Abstract
MicroRNAs (miRNAs) have been demonstrated to regulate the progression of numerous types of cancer, including acute myeloid leukemia (AML). Previous studies demonstrated that miR-1271-5p functions as a tumor suppressor; however, the roles of miR-1271-5p in AML remain unknown. In the present study, miR-1271-5p was significantly downregulated in AML tissues compared with normal tissues by reverse transcription-quantitative polymerase chain reaction analysis. Furthermore, the expression levels of miR-1271-5p in patients with AML may function as a prognostic marker. In addition, overexpression of miR-1271-5p significantly suppressed the proliferation and induced apoptosis of AML cells by Cell Counting kit-8 and fluorescence activated cell sorter assays; miR-1271-5p downregulation exhibited opposing effects. Additionally, transcription factor ZIC2 may be a direct target of miR-1271-5p in AML cells, which was demonstrated by a luciferase reporter assay and RNA pulldown assay. Overexpression of miR-1271-5p significantly reduced the mRNA and protein expression levels of ZIC2 in AML193 and OCI-AML2 cells by reverse transcription-quantitative polymerase chain reaction analysis and western blotting. Furthermore, an inverse correlation between miR-1271-5p and ZIC2 expression in AML samples was observed. In summary, ZIC2 was upregulated in AML tissues, and restoration of ZIC2 expression was able to promote the proliferation and reduce the apoptosis of AML cells transfected with miR-1271-5p mimics. The results of the present study demonstrated that miR-1271-5p inhibited the progression of AML by targeting ZIC2.
Keywords: microRNA-1271-5p, Zic family member 2, proliferation, apoptosis, acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous and malignant hematological tumor characterized by the accumulation of undifferentiated myeloid progenitor cells in the bone marrow and peripheral blood (1). At present, chemotherapy and stem cell transplantation are the principal therapeutic approaches for AML (2); however, the outcomes of patients with AML at advanced stage with metastasis and recurrence remain poor (3). Genetic mutations and aberrant expression levels of cancer-associated genes may induce AML (4,5). A previous study demonstrated that impaired expression of Wilms tumor 1, high meningioma 1 and ETS-related gene is associated with the clinical outcomes of AML (6). In addition, a number of microRNAs (miRNAs/miRs) have been reported to be involved in the progression of AML, including miR-19b and miR-375 (7,8); however, the underlying molecular mechanisms remain unknown.
miRNAs are a group of short noncoding RNAs with a length of ~21 nucleotides (9). A recent study has identified that miRNAs serve essential roles in regulating gene expression by binding to the target sequence and promoting the degradation of specific mRNAs (10). miRNAs regulate various biological processes in cancer cells, including cell differentiation, proliferation, metastasis and apoptosis (11,12). A variety of miRNAs are aberrantly expressed in various types of cancer, including AML (13,14). In AML, numerous miRNAs are dysregulated (15). Ding et al (16) identified that upregulated miR-130a may induce cell death in AML. Zhao et al (17) demonstrated that miR-144 is a potential non-invasive biomarker for patients with AML. Ma et al (14) identified that miR-362-5p is a novel prognostic predictor of cytogenically normal AML. Although the importance of miRNAs in AML has been identified, the underlying mechanisms remain unknown.
Recent studies have demonstrated that miR-1271-5p is involved in the pathogenesis of colorectal cancer and hepatocellular carcinoma (18,19); however, whether miR-1271-5p may regulate the progression of AML remains unknown. In the present study, miR-1271-5p inhibited the proliferation and induced apoptosis in AML via targeting zinc family member 2 (ZIC2), which suggested that miR-1271-5p may be a promising therapeutic target for the treatment of patients with AML.
Materials and methods
Clinical specimens
The peripheral blood samples (50 ml per sample) were obtained from patients with AML (n=35) and healthy donors (n=35) at The Third Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) between January 2014 and December 2016. The sex ratio of the patients and controls was ~1:1 and the patients were aged between 2 and 60 years old. Patients were diagnosed with AML according to the pathological and clinic features of the disease (20). Patients who received chemotherapy or radiotherapy prior to collection were excluded; all other patients with AML were included. Informed consent was obtained from each patient, and the present study was approved by the Institutional Ethics Committee of The Third Affiliated Hospital of Wenzhou Medical University. The patients with AML were divided into miR-1271-5p high expression and low expression groups according to the median value of miR-1271-5p expression.
Cell culture and transfection
AML cell lines (HL-60, Kasumi-1, AML193 and OCI-AML2) and a normal cell line derived from marrow stroma (HS-5) were purchased from The American Type Culture Collection (Manassas, VA, USA) and cultured in Eagle's minimum essential medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator containing 5% CO2.
The ZIC2 coding sequence was constructed in a pcDNA3 vector. For cell transfection, 5×105 AML cells were seeded in 6-well plates and cultured with Eagle's minimum rssential medium at 37°C overnight and subsequently transfected with 50 nM miR-1271-5p mimics (5′-CUUGGCACCUAGCAAGCACUCA-3′), inhibitors (5′-UGAGUGCUUGCUAGGUGCCAAG-3′) or the controls (5′-UCACAACCUCCUAGAAAGAGUAGA-3′) or pcDNA3-ZIC2 vector (1 µg; Invitrogen; Thermo Fisher Scientific, Inc.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. miR-1271-5p mimics, inhibitors and the controls were obtained from Applied Biosystems (Thermo Fisher Scientific, Inc.). At 48 h post-transfection, the effects of gene silencing or overexpressing were determined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Cell proliferation assay
In total, 2×103 AML193 and OCI-AML2 cells were transfected for 24 h and seeded in 96-well plates followed by incubation at 37°C for 1, 3 and 5 days. A total of 10 µl Cell Counting kit-8 (CCK-8) reagent (Beyotime Institute of Biotechnology, Haimen, China) was added to the medium and incubated for 2 h; the absorbance at 450 nm was measured using an ELISA reader (Molecular Devices, LLC; Sunnyvale, CA, USA).
Cell cycle and apoptosis analyses
For cell cycle analysis, 2×106 AML193 and OCI-AML2 cells were washed and resuspended in PBS solution containing 0.04 mg/ml propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.) and 100 mg/ml RNase (Invitrogen; Thermo Fisher Scientific, Inc.) for 15 min on ice. The samples were analyzed with a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) using the CELLQuest program 5.1 (BD Biosciences) followed by incubation at room temperature for 30 min. Modifit software 5.0 (Verity Software House, Inc., Topsham, ME, USA) was used to generate a histogram to determine the number of cells in each phase of the cell cycle.
For apoptosis analysis, 2×106 AML cells were transfected with miR-1271-5p mimics, inhibitors or controls for 24 h. Cells were collected by centrifugation at 2,500 × g for 2 min at 4°C. Apoptosis analysis was performed using an Annexin V-FITC Apoptosis Detection kit II (BD Biosciences) according to the manufacturer's protocol. Cells were analyzed by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences). The data were analyzed using Flowjo 7.6.1 (BD Biosciences).
RT-qPCR
Total RNA was extracted from cultured cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and cDNA was synthesized from total RNA with a PrimerScript RT Reagent kit (Takara Bio., Inc., Ostu, Japan) according to the manufacturer's protocol. The conditions were as follows: Incubation for 1 h at 42°C and inactivation at 70°C for 15 min. miRNA from total RNA was reverse transcribed using the Prime-Script miRNA cDNA Synthesis kit (Takara Bio., Inc.) according to the manufacturer's protocol. qPCR was performed using SYBR green Premix Ex Taq II (Takara Bio., Inc.) on a Step One Plus Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The PCR thermocycling conditions were as follows: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec. GAPDH and U6 were used as the endogenous controls for mRNA and miRNA expression levels irrespectively. Relative gene expression was calculated using the 2−∆∆Cq method (21). A total of three repeats were performed. The primer sequences were: U6 (forward, 5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse, 5′-ACGCTTCACGAATTTGCGTGTC-3′), GAPDH (forward, 5′-TCAACGACCACTTTGTCAAGCTCA-3′ and reverse, 5′-GCTGGTGGTCCAGGGGTCTTACT-3′), miR-1271-5p (forward, 5′-CTTGGCACCTAGCAAGCACTCA-3′ and reverse, 5′-GCGAGCACAGAATTAATACGAC-3′) and ZIC2 (forward, 5′-GTCCACACCTCCGATAAGCC-3′ and reverse, 5′-CTCATGGACCTTCATGTGCT-3′).
RNA pulldown assay
Purified synthesized RNAs (Invitrogen; Thermo Fisher Scientific, Inc.) were labeled using biotin with Pierce RNA 3′ End Desthiobiotinylation kit (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A positive control (biotin-labeled wild-type miR-1271-5p, biotin-miR-1271-5p) or negative controls (5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were incubated with AML cell lysates from 1×106 AML cells, isolated using lysis buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), at 4°C overnight. The Streptavidin magnetic beads (Sigma-Aldrich; Merck KGaA, Darmstadt) were added to the lysate and incubated at 4°C for 2 h. The RNA-enriched magnetic beads (Sigma-Aldrich; Merck KGaA) were collected and precipitated RNAs were eluted using elution buffer (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol, followed by RT-qPCR analysis as described above. Forkhead box protein K2 (FOXK2) was the positive control.
Western blotting
Proteins were extracted from AML cells using radioimmunoprecipitation lysis buffer (0.15 mM NaCl, 0.05 mM Tris-HCl, pH 7.5, 1% Triton, 0.1% SDS, 0.1% sodium deoxycholate and 1% NP40). The concentration of proteins was measured using the Bradford method. A total of 20 µg total protein extract was electrophoresed on 12% SDS-PAGE and transferred by electroblotting onto polyvinylidene fluoride membranes. The membranes were blocked for 1 h at 37°C in 5% (w/v) nonfat milk prior to incubation with appropriate dilutions of the primary antibodies against ZIC2 (1:2,000; cat. no. Z4525; Sigma-Aldrich; Merck KGaA) and GAPDH (1:2,000; cat. no. 5174; Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C overnight. The membranes were subsequently incubated with horseradish peroxidase-conjugated secondary antibody (1:5,000; cat. no. ab7090; Abcam, Cambridge, UK) at 25°C for 1 h. The signals were visualized using an enhanced chemiluminescence kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol.
Luciferase reporter assay
The potential targets of miR-1271-5p were predicted by TargetScan 7.0 (http://www.targetscan.org/vert_71/) and miRBase (http://www.mirbase.org/search.shtml). The sequence of ZIC3-3′UTR containing the wild-type or mutant potential binding site for miR1271-5p were inserted into the SpeI and HindIII sites in the pMIR-Report vector (Ambion; Thermo Fisher Scientific, Inc.) to generate the reporter plasmid. miR-1271-5p mimic (50 nM) or inhibitor, reporter plasmid (1 µg) and pRL-CMV Renilla luciferase reporter (1 ng; Ambion; Thermo Fisher Scientific, Inc.) were co-transfected into the cells using Lipofectamine® 2000 according to the manufacturer's protocol. Luciferase activity was assessed at 48 h post-transfection using a Dual-Luciferase® Reporter Assay (Promega Corporation, Madison, WI, USA), and the levels of firefly luciferase activity were normalized to that of Renilla luciferase.
Statistical analysis
All statistical analyses were performed using SPSS 20.0 (IBM Corp., Armonk, NY, USA). The results are expressed as the mean ± standard deviation. A Student's t-test or one-way analysis of variance followed by Tukey's post hoc test was used to analyze the difference in two or multiple groups. Pearson's correlation coefficient analysis was used to determine the correlations. The effect of miR-1271-5p expression on overall survival was analyzed by Kaplan-Meier analysis and a log-rank test. The experiments were repeated three times. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-1271-5p is downregulated in AML samples
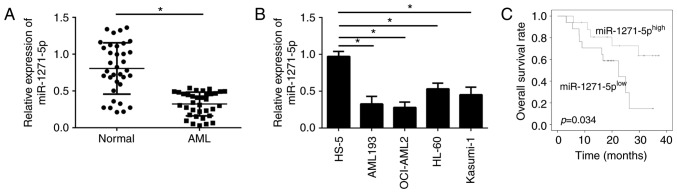
To examine the cellular functions of miR-1271-5p in AML, the expression levels of miR-1271-5p in the blood of patients with AML were evaluated by RT-qPCR. The expression levels of miR-1271-5p were significantly downregulated in AML samples compared with normal samples (P<0.05; Fig. 1A). In addition, miR-1271-5p expression levels were significantly reduced in AML cell lines compared with in the normal cell line HS-5 (P<0.05; Fig. 1B). The downregulation of miR-1271-5p in AML cells may be associated with the clinical outcomes of patients with AML. Furthermore, the AML samples were divided into two groups according to the mean value of miR-1271-5p expression levels. MiR-1271-5p expression above or below the mean value were considered as high or low expression, respectively. Kaplan-Meier analysis demonstrated that patients with AML with high miR-1271-5p expression exhibited improved survival rates (Fig. 1C). These data suggested that miR-1271-5p may be involved in the progression of AML.
Figure 1.
miR-1271-5p is downregulated in AML samples. (A) Average expression levels of miR-1271-5p in AML samples and normal tissues were analyzed were measured by RT-qPCR. (B) miR-1271-5p expression in AML cell lines was examined by RT-qPCR. (C) Median expression of miR-1271-5p was selected as the cutoff value; overall survival was determined by Kaplan-Meier analysis. Data are representative of three independent experiments and expressed as the mean ± standard deviation. *P<0.05 vs. the normal or HS-5 cells group. AML, acute myeloid leukemia; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-1271-5p inhibits AML cell proliferation and induces apoptosis
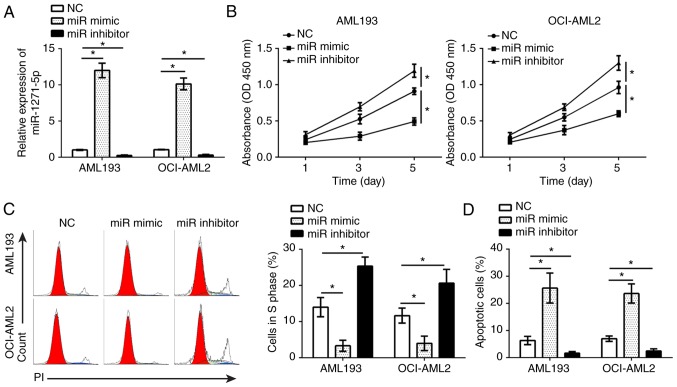
To determine the effects of miR-1271-5p on AML cells, gain- and loss-of-function studies were performed in AML193 and OCI-AML2 cells transfected with miR-1271-5p mimics or inhibitors. RT-qPCR analysis demonstrated that, compared with the control, miR-1271-5p was significantly up- or downregulated in AML193 and OCI-AML2 cells following transfection with mimics and inhibitors, respectively (P<0.05; Fig. 2A). A CCK-8 assay and flow cytometry were used to analyze the effects of miR-1271-5p on cell proliferation and the cell cycle. The results indicated that, compared with the control, miR-1271-5p overexpression significantly inhibited the proliferation of AML cells and reduced the number of cells in S phase, and vice versa (P<0.05; Fig. 2B and C). Additionally, flow cytometry indicated that, compared with the control, miR-1271-5p overexpression significantly promoted the apoptosis of AML cells, and vice versa (P<0.05; Fig. 2D). These results demonstrated that miR-1271-5p serves as a tumor suppressor in the progression of AML.
Figure 2.
miR-1271-5p inhibits AML cell proliferation and induces apoptosis. (A) Expression levels of miR-1271-5p in AML193 and OCI-AML2 cells transfected with miR-1271-5p inhibitor, mimic or NC were measured by reverse transcription-quantitative polymerase chain reaction. (B) Cell proliferation was determined in AML193 and OCI-AML2 cells transfected with miR-1271-5p inhibitor, mimic or NC by a Cell Counting kit-8 assay. (C) Cell cycle was analyzed in AML193 and AML2 cells transfected with miR-1271-5p inhibitor, mimic or NC by flow cytometry. (D) Cell apoptosis in AML193 and OCI-AML2 cells transfected with miR-1271-5p inhibitor, mimic or NC. Data are representative of three independent experiments and expressed as the mean ± standard deviation. *P<0.05 vs. the NC group. AML, acute myeloid leukemia; FACS, fluorescence activated cell sorter; miR, microRNA; NC, negative control.
ZIC2 is a target of miR-1271-5p in AML cells
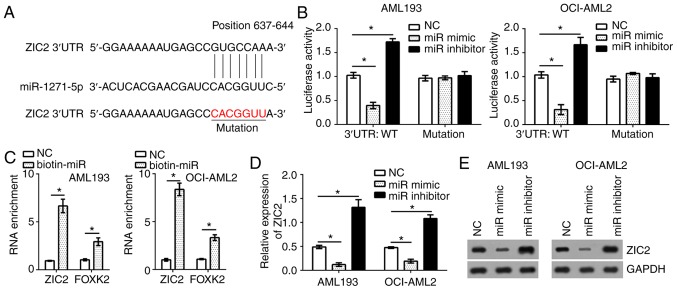
To identify the target genes of miR-1271-5p in AML cells, bioinformatics analysis was conducted using online software (TargetScan 7.0 and miRBase). The results identified that transcription factor ZIC2 was a possible target gene of miR-1271-5p. There was a potential binding site of miR-1271-5p in the 3′-untranslated region of ZIC2 mRNA (Fig. 3A). To determine the interaction between miR-1271-5p and ZIC2, a luciferase reporter assay was performed. The results indicated that, compared with the control, the luciferase activity of ZIC2 3′-UTR (wild-type, WT) in AML193 and OCI-AML2 cells were significantly reduced and increased by transfection with miR-1271-5p mimics and inhibitors, respectively (Fig. 3B); however, a mutation in the binding site of miR-1271-5p in ZIC2 3′-UTR abrogated the effects of miR-1271-5p on the luciferase activity (Fig. 3B). Furthermore, an RNA pulldown assay was performed using FOXK2 as a positive control (19). The results indicated that biotin-miR-1271-5p significantly promoted the precipitation of ZIC2 and FOXK2 mRNA in AML cells compared with the control (P<0.05; Fig. 3C), suggesting the direct interaction between miR-1271-5p and ZIC2. To further confirm whether ZIC2 was a target of miR-1271-5p in AML cells, the mRNA and protein expression levels of ZIC2 were evaluated by RT-qPCR and western blotting, respectively. Overexpression of miR-1271-5p significantly suppressed the mRNA expression levels of ZIC2 in AML193 and OCI-AML2 cells, and vice versa, compared with the control; a notable decrease in ZIC2 protein expression was observed in response to miR-1271-5p overexpression (P<0.05; Fig. 3D and E).
Figure 3.
ZIC2 is a target of miR-1271-5p. (A) ZIC2 was predicted as the target gene of miR-1271-5p. (B) Luciferase assay was utilized to detect ZIC2 3′-UTR (WT or mutation) activity. AML193 and OCI-AML2 cells were transfected with the ZIC2 3′-UTR (WT or mutation), and miR-1271-5p mimic or inhibitor; the luciferase activity was measured with a dual luciferase reporter assay kit. (C) RNA pulldown assay indicated that biotin-miR-1271-5p precipitated ZIC2 mRNA. FOXK2 served as a positive control. (D) mRNA levels of ZIC2 were analyzed by reverse transcription-quantitative polymerase chain reaction in AML193 and OCI-AML2 cells transfected with miR-1271-5p mimic or inhibitor. (E) Protein expression levels of ZIC2 were analyzed by western blotting in AML193 and OCI-AML2 cells transfected with miR-1271-5p mimic or inhibitor. Data are representative of three independent experiments and expressed as the mean ± standard deviation. *P<0.05 vs. the NC group. AML, acute myeloid leukemia; miR, microRNA; NC, negative control; UTR, untranslated region; FOXK2, forkhead box protein 2.
miR-1271-5p is negatively correlated with the expression levels of ZIC2 in AML samples
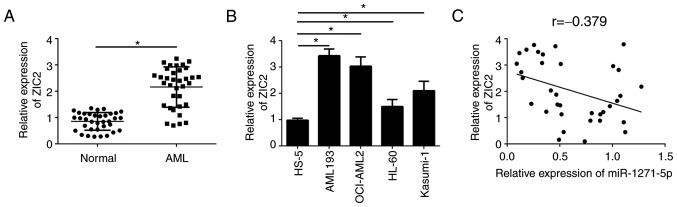
To further investigate the association between miR-1271-5p and ZIC2 in AML samples, the expression patterns of ZIC2 were analyzed by RT-qPCR. ZIC2 was significantly upregulated in AML tissues compared with normal samples (P<0.05; Fig. 4A). Consistently, ZIC2 was additionally upregulated in AML cell lines compared with the normal cell line, HS-5 (Fig. 4B). Furthermore, RT-qPCR analysis indicated that ZIC2 was negatively correlated with miR-1271-5p in AML samples (Fig. 4C). These results suggested that ZIC2 may be regulated by miR-1271-5p and serve an important role in AML.
Figure 4.
miRNA-1271-5p is negatively correlated with ZIC2 expression in AML samples. (A) ZIC2 mRNA expression in AML tissues was determined by RT-qPCR. (B) Relative expression levels of ZIC2 in AML cell lines were examined by RT-qPCR. (C) Correlation between ZIC2 mRNA and miR-1271-5p expression in AML samples. Data are representative of three independent experiments and expressed as the mean ± standard deviation. *P<0.05 vs. the normal group or HS-5 cells. AML, acute myeloid leukemia; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; ZIC2, Zic family member 2.
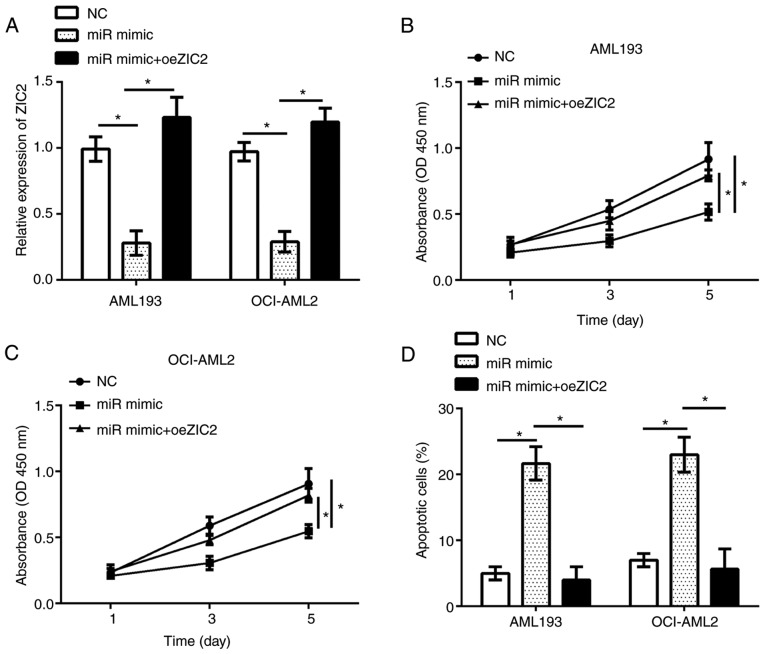
Restoration of ZIC2 reverses the effects of miR-1271-5p overexpression on the proliferation and apoptosis of AML cells
The functions of ZIC2 in AML remain unknown. The present study investigated the functions of ZIC2 and whether miR-1271-5p inhibits the progression of AML by regulating ZIC2; the expression of ZIC2 was restored in AML193 and OCI-AML2 cells transfected with miR-1271-5p. RT-qPCR analysis demonstrated that ZIC2 expression was significantly upregulated in AML193 and OCI-AML2 cells following transfection with an overexpression plasmid compared with the control (Fig. 5A). The results of CCK-8 and flow cytometry analyses indicated that, compared with the control, restoration of ZIC2 significantly rescued the proliferation and reduced apoptosis in AML193 and OCI-AML2 cells transfected with miR-1271-5p (Fig. 5B-D). In summary, these data suggested that miR-1271-5p regulated the progression of AML by targeting ZIC2.
Figure 5.
Restoration of ZIC2 reverses the effect of miRNA-1271-5p overexpression on AML cell proliferation and apoptosis. (A) Relative expression of ZIC2 in AML cells transfected with oeZIC2 was measured by reverse transcription-quantitative polymerase chain reaction. (B and C) Cell proliferation was determined by Cell Counting kit-8 assay in AML cells transfected with oeZIC2. (D) Cell apoptosis was analyzed by flow cytometry in AML cells transfected with indicated plasmids. Data are representative of three independent experiments and expressed as the mean ± standard deviation. *P<0.05 vs. the NC or miR-mimic group. AML, acute myeloid leukemia; miR, microRNA; NC, negative control; oeZIC2, Zic family member 2 overexpression plasmid.
Discussion
A recent study revealed that numerous miRNAs were aberrantly expressed in AML (22). For example, Tian et al (23) demonstrated that miR-494 was downregulated, and suppresses bone marrow stromal cell-mediated drug resistance in AML cells. Wang et al (12) demonstrated that miR-183 is upregulated and promotes cell proliferation by regulating programmed cell death 6 in pediatric AML. Yang et al (24) identified that downregulated miR-34c was associated with poor outcome in de novo AML. Jiang et al (25) observed that miR-22 is downregulated in AML and has a notable anti-tumor role with therapeutic potential. A recent study identified that miR-1271-5p suppressed the progression of colorectal cancer (18). Another previous study suggested that miR-1271-5p inhibited cell growth by targeting FOXK2 in hepatocellular carcinoma (19); however, the functions of miR-1271-5p in AML remain unknown. In the present study, the expression patterns of miR-1271-5p in AML samples and cell lines were investigated. The data indicated that miR-1271-5p was significantly downregulated in AML samples and cell lines; miR-1271-5p overexpression suppressed AML cell proliferation and induced apoptosis by directly targeting ZIC2.
ZIC2 is a member of the ZIC family of C2H2-type zinc finger proteins, which function as transcription factors and may regulate tissue specific gene expression (26). A recent study indicated that ZIC2 served oncogenic roles in human cancer, and regulated P21 protein-activated kinase 4 to promote the growth and metastasis of hepatocellular carcinoma (27). Marchini et al (28) demonstrated that ZIC2 exhibited features of an oncogene and its overexpression strongly correlated with the clinical course of epithelial ovarian cancer. Sakuma et al (29) identified that ZIC2 served as a prognostic marker in oral squamous cell carcinoma. Chan et al (30) demonstrated that ZIC2 synergistically promoted Hedgehog signaling via the nuclear retention of zinc finger protein GLI1 in cervical cancer cells. In addition, recent studies demonstrated that ZIC2 regulated the progression of nasopharyngeal carcinoma (31), liver cancer (32) and bladder cancer (33); however the functions of ZIC2 in AML remain unknown. In the present study, ZIC2 was identified as a direct target of miR-1271-5p in AML cells. Furthermore, ZIC2 was upregulated in AML samples and cell lines, which suggested that ZIC2 may serve oncogenic roles in AML. The expression level of miR-1271-5p was negatively correlated with that of ZIC2 in AML samples; however, ZIC2 mRNA may also be targeted by other miRNAs. The mechanisms underlying ZIC2-regulated proliferation and apoptosis in AML cell remain unknown. Wang et al (33) demonstrated that ZIC2 correlated with the phosphoinositide 3-kinase/protein kinase B signaling pathway to regulate cell proliferation and apoptosis in bladder cancer. Whether ZIC2 regulates AML cells in a similar manner requires further investigation.
In the present study, the interaction between miR-1271-5p and ZIC2 was identified. Overexpression of miR-1271-5p was associated with the reduced luciferase activity in AML cells transfected with WT-ZIC2 3′-UTR. In addition, the overexpression of miR-1271-5p markedly reduced the mRNA and protein expression levels of ZIC2 in AML193 and OCI-AML2 cells. Furthermore, an inverse correlation between the expression levels of miR-1271-5p and ZIC2 in AML samples was demonstrated by RT-qPCR. Additionally, restoration of ZIC2 reversed the effects of miR-1271-5p overexpression on the proliferation and apoptosis of AML cells.
In summary, the present study demonstrated that miR-1271-5p was downregulated in AML cells, which may regulate cell proliferation and apoptosis by targeting ZIC2. The findings additionally indicated ZIC2 as an oncogene in AML progression, suggesting that the miR-1271-5p/ZIC2 axis may be a promising therapeutic target for the treatment of AML.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XC and SY collected all the acute myeloid leukemia samples. XC, SY, JZ and MC performed the experiments. MC designed the present study and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
For the use of human samples, the present study was approved by the Institutional Ethics Committee of The Third Affiliated Hospital of Wenzhou Medical University. Written informed consent was obtained from all patients for the use of their clinical tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88:318–327. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- 2.Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127:53–61. doi: 10.1182/blood-2015-08-604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalaj M, Tavakkoli M, Stranahan AW, Park CY. Pathogenic microRNA's in myeloid malignancies. Front Genet. 2014;5:361. doi: 10.3389/fgene.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcucci G, Haferlach T, Döhner H. Molecular genetics of adult acute myeloid leukemia: Prognostic and therapeutic implications. J Clin Oncol. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 5.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 6.Baldus CD, Mrózek K, Marcucci G, Bloomfield CD. Clinical outcome of de novo acute myeloid leukaemia patients with normal cytogenetics is affected by molecular genetic alterations: A concise review. Br J Haematol. 2007;137:387–400. doi: 10.1111/j.1365-2141.2007.06566.x. [DOI] [PubMed] [Google Scholar]
- 7.Bi L, Zhou B, Li H, He L, Wang C, Wang Z, Zhu L, Chen M, Gao S. A novel miR-375-HOXB3-CDCA3/DNMT3B regulatory circuitry contributes to leukemogenesis in acute myeloid leukemia. BMC Cancer. 2018;18:182. doi: 10.1186/s12885-018-4097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang TJ, Lin J, Zhou JD, Li XX, Zhang W, Guo H, Xu ZJ, Yan Y, Ma JC, Qian J. High bone marrow miR-19b level predicts poor prognosis and disease recurrence in de novo acute myeloid leukemia. Gene. 2018;640:79–85. doi: 10.1016/j.gene.2017.10.034. [DOI] [PubMed] [Google Scholar]
- 9.Zhang S, Zhang Q, Shi G, Yin J. MiR-182-5p regulates BCL2L12 and BCL2 expression in acute myeloid leukemia as a potential therapeutic target. Biomed Pharmacother. 2018;97:1189–1194. doi: 10.1016/j.biopha.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Zebisch A, Hatzl S, Pichler M, Wölfler A, Sill H. Therapeutic resistance in acute myeloid leukemia: The role of non-coding RNAs. Int J Mol Sci. 2016;17(pii):E2080. doi: 10.3390/ijms17122080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan N, Zhang J, Cheng C, Zhang X, Feng J, Kong R. MicroRNA-384 represses the growth and invasion of non-small-cell lung cancer by targeting astrocyte elevated gene-1/Wnt signaling. Biomed Pharmacother. 2017;95:1331–1337. doi: 10.1016/j.biopha.2017.08.143. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Xue J, Kuang H, Zhou X, Liao L, Yin F. microRNA-1297 inhibits the growth and metastasis of colorectal cancer by suppressing cyclin D2 expression. DNA Cell Biol. 2017;36:991–999. doi: 10.1089/dna.2017.3829. [DOI] [PubMed] [Google Scholar]
- 13.You Y, Tan J, Gong Y, Dai H, Chen H, Xu X, Yang A, Zhang Y, Bie P. MicroRNA-216b-5p Functions as a Tumor-suppressive RNA by targeting TPT1 in pancreatic cancer cells. J Cancer. 2017;8:2854–2865. doi: 10.7150/jca.18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma QL, Wang JH, Yang M, Wang HP, Jin J. MiR-362-5p as a novel prognostic predictor of cytogenetically normal acute myeloid leukemia. J Transl Med. 2018;16:68. doi: 10.1186/s12967-018-1445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Zou Y, Lin L, Ma X, Chen H. Identification of serum miR-34a as a potential biomarker in acute myeloid leukemia. Cancer Biomark. 2018;22:799–805. doi: 10.3233/CBM-181381. [DOI] [PubMed] [Google Scholar]
- 16.Ding C, Chen SN, Macleod RAF, Drexler HG, Nagel S, Wu DP, Sun AN, Dai HP. MiR-130a is aberrantly overexpressed in adult acute myeloid leukemia with t(8;21) and its suppression induces AML cell death. Ups J Med Sci. 2018;123:19–27. doi: 10.1080/03009734.2018.1440037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q, Li J, Chen S, Shen K, Ai G, Dai X, Xie B, Shi Y, Jiang S, Feng J, Li W. Decreased miR-144 expression as a non-invasive biomarker for acute myeloid leukemia patients. Pharmazie. 2017;72:232–235. doi: 10.1691/ph.2017.6846. [DOI] [PubMed] [Google Scholar]
- 18.Wu Z, Hu Z, Han X, Li Z, Zhu Q, Wang Y, Zheng Q, Yan J. The BET-Bromodomain Inhibitor JQ1 synergized ABT-263 against colorectal cancer cells through suppressing c-Myc-induced miR-1271-5p expression. Biomed Pharmacother. 2017;95:1574–1579. doi: 10.1016/j.biopha.2017.09.087. [DOI] [PubMed] [Google Scholar]
- 19.Lin MF, Yang YF, Peng ZP, Zhang MF, Liang JY, Chen W, Liu XH, Zheng YL. FOXK2, regulted by miR-1271-5p, promotes cell growth and indicates unfavorable prognosis in hepatocellular carcinoma. Int J Biochem Cell Biol. 2017;88:155–161. doi: 10.1016/j.biocel.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Li J, Hu J, Zheng J, Zheng Z, Liu T, Lin Z, Lin M. Emodin enhances ATRA-induced differentiation and induces apoptosis in acute myeloid leukemia cells. Int J Oncol. 2014;45:2076–2084. doi: 10.3892/ijo.2014.2610. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Liao Q, Wang B, Li X, Jiang G. miRNAs in acute myeloid leukemia. Oncotarget. 2017;8:3666–3682. doi: 10.18632/oncotarget.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian C, Zheng G, Zhuang H, Li X, Hu D, Zhu L, Wang T, You MJ, Zhang Y. MicroRNA-494 activation suppresses bone marrow stromal cell-mediated drug resistance in acute myeloid leukemia cells. J Cell Physiol. 2017;232:1387–1395. doi: 10.1002/jcp.25628. [DOI] [PubMed] [Google Scholar]
- 24.Yang DQ, Zhou JD, Wang YX, Deng ZQ, Yang J, Yao DM, Qian Z, Yang L, Lin J, Qian J. Low miR-34c expression is associated with poor outcome in de novo acute myeloid leukemia. Int J Lab Hematol. 2017;39:42–50. doi: 10.1111/ijlh.12566. [DOI] [PubMed] [Google Scholar]
- 25.Jiang X, Hu C, Arnovitz S, Bugno J, Yu M, Zuo Z, Chen P, Huang H, Ulrich B, Gurbuxani S, et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat Commun. 2016;7:11452. doi: 10.1038/ncomms11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inaguma S, Ito H, Riku M, Ikeda H, Kasai K. Addiction of pancreatic cancer cells to zinc-finger transcription factor ZIC2. Oncotarget. 2015;6:28257–28268. doi: 10.18632/oncotarget.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu SX, Zhang CZ, Luo RZ, Wang CH, Liu LL, Fu J, Zhang L, Wang H, Xie D, Yun JP. Zic2 promotes tumor growth and metastasis via PAK4 in hepatocellular carcinoma. Cancer Lett. 2017;402:71–80. doi: 10.1016/j.canlet.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Marchini S, Poynor E, Barakat RR, Clivio L, Cinquini M, Fruscio R, Porcu L, Bussani C, D'Incalci M, Erba E, et al. The zinc finger gene ZIC2 has features of an oncogene and its overexpression correlates strongly with the clinical course of epithelial ovarian cancer. Clin Cancer Res. 2012;18:4313–4324. doi: 10.1158/1078-0432.CCR-12-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakuma K, Kasamatsu A, Yamatoji M, Yamano Y, Fushimi K, Iyoda M, Ogoshi K, Shinozuka K, Ogawara K, Shiiba M, et al. Expression status of Zic family member 2 as a prognostic marker for oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2010;136:553–559. doi: 10.1007/s00432-009-0689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan DW, Liu VW, Leung LY, Yao KM, Chan KK, Cheung AN, Ngan HY. Zic2 synergistically enhances Hedgehog signalling through nuclear retention of Gli1 in cervical cancer cells. J Pathol. 2011;225:525–534. doi: 10.1002/path.2901. [DOI] [PubMed] [Google Scholar]
- 31.Shen ZH, Zhao KM, Du T. HOXA10 promotes nasopharyngeal carcinoma cell proliferation and invasion via inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci. 2017;21:945–952. [PubMed] [Google Scholar]
- 32.Zhu P, Wang Y, He L, Huang G, Du Y, Zhang G, Yan X, Xia P, Ye B, Wang S, et al. ZIC2-dependent OCT4 activation drives self-renewal of human liver cancer stem cells. J Clin Invest. 2015;125:3795–3808. doi: 10.1172/JCI81979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Ma W, Liu Y. Long non-coding RNA HULC promotes bladder cancer cells proliferation but inhibits apoptosis via regulation of ZIC2 and PI3K/AKT signaling pathway. Cancer Biomark. 2017;20:425–434. doi: 10.3233/CBM-170188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.