Significance
This case study reveals that parasitic and symbiotic lifestyles affect the structure of essential molecular machineries of a living cell. We provide evidence that intracellular parasitism and endosymbiosis cause degeneration of the editing domains in aminoacyl-tRNA synthetases, a defect that is known to cause inaccurate translation of the genetic code. This finding suggests that most intracellular pathogens, including causative agents of human disease, have an unanticipated proteome diversity caused by inaccurate translation of the genetic code. Our finding may change current approaches to the study of proteomes of intracellular parasites, parasite–host interactions, and parasites’ sensitivity to drugs, which cause errors in transcription, translation, and protein folding.
Keywords: Muller’s ratchet, genome decay, mistranslation, aminoacyl-tRNA synthetases, quality control
Abstract
Intracellular organisms, such as obligate parasites and endosymbionts, typically possess small genomes due to continuous genome decay caused by an environment with alleviated natural selection. Previously, a few species with highly reduced genomes, including the intracellular pathogens Mycoplasma and Microsporidia, have been shown to carry degenerated editing domains in aminoacyl-tRNA synthetases. These defects in the protein synthesis machinery cause inaccurate translation of the genetic code, resulting in significant statistical errors in protein sequences that are thought to help parasites to escape immune response of a host. In this study we analyzed 10,423 complete bacterial genomes to assess conservation of the editing domains in tRNA synthetases, including LeuRS, IleRS, ValRS, ThrRS, AlaRS, and PheRS. We found that, while the editing domains remain intact in free-living species, they are degenerated in the overwhelming majority of host-restricted bacteria. Our work illustrates that massive genome erosion triggered by an intracellular lifestyle eradicates one of the most fundamental components of a living cell: the system responsible for proofreading of amino acid selection for protein synthesis. This finding suggests that inaccurate translation of the genetic code might be a general phenomenon among intercellular organisms with reduced genomes.
Evolutionary studies indicate that when an organism abandons its free-living lifestyle to become an intracellular entity, its genome undergoes a startling metamorphosis. Many formerly essential genes become degenerated, leading to loss of up to ∼95% of genes compared with genomes of closely related free-living species (1).
Early studies showed that genome reduction in host-restricted species is more than just a mere deletion of genes that become superfluous in the nutrient-rich environment of a host (2). Rather, these species tend to accumulate numerous deletions within essential genes, larger ratios between nonsynonymous and synonymous mutations, and other anomalies of genome structure (3–7). These trends of erosive evolution are thought to be driven by Muller’s ratchet, an evolutionary process by which genomes accumulate deleterious mutations in an irreversible manner that consequentially leads to mutational meltdown and genome decay (3–7). Muller’s ratchet states that in small asexual populations, due to genetic drift, there is a higher chance that the fittest individuals will fail to reproduce, while less fit individuals that carry slightly deleterious mutations will reproduce. If it happens repeatedly, the cumulative effect of deleterious mutations might result in massive gene loss, lower predicted thermal stability of proteins, and other forms of significant decline of cell efficiency (3–7). The most extreme cases of Muller’s ratchet are observed in organelles and in obligate intracellular organisms (8, 9). In these organisms, the genomes can be reduced up to ∼40 times smaller than the Escherichia coli genome, and their sequences might evolve at about a 10 times faster rate compared with free-living bacteria (3–6, 10, 11).
Previously, by exploring the translation machinery of pathogenic fungi Microsporidia (organisms with the smallest known eukaryotic genomes with a size of ∼2 Mb), we suggested that genomic erosion might alter one of the most fundamental properties of a living cell: its ability to accurately express the genetic code (12). We found that Microsporidia have a defect in the protein synthesis apparatus, namely degenerated editing domains in aminoacyl-tRNA synthetases (aaRSs) (12). Similar defects were found in other species with small genomes, including Mycoplasma (13) and some mitochondria (14–16). In all of these species, the editing domains are degenerated into poorly conserved and catalytically inactive protein segments. Editing domains of synthetases fulfill a quality-control process that ensures accurate selection of amino acids for protein synthesis. Because some aaRSs make occasional errors by the incorrect selection of structurally similar amino acids, they occasionally produce tRNAs that are charged with a noncognate amino acid (17–19). The editing domains hydrolyze erroneously charged aminoacyl-tRNA molecules and thereby help to maintain accurate translation of the genetic code (17–20). In prior studies, natural degeneration of the editing domains was shown to correspond to mistranslation of ∼0.5% of leucine and phenylalanine codons in Mycoplasma (13), and ∼5% of leucine codons in Microsporidia (12).
Our previous study of aaRSs in species with highly reduced genomes suggested that degenerated quality control and inaccurate protein synthesis represent inevitable consequences of genomic decay triggered by an intracellular lifestyle (12). In this study we examined whether editing deficiencies are a common phenomenon in highly reduced genomes. We accomplished this by assessing conservation of the editing domains of aaRSs in 10,423 complete bacterial genomes. We found that, while the editing sites are conserved in free-living species, they are degenerated in the overwhelming majority of obligate parasites and symbiotic bacteria, suggesting that most intracellular organisms translate the genetic code in an error-prone fashion.
Results
aaRS Editing Sites Are Mutated in ∼17% of Bacterial Species with Fully Sequenced Genomes.
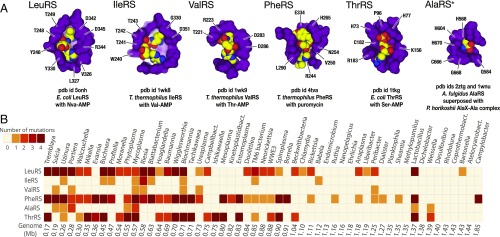
We first asked if the degeneration of aaRSs editing domains that was previously found in Microsporidia and Mycoplasma was present in other species. To answer this, conservation of the editing sites in aaRSs among complete bacterial genomes was analyzed. In total, 10,423 genomes corresponding to 2,277 bacterial species and their various strains were included in the analysis (Materials and Methods). In each of the aaRSs that have an editing domain (including LeuRS, IleRS, ValRS, PheRS, ThrRS, and AlaRS synthetases), we analyzed conservation of the active-site residues where side chains are either directly involved in substrate recognition or position water molecules for the nucleophilic attack (Materials and Methods and Fig. 1A). Multiple sequence alignments were used to verify the identity of these residues.
Fig. 1.
Editing sites in aaRSs are mutated in species with reduced genomes. (A) Fragments of crystal structures show the editing sites of aaRSs (shown in purple) in complex with mimics of misacylated tRNAs (shown as yellow, red, and blue balls). Labels point to the active site residues (E. coli numbering is used here and throughout the paper). (B) The heat map summarizes conservation of these active-site residues in the editing domains. Each square indicates the maximum number of editing-site mutations per species in a given genus. The complete list of species and identity of the editing-site mutations are shown in the supplementary data (https://figshare.com/articles/Supplementary_materials_for_Loss_of_protein_synthesis_quality_control_in_host-restricted_organisms_/7297499). The figure shows that, despite previously known high conservation of the editing sites across species, the editing sites are mutated in the majority of parasites and symbiotic bacteria with small genomes.
In agreement with previous observations (19), we found that editing-site residues remain conserved in ∼83% of bacterial species’ analyzed synthetases. However, the remaining ∼17% (387 species from 119 genera) have multiple mutations in the editing sites of at least one synthetase, and ∼5% of species (109 species from 43 genera) have multiples mutations in the editing sites of several aaRSs (Fig. 1B). As in the case of Mycoplasma, the editing-site mutations are found most frequently in threonine (ThrRS), leucine (LeuRS), and phenylalanine (PheRS) tRNA-synthetases. These editing-site mutations normally represent point mutations; but a small subset of genera—including Treponema, Borrelia, and Spirochaeta—contain species which completely lack the editing domain in PheRS. Overall, this finding illustrates that species with mutated editing sites are much more abundant in nature than was generally recognized (19).
aaRSs with Mutated Editing Sites Are Found Primarily in Organisms with Small Genomes.
In support of the Muller’s ratchet model (6), we found that species with mutated editing sites have highly reduced genomes and represent host-restricted organisms, such as endosymbionts or obligate intracellular parasites of animal and plant cells (Fig. 1B). Thus, 6.3% of species with genome sizes >3 Mb have editing-site mutations compared with 36% of species <3 Mb, while among bacteria with a genome size <1 Mb, the editing-site mutations are found in 88.3% of species (Fig. 1B).
Editing-Site Conservation Gradually Declines upon Genome Reduction.
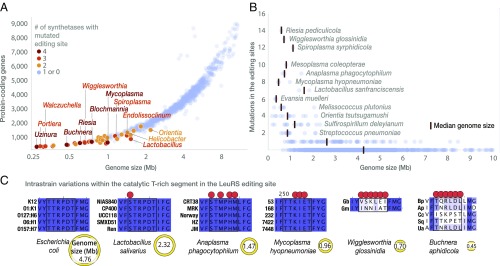
We next asked if the organisms with smaller genomes had a higher mutation load in the editing sites, as expected from the Muller’s ratchet (6, 7). Indeed, species with smaller genomes tend to have a higher number of synthetases with mutated editing sites: the average number of mutated synthetases is ascending from ∼0.06 in species with genomes 3 Mb and larger to ∼0.20 in species with genomes of 2–3 Mb, to ∼0.62 in species with genomes of 1–2 Mb to ∼1.58 in species with genomes <1 Mb (Fig. 2A). Furthermore, species with smaller genomes tend to have a higher total number of editing-site mutations: this number is gradually increasing from ∼0.09 in species with genomes 3 Mb and larger, ∼0.64 in species with genomes of 2–3 Mb, ∼1.89 in species with genomes of 1–2 Mb to 4.09 in species with genomes <1 Mb (Fig. 2B). Furthermore, in organisms with extremely small genomes (<0.7 Mb), the editing sites are no longer conserved even between strains of the same species, as exemplified by Buchnera aphidicola and Wigglesworthia glossinidia (Fig. 2C).
Fig. 2.
Species with reduced genomes have less-conserved editing sites. (A) The plot illustrates conservation of the editing sites in bacterial species with fully sequenced genomes. Each dot represents one bacterial species. For simplicity only one species per genus is shown; the complete list of bacterial species with mutated editing sites is shown in the supplementary data (https://figshare.com/articles/Supplementary_materials_for_Loss_of_protein_synthesis_quality_control_in_host-restricted_organisms_/7297499). As the genome size reduces, species carry more mutations in the editing sites. Thus, editing-site mutations in multiple aaRSs are highly uncommon in species with large genomes (above 2.5 Mb), whereas the majority of species with reduced genomes (below 1 Mb) have mutations in multiple synthetases. (B) The plot shows the total number of mutations in the editing sites of aaRSs per species. Upon transition from species with large genomes to species with reduced genomes, the editing sites accumulate an increasing number of mutations (total number of mutations in the editing sites of the six synthetases). (C) The panel illustrates conservation of the 242TTRPDT248 protein segment that is required for the LeuRS editing activity. Upon transition from free-living species (e.g., E. coli) to strict intracellular species with highly reduced genomes (W. glossinidia and B. aphidicola) this segment becomes less conserved. In the most extreme case, this segment is not conserved among different strains of the same species. Collectively, the figure shows that upon transition from large genomes (primarily found in free-living species) to reduced genomes (primarily found endosymbionts and obligate intracellular parasites), species accumulate more mutations per synthetase, as well as more mutated synthetases. Editing sites are transformed from nearly immutable protein segments into protein segments that are no longer conserved even between the closest groups of organisms, different strains of the same species.
Sequence Variations in the Editing Sites Suggest Loss of Enzymatic Activity.
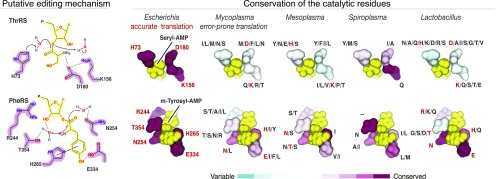
We next asked if editing-site mutations in the host-restricted species are comparable with those in the previously described editing-incompetent synthetases from Mycoplasma (13). In Mycoplasma, LeuRS and PheRS share two properties that underlie their editing incompetence: (i) their editing-site residues are not conserved even between closely related species of the genus; (ii) the editing-site residues that are directly involved in substrate recognition and coordination of water molecules for catalysis are replaced by residues of different size, charge or polarity, thereby impairing the chemistry of the editing reaction (13). We asked if these two properties are observed in the editing sites of other host-restricted species.
We analyzed mutations in the editing sites among bacterial genera for which genomes were sequenced for multiple species, including Mesoplasma, Spiroplasma, and Lactobacillus (Fig. 3). We focused on the editing sites of PheRS and ThrRS for which the roles of the editing-site residues in catalysis were determined earlier (Fig. 3) (21–23). We found that, similar to Mycoplasma, the editing sites of PheRS and ThrRS are not conserved within each of the analyzed genera. For example in Mesoplasma, Mesoplasma syrphidae and Mesoplasma chauliocola share as little as 12.5% sequence identity of the active-site residues of the editing domain (Fig. 3). In another example, Lactobacillus species have more than 20 combinations of residues in the ThrRS editing site, with some residues being represented by eight different amino acids (Fig. 3). Importantly, these variations in the editing sites include: (i) mutations of basic residues, which mediate tRNA binding, into small hydrophobic or negatively charged residues; and (ii) mutations of polar or hydroxyl-containing residues, which are involved in hydrogen bonding, into hydrophobic residues (Fig. 3). These sequence variations indicate that in species with small genomes the editing sites are degenerated into poorly conserved protein segments that most likely lack enzymatic activity.
Fig. 3.
Mutations in editing sites of aaRSs suggest loss of enzymatic activity. (Left) Mechanisms of the posttransfer editing by PheRS and ThrRS synthetases proposed by refs. 21–23. In PheRS synthetases, highly conserved editing-site residues Glu334 and Thr354 position water molecules for the nucleophilic attack, and Arg244 recruits the 3′-end of the tRNA molecule (here and throughout the paper we use E. coli numbering of amino acids). In the ThrRS, His73 and Lys156 coordinate water for the nucleophilic attack and the subsequent proton shuttle. (Right) Fragments of crystal structures show the editing sites of PheRS and ThrRS in complex with their substrates, seryl-AMP and m-Tyrosyl-AMP, respectively. The structures are colored by conservation of the active-site residues within a given genus. The letters next to each residue indicate identity of amino acids that are found in aminoacyl-tRNA sequences from a corresponding genus (for example, in Mycoplasma His32 is mutated to Ile, Leu, Met, Asn, or Ser). The figure illustrates that, similarly to Mycoplasma, many species with reduced genomes have highly variable editing domains where the residues that are required for catalysis stabilization of the CCA-end of the aminoacyl-tRNA, and positioning of the amino acid backbone in the editing site are replaced by amino acids of different size, charge, and hydrophobicity. Furthermore, these editing-site mutations are not conserved, even within species from the same genus. Mycoplasma, and many other host-restricted organisms, might lack residues required for the catalysis and substrate stabilization amino acid selection proofreading. This can result in partial or complete loss of the editing activity in these bacterial species.
The Editing Domains Appear to Be Either Editing-Competent or Editing-Null, but Never Promiscuous.
Past studies showed that the editing inactivation is not the most detrimental consequence of editing-site mutations (24). The most detrimental scenario is when the editing sites remain active but lose amino acid specificity, so that, in addition to hydrolysis of erroneously charge aminoacyl-tRNAs, the editing domains hydrolyze properly charged tRNAs. One illustrative example is LeuRS synthetase, in which the editing-site mutation T252A allows for hydrolysis of cognate Leu-tRNALeu. Consequently, the LeuRS mutant is not capable of producing Leu-tRNALeu (24).
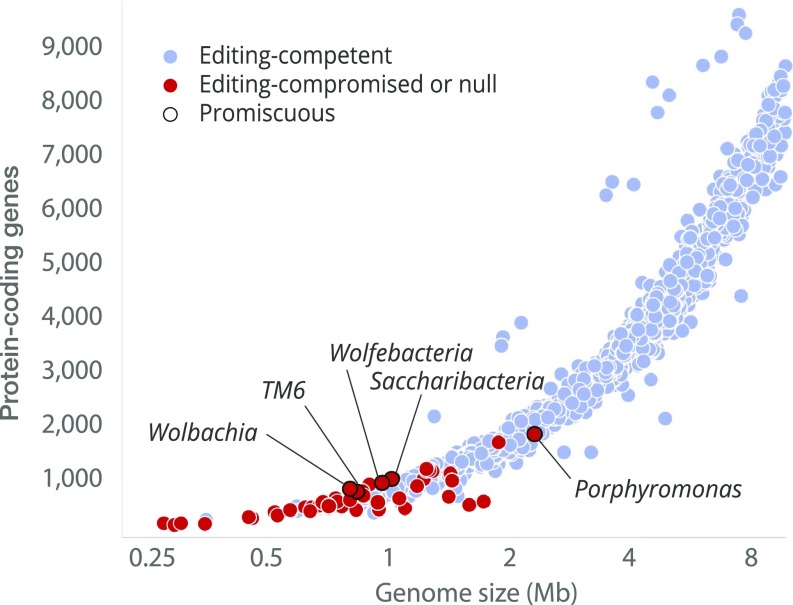
We asked if promiscuity-associated mutations can be found in species with unaltered catalytic residues in the editing domain of LeuRS synthetase (Fig. 4). We found that mutations associated with promiscuity are extremely rare and observed only in species with highly reduced genomes (Fig. 4). In total, only five species carried the T252A mutation: Porphyromonas gingivalis, TM6 bacterium, Saccharibacteria bacterium, Wolfebacteria bacterium, and Wolbachia endosymbiont. However, all of these bacteria carried additional mutations of catalytic residues in the editing domain, suggesting partial or complete loss of the editing activity (Fig. 4). This observation suggests that the editing domains remain either editing-competent or editing-null, but never promiscuous, which is consistent with the detrimental effect of promiscuity on tRNA synthetase ability to produce aminoacyl-tRNAs.
Fig. 4.
The editing domains appear to be either editing-competent or editing-null, but never promiscuous. The figure summarizes conservation of the active-site residues in the editing domain of LeuRS synthetase, with each dot corresponding to one bacterial species with a fully sequenced genome. Based on the active-site conservation, the species are divided into three groups: (i) species which LeuRS is predicted to be editing-competent due to consensus sequence in the editing site (in blue); (ii) species which LeuRS is predicted to be partially or fully editing-deficient due to mutations of the catalytic residues (in red); and (iii) species promiscuous toward cognate Leu-tRNALeu due to the mutation associated with promiscuity, T252A (E. coli LeuRS numbering; outlined in black). The figure shows that highly deleterious mutations that are associated with promiscuity are never observed among editing-competent synthetases, and are found exclusively in synthetases that are predicted to be editing-compromised or editing-null due to mutations of the catalytic residues of the editing center.
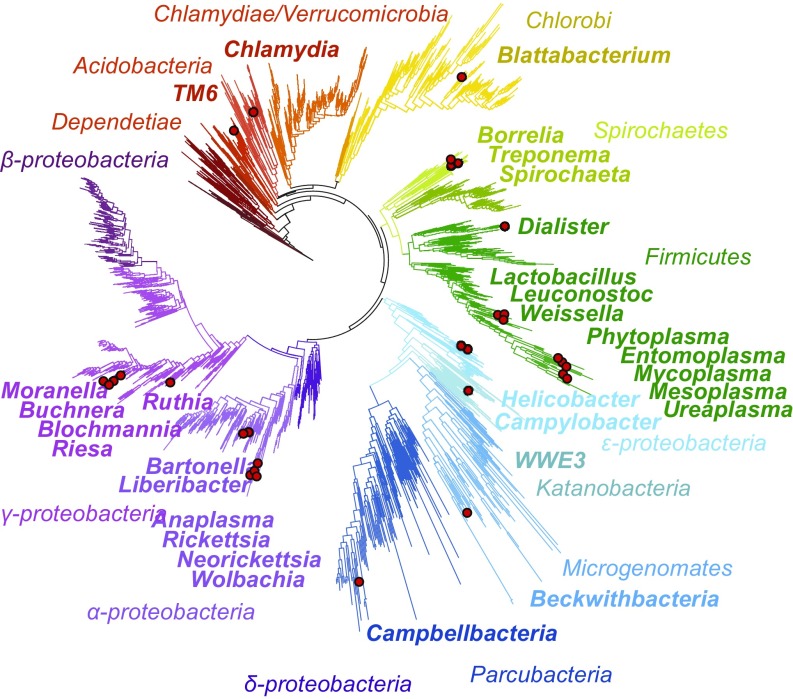
Species with Altered Editing Sites Are Not Closely Related but Are Found in All Major Branches of Bacteria.
We next asked if the editing-site mutations in a large fraction of bacterial species might be explained by a common evolutionary origin of these bacteria and their close relations to the Mycoplasma genus. To answer this, we mapped species with editing-site mutations on the bacterial tree of life (Fig. 5). We found that some of these species indeed belong to genera that are closely related to Mycoplasma, including Mesoplasma, Phytoplasma, Ureaplasma, and other pathogens from the Tenericutes phylum (Fig. 5). However, aside from Tenericutes, species with altered editing sites were found in nearly every major branch of bacterial phyla (Fig. 5). This finding indicates that degeneration of the editing sites is not limited to one narrow group of bacteria, but likely represents independent events among the diverse groups of bacteria.
Fig. 5.
Species with mutated editing domains are found among all major branches of bacteria. Red circles indicate bacterial genera with mutated editing sites of aaRSs. The figure illustrates that species with apparent editing defects can be found not only among close relatives of Mycoplasma (e.g., Phytoplasma, Entomoplasma, Mesoplasma, and other Firmicutes), but also in nearly every major branch of bacterial phyla.
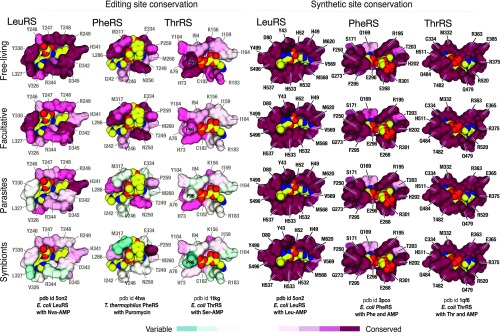
Synthetic Sites Remain Conserved Even in Species with Reduced Genomes.
We next asked if the alterations in editing sites are accompanied with comparable mutations in the synthetic sites of aaRSs. Synthetic sites are essential for life, due to their critical role in protein synthesis. We expected these sites to remain unaltered, even in species with drastically reduced genomes. To verify this, we analyzed conservation of the synthetic-site residues in four genera with differing lifestyles and average genome size (Fig. 6). We found that, unlike editing sites, the catalytic sites remain conserved across all branches of bacteria, including species with host-restricted lifestyles and extremely small genomes (Fig. 6). Thus, an apparent degeneration of aaRSs is limited to the quality-control editing sites, leaving the synthesis sites immutable.
Fig. 6.
Synthetic sites remain nearly invariant even in species with reduced genomes. The figure shows fragments of crystal structures in which aaRSs (shown as surface) are complexed with substrates (shown in yellow, red, and blue) that are bound to editing and synthetic sites. Active-site residues are colored by conservation. The figure illustrates gradual decline of the editing-site conservation upon transition from free-living to host-restricted species. At the same time, the synthetic sites remain highly conserved in all of the four groups of bacteria.
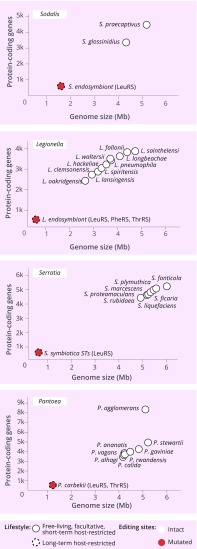
Editing-Site Degeneration Might Be Driven by Lifestyle Change from Free-Living to Host-Restricted.
Finally, we finally asked if we can observe editing-site degeneration in a closely related group of species whose members have highly distinct lifestyles. To date, there are four genera, Legionella, Pantoea, Serratia, and Sodalis, for which genome sequences are available for species with lifestyles varying from free-living to strictly host-restricted (Fig. 7). In the case of Legionella, human pathogens that cause a pneumonia-type Legionnaires’ disease, species are either free-living or facultative intracellular parasites and have relatively large genomes (∼2.7–5 Mb comprising ∼2,100–3,600 protein-coding genes). However, Legionella endosymbiont, a symbiont of Polyplax serrata, is a strictly intracellular organism whose genome comprises only ∼0.5 Mb base pairs and ∼400 protein-coding genes. Our analysis shows that, while the editing sites remain immutable in Legionella species with free-living or facultative symbiotic lifestyles, they are mutated in ThrRS, PheRS, and LeuRS in the L. endosymbiont (Fig. 7). The same pattern can be observed in Pantoea, Serratia, and Sodalis species, indicating that transition from a free-living to host-restricted lifestyle might be a factor that causes accumulation of editing-site mutations (Fig. 7).
Fig. 7.
Editing deficiency may be caused by transition from a free-living to host-restricted lifestyle. The diagrams illustrate conservation of the editing sites in four bacterial genera (Pantoea, Serratia, Sodalis, and Legionella) for which the full genome sequences are available for species with a different lifestyle, ranging from free-living to strictly host-restricted. The long-term host-restricted species S. endosymbiont, L. endosymbiont, S. symbiotica, and P. carbekii—which have highly reduced genomes (<1,000 protein-coding genes)—have mutated editing sites, while their free-living or facultative counterparts have conserved editing sites.
Discussion
This study revealed that the majority of host-restricted species with small genomes have accumulated defects in the protein synthesis machinery. Particularly, they lack catalytic residues in the aaRS editing domains that are required for accurate amino acid selection during protein synthesis. This finding suggests that most obligate intracellular parasites and endosymbiotic bacteria with highly reduced genomes translate genetic code in an error-prone manner, similarly to naturally editing-deficient Microsporidia (12) and Mycoplasma (13), and in engineered editing-deficient Saccharomyces cereviseae (25) and E. coli (26).
Notably, species with apparent defects in the editing sites include causative agents of human disease, such as ulcers (Helicobacteri pylori), scrub typhus (Orientia tsutsugamushi), hemorrhagic illness (Weisella ceti), sexually transmitted diseases (Ureaplasma parvum), Lyme disease (Borrelia afzelii), anaplasmosis (Rickettsia, Neorickettsia, Orientia, Ehrlichia, and Anaplasma species), and others. They also include Lactobacillus species, which are frequently used as probiotics to treat digestive problems. Discovery of editing defects in these clinically relevant species may help to uncover new aspects of their physiology, unanticipated proteome complexity, and unexplored strategies to disrupt pathogen–host interactions.
Our analysis indicates that the gradual decline of editing-site conservation might be driven by transition from a free-living to host-restricted lifestyle, reflecting overall genome erosion driven by Muller’s ratchet. This is especially evident when we compare species with different lifestyles but from the same genus (Fig. 6), or when we observe an accumulation of mutations in the editing sites across bacterial species with reduced genomes (Fig. 2). These findings indicate that long-term parasitism or endosymbiosis may have detrimental consequences for the integrity of the quality-control system that secures accurate translation of the genetic code. They also indicate that the accuracy of the genetic code translation might possibly gradually decline along with genome size reduction.
Our data indicate that different tRNA synthetases have different propensity to lose the editing function with the decrease of genome size (Fig. 1). While LeuRS, PheRS, and ThrRS have degenerated the editing sites in most intracellular organisms, other tRNA synthetases either remain conserved or carry fewer mutations in the editing sites, suggesting that the editing function of these synthetases (AlaRS, IleRS, and ValRS) is more important for maintaining the translation accuracy.
The question still remains: How do so many intracellular pathogens and endosymbiotic bacteria survive with defects in the editing domains? These domains have been long considered a highly conserved and essential system for accurate protein synthesis: mutations in these regions were shown to cause protein misfolding and genetic disorders in mammals (27–31); an impaired amino acid-starvation response in yeast (32); and growth defects, cell death, hypersensitivity to elevated amino acid levels, and mistranslation of up to 10% of codons that correspond to a defective synthetase in E. coli (33, 34).
One possible reason for the survival of host-restricted organisms may stem from the fact that these species show higher tolerance to protein aggregation due to elevated levels of chaperones, as exemplified by Buchnera and other primary endosymbionts that overexpress the GroEL chaperone (35–37). Although these higher levels of chaperones are thought to buffer accumulation of deleterious mutations caused by Muller’s ratchet (35–37), it seems plausible that they can prevent protein aggregation caused by mistranslation.
Another possible reason for survival might be related to the relatively ancient age of host-restricted species. Many intracellular symbionts and parasites are more than 50 My old (38), suggesting these species had time to adapt to a gradual erosion of editing sites or even use these defects to their benefit. In fact, beneficial use of mistranslation has been reported for several pathogens, including the parasitic fungi Candida albicans (25, 39, 40). While most species are thought to translate genes in a highly accurate manner, with roughly one error per 10,000 amino acids, C. albicans translates CUG codons ambiguously—either as leucine (94–97%) or as serine (3–6%)—causing frequent variations in protein sequence. This ambiguous translation leads to spectacular proteome diversity, with some proteins being expressed as more than 100,000 isoforms. These isoforms help C. albicans to escape immune recognition by the host (41, 42), suggesting that similar use of mistranslation might take place in host-restricted species with a degenerated editing system.
Although sequence degeneration in the editing sites is consistent with Muller’s ratchet, our data do not exclude other potential causes and outcomes of the editing loss. For example, while far fewer changes are seen in the synthetic active sites than in the editing sites (Fig. 6), it is possible that the small changes observed are sufficient to ameliorate the effects of lost editing. A case in point is the mitochondrial PheRS, which has completely lost the editing domain, but instead has far higher cognate versus noncognate substrate specificity than its bacterial counterpart with an intact editing domain (43). It is also possible that loss of the editing could have a net metabolic benefit in some cases. The editing reaction consumes ATP that might otherwise be used for other cellular processes, which might be critical for competitiveness under the nutrient-limited growth conditions experienced by many of the intracellular pathogens.
In sum, our work challenges previous assumptions about the level of accuracy of protein synthesis that is required to sustain life. It appears that, in an intracellular context, the pathogens and commensals can survive with reduced fidelity in protein synthesis. However, many questions about potential physiological roles of editing-site mutations still remain unresolved. It will be important to use mass spectrometry to verify our notion that the editing deficiency causes inaccurate protein synthesis in the majority of intracellular organisms. It will also be important to engineer parasites with restored editing domains and test how this will affect parasite growth, adaptation to stressors and interaction with a host. Perhaps the most important question is: Can we exploit this editing degeneration to manipulate protein synthesis accuracy and growth rate of host-restricted species? While additional research is needed to answer this question, our study demonstrates that genome reduction might eliminate or partially damage the quality-control pathway that is responsible for the accurate translation of the genetic code. Our work indicates that this damage might lead to a fundamental difference in how protein synthesis is carried out in free-living versus host-restricted species, raising a hope to exploit this difference for therapeutic purposes in the future.
Methods
Retrieving and Aligning Sequences of aaRSs.
To assess conservation of the editing domains in aaRSs, we analyzed bacterial species with fully sequenced genomes that were deposited to the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/genome/browse#!/overview/). The protein sequences were retrieved using a custom Python script from 10,423 bacterial genomes. aaRSs from genomes that were deposited as drafts were filtered for sequences with the proper protein name format, such as “Alanine-tRNA ligase (EC 6.1.1.7) (Alanyl-tRNA synthetase) (AlaRS)” for AlaRS. This allowed us to filter out truncated sequences, and sequences containing amino acids of unknown identity (denoted as “X”). The remaining sequences (∼8,400 sequences per aaRS, ∼2,400 bacterial species) were aligned using Clustal Omega with default settings (44). For each aaRS, the full list of species and sequences is shown. To simplify data visualization, we used custom Python scripts to reduce full datasets to datasets with only one sequence per one species (by removing multiple strains) and to datasets with only one sequence per genus (by removing multiple species).
Defining Active Site Residues in the Synthetic and Editing Sites of aaRSs.
The active sites residues were defined by using AreaImol (CCP4 suite) with default parameters and the solvent radius of 1.4 Å (45). For each synthetase, the program calculated area of protein–ligand interactions that were mediated by amino acid side chains of the editing site or the synthetic site. The crystal structure used in this studies were the following: for PheRS (22), ValRS (46), LeuRS (12, 47), IleRS (48), and ThrRS (21); the editing domain structures were determined in complex with a near-cognate amino acid bound to AMP, providing accurate information about the ligand-active site interactions. Because for AlaRS the crystal structures were determined in a vacant state, we defined editing-site residues based on previous biochemical studies, but used the structure of a homologous protein, AlaX, bound with near-cognate substrate serine to create an illustration shown in Fig. 1 (33, 49, 50). We have excluded ProRS synthetase from the analysis due to limited structural information about precise ligand–protein interaction, and due to complex genetics caused by isoforms and homologs in bacterial cells (51, 52). By doing so, we have selected only those editing-site residues in six aaRSs, with side chains that either: (i) directly bind the tRNA moiety of the substrate, (ii) directly bind the amino acid backbone of the substrate, (iii) or coordinate water molecules for the nucleophilic attack and the proton shuttle (Fig. 1).
Assessing Conservation of the Editing-Site Residues.
To assess conservation of the active-site residues, we cycled through the multiple sequence alignments and checked the identity of the active site residues in every sequence by using a custom Python script. The identity of these residues is listed. To color crystal structures by conservation, we used the ConSurf server (consurf.tau.ac.il/2016) with default settings for each synthetase (53).
Data Sharing.
All of the supplementary data may be downloaded as a zip-file at the following site: https://figshare.com/articles/Supplementary_materials_for_Loss_of_protein_synthesis_quality_control_in_host-restricted_organisms_/7297499.
Acknowledgments
We thank Kyle Hoffman, Hui Si Kwok, Jeffery Tharp, and Oscar Vargas-Rodriguez (all at Yale University) for critically reading the manuscript; and the reviewers for improving this study with their comments. This work was supported by National Institutes of Health Grant R35GM122560, and Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the Department of Energy Grant DE-FG02-98ER20311.
Footnotes
The authors declare no conflict of interest.
Data deposition: The supplementary data reported in this paper have been deposited in Figshare (https://figshare.com/articles/Supplementary_materials_for_Loss_of_protein_synthesis_quality_control_in_host-restricted_organisms_/7297499).
References
- 1.Casadevall A. Evolution of intracellular pathogens. Annu Rev Microbiol. 2008;62:19–33. doi: 10.1146/annurev.micro.61.080706.093305. [DOI] [PubMed] [Google Scholar]
- 2.Frank AC, Amiri H, Andersson SG. Genome deterioration: Loss of repeated sequences and accumulation of junk DNA. Genetica. 2002;115:1–12. doi: 10.1023/a:1016064511533. [DOI] [PubMed] [Google Scholar]
- 3.Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 4.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rispe C, Moran NA. Accumulation of deleterious mutations in endosymbionts: Muller’s ratchet with two levels of selection. Am Nat. 2000;156:425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- 6.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 7.Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 8.Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 9.Blanchard JL, Lynch M. Organellar genes: Why do they end up in the nucleus? Trends Genet. 2000;16:315–320. doi: 10.1016/s0168-9525(00)02053-9. [DOI] [PubMed] [Google Scholar]
- 10.Andersson SG, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 11.Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 12.Melnikov SV, et al. Error-prone protein synthesis in parasites with the smallest eukaryotic genome. Proc Natl Acad Sci USA. 2018;115:E6245–E6253. doi: 10.1073/pnas.1803208115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci USA. 2011;108:9378–9383. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lue SW, Kelley SO. An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry. 2005;44:3010–3016. doi: 10.1021/bi047901v. [DOI] [PubMed] [Google Scholar]
- 15.Roy H, Ling J, Alfonzo J, Ibba M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- 16.Ye Q, et al. Degenerate connective polypeptide 1 (CP1) domain from human mitochondrial leucyl-tRNA synthetase. J Biol Chem. 2015;290:24391–24402. doi: 10.1074/jbc.M115.672824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin L, Hale SP, Schimmel P. Aminoacylation error correction. Nature. 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 18.Nureki O, et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 19.Perona JJ, Gruic-Sovulj I. Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top Curr Chem. 2014;344:1–41. doi: 10.1007/128_2013_456. [DOI] [PubMed] [Google Scholar]
- 20.Döring V, et al. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 21.Dock-Bregeon AC, et al. Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol Cell. 2004;16:375–386. doi: 10.1016/j.molcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Tworowski D, Klipcan L, Peretz M, Moor N, Safro MG. Universal pathway for posttransfer editing reactions: Insights from the crystal structure of TtPheRS with puromycin. Proc Natl Acad Sci USA. 2015;112:3967–3972. doi: 10.1073/pnas.1414852112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling J, Roy H, Ibba M. Mechanism of tRNA-dependent editing in translational quality control. Proc Natl Acad Sci USA. 2007;104:72–77. doi: 10.1073/pnas.0606272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mursinna RS, Lincecum TL, Jr, Martinis SA. A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry. 2001;40:5376–5381. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]
- 25.Mohler K, Ibba M. Translational fidelity and mistranslation in the cellular response to stress. Nat Microbiol. 2017;2:17117. doi: 10.1038/nmicrobiol.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cvetesic N, et al. Proteome-wide measurement of non-canonical bacterial mistranslation by quantitative mass spectrometry of protein modifications. Sci Rep. 2016;6:28631. doi: 10.1038/srep28631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13:1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Schimmel P, Guo M. A tipping point for mistranslation and disease. Nat Struct Mol Biol. 2009;16:348–349. doi: 10.1038/nsmb0409-348. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci USA. 2014;111:17570–17575. doi: 10.1073/pnas.1420196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, et al. A human disease-causing point mutation in mitochondrial threonyl-tRNA synthetase induces both structural and functional defects. J Biol Chem. 2016;291:6507–6520. doi: 10.1074/jbc.M115.700849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohler K, et al. Editing of misaminoacylated tRNA controls the sensitivity of amino acid stress responses in Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45:3985–3996. doi: 10.1093/nar/gkx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beebe K, Ribas De Pouplana L, Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22:668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karkhanis VA, Mascarenhas AP, Martinis SA. Amino acid toxicities of Escherichia coli that are prevented by leucyl-tRNA synthetase amino acid editing. J Bacteriol. 2007;189:8765–8768. doi: 10.1128/JB.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fares MA, Ruiz-González MX, Moya A, Elena SF, Barrio E. Endosymbiotic bacteria: GroEL buffers against deleterious mutations. Nature. 2002;417:398. doi: 10.1038/417398a. [DOI] [PubMed] [Google Scholar]
- 36.Tomala K, Korona R. Molecular chaperones and selection against mutations. Biol Direct. 2008;3:5. doi: 10.1186/1745-6150-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shonhai A, Maier AG, Przyborski JM, Blatch GL. Intracellular protozoan parasites of humans: The role of molecular chaperones in development and pathogenesis. Protein Pept Lett. 2011;18:143–157. doi: 10.2174/092986611794475002. [DOI] [PubMed] [Google Scholar]
- 38.Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 39.Ribas de Pouplana L, Santos MA, Zhu JH, Farabaugh PJ, Javid B. Protein mistranslation: Friend or foe? Trends Biochem Sci. 2014;39:355–362. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Ling J, O’Donoghue P, Söll D. Genetic code flexibility in microorganisms: Novel mechanisms and impact on physiology. Nat Rev Microbiol. 2015;13:707–721. doi: 10.1038/nrmicro3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santos MA, Ueda T, Watanabe K, Tuite MF. The non-standard genetic code of Candida spp.: An evolving genetic code or a novel mechanism for adaptation? Mol Microbiol. 1997;26:423–431. doi: 10.1046/j.1365-2958.1997.5891961.x. [DOI] [PubMed] [Google Scholar]
- 42.Miranda I, et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio. 2013;4:e00285–e00313. doi: 10.1128/mBio.00285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds NM, et al. Cell-specific differences in the requirements for translation quality control. Proc Natl Acad Sci USA. 2010;107:4063–4068. doi: 10.1073/pnas.0909640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukunaga R, Yokoyama S. Structural basis for non-cognate amino acid discrimination by the valyl-tRNA synthetase editing domain. J Biol Chem. 2005;280:29937–29945. doi: 10.1074/jbc.M502668200. [DOI] [PubMed] [Google Scholar]
- 47.Palencia A, et al. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat Struct Mol Biol. 2012;19:677–684. doi: 10.1038/nsmb.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukunaga R, Yokoyama S. Structural basis for substrate recognition by the editing domain of isoleucyl-tRNA synthetase. J Mol Biol. 2006;359:901–912. doi: 10.1016/j.jmb.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 49.Sokabe M, Okada A, Yao M, Nakashima T, Tanaka I. Molecular basis of alanine discrimination in editing site. Proc Natl Acad Sci USA. 2005;102:11669–11674. doi: 10.1073/pnas.0502119102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naganuma M, Sekine S, Fukunaga R, Yokoyama S. Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization. Proc Natl Acad Sci USA. 2009;106:8489–8494. doi: 10.1073/pnas.0901572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crepin T, Yaremchuk A, Tukalo M, Cusack S. Structures of two bacterial prolyl-tRNA synthetases with and without a cis-editing domain. Structure. 2006;14:1511–1525. doi: 10.1016/j.str.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 52.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashkenazy H, et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]