Significance
Dissemination of chemical information throughout the cell is a fundamental biological process with clinical relevance. Pathological changes in local signaling enzyme activity are linked to diseases, including schizophrenia, Alzheimer’s disease, cardiac arrhythmias, and seizures. Mining of patient datasets has uncovered genetic variation in A-kinase anchoring proteins (AKAPs) that promotes mislocalization of protein kinase A (PKA). We investigate 42 SNPs in AKAPs that interrupt association with PKA to impact local cAMP signaling. The most detrimental variants are situated within the hydrophobic face of a conserved helical region on the AKAP that is essential for kinase anchoring. An unexpected outcome is the discovery of an alternative targeting mechanism for AKAPs that utilizes the intact PKA holoenzymes as cytoplasmic “anchors.”
Keywords: AKAPs, PKA, proximity labeling, kinase anchoring, nucleus
Abstract
A-kinase anchoring proteins (AKAPs) shape second-messenger signaling responses by constraining protein kinase A (PKA) at precise intracellular locations. A defining feature of AKAPs is a helical region that binds to regulatory subunits (RII) of PKA. Mining patient-derived databases has identified 42 nonsynonymous SNPs in the PKA-anchoring helices of five AKAPs. Solid-phase RII binding assays confirmed that 21 of these amino acid substitutions disrupt PKA anchoring. The most deleterious side-chain modifications are situated toward C-termini of AKAP helices. More extensive analysis was conducted on a valine-to-methionine variant in the PKA-anchoring helix of AKAP18. Molecular modeling indicates that additional density provided by methionine at position 282 in the AKAP18γ isoform deflects the pitch of the helical anchoring surface outward by 6.6°. Fluorescence polarization measurements show that this subtle topological change reduces RII-binding affinity 8.8-fold and impairs cAMP responsive potentiation of L-type Ca2+ currents in situ. Live-cell imaging of AKAP18γ V282M-GFP adducts led to the unexpected discovery that loss of PKA anchoring promotes nuclear accumulation of this polymorphic variant. Targeting proceeds via a mechanism whereby association with the PKA holoenzyme masks a polybasic nuclear localization signal on the anchoring protein. This led to the discovery of AKAP18ε: an exclusively nuclear isoform that lacks a PKA-anchoring helix. Enzyme-mediated proximity-proteomics reveal that compartment-selective variants of AKAP18 associate with distinct binding partners. Thus, naturally occurring PKA-anchoring-defective AKAP variants not only perturb dissemination of local second-messenger responses, but also may influence the intracellular distribution of certain AKAP18 isoforms.
The recent explosion of deep-sequencing data from diverse cohorts of patients reveals the impact of genetic variation within human populations (1). Nucleotide changes that accumulate over multiple generations are considered inert or “passenger” mutations that shape us as individuals. Conversely, acute genetic lesions that alter the activity of key enzymes or structural proteins are often linked to disease. Such pathological lesions are particularly prevalent in cell-signaling enzymes, such as ubiquitin ligases and protein kinases that guide the passage of chemical signals throughout the cell (2, 3). Consequently, mining of databases that chart these mutations can be combined with high-resolution structural analysis to inform the development of drugs that combat chronic disorders, such as Parkinson’s disease and various cancers (4).
Early examples of this precision-medicine approach include the identification of genetically modified forms of the BCR-Abl and B-RAF kinases that drive cancers, including chronic myelogenous leukemia, melanoma, and hepatocellular carcinoma (5, 6). The subsequent success of groundbreaking drugs, such as imatinib (Gleevec) and sorafenib, have transformed this innovative drug-discovery strategy into lucrative multinational ventures. The current kinase inhibitor landscape includes 243 small molecules in clinical trials for a variety of indications (7). However, manipulating complex cellular events is often more involved than simply targeting individual signaling enzymes. This is exemplified by evidence that many kinase cascades are constrained by scaffolding and anchoring proteins (8–10). These organizational elements ensure that signals are transmitted to precise sites within the cell (11). Hence, genetic lesions that disrupt protein kinase anchoring have been linked to various pathological responses and may represent untapped therapeutic targets (12–14).
A-kinase anchoring proteins (AKAPs) are prototypic kinase-targeting factors (15, 16). This structurally diverse group of proteins was initially discovered on the basis of solid-phase interaction with radiolabeled RII subunits of the cAMP-dependent protein kinase (PKA) (17). Subsequent interaction-cloning, proteomic, and bioinformatic approaches have identified 39 human genes that encode in excess of 60 AKAP isoforms (11) (Fig. 1A). AKAPs were originally named colloquially or on the basis of their molecular weight, but the systematic cataloging of anchoring proteins has been complicated by a HUGO nomenclature that only recognizes 15 AKAP genes (Fig. 1A). Nonetheless, a ubiquitous and defining characteristic of the AKAP family is a 14- to 18-residue helical region that binds with high affinity to PKA holoenzymes that are composed of a regulatory (R) subunit dimer and two catalytic (C) subunits (18). X-ray crystallographic analyses have shown that the hydrophobic face of this amphipathic helix slots into a binding groove formed by the docking and dimerization domain (D/D) of R subunits (19, 20). Negative-stain electron microscopy and 3D reconstructions show that intact hetero-pentameric AKAP-RII2-C2 assemblies adopt a range of flexible tripartite configurations (21, 22). This is because intrinsically disordered regions between the D/D domains and cAMP-binding sites on each regulatory subunit permit a 150- to 200-Å radius of motion to the associated catalytic subunits (21, 23). Such flexibility within anchored PKA holoenzyme complexes enables precise orientation of the catalytic subunit toward substrates (11, 24). Recent data advances this model, showing that intact, active, and anchored PKA holoenzymes are the functional signaling units inside cells (22). This “signaling island” concept radically changes the view of how AKAP complexes operate and indicates that protein phosphorylation is more regionally confined than previously appreciated (22).
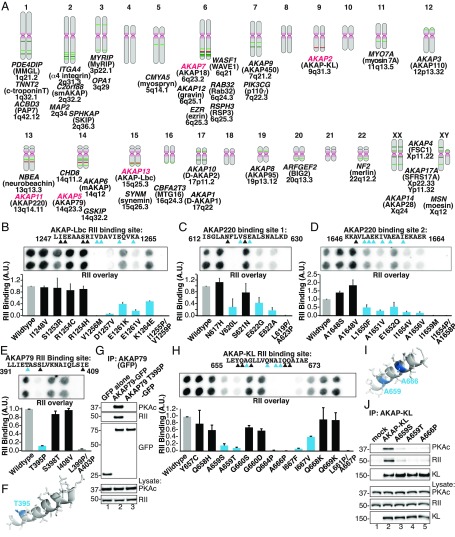
Fig. 1.
Identification and analysis of polymorphisms in PKA-anchoring domains of AKAPs. (A) Schematic of the human chromosomal locations for genes encoding 39 experimentally validated AKAPs. AKAPs harboring nonsynonymous SNPs in the PKA-anchoring helices that are investigated in this study are highlighted in red. (B–E and H) Peptide SPOT arrays containing WT and variant PKA binding helices were constructed and probed by RII overlay. (Upper) Solid-phase RII binding was assessed by far-Western blotting with biotin-RII and detected by neutravidin-HRP and enhanced chemoluminescence. (Lower) Densitometric analysis of RII binding was normalized to WT signal. (B) Peptide array and amalgamated densitometric data for analysis of the AKAP-Lbc (AKAP13) RII-binding site. Densitometric analysis of RII binding was normalized to WT signal. Variants that reduce binding to RII are highlighted in blue. (C) Screening and analysis of peptide arrays representing SNP sequences for PKA binding site 1 in AKAP220 (AKAP11). (D) Screening and analysis of peptide arrays representing SNP sequences for the PKA binding site 2 in AKAP220 (AKAP11). (E) Screening and analysis of peptide arrays representing SNP sequences for the PKA binding site in AKAP79 (AKAP5). (F) Molecular modeling of the AKAP79 amphipathic helix (PDB ID code 2H9R) showing threonine 395 at the second turn of the helix (blue). (G) Immune complexes of either WT AKAP79 or AKAP79 T395P were probed for coprecipitation of PKA RII and C subunits. Introduction of the helix-breaking proline residue disrupts the AKAP79–PKA interaction. (H) Screening and analysis of peptide arrays representing SNP sequences for the PKA binding in AKAPKL (AKAP2). (I) Modeling of the AKAP-KL amphipathic helix based on coordinates for AKAP-IS (PDB ID code 2IZX), showing the residues A659 and A666 (blue). (J) Immune complexes of either WT AKAP-KL or AKAP-KL A659S, A659T, or A666P variants were probed for coprecipitation of PKA RII and C subunits. Mutation of either position drastically impairs PKA binding. IP, immunoprecipitation
As our understanding of how signaling enzyme complexes operate becomes more refined, it is evident that perturbation of AKAP–PKA interactions is linked to disease (11, 14). Scrutiny of big datasets has uncovered genetic variation in AKAPs that correlate with susceptibility to diseases, such as schizophrenia, Alzheimer’s, certain cardiac arrhythmias, and the onset of febrile seizures (12, 14, 25, 26). In this report we systematically investigate the properties of polymorphic variants in AKAPs that elicit single amino acid changes in RII-anchoring helices. Unexpected outcomes of this endeavor include the discovery of an alternative AKAP-targeting mechanism that utilizes the intact PKA holoenzyme as a cytoplasmic anchor and the identification of an AKAP18 variant that does not function as an AKAP.
Results
SNPs in AKAP Helices Alter PKA Anchoring.
Genome-wide association surveys have identified point mutations and SNPs in genes encoding AKAPs that are linked to a variety of neurological and cardiac diseases (reviewed in ref. 14). The National Center for Biotechnology Information (NCBI) SNP (https://www.ncbi.nlm.nih.gov/SNP/index.html) and Variation Viewer databases (https://www.ncbi.nlm.nih.gov/variation/view/) were used to identify nonsynonymous nucleotide changes that introduce missense amino acids within the R subunit binding helices from a variety of AKAPs (Fig. 1A, red). This information was used to create peptide SPOT arrays that encompass native and variant PKA-binding sequences from five different anchoring proteins (Fig. 1A and SI Appendix, Table S1). Solid-phase RII-binding (RII-overlay) assessed whether these amino acid substitutions affected PKA anchoring (Fig. 1B). Peptides containing prolines were included in the peptide array as negative controls (SI Appendix, Table S1). Prior studies have shown that substitution with proline disrupts the structural integrity of PKA-anchoring helices (18, 27).
Densitometric analyses of multiple SPOT arrays revealed that amino acid substitution within PKA-anchoring helices influences RII interaction (Fig. 1 B–J). Nine naturally occurring variants in AKAP-Lbc (AKAP13) were tested (Fig. 1B). Single amino acid substitutions at four positions (R1245H, I1248V, S1253R, and R1254C) had no effect on RII-binding (Fig. 1B). However, three side-chain variants toward the C terminus of the PKA-anchoring helix (V1256M, D1257V, and E1261V) dramatically reduced solid-phase interaction with RII (Fig. 1B). Two other substitutions (E1261K and K1264E) had more modest effects (Fig. 1B).
Next we investigated SNPs in the vesicular and membrane-associated anchoring protein AKAP220 (AKAP11). This anchoring protein encodes two PKA-anchoring helices with different affinities for PKA holoenzymes (28, 29). Substitutions toward the C terminus of site 1 exhibited reduced affinity for RII, whereas amino acid changes throughout helix 2 adversely affected PKA anchoring (Fig. 1 C and D). Interestingly, substitution of serine or valine at position 1648 enhanced solid-phase binding of RII in site 2 of AKAP220 (Fig. 1D). This suggests that side chains that constrain the initial turns of helices are positive PKA-anchoring determinants.
Further support for this concept was provided by analysis of the neuronal anchoring protein AKAP79 (AKAP5) (30, 31). Three SNP variants in AKAP79 were investigated, but only substitution of Thr-395 with Pro affected RII-binding (Fig. 1E). Molecular modeling based upon the NMR structure of this region reveals that position 395 lies within the first turn of the anchoring helix (32) (Fig. 1F). Proline substitution prevents formation of the α-helix. Further validation was provided when this variant was introduced into full-length AKAP79 and expressed in HEK293 cells. Western blotting of WT AKAP79 immune complexes confirmed cofractionation of PKA regulatory and catalytic subunits (Fig. 1G, Upper three panels, lane 2). Cofractionation of PKA subunits was not detected in cells expressing the AKAP79-T395P variant (Fig. 1G, Upper three panels, lane 3).
AKAP-KLs (AKAP2) are a family of alternately spliced AKAP isoforms that organize kidney and lung second-messenger signaling events (33). AKAP-KL also sustains PKA regulation of lens aquaporin-0 water channels (34). Disruption of AKAP-KL–mediated PKA anchoring in the lens is implicated in pathological cataract formation (34). Peptide array analysis identified several missense variants of AKAP-KL that disrupted RII-binding (Fig. 1H). The most profound effects were observed in SNP variants that contained prolines at positions 664 and 666 in AKAP-KL. However, significant loss of PKA anchoring was also observed when positions 659 and 667 were replaced with bulkier side chains (Fig. 1H). We focused our biochemical analyses on two sites in the AKAP-KL helix that yielded severely impaired PKA-anchoring variants (Fig. 1I). Further validation of this result was provided when the A659S, A659T, and A666P variants were introduced into full-length AKAP-KL and expressed in HEK293 cells. Coprecipitation of PKA subunits was severely reduced or not detected in AKAP-KL immune complexes isolated from cells expressing each of the aforementioned SNP variants (Fig. 1J, Upper two panels, lanes 3–5). Control experiments with WT AKAP-KL immune complexes confirmed cofractionation of PKA regulatory and catalytic subunits (Fig. 1J, Upper two panels, lane 2). Collectively, the data presented in Fig. 1 show that naturally occurring variants on the hydrophobic face of RII-binding helices, and particularly those located in C-terminal turns, can have profound effects on PKA anchoring.
Characterization of AKAP18 Helix-Disrupting SNPs.
Alternative splicing of the AKAP7 gene drives tissue-specific expression of different AKAP18 protein products that range in size from 15 to 42 kDa (35–37) (Fig. 2 A and B). The low molecular weight α- and β-isoforms of AKAP18 couple PKA to a variety of ion channels in neurons, whereas the longer δ- and γ-isoforms have been implicated in modulation of excitation contraction coupling in the heart, water homeostasis in kidney collecting ducts, and roles in differentiated adipocytes (35, 38, 39) (Fig. 2B). Polymorphisms in AKAP18 have also been linked to an increased incidence of febrile seizures (26). The identification of a cluster of SNPs within the PKA-anchoring helix between residues 282 and 286 of AKAP18γ provided the impetus for more in-depth investigation (Fig. 2C). Peptide array analysis of SNP sequences at positions 282, 283, and 286 revealed that conservative side-chain substitutions disrupted RII-binding (Fig. 2C). Control experiments confirmed that PKA anchoring was also abolished in the context of the double-mutation AKAP18γ S278/L281P (Fig. 2C). In contrast, the AKAP18γ Arg280Ser (R280S) variant appeared to strengthen the interaction with RII (Fig. 2C). More stringent cell-based validation of AKAP18γ variants involved expression of the full-length proteins in HEK293 cells. Western blot analysis of AKAP18 immune complexes was used to detect cofractionation of PKA subunits (Fig. 2D, Upper panels). All variants at positions 282–286 of AKAP18γ were impaired in their ability to anchor PKA (Fig. 2D, lanes 4–6). In comparison, PKA subunits remained associated with the AKAP18γ R280S variant inside cells (Fig. 2D, lane 3). Secondary structure prediction algorithms place residue 280 of AKAP18γ on the hydrophilic face of the AKAP helix, a region that does not directly contact the RII dimer (40) (Fig. 2E). Conversely, side chains between positions 282–286 of AKAP18γ are located on the hydrophobic face of the helix (19, 40) (Fig. 2E). This substructure slots into a binding pocket formed by a four-helix bundle on the docking and dimerization domain of the R subunits (Fig. 2F, blue helix). The impaired binding in the V282M mutant provided us a unique opportunity to study this intermediate phenotype.
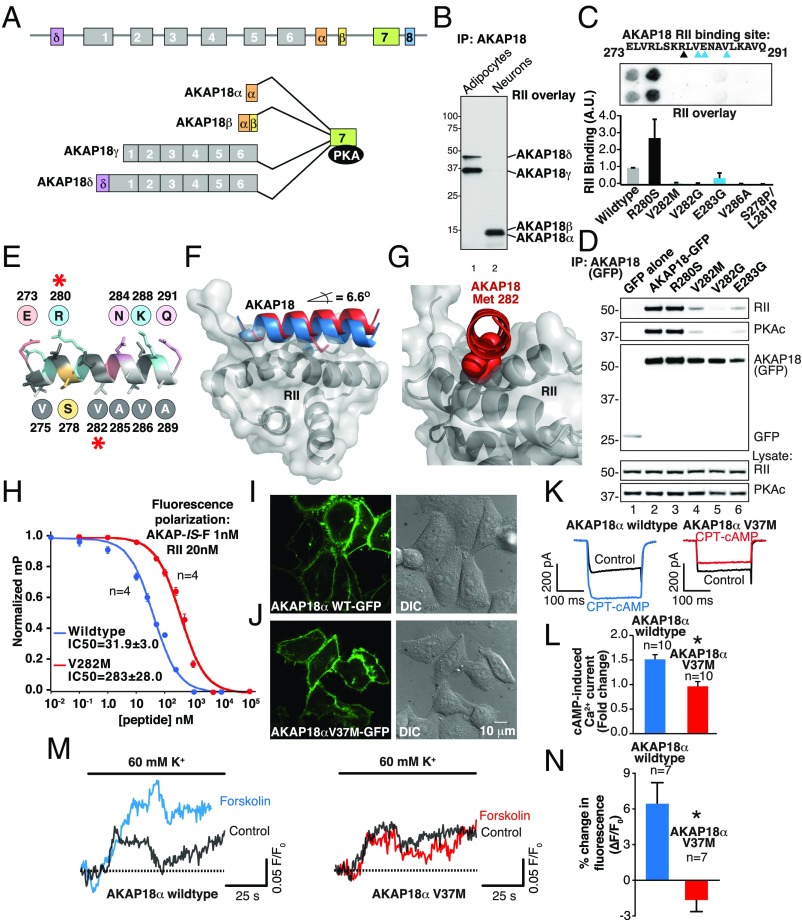
Fig. 2.
Polymorphisms in AKAP18 isoforms (AKAP7) alter PKA binding and regulation of ion channels. (A) Schematic of AKAP7 gene organization depicting exon–intron structure and alternatively spliced isoforms. (B) IP-RII overlay analysis show differential expression of distinct AKAP18 isoforms in 3T3-L1 adipocytes and cultured hippocampal neurons. (C) Peptide array and amalgamated densitometric data for analysis of the AKAP18 (AKAP7) RII-binding site. Densitometric analysis of RII binding was normalized to WT signal. Variants that reduce binding to RII are highlighted in blue. (D) AKAP18 SNP variants (denoted above each lane) were immunoprecipitated and copurification of PKA regulatory (Upper) and catalytic (Upper-mid) identified by Western blotting. Expression levels of each protein are indicated in the Lower three panels. (E) Model of AKAP18 PKA binding helix highlighting selected residues (red asterisk) on either face of the amphipathic helix. (F) Structural model shows WT AKAP18 helix (blue) and AKAP18 V282M helix (red) in complex with the RII D/D domain (gray). The V282M helix (red) is oriented at a 6.6° angle from WT. (G) En face and expanded view of the AKAP18 V282M helix shows displacement of the mutant peptide by bulky methionine side chain (red). (H) Fluorescence polarization data shows AKAP18 WT and V282M peptide displacement of a fluorescent AKAP-IS peptide from RII. The IC50 values for each are indicated. (I) Fluorescent and DIC images of HeLa cells expressing WT AKAP18α-GFP. (J) Fluorescent and DIC images of HeLa cells expressing AKAP18α V37M-GFP. (Scale bar: I–J, 10 μm.) (K) Whole-cell electrophysiological recording of cAMP-potentiated Ca2+ current in HEK293 cells expressing the L-type Ca2+ channel and either AKAP18α WT or the AKAP18α V37M variant. (L) Quantification of peak current potentiation. Data are presented as mean ± SEM. *P < 0.001 by unpaired Student’s t test. (M) Fluo4-AM measurement of cAMP-potentiated Ca2+ current in mouse aortic smooth muscle cells expressing either WT AKAP18α (blue) or the AKAP18α V37M variant (red). Calcium accumulation in response to depolarization was measured before and after brief forskolin stimulation. (N) Quantification of the change in fluorescence response to K+-induced depolarization. Data are presented as mean ± SEM. *P < 0.001 by unpaired Student’s t test.
Molecular modeling of the AKAP18–RIIα interface using PDB coordinates from published structures predicts that the V282M substitution generates a 6.6° upward displacement of the anchoring helix in relation to the binding pocket on RIIα (Fig. 2C, red helix). More detailed examination of the V282M–RIIα interface reveals that the added bulk and length of the methionine side-chain sterically hinders association with the AKAP-binding pocket on RIIα (Fig. 2G). In silico studies using the AutoDock program (41) computed a simulated free energy of binding of −4.54 kcal/mol (estimated Kd = 468 µM) for AKAP18 V282M compared with −10.58 kcal/mol (estimated Kd = 17.4 nM) for WT. Fluorescence polarization experiments calculated an IC50 of 283 ± 28 nM (n = 4) for AKAP18V282M peptide–RIIα interaction (Fig. 2H, red) compared with an IC50 of 32 ± 3 nM (n = 4) for the native sequence (Fig. 2H, blue). The differences in predicted and observed binding-affinity changes may be a consequence of AutoDock forcing all side chains into rigid conformations (41). Collectively, these data indicate that AKAP18 variants harboring the V282M substitution are severely impaired in their ability to anchor PKA holoenzymes.
Because the kinase-anchoring helix is conserved in most splice variants of AKAP18, we introduced the corresponding mutation (V37M) into the α-isoform. This membrane-associated anchoring protein mediates PKA-dependent modulation of ion channels in excitable cells (42). Fluorescent imaging confirmed that GFP-tagged AKAP18α V37M is retained at the plasma membrane when expressed in HEK293 cells (Fig. 2 I and J). Coexpression of AKAP18α V37M with L-type Ca2+ channel subunits permitted whole-cell electrophysiological analysis of local PKA modulation of calcium currents (36, 43). Stepwise depolarization of cells was used to generate a pulse of Ca2+ current. In cells expressing WT AKAP18α, application of cell-soluble cAMP elicits a 1.5-fold increase in Ca2+ current (n = 10) (Fig. 2 K and L, blue). This second-messenger response was abolished in cells expressing the AKAP18α V37M variant (Fig. 2 K and L, red).
Further substantiation in a more physiologically relevant system was possible by measuring depolarization-induced changes in Ca2+ flux in unpassaged cultured mouse aortic smooth-muscle cells expressing AKAP18α or AKAP18α V37M. Global Ca2+ influx through calcium channels was measured using the fluorescent calcium indicator dye acetoxymethyl-ester (AM) Fluo-4 (Fluo4-AM) upon depolarization with 60 mM potassium. In cells expressing AKAP18α, a brief pulse of forskolin to activate PKA enhanced Fluo4-AM fluorescence twofold (n = 7) (Fig. 2 M and N, blue). Importantly, this response was absent in cells expressing the AKAP18α V37M variant (Fig. 2 M and N, red). Taken together, these experiments suggest that naturally occurring variants in the RII-binding helix of AKAP18 isoforms uncouple cAMP-responsive events at the plasma membrane.
AKAP18 Long Isoforms Transit to the Nucleus in a PKA-Dependent Manner.
Long isoforms of AKAP18 encode targeting sequences that direct the anchoring protein to a variety of other subcellular locations (37, 44). For example, AKAP18γ and -δ are sequestered at the sarcoplasmic reticulum through protein–protein interactions with SERCA2 calcium reuptake channels, whereas in kidney collecting ducts, AKAP18δ is a component of intracellular vesicles (38, 39). However, these static targeting interactions do not fully reflect the complexity of AKAP18 retention in the cytoplasm. Surprisingly, application of a cell-soluble PKA-anchoring disruptor peptide (stHt-31) elicited the nuclear accumulation of AKAP18γ over a time course of 40 min (Fig. 3 A and B). In contrast, AKAP18γ remained cytoplasmic upon application of the control peptide stHt-31-P (Fig. 3C). These results raised the intriguing possibility of whether or not association with the PKA holoenzyme influences the cytoplasmic retention of AKAP18γ or -δ.
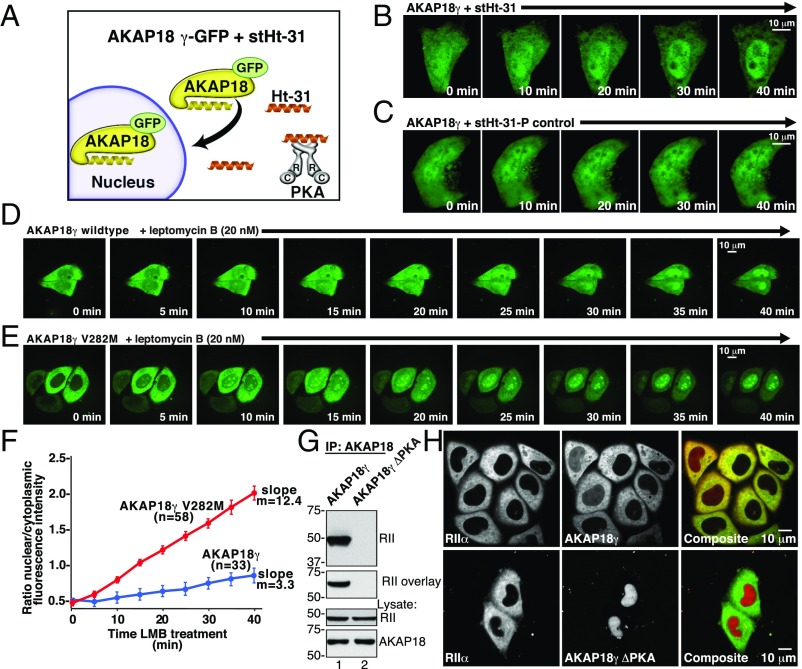
Fig. 3.
Association with the PKA holoenzyme is necessary for cytoplasmic retention of AKAP18 long forms. (A) Schematic depicting displacement of PKA from AKAP18γ by the cell-soluble PKA-anchoring disruptor peptide (stHt-31, orange) and the effect on subcellular targeting of AKAP18 long isoforms. Selected time points are shown from 0 to 40 min. (B and C) Live-cell imaging of cells expressing AKAP18γ-YFP following application of (B) stHt-31 or (C) the proline control peptide, stHt-31-P. (D and E) Live-cell imaging of cells expressing (D) WT AKAP18γ-GFP or (E) V282M AKAP18γ-GFP. Cells were treated with LMB and subcellular distribution was recorded over 40 min. (F) Amalgamated data from multiple experiments in D and E. Changes in the ratio of nuclear/cytoplasmic fluorescent signal WT AKAP18γ-GFP (n = 33 cells) and V282M AKAP18γ-GFP (n = 58 cells). (G) The PKA-binding deficient mutant AKAP18γ S278P/L281P (AKAP18γ ΔPKA) is unable coprecipitate RII or interact with it by solid-phase overlay. (H) Fluorescent confocal images of HeLa cells expressing RIIα-YFP (green) and (Lower) mCherry-AKAP18γ (red) or (Bottom) the PKA-deficient AKAP18γ mutant.
Leptomycin B (LMB) is a bacterial metabolite that blocks CRM1-dependent nuclear export in mammalian cells. This compound traps proteins that have the capacity to translocate into the nucleus (45). Hence, we reasoned that the diminished capacity of AKAP18γ V282M to anchor PKA would render this variant more prone to nuclear translocation. Live-cell imaging of HeLa cells treated with LMB (20 nM) monitored nuclear accumulation of GFP-tagged AKAP18γ adducts (Fig. 3 D and E). Nuclear accumulation rates were calculated by quantifying fluorescent intensity ratios between the nucleus and cytoplasm over a time course of 40 min (Fig. 3F). Minimal movement of the WT anchoring protein was detected (Fig. 3 D and F, blue) (n = 33 cells). In contrast, nuclear accumulation of AKAP18γ V282M was pronounced (Fig. 3 G and H, red) (n = 58 cells) and occurred at a rate that is 3.75 times faster than the WT anchoring protein (Fig. 3F). Considering these data in light of the experimental evidence presented in Fig. 2, we propose that a reduced affinity for PKA contributes to the transit of AKAP18γ V282M into the nucleus. Independent support for this hypothesis was provided by evidence that AKAP18γ S278P/L281P, a PKA binding-null mutant, was exclusively detected in the nucleus (Fig. 3 G and H). Control experiments confirmed that WT AKAP18γ associates with PKA, and is retained in the cytoplasm under these conditions (Fig. 3 G and H, Upper).
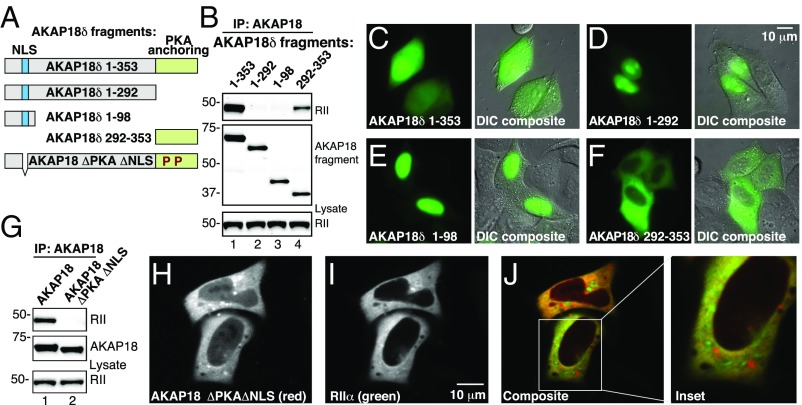
The long isoforms of AKAP18 (γ and δ) encode a putative nuclear localization signal (NLS) within exon 3 of the AKAP7 gene (46). Therefore, a potential explanation for the results in Fig. 3 is that PKA anchoring obscures this NLS. To test this hypothesis, a family of deletion constructs was generated where regions of the AKAP18δ were fused to YFP (Fig. 4A). In situ interaction with PKA was confirmed by coprecipitation of RII (Fig. 4B). The subcellular location of each YFP-tagged fragment was established by fluorescence microscopy (Fig. 4 C–F). Differential interference contrast (DIC) microscopy outlined cell contours (Fig. 4 C–F). These studies reveal that residues 1–98 of the anchoring protein are necessary and sufficient for nuclear targeting (Fig. 4E). To further demonstrate that the NLS is sufficient to direct AKAP18 into the nucleus in in the absence of PKA, we deleted the 8-amino acid KKRKKKRK sequence and introduced proline mutations that perturb the RII-binding helix (AKAP18γ ΔPKA ΔNLS) (Fig. 4 A and G, Top and Middle, lane 2). Fluorescent imaging revealed that both AKAP18γ ΔPKA ΔNLS and RIIα are retained within the cytoplasm (Fig. 4 H and I). Within an enlarged region of interest, it was apparent that the AKAP18γ ΔPKA ΔNLS (red) and RIIα (green) signals exhibit distinct but overlapping cytoplasmic distributions (Fig. 4J, Inset). This is consistent with the ability of RIIα to be incorporated into a spectrum of AKAP–PKA holoenzyme complexes dispersed throughout the cytoplasm (38, 47–50). Taken together, the results in Fig. 4 indicate that cytoplasmic retention of native AKAP18γ or -δ proceeds through a mechanism in which binding to the PKA holoenzyme masks a nuclear localization signal.
Fig. 4.
Long isoforms of AKAP18 contain a nuclear localization signal that may be masked by interaction with PKA. (A) Schematic showing fragments of YFP-tagged AKAP18δ isoforms that were used to map targeting regions within the anchoring protein. The first and last residue of each AKAP18 fragment is indicated. A polybasic NLS (blue) and PKA-anchoring regions (green) are identified. (B) IP of YFP-tagged AKAP18δ fragments. (Top) Western blot analysis of immune complexes shows PKA RII coprecipitating with full-length protein and the 292–353 fragment. (Middle) Loading control for AKAP18 fragments and (Bottom) loading controls for RIIα. (C–F) Fluorescent and DIC imaging detecting the differential subcellular distribution YFP-tagged AKAP18δ fragments. (G) Characterization of WT AKAP18γ and AKAP18γ ΔPKA ΔNLS mutant immune complexes. (Top) RII coprecipitates with the WT only. (Middle) Loading control for AKAP18. (Bottom) loading control for RIIα. (H) Fluorescent confocal image of the mCherry-tagged AKAP18γ ΔPKA ΔNLS mutant shows cytoplasmic retention. (I) RIIα-YFP is also cytoplasmic. (J) Single channel and composite images show both an overlapping and distinct cytoplasmic distribution of RII (green) and AKAP18γ ΔPKA ΔNLS (red). (Inset) Magnified image showing the regional distribution of RII and AKAP18.
Proximity Labeling and Proteomics Define Pools of AKAP18 Complexes.
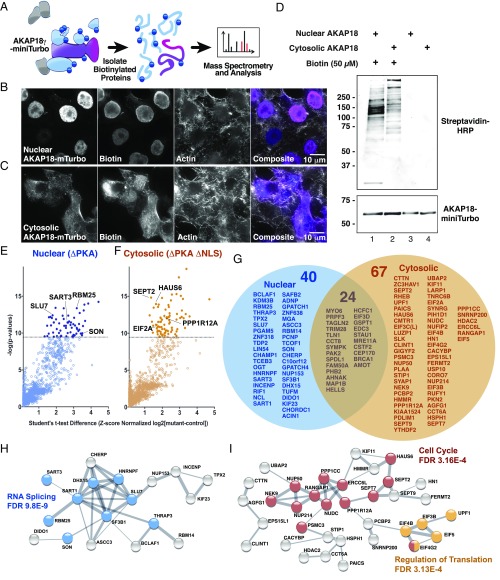
A corollary of these findings is that cytoplasmic and nuclear pools of AKAP18 interact with compartment-specific binding partners. To address this question, we tagged nuclear and cytoplasmic AKAP18 variants with miniTurbo, an engineered promiscuous ligase that permits local and efficient biotinylation of proteins within a radius of ∼5–10 nm (51). Tagged proteins are captured on NeutrAvidin beads and identified by MS (Fig. 5A). The intracellular location of nuclear and cytoplasmic AKAP18-miniTurbo was determined by immunofluorescence detection of a V5 tag (Fig. 5 B and C). In situ detection of biotinylated target proteins was assessed by immunofluorescence using NeutrAvidin-Rhodamine Red X dye (Fig. 5 B and C). Immunoblot analysis using streptavidin-HRP revealed distinct staining patterns for the nuclear and cytoplasmic pools of AKAP18-binding partners (Fig. 5D, Upper, lanes 1 and 2). Immunoblot detection of the AKAP18 miniTurbo constructs served as a loading control (Fig. 5D, Lower). MS identified 131 binding partners of AKAP18 (Fig. 5 E–G). Forty of these proteins preferentially interacted with the nuclear variant (Fig. 5G, blue), whereas 67 proteins bound only to the cytoplasmic form (Fig. 5G, orange). Network analysis suggests that nuclear AKAP18 isoforms associate with RNA splicing machinery (Fig. 5H). Cytoplasmic forms of AKAP18 interact with proteins involved in translational control and cell cycle progression (Fig. 5I). Taken together, these proximity-labeling results imply that AKAP18 isoforms associate with compartment-selective biding partners to participate in different cellular processes.
Fig. 5.
Proximity labeling and proteomic identification of compartment-selective AKAP18 interactors. (A) Schematic depicting the miniTurbo-based biotinylation/MS experiments. Biotin is shown as dark-blue circles. (Left) Upon biotin addition, miniTurbo biotinylates only proximal proteins. (Center) Biotinylated proteins are isolated with streptavidin pulldown and (Right) identified by MS. (B and C) Fluorescence microscopy depicting the subcellular location of (B) nuclear and (C) cytoplasmic AKAP18-miniTurbo variants and detection of compartment specific biotinylated proteins. (D) Western blot analysis of biotinylated proteins in cell lysates shows distinct and compartment-specific labeling patterns. (E and F) MS results plotted as log difference of experimental intensity minus control intensity plotted against the negative log of the P value. Selected nuclear (dark-blue) and cytoplasmic (tan) AKAP18 interacting partners are indicated. (G) Lists of statistically significant target proteins in nuclear and cytosolic preparations. Of 131 identified proteins, 40 were selectively associated with the nuclear variant, whereas 67 preferentially bound to the cytoplasmic variant. (H and I) STRING database-generated network depictions of compartment-specific proteins. Gene ontology process annotation and false-discovery rates (FDR) are listed for the highlighted nodes. Putative biological functions for each subnetwork are indicated.
Discovery of the Non-PKA–Anchoring AKAP18 Epsilon.
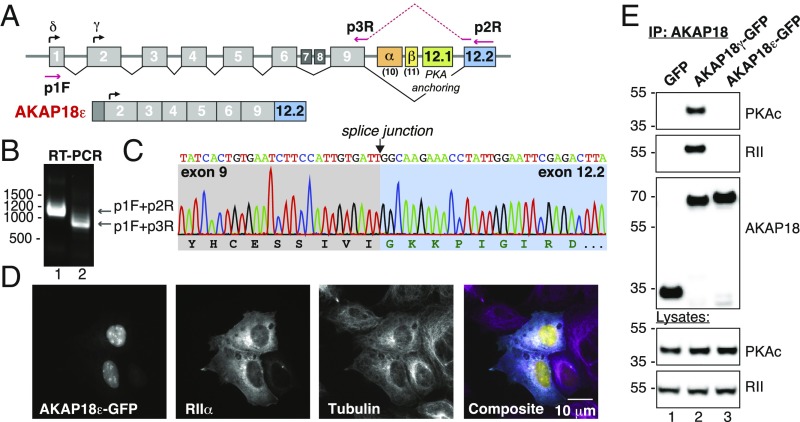
As outlined in Fig. 2A, numerous transcripts emanate from the AKAP7 gene. Scrutiny of the TCGA SpliceSeq database exposed a previously unidentified splicing event that bypasses the PKA-anchoring helix and replaces this region with a cryptic exon from the 3′UTR (Fig. 6A, blue). To amplify this transcript, we performed PCR on pooled human reverse-transcribed cDNA using specific primers (Fig. 6B, lane 1). PCR was also conducted with a 3′ primer that bridged sequences in exon 9 and exon 12.2 (p3R) (Fig. 6B, lane 2). Nucleotide sequencing of the cloned product revealed an ORF that encodes a non-PKA-anchoring isoform of 329 amino acids with an alternate C terminus (Fig. 6D, blue shading). We have named this variant AKAP18ε. When expressed in HEK293 cells, AKAP18ε−GFP is only detected in the nucleus (Fig. 6D). In contrast RIIα and α-tubulin are exclusively cytoplasmic (Fig. 6D). Biochemical characterization of AKAP18ε immune complexes isolated from HEK293 cells demonstrate that this isoform does not interact with either RIIα or Cα subunits (Fig. 6E, Top and Middle-mid panels, lane 3). Control experiments confirmed both PKA subunits were detected in AKAP18γ immune complexes (Fig. 6E, Top and Middle-mid panels, lane 2). Immunoblot detection of both PKA subunits in cell lysates served as loading controls (Fig. 6E, Bottom). Taken together these studies indicate that at least one nuclear isoform of AKAP18 does not function as a PKA-anchoring protein.
Fig. 6.
Identification of an AKAP18 isoform that lacks a PKA binding helix. (A) AKAP7 intron–exon map as adapted from TCGA SpliceSeq. The start sites for the δ- and γ-isoforms are noted, as are the α- and β-isoform-specific exons. The RII anchoring site (green) is encoded in exon 12.1 and the alternate 12.2 exon (blue) are presented. The locations of the primers used for RT-PCR are marked. (B) RT-PCR from pooled human cDNA samples using primers against predicted transcripts and novel exon junctions produces the expected products where intervening exons are spliced out. (C) Sequencing chromatogram of the exon 9–12.2 splice junction shows an in-frame protein sequence corresponding to a distinct C terminus. (D) Fluorescent confocal images showing nuclear localization of EmGFP-tagged AKAP18ε and cytoplasmic labeling of RIIα-V5 and α-tubulin. (E) Immune complexes of either WT AKAP18γ or AKAP18ε were immunoprecipitated and copurification of PKA catalytic (Upper) and RII (Upper-mid) subunits were identified by Western blotting. Expression levels of both AKAP18 forms and GFP are indicated in the Lower three panels.
Discussion
Genome-wide association metaanalyses have identified SNPs and point mutations in AKAPs from patients with neurological disorders, hypertension, cardiac arrhythmias, and certain cancers (14, 52, 53). In this report we investigate SNPs within exons that encode the PKA-binding domains of several AKAPs. These functionally conserved regions form amphipathic helices that bind PKA holoenzymes with high affinity (11). In total we screened 42 missense variants gleaned from a variety of patient-derived datasets. In vitro binding studies confirmed that half of these single amino acid substitutions disrupted PKA anchoring. Further scrutiny of AKAP-Lbc, AKAP220, and AKAP-KL sequences reveal that a majority of detrimental substitutions occur within the penultimate and final turns of the anchoring helix (Fig. 1). Structural studies corroborate that these hydrophobic side-chains interface with the D/D of the R subunits of PKA (19, 54).
Nonsynonymous SNPs in the AKAP7 gene have been indirectly linked to an increased incidence of febrile seizures, cardiovascular events, and autism spectrum disorders (14, 26, 55, 56). This provided an impetus for more extensive analysis of the valine-to-methionine variant in the PKA-anchoring helix. Molecular models presented in Fig. 2 F and G show that the extra molecular density of a methionine at position 282 in AKAP18γ deflects the helical pitch of the hydrophobic anchoring surface upward by 6.6°. This relatively modest topological flaw decreases the binding affinity for RII by 8.8-fold and impairs PKA-mediated potentiation of L-type Ca2+ currents, as shown in Fig. 2H. Because alternative splicing of the AKAP7 gene generates multiple isoforms, Met-282 polymorphic variants might negatively impact a certain local PKA signaling events. These include modulation of neuronal sodium channels, regulation of SERCA2-mediated calcium reuptake in the heart, and aquaporin-2–mediated control of water homeostasis in the kidney (38, 39, 57). The physiological significance to this concept is underscored by recent evidence that intact and anchored PKA holoenzymes are the predominant cytoplasmic signaling units and are sequestered within 200–400 Å of their preferred substrates (21, 22). Hence, AKAP variants that impact the intracellular targeting of PKA holoenzymes have the capacity to alter the dissemination of second-messenger signals.
Although polymorphic variation within AKAPs has the potential to uncouple local cAMP signaling, there is little information about the allelic frequency or prevalence in specific populations. In addition, scrutiny of the cBioPortal cancer genomics data interface does not reveal any clear pattern of AKAP-anchoring helix mutations that correlate with cancers. Perhaps the best-documented example of genetic variation comes from analysis of a nonsynonymous SNP that promotes an isoleucine to valine substitution at position 646 within the PKA-anchoring domain of D-AKAP-2 (AKAP10) (12, 58). This variant is associated with cardiac dysfunction in aging populations of European-American individuals that display a shortened electrocardiogram PR interval (12). The D-AKAP2 I646V variant is also associated with increased basal heart rate (58). Because basal heart rate correlates inversely with lifespan, the altered PKA-anchoring properties of D-AKAP2 may be linked to adverse pathological outcomes (59). However, the downstream signaling pathways affected by this modified kinase-anchoring event remain obscure. In vitro biochemical studies show that the D-AKAP2 646V variant binds to type I PKA holoenzyme with threefold higher affinity than D-AKAP2 646I, yet this variation has no effect on type II PKA anchoring (12). The physiological ramifications of these relatively small changes in affinity may be limited as D-AKAP2 exhibits a strong binding preference for type II PKA holoenzymes (60, 61). Furthermore, the porcine, murine, and chicken orthologs of D-AKAP2 have a valine at the corresponding position in the PKA-anchoring helix (12). While structural analyses of these D-AKAP2 orthologs are scant, it seems that the Ile to Val substitution can be tolerated with minimal effects on PKA anchoring. This raises the issue of whether or not silent variation in AKAP helices is a more general phenomenon. Data presented in Fig. 1F indicate that a silent change in the final turn of the AKAP79 helix (Ile-406 to Val) has no apparent effect on RII-binding. This may represent another example of evolutionary drift, as valine naturally occurs at this position in the mouse, rat, and macaque orthologs of AKAP79. Thus, malleability within amphipathic helices can accommodate structurally conserved variants or passenger mutations without any substantial impact on PKA anchoring.
AKAPs are sequestered within defined subcellular locations by specialized targeting motifs that interact with membranes or organelles (62–65). The α- and β-isoforms of AKAP18 were originally identified as membrane proximal lipid-modified PKA-anchoring proteins that facilitate cAMP-responsive modulation of ion channels (36, 66). In contrast the longer γ- and δ-isoforms of AKAP18 are retained within the cytoplasm through different targeting mechanisms (37, 39). Studies presented in Figs. 3–5 argue that interaction with the PKA holoenzyme influences the cytoplasmic distribution of AKAP18 long isoforms. This unique mode of AKAP targeting is supported by time-lapse imaging experiments in Fig. 3B, showing that application of PKA anchoring-disruptor peptides facilitates nuclear accumulation of AKAP18γ. This may be an AKAP18-selective phenomenon, because other anchoring proteins remain firmly attached to their targeting sites upon displacement of PKA. For example, delivery of anchoring inhibitor peptides has no effect on the mitochondrial location of D-AKAP-1 and OPA1 or the membrane tethering of AKAP79/150 and Gravin (47, 63, 67, 68). However, one discriminating feature may be the relatively small size of AKAP18, which is within the molecular dimensions of particles (30–60 kDa) that diffuse through the nuclear pore (69). With this in mind, we took advantage of the reduced PKA-anchoring affinity of the AKAP18γ V282M variant to show that the rate of nuclear accumulation is enhanced 3.8-fold compared with WT in the presence of LMB. Thus, paradoxically we can propose that intact PKA holoenzymes serve as a cytoplasmic anchor for certain long isoforms of AKAP18. Molecular explanations for this alternative targeting mechanism include the dimensions of the AKAP18-RII2-C2 pentameric complex (∼230,000 kDa), which is too large to transit the nuclear pore and that R subunit dimers are only detected in the cytoplasmic compartment (22). In contrast, free catalytic subunits only transit into the nucleus in response to pharmacological regimens that promote supraphysiological accumulation of cAMP or in adrenal adenomas where pathological Cα mutants are excluded from PKA holoenzymes (70–72).
Our demonstration of AKAP18γ nuclear shuttling and the subsequent discovery of AKAP18ε further expands our view of AKAP biology. For example, PKA anchoring contributes to the cytoplasmic retention of AKAP18 by masking a classic NLS that is located between residues 52–59. Furthermore, data in Fig. 6 characterizing the AKAP18ε isoform that lacks a PKA-anchoring helix, infer that this splice variant utilizes this NLS as its sole targeting signal. In both cases this would permit direct transport through the nuclear pore by targeting factors, such as Ran GTPase and importin (73). In addition, enzyme-catalyzed proximity-dependent proteomic screens substantiate that cytoplasmic and nuclear AKAP18s coordinate distinct interactomes. Network analyses presented in Fig. 5 H and I argue that cytoplasmic AKAP18 variants interface with RNA transport and translation machinery, whereas their nuclear counterparts facilitate signaling to spliceosomal components. Although individual nuclear functions for AKAP18γ, -δ, and -ε have yet to be ascribed, X-ray crystallography identified a common 2′-3′ phosphoesterase-like fold within each isoform (74, 75). One notable feature was the detection of 5′-AMP embedded deep within a grove of this shared domain. This degradation product of cAMP is coordinated by two His-X-Thr motifs that are hallmarks of the 2H-phosphoesterase–like fold (74). Postulated roles for this nucleotide–protein interaction include energy sensing or as an auxiliary nuclear localization element akin to ligand mediated targeting of steroid hormone receptors (74). Another important concept investigated in this study is that non-PKA–anchoring products can be transcribed from AKAP genes. On the surface, this notion may seem counterintuitive to the AKAP model (15, 76). However, omission of the PKA-anchoring helix could favor formation of AKAP18 macromolecular assemblies that respond to different activation signals (11, 43). This latter issue may have confounded analysis of AKAP18 knockout mice, as only an exon encoding the PKA-anchoring region was ablated from the AKAP7 gene (77).
Methods
Sequence Analysis and Variant Identification.
The variants described here were captured at several different points in time as new SNPs were deposited into the NCBI dbSNP and NCBI Variation Viewer databases.
Peptide Array Synthesis.
SPOT arrays were generated as described previously (60).
Live Cell Imaging.
HeLa cells expressing AKAP18-GFP constructs were washed in HBSS, mounted in a Ludin chamber (Life Imaging Services), and imaged using a Leica AS-MDW workstation. For nuclear trapping experiments, LMB (Calbiochem) was added to 20 nM and images were captured every 5 min for at least 90 min.
Structural Modeling with AutoDock.
AutoDock calculations were carried out using AutoDock Tools 1.5.6, according to the methods described in ref. 42.
Electrophysiology.
HEK293 cells were transfected with cDNAs encoding the α1c and β2a subunits of cardiac L-type Ca2+ channels (36) and WT or V37M AKAP18α-GFP. Whole-cell recordings were performed using an Axopatch 200B patch-clamp amplifier (Axon Instruments). L-type calcium channel currents were evoked step depolarization (200 ms) from a holding potential of –80 mV to +10 mV.
Calcium Imaging.
Male WT C57BL/6J mice of 5 to 8 wk of age were euthanized with sodium pentobarbital (250 mg/kg; intraperitoneally), as approved by the University of California, Davis Animal Care and Use Committee (protocol #20321). Mouse aorta were dissected, washed, and digested in DMEM solution containing 2.2 mg/mL of collagenase type 2 (Worthington). Unpassaged arterial myocytes were seeded on glass coverslips coated with laminin and kept in an incubator at 37 °C with 5% CO2 for 7–10 d before transfection. Transfected arterial myocytes were loaded with Fluo4-AM for 20 min at room temperature. Images were collected using an Andor spinning-disk confocal system coupled to an Olympus iX-81 inverted microscope equipped with an Olympus 40× oil immersion lens (NA = 0.75).
Proximity Labeling and Mass Spectrometry.
The cDNA for miniTurbo (51) was fused to 3′ end of AKAP18γ cDNAs and tested for expression in HEK293T cells. Transfected cells were incubated in media containing 50 µM biotin for 1 h. After lysate preparation and biotinylated target capture, proteins were reduced and alkylated digested using Lys-C (Wako) followed by further trypsin digestion. Peptides were analyzed by nanoLC-MS on an Orbitrap Fusion Lumos Tribrid Mass Spectrometer. Protein network prediction and gene ontology analysis were performed using STRING database version 10.5 (78). Networks were visualized using Cytoscape v3.6.1 software.
Complete methods can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Darren L. Beene and Laura Gabrovsek for technical assistance; current members of the J.D.S. laboratory for thoughtful ideas and critical discussions; and Melanie Milnes for administrative support. This work was supported by National Institutes of Health Grants 5R01DK105542 and 1R01DK119192 (to J.D.S.) and R01HL098200 and R01HL121059 (to M.F.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816614115/-/DCSupplemental.
References
- 1.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grabbe C, Dikic I. Cell biology. Going global on ubiquitin. Science. 2008;322:872–873. doi: 10.1126/science.1166845. [DOI] [PubMed] [Google Scholar]
- 4.Glusman G, et al. Mapping genetic variations to three-dimensional protein structures to enhance variant interpretation: A proposed framework. Genome Med. 2017;9:113. doi: 10.1186/s13073-017-0509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Druker BJ, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 7.Klaeger S, et al. The target landscape of clinical kinase drugs. Science. 2017;358:eaan4368. doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 9.Scott JD, Pawson T. Cell signaling in space and time: Where proteins come together and when they’re apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Good MC, Zalatan JG, Lim WA. Scaffold proteins: Hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langeberg LK, Scott JD. Signalling scaffolds and local organization of cellular behaviour. Nat Rev Mol Cell Biol. 2015;16:232–244. doi: 10.1038/nrm3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kammerer S, et al. Amino acid variant in the kinase binding domain of dual-specific A kinase-anchoring protein 2: A disease susceptibility polymorphism. Proc Natl Acad Sci USA. 2003;100:4066–4071. doi: 10.1073/pnas.2628028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esseltine JL, Scott JD. AKAP signaling complexes: Pointing towards the next generation of therapeutic targets? Trends Pharmacol Sci. 2013;34:648–655. doi: 10.1016/j.tips.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suryavanshi SV, Jadhav SM, McConnell BK. Polymorphisms/mutations in A-kinase anchoring proteins (AKAPs): Role in the cardiovascular system. J Cardiovasc Dev Dis. 2018;5:E7. doi: 10.3390/jcdd5010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott JD, Dessauer CW, Taskén K. Creating order from chaos: Cellular regulation by kinase anchoring. Annu Rev Pharmacol Toxicol. 2013;53:187–210. doi: 10.1146/annurev-pharmtox-011112-140204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musheshe N, Schmidt M, Zaccolo M. cAMP: From long-range second messenger to nanodomain signalling. Trends Pharmacol Sci. 2018;39:209–222. doi: 10.1016/j.tips.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Scott JD, Carr DW. Subcellular localization of the type II cAMP-dependent protein kinase. News Physiol Sci. 1992;7:143–148. [Google Scholar]
- 18.Carr DW, et al. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J Biol Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- 19.Gold MG, et al. Molecular basis of AKAP specificity for PKA regulatory subunits. Mol Cell. 2006;24:383–395. doi: 10.1016/j.molcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Kinderman FS, et al. A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol Cell. 2006;24:397–408. doi: 10.1016/j.molcel.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith FD, et al. Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. eLife. 2013;2:e01319. doi: 10.7554/eLife.01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith FD, et al. Local protein kinase A action proceeds through intact holoenzymes. Science. 2017;356:1288–1293. doi: 10.1126/science.aaj1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nygren PJ, et al. Intrinsic disorder within AKAP79 fine-tunes anchored phosphatase activity toward substrates and drug sensitivity. eLife. 2017;6:e30872. doi: 10.7554/eLife.30872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: Lessons from PKA. Nat Rev Mol Cell Biol. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logue MW, et al. Alzheimer Disease Genetics Consortium Two rare AKAP9 variants are associated with Alzheimer’s disease in African Americans. Alzheimers Dement. 2014;10:609–618.e611. doi: 10.1016/j.jalz.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabbout R, et al. A locus for simple pure febrile seizures maps to chromosome 6q22-q24. Brain. 2002;125:2668–2680. doi: 10.1093/brain/awf281. [DOI] [PubMed] [Google Scholar]
- 27.Means CK, et al. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc Natl Acad Sci USA. 2011;108:E1227–E1235. doi: 10.1073/pnas.1107182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whiting JL, et al. Protein kinase A opposes the phosphorylation-dependent recruitment of glycogen synthase kinase 3β to A-kinase anchoring protein 220. J Biol Chem. 2015;290:19445–19457. doi: 10.1074/jbc.M115.654822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiting JL, et al. AKAP220 manages apical actin networks that coordinate aquaporin-2 location and renal water reabsorption. Proc Natl Acad Sci USA. 2016;113:E4328–E4337. doi: 10.1073/pnas.1607745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carr DW, Stofko-Hahn RE, Fraser IDC, Cone RD, Scott JD. Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins. Characterization of AKAP 79. J Biol Chem. 1992;267:16816–16823. [PubMed] [Google Scholar]
- 31.Klauck TM, et al. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 32.Newlon MG, et al. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong F, Feldmesser M, Casadevall A, Rubin CS. Molecular characterization of a cDNA that encodes six isoforms of a novel murine A kinase anchor protein. J Biol Chem. 1998;273:6533–6541. doi: 10.1074/jbc.273.11.6533. [DOI] [PubMed] [Google Scholar]
- 34.Gold MG, et al. AKAP2 anchors PKA with aquaporin-0 to support ocular lens transparency. EMBO Mol Med. 2012;4:15–26. doi: 10.1002/emmm.201100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray PC, Tibbs VC, Catterall WA, Murphy BJ. Identification of a 15-kDa cAMP-dependent protein kinase-anchoring protein associated with skeletal muscle L-type calcium channels. J Biol Chem. 1997;272:6297–6302. doi: 10.1074/jbc.272.10.6297. [DOI] [PubMed] [Google Scholar]
- 36.Fraser ID, et al. A novel lipid-anchored A-kinase anchoring protein facilitates cAMP-responsive membrane events. EMBO J. 1998;17:2261–2272. doi: 10.1093/emboj/17.8.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trotter KW, et al. Alternative splicing regulates the subcellular localization of A-kinase anchoring protein 18 isoforms. J Cell Biol. 1999;147:1481–1492. doi: 10.1083/jcb.147.7.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lygren B, et al. AKAP complex regulates Ca2+ re-uptake into heart sarcoplasmic reticulum. EMBO Rep. 2007;8:1061–1067. doi: 10.1038/sj.embor.7401081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henn V, et al. Identification of a novel A-kinase anchoring protein 18 isoform and evidence for its role in the vasopressin-induced aquaporin-2 shuttle in renal principal cells. J Biol Chem. 2004;279:26654–26665. doi: 10.1074/jbc.M312835200. [DOI] [PubMed] [Google Scholar]
- 40.Götz F, et al. AKAP18:PKA-RIIα structure reveals crucial anchor points for recognition of regulatory subunits of PKA. Biochem J. 2016;473:1881–1894. doi: 10.1042/BCJ20160242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forli S, et al. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraser ID, Scott JD. Modulation of ion channels: A “current” view of AKAPs. Neuron. 1999;23:423–426. doi: 10.1016/s0896-6273(00)80795-3. [DOI] [PubMed] [Google Scholar]
- 43.Hoshi N, Langeberg LK, Gould CM, Newton AC, Scott JD. Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol Cell. 2010;37:541–550. doi: 10.1016/j.molcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson KR, Nicodemus-Johnson J, Carnegie GK, Danziger RS. Molecular evolution of A-kinase anchoring protein (AKAP)-7: Implications in comparative PKA compartmentalization. BMC Evol Biol. 2012;12:125. doi: 10.1186/1471-2148-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudo N, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown RL, August SL, Williams CJ, Moss SB. AKAP7gamma is a nuclear RI-binding AKAP. Biochem Biophys Res Commun. 2003;306:394–401. doi: 10.1016/s0006-291x(03)00982-3. [DOI] [PubMed] [Google Scholar]
- 47.Pidoux G, et al. Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J. 2011;30:4371–4386. doi: 10.1038/emboj.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diviani D, Langeberg LK, Doxsey SJ, Scott JD. Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr Biol. 2000;10:417–420. doi: 10.1016/s0960-9822(00)00422-x. [DOI] [PubMed] [Google Scholar]
- 49.Smith FD, et al. AKAP-Lbc enhances cyclic AMP control of the ERK1/2 cascade. Nat Cell Biol. 2010;12:1242–1249. doi: 10.1038/ncb2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marx SO, et al. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 51.Branon TC, et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reggi E, Diviani D. The role of A-kinase anchoring proteins in cancer development. Cell Signal. 2017;40:143–155. doi: 10.1016/j.cellsig.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Chen L, Kass RS, Dessauer CW. The A-kinase anchoring protein Yotiao facilitates complex formation between adenylyl cyclase type 9 and the IKs potassium channel in heart. J Biol Chem. 2012;287:29815–29824. doi: 10.1074/jbc.M112.380568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banky P, et al. Isoform-specific differences between the type Ialpha and IIalpha cyclic AMP-dependent protein kinase anchoring domains revealed by solution NMR. J Biol Chem. 2000;275:35146–35152. doi: 10.1074/jbc.M003961200. [DOI] [PubMed] [Google Scholar]
- 55.Ramirez AH, et al. Novel rare variants in congenital cardiac arrhythmia genes are frequent in drug-induced torsades de pointes. Pharmacogenomics J. 2013;13:325–329. doi: 10.1038/tpj.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poelmans G, Franke B, Pauls DL, Glennon JC, Buitelaar JK. AKAPs integrate genetic findings for autism spectrum disorders. Transl Psychiatry. 2013;3:e270. doi: 10.1038/tp.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Catterall WA. Ion channel voltage sensors: Structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tingley WG, et al. Gene-trapped mouse embryonic stem cell-derived cardiac myocytes and human genetics implicate AKAP10 in heart rhythm regulation. Proc Natl Acad Sci USA. 2007;104:8461–8466. doi: 10.1073/pnas.0610393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neumann SA, et al. AKAP10 (I646V) functional polymorphism predicts heart rate and heart rate variability in apparently healthy, middle-aged European-Americans. Psychophysiology. 2009;46:466–472. doi: 10.1111/j.1469-8986.2009.00802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herberg FW, Maleszka A, Eide T, Vossebein L, Tasken K. Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J Mol Biol. 2000;298:329–339. doi: 10.1006/jmbi.2000.3662. [DOI] [PubMed] [Google Scholar]
- 61.Alto NM, et al. Bioinformatic design of A-kinase anchoring protein-in silico: A potent and selective peptide antagonist of type II protein kinase A anchoring. Proc Natl Acad Sci USA. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dell’Acqua ML, Faux MC, Thorburn J, Thorburn A, Scott JD. Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4, 5-bisphosphate. EMBO J. 1998;17:2246–2260. doi: 10.1093/emboj/17.8.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J Biol Chem. 1997;272:8057–8064. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- 64.Hehnly H, et al. A mitotic kinase scaffold depleted in testicular seminomas impacts spindle orientation in germ line stem cells. eLife. 2015;4:e09384. doi: 10.7554/eLife.09384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dodge-Kafka KL, et al. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gray PC, et al. Primary structure and function of an A kinase anchoring protein associated with calcium channels. Neuron. 1998;20:1017–1026. doi: 10.1016/s0896-6273(00)80482-1. [DOI] [PubMed] [Google Scholar]
- 67.Nauert JB, Klauck TM, Langeberg LK, Scott JD. Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffold protein. Curr Biol. 1997;7:52–62. doi: 10.1016/s0960-9822(06)00027-3. [DOI] [PubMed] [Google Scholar]
- 68.Snyder EM, et al. Role for A kinase-anchoring proteins (AKAPS) in glutamate receptor trafficking and long term synaptic depression. J Biol Chem. 2005;280:16962–16968. doi: 10.1074/jbc.M409693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Timney BL, et al. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol. 2016;215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koschinski A, Zaccolo M. Activation of PKA in cell requires higher concentration of cAMP than in vitro: Implications for compartmentalization of cAMP signalling. Sci Rep. 2017;7:14090. doi: 10.1038/s41598-017-13021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berthon AS, Szarek E, Stratakis CA. PRKACA: The catalytic subunit of protein kinase A and adrenocortical tumors. Front Cell Dev Biol. 2015;3:26. doi: 10.3389/fcell.2015.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mo GC, et al. Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat Methods. 2017;14:427–434. doi: 10.1038/nmeth.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rout MP, Aitchison JD. The nuclear pore complex as a transport machine. J Biol Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 74.Gold MG, Smith FD, Scott JD, Barford D. AKAP18 contains a phosphoesterase domain that binds AMP. J Mol Biol. 2008;375:1329–1343. doi: 10.1016/j.jmb.2007.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bjerregaard-Andersen K, Østensen E, Scott JD, Taskén K, Morth JP. Malonate in the nucleotide-binding site traps human AKAP18γ/δ in a novel conformational state. Acta Crystallogr F Struct Biol Commun. 2016;72:591–597. doi: 10.1107/S2053230X16010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong W, Scott JD. AKAP signalling complexes: Focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 77.Jones BW, et al. Cardiomyocytes from AKAP7 knockout mice respond normally to adrenergic stimulation. Proc Natl Acad Sci USA. 2012;109:17099–17104. doi: 10.1073/pnas.1215219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Szklarczyk D, et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.