Significance
Eukaryotic chromosomes are organized into specialized, nucleoprotein domains that can regulate nuclear functions. Heterochromatin is a silenced chromatin region essential for maintaining genomic stability and driving developmental processes. Cell biological studies have indicated that heterochromatin is highly condensed, though the complicated structure and dynamics of heterochromatin have rendered it difficult to characterize. Here we reconstitute heterochromatin fibers with purified components and exploit a powerful combination of sedimentation velocity, molecular modeling, and atomic force microscopy to characterize the stoichiometry and condensed conformation of heterochromatin fibers.
Keywords: chromatin, heterochromatin, sedimentation, Sir
Abstract
Heterochromatin is a silenced chromatin region essential for maintaining genomic stability and driving developmental processes. The complicated structure and dynamics of heterochromatin have rendered it difficult to characterize. In budding yeast, heterochromatin assembly requires the SIR proteins—Sir3, believed to be the primary structural component of SIR heterochromatin, and the Sir2–4 complex, responsible for the targeted recruitment of SIR proteins and the deacetylation of lysine 16 of histone H4. Previously, we found that Sir3 binds but does not compact nucleosomal arrays. Here we reconstitute chromatin fibers with the complete complement of SIR proteins and use sedimentation velocity, molecular modeling, and atomic force microscopy to characterize the stoichiometry and conformation of SIR chromatin fibers. In contrast to fibers with Sir3 alone, our results demonstrate that SIR arrays are highly compact. Strikingly, the condensed structure of SIR heterochromatin fibers requires both the integrity of H4K16 and an interaction between Sir3 and Sir4. We propose a model in which a dimer of Sir3 bridges and stabilizes two adjacent nucleosomes, while a Sir2–4 heterotetramer interacts with Sir3 associated with a nucleosomal trimer, driving fiber compaction.
Eukaryotic cells regulate the accessibility of their genome to enzymatic processes by organizing it into two functionally distinct compartments, known as euchromatin and heterochromatin. Euchromatin consists of actively transcribed gene regions, whereas heterochromatin is refractory to external processes such as transcription and recombination (1). Heterochromatin organizes and protects centromeres and telomeres, guards against the spreading of transposons, and prevents aberrant homologous recombination within repetitive regions that can lead to chromosomal abnormalities (2–4). Additionally, heterochromatin formation is an essential developmental process that drives the differentiation and maintenance of cell types (2, 3, 5). Although heterochromatin carries a distinct subset of histone modifications and protein complexes, the mechanism by which heterochromatin maintains its silent state is poorly understood.
The most thoroughly characterized heterochromatin state exists in the budding yeast Saccharomyces cerevisiae, which requires the SIR proteins—Sir2, Sir3, and Sir4—for silencing (6, 7). The formation of SIR heterochromatin is believed to be a stepwise process in which a Sir2–4 complex is initially recruited to silencing regions via interactions between the Sir4 protein and DNA-binding factors such as Rap1, Orc1, and Abf1 (8–13). Sir2, an NAD+-dependent histone deacetylase, then deacetylates the H4 tail of an adjacent nucleosome at lysine 16 (H4K16), which promotes binding of the Sir3 protein to the nucleosome, in turn recruiting additional Sir2–4 complex (10, 14–16). As this cycle of deacetylation and binding continues, the SIR proteins spread away from the nucleation site, creating a silent, heterochromatin domain (17).
The importance of H4K16 to SIR heterochromatin was initially discovered when its mutation to glutamine (H4K16Q) was found to disrupt the repression of the silent mating loci, and compensatory mutations in the Sir3 protein were identified (18). The physical interaction between Sir3 and H4K16 has been explored at length both in vivo and in vitro (14, 19–21), with several crystal structures of a Sir3–nucleosome complex displaying an electronegative patch of Sir3 that forms a binding pocket for H4K16 (22–24). These data are consistent with previous biochemical data showing that high-affinity binding of Sir3 to histone peptides (25), mononucleosomes, and short nucleosomal arrays (26) is disrupted by H4K16 acetylation or glutamine substitution (9, 27). Surprisingly, a purified Sir2–3–4 complex binds with nearly equal affinity to acetylated nucleosomes or nucleosomes harboring H4K16Q (25, 27, 28). Furthermore, although all three SIR proteins can bind to H4K16Ac chromatin templates, the resulting Sir chromatin fibers do not block transcription unless H4K16 is deacetylated (25, 28). This suggests that although an interaction between modified H4K16 and SIR proteins is possible, the presence of H4K16Ac prevents the formation of a functional, repressive heterochromatin structure.
We previously assembled recombinant nucleosomal arrays with purified Sir3, and characterized their solution dynamics by sedimentation velocity analytical ultracentrifugation (SV-AUC) and structure by atomic force microscopy (AFM). We found that the Sir3 chromatin fibers remained quite extended compared with 30-nm fibers that were condensed with divalent cations, suggesting that SIR heterochromatin may not have a compact structure. Here, we characterize heterochromatin fibers reconstituted with all three SIR proteins by both SV-AUC and AFM. Notably, we have also adapted a grid-based modeling method, called 2D spectrum analysis (2DSA) (29), coupled with a genetic algorithm (GA) and Monte Carlo analysis (MC) (30, 31), to fit sedimentation and diffusion parameters to the SV-AUC data. These modeling methods have allowed determination of both the native molecular mass and shape parameters of SIR nucleosomal arrays, yielding estimates for Sir protein stoichiometries. Most importantly, the combination of results from SV-AUC and AFM provides strong evidence that the full complement of SIR proteins forms a nearly uniform, condensed chromatin fiber that requires the integrity of H4K16.
Results
Sir2–4 Binds to Both WT and H4K16Q Arrays.
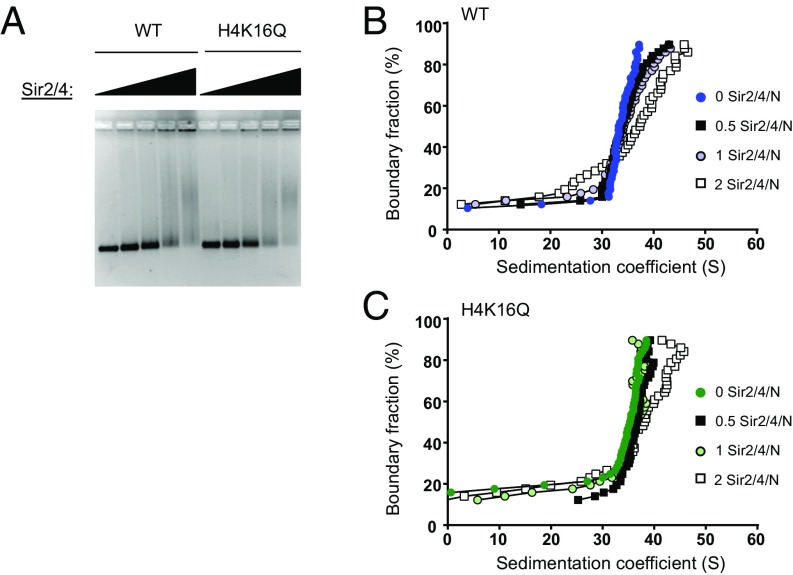
Previously, we established optimal ionic conditions (∼40 mM Na+) for the formation of Sir3 chromatin fibers that are highly sensitive to H4K16Q (21). Similar binding conditions yield Sir3 fibers that decrease nuclease accessibility of arrays and block early steps of recombinational DNA repair (20). To examine the binding of Sir2–4 to nucleosomal arrays under these same conditions, we assembled recombinant WT or H4K16Q nucleosomal arrays by salt dialysis using a DNA template containing 12 head-to-tail repeats of a 601-nucleosome positioning sequence (601-177-12). Binding of purified Sir2–4 complex was first analyzed by an electrophoretic mobility-shift assay (EMSA), in which increasing amounts of Sir2–4 complex were added to nucleosomal arrays and binding was monitored by the decrease in mobility on an agarose gel (Fig. 1A). Consistent with similar studies (27), Sir2–4 bound to WT and H4K16Q arrays at similar concentrations, with an apparent slight preference for H4K16Q (Fig. 1A). Next, Sir2–4 was titrated onto these nucleosomal arrays and interactions were monitored by sedimentation velocity analysis in an analytical ultracentrifuge. Sir2–4 bound maximally to arrays at a ratio of 1–2 Sir2–4 complexes per nucleosome (Fig. 1 A and B). Beyond a molar ratio of two Sir2–4 molecules per nucleosome, arrays became insoluble and were not used in sedimentation experiments. Notably, Sir2–4 appears to interact equally well with both WT and H4K16Q arrays, sedimenting at 33 to 38 S upon addition of two molecules of Sir2–4 per nucleosome. This contrasts with the sedimentation properties of Sir3 nucleosomal arrays that only assemble effectively with WT histones and sediment at 45 to 50 S (21) (see also Figs. 2A and 3A).
Fig. 1.
Sir2–4 complex binds both WT and H4K16Q arrays. (A) EMSA of Sir2–4 binding to WT and H4K16Q arrays. Sir2–4 titrations contain molar ratios of 0, 1, 2, 3, or 4 Sir2–4 per nucleosome. (B and C) vHW plots of Sir2–4 complex titrated onto WT and H4K16Q arrays. Numbers indicate the molar ratio of Sir2–4 complex per nucleosome.
Fig. 2.
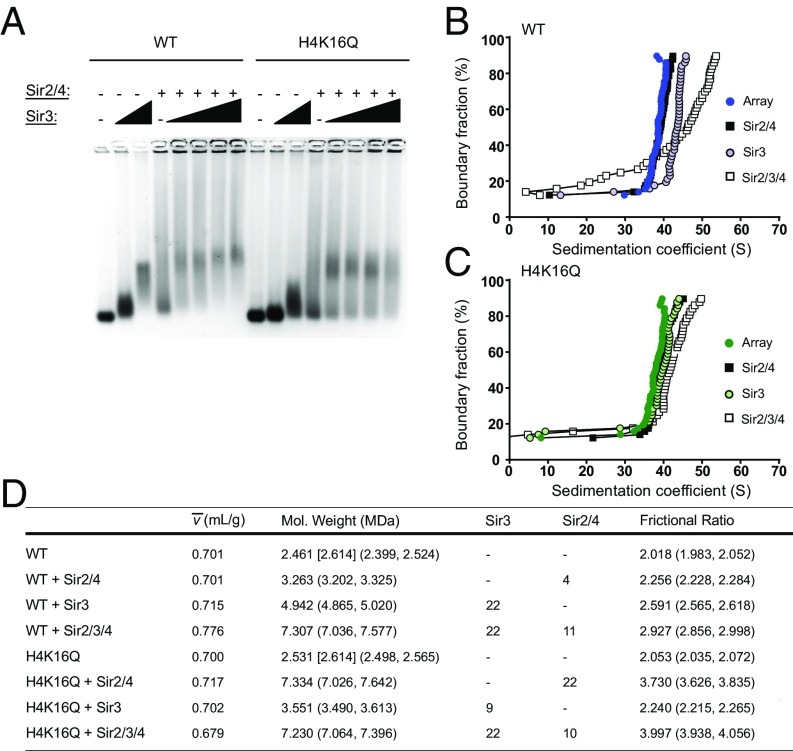
SIR interactions with WT and H4K16Q arrays are distinct. (A) EMSA of Sir3 titrated onto WT and H4K16Q arrays in the absence or presence of Sir2–4 complex. (B and C) vHW plots of Sir2–4, Sir3, and Sir3 and Sir2–4 complex added to WT and H4K16Q arrays. (D) 2DSA/GA-MC modeling results of the sedimentation data in B and C. Numbers in brackets represent expected molecular masses. Numbers in parentheses are 95% confidence intervals. Stoichiometries upon addition of Sir3 and Sir2–4 are speculative.
Fig. 3.
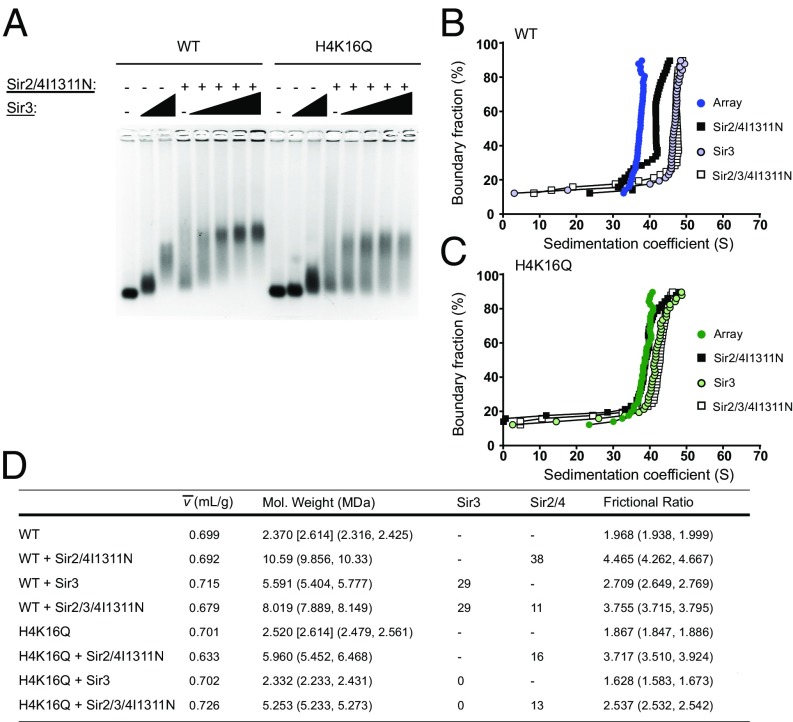
SIR-mediated compaction requires the interaction between Sir3 and Sir4. (A) EMSA of Sir3 titrated onto WT and H4K16Q arrays in the absence or presence of Sir2/4I1311N. (B and C) vHW plots of Sir3, Sir2–4I1311N, and Sir3 and Sir2–4I1311N complex added to WT and H4K16Q arrays. (D) 2DSA/GA-MC modeling results of the sedimentation data in B and C. Numbers in brackets represent expected molecular masses. Numbers in parentheses are 95% confidence intervals. Stoichiometries upon addition of Sir3 and Sir2–4 are speculative.
SIR Interactions with WT and H4K16Q Arrays Yield Fibers with Distinct Solution Dynamics.
To investigate the binding properties of arrays reconstituted with all three SIR proteins, increasing amounts of Sir3 were added to reactions that contained nucleosomal arrays and Sir2–4, and binding was first evaluated by EMSA (Fig. 2A). In isolation, Sir3 bound with much greater affinity to WT than H4K16Q arrays, as seen previously. However, titrating Sir3 in the presence of Sir2–4 led to the formation of a complex with similar mobility for both WT and H4K16Q arrays. Notably, this complex remained at constant mobility despite further additions of Sir3, suggesting that the ionic conditions that have been used to form a discrete Sir3 fiber also promote formation of a discrete, perhaps uniform, SIR heterochromatin fiber. This is in stark contrast to previous studies in different buffer conditions whereby addition of increasing concentrations of SIR proteins led to continual decreases in the mobility of EMSA species, suggestive of nonspecific DNA binding or aggregation (26, 27).
SIR heterochromatin fibers were next examined by SV-AUC (Fig. 2 B and C). As in Fig. 1, the addition of Sir2–4 onto WT and H4K16Q arrays led only to small changes in the sedimentation coefficient distribution. In contrast, addition of Sir3 to WT arrays led to a more substantial change in S (∼35 to 45 S), and addition of Sir3 did not shift H4K16Q arrays at all, consistent with binding to wild-type but not H4K16Q nucleosomes. Strikingly, addition of all three SIR proteins to WT arrays led to a large increase in the sedimentation coefficient distribution (∼50 S), whereas binding of all three SIR proteins to H4K16Q arrays led to a modest shift to ∼42 S. This result contrasts with the similar mobility of these fibers in the EMSA, and suggests that the binding of Sir proteins to WT and H4K16Q arrays may yield similar stoichiometries but distinct conformations in solution.
The sedimentation behavior of a macromolecule in an SV-AUC experiment is proportional to both its buoyant molecular mass and its frictional properties governed by its overall shape (29, 30, 32). Consequently, the observed SIR-induced changes in the S distribution of nucleosomal arrays in Fig. 2 could be due to an increased molecular mass, an altered conformation of the nucleosomal fiber, or a combination of both. To separate these parameters, we applied a set of modeling methods, 2DSA/GA-MC, which fit the sedimentation coefficient, partial concentration, molecular mass, and frictional ratio (f/f0) for solutes present in the experimental sample (21, 31, 33–35). Notably, an accurate determination of molecular mass by 2DSA/GA-MC analysis is dependent not only on the experimentally determined S and f/f0 values but also on the partial specific volume (). is the solvated volume of a macromolecule, defined in milliliters per gram, and is essential for describing the hydrodynamic behavior of molecules in solution (35–37). Consequently, we experimentally determined for each sample using a density contrast method we previously utilized in which samples are sedimented in three solvents containing either 0, 30, or 60% H218O (21). The resulting sedimentation coefficients were plotted as a function of solvent density, and the was calculated from the resulting plot (SI Appendix, Fig. S1).
The 2DSA/GA-MC analysis found that the molecular masses of both WT and H4K16Q arrays were ∼2.5 MDa, indicating the presence of ∼11 nucleosomes on the 12-mer templates (Fig. 2D and SI Appendix, Fig. S2). The addition of Sir3 to WT arrays led to an increase in molecular mass to 4.9 MDa, consistent with the binding of 22 molecules of Sir3 (113 kDa each), or a stoichiometry of two Sir3 molecules per nucleosome. In accordance with low levels of binding, the increase in molecular mass of H4K16Q arrays upon Sir3 addition indicated the presence of only ∼9 molecules of Sir3. In contrast, the addition of Sir2–4 complex to H4K16Q arrays led to an increase in molecular mass consistent with 22 complexes (of 215 MDa each) bound per array, whereas the increase in molecular mass on WT arrays reflected only four complexes bound per array. This difference may indicate Sir2–4 binding is more stable to H4K16Q nucleosomes, which mimic Sir2–4’s natural substrate, H4K16-acetyl, but which cannot be deacetylated by Sir2. This is consistent with a previous study which found Sir2–4 binds with greater affinity to acetylated nucleosomes in the absence of NAD+ (27). When the Sir2–4 complex was added with Sir3, the molecular mass of WT arrays increased to 7.3 MDa, which is consistent with the binding of 22 molecules of Sir3 and 11 molecules of Sir2–4 or a stoichiometry of two Sir3 molecules and one Sir2–4 per nucleosome. Interestingly, the addition of all three SIR proteins to H4K16Q arrays yielded a similar molecular mass (∼7.2 MDa), suggesting that the stoichiometry of SIR proteins is not sensitive to the integrity of H4K16.
Despite a nearly identical increase in molecular mass on addition of SIR proteins, WT arrays sedimented more rapidly than H4K16Q arrays. This suggests that WT arrays adopt a more compacted structure than H4K16Q arrays. This view is reflected in the frictional ratio, as the addition of Sir3 to wild-type arrays increased f/f0 from 2.0 to 2.6, and the further addition of Sir2–4 increased f/f0 to 2.9 (Fig. 2D and SI Appendix, Fig. S2). This suggests a more globular, asymmetric shape of the SIR chromatin fibers compared with nucleosomal arrays or arrays that contain only Sir3. In contrast, the addition of all three SIR proteins to H4K16Q arrays led to an increase of f/f0 from 2.1 to 4.0, indicative of a highly asymmetric, extended conformation. This interpretation is reinforced by the experimentally determined values for the partial specific volumes of these fibers, as the wild-type SIR fiber adopted a higher, protein-like value of 0.776 mL/g, whereas the of the SIR H4K16Q fiber remained a low, nucleic acid-like value of 0.679 mL/g, which could reflect the presence of exposed linker DNA in the structure (Fig. 2D and SI Appendix, Fig. S1).
The Compaction State of SIR Chromatin Requires a Sir3–Sir4 Interaction.
Sir4 contains a C-terminal coiled-coil domain that is required for both Sir4 dimerization and interaction with Sir3. Point mutations in this Sir4 domain have been identified (e.g., Sir4I1311N) that eliminate in vitro interactions between Sir4 and Sir3, and sir4I1311N disrupts recruitment of Sir3 to silencers in vivo (28, 36, 37). To address the contribution of this Sir4–Sir3 interaction to chromatin fiber assembly and dynamics, the Sir2–4 complex was purified from a yeast strain harboring the sir4I1311N allele, and the interactions of this complex with Sir3 and nucleosomal arrays were first analyzed by EMSA (Fig. 3A). On WT chromatin, the Sir2–Sir4I1311N complex appeared to bind well with Sir3 to form a chromatin fiber with a discrete mobility. However, binding of Sir3 and Sir2–Sir4I1311N was less effective on the H4K16Q arrays, and the complexes were more diffuse than complexes formed with wild-type Sir2–4, and perhaps less stable.
To address the possibility that loss of the Sir3–Sir4 interaction alters the solution dynamics of SIR chromatin fibers, WT and H4K16Q fibers bearing Sir4I1311N were analyzed by SV-AUC and 2DSA/GA-MC modeling (Fig. 3 B–D and SI Appendix, Figs. S3 and S4). Two results are apparent from the data. First, the Sir4I1311N substitution appeared to disrupt the stable binding of Sir3 to the H4K16Q arrays, with no additional increase in molecular mass detected upon Sir3 addition to H4K16Q arrays with Sir2–4 by 2DSA/GA-MC modeling (Fig. 3D and SI Appendix, Fig. S4). Second, on WT arrays the Sir3–Sir2–Sir4I1311N fibers had a molecular mass similar to that of a wild-type SIR chromatin fiber (∼8 MDa), suggesting a normal subunit stoichiometry. This result was surprising, as the Sir3–Sir2–Sir4I1311N fibers showed a similar sedimentation profile to fibers assembled with only Sir3 (Fig. 3B). These results are explained by the fact that the f/f0 ratio was increased dramatically by the Sir4I1311N substitution, from 2.9 to 3.8. Similarly, the value decreased from 0.715 for Sir3 arrays to 0.679 for Sir3–Sir2–Sir4I1311N arrays (Fig. 3D and SI Appendix, Fig. S3). Interestingly, an f/f0 of 3.8 and of 0.679 are nearly identical to the values for a SIR H4K16Q fiber, consistent with a more decondensed structure. These results indicate that the Sir4I1311N substitution does not disrupt the binding stoichiometry of SIR proteins to WT arrays but that the interaction between Sir3 and Sir4 is essential for organizing fibers into compact structures.
SIR Proteins Compact WT but Not H4K16Q Arrays.
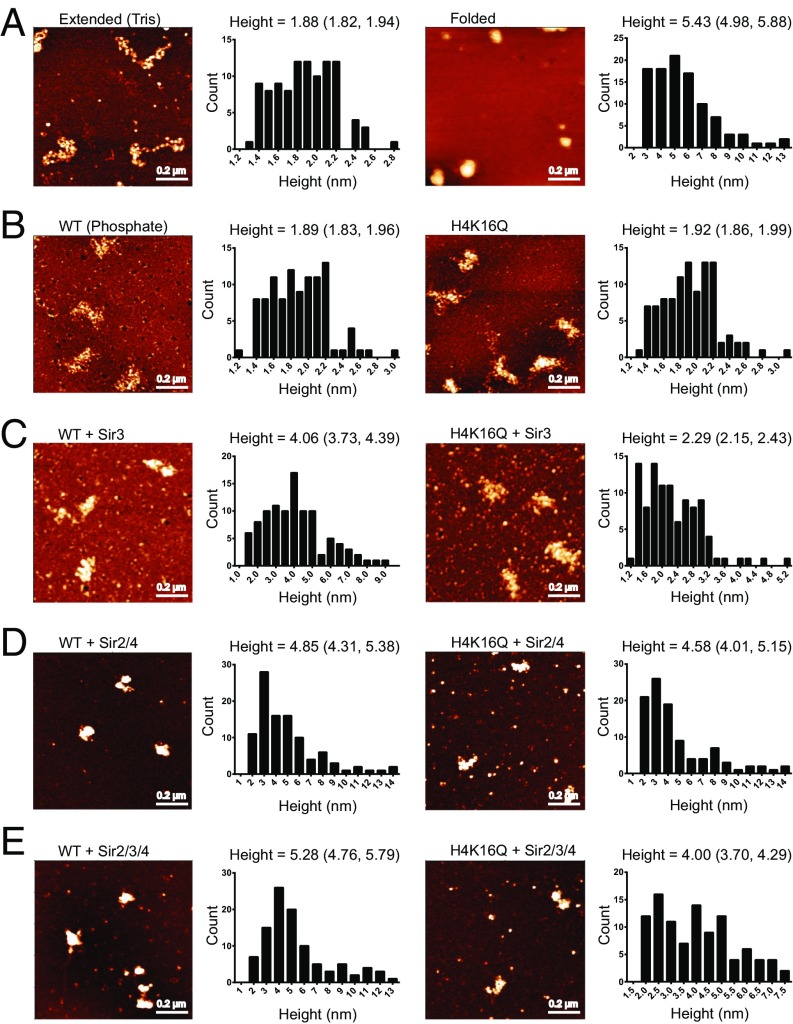
To directly observe the structural differences between SIR chromatin fibers, we visualized fibers by AFM. Given the large size of the SIR complex, we used nucleosomal array substrates reconstituted on DNA templates harboring 36 tandem copies of the 601-nucleosome positioning sequence, rather than the 12 copies used previously (Fig. 4). As an initial calibration of chromatin folding, WT arrays were initially imaged in low-salt buffer in the absence and presence of 1 mM MgCl2, which induces folding of extended fibers (−Mg2+) into condensed 30-nm fibers (+Mg2+) (38–40), with a proportional increase in height from 1.88 ± 0.31 to 5.43 ± 2.28 nm (Fig. 4A and SI Appendix, Table S1). In buffer containing moderate salt (Fig. 4B), both WT and H4K16Q arrays adopted a zigzag structure consistent with an intermediate folded state with a height of ∼1.9 nm (21, 41). The addition of Sir3 (Fig. 4 C and B) increased the height of WT arrays to 4.06 ± 1.67 nm, whereas addition of Sir3 to H4K16Q arrays increased the height only slightly to 2.29 ± 0.71 nm, corresponding to robust binding to WT but not H4K16Q arrays. Consistent with our previous study (21), Sir3 binding appeared to occlude linker DNA (as evidenced by the loss of a beads-on-a-string structure), but Sir3 fibers remained significantly decondensed compared with the 30-nm fibers shown in Fig. 4A. In contrast, addition of the full complement of SIR proteins to wild-type arrays led to formation of a more compacted structure, with the height increasing to 5.28 ± 2.61 nm (Fig. 4E and SI Appendix, Table S1). Notably, this structure is more compact than fibers formed with just the Sir2–4 complex (Fig. 4D). Although the height of the SIR chromatin fibers is similar to that of nucleosomal arrays compacted with Mg2+ (5.28 vs. 5.43 nm), a direct comparison of these two fibers is not suitable, given the large addition of protein mass to the SIR fibers. Interestingly, addition of all three SIR proteins to H4K16Q arrays increased the height to only 4.00 ± 1.50 nm, and these SIR fibers were clearly more extended and individual nucleosomes could still be identified, suggesting that while SIR proteins bound at similar levels to H4K16Q nucleosomes, they were unable to occlude linker DNA and form a compact structure.
Fig. 4.
SIR heterochromatin is compact. (A) AFM images of WT 601-177-36 arrays in low-salt Tris buffer in the absence (Left) or presence (Right) of 1 mM MgCl2. Histograms are of 100 individual measurements. Mean height and 95% confidence intervals are shown above. (B–E) WT and H4K16Q 601-177-36 arrays in sodium phosphate buffer incubated with the indicated SIR proteins.
Discussion
Heterochromatin fibers are believed to be highly compact structures that block DNA accessibility to DNA-binding transcription factors and components of the recombinational repair machinery (2, 3, 17). The in vitro assembly of budding yeast SIR chromatin fibers yields these expected functional properties—such fibers hinder access of nucleases, repress in vitro transcription by RNA polymerase II, and block early steps of in vitro recombinational repair (20, 25, 28). What has been limiting, however, is evidence that SIR chromatin fibers form condensed structures consistent with these repressive properties. Early studies from the Hansen and Woodcock groups suggested that the binding of Sir3 to nucleosomal arrays was sufficient to form highly compacted chromatin fibers, visualized by electron microscopy (EM) (42, 43). However, these studies were performed in buffers that lacked monovalent cations and, in these conditions, Sir3 bound equally well to free DNA, WT chromatin, and H4K16Q nucleosomal arrays (21, 42). Furthermore, binding of Sir3 to nucleosomal arrays in these conditions did not show saturation kinetics, suggesting that the “compact” structures may represent large aggregates. In contrast, work from the Moazed laboratory found that addition of the complete complement of Sir proteins to yeast nucleosomal arrays led to formation of long, highly extended (∼10-nm-diameter) chromatin filaments whose formation required NAD+-dependent histone deacetylation (19). In this case, however, fiber formation was performed in quite high monovalent-salt conditions (500 mM NaCl). And, finally, additional EM studies from the Moazed laboratory, performed in more moderate monovalent-salt conditions (e.g., 50 mM KCl) and with arrays assembled by an ATP-dependent assembly system, showed that SIR proteins can lead to formation of heterogeneous clumps of nucleosomes within arrays, exposing large regions of free DNA (28).
Here we have undertaken a solution analysis of the conformational dynamics of SIR chromatin fibers in buffers containing moderate levels of monovalent cations (e.g., 40 mM Na+). Using a combination of SV-AUC and molecular modeling, our results demonstrate formation of nearly homogeneous SIR chromatin fibers that are compact, not extended. The homogeneous nature of the reconstituted SIR chromatin fibers is illustrated by three experimental results: (i) Agarose gel electrophoresis shows a discrete protein complex that shows saturation binding properties; (ii) van Holde-Weischet (vHW) plots of sedimentation velocity data indicate a fairly uniform boundary of sedimenting species for SIR–nucleosomal array reconstitutions; and (iii) 2DSA/GA-MC analysis indicates primarily one major solute sedimenting in the SIR reconstitutions (SI Appendix, Fig. S2). The compact nature of SIR fibers is supported by the observation that such fibers exhibited a larger frictional ratio in solution, compared with either WT- or Sir3-containing arrays, consistent with a compact and asymmetric structure. Likewise, the WT SIR fiber adopted a higher, protein-like value for the partial specific volume, whereas the of the SIR–H4K16Q fiber remained a low, nucleic acid-like value, consistent with the ability of SIR proteins to form compact fibers only when H4K16 is intact. Furthermore, we found that this compact structure requires not only the integrity of H4K16 but also a physical interaction between Sir3 and Sir4.
Multiple Roles for Sir3–Sir4 Interactions.
Previous studies have shown that the Sir4I1311N substitution eliminates interactions between Sir3 and Sir4 in vitro and disrupts the recruitment of Sir3 to silencing domains in vivo (28, 36, 37). Furthermore, we found that Sir4I1311N disrupts the binding of Sir3 to H4K16Q nucleosomal arrays that harbor Sir2–4, consistent with a previous study showing a similar impact on Sir3 binding to arrays containing H4K16A (28). These results are all consistent with a recruitment role for the Sir3–Sir4 interaction surface (9, 37), and suggest that the presence of Sir3 on Sir2–4–bound chromatin lacking H4K16 is due to Sir3 binding to Sir4 and not to the H4 tail. Surprisingly, we found that the Sir4I1311N substitution eliminated the formation of compact SIR chromatin fibers, even though it had no apparent impact on the stoichiometry of SIR proteins on WT arrays. This suggests that interactions between Sir4 and Sir3 are key for the proper organization of SIR proteins on nucleosomal arrays, ensuring that Sir subunits are oriented in a manner that leads to a functional, condensed structure.
The Shared Role of Sir3 and Sir2–4 in Heterochromatin Formation and Function.
In vivo, overexpression of Sir3 can create extended, transcriptionally silent chromatin domains that are severely depleted of Sir2 and Sir4 (10, 44). Likewise, Sir3 addition to nucleosomal arrays is sufficient to inhibit early steps of recombinational repair, though this inhibition is strengthened by further addition of Sir2 and Sir4 (20). These data are consistent with the view that Sir3 is sufficient to create a partial, perhaps weaker form of heterochromatin fiber. How might the Sir2–4 complex reinforce the silencing properties of SIR heterochromatin and fold the chromatin fiber into an inaccessible, condensed structure? By adapting the 2DSA/GA-MC modeling algorithms, we have taken full advantage of analytical ultracentrifugation to describe both the native molecular mass and conformation of SIR chromatin fibers. Importantly, these studies provide experimental estimates of the stoichiometry of Sir proteins within condensed fibers, providing an opportunity for model development. Consistent with previous studies, our analyses support a model in which two copies of Sir3 bind to each nucleosome, with dimerization between Sir3 monomers forming an antiparallel bridge between adjacent nucleosomes (21, 45). Furthermore, the 2DSA/GA-MC analysis estimates a native molecular mass of SIR chromatin fibers consistent with one copy of Sir2–4 binding to each nucleosome.
Dimerization of Sir4 is essential for heterochromatin function in vivo (46), and Sir4 interacts with Sir3 (which also dimerizes)—why might Sir3 and Sir2–4 have different nucleosomal binding stoichiometries? We propose a model whereby a heterotetramer of Sir2–4 bridges the adjacent DNA linkers of a nucleosomal trimer by interacting with Sir3 (Fig. 5). This model suggests a stoichiometry of two Sir2–4 molecules per three nucleosomes, or 10 molecules of Sir2–4 per 11-mer array (Fig. 5). Importantly, this model is consistent with key roles for both Sir4 dimerization and Sir4–Sir3 interactions. This model is consistent with previous SIR stoichiometry estimates from Gasser and coworkers (26), who proposed a ratio of two SIR complexes per three nucleosomes. Furthermore, our model is nearly identical to a previously proposed model for a SIR chromatin fiber (47, 48). The resulting SIR chromatin fiber would be characterized by Sir3-dependent stabilization of nucleosomes, occlusion of DNA linkers, and Sir2–4–dependent compaction of nucleosomal arrays—all features predicted for repressive, heterochromatic structures.
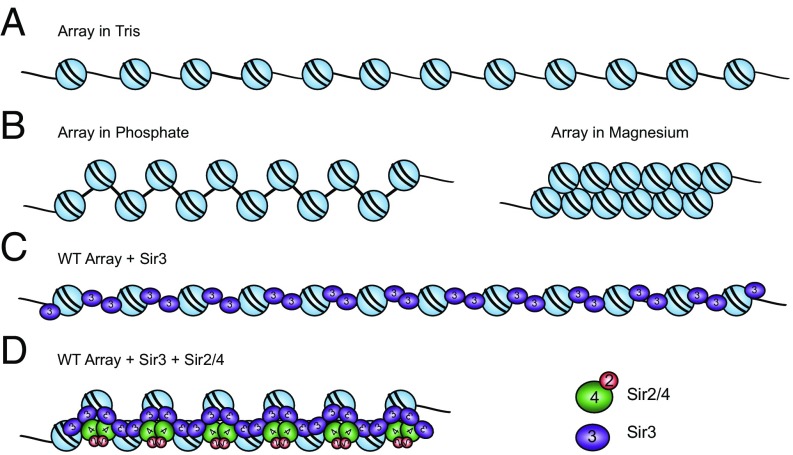
Fig. 5.
Model for a SIR chromatin fiber. (A) Diagram of a 12-mer array in low-salt Tris buffer. (B) Arrays in 20 mM phosphate buffer (pH 8.0; containing ∼40 mM Na+) are partially folded. Arrays in 1 mM MgCl2 buffer fold into 30-nm fibers. (C) Sir3 binds to arrays as a monomer, then subsequent dimerization via the Sir3 C-terminus bridges neighboring nucleosomes. (D) SIR proteins bind and condense WT arrays, though to a lesser extent than 30-nm fibers, with two molecules of Sir3 and likely one molecule of Sir2–4 per nucleosome.
Materials and Methods
FLAG-tagged Sir3 protein and tandem affinity purification (TAP)-tagged Sir2–4 complex were individually overexpressed and affinity-purified from yeast, as previously described (20). DNA templates, nucleosome reconstitution, EMSA, SV-AUC, and AFM were performed as previously described (21). Details are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the C.L.P. laboratory for helpful discussions. This work was supported by grants from the NIH (GM54096 to C.L.P; R01AI117839 to S.A.W.) and a grant from the National Cancer Institute (U54 CA143862 to S.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810647115/-/DCSupplemental.
References
- 1.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 3.Beisel C, Paro R. Silencing chromatin: Comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 4.Saksouk N, Simboeck E, Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin. 2015;8:3. doi: 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gartenberg MR, Smith JS. The nuts and bolts of transcriptionally silent chromatin in Saccharomyces cerevisiae. Genetics. 2016;203:1563–1599. doi: 10.1534/genetics.112.145243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diffley JF, Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989;246:1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- 9.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 10.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 11.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 12.Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by Rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- 13.Roy R, Meier B, McAinsh AD, Feldmann HM, Jackson SP. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J Biol Chem. 2004;279:86–94. doi: 10.1074/jbc.M306841200. [DOI] [PubMed] [Google Scholar]
- 14.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 15.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 16.Rusché LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 18.Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell. 2009;138:1109–1121. doi: 10.1016/j.cell.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swygert SG, et al. Solution-state conformation and stoichiometry of yeast Sir3 heterochromatin fibres. Nat Commun. 2014;5:4751. doi: 10.1038/ncomms5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnaudo N, et al. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nat Struct Mol Biol. 2013;20:1119–1121. doi: 10.1038/nsmb.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, et al. Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA. Proc Natl Acad Sci USA. 2013;110:8495–8500. doi: 10.1073/pnas.1300126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson A, Wu R, Peetz M, Gygi SP, Moazed D. Heterochromatic gene silencing by activator interference and a transcription elongation barrier. J Biol Chem. 2013;288:28771–28782. doi: 10.1074/jbc.M113.460071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martino F, et al. Reconstitution of yeast silent chromatin: Multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Oppikofer M, et al. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J. 2011;30:2610–2621. doi: 10.1038/emboj.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson A, et al. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell. 2009;35:769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demeler B. Methods for the design and analysis of sedimentation velocity and sedimentation equilibrium experiments with proteins. Curr Protoc Protein Sci. 2010;60:7.13.1–7.13.24. doi: 10.1002/0471140864.ps0713s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svedberg T, Pedersen KO, Bauer JH. The Ultracentrifuge. Clarendon; Oxford: 1940. [Google Scholar]
- 31.Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39:405–414. doi: 10.1007/s00249-009-0413-5. [DOI] [PubMed] [Google Scholar]
- 32.Hansen JC, Lebowitz J, Demeler B. Analytical ultracentrifugation of complex macromolecular systems. Biochemistry. 1994;33:13155–13163. doi: 10.1021/bi00249a001. [DOI] [PubMed] [Google Scholar]
- 33.Brookes EH, Demeler B. Gecco 2007: Genetic and Evolutionary Computation Conference. Vols 1–2. Association for Computing Memory; New York: 2007. Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments; pp. 361–368. [Google Scholar]
- 34.Demeler B, Brookes E. Monte Carlo analysis of sedimentation experiments. Colloid Polym Sci. 2008;286:129–137. [Google Scholar]
- 35.Gorbet G, et al. A parametrically constrained optimization method for fitting sedimentation velocity experiments. Biophys J. 2014;106:1741–1750. doi: 10.1016/j.bpj.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang JF, et al. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure. 2003;11:637–649. doi: 10.1016/s0969-2126(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 37.Rudner AD, Hall BE, Ellenberger T, Moazed D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol Cell Biol. 2005;25:4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Widom J. Physicochemical studies of the folding of the 100 Å nucleosome filament into the 300 Å filament. Cation dependence. J Mol Biol. 1986;190:411–424. doi: 10.1016/0022-2836(86)90012-4. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz PM, Hansen JC. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J Biol Chem. 1994;269:16284–16289. [PubMed] [Google Scholar]
- 41.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBryant SJ, Krause C, Woodcock CL, Hansen JC. The silent information regulator 3 protein, SIR3p, binds to chromatin fibers and assembles a hypercondensed chromatin architecture in the presence of salt. Mol Cell Biol. 2008;28:3563–3572. doi: 10.1128/MCB.01389-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adkins NL, et al. Role of nucleic acid binding in Sir3p-dependent interactions with chromatin fibers. Biochemistry. 2009;48:276–288. doi: 10.1021/bi801705g. [DOI] [PubMed] [Google Scholar]
- 44.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 45.Behrouzi R, et al. Heterochromatin assembly by interrupted Sir3 bridges across neighboring nucleosomes. eLife. 2016;5:e17556. doi: 10.7554/eLife.17556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy GA, et al. The Sir4 C-terminal coiled coil is required for telomeric and mating type silencing in Saccharomyces cerevisiae. J Mol Biol. 2003;334:769–780. doi: 10.1016/j.jmb.2003.09.066. [DOI] [PubMed] [Google Scholar]
- 47.Kueng S, Oppikofer M, Gasser SM. SIR proteins and the assembly of silent chromatin in budding yeast. Annu Rev Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- 48.Oppikofer M, Kueng S, Gasser SM. SIR-nucleosome interactions: Structure-function relationships in yeast silent chromatin. Gene. 2013;527:10–25. doi: 10.1016/j.gene.2013.05.088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.