Significance
Phylogenetic evidence suggests that a factor in the emergence of the ancestral eukaryotic cell may have been selection pressure resulting from invasion and proliferation of retroelements. Here we experimentally determine the effects of a retroelement invasion on genetically simple host organisms, and we demonstrate theoretically that the observed effects are sufficient to explain their observed rarity in bacteria. We also show that nonhomologous end-joining (NHEJ), a mechanism of DNA repair found in all extant eukaryotes, but only some bacteria, significantly enhances the efficiency of retrotransposition and the effects of retroelements on the host. We hypothesize that the interplay of NHEJ and retroelements may have played a previously unappreciated role in the evolution of advanced life.
Keywords: retroelements, LINE-1, introns, evolution, junk DNA
Abstract
Phylogenetic evidence suggests that the invasion and proliferation of retroelements, selfish mobile genetic elements that copy and paste themselves within a host genome, was one of the early evolutionary events in the emergence of eukaryotes. Here we test the effects of this event by determining the pressures retroelements exert on simple genomes. We transferred two retroelements, human LINE-1 and the bacterial group II intron Ll.LtrB, into bacteria, and find that both are functional and detrimental to growth. We find, surprisingly, that retroelement lethality and proliferation are enhanced by the ability to perform eukaryotic-like nonhomologous end-joining (NHEJ) DNA repair. We show that the only stable evolutionary consequence in simple cells is maintenance of retroelements in low numbers, suggesting how retrotransposition rates and costs in early eukaryotes could have been constrained to allow proliferation. Our results suggest that the interplay between NHEJ and retroelements may have played a fundamental and previously unappreciated role in facilitating the proliferation of retroelements, elements of which became the ancestors of the spliceosome components in eukaryotes.
The complexity of eukaryotes relative to bacteria and archaea is a consequence of the increased connectivity and plasticity of networks and interactions, rather than an increase in the amount of coding DNA (1). Such complexity is mediated by several mechanisms: one is the spliceosome, a complex molecular machine present in eukaryotes that operates on nascent mRNAs to generate mature transcripts. In some animals, for example, the spliceosome can generate multiple mRNAs through alternative splicings of a single primary transcript, allowing access to additional complexity without a concomitant increase in the amount of coding DNA. The spliceosome’s primary role is the removal of introns, intervening sequences that disrupt the coding regions of eukaryotic genes and make up, for example, ∼24–37% of the human genome (2). Conversely, bacteria and archaea lack a spliceosome, and intervening sequences are present only in limited numbers as retrotransposable elements called group II introns.
Group II introns are found in only ∼30% of sequenced bacterial species and are generally present in low copy numbers of ∼1–10 per individual in those species where they exist (3). Conversely, retroelements in eukaryotes are vastly more abundant. For example, retrotransposons in humans comprise another ∼45% of the genome in addition to introns and make up the majority of so-called “junk DNA” (2, 4). The human retroelement LINE-1 (or “L1”) alone makes up ∼17% of the genome, with ∼500,000 total integrants and ∼80–100 complete and active, or hot (L1H), copies per individual (5, 6). L1 activity contributes significantly to human genetic heterogeneity, disease, development, and evolution (7–10), and its known mechanisms of transposition show significant similarity to those of bacterial group II introns such as Ll.LtrB (11). This motivates their classification together as target-primed retrotransposons (12).
On the basis of manifold sequence, structural, and mechanistic similarities among bacterial group II introns, the spliceosome, eukaryotic spliceosomal introns, and autonomous eukaryotic retrotransposons, it has been hypothesized that an invasion of group II introns from an endosymbiotic eubacterial organelle contributed to the proliferation of introns within eukaryotic genomes before the last eukaryotic common ancestor (13, 14). If so, the resulting disruption to protein coding sequences could be alleviated by, among other contributing factors, consolidation of intron maturase splicing activity within the centralized spliceosome complex (3, 15) and the spatial decoupling of transcription and translation by a nuclear envelope (16, 17), although the order in which these developments occurred remains unclear. However, what enabled the proliferation of retroelements in eukaryotes and the evolutionary pressures and mechanisms limiting proliferation of retroelements in bacteria and archaea remain poorly understood and the subject of speculation (13, 18), particularly in light of the horizontal transfer of proliferative autonomous retroelements from humans to bacteria, as in the case of the recent transfer of L1 to the pathogen Neisseria gonorrhoeae (19).
To illuminate the changes in cellular machinery and tolerance of retroelements that would have been necessary to go from simple bacterial-like systems to eukaryotic ones, it would be important to understand precisely how retroelements may produce deleterious effects (20), what limits their activity in simple genomes, and what may have enabled their proliferation in eukaryotic genomes. To this end, we have constructed a bacterial version of L1 to quantitatively assess the function and effects of retroelement expression in the bacteria Escherichia coli and Bacillus subtilis, and we compare its effects with those of the bacterial group II intron Ll.LtrB. We find that L1 is functional in E. coli, successfully integrating into its genome. We demonstrate that retroelement expression is severely detrimental to both E. coli and B. subtilis, with wild-type B. subtilis in particular unable to tolerate any retroelement expression. We find that capacity of the host to perform nonhomologous end joining (NHEJ) repair of DNA double breaks increases retrotransposition rates by approximately three orders of magnitude, and that, surprisingly, NHEJ also strongly enhances bacterial sensitivity to the activity of retroelements. We show that these results demonstrate that retroelement activity generally leads to low copy numbers or extinction, as seen in bacteria and archaea, and that proliferation of retroelements in eukaryotes and subsequent addition of complexity to the eukaryotic genome may have been enabled by precise tuning of parameters, leading to suppression of growth defects and enhancement of integration efficiency.
Results
Description of Constructs.
To fully appreciate how human LINE-1 (L1) and bacterial Ll.LtrB molecularly affect their host genomes, we first review their remarkably similar mechanisms of action, likely evincing their shared evolutionary origin. L1 codes for the proteins ORF1p and ORF2p, and Ll.LtrB codes for LtrA. Although ORF1p is thought to bind transcribed L1 mRNA to prevent degradation, ORF2p and LtrA both contain endonuclease and reverse transcriptase domains facilitating replication of the retroelements into new chromosomal loci. After transcription and translation, each protein binds in cis to its encoding RNA, and the resulting ribonucleoprotein particle can then bind and cut a target DNA molecule, using the endonuclease domain. The mRNA 3′ end hybridizes with the cut DNA, which is used by the reverse transcriptase domain as a primer for target-primed reverse transcription (21). This generates a new cDNA copy of the retroelement at a nonspecific location in the genome, a process known as ectopic retrotransposition. L1 retrotransposition rates are poorly quantified in human somatic cells, and in E. coli, ectopic retrotransposition of Ll.LtrB occurs with a frequency of ∼1 per 109 exposed cells (11, 22, 23). In its native host, Lactococcus lactis, Ll.LtrB can also undergo a process called retrohoming, in which integration is targeted to a unique, specific site in the ltrB gene with ∼100% efficiency (11, 22, 23).
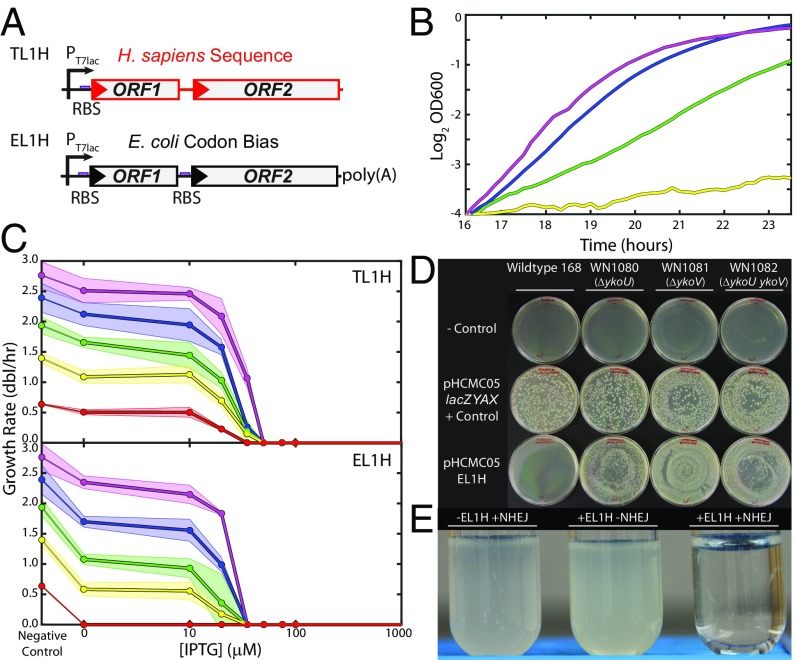
One author (T.E.K.) extracted the active or hot L1 element (L1H) #4–35 (5) from his own genome and modified it for tunable expression in E. coli. PCR was used to add a T7lac promoter at the 5′ end and a strong ribosomal binding site (RBS) to drive ORF1p expression (Fig. 1A, Top). The construct, named TL1H, was ligated into the plasmid pTKIP-neo (24, 25) and transformed into E. coli strain BL21(DE3). TL1H expression is tunable via addition of isopropyl β-d-1-thiogalactopyranoside (IPTG). We also synthesized de novo a version of L1H optimized for bacterial expression, EL1H (Fig. 2A). This construct uses E. coli codon bias, drives both ORF1 and ORF2 expression with consensus RBS sequences, and includes a ∼100-bp DNA-encoded poly-A tract at the 3′ end, a feature shown to enhance retrotransposition efficiency (26).
Fig. 1.
Bacterial L1 elements and effects on growth. (A) L1 constructs used in this study. (Top) TL1H has human sequence (indicated by red), and was modified for expression in E. coli using a bacterial T7lac promoter and a consensus Shine Dalgarno RBS driving ORF1. (Bottom) EL1H is driven by PT7lac and has consensus RBS for ORF1 and ORF2. EL1H has a 100-bp 3′ poly(A) tract and has E. coli codon bias (indicated by black). (B) L1 is detrimental to E. coli growth. Example growth curves for BL21(DE3) pTKIP-TL1H growing in M63 glucose medium including 0 (magenta), 10 μM (blue), 20 μM (green), and 35 μM (yellow) IPTG. (C) Growth response as a function of [IPTG] for BL21(DE3) pTKIP-TL1H (Top) and pTKIP-EL1H (Bottom) in various media; magenta, RDM glucose; blue, RDM glycerol; green, cAA glucose; yellow, M63 glucose; red, M63 glycerol. Growth rates were determined using the slope of the best fit regression of the initial linear portion of Log2(OD600) vs. time, as in B. Points are the average of three independent replicates, and shaded regions indicate the SD. (D) Wild-type B. subtilis cannot survive transformation with EL1H (first column), whereas NHEJ knockouts relieve sensitivity (second column: ΔykoU; third column ΔykoV; fourth column ΔykoU ΔykoV). First row: negative control (TE buffer only); second row: positive control (pHCMC05-lacZYAX); third row: pHCMC05-EL1H. We performed transformations in four independent replicates with identical results. (E) Example E. coli BL21(DE3) cultures in RDM glucose grown for 20 h. (Left) pTKIP, pUC57-NHEJ. (Middle) pTKIP-EL1H, pUC57. (Right) pTKIP-EL1H, pUC57-NHEJ. All cultures contain no IPTG and 100 ng/mL aTc.
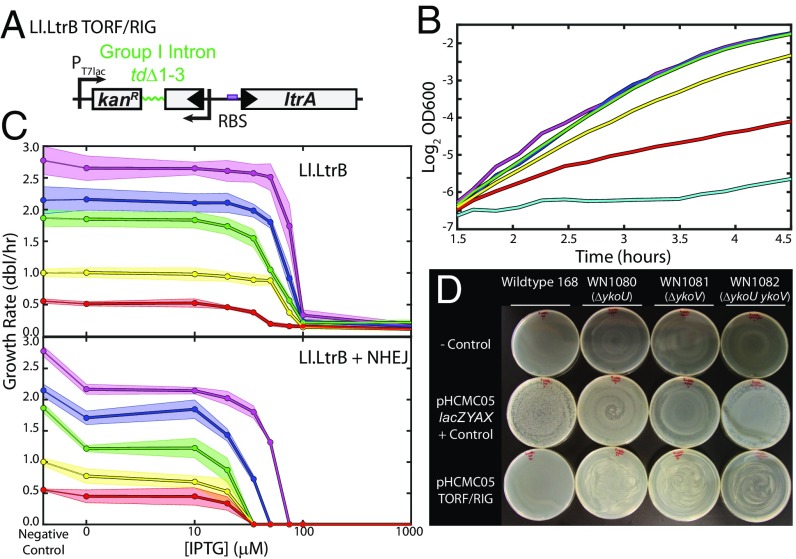
Fig. 2.
Effects of Ll.LtrB on bacterial growth. (A) The Ll.LtrB construct TORF/RIG. TORF/RIG drives the expression of the Ll.LtrB group II intron, with the ltrA coding sequence toward the 3′ end of the intron driven by a strong RBS. TORF/RIG includes a kanamycin resistance gene encoded in the opposite orientation whose coding sequence is disrupted by the group I intron tdΔ1–3 for determination of retrotransposition frequencies. (B) Expression of TORF/RIG is detrimental to E. coli growth. Example growth curves for BL21(DE3) pET-TORF/RIG growing in M63 glucose medium including 0 (magenta), 10 μM (blue), 20 μM (green), 35 μM (yellow), 50 μM (red), and 100 μM (cyan) IPTG. (C) Growth response as a function of [IPTG] for BL21(DE3) pET-TORF/RIG pZA31-tetR (Top) and pET-TORF/RIG pZA31-NHEJ (Bottom) in various media; magenta, RDM glucose; blue, RDM glycerol; green, cAA glucose; yellow, M63 glucose; red, M63 glycerol. Growth rates were determined using the slope of the best fit linear regression line of Log2(OD600) vs. time, as in B. Points are the average of three independent replicates, and shaded regions indicate the SD. (D) Wild-type B. subtilis cannot survive transformation with pHCMC05-TORF/RIG (first column), whereas NHEJ knockouts somewhat relieve sensitivity (second column: ΔykoU; third column: ΔykoV; fourth column: ΔykoU ΔykoV). First row: negative control (TE buffer only); second row: positive control (pHCMC05-lacZYAX); third row: pHCMC05-TORF/RIG. We performed transformations in four independent replicates with identical results.
Similarly, Ll.LtrB was transformed into E. coli BL21(DE3) on the plasmid pET-TORF/retromobility indicator gene (RIG), a kind gift of the Marlene Belfort laboratory (11, 27). pET-TORF/RIG uses the same pBR322 plasmid backbone as pTKIP, and Ll.LtrB is expressed from the same T7lac promoter as employed for L1 expression (Fig. 1B). Hence, expression levels of both L1 and Ll.LtrB are comparable between experiments in E. coli. In B. subtilis, we subcloned TORF/RIG and EL1H into the shuttle vector pHCMC05 under control of the IPTG-inducible hyper-spank promoter (28).
Effects of Retroelement Expression on Growth.
To assess the effects of L1 expression on bacteria, we first transformed pTKIP-TL1H/EL1H constructs into E. coli BL21(DE3), a strain that expresses T7 polymerase (29). A decrease in growth rate in response to increasing L1 expression is immediately apparent in cultures titrated with IPTG (Fig. 1 B and C). To test the generality of this effect, we next assessed the effects of L1 expression on B. subtilis. In contrast to E. coli, B. subtilis is a Gram-positive bacterium able to repair DNA double-strand breaks through a simple two-protein NHEJ system in a manner similar to eukaryotes (30). Hence, we hypothesized that B. subtilis would be more resistant to L1 and cleavage of DNA by ORF2p endonuclease than E. coli, which lacks capacity for NHEJ repair. Instead, we find the opposite: wild-type B. subtilis 168 cannot survive transformation with pHCMC05-EL1H (Fig. 1D). Conversely, we obtain high-yield transformation of EL1H into B. subtilis strains with knockouts of the individual NHEJ repair enzymes Ku (ykoV) or LigD (ykoU), as well as with both Ku and LigD knocked out (31). A Miller assay of expression level from the positive control plasmid pHCMC05-lacZYAX expressing E. coli’s metabolic lac enzymes from the hyper-spank promoter shows that expression is weak but leaky in B. subtilis (SI Appendix, Fig. S1). We conclude that wild-type B. subtilis is extremely sensitive to even low levels of L1H expression, and that this growth defect is enhanced by NHEJ repair.
We next cloned and expressed the B. subtilis NHEJ enzymes (BsKu and BsLigD) in E. coli under control of the aTc inducible PLtet01 promoter (32). We verified that BsKu and BsLigD were functional in E. coli by ensuring their ability to rescue strains where we induced the homing endonuclease I-SceI to create double-stranded chromosomal breaks at chromosomally integrated I-SceI recognition sites (SI Appendix, Fig. S2) (24, 25, 33). We then verified the enhancement of lethality of L1 by NHEJ by cotransformation of BL21(DE3) with plasmids expressing L1 and NHEJ enzymes. We find that even low leakage expression of EL1H without addition of IPTG is lethal to E. coli with concomitant induction of NHEJ enzymes with 100 ng/mL aTc (Fig. 1E).
To quantify the effect of L1 and Ll.LtrB RIG expression on E. coli growth, we measured the growth rate as a function of expression level by titration with IPTG and periodic measurement of optical density in a variety of growth media (Fig. 1 B and C for L1 and Fig. 2 B and C for Ll.LtrB). Even with no induction, leaky expression of L1 significantly reduces the growth rate relative to the parent strain carrying an empty plasmid, and complete growth arrest occurs at IPTG concentrations of 35–50 μM (Fig. 1C).
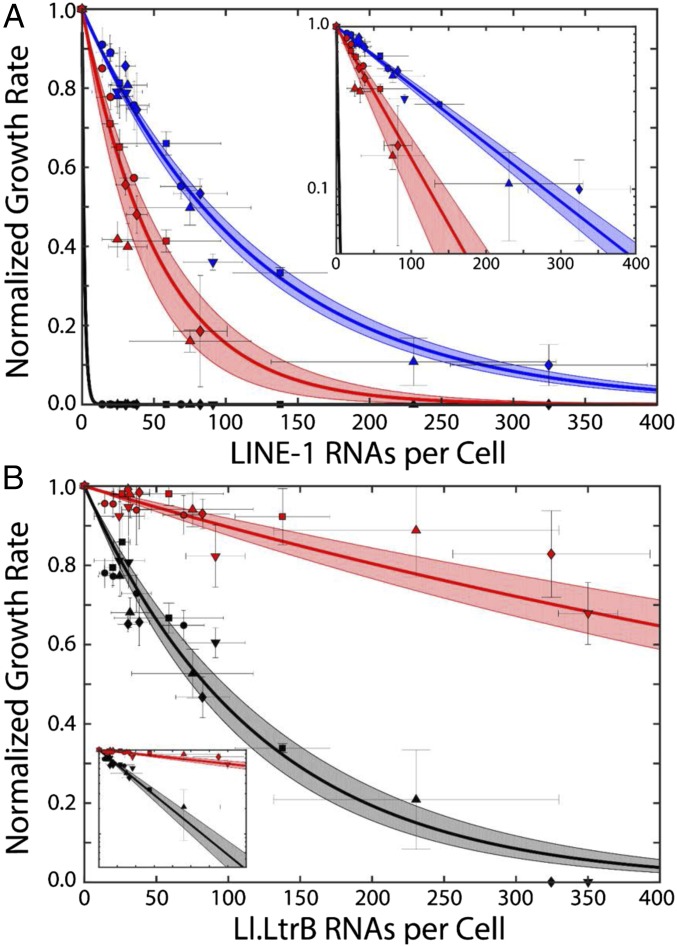
We measured the transcriptional response function of the T7lac promoter by qRT-PCR (SI Appendix, Fig. S3 A–D) of L1 mRNA extracted from bacteria grown at those IPTG concentrations at which cultures survive (SI Appendix, Fig. S3E). The resulting dose-responses as a function of L1 RNAs and Ll.LtrB RNAs per cell are shown in Fig. 3. In Fig. 3A, data from TL1H are plotted as blue points, EL1H as red points, and EL1H+NHEJ as black points. In Fig. 3B, data from Ll.LtrB are plotted as red points, and Ll.LtrB+NHEJ as black points. The normalized growth rate decreases exponentially with increasing numbers of retroelement RNAs, and growth conditions do not affect this response. Solid lines in Fig. 3 correspond to fits to the exponential function , where L is the average number of L1 or Ll.LtrB RNAs per cell and the parameter b quantifies the growth defect and sensitivity to retroelement expression. We find that, on average, each L1 transcript yields a decrease in E. coli’s growth rate of ∼0.83 ± 0.06% (TL1H) or 1.9 ± 0.6% (EL1H) in the absence of NHEJ, and ≥45 ± 1.6% with NHEJ. Each Ll.LtrB transcript reduces the growth rate by 0.11 ± 0.02% in the absence of NHEJ and 0.82 ± 0.11% with NHEJ. As might be expected because of the ability of LtrA maturase to excise Ll.LtrB from interrupted genes, the growth defect resulting from Ll.LtrB is weaker than that from L1.
Fig. 3.
Quantification of physiological effects of retroelement expression. (A) Normalized growth rate as a function of L1 expression on E. coli growth in a variety of media. ●, RDM glucose; ■, RDM glycerol; ◊, cAA glucose; ▲, M63 glucose; ▼, M63 glycerol. Blue points: TL1H; red points: EL1H; black points: EL1H and TL1H+NHEJ. Each point corresponds to the mean of three growth and four qRT-PCR measurements; error bars: SEM. Solid lines: fits to , yielding b = 0.0083 ± 0.0006 (TL1H), b = 0.019 ± 0.006 (EL1H), and b = 0.600 ± 0.031 (TL1H and EL1H+NHEJ). Fit errors are 95% CI (shaded regions). (Inset) Same, with log y axis. (B) Same as A, quantifying effects of pET-TORF/RIG pZA31-tetR (red) and pET-TORF/RIG pZA31-NHEJ (black). (Inset) Scales are identical to A. Exponential fits yield b = 0.0011 ± 0.0002 (−NHEJ), b = 0.0082 ± 0.0011 (+NHEJ).
The Ll.LtrB growth defect is also evident in plating assays to determine retrotransposition efficiency. Induction of Ll.LtrB expression with 100 µM IPTG reduces the number of viable colony forming units (cfus) per milliliter per OD by ∼10×. Simultaneous induction of Ll.LtrB with 100 µM IPTG and induction of NHEJ enzymes with 100 ng/mL anhydrotetracycline reduces viable cfus/OD/mL by ∼100×, whereas induction of expression of NHEJ enzymes alone has no detectable effect.
Finally, we attempted to transform Ll.LtrB into B. subtilis as the plasmid pHCMC05-TORF/RIG, with Ll.LtrB under control of the lacI-regulated hyper-spank promoter. As with L1, we find that wild-type B. subtilis 168 cannot survive transformation with Ll.LtrB, whereas knockouts for the NHEJ genes ykoU, ykoV, and both ykoU and ykoV are transformed with high yield (Fig. 2D).
L1 and Ll.LtrB Successfully Integrate in E. coli Chromosome.
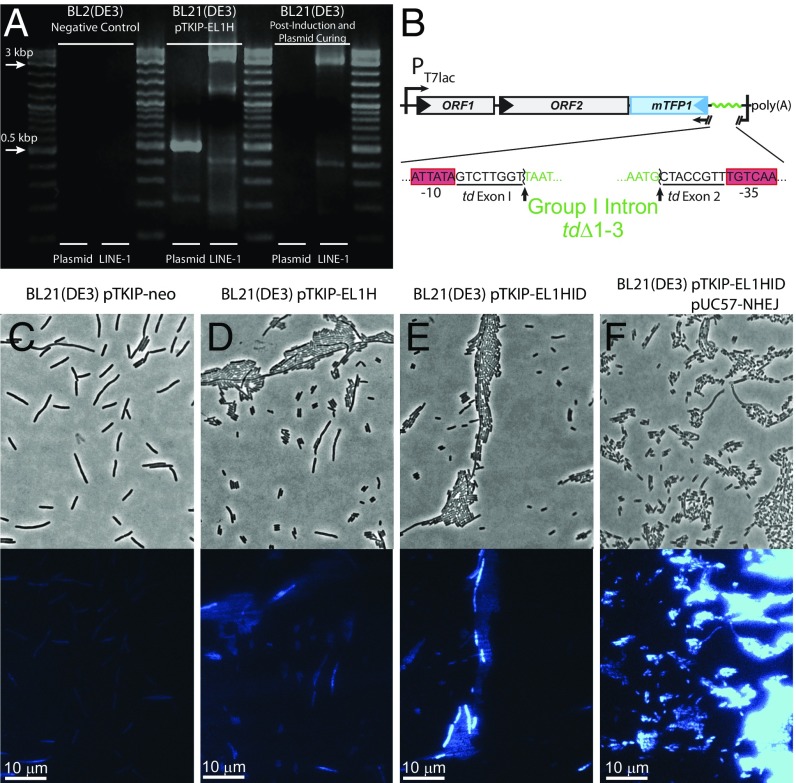
Several lines of evidence demonstrate that both Ll.LtrB and L1 successfully retrotranspose into the bacterial chromosome. E. coli carrying the pTKIP-EL1H plasmid was induced to express EL1H for several generations. Surviving cells were transformed with the plasmid pTKRED, which expresses the homing endonuclease I-SceI (24, 25, 33), to digest pTKIP-EL1H in vivo. Colony PCR and gel electrophoresis (Fig. 4A) show that cells no longer carrying pTKIP-EL1H still contain EL1H, demonstrating successful chromosomal integration. Colony PCR was also used to determine whether any surviving cells acquired the entire active EL1H sequence, using primers that amplified a 500-bp portion near the 5′ end. A positive signal was detected in 3 of 80 screened colonies, and was verified via sequencing (SI Appendix, Fig. S4).
Fig. 4.
L1 integrates into the E. coli genome. (A) Nonclonal colony PCR to detect EL1H (LINE-1 lanes) and pTKIP (plasmid lanes). (Left) BL21(DE3) negative control. (Middle) BL21(DE3) pTKIP-EL1H positive control. (Right) Strain post EL1H exposure and plasmid curing. (B) EL1HID, a construct for detecting successful retrotransposition of EL1H in individual cells by fluorescence. The integration detection cassette (ID) consists of mTFP1 with consensus σ70 promoter and RBS. −10 and −35 core promoter sequences are split by the group I intron tdΔ1–3 (sequences shown below). Upon successful retrotransposition, the cell fluoresces blue. (C–F) Phase contrast (Top) and fluorescence microscopy (Bottom) of induced (20 μM IPTG) (C) BL21(DE3) pTKIP-neo negative control, (D) BL21(DE3) pTKIP-EL1H, (E) BL21(DE3) pTKIP-EL1HID, and (F) BL21(DE3) pTKIP-EL1HID pUC57-NHEJ (0 IPTG, 5 ng/mL aTc).
As another phenotypic test, we synthesized the construct EL1HID (Fig. 4B) to report EL1H integration via fluorescence. EL1HID contains an mTFP1 gene expressed from a strong promoter whose −10 and −35 sequences are separated by the group I intron tdΔ1–3 (34). After transcription, tdΔ1–3 catalyzes its own excision from the transcribed mRNA, which reconstitutes the mTFP1 promoter, and allows expression of teal fluorescent protein on successful retrotransposition. When EL1HID was transformed into E. coli and weakly induced, ∼1% of cells exhibited a total fluorescence >10× brighter than any cells from control strains. With simultaneous weak induction of NHEJ enzymes, the fluorescent population increased to ∼80% (Fig. 4 C–F).
Using a similar RIG in Ll.LtrB (11), we found that NHEJ also enhances the rate of Ll.LtrB ectopic retrotransposition. The RIG is composed of a kanamycin resistance gene, the sequence of which is interrupted by tdΔ1–3 (Fig. 2A). After growing cultures of E. coli expressing Ll.LtrB and plating on selective media containing kanamycin, we determined the frequency of successful ectopic retrotransposition to be 3.0 ± 0.9 × 10−9, consistent with measurements by Coros et al. (11). For cells simultaneously expressing NHEJ enzymes, the efficiency increased approximately three orders of magnitude to 4.6 ± 0.4 × 10−6.
Discussion
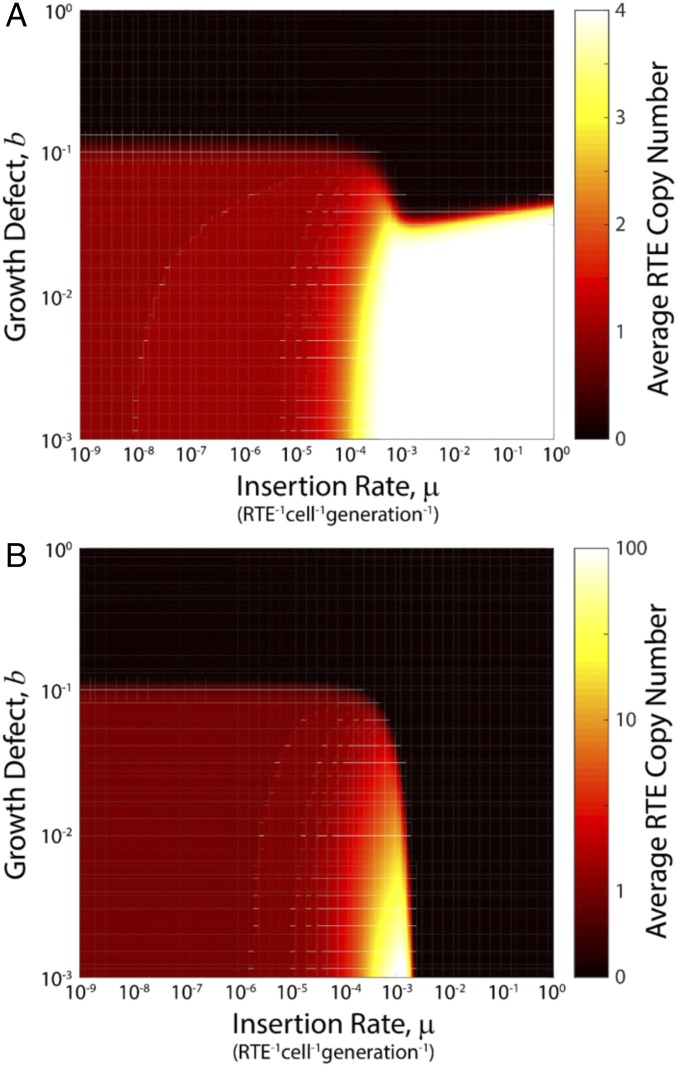
That both human L1H and bacterial Ll.LtrB expression results in exponential decrease in growth rate suggests a simple universal underlying mechanism: each retroelement mRNA transcript has a probability of integrating and disrupting essential genes affecting growth. In the simplest model of this type, the probability that a cell will survive is described by a binomial distribution with zero disruptive integration events, leading to an exponential decrease in growth rate with transcript number; including variable integration rates and physiological responses does not significantly affect the resulting behavior (SI Appendix, Supplementary Analysis). As a consequence, in bacteria, the growth defect is a monotonically increasing function of the integration rate. To further understand how retrotransposons will proliferate within a host genome, we constructed a simple model of retroelement activity, motivated by the existing body of work on retroelement activity (20, 35–41), and analyzed its dynamics (SI Appendix, Supplementary Analysis). Populations of asexually multiplying cells were simulated on the basis of measured integration rates and growth defects, and allowed to evolve over 10,000 generations. The resulting phase diagrams are shown in Fig. 5 for retrohoming (reflective boundary conditions) and retrotransposition (absorbing boundary conditions), respectively. We find that retrohoming generally leads to low but stable numbers of retroelements, whereas the parameters with which retrotransposition occurs must be finely tuned to achieve long-lived states with proliferation of retrotransposons in the host.
Fig. 5.
Phase diagram of retrotransposon dynamics. We simulated the model of retrotransposon dynamics, SI Appendix, Eq. 2.7 (SI Appendix, Supplementary Analysis), using a total system size [defined as the number of available empty sites in the environment plus (effective) number of individuals in the population] of Ω = 109, with an initial population of ψ1 = 0.1 and all other states empty. This initial state was allowed to evolve for 10,000 generations with Δ = 10−8 retrotransposon−1⋅cell−1⋅generation−1 and β = 10−2 cell−1⋅generation−1, at the conclusion of which we calculated the average number of retrotransposons per cell over the extant population. Results are shown for (A) reflecting boundary conditions with xmax = 4 and (B) absorbing boundary conditions with .
The phase portrait in Fig. 5B shows that there exists a small set of parameter values (low growth defect, b, of less than 0.01 and high integration rate, µ, of ∼10−3 retrotransposon−1⋅cell−1⋅generation−1), where retrotransposons can proliferate to high numbers. Coupling of the integration rate and growth defect implies that increases in the integration rate inexorably push bacteria toward the upper right of the phase diagram, and thus toward extinction. Hence, the bacterial phase space is highly constrained, and they are unlikely to be found within this small proliferative regime.
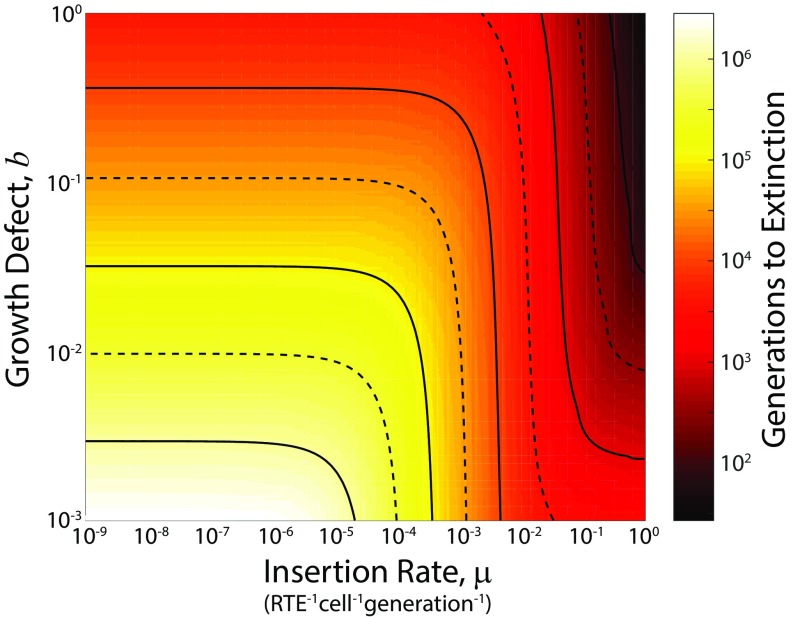
To demonstrate this, we performed simulations using absorbing boundary conditions across parameter values, and for each, we recorded the number of generations required for the retrotransposon to go extinct. The result is shown in Fig. 6. From this analysis, we see that the time required for a retrotransposon to go extinct can vary more than ∼7 orders of magnitude, depending on its dynamics and effects. For those parameter regimes corresponding to the aggressive autonomous retrotransposon L1 (b ≥ 10−2, µ ≥ 10−2 retrotransposon−1⋅cell−1⋅generation−1), extinction of retroelements is rapid, occurring in ∼100–10,000 generations. Conversely, parameter regimes corresponding to the group II intron Ll.LtrB (10−3 ≤ b ≤ 10−2, 10−9 ≤ µ ≤ 10−6 retrotransposon−1⋅cell−1⋅generation−1) can persist in low copy numbers (∼1 per cell) for millions to tens of millions of generations. We also see that the small parameter regime in which retrotransposons can proliferate to high copy numbers (b ≤ 10−2, µ ∼10−3 - 10−4 retrotransposon−1⋅cell−1⋅generation−1) persists for hundreds of thousands to millions of generations, and could be maintained longer with the inclusion of horizontal gene transfer.
Fig. 6.
Time to extinction of retrotransposons in a bacterial population. Simulations of the model SI Appendix, Eq. 2.7 (SI Appendix, Supplementary Analysis), with absorbing boundary condition at , system size of Ω = 109, Δ = 10−8 retrotransposon−1⋅cell−1⋅generation−1, β = 10−2 cell−1⋅generation−1 and initial population of ψ1 = 0.1 with all other states empty. Color indicates the number of generations required for the average number of retrotransposons per cell to drop below 1/Ω. Solid contour lines indicate major decade divisions; dashed contour lines indicate half-decade divisions.
Hence, this simple model suggests that for retroelements to proliferate to high numbers within asexual populations, the coupling of integration rate and growth defect must be weakened. In addition, increases in retrotransposition efficiency by NHEJ, present in all extant eukaryotes, must also be compensated for by suppression of the growth defect to enable proliferation. Indeed, it is hypothesized that many eukaryotic features arose specifically to mitigate the effects of retroelements (3, 13, 16, 17, 42, 43). For example, the nuclear membrane allows the spliceosome to complete intron excision before nuclear export and translation (16, 17). Furthermore, important spliceosomal components are derived from group II introns, and consolidation of splicing activity into the spliceosomal complex may facilitate efficient intron removal (3, 13). With the spliceosome, further complexity added to the eukaryotic genome by retroelements could then be exploited for benefit through, for example, alternative splicing by exon-skipping in some eukaryotes. In summary, proliferation of retroelements plays a dual role. On the one hand, group II introns create genome instability and negative physiological effects. On the other hand, by duplicating themselves, copies of group II introns are free to diversify and become the ancestors of both spliceosome and spliceosomal introns (13, 14).
We hypothesize that NHEJ enhances retrotransposition by directly joining the newly reverse-transcribed retroelement with the remaining free end of the endonuclease-induced break. Without NHEJ, this break can only be repaired through homologous recombination, generally leading to removal of the integrant and apparent low retrotransposition efficiencies, as observed in NHEJ-deficient E. coli. However, it is surprising that minimal, two-protein bacterial NHEJ systems interact with and enhance human L1 retrotransposition efficiency. Intriguingly, NHEJ proteins also heavily associate with telomeres and are required for proper telomere length regulation and end protection (44, 45). Furthermore, the reverse transcriptase activity of telomerase likely shares a common ancestor with group II introns, and in some organisms (e.g., Drosophila), telomere maintenance is performed by retroelements rather than telomerase (13). Combined with our results, we conjecture that NHEJ systems, together with retroelement proliferation, were implicated in the unexplained evolutionary transition from generally circular bacterial chromosomes to linear eukaryotic chromosomes (13, 42, 45).
Methods
Strains and Media.
Manipulation of constructs was performed with E. coli strain NEBTurbo (New England Biosciences). Experiments assaying effects of retroelement expression in E. coli were performed in the strain BL21(DE3). B. subtilis experiments were performed with strain 168, as well as ΔykoU (WN1080/BFS1845), ΔykoV (WN1081/BFS1846), and ΔykoU ΔykoV (WN1082/BFS1847) knockout strains (31).
Plasmid Construction.
See SI Appendix for descriptions of plasmid constructs.
B. subtilis Transformation.
B. subtilis transformation was performed as described in ref. 46, with modifications (SI Appendix, Supplementary Methods).
LacZ Measurements.
B. subtilis 168 pHCMC05-lacZYAX was inoculated into RDM glucose and, when OD600 of the culture reached ∼0.3–0.5, 0.5 mL culture was added to 0.5 mL Z-buffer + 0.1% SDS with 100 μL toluene. This mixture was vortexed and incubated in a 37 °C water bath for 30 min. The LacZ assay was then performed as previously described (SI Appendix, Fig. S1) (47, 48).
Growth Rate Determination.
Detailed methods of growth rate determination can be found in the SI Appendix.
Microscopy.
To perform fluorescence microscopy, 50 μL samples of culture were spread onto 1% agarose pads prepared on glass slides, covered with a #1.5 glass coverslip and imaged; see SI Appendix for details.
Quantitative RT-PCR.
Methods for qRT-PCR can be found in SI Appendix.
Ll.LtrB Retrotransposition Frequency Assays.
Retrotransposition efficiency of Ll.LtrB with and without NHEJ expression was determined by the protocol of ref. 11, with modifications; see SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Prof. Douglas Mitchell (University of Illinois Urbana–Champaign) for the gift of B. subtilis 168 and plasmids, Wayne L. Nicholson (University of Florida) for the gift of B. subtilis NHEJ knockout strains, and Marlene Belfort (University of Albany, State University of New York) for the gift of Ll.LtrB constructs and sequence information. This work was supported by the NSF Center for the Physics of Living Cells (Grant PHY 1430124), the Alfred P. Sloan Foundation (Award FG-2015-65532), and the Institute for Universal Biology, through partial support by the NASA Astrobiology Institute (NAI) under Cooperative Agreement No. NNA13AA91A issued through the Science Mission Directorate. G.L. is supported by the National Science Foundation Graduate Research Fellowship Program under Grant DGE-1144245. All work was reviewed and approved by the University of Illinois Urbana–Champaign Institutional Review Board and Institutional Biosafety Committee.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807709115/-/DCSupplemental.
References
- 1.Lynch M. The Origins of Genome Architecture. Sinauer Associates; Sunderland, MA: 2007. [Google Scholar]
- 2.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921, and erratum (2001) 412:565. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Lambowitz AM, Belfort M. Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol Spectr. 2015;3:MDNA3-0050-2014. doi: 10.1128/microbiolspec.MDNA3-0050-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson SR, et al. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr. 2015;3:MDNA3-0061-2014. doi: 10.1128/microbiolspec.MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodier JL. Retrotransposition in tumors and brains. Mob DNA. 2014;5:11. doi: 10.1186/1759-8753-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coros CJ, et al. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol Microbiol. 2005;56:509–524. doi: 10.1111/j.1365-2958.2005.04554.x. [DOI] [PubMed] [Google Scholar]
- 12.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novikova O, Belfort M. Mobile group II introns as ancestral eukaryotic elements. Trends Genet. 2017;33:773–783. doi: 10.1016/j.tig.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irimia M, Roy SW. Origin of spliceosomal introns and alternative splicing. Cold Spring Harb Perspect Biol. 2014;6:a016071. doi: 10.1101/cshperspect.a016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambowitz AM, Zimmerly S. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 17.Doolittle WF. The trouble with (group II) introns. Proc Natl Acad Sci USA. 2014;111:6536–6537. doi: 10.1073/pnas.1405174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeke JD. The unusual phylogenetic distribution of retrotransposons: A hypothesis. Genome Res. 2003;13:1975–1983. doi: 10.1101/gr.1392003. [DOI] [PubMed] [Google Scholar]
- 19.Anderson MT, Seifert HS. Opportunity and means: Horizontal gene transfer from the human host to a bacterial pathogen. MBio. 2011;2:e00005-11. doi: 10.1128/mBio.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iranzo J, Cuesta JA, Manrubia S, Katsnelson MI, Koonin EV. Disentangling the effects of selection and loss bias on gene dynamics. Proc Natl Acad Sci USA. 2017;114:E5616–E5624. doi: 10.1073/pnas.1704925114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 22.Cousineau B, Lawrence S, Smith D, Belfort M. Retrotransposition of a bacterial group II intron. Nature. 2000;404:1018–1021. doi: 10.1038/35010029. [DOI] [PubMed] [Google Scholar]
- 23.Ichiyanagi K, et al. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuhlman TE, Cox EC. Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res. 2010;38:e92. doi: 10.1093/nar/gkp1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tas H, Nguyen CT, Patel R, Kim NH, Kuhlman TE. An integrated system for precise genome modification in Escherichia coli. PLoS One. 2015;10:e0136963. doi: 10.1371/journal.pone.0136963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doucet AJ, Wilusz JE, Miyoshi T, Liu Y, Moran JV. A 3′ poly(A) tract is required for LINE-1 retrotransposition. Mol Cell. 2015;60:728–741. doi: 10.1016/j.molcel.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beauregard A, Chalamcharla VR, Piazza CL, Belfort M, Coros CJ. Bipolar localization of the group II intron Ll.LtrB is maintained in Escherichia coli deficient in nucleoid condensation, chromosome partitioning and DNA replication. Mol Microbiol. 2006;62:709–722. doi: 10.1111/j.1365-2958.2006.05419.x. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen HD, et al. Construction of plasmid-based expression vectors for Bacillus subtilis exhibiting full structural stability. Plasmid. 2005;54:241–248. doi: 10.1016/j.plasmid.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 30.Bowater R, Doherty AJ. Making ends meet: Repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2006;2:e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moeller R, et al. Role of DNA repair by nonhomologous-end joining in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV, and ionizing radiation. J Bacteriol. 2007;189:3306–3311. doi: 10.1128/JB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhlman TE, Cox EC. A place for everything: Chromosomal integration of large constructs. Bioeng Bugs. 2010;1:296–299. doi: 10.4161/bbug.1.4.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belfort M, Chandry PS, Pedersen-Lane J. Genetic delineation of functional components of the group I intron in the phage T4 td gene. Cold Spring Harb Symp Quant Biol. 1987;52:181–192. doi: 10.1101/sqb.1987.052.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Charlesworth B, Charlesworth D. The population dynamics of transposable elements. Genet Res. 1983;42:1–27. [Google Scholar]
- 36.Charlesworth B, Langley CH. The evolution of self-regulated transposition of transposable elements. Genetics. 1986;112:359–383. doi: 10.1093/genetics/112.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolgin ES, Charlesworth B. The fate of transposable elements in asexual populations. Genetics. 2006;174:817–827. doi: 10.1534/genetics.106.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langley CH, Brookfield JFY, Kaplan N. Transposable elements in mendelian populations. I. A theory. Genetics. 1983;104:457–471. doi: 10.1093/genetics/104.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brookfield JFY. The ecology of the genome: Mobile DNA elements and their hosts. Nat Rev Genet. 2005;6:128–136. doi: 10.1038/nrg1524. [DOI] [PubMed] [Google Scholar]
- 40.Hellen EHB, Brookfield JFY. Transposable element invasions. Mob Genet Elements. 2013;3:e23920. doi: 10.4161/mge.23920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch M, Bürger R, Butcher D, Gabriel W. The mutational meltdown in asexual populations. J Hered. 1993;84:339–344. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 42.Koonin EV. Viruses andd mobile elements as drivers of evolutionary transitions. Philos Trans R Soc B. 2016;371:20150442. doi: 10.1098/rstb.2015.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brodt A, Lurie-Weinberger MN, Gophna U. CRISPR loci reveal networks of gene exchange in archaea. Biol Direct. 2011;6:65. doi: 10.1186/1745-6150-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- 45.de Lange T. A loopy view of telomere evolution. Front Genet. 2015;6:321. doi: 10.3389/fgene.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sysoeva TA, et al. Structural characterization of the late competence protein ComFB from Bacillus subtilis. Biosci Rep. 2015;35:e00183. doi: 10.1042/BSR20140174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 48.Kuhlman T, Zhang Z, Saier MH, Jr, Hwa T. Combinatorial transcriptional control of the lactose operon of Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6043–6048. doi: 10.1073/pnas.0606717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.