 13.93684211 | 2.240613 |
| GO:0004222-metalloendopeptidase activity | 3 | 0.006336 | MMP9, MMP11, MMP1 | 23.58779693 | 5.938097 |
| GO:0003779-actin binding | 3 | 0.034758 | MYH7, ABLIM1, CEACAM1 | 9.587845513 | 28.8746 |
| GO:0005509-calcium ion binding | 4 | 0.038688 | MMP11, ATP2A1, MMP1, FAT1 | 4.956617485 | 31.61489 |
| KEGG_PATHWAY | |||||
| hsa04512: ECM-receptor interaction | 6 | 2.08×10−7 | FN1, LAMC2, SPP1, COL4A1, ITGA6, COL1A1 | 36.65782493 | 1.93×10−4 |
| hsa04510: Focal adhesion | 6 | 1.50×10−5 | FN1, LAMC2, SPP1, COL4A1, ITGA6, COL1A1 | 15.48170276 | 0.013864 |
| hsa04151: PI3K-Akt signaling pathway | 6 | 1.78×10−4 | FN1, LAMC2, SPP1, COL4A1, ITGA6, COL1A1 | 9.244147157 | 0.165019 |
| hsa05200: Pathways in cancer | 6 | 3.29×10−4 | MMP9, FN1, MMP1, LAMC2, COL4A1, ITGA6 | 8.115091016 | 0.304375 |
| hsa05222: Small cell lung cancer | 4 | 3.65×10−4 | FN1, LAMC2, COL4A1, ITGA6 | 25.01357466 | 0.33712 |
| hsa05146: Amoebiasis | 4 | 6.98×10−4 | FN1, LAMC2, COL4A1, COL1A1 | 20.05805515 | 0.644143 |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Construction and analysis of the PPI network are implemented via STRING based on hub genes
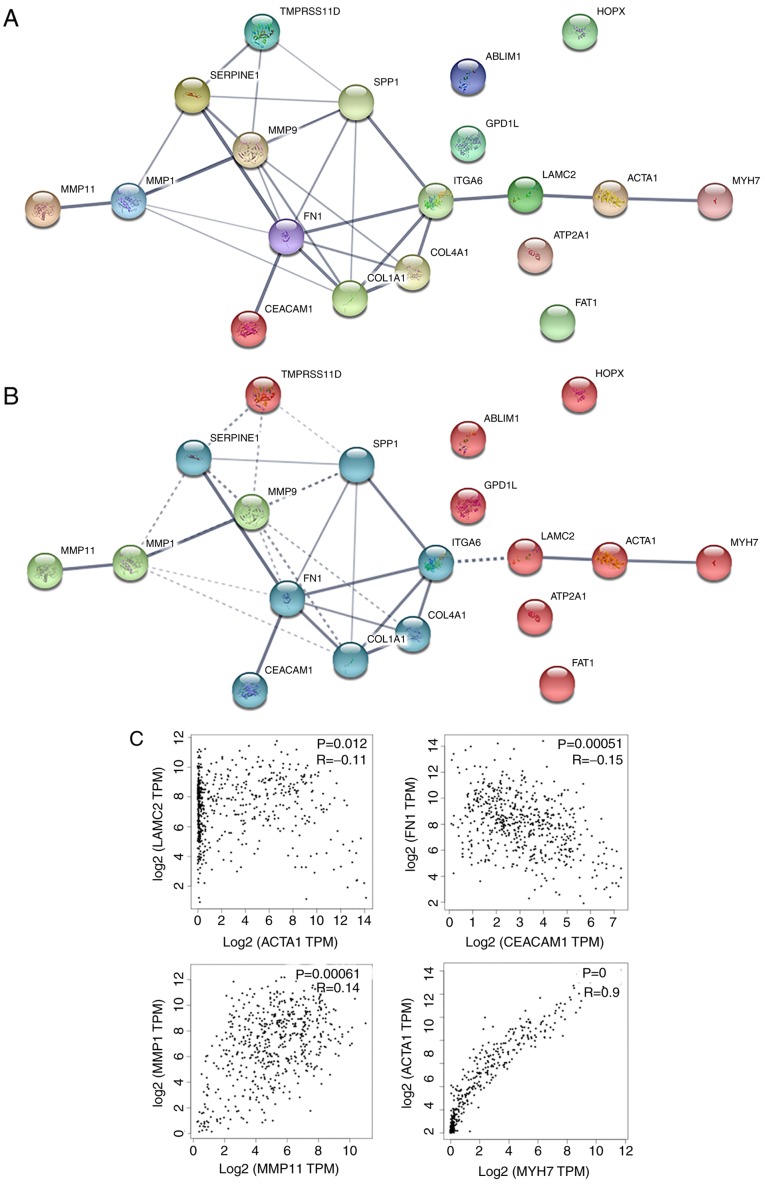
In order to analyze the interaction of hub genes, a PPI network was constructed using the online tool STRING. In total, 19 nodes and 29 edges were obtained (average node degree, 3.05; PPI enrichment P-value, 7.55×10−15; meaning of network edges, medium confidence score of 0.4), indicating a possibly high correlation among these hub genes (Fig. 4A).
Figure 4.
PPI networks of hub genes. (A) PPI network, and (B) k-means clustering of the PPI network are displayed. Nodes represent the proteins; colored nodes represent the query proteins and first layer of interactors; white nodes represent the second layer of interactors; edges represent the protein-protein associations; and the line thickness indicates the strength of data. (C) Associations among the expression of hub genes in the PPI networks produced using Gene Expression Profiling Interactive Analysis based on The Cancer Genome Atlas database (P≤0.05). PPI, protein-protein interaction.
To confirm the possibility of interactions in the PPI network, certain nodes of the PPI network were selected according to the following aspects: Edge thickness (line thickness indicates the strength of data support) (Fig. 4A), the results of the PPI clustering (k-means clustering; number of clustering=3; Fig. 4B) and the principle that the expression correlation means possible function interdependency. Correlation analysis of the gene expression levels were further verified using GEPIA, which is based on a large sample size from TCGA and GTEx databases. The selected correlations consisted of CEACAM1-FN1, MYH7-ACTA1, MMP11-MMP1 and ACTA1-LAMC2, and all the correlations were verified using GEPIA (Fig. 4C). The aforementioned evidence presumes that correlations among expression levels suggest that associations exist on the functional level, and it is further suggested that these genes may be associated with the development or progression of HNSCC.
Nine hub genes are positively or negatively associated with overall survival, and may be used as potential tumor markers and prognostic indicators
Based on the screening of tumor markers, overall survival analysis of hub genes was performed using GEPIA. The results suggested that nine hub genes (including SPP1, ITGA6, TMPRSS11D, MMP1, LAMC2, FAT1, ACTA1, SERPINE1 and CEACAM1) exhibited positive or negative associations with the overall survival of patients with HNSCC, and these hub genes may be used as potential tumor markers and prognostic indicators (Fig. 5). Nevertheless, the remaining hub genes had no effect on the prognosis of patients and may be involved in other processes of HNSCC development, which may be attributed to the interaction of a variety of factors in the occurrence of cancer. The nine hub genes validated by the online database GEPIA, correlated with the prognosis of patients. Furthermore, studies regarding the nine hub genes in HNSCC are limited according to literature retrieval in NCBI/PubMed (https://www.ncbi.nlm.nih.gov/pubmed; key words, hub gene and HNSCC, ignore language, publication data and article types). To the best of our knowledge, no previous studies regarding the effects of TMPRSS11D and ACTA1 in HNSCC have been performed to date.
Figure 5.
Association of hub gene expression with overall survival. Curves were produced using Gene Expression Profiling Interactive Analysis based on The Cancer Genome Atlas database (P≤0.05).
Discussion
Microarray and high-throughput sequencing technology provides valuable research information through profile screening of small sample sizes. Gene expression profile microarrays provide data from different samples, allowing for the identification of DEGs using bioinformatics analysis that can be applied in further studies (10,11,29). Microarray data are stored in public databases for sharing and re-mining, including GEO-datasets, TCGA and Oncomine (16,22,30). Through data mining and bioinformatics analysis, researchers are able to use public databases as valuable references for biological research (11,17,31).
In the present study, three data series associated with HNSCC (GSE83519, GSE58911 and GSE6631) were identified and then deep mining of genomics data was performed with the aid of bioinformatics software. The three included data series were based on paired samples and the target genes were identified as the common intersection of the three data series. The three data series examined in the present study were all derived from paired tumor and control samples of HNSCC patients, thus increasing the reliability of the study. Furthermore, the common DEGs identified demonstrated the same distribution tendency among the three data series. These common DEGs may be regarded as results from three experiment repetitions and an enlargement of the sample size, which increases the reliability of the gene expression trend. Comprehensive analysis of the above-mentioned databases confirmed the reliability of data with regard to experimental reproducibility and sample size. Furthermore, through screening and database validation, the intersection of the selected DEGs was found to include 19 mRNAs, which were defined as the hub genes and considered to be highly correlated with HNSCC. The 19 hub genes were differentially expressed in HNSCC simultaneously, suggesting that these genes may independently or dependently be involved in the occurrence or development of HNSCC. Thus, GO and KEGG pathway analyses were adopted to perform functional enrichment of hub genes, and these analyses categorized target genes in order to identify genes of interest. Functional enrichment analysis suggested that the highly correlated hub genes may participate in a variety of biological processes. Additionally, a PPI network was constructed based on the principle that proteins with associated expression levels may interact with one another. The PPI network and the network clustering results based on hub genes identified in the present study indicated that MMP11, CEACAM1, MYH7 and ACTA1 were located in the first layer of the network. These genes were suggested to be involved in the PPI network through higher binding, as indicated by the edge thickness, which included CEACAM1-FN1-SERPINE1, MYH7-ACTA1-LAMC2 and MMP11-MMP1-MMP9. In order to further confirm these associations, the correlations among connective nodes were verified based on large sample sizes using GEPIA, following the principle that proteins with associated expression levels may exhibit interactions, including CEACAM1-FN1, MYH7-ACTA1, MMP11-MMP1 and ACTA1-LAMC2. The results suggested that each protein in the primary line of the network exhibited a positive or negative correlation. The results further demonstrated that the hub genes participated in HNSCC through protein-protein interactions.
The bioinformatics analysis conducted in the present study provided important references for subsequent research; however, it also has certain limitations. For instance, genes that were differentially expressed among the three data sets were screened as hub genes, which suggested that these genes may have potential research value. However, useful information may be lost in the screening process. For example, TP53, a routine tumor suppressor, was screened out as it was only included in GSE83519, but not in other two databases. However, as a conventional tumor suppressor, TP53 has been demonstrated to be highly mutated and hold important biological significance in HNSCC (32).
The principle aim of present study was to screen a number of genes or proteins and provide a reference for clinical diagnosis, treatment and prognosis of HNSCC. In line with the purpose of screening tumor markers and prognostic indicators, analysis of the association of survival with the expression of the 19 hub genes was performed using GEPIA. The results indicated that nine of the hub genes (SPP1, ITGA6, TMPRSS11D, MMP1, LAMC2, FAT1, ACTA1, SERPINE1 and CEACAM1) were identified as significant prognostic indicators of HNSCC. To the best of our knowledge, few studies have investigated these nine hub genes in HNSCC according to literature retrieval using NCBI-PubMed. Previous studies have revealed that the expression level of SPP1 in the plasma is negatively associated with the survival time of the patients with HNSCC (33), and this was consistent with the results of the present study analysis; however, these results remain controversial; another study concerning HNSCC found no correlation between SPP1 expression level and prognosis of HNSCC (34). In addition, it has been reported that ITGA6 and LAMC2 promote the migration and invasion of HNSCC cells (35), and that MMP1 serves a role in the invasiveness of HNSCC (36). Furthermore, the mutation status of FAT1 is associated with the prognosis of HPV-negative HNSCC (37). A previous study also reported that TMPRSS11D expression was reduced in squamous cell carcinogenesis (38), whereas a different study revealed that the expression of this gene was significantly higher in non-small cell lung carcinoma tissues compared with that in adjacent normal tissues (39). Nevertheless, previous studies focusing on TMPRSS11D in HNSCC have not been identified. Notably, the analysis of the present study suggested that the expression level of TMPRSS11D was positively associated with the survival time. Thus, it is necessary to perform further research regarding the role of TMPRSS11D in HNSCC. A recent study reported that CEACAM1 promotes the growth of HNSCC tumors (40). However, to the best of our knowledge, no further studies on the underlying mechanisms have been performed, and there appears to be only one other study reporting that CEACAM1 is overexpressed in oral tumors and correlated with carcinogenesis (41). In addition, previous data analysis of colon cancer demonstrated that ACTA1 may be associated with the methylation status of the oncogenome (42), but no further research has been performed, particularly in HNSCC. Furthermore, a study reported that SERPINE1 promotes cell migration, and overexpression of SERPINE1 was revealed to be associated with poor survival in HNSCC (43). All of the aforementioned studies demonstrate that the mRNAs or proteins identified through bioinformatics analysis in the present study are involved in the biological process of HNSCC, which was in accordance with our hypothesis. Notably, no studies investigating TMPRSS11D and ACTA1 in HNSCC have been identified. Although the aforementioned literature analysis is only based on PubMed search, which is an authoritative and comprehensive database, it also reflects the research status of these genes to a certain extent. Therefore, the present study also provided a practical and feasible direction for further research.
Through rigorous statistical analysis, bioinformatics analysis is able to provide theoretical biological mechanisms and promote the development of biological research (13,15). However, further verification is required to confirm the results presented in the current study. In the initial analysis, the sample size of the three enrolled databases was small. If data from the TCGA or Oncomine databases, which contain more samples, are reasonably integrated in the initial analysis, the validity of the results of the present study will be increased.
In conclusion, in the current study, a novel set of core genes associated with HNSCC that may serve as potential biomarkers was identified based on a series of bioinformatics analyses and a large sample database. However, further validation is required to verify these results.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papilloma virus
- KDM5B
lysine (K)-specific demethylase 5B
- GNG7
guanine nucleotide-binding protein γ-7
- NCBI
National Center for Biotechnology Information
- GEO
Gene Expression Omnibus
- DEGs
differentially expressed genes
- TCGA
The Cancer Genome Atlas
- GEPIA
Gene Expression Profiling Interactive Analysis
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DAVID
The Database for Annotation Visualization and Integrated Discovery
- PPI
protein-protein interaction
Funding
The present study was funded by the Key Program of Hebei Natural Science Foundation (grant no. H2017206391) and the Project of Clinical Medical Talent Training and Basic Project Research Funded by Government [grant Hebei finance society (2017) no. 46].
Availability of data and materials
All data generated and analyzed during current study were extracted from public databases (NCBI-GEO, GEPIA, DAVID and STRING).
Authors' contributions
BSW conceived and designed the study; LZ contributed to the acquisition, analysis and interpretation of data, and was involved in drafting the manuscript; WWC and HC partially designed and revised the manuscript for important intellectual content; WNC and WXM participated in the data analysis and drafting of the manuscript; WG was involved in the data analysis and the production of figures.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Magnes T, Egle A, Greil R, Melchardt T. Update on squamous cell carcinoma of the head and neck: ASCO annual meeting. Memo. 2017;10:220–223. doi: 10.1007/s12254-017-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan H, Zhu G, She L, Wei M, Wang Y, Pi L, Chen C, Zhang D, Tan P, Chen J, et al. MiR-98 inhibits malignant progression via targeting MTDH in squamous cell carcinoma of the head and neck. Am J Cancer Res. 2017;7:2554–2565. [PMC free article] [PubMed] [Google Scholar]
- 4.Ardalan Khales S, Ebrahimi E, Jahanzad E, Ardalan Khales S, Forghanifard MM. MAML1 and TWIST1 co-overexpression promote invasion of head and neck squamous cell carcinoma. Asia Pac J Clin Oncol. 2018 Jan 15; doi: 10.1111/ajco.12843. (Epub ahead of print). doi: 10.1111/ajco.12843. [DOI] [PubMed] [Google Scholar]
- 5.Huang D, Qiu Y, Li G, Liu C, She L, Zhang D, Chen X, Zhu G, Zhang X, Tian Y, et al. KDM5B overexpression predicts a poor prognosis in patients with squamous cell carcinoma of the head and neck. J Cancer. 2018;9:198–204. doi: 10.7150/jca.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trivedi S, Mattos J, Gooding W, Godfrey TE, Ferris RL. Correlation of tumor marker expression with nodal disease burden in metastatic head and neck cancer. Otolaryngol Head Neck Surg. 2013;149:261–268. doi: 10.1177/0194599813486876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol. 2015;12:11–26. doi: 10.1038/nrclinonc.2014.192. [DOI] [PubMed] [Google Scholar]
- 8.Feng L, Wang R, Lian M, Ma H, He N, Liu H, Wang H, Fang J. Integrated analysis of long noncoding RNA and mRNA expression profile in advanced laryngeal squamous cell carcinoma. PLoS One. 2016;11:e0169232. doi: 10.1371/journal.pone.0169232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li CQ, Huang GW, Wu ZY, Xu YJ, Li XC, Xue YJ, Zhu Y, Zhao JM, Li M, Zhang J, et al. Integrative analyses of transcriptome sequencing identify novel functional lncRNAs in esophageal squamous cell carcinoma. Oncogenesis. 2017;6:e297. doi: 10.1038/oncsis.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seitz AK, Christensen LL, Christensen E, Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM, Palmfeldt J, Thomson M, et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep. 2017;7:395. doi: 10.1038/s41598-017-00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou AE, Ku J, Honda TK, Yu V, Kuo SZ, Zheng H, Xuan Y, Saad MA, Hinton A, Brumund KT, et al. Transcriptome sequencing uncovers novel long noncoding and small nucleolar RNAs dysregulated in head and neck squamous cell carcinoma. RNA. 2015;21:1122–1134. doi: 10.1261/rna.049262.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lena PG, Paz-Gallardo A, Paramio JM, Garcia-Escudero R. Clusterization in head and neck squamous carcinomas based on lncRNA expression: molecular and clinical correlates. Clin Epigenetics. 2017;9:36. doi: 10.1186/s13148-017-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129. doi: 10.1186/s12943-017-0696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong N, Khwaja SS, Baker CM, Gay HA, Thorstad WL, Daly MD, Lewis JS, Jr, Wang X. Prognostic microRNA signatures derived from The Cancer Genome Atlas for head and neck squamous cell carcinomas. Cancer Med. 2016;5:1619–1628. doi: 10.1002/cam4.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laukens K, Naulaerts S, Berghe WV. Bioinformatics approaches for the functional interpretation of protein lists: From ontology term enrichment to network analysis. Proteomics. 2015;15:981–996. doi: 10.1002/pmic.201400296. [DOI] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R, et al. NCBI GEO: Mining tens of millions of expression profiles-database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demokan S, Chuang AY, Chang X, Khan T, Smith IM, Pattani KM, Dasgupta S, Begum S, Khan Z, Liegeois NJ, et al. Identification of guanine nucleotide-binding protein γ-7 as an epigenetically silenced gene in head and neck cancer by gene expression profiling. Int J Oncol. 2013;42:1427–1436. doi: 10.3892/ijo.2013.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang GJ, Luo MS, Chen GP, Fu MY. MiRNA-mRNA crosstalk in laryngeal squamous cell carcinoma based on the TCGA database. Eur Arch Otorhinolaryngol. 2018;275:751–759. doi: 10.1007/s00405-018-4876-9. [DOI] [PubMed] [Google Scholar]
- 22.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Gene Ontology Consortium: Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao J, Huang JX, Lin M, Wu ZD, Yu H, Wang PC, Ye J, Chen P, Wu J, Zhao GJ. Microarray expression profile analysis of aberrant long non-coding RNAs in esophageal squamous cell carcinoma. Int J Oncol. 2016;48:2543–2557. doi: 10.3892/ijo.2016.3457. [DOI] [PubMed] [Google Scholar]
- 30.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le QT, Sutphin PD, Raychaudhuri S, Yu SC, Terris DJ, Lin HS, Lum B, Pinto HA, Koong AC, Giaccia AJ. Identification of osteopontin as a prognostic plasma marker for head and neck squamous cell carcinomas. Clin Cancer Res. 2003;9:59–67. [PubMed] [Google Scholar]
- 34.Lim AM, Rischin D, Fisher R, Cao H, Kwok K, Truong D, McArthur GA, Young RJ, Giaccia A, Peters L, et al. Prognostic significance of plasma osteopontin in patients with locoregionally advanced head and neck squamous cell carcinoma treated on TROG 02.02 phase III trial. Clin Cancer Res. 2012;18:301–307. doi: 10.1158/1078-0432.CCR-11-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinoshita T, Nohata N, Hanazawa T, Kikkawa N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumour-suppressive microRNA-29 s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer. 2013;109:2636–2645. doi: 10.1038/bjc.2013.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kidacki M, Lehman HL, Green MV, Warrick JI, Stairs DB. p120-catenin downregulation and PIK3CA mutations cooperate to induce invasion through MMP1 in HNSCC. Mol Cancer Res. 2017;15:1398–1409. doi: 10.1158/1541-7786.MCR-17-0108. [DOI] [PubMed] [Google Scholar]
- 37.Kim KT, Kim BS, Kim JH. Association between FAT1 mutation and overall survival in patients with human papillomavirus-negative head and neck squamous cell carcinoma. Head Neck. 2016;38(Suppl 1):E2021–E2029. doi: 10.1002/hed.24372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duhaime MJ, Page KO, Varela FA, Murray AS, Silverman ME, Zoratti GL, List K. Cell surface human airway trypsin-like protease is lost during squamous cell carcinogenesis. J Cell Physiol. 2016;231:1476–1483. doi: 10.1002/jcp.25173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao X, Tang Z, Huang F, Jin Q, Zhou X, Shi J. High TMPRSS11D protein expression predicts poor overall survival in non-small cell lung cancer. Oncotarget. 2017;8:12812–12819. doi: 10.18632/oncotarget.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tam K, Schoppy DW, Shin JH, Tay JK, Moreno-Nieves U, Mundy DC, Sunwoo JB. Assessing the impact of targeting CEACAM1 in head and neck squamous cell carcinoma. Otolaryngol Head Neck Surg. 2018;159:76–84. doi: 10.1177/0194599818756627. [DOI] [PubMed] [Google Scholar]
- 41.Wang FF, Guan BX, Yang JY, Wang HT, Zhou CJ. CEACAM1 is overexpressed in oral tumors and related to tumorigenesis. Med Mol Morphol. 2017;50:42–51. doi: 10.1007/s00795-016-0147-2. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Li H, Sun L, Wang Z, Xing C, Yuan Y. Aberrantly methylated-differentially expressed genes and pathways in colorectal cancer. Cancer Cell Int. 2017;17:75. doi: 10.1186/s12935-017-0444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavón MA, Arroyo-Solera I, Téllez-Gabriel M, León X, Virós D, López M, Gallardo A, Céspedes MV, Casanova I, López-Pousa A, et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget. 2015;6:29016–29033. doi: 10.18632/oncotarget.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and analyzed during current study were extracted from public databases (NCBI-GEO, GEPIA, DAVID and STRING).