Abstract
The adenosine 5′-triphosphate binding cassette subfamily B member (ABCB)11 gene is involved in bile transport, and mutations in this gene are associated with cholestasis and cholelithiasis. Therefore, the aim of the present study was to investigate the association between ABCB11 gene mutation and primary intrahepatic stone (PIS)s and to investigate the mechanism through which ABCB11 gene mutations affect the expression of the corresponding protein. Mutations of the ABCB11 gene in 443 PIS patients and 560 healthy participants were detected by exon sequencing. The expression levels of ABCB11 mRNA and bile salt export pump (BSEP) protein in the liver tissues of patients with PISs were measured by quantitative polymerase chain reaction and western blot analysis. The mutant plasmids constructed by site-directed mutagenesis of the human BSEP gene were transfected into human embryonic kidney 293 (293) cells and Madin-Darby canine kidney (MDCK) cells, and the expression and distribution of rs118109635 of BSEP was measured. There were two significant mutations in the ABCB11 gene of the PIS patients compared with the healthy population; a missense mutation, rs118109635 (P=0.025), and a synonymous mutation, rs497692 (P=0.006). The two mutations were associated with the occurrence of preoperative jaundice (P=0.026, and P=0.011, respectively). The expression levels of BSEP in PIS patients with the missense mutation rs118109635 was decreased, whereas its mRNA expression levels remained unchanged. In PIS patients with the synonymous mutation rs497692, the expression levels of ABCB11 were decreased at both the mRNA and protein level. It was also found that mutation A865V reduced the expression levels of BSEP in 293 cells at the cellular level; its distribution in MDCK cell membranes was decreased, whereas its mRNA levels remained unchanged. The mutated loci at rs118109635 and rs497692 of the ABCB11 gene were correlated with PISs, causing a decreased expression of BSEP and reduced distribution of the protein in the cell membrane. Therefore, mutations at rs118109635 and rs497692 of the ABCB11 gene may be risk factors for PISs.
Keywords: primary intrahepatic stone, adenosine 5′-triphosphate binding cassette subfamily B member 11, mutation, bile salt export pump, expression
Introduction
Primary intrahepatic stone (PIS) are rare among patients in Western countries (1,2). As reported in the literature, most PIS patients are Asian, and there is a high incidence of the disease in East Asia (3,4). Currently, PISs remain a common biliary system disease in Southwest China (5). Approximately 200–250 surgical procedures for PISs are performed in the Southwest Hospital, Third Military Medical University (Chongqing, China), each year.
Relapse and persistent progress of PISs to an advanced stage may result in biliary cirrhosis or intrahepatic cholangiocarcinoma (6), two of the most important causes of mortality due to benign biliary tract disease. At present, the etiology of PISs has not been fully elucidated; it is generally believed that chronic inflammation of the bile duct and bile stasis promote the formation of gallstones and that other factors such as bacterial infection, malnutrition and living environment may also be involved (7–9). The distribution of PIS cases in Eastern and Western countries and in the northern and southern parts of China differs notably, reflecting the strong racial and regional characteristics of PIS (10). Therefore, the possible role of genetic background in the pathogenesis of PISs is worthy of further investigation.
The protein encoded by the ATP binding cassette subfamily B member (ABCB)11 gene is known as the bile salt export pump (BSEP). Its primary function is to transport bile acid from the cell cavity to the bile duct and to facilitate cholesterol dissolution and inhibit its supersaturated crystallization (11–13). In a previous study of hepatolithiasis, it was found that the mRNA and protein expression levels of sodium/taurine co-aligners and cholesterol 7α hydroxylase, which are associated with bile salt and cholesterol metabolism, were significantly lower in hepatolithiasis patients than in normal controls (14). Therefore, this phenomenon needs to be investigated further.
It has been demonstrated that mutation of the ABCB11 gene is closely associated with cholestasis, including benign recurrent intrahepatic cholestasis 2 (15,16), progressive familial intrahepatic cholestasis 2 (17–19), and intrahepatic cholestasis of pregnancy (20–22), and bile duct stones often form in patients with BSEP deletion syndrome caused by ABCB11 gene mutations (23). In our previous study (5), two missense (rs2287617 and rs2287622) and one synonymous (rs3815675) mutations were detected in exons 9, 13 and 4, respectively. However, the distribution of allele frequencies for these three mutations was not significantly different between patients and controls. Conversely, one missense mutation (rs118109635) and two synonymous mutations (rs497692 and rs2287616) were also detected in exons 21, 24 and 9, respectively. The distribution of these allele frequencies were significantly different in PIS patients compared with those in the healthy group. The two mutations (rs118109635 and rs497692) in ABCB11 as well as decreased expression of its encoded protein BSEP, were closely associated with the recurrence of cholangitis and the level of jaundice of the PIS patients (5). The aim of the present study was to further verify the association between ABCB11 gene mutations and clinical morbidity in PIS patients on the basis of an expanded sample size. In addition, the present study focused on the observation of the effects of ABCB11 gene mutations on BSEP expression and on its distribution in the cell membrane. The mRNA and protein expression levels of the ABCB11 gene in the liver tissues of PIS patients were determined using quantitative polymerase chain reaction (PCR) and western blot analysis. To further investigate the effect of these mutations on the protein's expression and distribution, a wild-type plasmid containing all open reading frames of the human ABCB11 gene was constructed, and mutant plasmids were subsequently constructed by site-directed mutagenesis. After transfection of 293 cells with the mutant plasmids, changes in the expression of ABCB11 mRNA and cell membrane BSEP were detected. Subsequently, based on the polar characteristics of Madin-Darby canine kidney (MDCK) cells, the effect of the ABCB11 gene mutation on the distribution of BSEP at the cellular level was analyzed using immunofluorescence and laser confocal scanning microscopy.
Materials and methods
Clinical samples
A total of 443 patients (male, 134; female, 309; age range 17–81 years; male/female, 0.434) with PISs treated in the Southwest Hospital, Third Military Medical University (Chongqing, China) from December 2012 to May 2016 were included in the present study. Furthermore, 560 healthy individuals (male 245; female 315; age range 16–78 years; male/female, 0.778) were selected from the physical examination center of the same hospital as the control group. The inclusion criteria of the experimental group were confirmed diagnosis of PISs by imaging study; no prior liver resection, no history of chronic liver disease, no hepatitis A, B, or C virus or cytomegalovirus infection, no gallstones, and no tumors. The inclusion criteria of the control group were confirmed absence of PISs by imaging, no history of chronic liver disease, no hepatitis A, B, or C virus or cytomegalovirus infection, and no gallstones. Five milliliters of peripheral blood were collected from each of the above PIS patients and healthy participants, and the lymphocytes were immediately separated using human peripheral lymphocyte separation medium (Tianjin Hao Yang Biological Manufacturing Co., Ltd., Tianjin, China; http://www.tbdscience.com). The obtained samples were stored in a −80°C freezer.
Clinical data, including information on sex, age, height, body weight, indicators in the preoperative liver function test, preoperative serum jaundice, recurrent cholangitis, and stone type were collected (Tables I and II). Stone removal of the PIS patients was confirmed by abdominal ultrasound, abdominal CT, cholangiography and magnetic resonance imaging. Postoperatively, the patients were followed up every three months, and a stone found at follow-up was considered as a recurrence. The experiments were approved by the Ethics Committee of the Southwest Hospital of the Third Military Medical University of China. Written informed consent was obtained from all participants.
Table I.
Clinical data of patients with primary intrahepatic stones.
| Parameters | Patients, n | Median (range) |
|---|---|---|
| Sex, male/female | 134/309 | |
| Age, male/female (years) | 134/309 | 52 (17–81)/49 (17–78) |
| Height, male/female (cm) | 134/309 | 167 (155–187)/155 (144–176) |
| Weight, male/female (kg) | 134/309 | 60 (41–93)/51 (36–80) |
| Glutamic-pyruvic transaminase (IU/l) | 443 | 39.0 (4.9–587.0) |
| Glutamic oxalacetic transaminase (IU/l) | 443 | 38.0 (10.0–621.0) |
| Glutamyl transpeptidase (IU/l) | 443 | 165.0 (7.0–1968.0) |
| Alkaline phosphatase (IU/l) | 443 | 167.0 (31.0–1527.0) |
| Total protein (g/l) | 443 | 71.2 (36.2–87.2) |
| Albumin (g/l) | 443 | 40.0 (20.4–60.0) |
| Alobulin (g/l) | 443 | 31.2 (14.4–50.0) |
| Total bilirubin (µmol/l) | 443 | 16.4 (5.4–460.4) |
| Direct bilirubin (µmol/l) | 443 | 3.8 (0.4–271.9) |
| Indirect bilirubin (µmol/l) | 443 | 12.1 (3.7–196.6) |
| Total bile acid (µmol/l) | 443 | 5.1 (0.2–261.6) |
Table II.
Genotype distribution in groups of recurrence of cholangitis, preoperative jaundice, types of gallstone or recurrence of PIS.
| Recurrence of cholangitis | Preoperative jaundice | Types of gallstone | Recurrence of PIS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation (n) | Yes 309 (%) | No 134 (%) | P-value | Yes 143 (%) | No 300 (%) | P-value | CBS 420 (%) | CS 23 (%) | P-value | Yes 95 (%) | No 348 (%) | P-value |
| RS3815675T >C | ||||||||||||
| TT (n=218) | 154 (49.8) | 64 (47.8) | 0.222 | 71 (49.7) | 147 (49.0) | 0.895 | 209 (49.8) | 9 (39.1) | 0.426 | 46 (48.4) | 172 (49.4) | 0.266 |
| TC (n=179) | 128 (41.4) | 51 (38.1) | 56 (39.2) | 123 (41.0) | 169 (40.2) | 10 (43.5) | 43 (45.3) | 136 (39.1) | ||||
| CC (n=46) | 27 (8.7) | 19 (14.2) | 16 (11.1) | 30 (10.0) | 42 (10.0) | 4 (17.4) | 6 (6.3) | 40 (11.5) | ||||
| RS2287616T >C | ||||||||||||
| TT (n=210) | 148 (47.9) | 62 (46.3) | 0.143 | 68 (47.6) | 142 (47.3) | 0.998 | 201 (47.9) | 9 (39.1) | 0.493 | 48 (50.5) | 162 (46.6) | 0.303 |
| TC (n=186) | 134 (43.4) | 52 (38.8) | 60 (42.0) | 126 (42.0) | 176 (41.9) | 10 (43.5) | 41 (43.2) | 145 (41.7) | ||||
| CC (n=47) | 27 (8.7) | 20 (14.9) | 15 (10.5) | 32 (10.7) | 43 (10.2) | 4 (17.4) | 6 (6.3) | 41 (11.8) | ||||
| RS2287617G >A | ||||||||||||
| GG (n=440) | 308 (99.7) | 132 (98.5) | 0.168 | 141 (98.6) | 299 (99.7) | 0.510 | 418 (99.5) | 22 (95.7) | 0.369 | 92 (96.8) | 348 (100) | 0.009 |
| GA (n=3) | 1 (0.3) | 2 (1.5) | 2 (1.4) | 1 (0.3) | 2 (0.5) | 1 (4.3) | 3 (3.2) | 0 (0.0) | ||||
| AA (n=0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| RS2287622T >C | ||||||||||||
| TT (n=39) | 25 (8.1) | 14 (10.4) | 0.621 | 15 (10.5) | 24 (8.0) | 0.679 | 38 (9.0) | 1 (4.4) | 0.693 | 12 (12.6) | 27 (7.8) | 0.061 |
| TC (n=187) | 134 (43.4) | 53 (39.6) | 60 (42.0) | 127 (42.3) | 176 (41.9) | 11 (47.8) | 46 (48.4) | 141 (40.5) | ||||
| CC (n=217) | 150 (48.5) | 67 (50.0) | 68 (47.6) | 149 (49.7) | 206 (49.0) | 11 (47.8) | 37 (38.9) | 180 (51.7) | ||||
| RS118109635C>T | ||||||||||||
| CC (n=423) | 295 (95.5) | 128 (95.5) | 0.980 | 132 (92.3) | 291 (97.0) | 0.026 | 402 (95.7) | 21 (91.3) | 0.634 | 90 (95.5) | 333 (95.7) | 0.692 |
| CT (n=20) | 14 (4.5) | 6 (4.5) | 11 (7.7) | 9 (3.0) | 18 (4.3) | 2 (8.7) | 5 (5.3) | 15 (4.3) | ||||
| TT (n=0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| RS497692A >G | ||||||||||||
| AA (n=55) | 37 (12.0) | 18 (13.4) | 0.828 | 13 (9.1) | 42 (14.0) | 0.011 | 51 (12.1) | 4 (17.4) | 0.599 | 13 (13.7) | 42 (12.1) | 0.560 |
| AG (n=194) | 138 (44.7) | 56 (41.8) | 53 (37.1) | 141 (47.0) | 183 (43.6) | 11 (47.8) | 45 (47.4) | 149 (42.8) | ||||
| GG (n=194) | 134 (43.4) | 60 (44.8) | 77 (53.8) | 117 (39.0) | 186 (44.3) | 8 (34.8) | 37 (38.9) | 157 (45.1) | ||||
CBS, bilirubinate stones; CS, cholesterol stones; PIS, primary intrahepatic stones.
Genomic DNA extraction, primer design and ABCB11 gene exon sequencing
The Blood Genome DNA Extraction kit (Takara Bio, Inc., Otsu, Japan) was used to extract genomic DNA from the peripheral blood lymphocytes of patients and controls. In the 28 exon coding regions of the ABCB11 gene, the first exon does not participate in the coding of the protein, so Primer 6.0 primer design software (primerdesign.co.uk/home) was used to design 27 pairs of primers for the PCR amplification of the remaining exons (Table III). The PCR amplification was performed with the PrimeSTAR Max DNA Polymerase kit (Takara Bio, Inc.) and the cycling conditions were as follow: 30 cycles of 98°C for 10 sec, 55°C for 5 sec and 72°C for 30 sec. The amplified products were then sequenced using the ABI 3730XL Gene Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Table III.
Primer pairs for the amplification of the ABCB11 coding sequence.
| Exon | Forward primer | Reverse primer | Fragment length (bp) |
|---|---|---|---|
| ABCB11 | |||
| 2 | 5′-GGCTCTTTCAGGGAGTTATTAACC-3′ | 5′-ACTTGACCAGCTTGTCCTACTT-3′ | 335 |
| 3 | 5′-AGAGACAATATGAGCAGGAAGA-3′ | 5′-CTGCTTTGTGCCTTTGATATGA-3′ | 266 |
| 4 | 5′-TCTGTGAATCGCTAGTGAACCT-3′ | 5′-ACACCCACTGCCATAAATCAAC-3′ | 313 |
| 5 | 5′-ATACGAACTCTGCCACTCAATT-3′ | 5′-GTTAGATACCACTCCAGCTCAG-3′ | 492 |
| 6 | 5′-AATGTAATCTCTGGTGGCTTGA-3′ | 5′-TGTAGTTCTTAGGGCTTCTGAT-3′ | 262 |
| 7 | 5′-CTTAGTTCCCAAGAAGAGGCATT-3′ | 5′-CACACCAAATTGCAGTACCTTG-3′ | 487 |
| 8 | 5′-GAGAGGCTGTTAATGCTATCCA-3′ | 5′-TGTTGCTAACTGTACTCAGGAA-3′ | 410 |
| 9 | 5′-TCTTCCTCCTGTCAATGATGTTAC-3′ | 5′-ATTACTCTGCTTAGCTCCCTCTT-3′ | 412 |
| 10 | 5′-TGCTCTGTGTTTGCGATGATTT-3′ | 5′-TGTTTCCACAGACAGACTCCATA-3′ | 435 |
| 11 | 5′-TCTCTGCGTTAACATGGAAGAC-3′ | 5′-CAAGAGCGAAACTCCATCTCAA-3′ | 418 |
| 12 | 5′-GCAGAGATACGCCAAAGATGTT-3′ | 5′-AAGACACCTCCATTCCCTATTACT-3′ | 337 |
| 13 | 5′-CACAGACACCGAGTATCAACAC-3′ | 5′-CCAGGACAGTCTCAATGTATGC-3′ | 332 |
| 14 | 5′-TTTCTGCCCATTGGTCAAGTAT-3′ | 5′-CTCTTAGTTTCTCCCAGGAATGTA-3′ | 331 |
| 15 | 5′-GATCACTGTCAGAAGCCATCAA-3′ | 5′-TATCAACTACTCCCATCCCTCC-3′ | 336 |
| 16 | 5′-TCTAATGTCTGCACAGCCTATT-3′ | 5′-GTTGGGAGAACAGTGAGTATTGA-3′ | 441 |
| 17 | 5′-TTGCTACTTCTGATGGACTTCTC-3′ | 5′-AGGATTAGGACTACAGAGGACTC-3′ | 437 |
| 18 | 5′-AACTTGGACACCAGTTGATCCT-3′ | 5′-TAGTCTGACTTGAAACACTGCTAG-3′ | 300 |
| 19 | 5′-CCATATCCCATAGACATTTGAGGT-3′ | 5′-ATGAGAAGAAGAAAGCTAGTCCAG-3′ | 335 |
| 20 | 5′-CCACCAGAATGATACATTTCCTAC-3′ | 5′-TGAAGAGGGAGATGTTAGAGAA-3′ | 405 |
| 21 | 5′-GCAATGGGCTGTGTATCTCTTT-3′ | 5′-GTCAGTGTTAGAAGCAGTGGAA-3′ | 444 |
| 22 | 5′-TCTGAGACGGGTTGATTGCTTT-3′ | 5′-GCTTCCTTCAGTCTCTTCGTACTA-3′ | 331 |
| 23 | 5′-CCACTGAAATGTCACGAAAGGA-3′ | 5′-TGGAGACAGAAGAATACACAGAAG-3′ | 524 |
| 24 | 5′-ATGCTTGTTCAGTCCTCTTCTT-3′ | 5′-CCTGTGTCCATGTGTTCTGTT-3′ | 560 |
| 25 | 5′-TGAAGGTATCTCAAGCAGGGATT-3′ | 5′-AAAGTGAGTCTGGCAAAGCAAA-3′ | 397 |
| 26 | 5′-TTAGCCTTGGGATTGTTAGTCTG-3′ | 5′-CACTCTGGTCATTCTACTTCTCC-3′ | 402 |
| 27 | 5′-GAGGAGACCTTGACATGAGTTC-3′ | 5′-GGTTCCACAAAGTATTGCCAAT-3′ | 362 |
| 28 | 5′-GGATTGTTATTCAGGTCGTGTT-3′ | 5′-TTAGCTTGGATTCCGATGTAGG-3′ | 462 |
ABCB11, adenosine 5′-triphosphate binding cassette subfamily B member.
Extraction of mRNA and reverse transcription-quantitative PCR (RT-qPCR)
Total mRNA was extracted from the 200 mg freshly frozen liver tissue of the PIS patients and the control group using the Eastep Super Total RNA Extraction kit (Promega Corporation, Madison, WI, USA). PIS patient and control group liver tissue samples were cryopreserved in liquid nitrogen at the time of excision to facilitate RNA extraction and RT-qPCR analysis. The specific experimental procedure was conducted according to the manufacturer's protocol. DNA removal and reverse transcription were performed with the PrimeScript™ RT Reagent kit with gDNA Eraser (Takara Bio, Inc.) at 42°C for 2 min. Finally, the mRNA expression of the ABCB11 gene in the PIS patients was detected by RT-qPCR using SYBR Premix Ex Taq II (Takara Bio, Inc.) and a quantitative fluorescence PCR analyzer.
Total RNA was extracted from 293 cells 48 h after transfection with wild-type and mutant A856V plasmids using the Eastep Super Total RNA Extraction kit (Promega Corporation), followed by DNA removal and reverse transcription using the PrimeScript™ RT Reagent kit with gDNA Eraser (Takara Bio, Inc). The mRNA expression was detected by RT-qPCR using SYBR Premix Ex Taq II (Takara Bio, Inc.) and a quantitative fluorescence PCR analyzer.
The results of freshly frozen liver tissues and cells were normalized to the expression of β-actin. The primers for BSEP were forward primer, 5′-dTTGCCTTTGCCCAGTGCATCAT-3′ and reverse primer, 5′-dGGTTGTCGGTCCAGCAGTTGAA-3′ (BSEP), and those for β-actin were forward primer, 5′-dCCTGGCACCCAGCACAAT-3′ and reverse primer, 5′-dGGGCCGGACTCGTCATAC-3′. For the quantitative measurement of ABCB11 mRNA expression, RT-qPCR was performed using a CFX96 Real-Time system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with the following cycling conditions: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec (24).
Western blot analysis
Membrane proteins were extracted from the liver tissues of PIS patients and 293 cells transfected with wild-type and mutant A856V ABCB11, using the Mem-PER™ Plus Membrane Protein Extraction kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The Bicinchoninic Protein Quantification kit (CWBiotech, Beijing, China) was used for protein concentration determination. Equal amounts of protein (20 µg) were separated by 6% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA), which were blocked and incubated with primary antibodies overnight at 4°C. The primary antibodies used in this study included the following: Anti-BSEP (1:500; cat. no. 155421; Abcam, Cambridge, UK) and anti-Na/K-ATPase (1:1,000; cat. no. 58475; Abcam). The membranes were washed with Tris-buffered saline/Tween-20 and incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody for 2 h at room temperature. The SuperSignal West Femto Substrate Trial kit (Thermo Fisher Scientific, Inc.) was used for signal detection.
Plasmids and site-directed mutagenesis
The wild-type plasmid pEZ-M02-BSEP contains all the open reading frames (GeneCopoeia Inc., Rockville, MD, USA) of the ABCB11 gene. This plasmid was used as a template to obtain the A856V mutation by site-directed mutagenesis (25). The ABCB11 coding sequences of the above plasmids were verified by sequencing.
Cell culture and plasmid transfection
A total of 293 cells and MDCK cells obtained from the Cell Bank of the Chinese Academy of Sciences were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) with 1% penicillin-streptomycin solution (Beyotime Institute of Biotechnology, Haimen, China) and 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 humidified atmosphere. A total of 5×105 293 cells were seeded in 6-well plates and transiently transfected with 2,500 ng DNA and Lipofectamine® (Invitrogen; Thermo Fisher Scientific, Inc.) per well after 24 h at 37°C humidified atmosphere containing 5% CO2 when the cell confluence reached 80%. A total of 1×105 MDCK cells were seeded in 24-well plates and transiently transfected with 500 ng of DNA and Lipofectamine® (Invitrogen; Thermo Fisher Scientific, Inc.) per well after 24 h at 37°C humidified atmosphere containing 5% CO2 when the cell confluence reached 70%. The transfected cells were incubated at 37°C humidified atmosphere containing 5% CO2, and were harvested 48 h later for subsequent experiments.
Immunofluorescence and laser confocal microscopy
The MDCK cells were seeded on slides 24 hour at room temperature prior to transfection. Forty-eight hours after transfection, the cells were fixed with 2% paraformaldehyde for 10 min at room temperature and infiltrated with 1% Triton X-100 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 5 min at room temperature, followed by blocking with 10% goat serum (Gibco; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. Following rinsing with PBS, one group of 1×105 cells was incubated with rabbit anti-BSEP polyclonal antibody (1:100; cat. no. 155421; Abcam) and chicken anti-Na/K-ATPase monoclonal antibody (1:200; cat. no. 353; Abcam) overnight at 4°C, then washed with PBS and incubated with goat anti-rabbit Alexa Fluor 488 (1:200; cat. no. 150077; Abcam) and goat anti-chicken Alexa Fluor 647 (1:200; cat. no. 150171; Abcam) at room temperature for 2 h. Another group of 1×105 cells was incubated with rabbit anti-BSEP polyclonal antibody (1:100; cat. no. 155421) and mouse anti-calnexin polyclonal antibody (1:200; cat. no. 112995) overnight at 4°C, then washed with PBS and incubated with goat anti-rabbit Alexa Fluor 488 (1:200; cat. no. 150077) and goat anti-mouse Alexa Fluor 647 (1:200; cat. no. 150115; all Abcam) at room temperature for 2 h. Images of the cells were obtained using a Leica CTR 4000 laser scanning confocal microscope.
Statistical analysis
The genotype frequencies at all mutation loci were directly obtained by counting and the use of the Hardy-Weinberg equilibrium. The unconditional logistic regression model was used to evaluate the correlation between the ABCB11 gene mutations and PISs. The evaluation indicators included odds ratio (OR) and 95% confidence interval (CI); the odds ratio (OR) was corrected for sex, age and body mass index. The correlation between the genotype and the clinical data of the PIS patients was analyzed using the Chi-square test. All experimental data were analyzed by SPSS software (PASW Statistics 22, IBM Corp., Armonk, NY, USA) or Graphpad Prism (v6.0 for Windows; GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as the mean ± standard deviation of four independent experiments. Statistical analyses shown in the figures were performed using paired Student's t-tests or one-way analysis of variance with least significant difference post hoc test. P<0.05 was considered to indicate a statistically significant result.
Results
ABCB11 gene mutations may be associated with the pathogenesis of PISs
In the present study, peripheral blood lymphocytes were collected from 443 PIS patients and 560 healthy individuals for ABCB11 exon sequencing. Six mutations were detected, including three missense mutations and three synonymous mutations (Table IV). The missense mutation rs118109635 and the synonymous mutation rs497692 were detected within exons 21 and 24, of the ABCB11 gene, respectively. The gene frequency distributions of the two mutational loci in the PIS patients and the healthy control group differed significantly (P=0.025 and P=0.017, respectively). In addition, the missense mutations rs2287617 and rs2287622 and the synonymous mutations rs3815675 and rs2287616 were located in exons 9, 13, 4 and 9 of the ABCB11 gene, respectively. However, these four mutations showed no significant differences between PIS patients and healthy controls (P>0.05). These results indicated that there are two significant ABCB11 gene mutations in the PIS patients (rs118109635 and rs497692), consistent with previous findings (5).
Table IV.
Distribution of six mutations, and the association between the mutations and PIS.
| Mutation | Amino acid change | Genotype | Controls (%) n=560 | Cases (%) n=443 | Adjusted OR (95% CI) | P-value | P-value for HWE in control |
|---|---|---|---|---|---|---|---|
| s3815675 | Synonymous | TT | 280 (50.0) | 218 (49.2) | Ref. | Ref. | 0.308 |
| TC | 227 (40.5) | 179 (40.4) | 0.98 (0.73–1.30) | 0.877 | |||
| CC | 53 (9.5) | 46 (10.4) | 1.15 (0.72–1.83) | 0.569 | |||
| TC+TT | 507 (90.5) | 397 (89.6) | 0.86 (0.55–1.36) | 0.524 | |||
| rs2287616 | Synonymous | TT | 277 (49.4) | 210 (47.4) | Ref. | Ref. | 0.545 |
| TC | 225 (40.2) | 186 (42.0) | 1.06 (0.79–1.41) | 0.694 | |||
| CC | 58 (10.4) | 47 (10.6) | 1.08 (0.69–1.72) | 0.715 | |||
| TC+TT | 502 (89.6) | 396 (89.4) | 0.94 (0.61–1.46) | 0.793 | |||
| rs2287617 | R299k | GG | 555 (99.1) | 440 (99.3) | Ref. | Ref. | 0.943 |
| GA | 5 (0.9) | 3 (0.7) | 0.65 (0.14–3.06) | 0.582 | |||
| AA | 0 (0.0) | 0 (0.0) | NA | NA | |||
| GA+AA | 5 (0.9) | 3 (0.7) | 0.65 (0.14–3.06) | 0.582 | |||
| rs2287622 | V444A | TT | 50 (8.9) | 39 (8.8) | Ref. | Ref. | 0.886 |
| TC | 237 (42.3) | 187 (42.2) | 0.91 (0.55–1.51) | 0.709 | |||
| CC | 273 (48.8) | 217 (49.0) | 0.92 (0.56–1.52) | 0.750 | |||
| TC+TT | 287 (51.2) | 226 (51.0) | 1.00 (0.76–1.31) | 0.992 | |||
| rs118109635 | A865V | CC | 551 (98.4) | 423 (95.5) | Ref. | Ref. | 0.627 |
| CT | 9 (1.6) | 20 (4.5) | 2.67 (1.13–6.31) | 0.025 | |||
| TT | 0 (0.0) | 0 (0.0) | NA | NA | |||
| CT+TT | 9 (1.6) | 20 (4.5) | 2.67 (1.13–6.31) | 0.025 | |||
| rs497692 | Synonymous | AA | 106 (18.9) | 55 (12.4) | Ref. | Ref. | 0.549 |
| AG | 249 (44.5) | 194 (43.8) | 1.44 (0.96–2.17) | 0.079 | |||
| GG | 205 (36.6) | 194 (43.8) | 1.79 (1.18–2.70) | 0.006 | |||
| AG+GG | 454 (81.1) | 388 (87.6) | 1.60 (1.08–2.34) | 0.017 |
CI, confidence interval; HWE, HardyWeinberg Equilibrium; NA, not available; OR, odds ratio; PIS, primary intrahepatic stones; Ref., reference.
Correlation of ABCB11 gene mutations and clinical characteristics of PIS patients
The analysis of the ABCB11 gene mutations and the relevant clinical data for the PIS patients in the present study (Table II) showed that the genotype of the rs118109635 mutation (CT genotype) of the ABCB11 gene was correlated with preoperative jaundice (P=0.026) and that the homozygous genotype (GG) and the heterozygous genotype (GA) caused by mutations at the rs497692 locus in the PIS patients in the present study were associated with preoperative jaundice (P=0.011). In addition, the mutation at the rs2287617 locus of the ABCB11 gene was associated with the recurrence of PISs (P=0.009). Therefore, the rs118109635 and rs497692 mutations of the ABCB11 gene are closely associated with the clinical characteristics of PISs.
Effects of ABCB11 gene mutations in PIS patients on the expression of ABCB11 mRNA and its encoded protein
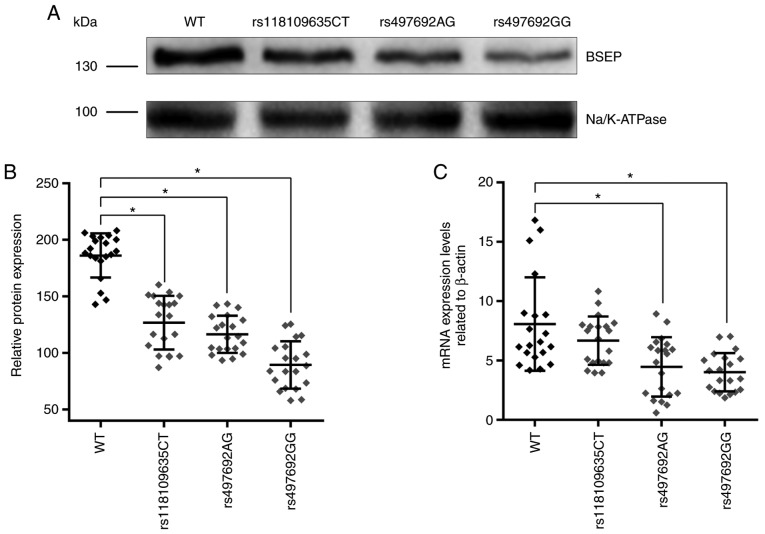
The expression of ABCB11 mRNA in the liver tissues of PIS patients with different genotypes was measured by RT-qPCR. The results showed that whereas the mRNA expression levels of the mutant rs497692 were decreased, the rs118109635 mutation had no significant impact on ABCB11 mRNA expression levels (Fig. 1C). The expression of BSEP in the liver tissue of PIS patients with different genotypes was detected by western blot analysis; BSEP was detected as a 140–150 kDa band (Fig. 1A). The results revealed that the expression of BSEP was significantly decreased in patients with the ABCB11 gene mutations rs118109635 and rs497692 (Fig. 1A and B). Thus, the ABCB11 gene mutations (rs118109635 and rs497692) may affect the transcription and translation of the ABCB11 gene.
Figure 1.
Detection of mRNA and BSEP protein expression levels in PIS patients with different ABCB11 genotypes. (A) Representative western blot analysis using the liver tissues of PIS patients with different genotypes and (B) quantification of BSEP expression levels. (C) RT-qPCR was performed to measure the ABCB11 mRNA expression levels in the liver tissues of PIS patients with different genotypes. Data are presented as the mean ± standard deviation (n=20; *P<0.05). BSEP, bile salt export pump; PIS, primary intrahepatic stone; WT, wild type.
Expression of mutated BSEP on the 293 cell membrane
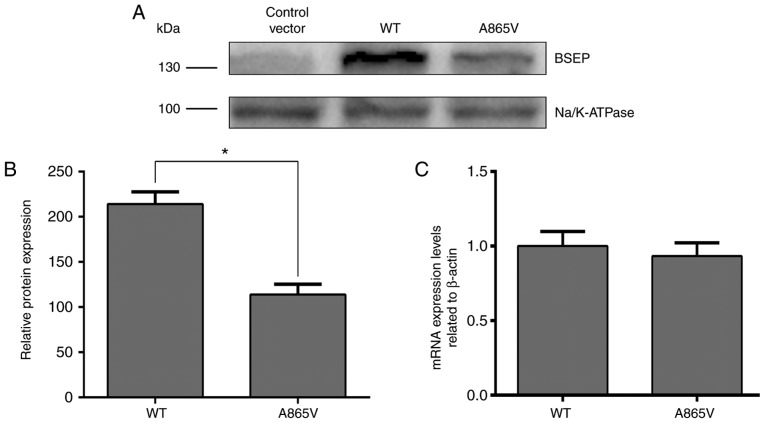
The aim of this experiment was to investigate the effect of ABCB11 gene mutation on BSEP expression and its subcellular localization. Therefore, 293 and MDCK cell lines were used, albeit neither express BSEP protein, as experimental cells, and investigated the effect of ABCB11 gene mutations on protein expression and subcellular localization by transfection of ABCB11 mutant plasmid and wild-type plasmid. Due to the high expression of BSEP protein in liver cell lines, it may interfere with the observation of cell transfection experiment results. The 293 and MDCK cells are polarized cells commonly used to study trafficking of membrane proteins (17,23). The transfection efficiency in MDCK cells was quite low (25). Thus, to determine the effects of ABCB11 mutations on BSEP expression, 293 cells were used. Plasmids containing wild-type and mutated BSEP cDNA were constructed and transfected into 293 cells (25) to determine the effects of the A865V mutation on BSEP expression. The expression of BSEP in the A865V mutant measured by western blot analysis was significantly lower than that in wild-type 293 cells (Fig. 2A and B). Although the A865V mutation had no apparent effect on BSEP mRNA expression levels (Fig. 2C), the decrease in BSEP protein expression levels may be associated with the degradation of BSEP due to its retention in the endoplasmic reticulum as a result of a change in its three-dimensional structure (17). Thus, the above results indicated that the A865V mutation downregulated the translocation of the BSEP protein.
Figure 2.
Effect of ABCB11 gene missense mutations on BSEP expression. (A) 293 cells were transfected with wild-type or mutant ABCB11 plasmids and the expression of BSEP protein was detected by western blotting. ‘Control’ represents the empty plasmid vector and (B) quantification of protein expression levels. (C) Total RNA was extracted from 293 cells 48 h after transfection with the wild-type and mutant ABCB11 plasmids, and RT-qPCR was performed to measure the ABCB11 mRNA expression levels. Data are presented as the mean ± standard deviation from 4 independent experiments (*P<0.05). BSEP, bile salt export pump; PIS, primary intrahepatic stone; WT, wild type.
Subcellular localization of wild-type and mutant BSEP
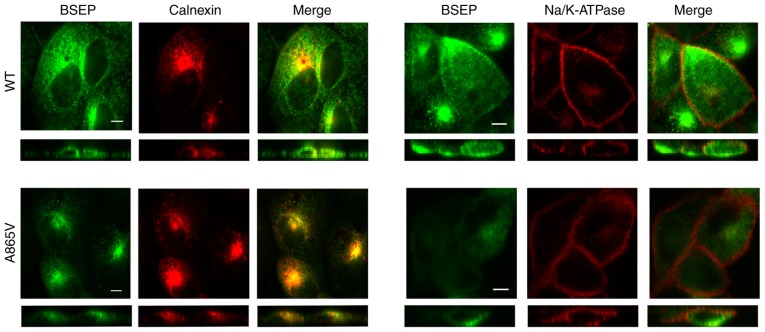
The cells used in this experiment needed to undergo multiple antibody incubation and PBS washing steps. The 293 cells have poor adhesion and can easily be detached from the slide during multiple incubation and washing (25). Thus, the impact of ABCB11 mutations against BSEP subcellular localization was assessed in MDCK cells. MDCK cells were transiently transfected with wild-type and mutant plasmids, and the expressed proteins were labeled and stained with antibodies to BSEP, calnexin and Na/K-ATPase to determine the subcellular location of BSEP by immunofluorescence confocal microscopy. Calnexin and Na/K-ATPase were used as markers for the endoplasmic reticulum and the cell membrane, respectively. Results demonstrated that wild-type BSEP was distributed inside the cells and in the cell membrane, whereas the A856V mutant BSEP was mainly distributed inside the cells and showed significantly reduced distribution in the cell membrane (Fig. 3). Thus, the A856V mutation affected the subcellular localization of BSEP.
Figure 3.
Analysis of wild-type and mutant BSEP distribution by immunofluorescence. The resulting distribution of fluorescence was observed by laser confocal microscopy. Green represents BSEP and red represents calnexin (left) and Na/K-ATPase (right) and yellow represents co-localization of the two proteins. In each panel, an X-Y image (upper) and an X-Z image (lower) are shown. The data shown were obtained from four independent experiments. Scale bar, 5 µm.
Discussion
The present study demonstrated that two mutations in the ABCB11 gene, rs118109635 (C>T) and rs497692 (A>G), are associated with the pathogenesis of PISs, and these mutations are closely associated with the occurrence of preoperative jaundice in patients with PISs. Compared with the wild-type protein, the expression of BSEP in the liver tissues of patients with mutations (rs118109635 and rs497692) was significantly decreased, and the decrease in protein expression levels was evident is the rs497692 homozygous genotype. Although the ABCB11 mRNA expression levels of the mutant rs497692 heterozygote and homozygous genotypes were both decreased, the rs118109635 mutation had no significant effect on mRNA expression. The in vitro experiments showed that the A865V mutation significantly decreased BSEP protein expression levels in the cell membrane, indicating that the A865V mutation may affect the subcellular localization of BSEP.
According to existing reports, the incidence of PISs is high in East Asia, and its incidence in Southern China is significantly higher than in Northern China; thus, it is believed that the incidence of PISs displays very strong regional and ethnic differences (1). The identified mutations in the ABCB11 gene lead to defects in BSEP protein expression that can cause changes in bile salt content (26), and it has been shown that mutations in this gene are associated with the pathogenesis of cholestasis (27–29). Therefore, the role of ABCB11 gene mutations in the pathogenesis of PISs was investigated. In the present study, ABCB11 gene exon sequencing was performed on a large sample of 443 cases of PIS patients and 560 healthy individuals. Two mutations associated with the pathogenesis of PISs: The missense mutation rs118109635 and the synonymous mutation rs497692. This result is consistent with the findings reported in a previous study (5). Additionally, analysis of the relevant clinical data showed that the ABCB11 gene mutations rs118109635 and rs497692 were correlated with the occurrence of preoperative jaundice in PIS patients. Therefore, the role of ABCB11 gene mutations in the pathogenesis of PISs is worthy of further investigation.
To further clarify the association between mutations rs118109635 and rs497692 and the pathogenesis of PISs, the mRNA expression levels of ABCB11 in the liver tissue of the PIS patients was measured. The results showed that the missense mutation rs118109635 had no significant impact on ABCB11 mRNA expression, but western blot analysis showed that BSEP expression levels were significantly reduced in patients with this mutation. Western blot analysis and confocal immunofluorescence microscopy experiments performed in vitro confirmed that mutation A865V (rs118109635) significantly decreased BSEP expression levels at the subcellular levels, with no significant change in the ABCB11 mRNA expression levels. Thus, the rs118109635 mutation downregulated the expression of the transmembrane transporter BSEP in the cell membrane. Previous studies of ABCB11 gene mutations confirmed that ABCB11 gene missense mutations change the amino acid sequence of the BSEP protein and alter its three-dimensional structure (30). Most misfolded proteins are degraded by proteases in the endoplasmic reticulum, ultimately leading to reduced protein expression levels of BSEP in the cell membranes (17). Therefore, the missense mutation rs118109635 may be involved in the pathogenesis of PISs due to its effect on the expression of BSEP; this hypothesis awaits further confirmation.
Furthermore, to determine the role of mutation rs497692 in the pathogenesis of PISs, PIS clinical specimens were analyzed by RT-qPCR. The results showed that the expression mRNA expression levels of rs497692 was decreased and that the expression levels of mutant BSEP associated with the heterozygote and homozygous genotypes were significantly decreased compared to those of the wild-type gene and protein. The relevant studies demonstrated that mutation rs497692 of the ABCB11 gene is a synonymous mutation that can change gene splicing by exon skipping, thus affecting the expression of mature mRNA and ultimately leading to a decrease in the cellular level of the encoded protein (30). Thus, mutation rs497692 may serve an important role in the pathogenesis of PISs by altering the transcription level of the ABCB11 gene.
A previous study of BSEP transport function showed no change in the bile transport activity of individual BSEP molecules in the cell membrane; instead, the decrease in BSEP transport function observed in the mutant is due to decreased molecular distribution of BSEP on the membrane, resulting in overall reduced bile transport by BSEP (17). Therefore, the rs118109635 and rs497692 mutations decreased BSEP expression hepatocyte apical and bile duct membrane by different mechanisms, leading to reduced bile transport capacity in PIS patients, changes in bile composition, and stone formation. In addition, in vitro experiments have shown that administration of 4-butyl benzene increases the membrane expression of BSEP protein transcribed from the mutated ABCB11 gene in the cell membrane (31). These results may be beneficial in the treatment of PIS patients with reduced BSEP expression caused by the identified ABCB11 gene mutation.
The present study has limitations. The regulatory mechanism of ABCB11 gene mutation on mRNA and BSEP protein expression levels need to be studied further, and at the same time, ABCB11 mutated rat models need to be used in order to further define the biological function of these mutations, and determine the specific role of ABCB11 gene mutations in primary intrahepatic stone.
In conclusion, the present study confirmed that the ABCB11 gene in patients with PISs displayed two disease-associated mutations, rs118109635 and rs497692. The rs497692 and rs118109635 mutations affected transcription and translation of the ABCB11 gene and downregulated BSEP expression in the cell membrane, possibly affecting the transport of specific substrates by BSEP and eventually causing changes in bile composition that serve an important role in the pathogenesis of PISs. Therefore, mutations rsB8109635 and rs497692 of the ABCB11 gene may be risk factors for PISs that can provide a reliable basis for improvement of the clinical treatment of PIS patients in the future.
Acknowledgements
The authors would like to thank The Institute of Hepatobiliary Surgery, Southwest Hospital, Third Military Medical University (Army Medical University), Chongqing, China, for providing laboratory facilities.
Glossary
Abbreviations
- BSEP
bile salt export pump
- BRIC2
benign recurrent intrahepatic cholestasis type 2
- CBS
calcium bilirubinate stones
- CI
confidence interval
- CS
cholesterol stones
- 293
human embryonic kidney 293
- HWE
Hardy-Weinberg Equilibrium
- ICP
intrahepatic cholestasis of pregnancy
- MDCK
Madin-Darby canine kidney
- PFIC2
progressive familial intrahepatic cholestasis type 2
- OR
odds ratio
- PIS
primary intrahepatic stone
- SNP
single nucleotide polymorphisms
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81270535) and Southwest Hospital (grant no. SWH2016JCYB-10).
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
YH, LG and SP designed the study. LG and YH conducted the experiments and drafted the manuscript. LG, SP, JC, JB and PJ were involved in the statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The experiments were approved by the Ethics Committee of the Southwest Hospital of the Third Military Medical University of China. Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Clemente G, De Rose AM, Murri R, Ardito F, Nuzzo G, Giuliante F. Liver resection for primary intrahepatic stones: Focus on postoperative infectious complications. World J Surg. 2016;40:433–439. doi: 10.1007/s00268-015-3227-x. [DOI] [PubMed] [Google Scholar]
- 2.Li SQ, Hua YP, Shen SL, Hu WJ, Peng BG, Liang LJ. Segmental bile duct-targeted liver resection for right-sided intrahepatic stones. Medicine (Baltimore) 2015;94:e1158. doi: 10.1097/MD.0000000000001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catena M, Aldrighetti L, Finazzi R, Arzu G, Arru M, Pulitanò C, Ferla G. Treatment of non-endemic hepatolithiasis in a Western country. The role of hepatic resection. Ann R Coll Surg Engl. 2006;88:383–389. doi: 10.1308/003588406X98711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama F, Soloway RD, Nakama T, Miyazaki K, Ichimiya H, Sheen PC, Ker CG, Ong GB, Choi TK, Boey J, et al. Hepatolithiasis in East Asia. Retrospective study. Dig Dis Sci. 1986;31:21–26. doi: 10.1007/BF01347905. [DOI] [PubMed] [Google Scholar]
- 5.Pan S, Li X, Jiang P, Jiang Y, Shuai L, He Y, Li Z. Variations of ABCB4 and ABCB11 genes are associated with primary intrahepatic stones. Mol Med Rep. 2015;11:434–446. doi: 10.3892/mmr.2014.2645. [DOI] [PubMed] [Google Scholar]
- 6.Meng ZW, Han SH, Zhu JH, Zhou LY, Chen YL. Risk factors for cholangiocarcinoma after initial hepatectomy for intrahepatic stones. World J Surg. 2017;41:835–843. doi: 10.1007/s00268-016-3752-2. [DOI] [PubMed] [Google Scholar]
- 7.Cetta FM. Bile infection documented as initial event in the pathogenesis of brown pigment biliary stones. Hepatology. 1986;6:482–489. doi: 10.1002/hep.1840060327. [DOI] [PubMed] [Google Scholar]
- 8.Sakpal SV, Babel N, Chamberlain RS. Surgical management of hepatolithiasis. HPB (Oxford) 2009;11:194–202. doi: 10.1111/j.1477-2574.2009.00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tazuma S. Gallstone disease: Epidemiology, pathogenesis, and classification of biliary stones (common bile duct and intrahepatic) Best Pract Res Clin Gastroenterol. 2006;20:1075–1083. doi: 10.1016/j.bpg.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Meier Y, Pauli-Magnus C, Zanger UM, Klein K, Schaeffeler E, Nussler AK, Nussler N, Eichelbaum M, Meier PJ, Stieger B. Interindividual variability of canalicular ATP-binding-cassette (ABC)-transporter expression in human liver. Hepatology. 2006;44:62–74. doi: 10.1002/hep.21214. [DOI] [PubMed] [Google Scholar]
- 11.Kubitz R, Dröge C, Kluge S, Stindt J, Häussinger D. Genetic variations of bile salt transporters. Drug Discov Today Technol. 2014;12:e55–e67. doi: 10.1016/j.ddtec.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Kubitz R, Dröge C, Stindt J, Weissenberger K, Häussinger D. The bile salt export pump (BSEP) in health and disease. Clin Res Hepatol Gastroenterol. 2012;36:536–553. doi: 10.1016/j.clinre.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Wang J, Tang S, Zhang Y, Lv X, Wu S, Yang Z, Xia Y, Chen D, Zhan S. Role of polymorphic bile salt export pump (BSEP, ABCB11) transporters in anti-tuberculosis drug-induced liver injury in a Chinese cohort. Sci Rep. 2016;6:27750. doi: 10.1038/srep27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chai J, He Y, Cai SY, Jiang Z, Wang H, Li Q, Chen L, Peng Z, He X, Wu X, et al. Elevated hepatic multidrug resistance-associated protein 3/ATP-binding cassette subfamily C 3 expression in human obstructive cholestasis is mediated through tumor necrosis factor alpha and c-Jun NH2-terminal kinase/stress-activated protein kinase-signaling pathway. Hepatology. 2012;55:1485–1494. doi: 10.1002/hep.24801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beausejour Y, Alvarez F, Beaulieu M, Bilodeau M. Description of two new ABCB11 mutations responsible for type 2 benign recurrent intrahepatic cholestasis in a French-Canadian family. Can J Gastroenterol. 2011;25:311–314. doi: 10.1155/2011/534918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi H, Naoi S, Hirose Y, Matsuzaka Y, Tanikawa K, Igarashi K, Nagasaka H, Kage M, Inui A, Kusuhara H. Successful treatment with 4-phenylbutyrate in a patient with benign recurrent intrahepatic cholestasis type 2 refractory to biliary drainage and bilirubin absorption. Hepatol Res. 2016;46:192–200. doi: 10.1111/hepr.12561. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi H, Takada T, Suzuki H, Akita H, Sugiyama Y. Two common PFIC2 mutations are associated with the impaired membrane trafficking of BSEP/ABCB11. Hepatology. 2005;41:916–924. doi: 10.1002/hep.20627. [DOI] [PubMed] [Google Scholar]
- 18.Waisbourd-Zinman O, Surrey LF, Schwartz AE, Russo PA, Wen J. A Rare BSEP mutation associated with a mild form of progressive familial intrahepatic cholestasis type 2. Ann Hepatol. 2017;16:465–468. doi: 10.5604/01.3001.0009.8604. [DOI] [PubMed] [Google Scholar]
- 19.Keitel V, Burdelski M, Warskulat U, Kühlkamp T, Keppler D, Häussinger D, Kubitz R. Expression and localization of hepatobiliary transport proteins in progressive familial intrahepatic cholestasis. Hepatology. 2005;41:1160–1172. doi: 10.1002/hep.20682. [DOI] [PubMed] [Google Scholar]
- 20.Anzivino C, Odoardi MR, Meschiari E, Baldelli E, Facchinetti F, Neri I, Ruggiero G, Zampino R, Bertolotti M, Loria P, Carulli L. ABCB4 and ABCB11 mutations in intrahepatic cholestasis of pregnancy in an Italian population. Dig Liver Dis. 2013;45:226–232. doi: 10.1016/j.dld.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Keitel V, Vogt C, Häussinger D, Kubitz R. Combined mutations of canalicular transporter proteins cause severe intrahepatic cholestasis of pregnancy. Gastroenterology. 2006;131:624–629. doi: 10.1053/j.gastro.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Lim TY, Coltart I, Foskett P, Thompson R, Strautnieks S, Penna L, Williamson C, Miquel R, Heneghan MA. Donor transmitted mutation of the ABCB11 gene and ensuing intrahepatic cholestasis of pregnancy in a liver transplant recipient. Liver Transpl. 2017;23:1229–1232. doi: 10.1002/lt.24776. [DOI] [PubMed] [Google Scholar]
- 23.Lam P, Pearson CL, Soroka CJ, Xu S, Mennone A, Boyer JL. Levels of plasma membrane expression in progressive and benign mutations of the bile salt export pump (Bsep/Abcb11) correlate with severity of cholestatic diseases. Am J Physiol Cell Physiol. 2007;293:C1709–C1716. doi: 10.1152/ajpcell.00327.2007. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Chen K, Jiang P, Zhang X, Li X, Li Z. CD44v/CD44s expression patterns are associated with the survival of pancreatic carcinoma patients. Diagn Pathol. 2014;9:79. doi: 10.1186/1746-1596-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordo-Gilart R, Andueza S, Hierro L, Martínez-Fernández P, D'Agostino D, Jara P, Alvarez L. Functional analysis of ABCB4 mutations relates clinical outcomes of progressive familial intrahepatic cholestasis type 3 to the degree of MDR3 floppase activity. Gut. 2015;64:147–155. doi: 10.1136/gutjnl-2014-306896. [DOI] [PubMed] [Google Scholar]
- 26.Wang R, Chen HL, Liu L, Sheps JA, Phillips MJ, Ling V. Compensatory role of P-glycoproteins in knockout mice lacking the bile salt export pump. Hepatology. 2009;50:948–956. doi: 10.1002/hep.23089. [DOI] [PubMed] [Google Scholar]
- 27.Hu G, He P, Liu Z, Chen Q, Zheng B, Zhang Q. Diagnosis of ABCB11 gene mutations in children with intrahepatic cholestasis using high resolution melting analysis and direct sequencing. Mol Med Rep. 2014;10:1264–1274. doi: 10.3892/mmr.2014.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JS, Ko JS, Seo JK, Moon JS, Park SS. Clinical and ABCB11 profiles in Korean infants with progressive familial intrahepatic cholestasis. World J Gastroenterol. 2016;22:4901–4907. doi: 10.3748/wjg.v22.i20.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai Y, Xie Y, Tang CW. Compound heterozygous mutations of ABCB11 responsible for benign recurrent intrahepatic cholestasis. J Dig Dis. 2015;16:299–302. doi: 10.1111/1751-2980.12227. [DOI] [PubMed] [Google Scholar]
- 30.Byrne JA, Strautnieks SS, Ihrke G, Pagani F, Knisely AS, Linton KJ, Mieli-Vergani G, Thompson RJ. Missense mutations and single nucleotide polymorphisms in ABCB11 impair bile salt export pump processing and function or disrupt pre-messenger RNA splicing. Hepatology. 2009;49:553–567. doi: 10.1002/hep.22683. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi H, Sugiyama Y. 4-phenylbutyrate enhances the cell surface expression and the transport capacity of wild-type and mutated bile salt export pumps. Hepatology. 2007;45:1506–1516. doi: 10.1002/hep.21630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.