Graphical abstract
Keywords: ZnO-NP, S. cerevisiae, 2DE, 1H NMR, Metabolomics, qRT-PCR
Highlights
-
•
Untargeted proteomic and metabolic approaches provide complete toxicity assessment.
-
•
ZnO-NPs de-regulate the proteome and metabolome of S. cerevisiae.
-
•
ZnO-NPs affect the key metabolites of central metabolic pathways.
-
•
Protein and/or metabolite can be used as biomarker specific to the ZnO-NPs induced toxicity.
Abstract
As zinc oxide nanoparticles are being increasingly used in various applications, it is important to assess their potential toxic implications. Stress responses and adaptations are primarily controlled by modulation in cellular proteins (enzyme) and concentration of metabolites. To date proteomics or metabolomics applications in nanotoxicity assessment have been applied to a restricted extent. Here we utilized 2DE and 1H NMR based proteomics and metabolomics respectively to delineate the toxicity mechanism of zinc oxide nanoparticles (ZnO-NPs) in budding yeast S. cerevisiae. We found that the physiological and metabolic processes were altered in the S. cerevisiae upon ZnO-NPs exposure. Almost 40% proteins were down-regulated in ZnO-NPs (10 mg L−1) exposed cell as compared to control. Metabolomics and system biology based pathway analysis, revealed that ZnO-NPs repressed a wide range of key metabolites involved in central carbon metabolism, cofactors synthesis, amino acid and fatty acid biosynthesis, purines and pyrimidines, nucleoside and nucleotide biosynthetic pathways. These metabolic changes may be associated with the energy metabolism, antioxidation, DNA and protein damage and membrane stability. We concluded that untargeted proteomic and metabolic approaches provide more complete measurements and suggest probable molecular mechanisms of nanomaterials toxicity.
1. Introduction
Among engineered nanomaterials, zinc oxide nanoparticles (ZnO-NPs) are the most common industrially produced nanomaterials. They gained much interest due to their applications in diverse fields ranging from sunscreens, to ceramics, superconductors and optoelectronic devices [1]. Owing to their extensive use and increased environmental and occupational exposure ZnO-NPs have become an important compound for public health issues therefore, an appropriate risk assessment for this compound is necessary. For these reasons, in the past few years several researches have been conducted [[2], [3], [4], [5]]. The accumulating research evidences documented that exposure to ZnO-NPs can be toxic to biological system ranging from prokaryotes to higher eukaryotes including humans [[2], [3], [4], [5]]. Among the eukaryotic microorganisms, the budding yeast Saccharomyces cerevisiae (S. cerevisiae) represents a well-accepted and extensively used eukaryotic model system for basic and applied studies because its fundamental cellular processes and pathways are similar to those of higher eukaryotic organisms including humans [6]. Number of characteristics features and availability of yeast metabolome database (YMDB) makes it an excellent organism to carry out functional toxicological and metabolomic studies [7]. Several advantages are recognised by exploiting yeast cells, such as to dissect unknown mechanisms of action, to identify potential cellular targets and to characterize novel molecular biomarkers [8,9]. Although, few nanotoxicity studies have been done on this model organism [4,10], but the proteomic, metabolomic and systems biology based studies are scarce. In our previous research, we draw a conclusive mechanism(s) underlying the nanotoxicity of ZnO-NPs [11]. Although several toxicity pathways are reported but the studies explicitly dealing with the proteomic and metabolomic variations upon ZnO-NPs exposure to S. cerevisiae, have gained relatively little attention.
In order to gain more information on the mechanism of nanotoxicity there is an urgent need to apply metabolomics in the field of nanotoxicology and in its integration with molecular profiling at protein level therefore, more studies are necessary in these directions [12,13]. We utilized proteomic and metabolomics to present a multi-omics approach for better understanding the nanotoxicity. Briefly, we investigated the variation in the ZnO-NPs exposed budding yeast S. cerevisiae at the proteome and the metabolome level. Qualitative data of de-regulated proteins obtained using two-dimensional gel electrophoresis (2DE) and quantitative data of de-regulated metabolites gained by utilizing 1H-NMR were interpreted using systems biology analysis. The aim of this study is to understand the biological processes that are modulated upon interaction of ZnO-NPs with living systems. This research contributing towards a detailed understanding of the biochemical mechanism(s) triggered in the yeast upon ZnO-NPs exposure. The knowledge gained based on the information of networks, processes, and pathways modulated can be used to advance the design of nanoparticles and/or to discover definite biomarkers of nanotoxicity.
2. Material and Methods2.1Yeast G
2.1. Yeast growth and ZnO-NPs treatment
S. cerevisiae, BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) cells were pre-cultured in liquid YPD medium. The overnight grown culture inoculated in 300 ml (in duplicate) YPD medium (OD600nm 0.2), the cells were grown until mid-exponential growth phase (OD600nm 1.3–1.5). Cells were harvested by centrifugation and thoroughly washed thrice with sterile deionized water (DI) water. The cell pellets were resuspended and diluted using 300 ml sterile DI water. On the basis of our previous cytotoxicity test, the experimental test concentrations of ZnO-NPs (10 mg L−1) was selected for the assessment of metabolic toxicity [11]. Under sterile conditions, water suspended yeast cells were equally divided into two flasks one left untreated (control) and other used for ZnO-NPs treatment in duplicates. Cells were incubated for three hours (200 rpm, 30 °C) for toxicity evaluation in an incubator shaker. After exposure, the 100 ml control and treated cultures and rest 50 ml cultures were used for the metabolite and protein extraction respectively.
2.2. Protein extraction and Western blotting
Fifty ml culture was used for protein performed by a previously described method with desired modifications [11,14]. Briefly, control and ZnO-NPs treated cells were harvested and instantaneously frozen at −80 °C. Frozen cells were thoroughly washed by three times with sterile DI water subsequently cell pellet were resuspended in 400 μl extraction buffer [50 mM Tris−HCl (pH 7.5); NaCl 200 mM; Triton X-100 0.1%; Glycerol 5%; EDTA 1 mM; DTT 5 mM; PMSF 0.5 mM]. Protease inhibitor cocktail (Sigma-Aldrich) and glass beads (acid washed; 0.5 mm; Sigma-Aldrich) were added to the cell suspension. Cells were disrupted by vigorous vortexing eight times for 40 s (samples were cooled on ice for 40 s in between the vortex steps). Remove all the glass beads from the cell extract and transferred cell extract to a fresh microcentrifuge tube and centrifuged at 12,000 for 10 min at 4 °C. The supernatant was recovered completely by transferring to a fresh microcentrifuge tube (fraction 1). The insoluble fractions were again suspended in 300 μl extraction buffer mix cell suspension by thorough vortexing and pipetting up and down with a 200 μl pipette tip. The sample was boiled for 5 min and immediately cooled on ice for 5 min. After centrifugation for 10 min (12,000 RPM, 4 °C), the supernatant (fraction 2) was then transferred to a fresh microcentrifuge tube and mixed with fraction 1. Consequently, add 100 μl of DNase and RNase solution [1% (w/v) DNase I, 0.25% (w/v) RNase A, 50 mM MgCl2, 0.5 M Tris−HCl, pH 7.0] and the combined fractions were incubated on ice for 20 min. The mixed protein extract was then purified by using a 2-D Clean-Up Kit (GE Healthcare, Uppsala, Sweden), and the purified protein sample was dissolved in rehydration solution [8 M urea; 4% (w/v) CHAPS; 5.4 mg/ml dithiothreitol; 0.002% (w/v) bromophenol blue] supplemented with 2% (v/v) 3–10 IPG buffer (GE Healthcare). Total protein concentration was determined by Bradford method. Dissolved protein samples were stored at −80 °C before proteomic assays. The equal concentrations of protein extracts were separated by electrophoresis on SDS-polyacrylamide gel. Immunoblotting analysis of protein extracts was performed as described previously [11]. Blots were incubated (1 h) with primary antibodies anti-Tbp (polyclonal antibodies against recombinant yeast TBPp) and anti-Rap1 (polyclonal antibodies against recombinant Rap1p). After that exposed to the IRDyeH 800CW anti-rabbit IgG secondary antibody for 1 h. Blots were scanned with an Odyssey Infrared Imager (LI−CORH Biosciences).
2.3. 2-D gel electrophoresis (2DE)
2DE experiment was performed by a standard protocol described elsewhere [14]. For the first dimension (isoelectric focussing; IEF) 90 μg (make up the total volume with rehydration buffer to 125 μl) protein samples were loaded on a 7 cm Immobiline Dry-Strip pH 3–10 (GE Healthcare) avoiding air bubbles between protein samples and strips, and the IPG strips were rehydrated overnight at room temperature. IEF was performed with Ettan IPGphore 3 system (GE Healthcare) at 20 °C with a 4 step gradient program: 100 V for 60 min (0.25 kV h), 500 V for 60 min, 1000 V for 60 min, 5000 V for 180 min and finally at 5000 V for a total running of 40 kV h. Prior to the second dimension (SDS-PAGE), the IPG strips were equilibrated by incubating for 15 min in equilibration buffer [75 mM Tris−HCl (pH 8.8); 6 M urea; 30% (v/v) glycerol; 2% (w/v) SDS; 0.002% (w/v) bromophenol blue] containing 1% (w/v) dithiothreitol, followed by 15 min incubation in equilibration buffer containing 2.5% (w/v) iodoacetamide. Second-dimension electrophoresis was performed in miniVE vertical electrophoresis system (Amersham Biosciences). The IPG strips were placed on top of 12.0% polyacrylamide gels and sealed with agarose solution (0.5% w/v) containing bromophenol blue. The vertical gels were run at 10 mA per gel until the bromophenol blue had migrated to the bottom of the gel. Proteins were visualized using colloidal coomassie blue (CBB G-250) staining. The CBB stained gels were scanned by A3 Epson Transparency Unit (Epson).
2.4. Image analysis
The scanned gels for protein spots were analyzed by ImageMaster™ 2D Platinum software (GE Healthcare) for background subtraction, spot detection, quantification and spot comparison between control and treated gels. Quantification of the protein spots were calculated as the volumes (intensity × mm2) of the spots. The relative abundance (control vs. treated) of the spot volume at the desired time was compared using Student’s t-test. P values >0.05 were considered statistically significant.
2.5. Metabolite Extraction and 1H NMR sample preparation
Extraction of metabolites was done as described previously [15]. Briefly, the cells were collected by cenrifugation, washed with 100 ml of ice-cold water and instantaneously frozen at −80 °C. Frozen cells were carefully washed thrice with 10 ml ice-cold water. Then cell pellets were resuspended in 4 ml 75% EtOH and an equal volume of glass beads; metabolites were extracted by vigorous vortexing 10 times for 35 s with 40 s breaks in ice. Subsequently, samples were centrifuged for 5 min at 2000 x g to remove cell debris and glass beads from the extract and equal volumes of the supernatant containing metabolites were evaporated under vacuum. Aqueous samples were dissolved in 600 μl of deuterated phosphate buff ;er (Na2DPO4 100 mM, pD 7.5) in D2O with 0.1 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) as an internal standard for chemical shift and concentration.
2.6. 1H NMR experiments and spectra pre-processing
Spectra were recorded in an AVANCE- III 700 MHz FT NMR spectrometer (Ascend™, Bruker), in a DQD mode at a frequency of 4323.96 Hz 5-mm QCI cryoprobe. Bruker pulse sequences were acquired with 128 free induction decays (FIDs) and 64 k data points over a spectral width of 20 ppm. 512 scans were used with a relaxation delay of 5 s. The FID values were multiplied by an exponential function with a 0.3 Hz line broadening factor. Spectra were automatically processed for the phase and base correction and after that referenced using the Topspin (AU program apk0.NOE) library. 1H NMR spectra were pre-processed with Topspin 3.5 pl 7 software (Bruker) and normalised to the sum of total spectrum intensity to minimise the effect of the differences in sample concentration. The regions corresponding to water and DSS were excluded during the normalisation and processing. For metabolites quantification, we exploited the algorithm called GSD (global spectrum deconvolution). Overlapping regions were deconvoluted and absolute quantification also performed for metabolites with resonances in crowded spectral areas [16]. For each compound, the mean value of the different assigned signals was determined.
2.7. Metabolite identification and quantification
Metabolite assignment was performed by a detailed, targeted metabolite profiling analysis of the 1H NMR signals using the YMDB [7]. Each spectral intensity data set was normalized to the assigned chemical compounds according to total sum of the spectral regions, converted to tab-delimited text (.txt) and comma-separated values (.csv) imported into Web-based MetaboAnalystv3.5 software to carry out normalization, scaling and multivariate analysis. Significance was determined with a p-value threshold (<0.05). Metabolites levels were normalized using the log2 function and then, mean centering and pare to scaling was applied for all PCA and heatmap by MetaboAnalystv3.5 [17].
2.8. Isolation of total RNA and real-time quantitative PCR (qPCR)
RNA extraction, cDNA preparation and the qPCR experiments were performed by following the previously described methods with desired modifications [18]. Briefly, the exponentially growing cells were harvested (OD600 = 1.5) from each control and ZnO-NPs treated samples. Total RNA was extracted by a heat/freeze phenol method and treated with a TURBO DNA-free™ Kit (Invitrogen™), thereafter quantified in a NanoDrop 1000 Spectrophotometer (Thermo Scientific) for 260/280 ratio (i.e., 1.94) and its integrity was checked on 2% agarose gel. Genomic DNA contamination was checked by PCR on a total RNA template using primers targeting the ACT gene. For the synthesis of cDNA equal amount (1 μg) of purified RNA from each control and treated samples were used. cDNA was synthesized by a high capacity RNA-to-cDNA kit (Bio-Rad Laboratories Pvt. Ltd., Gurgaon, India) following the manufacturer’s instructions. Three technical replicates were performed for minimum three biological replicates using equal concentration of synthesized cDNA from every control and ZnO-NPs treated samples. Gene information and the primers used for real time PCR in this study were listed in Table 1. Real-time PCR experiments were performed using 2XSYBR green master mixes (ABI) in an ABI-7300 RT-PCR with Sequence Detection System v1.4 (Applied Biosystems, CA). Thermal cycler conditions included initial denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 58 °C for 20 s, and elongation at 72 °C for 20 s. Melting curve analysis was performed for each primer pair, and relative changes in expression levels between control and treated samples were calculated by using the 2-ΔΔCT method. The relative expression fold change were calculated using the actin (ACT1) housekeeping gene as control, data represent the means ± standard errors of the results of three independent experiments.
Table 1.
List of primers and genes information used in this study.
| S.No | Genes | Oligo Sequences | Description |
|---|---|---|---|
| 1 | ACT1 | F-CCTTCTGTTTTGGGTTTGGA R-CGGTGATTTCCTTTTGCATT |
Actin; structural protein involved in cell polarization, endocytosis, and other cytoskeletal functions. |
| 2 | CRD1 | F-TACGGCCTGAAAACCATTGC R-CATGCCACACGACCAGGATA |
Cardiolipin synthase; produces cardiolipin, which is a phospholipid of the mitochondrial inner membrane that is required for normal mitochondrial membrane potential and function and for correct integration of membrane-multispanning proteins into the mitochondrial outer membrane; required to maintain tubular mitochondrial morphology and functions in mitochondrial fusion; also required for normal vacuolar ion homeostasis. |
| 3 | PSD1 | F-TACCGCTGAATGCGATGTCT R-AACGTCTTCGCCTTGTGCTA |
Phosphatidylserine decarboxylase; involved in phosphatidylcholine biosynthesis, positive regulation of protein processing, integral component of mitochondrial inner membrane; regulates mitochondrial fusion and morphology by affecting lipid mixing in the mitochondrial membrane. |
| 4 | ACO1 | F-ATGTTATGGCAGGTCGTCCA R-ACCCATACCAGTAGCGGAGA |
Aconitase; required for the tricarboxylic acid (TCA) cycle and also independently required for mitochondrial genome maintenance; component of the mitochondrial nucleoid; mutation leads to glutamate auxotrophy; human homolog ACO2 can complement yeast null mutant |
| 5 | SOD1 | F-GTGTCTCTGCTGGTCCTCAC R-GGATAACGACGCTTCTGCCT |
SuperOxide Dismutase- Cytosolic copper-zinc superoxide dismutase; detoxifies superoxide; stabilizes Yck1p and Yck2p kinases in glucose to repress respiration; phosphorylated by Dun1p, enters nucleus under oxidative stress to promote transcription of stress response genes; localized to the nucleus, cytosol, and mitochondrial intermembrane space. |
| 6 | SOD2 | F-GCATTACACCAAGCACCATC R-GAGCCAGGTTTTCCCAGAAT |
SuperOxide Dismutase- Mitochondrial manganese superoxide dismutase; protects cells against oxygen toxicity and oxidative stress; human mitochondrial SOD2 can complement a yeast null mutant and human cytoplasmic SOD1 can also complement when targeted to the mitochondrial matrix. |
| 7 | INO2 | F-AACACACGAGCTCGGCATAA R-CTCAACAACCCCGAACTGGA |
INOsitol requiring- Transcription factor; component of the heteromeric Ino2p/Ino4p basic helix-loop-helix transcription activator that binds inositol/choline-responsive elements (ICREs), required for derepression of phospholipid biosynthetic genes in response to inositol depletion; involved in diauxic shift. |
| 8 | INO4 | F-CAACCCCAGGAAAGTCGGTC R-CCATCACCCAGCTCCCAAAT |
INOsitol requiring- Transcription factor involved in phospholipid synthesis; required for derepression of inositol-choline-regulated genes involved in phospholipid synthesis; forms a complex, with Ino2p, that binds the inositol-choline-responsive element through a basic helix-loop-helix domain. |
| 9 | CHO2 | F-CGCATCGCGTGGTTAAAGAT R-CGGACACCAGAAGAACCTTGT |
CHOline requiring- Phosphatidylethanolamine methyltransferase (PEMT); catalyzes the first step in the conversion of phosphatidylethanolamine to phosphatidylcholine during the methylation pathway of phosphatidylcholine biosynthesis. |
| 10 | YAP1 | F-TACACGTGATGGCGAGGATA R-CCACTTCATTTTGCTGCTGA |
Yeast AP-1- Basic leucine zipper (bZIP) transcription factor; required for oxidative stress tolerance; activated by H2O2 through the multistep formation of disulfide bonds and transit from the cytoplasm to the nucleus; Yap1p is degraded in the nucleus after the oxidative stress has passed; relative distribution to the nucleus increases upon DNA replication stress. |
| 11 | KGD1 | F-GGTTAACTGCTGCCTGGGAA R-CCCAATCGATGCCTTCACCT |
Alpha-KetoGlutarate Dehydrogenase- Subunit of the mitochondrial alpha-ketoglutarate dehydrogenase complex; catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA. |
3. Results and discussion
3.1. ZnO-NPs cause de-regulation in protein expression
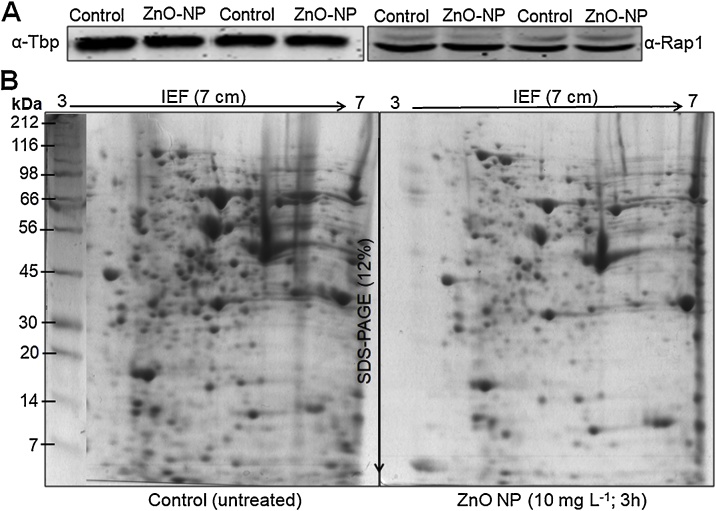
To maintain the equilibrium between essential and non essential levels of certain toxic compounds, eukaryotic cells utilize complicated mechanisms to regulate its cellular uptake, sequestration to sub-cellular organelles and complexes, as well as detoxification [19]. Since metals nanoparticles cannot be degraded or modified like other toxic organic compounds they remains in cells and interfere with cellular homeostatic pathways and can induce oxidative stress, alteration of enzyme and protein function, lipid peroxidation, and DNA damage [20]. In general metals and metalloids have the capacity to bind to proteins, often via thiol groups of cysteine residues, and inhibit enzyme function by inducing improper protein folding [21]. Most metals are weak mutagens and do not causes direct DNA damage however, they may trigger genotoxicity by interfering with DNA repair molecules and mechanisms [22]. Many metals including Zn have the capacity to influence membrane fluidity, which in turn could contribute to their toxicity [23]. Cellular response of S. cerevisiae at different “-omics” levels to various environmental stress conditions has been comprehensively studied but this organism is poorly utilized to study the nanomaterials toxicity [8,9]. Earlier, it was established that despite, the rigid cell wall of yeast, ZnO-NPs induce mechanical damage by breaking the cell walls and get internalized inside the yeast cells with the help of endocytosis [4]. This may trigger several cellular and sub-cellular responses such as oxidative stress by generating radical oxygen species (ROS), inflammatory & immunological responses, genotoxicity and ultimately cell death [[2], [3], [4], [5]]. It is very well established that metal nanoparticles impose oxidative stress thus induce defects in mitochondrial and ER enzymes [10,[24], [25], [26]]. The combined proteome and metabolome analysis and its integration with systems biology offers an opportunity to improve our knowledge about the toxic effects and mechanisms induced by NPs [12,27]. S. cerevisiae has the ability to efficiently respond to a particular stress, they developed several mechanisms for adaptation and thus survival [28,29]. Stress can trigger signalling cascades, regulation in mRNA levels and consequently induce significant changes in cell metabolic pathways and metabolite levels by adjusting protein/enzyme levels [29,30]. In a comparative proteome and metabolome analysis, it is essential that the protein concentration must be equal in control (untreated) and treated (ZnO-NPs) cells. Therefore, we performed western blotting using anti-TBP and -Rap1 as reference proteins. Western blots showed equal band intensity in both the control (untreated) and ZnO-NPs (10 mg L−1) treated protein samples (Fig. 1A). Being assured, we analyzed the relative proteomic changes in the control and treated cell. The 2DE image gels analysis showed a total of 650 and 425 protein spots in the control and ZnO-NPs treated samples, respectively with an average percentage of matched spots across gels of 94% (Fig.1B). Among the protein spots detected, 46 protein spots were found to be down-regulated and 20 spots were up-regulated by at least 2-fold in ZnO-NPs treated cells. A total number of 66 protein spots were differentially expressed and significant (p < 0.05) between the control and treated groups. Although we did not further characterize these proteins but qualitative 2DE gels showed a significant difference in the protein profile of control and ZnO-NPs exposed cells.
Fig. 1.
Proteomic analysis of the S. cerevisiae BY4741 cells exposed to ZnO-NPs. (A) Western blot (B) 2DE gel images showing protein spots in control and ZnO-NPs (10 mg L−1) treated cells for 3 h. Protein ladder (kDa).
3.2. ZnO-NPs cause de-regulation in metabolites
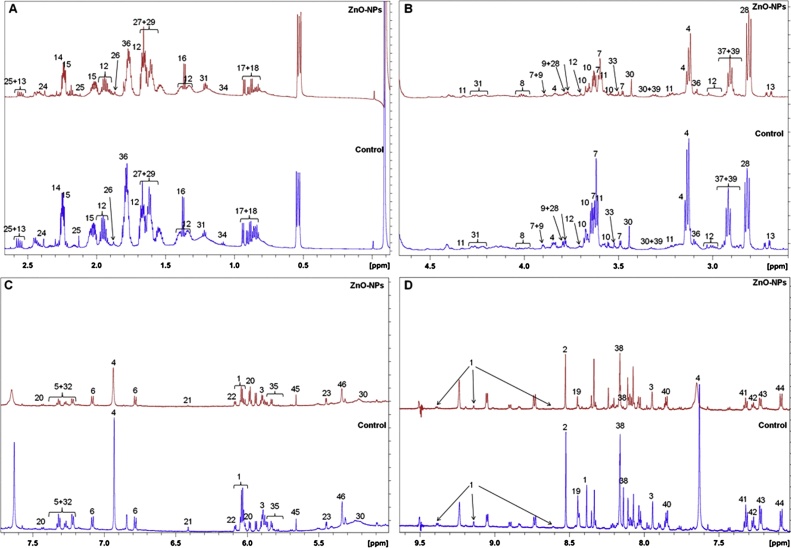
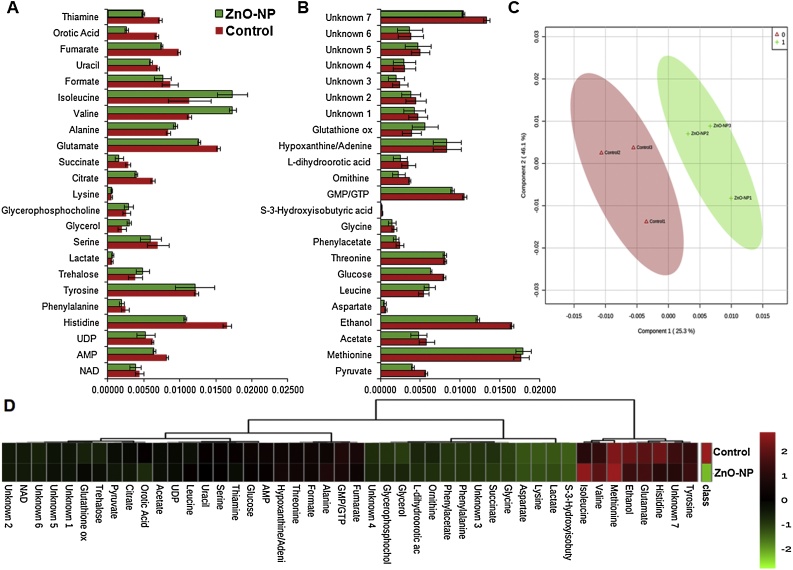
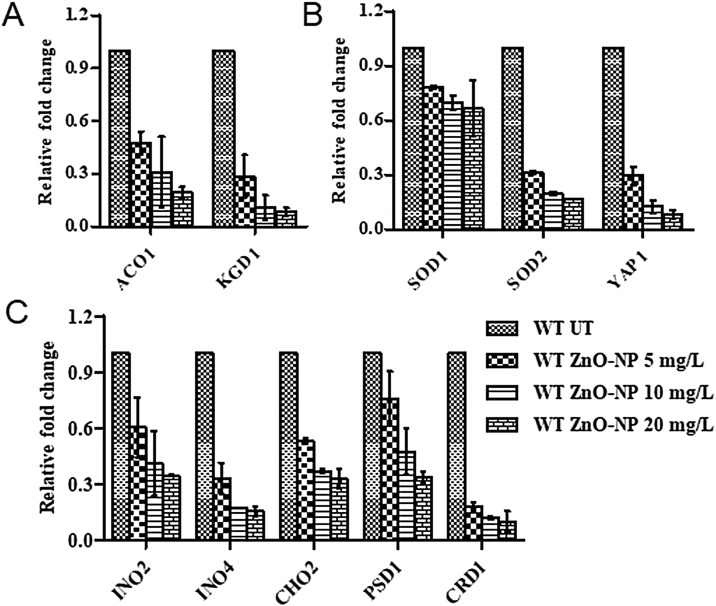
Proteome analysis results motivated us to analyse the de-regulation in the metabolites. Untargeted 1H NMR analysis approach has been utilized to evaluate the metabolic changes. Due to the complex 1H NMR experimental set-up, we have chosen the most effective (EC50) concentration of ZnO-NPs (i.e., 10 mg L−1). To partly overcome the challenge of complex metabolomics data analysis, integrated metabolomics approaches were used to achieve insights into molecular pathways from identified metabolite annotations. The chemical shifts obtained after statistical analysis were submitted to the public database search for metabolite annotations and identification, we used the standard reporting system, described previously [16]. According to the chemical shifts data and coupling patterns, we identified 46 metabolites (including seven unknown metabolites) from the whole S. cerevisiae cell extract (Fig. 2). Chemical shifts of signals were assigned to the metabolites in the form of amino acids, organic acids, fatty acids, carbohydrates, and nucleotide derivatives. Results show that ZnO-NPs had a significant impact on the deregulation pattern (p < 0.05) of metabolites in the yeast cell. To our surprise, most of the annotated metabolites were significantly down-regulated and few of them were up-regulated. Metabolic pathway analysis revealed that ZnO-NPs repressed a wide range of key metabolites involved in central metabolism, including cofactors synthesis, amino acid & organic acid biosynthesis, purines & pyrimidines and nucleoside & nucleotide biosynthetic pathways. These metabolites are classified by their chemical nature and functions associated with the metabolic pathways. Relative concentrations of the annotated metabolites and the parameters used for its calculation are presented in the supplementary Table S1. List of metabolites with their respective chemical shifts, de-regulation and YMDB, KEGG ID is presented in Table 2 and the main variations are summarized in the form of a bar graph (Fig. 3 A, B). To visualize the differences between control and ZnO-NPs treated S. cerevisiae, the 46 identified metabolites were normalized and analyzed by Partial Least Squares Discriminant Analysis (PLS-DA) using metaboanalyst which is a supervised clustering method to maximize the separation between groups. The score plot shows that cells exposed to ZnO-NPs are clearly separated along the first principal axis (PC1), which explained 25.3% of the total variability (Fig. 3C) indicating that ZnO-NPs considerably altered the metabolic profiles of the yeast cell. The heat map obtained from the selected metabolites indicates that some metabolites are down-regulated while others are up-regulated upon ZnO-NPs exposure (Fig. 3D). To further validate these results we have checked the transcript level changes in few selected genes associated with the TCA cycle (ACO1 and KDG1) (Fig. 4A), oxidative pathway (SOD1, SOD2 and YAP1) (Fig. 4B), and lipid biosynthetic pathways (INO2, INO4, CHO1, PSD1 and CRD1) (Fig. 4C). We found that all the selected genes were down-regulated upon ZnO-NPs exposure. These observations are in favour of metabolic data and thus indicate that the ZnO-NPs impose toxicity by perturbing the mitochondria, ER and lipid homeostasis.
Fig. 2.
1H NMR spectra showing chemical shifts. Different peaks marked as numbers represents different metabolites.
Table 2.
1H NMR spectroscopic (chemical shift) data of the assigned metabolites. YMDB IDs, KEGG IDs and respective cellular functions are provided.
| Peak No. | Metabolites | Chemical Shifts (ppm) | Regulation | YMDB ID | KEGG ID | Cellular Functions |
|---|---|---|---|---|---|---|
| 1 | NAD+ | 9.33 (s) 9.15 (d) 8.83 (d) 8.42 (s) 8.19 (m) 6.13 (d) 6.08 (d) 6.02 (d) | Down | YMDB00110 | C00003 | Coenzyme in redox reactions |
| 2 | AMP | 8.6 (s) 8.17 (s) | Down | YMDB00058 | C00020 | Maintains cellular energy homeostasis. as well as in signal transduction as cyclic adenosine monophosphate (cAMP) |
| 3 | UDP | 7.95 (d) | Down | YMDB00307 | C00015 | Important extracellular signaling molecule |
| 4 | Histidine | 7.8 (s) 7.05 (s) 3.96 (dd) 3.22 (dd) 3.12 (dd) | Down | YMDB00369 | C00135 | Often found in enzyme active sites, where the chemistry of the imidazole ring of histidine makes it a nucleophile and a good acid/base catalyzer. Its biosynthesis is inherently linked to the pathways of nucleotide formation |
| 5 | Phenylalanine | 7.42 (m) 7.36 (m) 7.32 (d) 3.97 (dd) 3.29 (dd) 3.12 (dd) | Down | YMDB00304 | C00079 | Incorporated into polypeptide chains, production of tyrosine via the tetrahydrobiopterin-requiring phenylalanine hydroxylase and conversion to a fusel alcohol |
| 6 | Tyrosine | 7.18 (d) 6.89 (d) 3.97 (dd) 3.13 (dd) 3.02 (dd) | No | YMDB00364 | C00082 | Converted to NAD+ |
| 7 | Trehalose | 5.18 (d) 3.85 (m) 3.75 (dd) 3.64 (dd) 3.44 (t) | Up | YMDB00008 | C01083 | Implicated in anhydrobiosis - the ability of plants and animals to withstand prolonged periods of desiccation |
| 8 | Lactate | 4.11 (dd) 1.32 (d) | Up | YMDB00247 | C00186 | Alternative byproduct in anaerobic respiration |
| 9 | Serine | 3.94 (m) 3.83 (dd) | Down | YMDB00112 | C00065 | Participates in the biosynthesis of purines and pyrimidines. It is the precursor to several amino acids including glycine, cysteine, and tryptophan. It is also the precursor to numerous other metabolites, including sphingolipids and folate, which is the principal donor of one-carbon fragments in lipid biosynthesis |
| 10 | Glycerol | 3.77 (m) 3.65 (dd) 3.55 (dd) | Up | YMDB00283 | C00116 | Important component of triglycerides and phospholipids |
| 11 | Glycerophospho-choline | 4.31 (m) 3.6 (dd) 3.22 (s) | Up | YMDB00309 | C00670 | A choline derivative and one of the two major forms of choline storage (along with phosphocholine) in the cytosol. Important component of lipid metabolism |
| 12 | Lysine | 3.7 (m) 3.00 (t) 1.87 (m) 1.71 (m) 1.45 (m) | No | YMDB00330 | C00047 | Important in nitrogen metabolism, converted to Acetyl CoA |
| 13 | Citrate | 2.64 (d) 2.52 (d) | Down | YMDB00086 | C00158 | Component of the citric acid cycle |
| 14 | Succinate | 2.39 (s) | Down | YMDB00338 | C00042 | Component of the citric acid cycle |
| 15 | Glutamate | 3.74 (dd) 2.34 (td) 2.05 (m) | Down | YMDB00271 | C00025 | Enter the Krebs cycle for energy metabolism, and be converted into glutamine, which is one of the key players in nitrogen metabolism |
| 16 | Alanine | 1.47 (d) | Up | YMDB00154 | C00041 | Tightly coupled to metabolic pathways such as glycolysis, gluconeogenesis, and the citric acid cycle. It also arises together with lactate and generates glucose from protein degradation via the alanine cycle. Alanine's catabolic pathway directly produces pyruvate |
| 17 | Valine | 1.03 (d) 0.98 (d) | Up | YMDB00152 | C00183 | Essential amino acid |
| 18 | Isoleucine | 1.00 (d) 0.94 (t) | Up | YMDB00038 | C00407 | Essential amino acid |
| 19 | Formate | 8.44 (s) | Down | YMDB00101 | C00058 | Intermediate in TCA cycle |
| 20 | Uracil | 7.53 (d) 5.79 (d) | Down | YMDB00098 | C00106 | Involved in pyrimidine and beta-alanine metabolism Associated in pantothenate and CoA biosynthesis |
| 21 | Fumarate | 6.4 (s) | Down | YMDB00101 | C00122 | Intermediate in TCA cycle |
| 22 | Orotic acid | 6.18 (s) | Down | YMDB00405 | C00295 | Intermediate in the uridine-5'-phosphate (UMP) biosynthesis pathway. |
| 23 | Thiamine derivate | 5.46 (s) | Down | YMDB00220 | C00378 | Plays a key role in intracellular glucose metabolism |
| 24 | Pyruvate | 2.36 (s) | Down | YMDB00175 | C00022 | Used to construct alanine and be converted into ethanol. Supplies energy to living cells through the citric acid cycle when oxygen is present (aerobic respiration), and alternatively ferments to produce lactate in absence of oxygen |
| 25 | Methionine | 2.63 (t) 2.12 (s) | No | YMDB00318 | C00073 | Incorporate into polypeptide chains, and use in the production of alpha-ketobutyrate and cysteine via SAM (S-adenosylmethionine). The transulfuration reactions that produce cysteine from homocysteine and serine also produce alpha-ketobutyrate, the latter being converted first to propionyl-CoA and then via a 3-step process to succinyl-CoA |
| 26 | Acetate | 1.91 (s) | Down | YMDB00056 | C00033 | Utilized by organisms in the form of acetyl coenzyme A. |
| 27 | Ethanol | 3.65 (q) 1.71 (t) | Down | YMDB00883 | C00469 | Involved in Glycolysis/Gluconeogenesis and pyruvate metabolism |
| 28 | Aspartate | 3.88 (dd) 2.80 (dd) | Up | YMDB00896 | C00049 | Precursor to several amino acids, including methionine, threonine, isoleucine, and lysine. Asparagine is derived from aspartate via transamidation. Aspartate is also a metabolite in the urea cycle and participates in gluconeogenesis. Involved in biosynthesis of inosine, the precursor to the purine bases |
| 29 | Leucine | 3.71 (m) 1.69 (m) 0.95 (t) | Up | YMDB00387 | C00123 | Products of its breakdown are acetyl-CoA and acetoacetate, it is one of the two exclusively ketogenic amino acids lysine being the other one |
| 30 | Glucose | 5.22 (d) 4.64 (d) 3.89 (dd) 3.83 (m) 3.73 (m) 3.52 (dd) 3.46 (m) 3.40 (td) 3.23 (dd) | Down | YMDB00273 | C00031 | Primary source of energy |
| 31 | Threonine | 4.24 (m) 1.31 (d) | No | YMDB00214 | C00188 | Yields ketogenic and glucogenic byproducts |
| 32 | Phenylacetate | 7.38 (m) 7.30 (m) 3.52 (s) | Up | YMDB00838 | C00548 | |
| 33 | Glycine | 3.55 (s) | Down | YMDB00016 | C00037 | Involved in glutathione and nitrogen metabolism |
| 34 | S-3-Hydroxy isobutyric acid |
1.06 (d) | Up | YMDB00337 | C01188 | Intermediate in the metabolic pathways of valine and thymine amino acids |
| 35 | GMP/GTP | 5.93 (d, J = 5.3 Hz) | Down | YMDB00261/ YMDB00558 | C00144/ C00044 | Intermediate in purine metabolism |
| 36 | Ornithine | 1.65-2.00 (4 H); 3.04 (t, 2 H) | Down | YMDB00353 | C00077 | Non-proteinogenic amino acid that plays a role in the urea cycle. Aalso a precursor of citrulline and arginine |
| 37 | L-dihydroorotic acid | 2.76 (d); 2.81 (d) | Down | YMDB00396 | C00337 | An intermediate in the uridine-5'-phosphate biosynthesis pathway |
| 38 | Hypoxanthine/ Adenine | 8.18 (s); 8.20 (s) | Down | YMDB00555 | C00262 | Naturally occurring purine derivative and a reaction intermediate in the metabolism of adenosine and in the formation of nucleic acids by the salvage pathway. Also a spontaneous deamination product of adenine |
| 39 | Glutathione ox | 3.30 (dd) 2.96 (dd) | Down | YMDB00057 | C00127 | It is an antioxidant and a coenzyme in various enzymatic reactions. It is found almost exclusively in its reduced form |
| 40 | Unknown 1 | 7.89 (dd) | Down | |||
| 41 | Unknown 2 | 7.3 (dd) | Down | |||
| 42 | Unknown 3 | 7.2 (d) | Down | |||
| 43 | Unknown 4 | 7.02 (d) | Down | |||
| 44 | Unknown 5 | 7.01 (d) | No | |||
| 45 | Unknown 6 | 5.67 (s) | No | |||
| 46 | Unknown 7 | 5.31 (s) | Down |
Fig. 3.
Deregulation in the metabolome of S. cerevisiae BY4741 cells exposed to ZnO-NPs (10 mg L−1) for 3 h. (A, B) Relative concentration of different annotated metabolites (C) Scores plot (PC1 vs PC2) of partial least-squares-discriminant analysis (PLS-DA) of metabolites (D) Heat map generated by hierarchical cluster analysis of 1H NMR data. Centroid method and euclidean distance were considered for clustering analysis. Data was log2 transformed and row scaled prior to cluster analysis. Red and green bars indicate a significant decrease and increase in the metabolite content. The dendrogram reveal the relationships between control and ZnO-NP treated samples based on the abundance of metabolites. All individual samples (including replicates) were included.
Fig. 4.
RT-qPCR analysis to determine relative transcript level expression of selected genes of (A) oxidative stress (SOD1, SOD2 and YAP1), TCA cycle (ACO1 and KDG1) and lipid biosynthetic (INO2, INO4, CHO2, PSD1 and CRD1) pathways. Results are mean ± SEM from three independent experiments. * indicate the P ≤ 0.05 compared to control cells (Student’s t-test).
Mitochondria are essential bioenergetic organelles participate in several vital processes and fulfil the energy requirements of the cell. Metabolite profiling presented in this study showed improper mitochondrial functioning as evidenced by diminished biosynthesis of various metabolites of key mitochondrial processes i.e., glycolysis, the tricarboxylic acid (TCA) cycle, and the pentose phosphate pathway, indicating that ZnO-NPs affect the central carbon metabolism (CCM). The CCM network fulfils energy demand of the cell and is important for providing synthetic precursors for other primary and secondary metabolites. Thus, the regulation of CCM is important for the regulation and coordination of a wide range of metabolic reactions and pathways. Under stress conditions, the CCM and other metabolic pathways can be altered and these metabolic changes have been studied by using different approaches [31,32]. In aerobic metabolism, ROS are formed as a natural by-product, but in excess they can chemically modify various macromolecules and cause damage to vital organelles (mitochondria, ER, lysosomes etc.) [33,34]. Earlier it was suggested that zinc ions produced by dissolved ZnO-NPs affect the TCA cycle by repressing the enzymes and resulting in decreased levels of citrate, succinate and α-ketoglutarate [35]. Another study on rat lung confirmed that ZnO-NPs down-regulate the key enzymes of glycolysis the tricarboxylic acid (TCA) cycle and the electron transport chain, resulting in diminished ATP generation and ultimately cell death [27]. Our results also showed decreased levels in the citrate and succinate this might affect the aerobic metabolism and the TCA cycle thus inhibits ATP production. Succinic acid is an important metabolite in the TCA cycle for energy generation, and it also participates in the GABA pathway involved in ROS stress alleviation [36]. Decreased levels of glucose and acetate while increased levels of lactate and alanine indicates that glucose can be changed into lactate or alanine in order to supply enough energy for survival. Result might entail the disturbance of aerobic metabolism and TCA cycle. Moreover, acetate is also viewed as an alternative energy source [37]. These alterations in metabolites provide evidence that there was induction of central elements of the oxidative stress response and the basic detoxification process against short-term ZnO-NPs exposure.
We did not observe any significant change in tyrosine, lysine and methionine while branched-chain amino acids (isoleucine, leucine and valine) and alanine were up-regulated. These essential amino acids are involved in the energetic pathways of eukaryotic organisms and their increased levels may be attributed to protein breakdown. Previous results also showed that branched-chain amino acids were increased in mouse model upon acrolein-induced lung injury this might be due do the result of the utilization of these amino acids as energy sources [38]. Another stress indicator found was the accumulation of trehalose. The biosynthesis of trehalose is considered as a cellular defence mechanism against ROS accumulation and other stressful conditions [12], the protective role of this carbohydrate against ROS has been confirmed in cell devoid of SOD [39]. Glycerol is a protective agent synthesized by yeast cells against environmental stress, which can function in maintaining the redox homeostasis [40]. The increased glycerol biosynthesis rate thus indicates the alteration in stress defence of the cells against ZnO-NPs toxicity. One of the most important metabolites found up-regulated is glycerophosphocholine, an important member of eukaryotic cellular membrane components. It is involved in the membrane choline phospholipid metabolism pathway, where phosphatidylcholine can be converted to glycerophosphocholine, which can be reconverted to phosphatidylcholine. Cells consume glycerophosphocholine to generate phosphatidylcholine for maintaining cell membrane integrity [41,42]. Based on this we can propose a probable mechanism of membrane component maintenance upon ZnO-NPs exposure. Our finding is in parallel to the result of a research conducted in the rat lung upon acute inhalation of ZnO particles [27].
Protein synthesis is crucial for cellular stress response and adaptation. The basic amino acids are the building blocks of protein. During metabolic profiling several amino acids were de-regulated. Among them, threonine, histidine, phenylalanine, glutamate and glycine were down-regulated. The decrease in glycine and oxidised glutathione is reported that might have been a protective response to oxidative stress. Glycine is one of the precursor amino acids that can synthesize glutathione, which is an important antioxidant that provides redox defence against inflammation and oxidative stress induced by metal stress [43,44]. Nucleotides are crucial players in a huge number of diverse cellular processes. Nucleic acids are made up of purines and pyrimidines, and ATP is the central cellular energy supplier. The GMP/GTP and modified nucleotides for eg. cyclic AMP are signalling molecules. Finally, nucleotides are incorporated in cofactors (e.g., NAD and coenzyme A) and serve as precursors (e.g., UDP-glucose and GDP-mannose). In this study, all the above metabolites were down-regulated after ZnO-NPs exposure. Purine and pyrimidine synthesis occurs through distinct metabolic pathways, and these are highly conserved among both prokaryotic and eukaryotic species. The purine and pyrimidine pathways ensure net synthesis of nucleotides but also allow interconversion of nucleobases, nucleosides, and nucleotides, thus permitting a proper balance of the final products to be achieved. Formate is a possible alternative single-carbon source for the production of N5, N10-methylene-tetrahydrofolate, which is necessary for the biosynthesis of nucleic acid. It might be a possibility that the under stressed condition cells utilize more amount of formate to produce purine nucleotides thus reflect low formate level and might indicate that ZnO-NPs cause DNA damage.
4. Conclusion
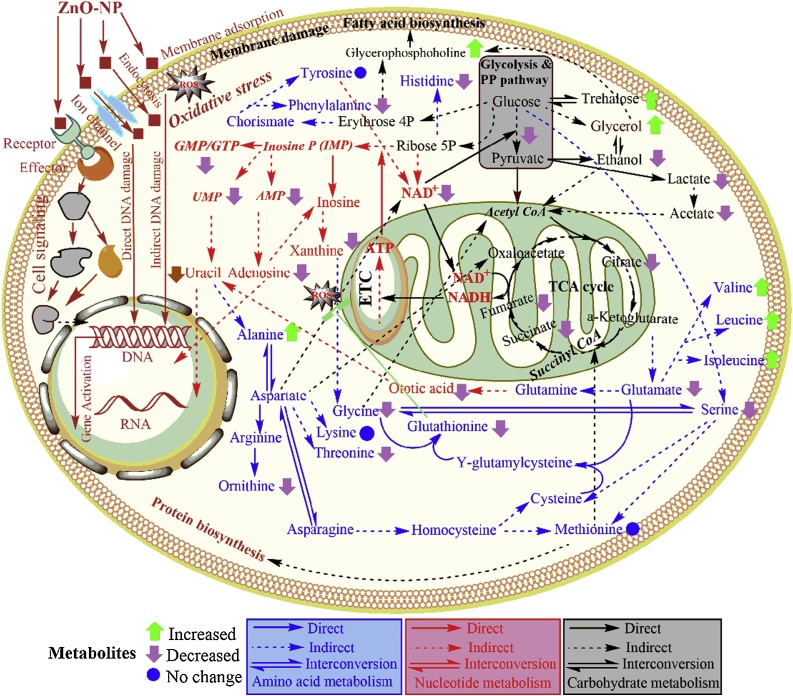
In summary, our results indicate that ZnO-NPs induced physiological and metabolic toxicity can be characterized by de-regulation of proteins and metabolites. An overview of the main interconnections for the de-regulated metabolites in S. cerevisiae is shown in Fig. 5. Taken together, it is clear that under stress conditions the enzymes of carbon, amino acids, lipid and nucleotide metabolism play important roles in maintaining optimal metabolite concentrations and eventually global metabolic homeostasis. On the basis of proteome and metabolome analysis we can hypothesise that ZnO-NPs affect physiology and metabolism of S. cerevisiae. In general the toxicity mechanisms showed decline in the growth-related processes such as carbohydrate metabolism and protein biosynthesis including the repression of genes of antioxidative and lipid metabolic pathways.
Fig. 5.
Integrative overview of the metabolic alteration based on the assigned metabolites occurring under ZnO-NP stress in S. cerevisiae. Amino acids, nucleotide and carbohydrate metabolic pathways are highlighted in blue, red and black colours respectively. The metabolic scheme was based on information gathered in the KEGG pathway database (http://www.genome.jp/kegg/pathway.html) and in yeast biochemical pathway database (http://pathway.yeastgenome.org/).
Conflicts of interest
The authors declare that they have no conflict of interest.
Funding
This work was supported by a project grant by SERB- DST New Delhi India (File No. PDF/000200).
Transparency document
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jog.2018.11.004.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Djurišić A.B., Chen X., Leung Y.H., Ng A.M.C. ZnO nanostructures: growth, properties and applications. J. Mater. Chem. 2012;22:6526–6535. [Google Scholar]
- 2.Sirelkhatim A., Mahmud S., Seeni A., Kaus N.H.M., Ann L.C., Bakhori S.K.M., Hasan H., Mohamad D. Review on zinc oxide nanoparticles: antibacterial activity and toxicity mechanism. Nano-Micro Lett. 2015;7:219–242. doi: 10.1007/s40820-015-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung Y.H., Xu X., Ma A.P., Liu F., Ng A.M., Shen Z., Gethings L.A., Guo M.Y., Djurišić A.B., Lee P.K. Toxicity of ZnO and TiO2 to Escherichia coli cells. Sci. Rep. 2016;6:35243. doi: 10.1038/srep35243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W., Bao S., Fang T. The neglected nano-specific toxicity of ZnO nanoparticles in the yeast Saccharomyces cerevisiae. Sci. Rep. 2016;6 doi: 10.1038/srep24839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condello M., De Berardis B., Ammendolia M.G., Barone F., Condello G., Degan P., Meschini S. ZnO nanoparticle tracking from uptake to genotoxic damage in human colon carcinoma cells. Toxicol. Vitr. 2016;35:169–179. doi: 10.1016/j.tiv.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Duina A.A., Miller M.E., Keeney J.B. Budding yeast for budding geneticists: a primer on the Saccharomyces cerevisiae model system. Genetics. 2014;197:33–48. doi: 10.1534/genetics.114.163188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jewison T., Knox C., Neveu V., Djoumbou Y., Guo A.C., Lee J., Liu P., Mandal R., Krishnamurthy R., Sinelnikov I. YMDB: the yeast metabolome database. Nucleic Acids Res. 2011;40:D815–D820. doi: 10.1093/nar/gkr916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.dos Santos S.C., Teixeira M.C., Cabrito T.R., Sá-Correia I. Yeast toxicogenomics: genome-wide responses to chemical stresses with impact in environmental health, pharmacology, and biotechnology. Front. Genet. 2012;3:63. doi: 10.3389/fgene.2012.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braconi D., Bernardini G., Santucci A. Saccharomyces cerevisiae as a model in ecotoxicological studies: a post-genomics perspective. J. Proteomics. 2016;137:19–34. doi: 10.1016/j.jprot.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Kasemets K., Ivask A., Dubourguier H.-C., Kahru A. Toxicity of nanoparticles of ZnO, CuO and TiO 2 to yeast Saccharomyces cerevisiae. Toxicol. Vitr. 2009;23:1116–1122. doi: 10.1016/j.tiv.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Babele P.K., Thakre P.K., Kumawat R., Tomar R.S. Zinc oxide nanoparticles induce toxicity by affecting cell wall integrity pathway, mitochondrial function and lipid homeostasis in Saccharomyces cerevisiae. Chemosphere. 2018;213:65–75. doi: 10.1016/j.chemosphere.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 12.Gioria S., Lobo Vicente J., Barboro P., La Spina R., Tomasi G., Urbán P., Kinsner-Ovaskainen A., François R., Chassaigne H. A combined proteomics and metabolomics approach to assess the effects of gold nanoparticles in vitro. Nanotoxicology. 2016;10:736–748. doi: 10.3109/17435390.2015.1121412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farres M., Pina B., Tauler R. LC-MS based metabolomics and chemometrics study of the toxic effects of copper on Saccharomyces cerevisiae. Metallomics. 2016;8:790–798. doi: 10.1039/c6mt00021e. [DOI] [PubMed] [Google Scholar]
- 14.Jun H., Kieselbach T., Jönsson L.J. Comparative proteome analysis of Saccharomyces cerevisiae: a global overview of in vivo targets of the yeast activator protein 1. BMC Genomics. 2012;13:1. doi: 10.1186/1471-2164-13-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Airoldi C., Tripodi F., Guzzi C., Nicastro R., Coccetti P. NMR analysis of budding yeast metabolomics: a rapid method for sample preparation. Mol. Biosyst. 2015;11:379–383. doi: 10.1039/c4mb00452c. [DOI] [PubMed] [Google Scholar]
- 16.Puig-Castellví F., Alfonso I., Piña B., Tauler R. 1H NMR metabolomic study of auxotrophic starvation in yeast using multivariate curve resolution-alternating least squares for pathway analysis. Sci. Rep. 2016;6:30982. doi: 10.1038/srep30982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia J., Sinelnikov I.V., Han B., Wishart D.S. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sariki S.K., Sahu P.K., Golla U., Singh V., Azad G.K., Tomar R.S. Sen1, the homolog of human Senataxin, is critical for cell survival through regulation of redox homeostasis, mitochondrial function, and the TOR pathway in Saccharomyces cerevisiae. FEBS J. 2016;283:4056–4083. doi: 10.1111/febs.13917. [DOI] [PubMed] [Google Scholar]
- 19.Wysocki R., Tamás M.J. How Saccharomyces cerevisiae copes with toxic metals and metalloids. FEMS Microbiol. Rev. 2010;34:925–951. doi: 10.1111/j.1574-6976.2010.00217.x. [DOI] [PubMed] [Google Scholar]
- 20.Hosiner D., Gerber S., Lichtenberg-Frate H., Glaser W., Schüller C., Klipp E. Impact of acute metal stress in Saccharomyces cerevisiae. PLoS One. 2014;9 doi: 10.1371/journal.pone.0083330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S.K., Goloubinoff P., Christen P. Heavy metal ions are potent inhibitors of protein folding. Biochem. Biophys. Res. Commun. 2008;372:341–345. doi: 10.1016/j.bbrc.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 22.Beyersmann D., Hartwig A. Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Arch. Toxicol. 2008;82:493. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- 23.García J.J., Martinez-Ballarin E., Millan-Plano S., Allue J., Albendea C., Fuentes L., Escanero J. Effects of trace elements on membrane fluidity. J. Trace Elem. Med. Biol. 2005;19:19–22. doi: 10.1016/j.jtemb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Sharma V., Anderson D., Dhawan A. Zinc oxide nanoparticles induce oxidative DNA damage and ROS-triggered mitochondria mediated apoptosis in human liver cells (HepG2) Apoptosis. 2012;17:852–870. doi: 10.1007/s10495-012-0705-6. [DOI] [PubMed] [Google Scholar]
- 25.Huerta-García E., Pérez-Arizti J.A., Márquez-Ramírez S.G., Delgado-Buenrostro N.L., Chirino Y.I., Iglesias G.G., López-Marure R. Titanium dioxide nanoparticles induce strong oxidative stress and mitochondrial damage in glial cells. Free Radic. Biol. Med. 2014;73:84–94. doi: 10.1016/j.freeradbiomed.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Park E.-J., Choi D.-H., Kim Y., Lee E.-W., Song J., Cho M.-H., Kim J.-H., Kim S.-W. Magnetic iron oxide nanoparticles induce autophagy preceding apoptosis through mitochondrial damage and ER stress in RAW264. 7 cells. Toxicol. Vitr. 2014;28:1402–1412. doi: 10.1016/j.tiv.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Lee S.-H., Wang T.-Y., Hong J.-H., Cheng T.-J., Lin C.-Y. NMR-based metabolomics to determine acute inhalation effects of nano-and fine-sized ZnO particles in the rat lung. Nanotoxicology. 2016;10:924–934. doi: 10.3109/17435390.2016.1144825. [DOI] [PubMed] [Google Scholar]
- 28.Morano K.A., Grant C.M., Moye-Rowley W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taymaz-Nikerel H., Cankorur-Cetinkaya A., Kirdar B. Genome-wide transcriptional response of Saccharomyces cerevisiae to stress-induced perturbations. Front. Bioeng. Biotechnol. 2016;4:17. doi: 10.3389/fbioe.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minic Z. Proteomic studies of the effects of different stress conditions on central carbon metabolism in microorganisms. J. Proteomics Bioinform. 2015;8:80. [Google Scholar]
- 31.Wiebe M.G., Rintala E., Tamminen A., Simolin H., Salusjärvi L., Toivari M., Kokkonen J.T., Kiuru J., Ketola R.A., Jouhten P. Central carbon metabolism of Saccharomyces cerevisiae in anaerobic, oxygen-limited and fully aerobic steady-state conditions and following a shift to anaerobic conditions. FEMS Yeast Res. 2007;8:140–154. doi: 10.1111/j.1567-1364.2007.00234.x. [DOI] [PubMed] [Google Scholar]
- 32.Guo W., Chen Y., Wei N., Feng X. Investigate the metabolic reprogramming of Saccharomyces cerevisiae for enhanced resistance to mixed fermentation inhibitors via 13C metabolic flux analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrero E., Ros J., Bellí G., Cabiscol E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta (BBA)-Gen. Sub. 2008;1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Santo A., Zhu H., Li Y.R. Free radicals: from health to disease. React. Oxyg. Species. 2016;2:245–263. [Google Scholar]
- 35.Yan G., Huang Y., Bu Q., Lv L., Deng P., Zhou J., Wang Y., Yang Y., Liu Q., Cen X. Zinc oxide nanoparticles cause nephrotoxicity and kidney metabolism alterations in rats, Journal of Environmental Science and Health. Part A. 2012;47:577–588. doi: 10.1080/10934529.2012.650576. [DOI] [PubMed] [Google Scholar]
- 36.Cao J., Barbosa J.M., Singh N.K., Locy R.D. GABA shunt mediates thermotolerance in Saccharomyces cerevisiae by reducing reactive oxygen production. Yeast. 2013;30:129–144. doi: 10.1002/yea.2948. [DOI] [PubMed] [Google Scholar]
- 37.Jiang L., Gulanski B.I., De Feyter H.M., Weinzimer S.A., Pittman B., Guidone E., Koretski J., Harman S., Petrakis I.L., Krystal J.H. Increased brain uptake and oxidation of acetate in heavy drinkers. J. Clin. Invest. 2013;123:1605–1614. doi: 10.1172/JCI65153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabisiak J.P., Medvedovic M., Alexander D.C., McDunn J.E., Concel V.J., Bein K., Jang A.S., Berndt A., Vuga L.J., Brant K.A. Integrative metabolome and transcriptome profiling reveals discordant energetic stress between mouse strains with differential sensitivity to acrolein‐induced acute lung injury. Mol. Nutr. Food Res. 2011;55:1423–1434. doi: 10.1002/mnfr.201100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Jesus Pereira E., Panek A.D., Eleutherio E.C.A. Protection against oxidation during dehydration of yeast. Cell Stress Chaperones. 2003;8:120–124. doi: 10.1379/1466-1268(2003)008<0120:paoddo>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paget C.M., Schwartz J.M., Delneri D. Environmental systems biology of cold‐tolerant phenotype in Saccharomyces species adapted to grow at different temperatures. Mol. Ecol. 2014;23:5241–5257. doi: 10.1111/mec.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu J.Z., Rommereim D.N., Minard K.R., Woodstock A., Harrer B.J., Wind R.A., Phipps R.P., Sime P.J. Metabolomics in lung inflammation: a high-resolution 1H NMR study of mice exposedto silica dust. Toxicol. Mech. Methods. 2008;18:385–398. doi: 10.1080/15376510701611032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatem E., Berthonaud V., Dardalhon M., Lagniel G., Baudouin-Cornu P., Huang M.-E., Labarre J., Chédin S. Glutathione is essential to preserve nuclear function and cell survival under oxidative stress. Free Radic. Biol. Med. 2014;67:103–114. doi: 10.1016/j.freeradbiomed.2013.10.807. [DOI] [PubMed] [Google Scholar]
- 44.Ilyas S., Rehman A. Oxidative stress, glutathione level and antioxidant response to heavy metals in multi-resistant pathogen, Candida tropicalis. Environ. Monit. Assess. 2015;187:4115. doi: 10.1007/s10661-014-4115-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.