Abstract
We describe a comprehensive method for imaging and analysis of local protein dynamics at single sites of exocytosis in living cultured endocrine cells. This method is well suited to quantitatively map the complex dynamics of individual molecules at single sites of vesicle fusion in live cells.
Keywords: TIRF, exocytosis, live-cell imaging, vesicle fusion, image analysis, endocrine cells
1. Introduction
Exocytosis, the regulated secretion of molecules from cells is critical to many aspects of eukaryotic physiology [1]. During exocytosis, materials packaged into the lumen of secretory vesicles are released into the external environment when the vesicle membrane and plasma membrane fuse. Examples of well-studied exocytic systems are neurotransmitter release at neuronal synapses [2,3], adrenaline and insulin release from neuroendocrine and endocrine cells [4–7], acrosome and cortical granule fusion during fertilization [8,9], acetylcholine stimulation of muscle [10], and many more physiological processes.
The process of exocytosis is driven by a molecular complex that includes the proteins Syntaxin1a, SNAP-25, and VAMP [11]. These three SNARE proteins are thought to assemble between the vesicle and plasma membrane, forming a four helix bundle that zippers together and pulls both membranes close to one another. Additional factors including complexin, munc18, munc13, CAPS, tomosyn, and others are thought to regulate SNARE complex assembly in distinct cellular systems [12–15,11,16,17]. Physiologically, membrane fusion is triggered by increases in cytoplasmic calcium concentrations, which is sensed by the protein synaptotagmin [18,19]. Additional proteins including Rab GTPases and their effectors help direct vesicles to the plasma membrane and tether them close to sites of fusion [20–22]. While we understand a great deal about the genetics, biochemistry, structures, and mechanisms of many of these factors and their possible roles in exocytosis, it is still unclear which of these molecules are present at sites of fusion before, during, and after exocytosis. To address this central question, we visualize exocytic events using fluorescent proteins to directly interrogate the temporal dynamics of individual molecules at single sites of exocytosis in living cells.
Here, we present detailed methods for two-color through-the-objective total internal reflection fluorescence (TIRF) microscopy (also known as evanescent field microscopy) to visualize exocytosis in living cells [23,24]. TIRF illumination provides an exponentially-decaying fluorescence excitation field around one hundred nanometers above the coverslip/liquid interface [4]. This illumination depth is ideally suited to visualize with high contrast fluorescently-labeled 50–300 nm diameter vesicles arriving at, and fusing with, the plasma membrane at the bottom surface of an adherent cell [4]. In objective-based TIRF, adherent cells can be maintained in a bath solution and locally perfused to acutely stimulate exocytosis (Figure 1).
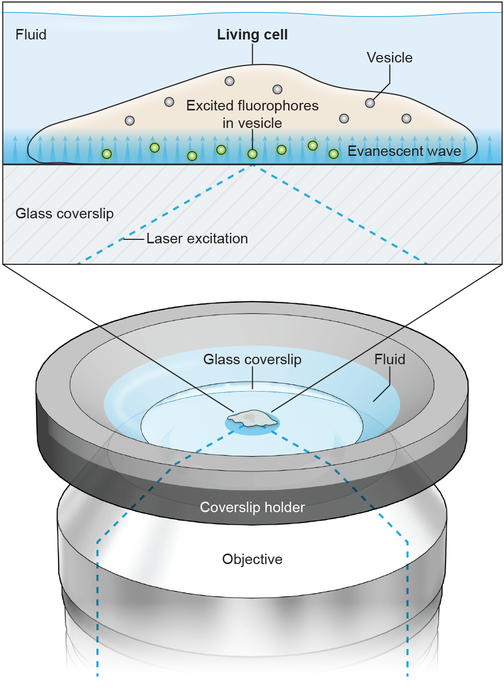
Figure 1.
Diagram of imaging geometry and a cell under TIRF illumination. A coverslip with living cells grown on it is mounted into a coverslip holder with fluid bathing the cells. From below, the excitation laser is directed through the imaging objective and its angle of incidence with the glass coverslip adjusted to achieve total internal reflection. This produces an evanescent illumination field, which decays exponentially, at the glass-water interface to illuminate fluorophores inside vesicles in the cell. With a low-profile coverslip holder, a perfusion pipette can be positioned above the cell using a micromanipulator to locally superfuse cells with stimulation solution.
By visualizing exocytosis in two colors using TIRF, we identify the individual and average behavior of molecules at single sites of exocytosis before, during, and after fusion [5,25]. This approach is valuable because it provides direct dynamic information about the local behavior of individual molecules, which in turn helps to interpret biochemical data and build mechanistic models to describe the regulation of the exocytic fusion machinery in mammalian cells [26,27].
2. Materials:
2.1. General cell culture and imaging
30% hydrogen peroxide solution
27–30% ammonium hydroxide solution
25 mm #1.5 glass coverslips
Ceramic or Teflon coverslip holder (Thomas Scientific)
Sharp tweezers for coverslip handling, long-handled tweezers or tongs for handling coverslip holders
Two 2 L glass beakers
100% ethanol
Hotplate
Fume hood
Poly-L-lysine solution (0.01%)
RPMI media without phenol red: 10% FBS, 1% pen/strep, 11.1 mM glucose, 2 mM glutamine, 1 mM pyruvate, 10 mM HEPES, 50 μM beta-mercaptoethanol
Dulbecco’s phosphate buffered saline
0.25% trypsin-EDTA solution
6-well tissue culture plates
T25 or T75 tissue culture flasks
Lipofectamine 2000
Immersion oil (NA: 1.518)
FluoSphere 0.1 μm yellow-green beads (Thermo Fisher Scientific)
Micro-perfusion system (ALA Scientific μFLow-8 with 100 μm diameter tip Quartz manifold)
Coverslip chamber (Warner Instruments Series 40 or Thermo Fisher Scientific AttoFluor)
Micromanipulator platform (ALA Scientific MT-75 series or similar)
Coarse micromanipulator (ALA Scientific MM-3 series or similar) with small manipulator mounted on end (ALA Scientific YOU-2 or similar)
Aspirating system (ALA Scientific VWK or similar)
Imaging Buffer (IB): 130 mM NaCl, 2.8 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 10 mM glucose, pH 7.4 (titrated with NaOH)
Ionomycin Buffer: IB with 10 μM ionomycin added
Stimulation Buffer: 50 mM NaCl, 105 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 1 mM NaH2PO4, pH 7.4 (titrated with KOH)
2.2. TIRF microscope setup
The microscope setup described here is specifically designed for two-color TIRF microscopy optimized for visualizing GFP and mCherry fluorescent proteins. Our system is built around an inverted microscope configured for through-the-objective TIRF. Fluorescence is excited by lasers at 488 nm and 561 nm, and lasers are combined and controlled with an acousto-optic tunable filter (AOTF), and directed into the back port of the microscope via optics for changing TIRF angle, size, and collimation. This microscope uses the Olympus ZDC2 drift correction system which is based on far-red light reflection from the glass/liquid interface on the coverslip. Optical filters and dichroics for excitation light control are bright-line full multiband LF405/488/561/635 filters (Semrock).
TIRF is achieved through an Olympus 100X 1.45 NA oil immersion objective. The resulting emission is divided by an image splitter’s dichroic (565DCXR) and projected side-by-side through 525Q/50 and 605Q/55 emission filters onto the chip of a back-illuminated EMCCD camera. Images are acquired using Andor IQ2 software.
Inverted microscope equipped with ZDC correction system (Olympus IX-81)
Sapphire 488 nm (Coherent) and 561 nm (Melles Griot LD-561–20A) lasers
Laser combiner system and AOTF (Andor, 400 Series)
100X 1.45 NA oil-immersion PlanApo objective (Olympus)
DualView image splitter (Photometrics DV2)
iXon Ultra 897 EM-CCD (Andor DU 897)
Software (Andor iQ2)
3. Methods:
3.1. Coverslip preparation:
Coverslips must be thoroughly cleaned and sterilized with ethanol prior to use. Our cleaning protocol is adapted from the first step of the classic RCA etch protocol [28]. This protocol removes organic contaminants, substantially reduces background fluorescence on the glass, and imparts partial charge to the glass to help surface-additives adhere.
Add coverslips to a ceramic or Teflon coverslip holder.
Place coverslip holders in the bottom of a 2 L glass beaker and add 300 mL water (see NOTE 1).
Add 60 mL of 30% hydrogen peroxide to the beaker and move into a fume hood.
Add 60 mL of 27% ammonium hydroxide to the beaker. Move the beaker to a hotplate and turn to high. Wait for five minutes and check for gentle bubbling in the beaker (approximately 50°C). After gentle bubbling begins, incubate the coverslips for 15 minutes. At the end of the incubation, the solution should be vigorously bubbling at 80–90°C. CAUTION: the fumes from the beaker are caustic and must not be inhaled.
Remaining in the hood, remove the beaker from the hotplate. Using long handled tweezers or wire tongs transfer the coverslip holders to a 2 L beaker filled with 1 L of water. Next transfer coverslip holders to smaller containers of 100 % ethanol for long-term storage.
Depending on the cell type, coverslips may need to be coated so that cells will adhere properly. Common surface additives are poly-L-lysine (PLL), poly-D-lysine (PDL), collagen, fibronectin, and others. For INS-1 cells, we briefly coat the coverslips with PLL immediately prior to adding cells. Remove the coverslips from ethanol inside a biological safety cabinet under sterile conditions. Allow the coverslips to air-dry entirely and transfer one coverslip to each well of a six-well plate. Add approximately 100 – 200 μL of PLL solution to each coverslip. It is not necessary to completely cover the coverslip.
Incubate ten minutes at room temperature and then aspirate the PLL solution from the coverslip. Wash coverslips twice with 2 mL media and then cover with 2 mL media in preparation for cell addition. It is important to rinse away all of the unbound PLL.
Pre-coated coverslips are also available from several companies. We have used Neuvitro brand (# GG-25–1.5) coverslips with a variety of coatings (poly-D-lysine, poly-L-lysine, fibronection, collagen).
3.2. Cell culture and transfection:
We study the exocytosis of dense-core vesicles from INS-1/832–13 cells, derived from a rat insulinoma of the pancreas [29]. Other cells that can be used with slight modification to this protocol are rat pheochromocytoma PC12 cells originally derived from the adrenal gland [5]. Cells are passaged using routine tissue culture methods in T-75 plastic tissue culture flasks until approximately passage number 80, after which the culture is discarded.
-
1.
After rinsing cells in DPBS, trypsinizing, and pelleting, cells are re-suspended in media and added dropwise to coverslips in six-well plates. Ideally cells should be plated densely enough to be healthy (INS-1 cells, for example, do not thrive at low density) but not so dense that it will be difficult later to identify single cells in the microscopic field-of-view.
-
2.
After plating, allow cells to rest overnight in a tissue-culture incubator.
-
3.
Transfect the cells using your transfection method of choice one day post-plating. For many transfection methods, maximum transfection efficiency is achieved with freshly-plated cells. We routinely obtain ~10–20% transfection efficiency using Lipofectamine 2000 with INS-1 cells. Nucleofection with Lonza Kit V and protocol T-020 produces much higher transfection efficiency (>50%).
-
4.
Cells must be transfected with a vesicle cargo marker. For INS-1 cells in our system, we transfect with NPY-GFP using 1 μg of DNA per coverslip (Addgene plasmid #74629).
-
a.
Choice of cargo marker will be dependent on the cell line and experiment. In general, useful cargoes are specifically packaged into or associated with secretory vesicles. More general markers of vesicles, such as Rab proteins or VAMPs, often have high levels of diffuse background.
-
b.
Other common cargo markers used to visualize dense-core vesicles are tissue plasminogen activator (tPA), phogrin, and chromogranin. TPA can be experimentally useful because after fusion it is released from the vesicle lumen very slowly, allowing for visualization of much longer timescale exocytic events and mechanisms (control of cargo release, for example).
-
5.
Vesicles can be labeled in any convenient color, though probes must be sensitive to the pH changes that vesicles experience during fusion to robustly detect exocytic events. The lumen of most secretory vesicles is acidic (~5.5 pH) and will quench GFP, pHluorin, and other pH-sensitive fluorophores. Upon membrane fusion, the acidic lumen of the vesicle is neutralized by exposure to the extracellular buffer, leading to a sudden and dramatic brightening in the fluorescent signal from the vesicle followed by its loss as the cargo exits the vesicle. This fluorescence behavior is a hallmark of vesicle fusion (Figure 2B
-
C).
In the absence of pH-sensitivity, exocytosis will manifest as merely a loss of vesicle signal, which could also be caused by vesicle diffusion out of the TIRF field or photobleaching. Probes tagged with mCherry or ECFP which have lower pKa values exhibit this behavior. In most experiments, we co-transfect cells with a second protein-of-interest tagged with a red fluorescent protein such as mCherry or mRFP. Thus if desired, perform co-transfection by adding 1 μg of DNA of a second construct in addition to NPY-GFP to each coverslip.
-
6.
After transfection, allow cells to rest overnight. Cells can be imaged one to two days post transfection. At short timescales post transfection (~6 hours), low expression levels of the transgenic protein can be visualized if overexpression is a concern.
Figure 2:
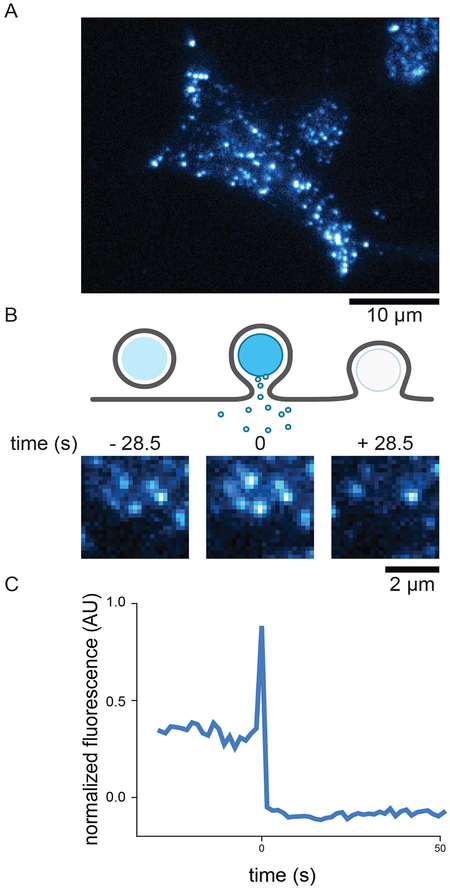
Representative images of labeled dense-core vesicles in INS-1 cells visualized with TIRF microscopy. A) A TIRF micrograph of an INS-1 cells transfected with NPY-GFP. Diffraction limited spots are vesicles resting near the bottom cell membrane within the evanescent illumination field. The contrast of the image has intentionally been adjusted much higher than normal for print clarity which causes the diffuse background within the cell body. B) A cartoon of exocytosis, shown as a side view of a vesicle resting near the membrane. Upon exocytosis the vesicular and plasma membranes fuse to release cargo. After exocytosis, vesicle fluorescence has been lost. Below the cartoon are representative images of a single dense-core vesicle undergoing exocytosis. The vesicle is present prior to fusion (left frame, vesicle at center), its intensity increases dramatically upon fusion (middle frame), and is then lost rapidly (right frame). D) Fluorescence intensity trajectory extracted from the image sequence in B. The intensity is background subtracted and normalized as described in the text
3.3. Microscope and sample preparation:
The following protocol is for an imaging experiment to visualize NPY-GFP labeled vesicles and a mCherry-tagged protein-of-interest using a two-color TIRF microscope, as described in the Materials section. The protocol is broadly applicable to imaging in any other one or two-color modes with different fluorophores, with necessary changes in filters and imaging settings.
Before each round of imaging experiments, the microscope imaging channels must be aligned and the TIRF field calibrated and evaluated for uniformity. Alignment is performed by imaging 100 nm fluorescent beads with 488 nm illumination in TIRF such that the beads are visible in both the green (direct emission) and red (bleed-through signal) channels. These alignment images are essential for processing the data and must be collected prior to each experimental session. We collect these images before each set of experiments to ensure that no mechanical drift in the imaging system is changing the channel alignment.
Place a 25 mm coverslip in a coverslip chamber (see NOTE 2). The coverslip need not be cleaned. After securing the coverslip, before adding buffer, briefly rub the coverslip with a forefinger, which helps beads stick to the glass. Add 500 μL IB to the coverslip followed by 5 μL diluted fluorescent beads (diluted 1:1000). Transfer bead-coated coverslip to the microscope.
Using the 488 nm laser, focus on the beads stuck to the coverslip. For longer timescale imaging experiments (more than a few minutes) it is useful to employ an auto-focus or drift-correction device. Autofocus devices can usually be calibrated using a bead sample, disengaged to replace the bead sample with a cell sample, and re-engaged to quickly find and keep focus.
Adjust the TIRF angle as necessary to produce shallow evanescent illumination. In epi-illumination unbound beads will be visible diffusing freely and briefly contacting the coverslip, while in TIRF illumination, only beads stuck to the coverslip will be visible as immobile spots. Adjust imaging parameters such that the emission in the green channel is in the dynamic range of the camera.
Acquire at least three bead images from different fields-of-view using 488 nm illumination. Ideally the field-of-view should contain at least 20–30 beads but not be so dense that picking out individual beads is difficult.
The bead sample is also useful to verify even TIRF illumination over the entire field-of-view, which will be conspicuous in areas of high bead density.
Prepare the perfusion apparatus (see NOTE 3). Load at least one channel of the perfusion system with IB and one channel with stimulation solution (ionomycin or stimulation buffer, as desired). Check nitrogen pressure and ensure the perfusion apparatus is not clogged by verifying solution flow from the tip of the perfusion manifold from both the IB and stimulation channels.
3.4. Imaging and Stimulation
Exocytosis can be triggered with a variety of stimuli depending on the cell type and secretion process of interest. We trigger secretion by brief (5 seconds) local superfusion of cells in the field-of-view, which applies a very small volume of stimulation solution. Local superfusion prevents bulk depolarization of the coverslip and allows for multiple rounds of stimulation and imaging in different fields-of-view over the same coverslip.
Rinse coverslip with cells three times in IB before placing in coverslip chamber. Rinsing can be easily accomplished by moving the coverslip sequentially through three small dishes filled with IB. Load the coverslip into a coverslip chamber and cover cells with 500 μL IB and place on microscope. Engage the auto-focus device and verify the cells are in focus with brightfield illumination.
Using the microscope eyepieces and the micromanipulator controlling the perfusion pipette, position the perfusion tip as near as possible to the focal plane of the cells and to the side of the field-of-view (see NOTE 4). After positioning the perfusion tip, perfuse with IB briefly to ensure no air was trapped in the tip of the perfusion pipette and blow any debris off. Move the stage in the x-y plane in all directions to ensure that the tip is not touching cells or the coverslip.
If necessary, add an aspirating pipette to the microscope stage insert, positioning the tip just over the desired buffer level in the coverslip (see NOTE 5). The aspirating pipette setup is used to maintain constant buffer level in the coverslip chamber after addition of IB for sample rinsing.
Switch to fluorescent illumination and scan the coverslip for a cell suitable for imaging. High framerate of 10/s enables easy screening for cells. Ideal cells expressing the vesicle marker show well defined, diffraction limited vesicles in the TIRF illumination field (Figure 2A).
If sample has been co-transfected for two-color imaging, verify that the cell is expressing the second construct.
Adjust imaging parameters (laser power, most commonly, or EM gain and exposure time as well) to ensure that the maximum intensity from both the green and red channels is within the dynamic range of the camera.
-
Begin acquiring images, alternating between the green and red channels, for at least 20 frames before perfusing the cells with a stimulation solution. To visualize DCV exocytosis in INS-1 cells using NPY-GFP, we typically use a camera exposure time of 500 ms in both green and red channels, followed by a 500 ms wait, for a total framerate in both channels of 0.67/s. Lower exposure times are, however, possible.
-
a.
At lower framerates of 0.67/s, longer movies and more events can be captured at the expense of time resolution. Conversely, high framerates of 10/s or more offer improved time resolution but typically yield fewer events and less total observation time due to photobleaching.
-
a.
Trigger perfusion for 5 seconds. Perfusion can be triggered manually or automatically via software control. The exact timing of perfusion does not matter greatly as exocytic events will be time aligned later to the moment of exocytosis.
After the images have been acquired, rinse the coverslip with 3–5 mL of IB to remove residual stimulation solution. Rinsing can be performed by manually adding IB to the coverslip or with larger volume perfusion systems.
After the experiment, save movies as TIFF stacks for further processing.
3.5. Image Analysis
Our analysis pipeline uses custom written MATLAB scripts to handle TIFF stacks and process image data. The same analysis can be performed in a variety of programming languages or manually with image processing software such as Metamorph or ImageJ. This analysis pipeline assumes a camera-face of 512×512 pixels with DualView setup, as described in the Materials section.
Split: Split the raw TIFF stacks into green and red channels by extracting the appropriate frames from the movie and rewriting new TIFF stacks for each channel. These new movies will be half the length of the original acquisition.
Transform: Using an image of beads acquired before the experiment, map the coordinates of at least six beads from their fluorescence in the green and red channels. Use these coordinates to spatially transform the red channel images onto the same coordinate plane as the green image. This processing step should yield a new red channel movie that can be superimposed on the green channel movie to provide perfect spatial overlap between the two channels (see NOTE 6). Bead images are used as fiducials to account for alignment differences between the green and red illumination paths and DualView image splitting.
Identify: Next exocytic events should be identified by visual inspection of the green channel movies. The rubric for identifying exocytic events will vary depending on the vesicle cargo marker that is used, generally however, events should come from single diffraction limited spots of fluorescence that were stably present at the plasma membrane prior to fusion. Most cargo markers useful to detect vesicle fusion will increase in fluorescence intensity upon membrane fusion, and this dramatic brightening of the fluorescence spot is the marker of an exocytic event (Figure 2B).
Extract: Extract the average pixel intensity of a 3×3 pixel region centered at the exocytic event coordinates (Fcenter) over a suitable time interval from the green and red channels. Again the time interval will depend on a variety of factors: the vesicle luminal marker used, the process of interest in the experiment (for example pre-fusion states, the moment of fusion itself, cargo release kinetic post-fusion). The fluorescence intensity should be background subtracted using local cellular background fluorescence (see NOTE 7).
- Normalize: Normalize trajectories before averaging to account for differences in brightness from cell-to-cell due to biological or experimental variability. Again there are a variety of methods available, but we normalize each trajectory from 0 to 1 according to:
where Fmin is the minimum fluorescence intensity over the Fcenter background-subtracted trace and Fmax is the maximum. Fsurround is the background subtraction method used here, where Fsurround is simply the mean pixel intensity in a 25 pixel box around Fcenter. An example is shown in Figure 2C. Time-align: Time-align fluorescence intensity trajectories to generate average trajectories. Trajectories should be aligned to a moment near fusion, and the fluorescence signal corresponding to this will depend on the vesicle marker. For NPY-GFP, which is our probe of choice for measuring INS-1 dense-core vesicle fusion, the marker is reasonably fluorescent in the vesicle lumen and decays sharply after fusion. We align to the timeframe of maximum intensity decrease, usually occurring in one step at our framerate, as this is the most robust feature of all intensity trajectories independent of how bright the vesicle might be prior to fusion. Probes that are dim in the unfused vesicle lumen (tPA-GFP in INS-1 cells, or pHluorin constructs) can be aligned to the timeframe corresponding to the maximum intensity increase, which should be close in time to when the vesicle fuses. Our analysis pipeline employs a script that detects the maximum intensity decrease or increase in a window around an estimated time of fusion provided by the user when selecting the exocytic events. After time-alignment, trajectories can be averaged and plotted.
4. NOTES:
A round 2 L glass beaker can accommodate four or five of the Thomson ceramic coverslip holders. If more or less coverslips are to be cleaned in one batch, or a different sized beaker used, adjust the total volume of cleaning solution to ensure coverage of the coverslips. Maintain a ratio of water: hydrogen peroxide:ammonium hydroxide at 5:1:1.
For imaging stimulated exocytosis, we recommend coverslip chambers from Warner Instruments. These low-profile coverslip chambers occlude a minimal amount of the coverslip and allow for easy access for perfusion, rinsing, and aspirating pipettes. We add custom-cut Parafilm o-rings, approximately the same size as the polycarbonate component of the chamber, which we place over the coverslip after seating it in the aluminum lower component of the chamber. The upper polycarbonate component is then pressed down over the Parafilm ring, which helps to ensure a tight seal and prevent liquid leakage. We have also used stainless steel Attofluor chambers from Thermo Scientific (# A7816), which have great leak protection but occlude more of the coverslip and have a higher profile.
The quartz perfusion manifolds from ALA Scientific are much more forgiving and reproducible than glass perfusion pipettes. Care should be taken to clean the perfusion system after each use. Channels used for perfusion must be thoroughly rinsed with water after each experiment or they will clog. Long-term storage in 20% ethanol is recommended.
Proper positioning of the perfusion pipette tip is tricky. The tip must be “found” in the field-of-view by looking for the shadow that it casts in the brightfield illumination light. Begin by using the micromanipulator to position the shaft of the perfusion pipette generally over the objective lens. Using the eyepieces and micromanipulator, move the perfusion tip back-and-forth in the y-plane (where the y-plane is parallel to the microscope eyepieces) and look for a dark shadow moving through the field-of-view. If no shadow can be found, move the perfusion pipette further away from the objective lens and try again. Once the shadow of the tip is located, center it in the field of view and begin using the micromanipulator controls to slowly step the tip down. After each movement downwards, re-center the shadow in the field-of-view. Eventually, the shadow will resolve into the perfusion pipette and the tip will become sharply focused. The ALA quartz perfusion manifold appears somewhat transparent when in the focal plane, which is a good sign the tip is close enough to the coverslip.
We use a vacuum pump-driven aspirating system from ALA Scientific. Attached to the aspirating line is a large gauge blunt needle bent downward with a magnet glued to the needle base. The bend of the needle can be changed to suit any particular stage insert or coverslip chamber. The magnet is used to clamp the pipette to our microscope stage insert to position the pipette tip over the coverslip.
A good method for validating a transformation method is to acquire a movie of stationary beads, which can then be split and transformed to verify complete overlap of bead fluorescence from each channel.
There are a variety of ways to implement a background subtraction and we recommend experimenting with several to identify what is most relevant to the particular experiment and imaging probes being used. For example, high background levels that change in response to fusion, as happens with membrane-bound vesicle lumen markers such as VAMP, can critically confound analysis.
>5. References
- 1.Jahn R, Lang T, Sudhof TC (2003) Membrane fusion. Cell 112 (4):519–533 [DOI] [PubMed] [Google Scholar]
- 2.Sudhof TC (2013) Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron 80 (3):675–690. doi: 10.1016/j.neuron.2013.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zenisek D, Steyer JA, Almers W (2000) Transport, capture and exocytosis of single synaptic vesicles at active zones. Nature 406 (6798):849–854. doi: 10.1038/35022500 [DOI] [PubMed] [Google Scholar]
- 4.Steyer JA, Almers W (1999) Tracking single secretory granules in live chromaffin cells by evanescent-field fluorescence microscopy. Biophysical journal 76 (4):2262–2271. doi: 10.1016/S0006-3495(99)77382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taraska JW, Perrais D, Ohara-Imaizumi M, Nagamatsu S, Almers W (2003) Secretory granules are recaptured largely intact after stimulated exocytosis in cultured endocrine cells. Proceedings of the National Academy of Sciences of the United States of America 100 (4):2070–2075. doi: 10.1073/pnas.0337526100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuboi T, Rutter GA (2003) Insulin secretion by ‘kiss-and-run’ exocytosis in clonal pancreatic islet beta-cells. Biochem Soc Trans 31 (Pt 4):833–836. doi: 10.1042/10.1.1.672.7754 [DOI] [PubMed] [Google Scholar]
- 7.Gandasi NR, Barg S (2014) Contact-induced clustering of syntaxin and munc18 docks secretory granules at the exocytosis site. Nature communications 5:3914. doi: 10.1038/ncomms4914 [DOI] [PubMed] [Google Scholar]
- 8.Tomes CN, Michaut M, De Blas G, Visconti P, Matti U, Mayorga LS (2002) SNARE complex assembly is required for human sperm acrosome reaction. Dev Biol 243 (2):326–338. doi: 10.1006/dbio.2002.0567 [DOI] [PubMed] [Google Scholar]
- 9.Ramalho-Santos J, Schatten G, Moreno RD (2002) Control of membrane fusion during spermiogenesis and the acrosome reaction. Biology of reproduction 67 (4):1043–1051 [DOI] [PubMed] [Google Scholar]
- 10.Heuser JE, Reese TS (1973) Evidence for recycling of synaptic vesicle membrane during transmitter release at the frog neuromuscular junction. The Journal of cell biology 57 (2):315–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jahn R, Fasshauer D (2012) Molecular machines governing exocytosis of synaptic vesicles. Nature 490 (7419):201–207. doi: 10.1038/nature11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brose N, Hofmann K, Hata Y, Sudhof TC (1995) Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. The Journal of biological chemistry 270 (42):25273–25280 [DOI] [PubMed] [Google Scholar]
- 13.Fujita Y, Shirataki H, Sakisaka T, Asakura T, Ohya T, Kotani H, Yokoyama S, Nishioka H, Matsuura Y, Mizoguchi A, Scheller RH, Takai Y (1998) Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron 20 (5):905–915 [DOI] [PubMed] [Google Scholar]
- 14.Hata Y, Slaughter CA, Sudhof TC (1993) Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature 366 (6453):347–351. doi: 10.1038/366347a0 [DOI] [PubMed] [Google Scholar]
- 15.Hatsuzawa K, Lang T, Fasshauer D, Bruns D, Jahn R (2003) The R-SNARE motif of tomosyn forms SNARE core complexes with syntaxin 1 and SNAP-25 and down-regulates exocytosis. The Journal of biological chemistry 278 (33):31159–31166. doi: 10.1074/jbc.M305500200 [DOI] [PubMed] [Google Scholar]
- 16.Loyet KM, Kowalchyk JA, Chaudhary A, Chen J, Prestwich GD, Martin TF (1998) Specific binding of phosphatidylinositol 4,5-bisphosphate to calcium-dependent activator protein for secretion (CAPS), a potential phosphoinositide effector protein for regulated exocytosis. The Journal of biological chemistry 273 (14):8337–8343 [DOI] [PubMed] [Google Scholar]
- 17.Voets T, Toonen RF, Brian EC, de Wit H, Moser T, Rettig J, Sudhof TC, Neher E, Verhage M (2001) Munc18–1 promotes large dense-core vesicle docking. Neuron 31 (4):581–591 [DOI] [PubMed] [Google Scholar]
- 18.Chapman ER (2008) How does synaptotagmin trigger neurotransmitter release? Annual review of biochemistry 77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135 [DOI] [PubMed] [Google Scholar]
- 19.Wang CT, Grishanin R, Earles CA, Chang PY, Martin TF, Chapman ER, Jackson MB (2001) Synaptotagmin modulation of fusion pore kinetics in regulated exocytosis of dense-core vesicles. Science 294 (5544):1111–1115. doi: 10.1126/science.1064002 [DOI] [PubMed] [Google Scholar]
- 20.Fukuda M (2008) Regulation of secretory vesicle traffic by Rab small GTPases. Cellular and molecular life sciences : CMLS 65 (18):2801–2813. doi: 10.1007/s00018-008-8351-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geppert M, Goda Y, Stevens CF, Sudhof TC (1997) The small GTP-binding protein Rab3A regulates a late step in synaptic vesicle fusion. Nature 387 (6635):810–814. doi: 10.1038/42954 [DOI] [PubMed] [Google Scholar]
- 22.Wada K, Mizoguchi A, Kaibuchi K, Shirataki H, Ide C, Takai Y (1994) Localization of rabphilin-3A, a putative target protein for Rab3A, at the sites of Ca(2+)-dependent exocytosis in PC12 cells. Biochemical and biophysical research communications 198 (1):158–165. doi: 10.1006/bbrc.1994.1023 [DOI] [PubMed] [Google Scholar]
- 23.Axelrod D (1981) Cell-substrate contacts illuminated by total internal reflection fluorescence. The Journal of cell biology 89 (1):141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Axelrod D, Burghardt TP, Thompson NL (1984) Total internal reflection fluorescence. Annual review of biophysics and bioengineering 13:247–268. doi: 10.1146/annurev.bb.13.060184.001335 [DOI] [PubMed] [Google Scholar]
- 25.Sochacki KA, Larson BT, Sengupta DC, Daniels MP, Shtengel G, Hess HF, Taraska JW (2012) Imaging the post-fusion release and capture of a vesicle membrane protein. Nature communications 3:1154. doi: 10.1038/ncomms2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merrifield CJ, Feldman ME, Wan L, Almers W (2002) Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol 4 (9):691–698. doi: 10.1038/ncb837 [DOI] [PubMed] [Google Scholar]
- 27.Taylor MJ, Perrais D, Merrifield CJ (2011) A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS biology 9 (3):e1000604. doi: 10.1371/journal.pbio.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kern W (1990) The Evolution of Silicon-Wafer Cleaning Technology. J Electrochem Soc 137 (6):1887–1892 [Google Scholar]
- 29.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB (2000) Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes 49 (3):424–430 [DOI] [PubMed] [Google Scholar]