Abstract
Motivation
In the study of 3D RNA structure, information about non-canonical interactions between nucleobases is increasingly important. Specialized databases support investigation of this issue based on experimental data, and several programs can annotate non-canonical base pairs in the RNA 3D structure. However, predicting the extended RNA secondary structure which describes both canonical and non-canonical interactions remains difficult.
Results
Here, we present RNAvista that allows predicting an extended RNA secondary structure from sequence or from the list enumerating canonical base pairs only. RNAvista is implemented as a publicly available webserver with user-friendly interface. It runs on all major web browsers.
Availability and implementation
1 Introduction
Full understanding of RNA-mediated biology requires knowledge of RNA structure, which is divided into three levels of organization: primary (nucleotide sequence), secondary and tertiary. Unlike proteins, RNA can act in an unstructured form (e.g. codons must be unpaired from mRNA self-structure in order to be translated by pairing with tRNAs) and some conserved sequence motifs can be detected based on RNA sequence itself. Nevertheless, the most functional motifs (involved in protein binding and cellular processes regulation) have a structural context and are related to secondary structure patterns. Their structural similarity also arises in the absence of significant sequence identity (Pietrosanto et al., 2016). Structural motifs can even encode a stronger functional signal than sequence ones. In general, knowing RNA secondary structure reveals essential constraints governing the molecule’s physical properties and function (Pietrosanto et al., 2016; Rybarczyk et al., 2016). At a fundamental level, RNA secondary structure consists of base-paired and unpaired nucleotides from which arise such structural elements as helical stems and single-stranded regions (hairpins, bulges, internal loops and n-way junctions). Base pairs are either canonical (Watson-Crick or wobble base pairs) or non-canonical (formed by edge-to-edge hydrogen bonding interactions between the bases) (Leontis and Westhof, 2001). Non-canonical ones play an important role, e.g. in base-specific interactions with proteins or ligands. Taking them into account is also essential to make the RNA 3D structure modeling more reliable and accurate (Halder and Bhattacharyya, 2013).
Experimental determination of RNA secondary structure is a laborious and expensive task (Weeks, 2010). Thus, its computational assessment via 3D structure-based annotation or sequence-based prediction is an attractive alternative. Among over 50 methods developed for the latter purpose, only seven can predict extended RNA secondary structure containing both canonical and non-canonical base pairs (Dallaire and Major, 2016; Honer zu Siederdissen et al., 2011; Parisien and Major, 2008; Pietrosanto et al., 2016; Rybarczyk et al., 2015; Sloma and Mathews, 2017; Weinreb et al., 2016). The remaining ones handle canonical base pairs only. In the case of non-canonical pairs, an annotation problem seems to be better explored. Following this observation, in (Rybarczyk et al., 2015), we have introduced our own methodology to predict extended RNA secondary structure. It leads through RNA 3D structure prediction from sequence, followed by extended secondary structure annotation. Initially, we proposed to apply RNAComposer (Antczak et al., 2016; Popenda et al., 2012) in the first step, and RNApdbee (Antczak et al., 2014; Zok et al., 2018) in the following step. They had to be executed one by one, with all parameters set by the user separately in each step. Here, we present the RNAvista webserver that facilitates the use of our approach by integrating specialized versions of RNAComposer and RNApdbee’s engines in a fully automated computational pipeline.
2 Materials and methods
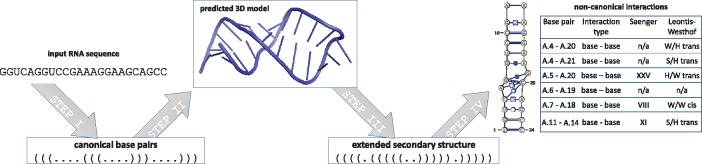
The RNAvista webserver assesses extended secondary structure of RNA from a given sequence or canonical secondary structure. It was built based on the following four-step procedure (Fig. 1): (i) prediction of canonical interactions, (ii) prediction of the tertiary structure, (iii) annotation of extended secondary structure and (iv) output data encoding and visualization.
Fig. 1.
Consecutive steps in the RNAvista workflow
The first step is optional. It runs when the user inputs RNA sequence only and is skipped if canonical base pairs have been defined in dot-bracket notation (DBN). Six algorithms, CentroidFold (Sato et al., 2009), ContextFold (Zakov et al., 2011), CONTRAfold (Do et al., 2006), IPknot (Sato et al., 2011), RNAfold (Hofacker et al., 1994) and RNAstructure (Reuter and Mathews, 2010), are incorporated in the RNAvista webserver to perform this computational step and the user can decide which one to apply. In the second step, the RNAComposer method (Popenda et al., 2012) is used to predict the RNA 3D model from canonical secondary structure. The model is built based on 3D structure elements derived from experimentally determined RNA structures that often include non-canonical and pseudoknot interactions. Thus, at this step, the structure is enriched with non-canonical base pair data. Next, the extended secondary structure is derived from the predicted RNA 3D model, and non-canonical interactions are classified according to both Saenger (Saenger, 1984) and Leontis-Westhof (Leontis and Westhof, 2001) nomenclatures. These tasks are performed by RNApdbee method (Antczak et al., 2014). Optionally, the user can choose which built-in procedures of RNApdbee, RNAView (Yang et al., 2003), MC-Annotate (Gendron et al., 2001) or 3DNA/DSSR (Lu and Olson, 2003), should be applied in the annotation process. Finally, the output structure is saved in text formats (DBN – dot-bracket notation, BPSEQ and CT – connect) and visualized. Non-canonical base pairs are graphically annotated using Leontis-Westhof pictograms.
2.1 Input and output description
In the simplest usage scenario, the user should input an RNA sequence (up to 500 nts long) in FASTA format and click the Run button. If the user has knowledge of possible canonical secondary structure, it can be introduced at the input in extended DBN. Input data can be typed in directly to the edit box or loaded from a local file. Three examples are available to facilitate familiarization with the system.
RNAvista allows to set options of intermediate processing steps. An option panel, displayed on clicking Show advanced options, enables to select: (i) one of six algorithms for canonical base pair prediction (default: CentroidFold), (ii) one of three methods that derive extended secondary structure from 3D model (default: 3DNA/DSSR with Analyze helices option; Lu and Olson, 2003), (iii) one of two algorithms for resolving 2D structure topology (default: Hybrid Algorithm; Antczak et al., 2018).
Output data includes: (i) predicted secondary structure in graphical view (with non-canonical base pairs annotated), DBN, BPSEQ and CT formats, (ii) the list of non-canonical base pairs with their classification, (iii) view of the corresponding 3D structure and (iv) log files regarding intermediate processing steps. The data are presented on the result page and can be downloaded to a local drive.
3 Results
In our previous work (Rybarczyk et al., 2016), we have conducted large-scale tests aimed to verify the accuracy of the results generated by the pipeline integrating RNAComposer and RNApdbee (now implemented in RNAvista webserver). Using data from RNA STRAND (Andronescu et al., 2008), we executed one prediction experiment based on RNA sequence only and the second starting from canonical secondary structure. The input dataset was divided into size-wise subsets. The results showed that—depending on the input sequence length—the percentage of correctly predicted non-canonical base pairs ranged between 30.64 and 57.57% (for sequence-based prediction), and 49.91–70.51% (for secondary structure-based prediction) in comparison to the reference structure. These results are also true for RNAvista webserver.
Here, we additionally decided to estimate the accuracy of predicting and annotating recurrent RNA motifs known to be defined by non-canonical interactions only. We have run RNAvista to predict the secondary structures of seven featured motifs from RNA 3D Motif Atlas (Petrov et al., 2013): K-turn, T-loop, C-loop, Sarcin, GNRA, Double sheared and Triple sheared. 12 PDB-deposited RNA 3D structures carrying these modules have been selected for the experiment. We have executed RNAvista in both modes with the default settings (3DNA/DSSR, Hybrid Algorithm) to predict whole structures of selected 12 RNAs. Next, for every recurrent motif shelled out of the predicted RNA model, we compared its extended secondary structure generated by RNAvista to the reference one, and we calculated positive predictive value (PPV), true positive rate (sensitivity, TPR), and Matthews correlation coefficient (MCC). PPV, TPR and MCC values were computed for the analyzed motifs exclusively, thus, considering non-canonical interactions only. In the sequence-based mode (Table 1), RNAvista was tested with every incorporated method dedicated to canonical secondary structure prediction. One can see that the first step of the computational pipeline profoundly influences the results. An accurate structure defined by canonical interactions significantly contributes to obtaining a precise extended secondary structure (Table 2). Additionally, the results reveal the advantage of CentroidFold (Sato et al., 2009), the default algorithm of RNAvista, over the other methods.
Table 1.
The accuracy of non-canonical interactions within recurrent RNA motifs predicted by RNAvista from the sequence (best values in bold)
| Motif | PDB ID: | CentroidFold |
ContextFold |
CONTRAFold |
IPknot |
RNAFold |
RNAstructure |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chain | PPV | TPR | MCC | PPV | TPR | MCC | PPV | TPR | MCC | PPV | TPR | MCC | PPV | TPR | MCC | PPV | TPR | MCC | |
| T-loop | 1J1U: B | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 4P5J | 1.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 1.000 | 1.000 | 1.000 | |
| Sarcin | 1JBR: D | 0.714 | 0.833 | 0.772 | 0.429 | 0.500 | 0.463 | 0.571 | 0.667 | 0.617 | 0.429 | 0.500 | 0.463 | 0.143 | 0.250 | 0.189 | 0.286 | 0.500 | 0.378 |
| 1Q93: B | 1.000 | 1.000 | 1.000 | 0.143 | 0.250 | 0.189 | 1.000 | 1.000 | 1.000 | 0.243 | 0.333 | 0.218 | 0.429 | 0.600 | 0.507 | 0.143 | 0.250 | 0.189 | |
| GNRA | 1JID: B | 1.000 | 1.000 | 1.000 | 1.000 | 0.500 | 0.707 | 1.000 | 0.500 | 0.707 | 1.000 | 0.500 | 0.707 | 1.000 | 0.500 | 0.707 | 1.000 | 0.500 | 0.707 |
| 1Q93: B | 1.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 1.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 1.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | |
| C-loop | 4JRC: A | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 5B2Q: B | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| K-turn | 5FJC: A | 0.000 | 0.000 | 0.000 | 0.200 | 0.333 | 0.258 | 0.200 | 0.250 | 0.224 | 0.200 | 0.333 | 0.258 | 0.200 | 0.333 | 0.258 | 0.200 | 0.333 | 0.258 |
| 4QVI: B | 0.600 | 1.000 | 0.775 | 1.000 | 1.000 | 1.000 | 0.400 | 0.500 | 0.447 | 0.400 | 1.000 | 0.632 | 0.200 | 0.500 | 0.316 | 0.200 | 0.500 | 0.316 | |
| Double sheared | 5AOX: F | 0.500 | 0.500 | 0.500 | 0.000 | 0.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 1MMS: C | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Triple sheared | 4GMA | 0.000 | 0.000 | 0.000 | 1.000 | 1.000 | 1.000 | 0.333 | 0.667 | 0.471 | 0.333 | 1.000 | 0.577 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Table 2.
The accuracy of non-canonical interactions within recurrent RNA motifs predicted by RNAvista from canonical secondary structure
| Motif | PDB ID: Chain | Chain: Motif size [nts] | PPV | TPR | MCC |
|---|---|---|---|---|---|
| T-loop | 1J1U: B | 74: 9 | 1.000 | 1.000 | 1.000 |
| 4P5J | 86: 9 | 1.000 | 1.000 | 1.000 | |
| Sarcin | 1JBR: D | 31: 15 | 0.714 | 0.833 | 0.772 |
| 1Q93: B | 27: 15 | 1.000 | 1.000 | 1.000 | |
| GNRA | 1JID: B | 29: 6 | 1.000 | 0.500 | 0.707 |
| 1Q93: B | 27: 6 | 1.000 | 1.000 | 1.000 | |
| C-loop | 4JRC: A | 57: 7 | 0.000 | 0.000 | 0.000 |
| 5B2Q: B | 94: 7 | 1.000 | 1.000 | 1.000 | |
| K-turn | 5FJC: A | 94: 12 | 1.000 | 1.000 | 1.000 |
| 4QVI: B | 81: 12 | 1.000 | 1.000 | 1.000 | |
| Double sheared | 5AOX: F | 87: 8 | 0.500 | 1.000 | 0.707 |
| 1MMS: C | 58: 8 | 0.667 | 1.000 | 0.816 | |
| Triple sheared | 4GMA | 210: 12 | 1.000 | 1.000 | 1.000 |
4 Conclusions
We presented RNAvista, the first webserver to predict extended RNA secondary structure (including non-canonical base pairs) from sequence or canonical secondary structure. We believe RNAvista can contribute to better understanding of RNA structure and improve its full description.
Funding
National Science Centre, Poland [2016/23/B/ST6/03931], Faculty of Computing, Poznan University of Technology, Poland [09/91/DSPB/0649], and the Institute of Bioorganic Chemistry, Polish Academy of Sciences.
Conflict of Interest: none declared.
References
- Andronescu M., et al. (2008) RNA STRAND: the RNA secondary structure and statistical analysis database. BMC Bioinformatics, 9, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antczak M., et al. (2014) RNApdbee–a webserver to derive secondary structures from pdb files of knotted and unknotted RNAs. Nucleic Acids Res., 42, W368–W372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antczak M., et al. (2016) New functionality of RNAComposer: an application to shape the axis of miR160 precursor structure. Acta Biochim. Pol., 63, 737–744. [DOI] [PubMed] [Google Scholar]
- Antczak M., et al. (2018) New algorithms to represent complex pseudoknotted RNA structures in dot-bracket notation. Bioinformatics, 34, 1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallaire P., Major F. (2016) Exploring alternative RNA structure sets using MC-flashfold and db2cm. Methods Mol. Biol., 1490, 237–251. [DOI] [PubMed] [Google Scholar]
- Do C., et al. (2006) CONTRAfold: rNA secondary structure prediction without energy-based models. Bioinformatics, 22, e90–e98. [DOI] [PubMed] [Google Scholar]
- Gendron P., et al. (2001) Quantitative analysis of nucleic acid three-dimensional structures. J. Mol. Biol., 308, 919–936. [DOI] [PubMed] [Google Scholar]
- Halder S., Bhattacharyya D. (2013) RNA structure and dynamics: a base pairing perspective. Prog. Biophys. Mol. Biol., 113, 264–283. [DOI] [PubMed] [Google Scholar]
- Hofacker I., et al. (1994) Fast folding and comparison of RNA secondary structures. Monatshefte Für Chem. Chem. Mon., 125, 167–188. [Google Scholar]
- Honer zu Siederdissen C., et al. (2011) A folding algorithm for extended RNA secondary structures. Bioinformatics, 27, i129–i136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N., Westhof E. (2001) Geometric nomenclature and classification of RNA base pairs. RNA, 7, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Olson W. (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res., 31, 5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M., Major F. (2008) The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature, 452, 51–55. [DOI] [PubMed] [Google Scholar]
- Petrov A., et al. (2013) Automated classification of RNA 3D motifs and the RNA 3D Motif Atlas. RNA, 19, 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrosanto M., et al. (2016) A novel method for the identification of conserved structural patterns in RNA: from small scale to high-throughput applications. Nucleic Acids Res., 44, 8600–8609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M., et al. (2012) Automated 3D structure composition for large RNAs. Nucleic Acids Res., 40, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter J., Mathews D. (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybarczyk A., et al. (2015) New in silico approach to assessing RNA secondary structures with non-canonical base pairs. BMC Bioinformatics, 2, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybarczyk A., et al. (2016) Computational prediction of non-enzymatic RNA degradation patterns. Acta Biochim. Pol., 63, 745–751. [DOI] [PubMed] [Google Scholar]
- Saenger W. (1984). Principles of Nucliec Acid Structure. Springer-Verlag, Berlin. [Google Scholar]
- Sato K., et al. (2009) CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res., 37, W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., et al. (2011) IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics, 27, i85–i93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloma M., Mathews D. (2017) Base pair probability estimates improve the prediction accuracy of RNA non-canonical base pairs. PLoS Comput. Biol., 13, e1005827–e1008609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K. (2010) Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol., 20, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C., et al. (2016) 3D RNA and functional interactions from evolutionary couplings. Cell, 165, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., et al. (2003) Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res., 31, 3450–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakov S., et al. (2011) Rich parameterization improves RNA structure prediction. J. Comput. Biol., 18, 1525–1542. [DOI] [PubMed] [Google Scholar]
- Zok T., et al. (2018) RNApdbee 2.0: multifunctional tool for RNA structure annotation. Nucleic Acids Res., 46, W30–W35. [DOI] [PMC free article] [PubMed] [Google Scholar]