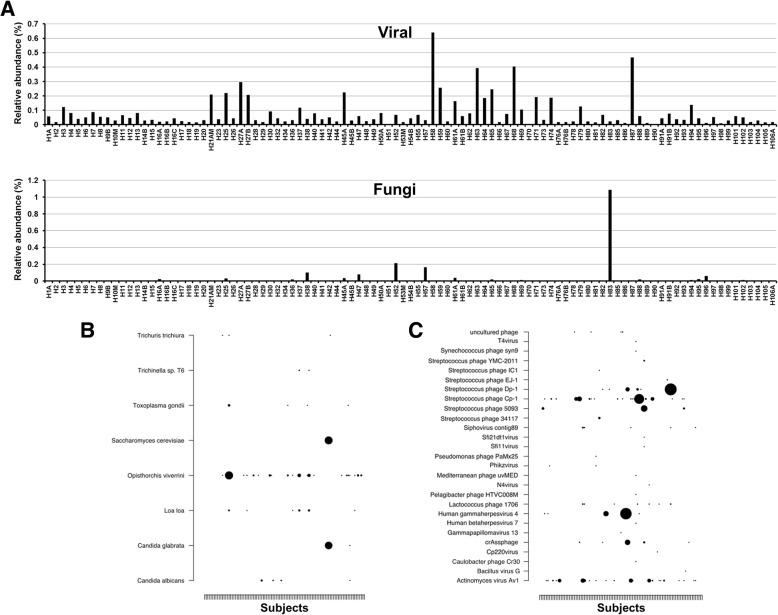
Fig. 5.
Presence of non-bacterial microbial taxa within the esophageal microbiome. a Relative abundance of viruses and fungi within the esophageal microbiome of each subject. Relative abundances were calculated using taxonomy arising from MEGAN6. This was used due to its capacity to detect microbial eukaryotes. b Relative abundances of specific eukaryotic and c viral taxa within each subject arising from the MEGAN6 analysis. Size of circle signifies the relative abundance levels of the organism. Size of circles ranges from 0.0094 (smallest) to 0.64% (largest) for viruses and 0.0097–1.08% for eukaryotes. Subjects are ordered by time of recruitment (left to right). The presence of Candida spp. and Saccharomyces were confirmed using 18S amplicon sequencing. We could not confirm the detection of Trichuris, Trichinella, and Loa loa; thus, these identifications should be taken with caution as they most probably arose from misclassifications within the metagenomic annotation process. No association with any of the clinical metadata was found