Abstract
With a 5-year survival rate of only 8%, pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-associated mortality worldwide. Unfortunately, even following radical surgery, patient outcomes remain poor. Emerging as a new class of biomarkers in human cancer, microRNAs (miRNAs/miRs) have been reported to have various tumor suppressor and oncogenic functions. In the present study, miRNA expression profiles of patients with PDAC and corresponding clinical data with survival profiles were obtained from The Cancer Genome Atlas database. A co-expression network was constructed to detect the modules significantly associated with clinical features by weighted gene co-expression network analysis. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed on the hub miRNAs in the module of interest for functional annotation. A prognosis model consisting of hub miRNAs was generated using the R package ‘rbsurv’ and validated in survival analysis. The expression data of 523 miRNAs in 124 patients with PDAC were analyzed in a co-expression network. The turquoise module containing 131 miRNAs was identified to be associated with pathological T stage (cor=−0.21; P=0.02). The 39 hub miRNAs of the turquoise module were then detected using the ‘networkScreening’ function in R. These miRNAs were predominantly involved in biological processes including ‘regulation of transcription’, ‘apoptotic process’, ‘TGF-β receptor signaling pathway’, ‘Ras protein signal transduction’ and significantly enriched in ‘cell cycle’, ‘adherens junction’, ‘FoxO’, ‘Hippo’ and ‘PI3K-Akt signaling’ pathways. A prognostic signature consisting of four hub miRNAs (miR-1197, miR-218-2, miR-889 and miR-487a) associated with pathological T stage was identified to stratify the patients with early-stage PDAC into high and low risk groups. The signature may serve as a potential prognostic biomarker for patients with early-stage PDAC who undergo radical resection.
Keywords: pancreatic ductal adenocarcinoma, The Cancer Genome Atlas, weighted gene co-expression network analysis, prognostic
Introduction
Pancreatic ductal adenocarcinoma (PDAC), which accounts for ~80% of all pancreatic tumors, is an aggressive malignancy and the fourth leading cause of cancer-associated mortality worldwide (1,2). With the rising pancreatic cancer incidence rates, the American Cancer Society estimates that 55,440 new cases will occur in the United States with 44,330 PDAC-associated deaths in 2018 (3). Treatment of pancreatic cancer consists of surgery, radiotherapy, chemotherapy and palliative care. Appropriate treatments are selected depending on disease stage. Compared with other treatments, surgical resection is the best curative option to significantly prolong patient survival (4). Unfortunately, numerous studies have reported that prognosis remains poor following radical surgical resection, with a median survival of 10–20 months and 5-year survival rates of only 10–25% in patients who receive pancreaticoduodenectomy (5–11); the survival rates are even lower for patients who receive pancreatectomy (12,13). For patients who undergo radical resection, gemcitabine-based adjuvant chemotherapy is currently the standard treatment following surgery. If the patients exhibit nodal involvement or microscopic residual disease following resection, radiotherapy is recommended (14). Despite substantial advancements in screening, diagnosis and treatment, PDAC has an extremely dismal prognosis, with a median survival of 2–8 months and a 5-year survival rate as low as 8% (15). Using prognostic biomarkers to select the most suitable adjuvant therapies for patients may improve clinical outcomes and reduce the toxicity caused by ineffective therapies.
As a class of single-stranded small non-coding RNAs (typically 14–25 nucleotides), microRNAs (miRNAs/miRs) suppress protein translation at the post-transcriptional level via translational inhibition or degradation of their target mRNAs, and subsequently affect various biological processes (16–18). miRNA dysregulation has been reported in a range of human malignancies, and identified as an important mechanism that regulates cancer-associated genes and signaling pathways (19–22). Recently, the vital roles of miRNAs in PDAC pathogenesis have been revealed. Certain miRNAs, which are potentially oncogenic, are upregulated in PDAC tissues compared with normal tissues, including miR-10b, miR-17 and miR-21, whereas several potentially tumor-suppressive miRNAs, including miR-26, miR-34a and miR-96, are downregulated (23). For example, the putative onco-miR, miR-155, enhances reactive oxygen species-induced proliferation by targeting forkhead box O (FoxO)3a and GTPase KRas, and also promotes tumor growth via tumor protein p53-inducible nuclear protein 1 in PDAC (24,25). The overexpression of miR-214 decreases the sensitivity of PDAC tumor cells to gemcitabine by inhibiting inhibitor of growth family member 4 (26). Furthermore, the association of certain aberrantly expressed miRNAs with the clinical outcomes of patients with PDAC has been reported in numerous previous studies (27–30). For example, elevated miR-21 expression levels are significantly correlated with worse overall survival and progression-free survival in patients with PDAC (31). Additionally, downregulation of miR-506 leads to increased sphingosine kinase 1 expression, which is also associated with shorter survival in patients with PDAC (32).
Based on the potential of miRNAs to be effective biomarkers, further in-depth analysis of the association between miRNAs and PDAC prognosis is important. Traditional biological research focuses on the specific functions and characteristics of individual genes, transcripts and proteins, which can only explain part of a biological system. Weighted gene co-expression network analysis (WGCNA) is an effective systems biology method used to assign highly co-expressed genes into modules using unsupervised hierarchical clustering and assess the association of modules with clinical features, allowing the identification of key genes in a module for further analysis according to intra-modular connectivity and gene significance (33).
In the present study, WGCNA was used to analyze miRNAs to detect the potential key miRNAs associated with the prognosis of patients with early-stage PDAC that underwent radical resection. These results may produce further useful information for the assessment of PDAC prognosis.
Materials and methods
Data collection and processing
miRNA sequencing data and clinical information, including survival profiles, of patients with PDAC were obtained from The Cancer Genome Atlas (TCGA; cancergenome.nih.gov/). Datasets with the following criteria were used: i) The histological type was pancreatic ductal adenocarcinoma; ii) the pathological stage was stage I or II, according to the standard of American Joint Committee on Cancer (7th edition) (34); iii) patients underwent radical resection, including the Whipple procedure, distal or total pancreatectomy; and iv) survival data were available. Consequently, level three miRNAseq data generated using the Illumina HiSeq platform (Illumina, Inc., San Diego, CA, USA) for 124 patients with PDAC and the corresponding clinical data were available for analysis. The variance in expression of each miRNA in each sample was calculated and ranked using the quartile method. miRNAs with expression variance less than the median variance were removed. Subsequently, the ‘goodSamplesGenes’ function in the R package ‘WGCNA’ (R version 3.3.3) was used to remove miRNAs with verbose <3 for excessive missing values and identification of outlier samples (35). Additionally, the clustering analysis of samples was performed using the ‘hclust’ function (method ‘average’) in R with an appropriate cut-off value. Further approval of the data and analysis in the present study was not required by the TCGA ethics committee. Meanwhile, miRNA sequencing data with relevant clinical data were searched on the Gene Expression Ominbus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) to further validate our results (36).
Weighted gene co-expression network analysis
Based on the filtered miRNA expression data, the scale-free gene modules of co-expression were constructed by WGCNA (35). To ensure the reliability of the co-expression network, hierarchical clustering was performed based on Euclidean distance to detect and remove sample outliers. The Pearson correlation coefficient between all input miRNAs was calculated for converting expression data into correlation matrices. An adequate soft-threshold power that met the scale-free topology criterion was selected for transforming the former correlation matrix into an adjacency matrix, which was subsequently converted into a Topological Overlap Matrix (TOM) using the ‘TOMsimilarity’ function in R (33,37). TOM-based dissimilarity was computed as measure distance, and a miRNA clustering tree (dendrogram) and module colors were obtained. In the clustering dendrogram, the minimum module size and cut height were separately set to 30 and 0.25, respectively; thus, modules below these values were merged into new modules.
Analysis of association between modules and clinical characteristics
The association between modules and clinical characteristics (age, gender, pathological stage, pathological T stage, pathological N stage, histological grade, type of surgery performed, radical resection, number of lymph nodes, radiation therapy, targeted molecular therapy, history of alcohol consumption and smoking history) were estimated by Pearson's correlation tests for the phenotype (clinical characteristics) and module eigengene. P<0.05 was considered to indicate statistical significance (38). The association of individual miRNAs with clinical characteristics was quantified by Gene Significance (GS), while the correlation of the miRNA expression profile with module eigengenes was weighted by Module Membership (MM) (39).
Identification of hub miRNAs and functional annotation
Hub miRNAs are highly connected intra-modular miRNAs that determine the characteristics of the module to a certain extent (40,41). The module eigengene function was used for calculating the eigengene, which represents the whole module miRNA expression level. Subsequently, the ‘signedKME’ function was used to calculate the distance between a gene and a module eigengene. Screening based on GS and MM was used to detect the hub miRNAs in specified modules. The corrected miRNAs with q-weighted P<0.01 were selected as hub miRNAs (35). For visualizing the network that consists of the hub miRNAs in the turquoise module, Cytoscape software (version 3.6.1; http://cytoscape.org) was used, which is an open source software platform used primarily for visualization of biological pathways and molecular interactions (42).
For interpreting the biological function of the hub miRNAs, target genes were predicted using miRWalk 2.0 (zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html), miRDB (www.mirdb.org), TargetScanHuman 7.2 (www.targetscan.org/vert_71/), miRanda (34.236.212.39/microrna/home.do), miRTarBase 7.0 (mirtarbase.mbc.nctu.edu.tw/php/index.php) and PicTar (pictar.mdc-berlin.de) (43–48). Genes common to four of these six databases were selected as target genes and used in further analysis. The list of target genes was then uploaded to the Database For Annotation, Visualization And Integrated Discovery (DAVID) bioinformatics resource (version 6.8; http://david.ncifcrf.gov) for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (49).
Prognostic model construction and survival analysis
After dichotomizing the data around the median expression of each hub miRNAs, the R packages ‘survival’ and ‘survminer’ (package version 0.4.2) (https://cran.r-project.org/web/packages/survminer/index.html) were used for the single hub miRNA survival analysis and constructing the survival curves. Subsequently, the ‘rbsurv’ function in R was applied to develop models that utilized the partial likelihood of the Cox model to select survival-associated miRNAs by employing forward selection, generating a series of gene models. One of these models, which was validated to have significant importance for survival analysis via the ‘ggsurvplot’ function contained in the ‘survminer’ package, was the optimal model (50,51). Meanwhile, the expression of the miRNAs for each sample in this model was visualized with the ‘pheatmap’ package in R (https://cran.r-project.org/web/packages/pheatmap/index.html) (52). For all the survival analysis, overall survival (OS) was determined as survival endpoints, the hazard ratio (HR) was calculated via a Cox regression model and Kaplan-Meier curves were compared by the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Study population and sequencing data
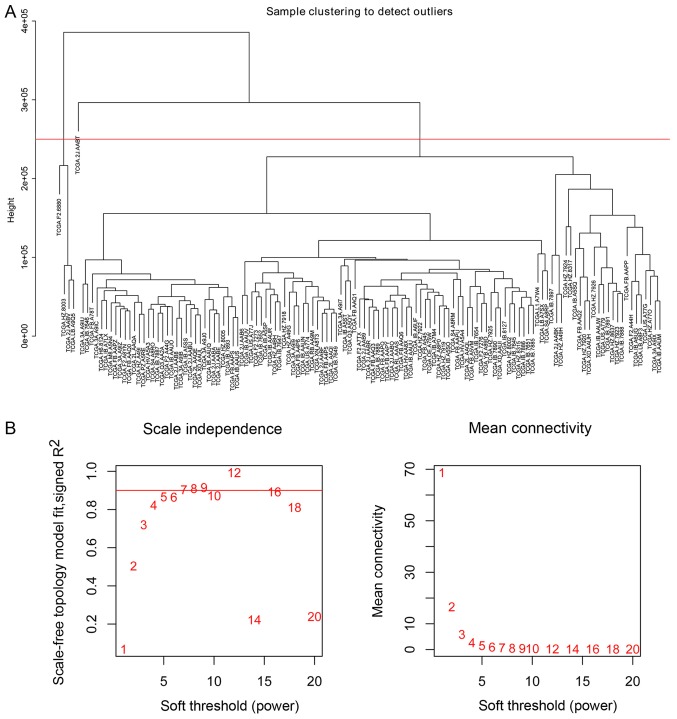
The clinical characteristics with survival profiles of 185 patients with pancreatic adenocarcinoma and a miRNA sequencing dataset from 178 patients were downloaded from TCGA. A total of 129 patients diagnosed with early-stage PDAC who underwent radical resection met the inclusion criteria of the present study (Table I). It should be noted that five patients were removed due to detection as sample outliers by clustering analysis prior to construction of the co-expression network in the subsequent analysis. Using miRNA sequencing data, 523 miRNAs for each sample were filtered into further analysis according to the ranked expression variance. Following testing using the ‘goodSamplesGenes’ function, the expression of these selected 523 miRNAs in 129 patients with PDAC were implemented into hierarchical clustering analysis (Fig. 1A). Consequently, five outlier samples whose leaf node height was significantly higher than other samples and deviated from the main cluster obviously in the dendrogram were identified. To avoid effects on the subsequent analysis, a height cut at a certain value ranged from about 2.2e+05 to 2.9e+05 was utilized to remove these samples automatically. The expression data of 523 miRNAs in 124 patients with PDAC was used for construction of the co-expression network.
Table I.
Clinical characteristics of patients with pancreatic ductal adenocarcinoma (n=124).
| Variable | Number of cases (%) |
|---|---|
| Age (years) | |
| <60 | 40 (32.3) |
| ≥60 | 84 (67.7) |
| Sex | |
| Female | 55 (44.4) |
| Male | 69 (55.6) |
| Pathological stage | |
| I | 10 (8.1) |
| II | 114 (91.9) |
| Pathological T stage | |
| T1/T2 | 18 (14.5) |
| T3 | 106 (85.5) |
| Pathological N stage | |
| N0 | 28 (22.6) |
| N1 | 95 (76.6) |
| NX | 1 (0.8) |
| Histological grade | |
| 1 | 16 (12.9) |
| 2 | 71 (57.2) |
| 3/4 | 37 (29.8) |
| Type of surgery performed | |
| Whipple | 107 (86.3) |
| Distal/total pancreatectomy | 17 (13.7) |
| Residual tumor | |
| R0 | 72 (58.1) |
| R1/R2 | 44 (35.5) |
| RX | 8 (6.4) |
| Number of lymph nodes | |
| 0 | 27 (21.8) |
| ≥1 | 97 (78.2) |
| Radiation therapy | |
| No | 80 (64.5) |
| Yes | 34 (27.4) |
| NA | 10 (8.1) |
| Targeted molecular therapy | |
| No | 34 (27.4) |
| Yes | 61 (49.2) |
| NA | 29 (23.4) |
| Alcohol consumption history | |
| No | 46 (37.1) |
| Yes | 71 (57.3) |
| NA | 7 (5.6) |
| Tobacco smoking history | |
| <3 | 63 (50.8) |
| ≥3 | 38 (30.6) |
| NA | 23 (18.6) |
NA, not available.
Figure 1.
Sample clustering to detect outliers and analysis of network topology for various soft threshold powers. (A) Cluster dendrogram of 129 samples based on their Euclidean distance. The red line represents the cut-off of data filtering in the step of data processing. Five sample outliers were removed. (B) Analysis of the scale-free fit index and the mean connectivity for various soft-thresholding powers (β). The red line represents the cut-off value (R2=0.9).
Co-expression network analysis
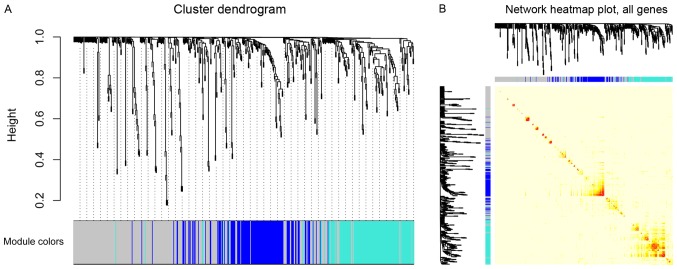
The ‘PickSoftThreshold’ function in the WGCNA package was used to estimate the suitable soft threshold power, which was set as five (Fig. 1B). Three modules were identified by a one-step network construction method using the ‘blockwiseModules’ function in R (minModuleSize, 30; mergeCutHeight, 0.25) containing blue, turquoise and grey modules. Because module identification does not employ prior biological knowledge about the miRNA, the biological meaning of each module is initially unknown and hence the module were assigned a color label (blue, turquoise and grey). Notably, miRNAs that failed to be classified into a module were assigned to the grey module (Fig. 2A). The number of miRNAs in each module was 131 in blue, 131 in turquoise and 261 in grey. The co-expression network was visualized as a TOM plot consisting of a hierarchical clustering dendrogram and TOM matrix (Fig. 2B).
Figure 2.
Clustering dendrogram and heatmap plot of miRNAs. (A) miRNA clustering dendrogram obtained by hierarchical clustering of TOM-based dissimilarity with the corresponding module colors indicated by the color row. Each colored row represents color-coded module which contains a group of highly connected miRNAs. As a result, two co-expression modules were constructed and shown as blue and turquoise. (B) Heatmap plot of the topological overlap matrix. In the heatmap, rows and columns correspond to single miRNAs, light colors represent low topological overlap, while progressively darker orange and red colors represent higher topological overlap. Blocks of darker colors along the diagonal represent the modules. The corresponding miRNA dendrogram and module assignment are shown on the left and top. TOM, Topological Overlap Matrix; miRNA, microRNA.
Association of modules with clinical characteristics
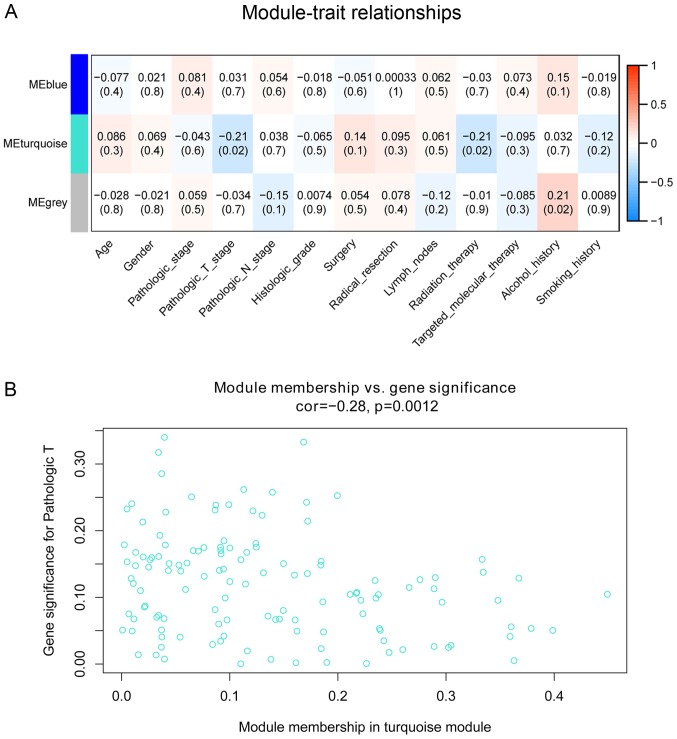
The associations of co-expression network modules with the clinical characteristics of the patients with PDAC are illustrated as heatmaps of module-trait correlation (Fig. 3A). The turquoise module was significantly associated with pathological T stage (cor=−0.21; P=0.02) and radiation therapy (cor=−0.21; P=0.02). A scatterplot of GS vs. MM in the turquoise module was generated. This analysis revealed a highly significant association between the turquoise module and pathological T stage (cor=−0.21; P=0.0012; Fig. 3B); however, radiotherapy was not significantly associated with the turquoise module in this analysis (cor=0.035; P=0.69).
Figure 3.
Association between module and clinical characteristics. (A) Module trait relationships. Each row corresponds to a module eigengene, column to a feature. Each cell contains the corresponding correlation and P-value. The table is color-coded by correlation according to the color legend. The correlations between ME turquoise with pathological T stage (cor=−0.21; P=0.02) and radiation therapy (cor=−0.21; P=0.02) were significant. (B) A scatterplot of Gene Significance for pathological T stage vs. Module Membership in the turquoise module. ME, module eigengene; cor, correlation coefficient.
Hub miRNA detection and functional analysis
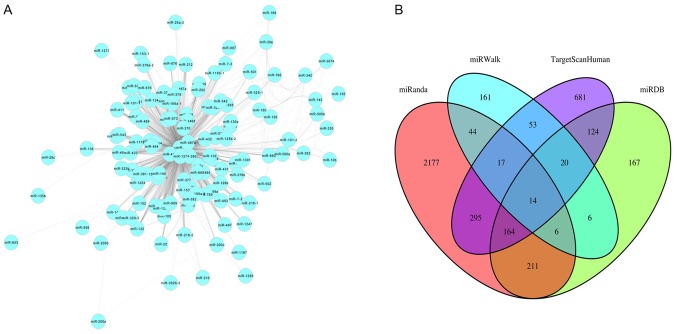
Cytoscape software (version 3.6.1; http://cytoscape.org) was used to visualize the miRNAs in the co-expression network turquoise module based on topological overlap of miRNAs (Fig. 4A). Nodes represent miRNAs and edges represent connectivity. The more edges the node is connected with, the higher intra-module connectivity the node possesses, and the more likely it is to be a hub miRNA. Through function ‘networkScreening’ based on GS and MM, 39 hub miRNAs were identified in the turquoise module (Table II). TargetScanHuman, miRWalk, miRDB, miRanda, miRTarBase and PicTar were used to identify target genes of the miRNAs. Genes common to four of these six databases were selected as target genes. Overlapping target genes for each miRNA from four web tools were screened out using Venn diagrams in order to reduce the false positive rate, such as for miR-140 (Fig. 4B). In total, 404 genes were identified as targeted genes of the 39 hub miRNAs in the turquoise module.
Figure 4.
Hub miRNA detection and functional analysis. (A) Visualization of the turquoise module. The turquoise module assigned into 131 miRNAs and was visualized by Cytoscape software. The turquoise nodes represent miRNAs, the edges represent the connectivity between two unspecified miRs. (B) Venn diagram for miR-140 targets. The overlapping target genes were predicted using four of six web tools (TargetScanHuman, miRWalk, miRDB, miRanda, miRTarBase and PicTar). There were 14 commonly identified genes in the four web tools, which were considered as target genes for miR-140. miR, microRNA.
Table II.
Hub miRs associated with pathological T stage in the turquoise module.
| Module | Hub miRNAs | q-weighted P-value |
|---|---|---|
| Turquoise | miR-376c, miR-379, miR-654, miR-873, miR-889, miR-487b, miR-323, miR-410, miR-204, miR-127, miR-758, miR-487a, miR-432, miR-154, miR-381, miR-431, miR-376a-1, miR-409, miR-377, miR-543, miR-370, miR-433, miR-375, miR-382, miR-1468, miR-551b, miR-539, miR-129-1, miR-1224, miR-129-2, miR-369, miR-1179, miR-496, miR-212, miR-1197, miR-218-2, miR-134, miR-411, miR-140 | <0.01 |
The 39 hub miRNAs were detected using the ‘networkScreening function’ in R based on Gene Significance and Module Membership. miR, microRNA.
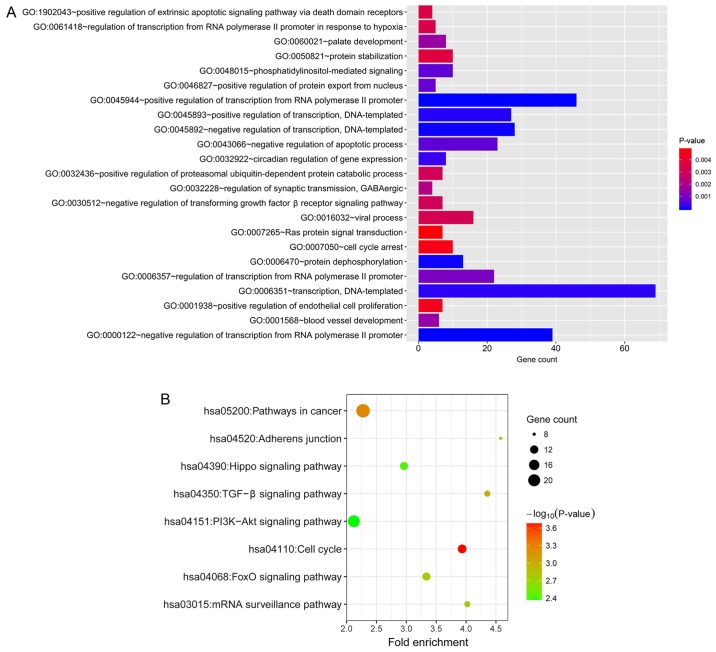
For further insights into the biological relevance of the target genes, GO and KEGG pathway enrichment analysis were performed using the data uploaded to DAVID. The GO biological process that was most enriched was ‘negative regulation of transcription from RNA polymerase II promoter’ (GO:0000122; Fig. 5A). The KEGG pathways that were most enriched were ‘cell cycle’ (hsa04110), ‘pathways in cancer’ (hsa05200), ‘TGF-β signaling pathway’ (hsa04350), ‘FoxO signaling pathway’ (hsa04068), ‘mRNA surveillance pathway’ (hsa03015), ‘adherens junction’ (hsa04520), ‘Hippo signaling pathway’ (hsa04390), ‘PI3K-Akt signaling pathway’ (hsa04151; Fig. 5B).
Figure 5.
GO and KEGG analysis. (A) Significantly enriched GO biological processes of target genes. (B) Significantly enriched KEGG pathways of target genes. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Prognostic model construction and survival analysis
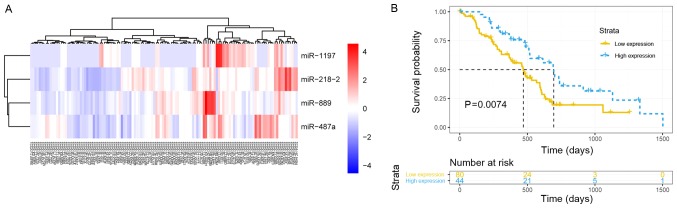
To explore the association of the 39 hub miRNAs with the prognosis of the patients with PDAC and construct a prognostic model, the expression data of hub miRNAs and corresponding survival profiles were analyzed in R software using the ‘rbsurv’ function. A model consisting of four hub miRNAs (miR-1197, miR-218-2, miR-889 and miR-487a) associated with pathological T staging was built and validated by survival analysis. The patients with PDAC were successfully stratified into high and low signature expression groups using the ‘pheatmap’ function (agglomeration method, ward. D2) in R (Fig. 6A). The median overall survival was 695 vs. 470 days in the high and low miR signature expression groups, respectively (P=0.0074; Fig. 6B). The results demonstrated that the higher expression of the signature, the longer the OS of the patients with early-stage PDAC who underwent radical resection.
Figure 6.
Identification of a four-miR signature. (A) Heatmap of the four-miRNA signature expression. Patients with early-stage pancreatic ductal adenocarcinoma were stratified into low- and high-expression groups by unsupervised hierarchical clustering. Blue represents low expression values and red represents high expression values. (B) Survival curve displaying the difference of survival rate between the low- and high-expression groups divided by the expression of the four-miRNA signature. miR, microRNA.
Discussion
As one of the most fatal cancers worldwide, treatment of PDAC faces therapeutic challenges due to an increasing incidence rate and insufficient diagnosis at the early stage (3). Although patients who receive radical resection have a relatively good clinical outcome, <20% of patients are eligible for surgery (53). Furthermore, low sensitivity to adjuvant therapy and substantial toxicity are major problems (4,54). Identification of reliable biomarkers to screen for patients who will benefit from surgery is ongoing (55). Advancements in algorithms have allowed the use of systems biology methods to explore the mechanisms of tumorigenesis and development in depth (56).
In the present study, WGCNA was performed using miRNA expression data and corresponding clinical information of 124 patients with PDAC. An association between the turquoise module and pathological T stage was detected in the co-expression network. The pathological T stage indicates whether the adjacent tissues have been invaded by the tumor and what the original tumor size was, and was previously identified as an independent prognostic factor for patients with PDAC who underwent resection in previous studies (57,58); however, the mechanisms by which miRNAs affect the pathological T stage remains unknown. In the present study, 39 hub miRNAs that represented the features of the turquoise module were selected to predict target genes for enrichment analysis. In GO enrichment analysis, ‘negative regulation of transcription from RNA polymerase II promoter’ was the most significantly enriched biological process, while the other enriched processes included ‘regulation of transcription’, ‘negative regulation of apoptotic process’, ‘TGF-β receptor signaling pathway’, ‘positive regulation of endothelial cell proliferation’, ‘cell cycle arrest’ and ‘Ras protein signal transduction’.
The proliferation and growth of tumor cells is regulated by various factors, including abnormalities in growth promotion and inhibitory signaling, increased telomerase activity, reduced apoptosis, continuous tumor angiogenesis and self-renewal of cancer stem cells (59). The functions of the majority of oncogenes and tumor suppressor genes are associated with the cell cycle. Gene mutations may cause dysregulation of the cell cycle, resulting in uncontrolled growth via excessive proliferation and decreased apoptosis (60). In addition, tumor growth requires delivery of oxygen and nutrients via blood vessels. Vascular endothelial growth factor (VEGF) is secreted by tumor cells and acts directly on receptors on the surface of endothelial cells to induce their proliferation and activation to form new blood vessels (61). Transforming growth factor-β (TGF-β) is a multifunctional cytokine. In early-stage pancreatic cancer, cell growth is inhibited by activation of TGF-β receptor signaling, which increases Smad3 phosphorylation and nuclear translocation; in advanced tumors, the activation of TGF-β signaling can promote tumor vascularization and metastasis by targeting VEGFA, and increasing fibrosis in the pancreatic tissue (62–64). Ras-Raf-MEK 1-mitogen activated protein kinase signaling, a receptor tyrosine kinase-mediated signaling pathway, serves an important role in the regulation of cell proliferation and survival. As an upstream mediator of this pathway, abnormal activation of Ras signaling may lead to unrestrained cell proliferation, and is associated with poor prognosis in PDAC (65).
In KEGG pathway analysis, the most significantly enriched pathway was ‘cell cycle’, and other significant cancer-associated pathways included ‘TGF-β signaling pathway’, ‘FoxO signaling pathway’, ‘mRNA surveillance pathway’, ‘adherens junction’, ‘Hippo signaling pathway’ and ‘PI3K-Akt signaling pathway’. It is well established that cell cycle disorder can cause tumorigenesis (66). Additionally, TGF-β signaling promotes cell proliferation, angiogenesis and the self-renewal of tumor stem cells. FoxOs, as a subfamily of the fork head transcription factor family, have confirmed roles in tumor cell differentiation, proliferation, apoptosis, DNA repair and damage, and also act as mediators of oxidative stress (67). The translation of mRNA transcripts into proteins is an essential part of the central dogma of molecular biology. However, fidelity errors are occasionally present in the mRNA molecules, which may result in errors in the translated protein. The mRNA surveillance pathway is a quality control mechanism that detects and degrades abnormal mRNA molecules (68).
Adherens junctions are created by homotypic interactions between the extracellular domains of cadherin proteins to join epithelial cells together, and alterations in cell adhesion through adherens junctions may mediate tumor cell invasion and migration in pancreatic cancer (57). Hippo signaling is a highly conserved serine/threonine kinase cascade involved in the control of cell proliferation and apoptosis to regulate organ size (69). Phosphoinositide 3-kinase (PI3K)-protein kinase B (Akt) signaling, which is one of the most commonly dysregulated signaling pathways in cancer, can be triggered to cause continuous activation of Akt, which ultimately inhibits tumor cell apoptosis. In PDAC, mechanistic target of rapamycin kinase (mTOR), a key kinase downstream of PI3K-Akt, plays key roles in the renewal of cancer stem cells, and resistance to radiotherapy or chemotherapy (70).
Based on the current enrichment analysis, 39 hub miRNAs in the turquoise module were identified to be enriched in the biological processes and pathways involved in tumor development and progression, which may be involved in the negative correlation detected between miRNA expression and pathological T stage.
By utilizing prognostic biomarkers, PDAC patients could be divided into different subgroups and received individualized adjuvant therapy, which might improve clinical outcomes (71). However, a single miRNA is less specific and sensitive than a panel of miRNAs (72,73). With data on miRNAs as prognostic biomarkers accumulating, inconsistencies among study results have arisen. Though factors might ascribe to discrepancies of lab protocols, measurement platforms, samples size and ethnicity, the credibility of the predictive value of a single miRNA is dubious. Besides, focusing on the target protein of a single miRNA typically results in researchers overlooking the pathways involved in complex mechanisms such as drug metabolism and treatment resistance, which could be simultaneously affected by certain prognosis-associated miRNAs (74). Furthermore, some studies have proven that it is more effective to predict prognosis with a panel than a single miRNA by statistical analysis (73,75).
To explore the association between the 39 hub miRNAs and PDAC prognosis, a predictive model was constructed to identify a prognostic signature composed of miR-218-2, miR-889, miR-487a and miR-1197. The previous study by Guan et al (76) reported that miR-218 acts as an inhibitor of tumor angiogenesis by targeting rapamycin-insensitive companion of mTOR, an mTOR component, in prostate cancer (76). In a study by Zhu et al (77), miR-218-5p was demonstrated to suppress the cell proliferation and migration of non-small-cell lung cancer via epidermal growth factor receptor (77). Although miR-218, which is transcribed from two loci located at chromosome 5q35.1 (miR-218-2) and 4p15.31 (miR-218-1), has been considered as an anti-oncogene in various types of cancer, miR-218-2 has opposing roles in different types of cancer. In glioblastoma, miR-218-2 promotes cell proliferation, migration and invasion by targeting cell division cycle 27 (78); whereas in thyroid cancer, co-expression of miR-218-2 with its host gene, slit guidance ligand 3, exerts a tumor-suppressive effect (79). Xie et al (80) reported that miR-889 downregulation by histone deacetylase enhances the cytotoxicity of natural killer cells in hepatocellular carcinoma. Xu et al (81) observed that miR-889 promotes the transformation of esophageal cancer cells from the G1 to S phase; furthermore, miR-889 may induce epithelial-to-mesenchymal transition (EMT) via the Akt-mTOR and Wnt pathways (81). In a study of advanced colorectal cancer, Molina-Pinelo et al (82) reported that the reduced expression of miR-889 is significantly associated with improved overall survival and progression-free survival in patients who received chemotherapy (82). Chang et al (83) revealed that miR-487a has the ability to promote hepatocellular carcinoma metastasis and proliferation by targeting phosphoinositide-3-kinase regulatory subunit 1 and sprouty-related EVH1 domain-containing 2 mRNA (83). Additionally, miR-487a was demonstrated to induce cell invasion and migration of lung adenocarcinoma by regulating migration-associated genes AXL receptor tyrosine kinase and ovo-like zinc finger 2 (84). In breast cancer cells, EMT induced by TGF-β1 may be restricted by overexpression of the membrane-associated guanylate kinase WW and PDZ domain-containing 2 gene, which is regulated by miR-487a (85). Only a limited number of studies have investigated miR-1197 (86,87). The effects of these miRNAs on tumor development and progression are inconsistent with each other and vary among different types of cancer (78,79). The role of each miRNA in biological processes and the expression level should be considered when interpreting study results among varying cancer types. Thus, comprehensive consideration and further study are required.
Certain limitations of the present study should be considered. As the results were based on single-data source, Gene Expression Omnibus datasets were searched for further validation. Unfortunately, no data with full survival profiles were available for analysis. Additionally, the conclusions made are based on bioinformatics data only, lacking of independent external validation with experimental and clinical data. Although the accuracy of this bioinformatics method has been proven, a realistic assessment of the performance of the panel will be warranted in the future.
Despite these limitations, the present study identified 39 hub miRNAs associated with pathological T stage via WGCNA. A four-miRNA signature associated with pathological T stage was identified and validated for association with the prognosis of PDAC, which may serve as a novel biomarker for prognosis prediction and to improve clinical outcome in patients with PDAC. The prognostic signature consisted of four hub miRNAs (miR-1197, miR-218-2, miR-889 and miR-487a) associated with pathological T stage, and expression of the signature miRs was significantly associated with survival. These findings suggest that the increased expression of the signature in early-stage PDAC patients who underwent radical resection surgery may be an indicator of improved overall survival.
Acknowledgements
Not applicable.
Funding
The present study was supported by People's Liberation Army General Hospital Medical Big Data R&D Project (grant no. 2017MBD-013).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LY and JW performed data analyses and wrote the manuscript. FZ, JZ, HT and XZ contributed significantly in data analyses and manuscript revision. YH conceived and designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kamposioras K, Papadopoulos V. Pancreatic adenocarcinoma treatment. Anticipating better results. https://www.esmo.org/Career-Development/Young-Oncologists-Corner/Journal-Club/Pancreatic-Adenocarcinoma-Treatment.-Anticipating-Better-Results. [Nov 27;2012 ];European Society for Medical Oncology, Lugano. 2012
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 5.Bakkevold KE, Arnesjø B, Dahl O, Kambestad B. Adjuvant combination chemotherapy (AMF) following radical resection of carcinoma of the pancreas and papilla of Vater-results of a controlled, prospective, randomised multicentre study. Eur J Cancer. 1993;29:698–703. doi: 10.1016/S0959-8049(05)80349-1. [DOI] [PubMed] [Google Scholar]
- 6.Kuhlmann KF, de Castro SM, Wesseling JG, ten Kate FJ, Offerhaus GJ, Busch OR, van Gulik TM, Obertop H, Gouma DJ. Surgical treatment of pancreatic adenocarcinoma: Actual survival and prognostic factors in 343 patients. Eur J Cancer. 2004;40:549–558. doi: 10.1016/j.ejca.2003.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Tsao JI, Rossi RL, Lowell JA. Pylorus-preserving pancreatoduodenectomy. Is it an adequate cancer operation. Arch Surg. 1994;129:405–405. doi: 10.1001/archsurg.1994.01420280081010. [DOI] [PubMed] [Google Scholar]
- 8.Yeo CJ, Cameron JL, Lillemoe KD, Sitzmann JV, Hruban RH, Goodman SN, Dooley WC, Coleman J, Pitt HA. Pancreaticoduodenectomy for cancer of the head of the pancreas. 201 patients. Ann Surg. 1995;221:721–731. doi: 10.1097/00000658-199506000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeo CJ, Cameron JL, Sohn TA, Lillemoe KD, Pitt HA, Talamini MA, Hruban RH, Ord SE, Sauter PK, Coleman J, et al. Six hundred fifty consecutive pancreaticoduodenectomies in the 1990s: Pathology, complications, and outcomes. Ann Surg. 1997;226:248–257. doi: 10.1097/00000658-199709000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millikan KW, Deziel DJ, Silverstein JC, Kanjo TM, Christein JD, Doolas A, Prinz RA. Prognostic factors associated with resectable adenocarcinoma of the head of the pancreas. Am Surg. 1999;65:618–623. [PubMed] [Google Scholar]
- 11.Benassai G, Mastrorilli M, Quarto G, Cappiello A, Giani U, Mosella G. Survival after pancreaticoduodenectomy for ductal adenocarcinoma of the head of the pancreas. Chir Ital. 2000;52:263–270. [PubMed] [Google Scholar]
- 12.Dalton RR, Sarr MG, van Heerden JA, Colby TV. Carcinoma of the body and tail of the pancreas: Is curative resection justified. Surgery. 1992;111:489–494. [PubMed] [Google Scholar]
- 13.Johnson CD, Schwall G, Flechtenmacher J, Trede M. Resection for adenocarcinoma of the body and tail of the pancreas. Br J Surg. 2010;80:1177–1179. doi: 10.1002/bjs.1800800937. [DOI] [PubMed] [Google Scholar]
- 14.Khorana AA, Mangu PB, Katz MHG. Potentially curable pancreatic cancer: American society of clinical oncology clinical practice guideline. J Oncol Prac. 2017;13:388–391. doi: 10.1200/JOP.2017.023044. [DOI] [PubMed] [Google Scholar]
- 15.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 16.Suárez Y, Sessa WC. microRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP, Chen CZ. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nature Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 19.Mcguire A, Brown JAL, Kerin MJ. Metastatic breast cancer: The potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145–155. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maura F, Cutrona G, Mosca L, Matis S, Lionetti M, Fabris S, Agnelli L, Colombo M, Massucco C, Ferracin M, et al. Association between gene and miRNA expression profiles and stereotyped subset #4 B-cell receptor in chronic lymphocytic leukemia. Leuk Lymphoma. 2015;56:3150–3158. doi: 10.3109/10428194.2015.1028051. [DOI] [PubMed] [Google Scholar]
- 21.Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen J, Zhang Y, Lai P, Fan X, Zhou X, et al. miR-483-5p promotes invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1 and ALCAM. Cancer Res. 2014;74:3031–3042. doi: 10.1158/0008-5472.CAN-13-2193. [DOI] [PubMed] [Google Scholar]
- 22.Singh S, Chitkara D, Kumar V, Behrman SW, Mahato RI. miRNA profiling in pancreatic cancer and restoration of chemosensitivity. Cancer Lett. 2013;334:211–220. doi: 10.1016/j.canlet.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Wald P, Liu XS, Pettit C, Dillhoff M, Manilchuk A, Schmidt C, Wuthrick E, Chen W, Williams TM. Prognostic value of microRNA expression levels in pancreatic adenocarcinoma: A review of the literature. Oncotarget. 2017;8:73345–73361. doi: 10.18632/oncotarget.20277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Zhu CF, Ma MZ, Chen G, Song M, Zeng ZL, Lu WH, Yang J, Wen S, Chiao PJ, et al. Micro-RNA-155 is induced by K-Ras oncogenic signal and promotes ROS stress in pancreatic cancer. Oncotarget. 2015;6:21148–21158. doi: 10.18632/oncotarget.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giulietti M, Occhipinti G, Principato G, Piva F. Identification of candidate miRNA biomarkers for pancreatic ductal adenocarcinoma by weighted gene co-expression network analysis. Cell Oncol (Dordr) 2017;40:1–12. doi: 10.1007/s13402-017-0315-y. [DOI] [PubMed] [Google Scholar]
- 28.Dhayat SA, Abdeen B, Köhler G, Senninger N, Haier J, Mardin WA. MicroRNA-100 and microRNA-21 as markers of survival and chemotherapy response in pancreatic ductal adenocarcinoma UICC stage II. Clin Epigenetics. 2015;7:132. doi: 10.1186/s13148-015-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH, et al. Identification of MicroRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5:e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang C, Yu XJ, Guo XZ, Sun MH, Wang Z, Song Y, Ni QX, Li HY, Mukaida N, Li YY. MicroRNA-33a-mediated downregulation of Pim-3 kinase expression renders human pancreatic cancer cells sensitivity to gemcitabine. Oncotarget. 2015;6:14440–14455. doi: 10.18632/oncotarget.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan K, Cunningham D, Peckitt C, Barton S, Tait D, Hawkins M, Watkins D, Starling N, Rao S, Begum R, et al. miR-21 expression and clinical outcome in locally advanced pancreatic cancer: Exploratory analysis of the pancreatic cancer Erbitux, radiotherapy and UFT (PERU) trial. Oncotarget. 2016;7:12672–12681. doi: 10.18632/oncotarget.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Wu H, Li W, Yin L, Guo S, Xu X, Ouyang Y, Zhao Z, Liu S, Tian Y, et al. Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene. 2016;35:5501–5514. doi: 10.1038/onc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]
- 34.Cuccurullo V. AJCC cancer staging handbook: From the AJCC cancer staging manual (7th edition) European J Nuclear Med Mol Imag. 2011;38:408. doi: 10.1007/s00259-010-1693-9. [DOI] [Google Scholar]
- 35.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams S. Pearson's correlation coefficient. N Z Med J. 1996;109:38. [PubMed] [Google Scholar]
- 39.Shi K, Bing ZT, Cao GQ, Guo L, Cao YN, Jiang HO, Zhang MX. Identify the signature genes for diagnose of uveal melanoma by weight gene co-expression network analysis. Int J Ophthalmol. 2015;8:269–274. doi: 10.3980/j.issn.2222-3959.2015.02.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 41.Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23:375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. ELife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doron B, Manda W, Aaron G, Marks DS, Chris S. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36:149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 49.Jiao X, Sherman BT, Huang DW, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freije WA, Castrovargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 51.Cho HJ, Yu A, Kim S, Kang J, Hong S-M. Robust likelihood-based survival modeling with microarray sata. J Stat Softw. 2009;29:1–16. doi: 10.18637/jss.v029.i01. [DOI] [Google Scholar]
- 52.Kolde R. Pheatmap: Pretty Heatmaps. https://cran.r-project.org/web/packages/pheatmap/index.html. [May 19;2018 ]; Version 1.0.10.
- 53.Bukki J. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2139–2140. doi: 10.1056/NEJMc1412266. [DOI] [PubMed] [Google Scholar]
- 54.Guo S, Fesler A, Wang H, Ju J. microRNA based prognostic biomarkers in pancreatic Cancer. Biomark Res. 2018;6:18. doi: 10.1186/s40364-018-0131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu IS, Cheung WY. A Contemporary review of the treatment landscape and the role of predictive and prognostic biomarkers in pancreatic adenocarcinoma. Can J Gastroenterol Hepatol. 2018;2018:1863535. doi: 10.1155/2018/1863535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Li YX, Li YY. Differential rebgulatory analysis based on coexpression network in cancer research. Biomed Res Int. 2016;2016:4241293. doi: 10.1155/2016/4241293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiang KC, Yeh CN, Ueng SH, Hsu JT, Yeh TS, Jan YY, Hwang TL, Chen MF. Clinicodemographic aspect of resectable pancreatic cancer and prognostic factors for resectable cancer. World J Surg Oncol. 2012;10:77. doi: 10.1186/1477-7819-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto T, Yagi S, Kinoshita H, Sakamoto Y, Okada K, Uryuhara K, Morimoto T, Kaihara S, Hosotani R. Long-term survival after resection of pancreatic cancer: A single-center retrospective analysis. World J Gastroenterol. 2015;21:262–268. doi: 10.3748/wjg.v21.i1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanahan D, Robert RA. Weinberg: Hallmarks of cancer: The next generation. Cell. 2011;144:644–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 60.Ho A, Dowdy SF. Regulation of G(1) cell-cycle progression by oncogenes and tumor suppressor genes. Curr Opin Genet Dev. 2002;12:47–52. doi: 10.1016/S0959-437X(01)00263-5. [DOI] [PubMed] [Google Scholar]
- 61.Holmes K, Roberts OL, Thomas AM, Cross MJ. Vascular endothelial growth factor receptor-2: Structure, function, intracellular signalling and therapeutic inhibition. Cell Signal. 2007;19:2003–2012. doi: 10.1016/j.cellsig.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 62.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H, Liu C, Kong Y, Huang H, Wang C, Zhang H. TGFβ signaling in pancreatic ductal adenocarcinoma. Tumor Biol. 2015;36:1613–1618. doi: 10.1007/s13277-014-2757-4. [DOI] [PubMed] [Google Scholar]
- 64.Katz LH, Likhter M, Jogunoori W, Belkin M, Ohshiro K, Mishra L. TGF-β signaling in liver and gastrointestinal cancers. Cancer Lett. 2016;379:166–172. doi: 10.1016/j.canlet.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonckheere N, Vasseur R, Van SI. The cornerstone K-RAS mutation in pancreatic adenocarcinoma: From cell signaling network, target genes, biological processes to therapeutic targeting. Crit Rev Oncol Hematol. 2017;111:7–19. doi: 10.1016/j.critrevonc.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 66.Mikhail V. Blagosklonny: Cell immortality and hallmarks of cancer. Cell Cycle. 2003;2:296–299. [PubMed] [Google Scholar]
- 67.Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. 2017;13:815–827. doi: 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore MJ. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 69.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/S0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 70.Dogan F, Biray AC. Correlation between telomerase and mTOR pathway in cancer stem cells. Gene. 2018;641:235–239. doi: 10.1016/j.gene.2017.09.072. [DOI] [PubMed] [Google Scholar]
- 71.Frampton AE, Krell J, Jamieson NB, Gall TMH, Giovannetti E, Funel N, Mato Prado M, Krell D, Habib NA, Castellano L, et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: A meta-analysis. Eur J Cancer. 2015;51:1389–1404. doi: 10.1016/j.ejca.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Zhou X, Wang X, Huang Z, Xu L, Zhu W, Liu P. An ER-associated miRNA signature predicts prognosis in ER-positive breast cancer. J Exp Clin Cancer Res. 2014;33:94. doi: 10.1186/s13046-014-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 74.Zhou X, Huang Z, Xu L, Zhu M, Zhang L, Zhang H, Wang X, Li H, Zhu W, Shu Y, Liu P. A panel of 13-miRNA signature as a potential biomarker for predicting survival in pancreatic cancer. Oncotarget. 2016;7:69616–69624. doi: 10.18632/oncotarget.11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155: 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 76.Guan B, Wu K, Zeng J, Xu S, Mu L, Yang G, Wang K, Ma Z, Tian J, Shi Q, et al. Tumor-suppressive microRNA-218 inhibits tumor angiogenesis via targeting the mTOR component RICTOR in prostate cancer. Oncotarget. 2017;8:8162–8172. doi: 10.18632/oncotarget.14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu K, Ding H, Wang W, Liao Z, Fu Z, Hong Y, Zhou Y, Zhang CY, Chen X. Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget. 2016;7:28075–28085. doi: 10.18632/oncotarget.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feng Z, Zhang L, Zhou J, Zhou S, Li L, Guo X, Feng G, Ma Z, Huang W, Huang F. mir-218-2 promotes glioblastomas growth, invasion and drug resistance by targeting CDC27. Oncotarget. 2017;8:6304–6318. doi: 10.18632/oncotarget.13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guan H, Wei G, Wu J, Fang D, Liao Z, Xiao H, Li M, Li Y. Down-regulation of miR-218-2 and its host gene SLIT3 cooperate to promote invasion and progression of thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1334–E1344. doi: 10.1210/jc.2013-1053. [DOI] [PubMed] [Google Scholar]
- 80.Xie H, Zhang Q, Zhou H, Zhou J, Zhang J, Jiang Y, Wang J, Meng X, Zeng L, Jiang X. microRNA-889 is downregulated by histone deacetylase inhibitors and confers resistance to natural killer cytotoxicity in hepatocellular carcinoma cells. Cytotechnology. 2018;70:513–521. doi: 10.1007/s10616-017-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu Y, He J, Wang Y, Zhu X, Pan Q, Xie Q, Sun F. miR-889 promotes proliferation of esophageal squamous cell carcinomas through DAB2IP. FEBS Lett. 2015;589:1127–1135. doi: 10.1016/j.febslet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 82.Molina-Pinelo S, Carnero A, Rivera F, Estevez-Garcia P, Bozada JM, Limon ML, Benavent M, Gomez J, Pastor MD, Chaves M, et al. MiR-107 and miR-99a-3p predict chemotherapy response in patients with advanced colorectal cancer. BMC Cancer. 2014;14:656. doi: 10.1186/1471-2407-14-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang RM, Xiao S, Lei X, Yang H, Fang F, Yang LY. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. 2016;23:2593–2604. doi: 10.1158/1078-0432.CCR-16-0851. [DOI] [PubMed] [Google Scholar]
- 84.Gonzalez-Vallinas M, Rodriguez-Paredes M, Albrecht M, Sticht C, Stichel D, Gutekunst J, Pitea A, Sass S, Sánchez-Rivera FJ, Bermejo JL, et al. Epigenetically regulated chromosome 14q32 miRNA cluster induces metastasis and predicts poor prognosis in lung adenocarcinoma patients. Mol Cancer Res. 2018;16:390–402. doi: 10.1158/1541-7786.MCR-17-0334. [DOI] [PubMed] [Google Scholar]
- 85.Ma M, He M, Jiang Q, Yan Y, Guan S, Zhang J, Yu Z, Chen Q, Sun M, Yao W, et al. MiR-487a promotes TGF-β1-induced EMT, the migration and invasion of breast cancer cells by directly targeting MAGI2. Int J Biol Sci. 2016;12:397–408. doi: 10.7150/ijbs.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stylli SS, Adamides AA, Koldej RM, Luwor RB, Ritchie DS, Ziogas J, Kaye AH. miRNA expression profiling of cerebrospinal fluid in patients with aneurysmal subarachnoid hemorrhage. J Neurosurg. 2017;126:1131–1139. doi: 10.3171/2016.1.JNS151454. [DOI] [PubMed] [Google Scholar]
- 87.Fisher AJ, Cipolla E, Varre A, Gu H, Mickler EA, Vittal R. Potential mechanisms underlying TGF-β-mediated complement activation in lung fibrosis. Cell Mol Med Open Access. 2017;3:14. doi: 10.21767/2573-5365.100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.