Abstract
Background
Ocular melanoma is a rare but often deadly malignancy that arises in the uvea (commonest primary site), conjunctiva or the orbit. Primary orbital melanoma (POM) is exceedingly rare, with approximately 60 cases reported to date. Despite recent advances in our understanding of the genetics of primary uveal and conjunctival melanomas, this information is lacking for POM.
Methods
DNA was extracted from 12 POM tissues, with matched germline DNA (where available). MLPA was conducted to detect chromosomal alterations and Sanger sequencing used to identify point mutations in candidate melanoma driver genes (BRAF, NRAS, KRAS, GNA11, GNAQ), and other genes implicated in melanoma prognosis (EIF1AX, SF3B1). Immunohistochemistry was performed to analyse BAP1 nuclear expression.
Results
MLPA detected copy number alterations in chromosomes 1p, 3, 6 and 8. Sequencing of melanoma driver genes revealed GNAQ (p.Q209L) mutations in two samples; although it is possible that these samples represent extraocular spread of an occult uveal melanoma. A recurrent mutation in SF3B1 (p.R625H) was observed in indolent, but not aggressive, tumours; a mutation in EIF1AX (p.N4S) was detected in one patient with non-aggressive disease.
Conclusions
EIF1AX and SF3B1 mutations appear have a role in determining the clinical course of POM and detection of these changes could have clinical significance. Further in depth analysis of this rare group using differing ‘omic technologies will provide novel insights into tumour pathogenesis.
Keywords: Orbit, Melanoma, Primary, SF3B1, EIF1AX
Background
Ocular melanoma is a rare, generally lethal, malignancy that can arise in the uveal tract, the conjunctiva, or the ocular adnexa (eyelid or orbit). Orbital melanoma occurs either as primary disease, as secondary disease (local invasion from an ocular or sinonasal primary tumour), or as a metastasis from the contralateral eye or from the skin. Melanoma accounts for 5–20% of metastatic and secondary orbital malignancies, but only a very small proportion of primary orbital neoplasia [1–3]. Primary orbital melanoma (POM) is extremely rare, with approximately 60 cases reported to date. It is possible that POM arise from melanocytic cells lining the leptomeninges or ciliary nerves, or from ectopic intraorbital nests of melanocytes [4]. POM can occur de novo, but it is sometimes reported in association with pigmentary changes of the periocular tissues – such as naevus of Ota, blue cellular naevus or oculo-dermal melanosis [5]. The prognosis of POM appears to be quite variable. Generally, the disease is considered to have a very poor prognosis, but there appears to be a subset of patients who have long-survival [6, 7].
The genetic aberrations that drive cutaneous melanoma (CM) and uveal melanoma (UM) are well-described and quite distinct from each other. Mutations in the RAS pathway are found in > 75% of CM and have also been described in conjunctival melanoma, whereas mutations in GNAQ and GNA11 are found in ~ 85% of UM [8, 9]. Several genes have also recently been implicated in UM prognosis, such as BAP1, SF3B1 and EIF1AX. There are no data, however, describing the genetic alterations found in POM. In this study, we examined POM for alterations in candidate melanoma driver genes (BRAF, NRAS, KRAS, GNA11, GNAQ) and genes implicated in UM prognosis (EIF1AX, SF3B1, BAP1).
Methods
This study received ethics approval from Moorfields Eye Hospital Biobank ethics board (15/SW/0104). Informed, written consent was obtained from patients and research adhered to the tenets of the Declaration of Helsinki. Twelve tumour DNA samples from 11 patients were included in the study; the demographics and clinical course (including disease presentation, occurrence of metastasis and overall survival) of the patients has been previously published but is also summarised in Table 1 [7]. Prognosis was defined as poor (survival of < 6 months with or without metastasis), intermediate (survival > 6 months with local or systemic spread of disease) or good (survival > 6 months without spread; or very late recurrence of disease (> 10 years)).
Table 1.
Summary of clinical characteristics of 11 patients with primary orbital melanoma. “RT” denotes fractionated external beam radiotherapy. † indicates a deceased patient
| Case no. | Gender | Age at onset (years) | Side | Primary treatment of orbital disease | Orbital progression | Time orbital treatment to orbital recurrence (months) | Systemic progression | Time orbit to systemic disease (months) | Systemic therapy | Survival (months) | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 81 | R | Debulking | N | – | Y | 0 | 3† | Too unwell for adjuvant radiotherapy | |
| 2 | M | 40 | L | Exenteration | N | – | Y | 0 | 4† | Too unwell for adjuvant radiotherapy | |
| 4 | F | 58 | L | Exenteration | Unknown | – | Unknown | – | 25 | Patient declined adjuvant radiotherapy & active follow up | |
| 5 | M | 45 | L | Debulking + RT | Y | 6 | N | – | 37 | Exenteration after orbital progression | |
| 6 | F | 84 | R | Exenteration + RT | N | – | Y | 12 | Palliative RT for bone metastases | 18† | Conjunctival melanosis |
| 8 | M | 45 | L | Debulking + RT | Y | 7 | Y | 45 | Liver resection | 91 | Nevus of Ota Exenteration after orbital progression |
| 9 | F | 47 | L | Debulking + RT | Y | 161 | Y | 168 | Nil active | 174† | Conjunctival nevus Late exenteration |
| 10 | M | 46 | R | Debulking +RT | N | – | N | – | 26 | ||
| 11 | F | 43 | L | Debulking + RT | N | – | N | – | 188 | West African | |
| 12 | F | 70 | L | Debulking +RT | N | – | N | – | 22 | ||
| 13 | M | 55 | R | Debulking + RT | Y | 2 | Y | 5 | Immunotherapy | 35 | Immunotherapy for progressive systemic and orbital disease |
DNA extraction
Tumour DNA was extracted from surgical samples as previously described [10]. Germline DNA was extracted from peripheral blood using standard phenol-based method.
MLPA
MLPA was undertaken using the SALSA MLPA P027-C1 kit (MRC Holland, The Netherlands) according to manufacturer’s instructions and previously published methods [10], to detect copy number alterations of chromosomes 1p, 3, 6 and 8. Classification of chromosomes was based on the dosage quotients generated and ≥ 75% probes on a particular chromosome being classified as loss, normal or gain as previously described [11]. Chromosomes were considered ‘unclassified’ when the criteria could not be met.
Sanger sequencing
PCR was performed using custom primer sequences to target regions (Table 2) using PCR Master Mix 2X, ThermoFisher Scientific UK (cycle conditions available upon request). Sanger sequencing was performed using BigDye Terminator v3.1 on ABI3730 using a standard protocol (both Applied Biosystems, UK). Sequence data was analysed on Seqman Pro (DNASTAR, USA). Identified changes were validated in a second PCR and sequencing reaction. Validated mutations were assayed in germline genomic DNA from the patient, to confirm that they were a somatic mutation.
Table 2.
Regions of interest assayed and primer sequences used. The commonly reported mutations in the exons of interest were taken from individual gene reports on COSMIC database [13]
| Gene | Exon | Forward primer | Reverse primer | Previously reported mutations | Implicated in |
|---|---|---|---|---|---|
| BRAF | 11 | CTGTTTGGCTTGACTTGAC | CATATCCTATTATGACTTG | Minor mutation hotspots at codons 466 and 469 | Driver mutation, cutaneous melanoma |
| 15 | CTGATAGGAAAATGAGAT | CAGCAGCATCTCAGGGCC | p.V600E | ||
| NRAS | 2 | GTGGCTCGCCAATTAAC | CCGACAAGTGAGAGAC | Mutation hotspots at codons 12 and 13 | Driver mutation, cutaneous melanoma |
| 3 | GCATTGCATTCCCTGTGG | GAACACAAAGATCATCC | p.Q61R, p.Q61L | ||
| KRAS | 2 | GATAGTGTATTAACC | AACCTTTATCTGTATC | Mutation hotspots at codons 12 and 13 | Driver mutation,cutaneous melanoma |
| 3 | GTGCACTGTAATAATCT | AATTACTCCTTAATGTC | p.Q61R, p.Q61H | ||
| GNAQ | 4 | CTTTCCGTAGACAGCTTTG | GTACTCAAGGCATAAAAG | p. R183Q | Driver mutation, uveal melanoma |
| 5 | GCTATATTTATGTTGAC | CTATCATTTACTTGTATC | p.Q209L, p.Q209R, p.Q209P | ||
| GNA11 | 4 | GCAGCCGGCCTGAGCA | ACACACACTGAGGATG | p. R183C | Driver mutation, uveal melanoma |
| 5 | GCCAGGTGGCTGAGT | GCAGGGCCTTACTGG | p.Q209L, p.Q209P | ||
| SF3B1 | 14 | GGCCGAGAGATCATTTCTAAT | AAGAAGGGCAATAAAGAAGGA | Mutation hotspot at codon 625, and multiple other hotspots | Prognosticating mutation in uveal melanoma, possible driver mutation |
| EIF1AX | 1 | GCCACGCCTGCGTCATAAAG | CGAGCTCAGAGTCGCGTGTG | Mutation hotspots at codons 2,3,4,6 | Prognosticating mutation in uveal melanoma, possible driver mutation |
| 2 | AAAGGAAATTCCAAGAAGGGTAGGG | TAATCGTGCCACCACACTTCACC | Mutation hotspots at codons 13, 15 |
BAP1 immunohistochemistry
BAP1 protein expression was examined in 4 μm sections of each POM case using a mouse anti-BAP1 antibody (sc-28,383, Santa Cruz, Insight Biotechnology Ltd., Middlesex, UK) and the DAKO Envision FLEX Kit, according to previously published methods [12]. POM were classified as either BAP1 positive or BAP1 negative according to whether the tumour cells displayed nuclear staining or not, respectively.
Results
Copy number changes in chromosomes 6 and 8 are frequently observed in POM
MLPA was performed to investigate chromosomal changes within POM. Copy number alterations were obtained for 7/11 tumour samples (Table 3). Four tumours were reported as disomy 3 and three cases had unclassifiable chromosome 3 status. The most frequent change was gain of chromosome 6p, in 5/7 cases. Polysomy 8q was reported in four cases; gain of chromosome 1p was also observed (2/7 cases). It was not possible to correlate chromosomal alterations with outcome because of the small number of patients for whom data were available.
Table 3.
MLPA, gene sequencing and BAP1 IHC in eleven primary orbital melanoma cases. The case number refers to the clinical histories previously published [7]. G gain, U unclassified, N normal copy normal, L loss; − reference sequence, + heterozygous mutation observed
| Case ID | BAP1 IHC | MLPA result - chromosome | SF3B1 (R625H) | EIF1AX (N4S) | GNAQ (Q209L) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1p | 3 | 6p | 6q | 8p | 8q | |||||
| 1 | Positive | G | U | G | N | N | G | – | – | – |
| 2 | Positive | MLPA QC fail | – | – | – | |||||
| 4 | Negative | N | N | G | N | N | G | + | – | + |
| 5 | Positive | MLPA QC fail | – | – | + | |||||
| 6 | NA | N | N | N | N | N | N | – | – | – |
| 8 | Positive | N | U | G | N | N | N | – | – | – |
| 9 | Positive | G | N | G | L | L | G | + | – | – |
| 10 | Positive | N | N | G | N | N | G | – | + | – |
| 11 | Positive | U | U | U | G | G | N | + | – | – |
| 12 | Positive | MLPA QC fail | + | – | – | |||||
| 13 | Negative | MLPA QC fail | – | – | – | |||||
Analysis of driver mutations in POM
Sequencing of mutation hotspots in five melanoma driver genes (BRAF, NRAS, KRAS, GNAQ, GNA11) did not reveal any changes in 9/11 patients. In two patients (numbers 4 and 5), a change in GNAQ was found: c.A626T, p.Q209L (Fig. 1a). GNAQ mutations account for approximately 50% of UM driver changes, and p.Q209L is found in 33% of GNAQ mutant tumours [13]. It was considered, therefore, whether these two samples could represent an extraocular manifestation of a primary UM, with an occult uveal source. Review of the available imaging did not reveal any possible uveal source. In light of this, the histology slides were re-reviewed by a second expert pathologist (PL, blinded to genetic findings). It was observed that patient 4 had some small regions of increased pigmentation in the choroid that might represent a uveal source of the tumour, although it was felt more likely that these were insignificant findings given the benign appearance and size of the lesions (Fig. 2). In the case of patient 5, the exenteration sample was not available for histological analysis and, so, although there was no evidence of choroidal involvement in the samples examined, it remains impossible to fully exclude an occult uveal source.
Fig. 1.
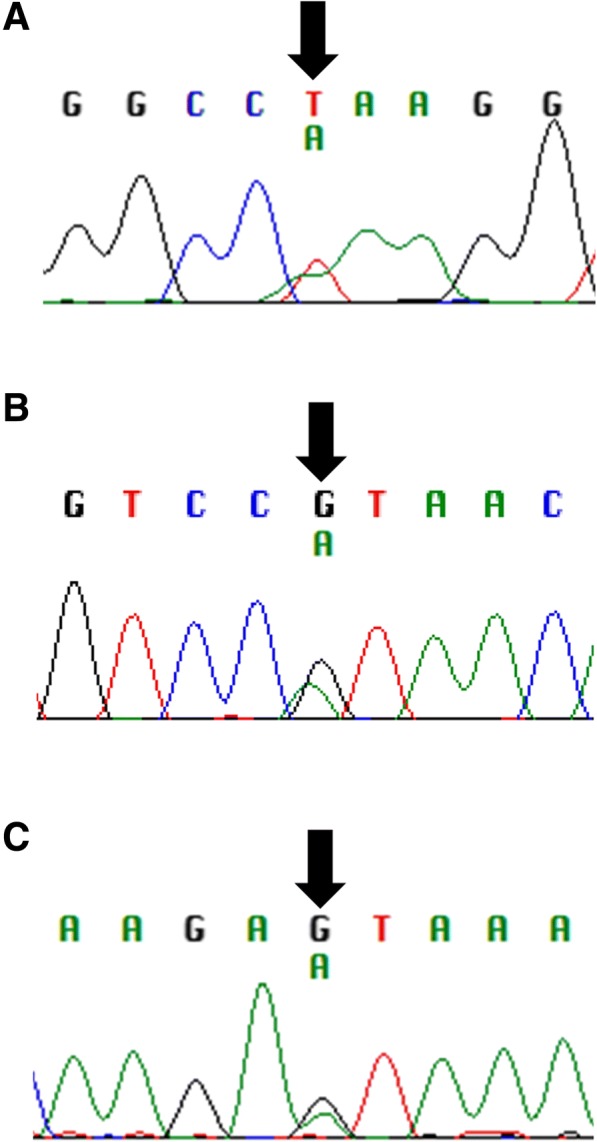
Point mutations identified in tumour tissue of primary orbital melanoma. a Two cases (patients 4 and 5) harboured a heterozygous mutation in GNAQ (c.A626T, p.Q209L). b A recurrent heterozygous mutation in SF3B1 (c.G1874A, p.R625H) was identified in four patients with favourable prognoses (cases 4, 9, 11, 12). c One patient, case 10, carried a heterozygous change in EIF1AX (c.A11T, p.N4S)
Fig. 2.
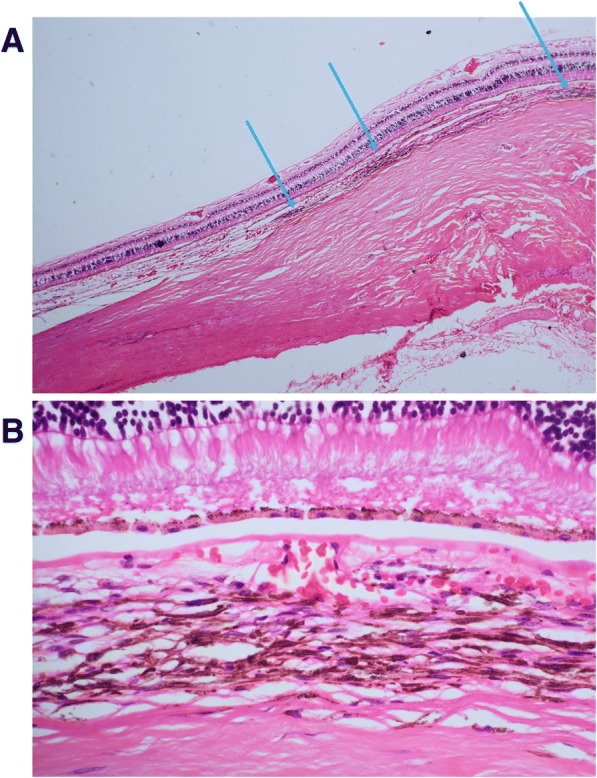
a H&E staining of serial slices of exenteration specimen revealed several small regions of increased choroidal melanosis in case 4 (arrowed). b Representative image of region with increased pigmentation, showing that the pigmented cells were not malignant in appearance
Identification of a recurrent change in SF3B1
Sequencing of exon 14 of SF3B1 revealed that four patients (cases 4, 9, 11, 12) harboured the same mutation; c.G1874A, p.R625H (Fig. 1b). One patient (case 10) was found to harbour a change in exon 1 of EIF1AX; c.A11T, p.N4S (Fig. 1c). These changes were not present in the germline DNA of the patients.
It was noted that the patients harbouring the recurrent SF3B1 mutation all had highly favourable prognosis (Table 4). The mean survival to-date of those patients harbouring the SF3B1 mutation is 104.8 months (range = 22–188 months), whilst the mean survival of patients without the SF3B1 mutation is 29.8 months (range = 3–91 months). It is not possible, however, to assess the significance of these differences due to the small sample size. Furthermore, it should be noted that of the seven patients with systemic and/or orbital progression of disease, only one carried the SF3B1 mutation – and this patient remained without disease progression for over 13 years prior to sudden recurrence, deterioration and death. The SF3B1 mutation was found in both the first tumour biopsy sample (disease which remained quiescent for 13 years) and the recurrent tumour sample.
Table 4.
Correlation between prognosis in patients harbouring SF3B1 and EIF1AX changes versus wildtype patients. Deceased patients indicated by †; survival correct to January 2018. The two patients highlighted in grey might represent secondary orbital melanoma with an occult uveal origin
| Case no. | Prognosis group | Orbital progression | Systemic progression | Survival (months) | SF3B1 p.R625H | EIF1AX p.N4S |
|---|---|---|---|---|---|---|
| 1 | Poor | No | Yes | 3† | ||
| 2 | No | Yes | 4† | |||
| 13 | Intermediate | Yes | Yes | 25 | ||
| 5 | Yes | No | 37 | |||
| 6 | No | Yes | 18† | |||
| 8 | Yes | Yes | 91 | |||
| 9 | Good | Yes (after 13 years) | Yes (after 13 years) | 174† | YES | |
| 10 | No | No | 26 | YES | ||
| 11 | No | No | 188 | YES | ||
| 12 | No | No | 22 | YES | ||
| 4 | Unknown | Unknown | 35 | YES |
BAP1 immunohistochemistry
BAP1 IHC was possible in 10 cases, a single case had no remaining available tumour material. Nuclear BAP1 (nBAP1) expression was noted in the tumour cells in eight cases and was absent in two (Fig. 3). Of the two nBAP1 negative cases, one showed disomy of chromosome 3 and the presence of SF3B1 and GNAQ mutations, whilst in the second case MLPA had not yielded any results and no mutations were detected.
Fig. 3.
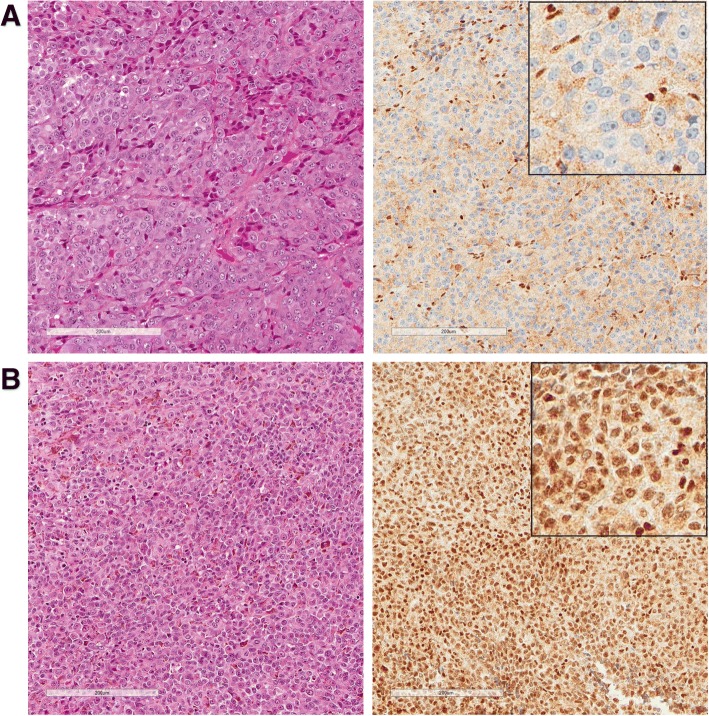
Representative images of BAP1 immunohistochemistry. a H&E and nuclear BAP1 (nBAP1) negative tumour (Case ID 13), with normal staining found in admixed lymphocytes. b H&E and nBAP1 positive tumour (Case ID 12). Insets show nBAP1 staining at ×40 magnification
Discussion
In this study we examined the largest reported clinical series of POM for mutations implicated in melanoma, giving the first report of genetic alterations in this rare tumour. It was demonstrated that POM appears to be genetically distinct to CM, but shares some overlapping features with UM; however, a distinct driver mutation might exist in at least some cases of POM. Furthermore, mutations in SF3B1 and EIF1AX might influence prognosis. POM is an extremely rare tumour, with approximately 60 cases reported to-date in the literature. One striking feature is the highly variable prognosis – whilst most reported cases have a dismal prognosis, a subset of patients appear to follow a relatively benign course [6, 7]. This feature was certainly observed in the patient cohort used in this study – some patients had aggressive primary disease with widespread systemic involvement and rapid deterioration; whereas other patients survived for more than a decade [7].
POM were not associated with monosomy 3 in the small number of tested samples, which is a frequent chromosomal alteration in UM associated with poor prognosis [14]. In addition, although polysomy 8q was observed in 4/7 POM in this cohort, all were associated with patients who had a good prognosis, which again is contrary to its association with a poor outcome in UM. These data would suggest that chromosomal alterations in POM do not follow a similar pattern to that observed in UM, although testing of more cases will be important. Similarly, loss of nBAP1, which is associated with a poor prognosis and monosomy of chromosome 3 in UM [12, 15, 16] was noted in only two cases and was neither associated with a poor outcome nor with monosomy of chromosome 3.
Mutations in genes of the MAPK pathway are known to drive the majority of cases of CM. Mutations in BRAF account for the majority of driver mutations (up to 60%) [17]. However, mutations in NRAS and KRAS are also relatively frequent (13–25 and 2%, respectively) [18]. Screening for mutations in these genes did not, however, reveal any changes in our POM cohort. Mutations in two Gq alpha sub-unit genes, which interact indirectly with the MAPK pathway, are found frequently in UM: GNAQ (~ 45%) and GNA11 (~ 35%) [19, 20]. In our cohort, two cases (patients 4 and 5) were found to carry a heterozygous mutation in GNAQ (p.Q209L). This change is the most frequently observed change in GNAQ mutant UM, occurring in one third of such cases [21]. This dominant-negative mutation alters the catalytic (GTPase) domain of GNAQ and results in a constitutively active protein [22]. Expression of GNAQ p.Q209L in mice resulted in malignant transformation of melanocytes and increased signalling through the MAPK pathway [21]. It is often speculated whether POM cases are true primary disease, or whether an occult uveal source might be present. It was considered necessary, therefore, to assess whether these two cases could be occult uveal tumours, given the frequency of GNAQ p.Q209L in UM. The imaging of both cases was reviewed, and a uveal source could not be discerned. The tumour histology of the cases was re-reviewed by an expert pathologist, without knowledge of the genetic findings. In one case harbouring the GNAQ change, there was evidence of very minor choroidal pigmentary changes which could represent a possible uveal source; in the other case, there was insufficient biopsy material to fully exclude a uveal source. It is possible, therefore, that these two cases might indeed have an occult uveal source. It is also plausible, however, that these are POM that share common genetic features with UM. Mutations in other G protein genes, such as CYSLTR2 and PLCB4 have been reported in a small number of UM patients and these could be sequenced in our cohort to extend the study further [23, 24].
Next, it was considered whether mutations in SF3B1 or EIF1AX contributed to the pathogenesis of disease and/or variable prognosis seen within our cohort. Mutually exclusive changes in both genes were found (which might associate with good prognosis), highlighting another feature that overlaps with the genetic landscape of UM. Sequencing of exon 14 of SF3B1 revealed a recurrent heterozygous mutation (c.G1874A, p.R625H) in four patients – cases 4, 9, 11 and 12. It was noted that all four of these patients had a favourable outcome, with a mean survival to-date of 104.8 months (22–188 months). Three of these patients have not had any local or systemic progression or recurrence of disease. One patient – case 9 – harbouring the SF3B1 mutation had systemic progression and has since died, however, this was after a remarkable disease-free period of 13 years. The presence of the SF3B1 mutation was confirmed in both the primary and recurrent tumour of this individual. In contrast, no patients with poor survival, or early local/systemic progression, carried the SF3B1 change. This would suggest that the observed mutation in SF3B1 confers a favourable prognosis in POM.
SF3B1 encodes splicing factor 3b, subunit 1 protein, which is a component of the splicing factor 3b protein complex. This complex is part of the spliceosome, the macromolecular structure responsible for transcriptional mRNA processing. Mutations in SF3B1 have been implicated as a common driver mutation in myelodysplatic syndromes (MDS), myelofibrosis and chronic myeloid leukaemia [25, 26]. In MDS, SF3B1 mutations have been associated with favourable overall survival and a lower likelihood of transformation into acute leukaemia [27, 28]. More recently, mutations in SF3B1 (particularly at codon 625) have been identified in various pigmented tumours, including UM [29], mucosal melanoma [30], leptomeningeal melanoma [31] and blue naevi-like cutaneous melanoma [32]. They are, however, rare in CM [33, 34]. As with MDS, SF3B1 mutations confer a favourable prognosis in UM, with lower age of onset and concurrent disomy 3 [35]. However, it should be noted that SF3B1 mutant UM are reported to give rise to late metastasis (median 8.2 years after initial diagnosis) [36]. Furthermore, The Cancer Genome Atlas project reported that UM cases with SF3B1 mutations have an intermediate prognosis [9]. One of our patients harbouring the change (case 9) did indeed have late onset metastasis to the brain (13 years after diagnosis). It is also interesting to note that secondary melanoma within the orbit was noted to have frequent late recurrence, and the incidence of SF3B1 in these tumours could be studied [37].
One patient, case 10, was found to harbour a heterozygous mutation in exon 1 of EIF1AX (c.A11T, p.N4S). This patient also had a highly favourable prognosis. EIF1AX, located on the X chromosome, encodes the eukaryotic translation initiation factor 1A protein. This factor is essential in the initiation phase of translation, through interaction with tRNA and the ribosome [38]. Mutations in exon 1 or 2 of this gene have been frequently reported in UM [39], but also rarer melanoma types including blue-nevus associated melanoma and leptomeningeal melanoma [33, 34]. EIF1AX mutations are associated with good prognosis disomy 3 UM, and usually occur in non-metastatic cases [39–41]. This is in agreement with the patient reported here, who displayed all of these features to-date with follow up of over 2 years. It is highly plausible, therefore, that mutations in EIF1AX also correspond to good prognosis in POM.
It must be considered whether the changes seen in SF3B1 and EIF1AX are the driver mutation of POM, or a secondary change in the tumour. It is thought that mutations in SF3B1 arise as a second genetic change in UM and blue nevus-like melanoma, after initial mutation in GNAQ or GNA11; however, SF3B1 changes can be driver mutations in MDS [25, 34]. Indeed, in UM, mutations in either gene (SF3B1 or EIF1AX) are almost never seen in the absence of a GNAQ or GNA11 mutation [13]. Whole exome or whole genome sequencing of these tumours will be necessary to elucidate the full spectrum of genetic mutations in POM. A further consideration is the role of underlying precursor or premalignant lesions in the genetic aetiology of POM. The tumour can arise following malignant transformation of pigmentary changes of the periocular tissues – such as naevus of Ota, blue cellular naevus or oculo-dermal melanosis [5]. This was the case in three individuals studied here (Cases 6, 8 and 9; in addition, minor increased choroidal pigmentation was observed on re-examination of Case 4. It would be interesting to study the genetic aberrations in the premalignant tissue, as this might shed light on the temporal occurrence of sequential mutations and their role in pathogenesis.
Conclusions
In summary, we have provided the first genetic insights into POM. We have shown that – in contrast to CM and UM – mutations in the tested MAPK pathway genes or Gq alpha subunit genes are not implicated in the majority of POM. Furthermore, we have shown that mutations in either SF3B1 or EIF1AX are likely to be associated with a favourable prognosis in POM cases. Testing for these changes in the clinical setting might allow better prognostication for patients. However, it is possible that SF3B1 mutations are associated with late recurrence of disease, and so long-term follow-up and high index of suspicion should be maintained in all cases of orbital melanoma. Additional ‘omic technologies, such as whole genome sequencing, should be used to explore whether there are other driver mutations involved in the development of these rare tumours.
Acknowledgements
The authors would like to thank all of the patients for taking part in this study, and Lucretia Medard for assistance with processing of pathology samples. The authors are also grateful to Prof. Alex Blakemore for allowing use of laboratory space.
Funding
The project received funding from Melanoma Focus (UK charity no. 1124716), British Association of Dermatology and NIHR Imperial Biomedical Research Centre through the Academic Foundation Programme.
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
Abbreviations
- CM
Cutaneous melanoma
- IHC
Immunohistochemistry
- MAPK
Mitogen-activated protein kinase
- MDS
Myelodysplastic syndrome(s)
- MLPA
Multiplex ligation dependent probe analysis
- PCR
Polymerase chain reaction
- POM
Primary orbital melanoma
- UM
Uveal melanoma
Authors’ contributions
Study design- AMR, CNJ, SEC, GER; patient recruitment and consent: DHV, GER; bench work: AMR, RL, UKR, HK, ST; histological assessment: PJL, SEC; analysis of data: AMR, HK, SEC; writing and review of manuscript: AMR, HK, SEC, GER. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study received ethics approval from Moorfields Eye Hospital Biobank ethics board (15/SW/0104). Informed, written consent was obtained from patients and research adhered to the tenets of the Declaration of Helsinki.
Consent for publication
Consent to publish data was included in the research consent process.
Competing interests
The authors have no competing interests to declare.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anna M. Rose, Email: anna.rose@ucl.ac.uk
Rong Luo, Email: seraphina.rong13@imperial.ac.uk.
Utsav K. Radia, Email: utsav.radia13@imperial.ac.uk
Helen Kalirai, Email: H.Kalirai@liverpool.ac.uk.
Sophie Thornton, Email: Sophie.Thornton@liverpool.ac.uk.
Philip J. Luthert, Email: p.luthert@ucl.ac.uk
Channa N. Jayasena, Email: c.jayasena@imperial.ac.uk
David H. Verity, Email: veritydh@aol.com
Sarah E. Coupland, Email: S.E.Coupland@liverpool.ac.uk
Geoffrey E. Rose, Phone: 020 7566 2034, Email: geoff.rose@moorfields.nhs.uk
References
- 1.Valenzuela AA, Archibald CW, Fleming B, Ong L, O’Donnell B, Crompton J, Selva D, McNab AA, Sullivan TJ. Orbital metastasis: clinical features, management and outcome. Orbit. 2009;28:153–159. doi: 10.1080/01676830902897470. [DOI] [PubMed] [Google Scholar]
- 2.Zografos L, Ducrey N, Beati D, Schalenbourg A, Spahn B, Balmer A, et al. Metastatic melanoma in the eye and orbit. Ophthalmology. 2003;110(11):2245–2256. doi: 10.1016/j.ophtha.2003.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad SM, Esmaeli B. Metastatic tumours of the orbit and ocular adnexa. Curr Opin Ophthalmol. 2007;18(5):405–413. doi: 10.1097/ICU.0b013e3282c5077c. [DOI] [PubMed] [Google Scholar]
- 4.Rootman J (editor). Diseases of the Orbit: a multidisciplinary approach. 2nd ed. Philadelphia: Lippincott Williams & Wilkins (2003). p. 53–84.
- 5.Henry HL, Chan MB, Kono T. Nevus of Ota: clinical aspects and management. Skin Med. 2003;2:89–96. doi: 10.1111/j.1540-9740.2003.01706.x. [DOI] [PubMed] [Google Scholar]
- 6.Henderson JW. Orbital Tumors. 3rd ed. p. 278.
- 7.Rose AM, Luthert PJ, Jayasena CN, Verity DH, Rose GE. Primary orbital melanoma: presentation, treatment, and long-term outcomes for 13 patients. Front Oncol. 2017;7:316. doi: 10.3389/fonc.2017.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cicenas J, Tamosaitis L, Kvederaviciute K, Tarvydas R, Staniute G, Kalyan K, Meskinyte-Kausiliene E, Stankevicius V, Valius M. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Med Oncol. 2017;34:26. doi: 10.1007/s12032-016-0879-9. [DOI] [PubMed] [Google Scholar]
- 9.Robertson AG, Shih J, Yau C, et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell. 2017;32:204–220.e15. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lake SL, Coupland SE, Taktak AF, Damato BE. Whole-genome microarray detects deletions and loss of heterozygosity of chromosome 3 occurring exclusively in metastasizing uveal melanoma. Invest Ophthalmol Vis Sci. 2010;51(10):4884–4891. doi: 10.1167/iovs.09-5083. [DOI] [PubMed] [Google Scholar]
- 11.Caines R, Eleuteri A, Kalirai H, et al. Cluster analysis of multiplex ligation-dependent probe amplification data in choroidal melanoma. Mol Vis. 2015;21:1–11. [PMC free article] [PubMed] [Google Scholar]
- 12.Farquhar N, Thornton S, Coupland SE, et al. Patterns of BAP1 protein expression provide insights into prognostic significance and the biology of uveal melanoma. J Pathol Clin Res. 2017;4(1):26–38. doi: 10.1002/cjp2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.COSMIC Catalogue of Somatic Mutations in Cancer. Available at: http://cancer.sanger.ac.uk/cosmic. Accessed Dec 2018.
- 14.Damato B, Coupland SE. Translating uveal melanoma cytogenetics into clinical care. Arch Ophthalmol. 2009;127(4):423–429. doi: 10.1001/archophthalmol.2009.40. [DOI] [PubMed] [Google Scholar]
- 15.van de Nes JA, Nelles J, Kreis S, et al. Comparing the prognostic value of BAP1 mutation pattern, chromosome 3 status, and BAP1 immunohistochemistry in uveal melanoma. Am J Surg Pathol. 2016;40(6):796–805. doi: 10.1097/PAS.0000000000000645. [DOI] [PubMed] [Google Scholar]
- 16.Koopmans AE, Verdijk RM, Brouwer RW, et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod Pathol. 2014;27:1321–1330. doi: 10.1038/modpathol.2014.43. [DOI] [PubMed] [Google Scholar]
- 17.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Bröcker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 18.Cicenas J, Tamosaitis L, Kvederaviciute K, Tarvydas R, Staniute G, Kalyan K, Meskinyte-Kausiliene E, Stankevicius V, Valius M. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Med Oncol. 2017;34(2):26. doi: 10.1007/s12032-016-0879-9. [DOI] [PubMed] [Google Scholar]
- 19.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457(7229):599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.GNAQ, MyCancerGenome Database. Available at: https://www.mycancergenome.org/content/disease/melanoma/gnaq/100/. Accessed Mar 2018.
- 22.Kalinec G, Nazarali AJ, Hermouet S, Xu N, Gutkind JS. Mutated alpha subunit of the Gq protein induces malignant transformation in NIH 3T3 cells. Mol Cell Biol. 1992;12(10):4687–4693. doi: 10.1128/MCB.12.10.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore AR, Ceraudo E, Sher JJ, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48(6):675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson P, Aoude LG, Wadt K, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7(4):4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 26.Patel BJ, Przychodzen B, Thota S, et al. Genomic determinants of chronic myelomonocytic leukemia. Leukemia. 2017;31(12):2815–2823. doi: 10.1038/leu.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cazzola M, Della Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122(25):4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malcovati L, Papaemmanuil E, Bowen DT, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118(24):6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45(2):133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hintzsche JD, Gorden NT, Amato CM, Kim J, Wuensch KE, Robinson SE, Applegate AJ, Couts KL, Medina TM, Wells KR, Wisell JA, McCarter MD, Box NF, Shellman YG, Gonzalez RC, Lewis KD, Tentler JJ, Tan AC, Robinson WA. Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma. Melanoma Res. 2017;27(3):189–199. doi: 10.1097/CMR.0000000000000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Küsters-Vandevelde HV, Creytens D, van Engen-van Grunsven AC, Jeunink M, Winnepenninckx V, Groenen PJ, Küsters B, Wesseling P, Blokx WA, Prinsen CF. SF3B1 and EIF1AX mutations occur in primary leptomeningeal melanocytic neoplasms; yet another similarity to uveal melanomas. Acta Neuropathol Commun. 2016;4:5. doi: 10.1186/s40478-016-0272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griewank KG, Müller H, Jackett LA, et al. SF3B1 and BAP1 mutations in blue nevus-like melanoma. Mod Pathol. 2017;30(7):928–939. doi: 10.1038/modpathol.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schilling B, Bielefeld N, Sucker A, Hillen U, Zimmer L, Schadendorf D, Zeschnigk M, Griewank KG. Lack of SF3B1 R625 mutations in cutaneous melanoma. Diagn Pathol. 2013;8:87. doi: 10.1186/1746-1596-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong Y, Krauthammer M, Halaban R. Rare SF3B1 R625 mutations in cutaneous melanoma. Melanoma Res. 2014;24(4):332–334. doi: 10.1097/CMR.0000000000000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helgadottir H, Höiom V. The genetics of uveal melanoma: current insights. Appl Clin Genet. 2016;9:147–155. doi: 10.2147/TACG.S69210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yavuzyigitoglu S, Koopmans AE, Verdijk RM, et al. Uveal melanomas with SF3B1 mutations: a distinct subclass associated with late-onset metastases. Ophthalmology. 2016;123(5):1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 37.Rose AM, Cowen S, Jayasena CN, Verity DH, Rose GE. Presentation, treatment, and prognosis of secondary melanoma within the orbit. Front Oncol. 2017;7:125. doi: 10.3389/fonc.2017.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhuri J, Si K, Maitra U. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J Biol Chem. 1997;272(12):7883–7891. doi: 10.1074/jbc.272.12.7883. [DOI] [PubMed] [Google Scholar]
- 39.Martin M, Masshofer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45(8):933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decatur CL, Ong E, Garg N, Anbunathan H, Bowcock AM, Field MG, Harbour JW. Driver mutations in uveal melanoma: associations with gene expression profile and patient outcomes. JAMA Ophthalmol. 2016;134(7):728–733. doi: 10.1001/jamaophthalmol.2016.0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ewens KG, Kanetsky PA, Richards-Yutz J, et al. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Invest Ophthalmol Vis Sci. 2014;55(8):5160–5167. doi: 10.1167/iovs.14-14550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.