Abstract
Motivation
Single nucleotide polymorphism (SNP) array is the most widely used platform to assess somatic copy number variations (CNVs) in cancer studies. Many SNP data-based CNV callers are available, however, the false positive rates from automated calling are commonly high, and reported breakpoints can be inaccurate. Manual review for each reported CNV by visualizing the SNP data is important, but is challenging for users lacking computational experience. To address this, we present a Shiny/R application ShinyCNV, an interactive graphical user interface to view and annotate CNVs.
Results
With this application, normalized SNP data, which includes log R ratio (LRR) and B allele frequency, can be plotted against the reported CNVs, and users can visually check the reliability of CNVs per se or adjust the incorrectly assigned breakpoints. Further, the interactive LRR spectrum panel within ShinyCNV can facilitate the process to identify commonly affected CNV regions from a group of samples, and to visually check if important focal gains/losses are missing from reported CNVs. ShinyCNV is designed to be intuitive for cancer researchers and can be easily installed for either personal use or deployed on servers to provide online service.
Availability and implementation
ShinyCNV and the tutorial are freely available from https://github.com/gzhmat/ShinyCNV.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Somatic copy number variations (CNVs) are important in cancer initiation, progression and relapse (Caren et al., 2010; Mullighan et al., 2007; Mullighan et al., 2009; Weir et al., 2007). In cancer genomic studies, high-density single nucleotide polymorphism (SNP) array has been proven a more systematic and sensitive platform in identifying CNVs compared with conventional cytogenetic karyotyping or fluorescence in situ hybridization approaches, especially for focal copy number gains and losses (Mullighan et al., 2007; Zhang et al., 2016). However, the segments detected from various CNV detection software are often false positive or with inaccurate breakpoints due to low quality of SNP data, CNV complexity and/or insufficiently trained segmentation algorithms. To ensure high-quality results, manual review may be required to compare the normalized SNP data and reported CNVs. Several CNV analysis tools have permitted this visual comparison (Li, 2008; Li et al., 2008; Sun et al., 2009), but to our knowledge, none are specifically designed to facilitate this process in a semi-automated manner that allows simple adjustment of breakpoint following data inspection.
Here we present ShinyCNV, an interactive Shiny/R application which takes CNV regions reported from external tools and normalized SNP data (either from Illumina or Affymetrix platform) as input, to help users identify reliable CNVs and adjust inaccurate segment boundaries simply and intuitively.
2 Implementation
ShinyCNV is designed to visualize and annotate CNV analysis result efficiently for users with limited experience in programing. To achieve this, this application is developed based on the widely used R environment, and the relied extra packages will be installed automatically during the first run. In general, ShinyCNV is developed by wrapping up the graphics and data-table processing functions in R packages, and the interactive features are provided from Shiny R package.
To accommodate the challenge of presenting millions of SNP probes from dozens of samples on an interactive platform, the visualization is constructed based on the basic R graphic toolkit, and the data tables are mainly processed by ‘data.table’ and ‘dplyr’ R packages due to their high efficiency. The normalized SNP intensity is further sampled and smoothed to grant users with interactive and responsive experience, while the visual sensitivity for checking CNVs is maintained as much as possible.
3 Results
To access ShinyCNV, users may download the package including R scripts and reference files from https://github.com/gzhmat/ShinyCNV to their personal computer due to the relatively large size of input SNP data.
3.1 Input data
Two types of input datasets are required to run ShinyCNV:
3.1.1 Normalized SNP data
Currently, Illumina Human Infinium Genotyping array and Affymetrix Genome-Wide SNP6.0 are the two most widely used SNP platforms in cancer research, and both types of data can be accepted by ShinyCNV after normalization.
For the Infinium array, GenomeStudio (Illumina) is freely available to perform normalization based on a set of reference samples (e.g. HapMap samples). Through normalization, two quantitative metrics are generated: one is B allele frequency (BAF), which is interpolated from three known genotype clusters AA (with 0 B allele), AB (with 50% B allele) and BB (with 100% B allele); the other is log R ratio [LRR, log2(Robserved/Rexpected)], where Rexpected is interpolated from the observed allelic ratio compared to the reference genotype clusters (Peiffer et al., 2006; Wang et al., 2007). For tumor samples observed with high hyperdiploidy (chromosome number ≥51) or hypodiploidy (chromosome number ≤39), it would be beneficial to use internal reference chromosomes (copy number is two) to perform normalization (Pounds et al., 2009). A detailed guide on how to prepare normalized data is available from PennCNV’s website http://penncnv.openbioinformatics.org/. System memory required for loading the dataset is linearly correlated with the sample size (Supplementary Fig. S1).
Although the concept of using BAF and LRR in CNV detection is proposed by analyzing SNP data from Infinium platform, the Affymetrix SNP6.0 data can also be converted into this format and a tutorial is available from PennCNV’s website too.
3.1.2 Reported CNV regions
To detect initial CNV regions from SNP data, users are open to any CNV callers as long as the segment breakpoint and estimated copy number are available (Supplementary Table S1). For demonstration, we used OncoSNP (Yau et al., 2010) due to its features in detecting somatic CNV by integrating tumor and matched normal control data, and estimating normal sample contamination and intra-tumor heterogeneity etc. More importantly, the input SNP data for OncoSNP is in the same format for ShinyCNV.
3.2 ShinyCNV panels
For demonstration, we used SNP dataset from Illumina Infinium Omni2.5-exome8-v1.3 platform (2 561 003 probes per samples) to assess somatic CNVs in six cases with leukemia.
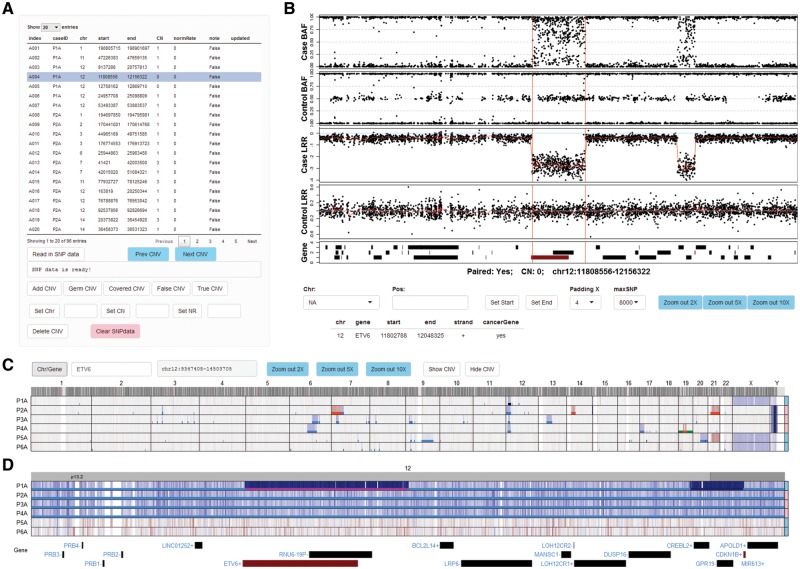
After uploading the CNV segments into the CNV table panel of ShinyCNV (Fig. 1A), normalized SNP data can be imported conveniently after pointing out the data directory. Through clicking the buttons from CNV table panel, users can perform the following editing: add/delete CNV, annotate CNV (as germline, covered in larger CNV regions, false positive or true somatic) and update CNV (change chromosome, copy number or norm sample contamination rate). Meanwhile, the boundaries of selected CNV in CNV table will be shown in BAF/LRR panel together with normalized SNP intensity for visual check (Fig. 1B). The most common use of BAF/LRR panel is to zoom into the potential breakpoint to update the CNV boundary by mouse-clicking, which is critical if the breakpoint is close to key cancer genes. With CNV table and interactive BAF/LRR figure, it is sufficient to check the reliability of the reported CNVs and correct their breakpoints.
Fig. 1.
Viewing and annotation of CNVs by ShinyCNV. A, Table of CNVs. This table is imported from reported CNVs by OncoSNP (Yau et al., 2010): index, user defined CNV identifier; CN, copy number; normRate (NR), normal sample contamination rate, estimated by OncoSNP and can be marked as 0 if unknown. The CNVs are noted as ‘False’ initially and need manual review. Any modification of this table would trigger update to a backup file. B, B allele frequency (BAF) and log R ratio (LRR) of each probe for case and matched control samples. X axis shows the chromosome position. For LRR, the two-copy regions should be around 0, which means normal, and the higher and lower LRR intensity indicates copy number gain and loss, respectively. For BAF, the SNPs should be clustered along Y axis at 0, 0.5 and 1 for two-copy regions, while other types of clustering would indicate copy number changes. When a CNV region is selected, BAF and LRR around this region will be plotted for case and control samples, with the reported CNV region marked by two vertical red lines. RefSeq genes are shown at the bottom panel, and the ones reported as consensus cancer genes according to COSMIC database are in red. Detailed gene information can be seen by clicking the gene bars. For CNVs reported with incorrect breakpoints, users can click the BAF/LRR figure (with SNPs around) and the nearest SNP’s chromosome coordination will be shown in ‘Pos:’ input box, and then the selected CNV’s start or end position can be updated by clicking ‘Set Start’ or ‘Set End’. Drop-list ‘Padding X’ is provided to adjust the length of padding regions around the CNV; ‘maxSNP’ is available to customize the maximum number of SNPs shown in each BAF/LRR panel. The figure can be zoomed in by mouse-swiping in the panel, and zoom out by clicking the ‘Zoom out *X’ buttons. C, Normalized SNP intensity (LRR) spectrum for all the analyzed cases. Chromosomes are labeled on the top and case IDs are at the left side. Cases’ genders are annotated at the right side, with pink for female and skyblue for male. Probe intensity is corresponding to DNA copy, which is shown in blue and red to indicate potential copy number loss and gain, respectively. Two buttons are provided to show (‘Show CNV’) or hide (‘Hide CNV’) the imported CNVs in different colors: copy-neutral loss of heterozygosity in green, one copy loss in light blue, two-copy loss (0 copy left) in dark blue, one copy gain in red and two or more copy gain in dark red. The selected CNV is highlighted in magenta as shown in Figure 1D. D, Zoomed-in view of a cancer gene ETV6. Users can zoom in to specific chromosome, region and gene through the input box ‘Chr/Gene’. Mouse-swiping zoom in and button supported zoom out are only available within chromosome range. An online tutorial is available at https://github.com/gzhmat/ShinyCNV
To check CNV spectrum across different samples, whole genome level SNP intensity (here is LRR) is shown at the bottom of ShinyCNV immediately after SNP data imported (Fig. 1C). Users can navigate to any chromosome, gene or genomic region through input box. More conveniently, genomic regions can be zoomed in by mouse-swiping in the figure region to show finer details (Fig. 1D). This panel is not only useful for identifying commonly affected CNV regions, but also for checking if important focal gain/loss is missing from reported CNVs.
4 Conclusion and outlook
ShinyCNV is designed with a clear purpose to visualize and annotate CNVs for users with limited experience in programing. With basic R environment and internet connection, this user-friendly Shiny application will be automatically set up and intuitively applied to visually review the reported CNVs. Since Shiny package was designed to build interactive web applications, it is straightforward to deploy ShinyCNV on servers to provide online service for users.
For now, the default parameters are optimized and encoded in the script to speed up the visualization process for most researchers, while the following versions will have more functionalities and front-end widgets to meet customized requirements based on feedbacks from users. More recently, applying whole genome sequence data to detect CNVs is becoming increasingly common, and ShinyCNV can also be adapted for CNV viewing and annotating once the normalized sequence depth (as LRR) and variant allele frequency (as BAF) are properly prepared.
Supplementary Material
Acknowledgements
We thank the staff of the Biorepository, and the Hartwell Center for Bioinformatics and Biotechnology of St. Jude Children’s Research Hospital.
Funding
This work was supported by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, American Society of Hematology Scholar Award (to Z.G.), The Leukemia & Lymphoma Society's Career Development Program Special Fellow (to Z.G.), St. Baldrick’s Foundation Robert J. Arceci Innovation Award (to C.G.M), National Cancer Institute (NCI) Outstanding Investigator Award R35 CA197695 (to C.G.M.), National Institute of General Medical Sciences (NIGMS) P50 GM115279 (to C.G.M) and the NCI grants P30 CA021765 (St. Jude Cancer Center Support Grant).
Conflict of Interest: none declared.
References
- Caren H. et al. (2010) High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc. Natl. Acad. Sci. USA, 107, 4323–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. (2008) Automating dChip: toward reproducible sharing of microarray data analysis. BMC Bioinformatics, 9, 231.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. (2008) Major copy proportion analysis of tumor samples using SNP arrays. BMC Bioinformatics, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C.G. et al. (2007) Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature, 446, 758–764. [DOI] [PubMed] [Google Scholar]
- Mullighan C.G. et al. (2009) Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med., 360, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer D.A. (2006) High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res., 16, 1136–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounds S. et al. (2009) Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics, 25, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W. et al. (2009) Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res., 37, 5365–5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. (2007) PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res., 17, 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B.A. et al. (2007) Characterizing the cancer genome in lung adenocarcinoma. Nature, 450, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau C. et al. (2010) A statistical approach for detecting genomic aberrations in heterogeneous tumor samples from single nucleotide polymorphism genotyping data. Genome Biol., 11, R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. et al. (2016) Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet., 48, 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.