Abstract
Introduction:
The development and application of new molecular diagnostic assays based on next-generation sequencing and proteomics require improved methodologies for procurement of target cells from histological sections. Laser microdissection can successfully isolate distinct cells from tissue specimens based on visual selection for many research and clinical applications. However, this can be a daunting task when a large number of cells are required for molecular analysis or when a sizeable number of specimens need to be evaluated.
Materials and Methods:
To improve the efficiency of the cellular identification process, we describe a microdissection workflow that leverages recently developed and open source image analysis algorithms referred to as computer-aided laser dissection (CALD). CALD permits a computer algorithm to identify the cells of interest and drive the dissection process.
Results:
We describe several “use cases” that demonstrate the integration of image analytic tools probabilistic pairwise Markov model, ImageJ, spatially invariant vector quantization (SIVQ), and eSeg onto the ThermoFisher Scientific ArcturusXT and Leica LMD7000 microdissection platforms.
Conclusions:
The CALD methodology demonstrates the integration of image analysis tools with the microdissection workflow and shows the potential impact to clinical and life science applications.
Keywords: Image analysis, microdissection, probabilistic pairwise Markov model, SIVQ
INTRODUCTION
Laser microdissection facilitates procurement of a phenotypically-defined cell population(s) from a tissue section or cytology slide. Currently, there are four commercially available microdissection devices and each instrument employs a slightly different mechanism for dissection.[1] The ThermoFisher Scientific ArcturusXT instrument is a dual laser system with an infrared (IR) laser to capture single cell targets onto a film and an ultraviolet (UV) laser to cut out large tissue areas. Alternatively, the Leica LMD7000 device employs only an UV laser to cut out target areas in the tissue, dropping the isolated area by gravity into a capture vessel. While both the ThermoFisher Scientific and Leica equipment have optional rudimentary image processing packages (AutoScan and AVC, respectively) to aid the microdissection process, these tools have had limited utility. Regardless of the method or machine used, laser dissection has significantly impacted the biomedical and life science research fields by highlighting the complex and heterogeneous pathophysiology that exists within tissues and providing a new tool for interrogation. To date, there are over 4000 peer-reviewed publications in the literature using laser dissection covering a range of topics, including the tumor microenvironment, neuroscience and brain studies, plant genomics, and analysis of infectious organisms and host response.[2,3,4,5,6,7,8]
However, the time and labor required to identify and isolate specific cells at high precision is a tedious user-driven process across all microdissection instruments and is a major drawback of the technology when high-throughput capability is required.
To alleviate this limitation, we turned to the field of digital pathology whole-slide imaging (WSI) to facilitate and streamline the microdissection process. Advances in digital imaging and pattern recognition algorithms improve the speed and accuracy of identifying features for the pathologists;[9,10,11,12] which can then be adopted to aid in the cellular identification step for the microdissection workflow. Although image-processing applications in pathology date back to the 1950s,[13] there are only a few applications that are routinely used today for patient care, and the only one that has received FDA approval, IHC quantification for clinical use (Leica Biosystems Imaging, Inc.). However, the current and newly developing analytic tools can quantify and classify histological regions of interests (ROI) more effectively than in the past, and these methods hold great potential to aid investigators, pathologists, and other clinicians in a variety of laboratory-based studies and diagnostic applications.[14,15,16,17,18,19,20] In the present manuscript, we describe an algorithm agnostic framework that incorporates computer algorithms (either individually or multiple algorithms in a cascade fashion) into the microdissection workflow [Figure 1], using both the ThermoFisher Scientific ArcturusXT and Leica LMD7000 dissection platforms for specific use cases.
Figure 1.

Schematic representation of the computer-aided laser dissection workflow
MATERIALS AND METHODS
Tissue specimens
The prostate study was performed under a University of Michigan IRB approved protocol and a material transfer agreement with the NIH. Formalin-fixed, paraffin-embedded (FFPE) tissue/cell block specimens were sectioned at a thickness of 5–7 μ. All specimens were anonymized. The mouse kidney study was performed under the University of Arkansas for Medical Sciences IRB approved protocol. For additional information regarding specimen handling, processing and sectioning, please see Hanson et al. and Eberle et al.[21,22]
Histology slides
Three types of slides were used for the use cases. For the ArcturusXT experiments, positive charged glass (Superfrost Plus microscope slides, Thermo Scientific); Arcturus PEN Membrane Glass Slides (ThermoFisher Scientific); or Arcturus PEN Membrane Frame Slides (ThermoFisher Scientific). For the Leica LMD7000, Frame Slides with PET membrane, 1.4 μm (Leica Microsystems); or glass slides with membrane, 1.4 μm (Leica Microsystems).
Specimen staining
Staining was performed as described previously for hematoxylin and eosin staining.[1]
For immunohistochemical staining, sections were dewaxed and rehydrated through xylenes and graded alcohols. Heat-induced epitope retrieval (HIER) was performed using heated citrate buffer (pH 6.0) (BioGenex). The primary antibody to prostate-specific antigen (PSA) was diluted 1:50 in antibody diluent and incubated on the tissue for 30 min at room temperature. The prediluted secondary antibody (DAKO Envision + Dual Link) was incubated on the tissue for 30 min at room temperature. DAB solution was placed on the tissue for 5–10 min and the intensity of staining was observed and stopped by placing the tissue in di H2O. After staining, the tissue was dehydrated through graded alcohols and xylenes.
Imaging
Xylenes (Sigma Aldrich) or ethanol was used as a pseudo-coverslip to improve image quality by alleviating the refractive index mismatch between dry samples and objective lens on the ThermoFisher Scientific ArcturusXT device. Signature Series Cover Glass; 24 mm × 50 mm (Thermo Scientific) was placed on the tissue with xylenes or ethanol to improve the image quality. After the slide was imaged, the coverslip was removed and placed back on the stage for dissection maintaining the original orientation.
Instrumentation
ArcturusXT™ – IR and UV-cutting laser capture microdissection (LCM) instrument. Arcturus CapSure Macro LCM Caps (ThermoFisher Scientific,), QIAamp DNA Micro Kit (Qiagen) were utilized with the ArcturusXT™ Laser Microdissection Instrument (ThermoFisher Scientific) attached to a personal computer.
Leica LMD7000 – UV laser microdissection (LMD) device with max pulse energy 120 uJ, pulse frequency 10–5000 Hz, Wavelength 349 nm.
IHC – Handy Steamer (Black and Decker) was used for HIER. Leica LMD7000 Laser Microdissection System (Leica Microsystems).
Software
AutoScanXT – (ThermoFisher Scientific) An image analysis program available as an add-on to the standard ArcturusXT system. AutoScanXT allows the user to identify targets of interest versus background to enable a semi-automated microdissection experience and more precise dissections. AutoscanXT was used in the SIVQ workflow as previously described.[23]
Probabilistic pairwise Markov model (PPMM) – An algorithm that was used in the present study to identify prostate cancer on histologic images.[24,25] For access to the PPMM algorithm, please contact Dr. Anant Madabhushi (anant.madabhushi@case.edu).
ImageJ (Version 1.47g, National Institutes of Health, https://imagej.nih.gov/ij/index.html) – Images of stained cells acquired using the ArcturusXT microscope were color inverted so that the stained cells appeared bright in a dark background by using the “edit-> invert” function. The inverted images were converted to binary masks representative of the shapes of the cells with an appropriate threshold selected manually by using the “adjust-> threshold” function. The binary masks were saved as a jpeg file followed by editing the header information containing the spatial coordinates of the mask images using a Hex editor software HxD (available at http://www.download.com) so that the modified files could be imported and read by the AutoScanXT software for segmentation-guided LCM. The segmentation process was streamlined by scripting the steps described above into an ImageJ macro program and executing the script using “plugins->compile and run” function in ImageJ. (http://rsbweb.nih.gov/ij/download.html).
Spatially invariant vector quantization (SIVQ) – A pattern recognition algorithm useful for a variety of image recognition tasks and formats (remote sensing, radiology, and others).[23,25,26,27,28,29] This tool suite is publicly available for academic collaborations. For access to the SIVQ algorithm, please contact Dr. Ulysses Balis (Ulysses@med.umich.edu).
Extensible Segmentor (eSeg) – An image segmentation software system currently under development was used in the present study. For more information and access contact Dr. Donald Johann (don.johann@gmail.com).
RESULTS
The goal of the present study was to evaluate the integration of image analysis algorithms (both open-source and proprietary; utilized either individually or in a cascade) to the ThermoFisher Scientific ArcturusXT and Leica LMD7000 microdissection platforms. In the five use cases described below, we show how such algorithms can aid the speed of the cellular identification and targeting steps in the microdissection workflow.
Use Case #1: Integration of probabilistic pairwise markov model into the ArcturusXT laser capture microdissection workflow
In the first example, we evaluated the use of the PPMM[24] algorithm for prostate cancer detection, which exploits the presence of small, abnormally shaped glands with narrow lumens within the tumor; a hallmark characteristic of prostate cancer. PPMM identifies cancerous regions by evaluating the white spaces within the lumens analyzing their size, shape, and proximity to other abnormal glands.[24] The PPMM was originally created to analyze high-resolution WSI data sets.[24] However, for this study, it was adapted to work with lower resolution jpeg images generated by the ArcturusXT instrument. The ArcturusXT captures images in the jpeg format affecting the image quality and resolution, which needs to be considered and factored into future algorithm development efforts. Moreover, the add-on software (AutoScan) acts as a “bridge” for image analysis programs and needs to be addressed for successful incorporation of PPMM.
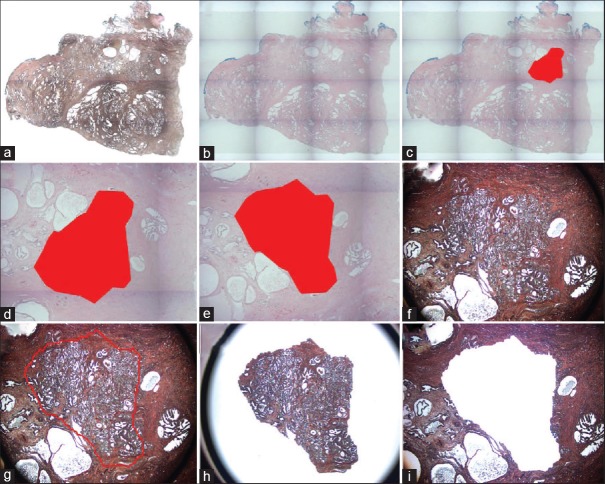
Tissue sections from a human prostatectomy tissue block containing a nodule of prostate cancer (Gleason Grade 3+3) (also employed in Use Case #2 and #3) were cut onto glass or membrane slides and stained with hematoxylin and eosin (H&E). After H&E staining, a tiled ×2 magnification image was taken on the ArcturusXT [Figure 2a]. Xylenes were added to the specimen to provide a pseudo-coverslip [Figure 2b] allowing for image analysis. The regions that were identified as “cancer” by the PPMM algorithm were annotated in red [Figure 2c]. This annotation was then used as a guide for the microdissection process.
Figure 2.
Probabilistic pairwise markov model-laser capture microdissection (a) A low power image of an uncoverslipped Haemotoxylin and Eosin stained human prostate tissue section. (b) A low power image of the Haemotoxylin and Eosin stained tissue with the pseudo-coverslip. (c) Low power image of Probabilistic pairwise Markov model -identified tumor region annotated in red. (d) High power view of the Probabilistic pairwise Markov model area. (e) Pseudo-coverslipped image is rotated to fit. (f) Uncoverslipped sample at high power. (g) This image was imported into AutoScanXT and dissected by the ArcturusXT. (h) Laser capture microdissection cap showing area that was dissected. (i) The remaining tissue in the section is shown
Unlike in our previous study,[23] where an image of a single field of view (FOV) was directly imported into the ArcturusXT AutoScan software after the x, y positional coordinates were re-embedded into the image file, this was not possible using tiled images for two reasons. First, tiled images captured on the ArcturusXT do not contain positional x, y coordinates, and second, there is a memory limitation (up to 80MB; personal communication) on AutoScan that does not allow whole slide tiled images to be imported and analyzed. Therefore, the annotated image was cropped [Figure 2d]. To dissect a large area of tissue as that identified by the PPMM algorithm, it is ideal to section the tissue onto a metal-framed membrane slide. Thus, a serial section from the same tissue block was cut onto a metal-framed membrane slide for the dissection. The cropped PPMM annotated image had to be flipped vertically to match the orientation of the tissue on the membrane slide [Figure 2e].
The correctly oriented, annotated image was then imported into the AutoScan software on the ArcturusXT and the LCM cap was placed on the ROI [Figure 2f] and a dissection map was drawn using a freehand drawing tool based on the annotation as described previously.[23] Due to the lack of x, y coordinates, the default placement of the annotation map for the AutoScan software was into the upper left corner of the slide stage. Therefore, the annotation was selected, copied, and then pasted where the original image was taken, using the original PPMM analyzed image as a guide [Figure 2g]. The IR laser spots (which tack the tissue to the cap) could not be automatically placed, so the microdissection map was “nicked” by erasing a small portion of the outline and then the gap was redrawn by the user, automatically placing the appropriate IR spots.
As expected, the precision of the algorithm was weaker than previously reported on digital WSIs, due to the low resolution of the jpeg image created by the ArcturusXT. However, the tumor cells were identified and successfully microdissected [Figure 2h and i]. Note that annotation regions cannot exceed a 6 mm diameter area due to the limit of the LCM capsize.
Use Case #2: Integration of ImageJ with the ArcturusXT laser capture microdissection workflow
The second algorithm uses the open-source image analysis software suite ImageJ. Here we evaluated the identification of prostate glands stained via immunohistochemistry (IHC), using threshold-based segmentation.
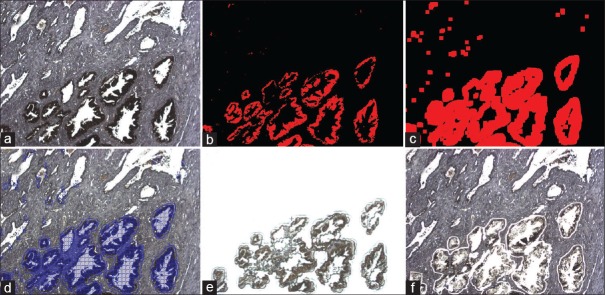
A whole mount human prostate tissue section was immunohistochemically (IHC) stained for PSA, which labeled the prostate epithelial glands [Figure 3a]. The stained section was then imaged on the ArcturusXT and a ×10 FOV was captured. A segmentation algorithm was written in ImageJ to identify the stained glands.
Figure 3.
ImageJ (a) Uncoverlipped immuno-stained human prostate tissue. (b) ImageJ binary mask for the DAB stained areas. (c) ImageJ dilation tool to increase coverage. (d) ImageJ imported into AutoScan software on the ArcturusXT. (e) Image of the laser capture microdissection cap after dissection. (f) Tissue that remains behind after laser capture microdissection
The first step in developing the algorithm was the generation of a binary mask image where the corresponding pixels containing the identified glands were masked with opaque red annotation [Figure 3b]. The mask image was then further processed by the dilation function in ImageJ to fully cover the corresponding area [Figure 3c]. Next, the x, y positional coordinates of the original IHC image were copied to the header of the mask image file, which was subsequently imported back into the AutoScan program. AutoScan then drew a dissection map, and the IHC stained prostate glands [Figure 3d] were microdissected according to the locations of the annotation mask [Figure 3e and f]. Using this approach, the IHC stained tissue was successfully procured.
Use Case #3: The integration of SIVQ with probabilistic pairwise markov model for the ArcturusXT laser capture microdissection workflow
A recently described digital pathology algorithm is Spatially Invariant Vector Quantization (SIVQ),[26,27,28,29,30] a user-friendly pattern recognition method applicable to a wide variety of matching tasks for pathology images. SIVQ was designed such that the user simply “clicks” on a tissue/cellular or sub-cellular feature of interest within the image and the algorithm then quickly (30–40 s) scans it for similar targets based on morphologic features. The method is typically employed in an iterative manner to continually improve the match characteristics as the operator gains experience in selecting predicate features of interest. We have previously demonstrated a proof of principle integration of SIVQ into the LCM workflow[23,31] and showed an algorithmic cascade approach for identifying prostate tumor epithelium by combining PPMM with SIVQ.[30] In this report, we further assessed the potential implementation of the PPMM-SIVQ cascade onto the ArcturusXT device for microdissection.
The third use case involved microdissection using an algorithmic cascade for the specific identification of tumor cells in a heterogeneous prostate tissue section. Similar to use case #2, PPMM was initially used to identify the cancerous region(s) in the tissue section; however, SIVQ was subsequently used to augment the process and identify epithelial cells within the PPMM- identified tumor region.
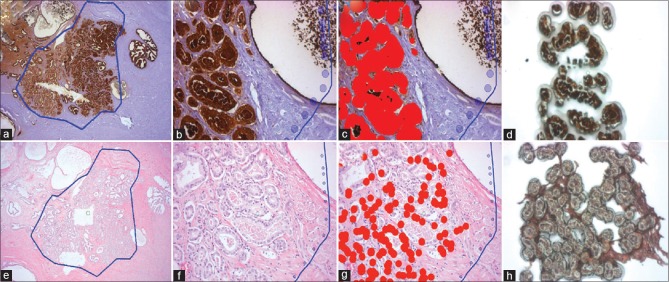
A recut tissue section from Use Case #1 was stained for PSA. The PPMM annotation from Use Case #1 was then imported as described above (the annotation from the PPMM analysis was cropped, but not flipped because the IHC staining had the correct orientation) [Figure 4a]. An image was then taken from within the PPMM annotation and exported for SIVQ analysis [Figure 4b]. A ring vector predicate image was captured from the IHC stained area and used to identify the other stained regions [Figure 4c]. The SIVQ analysis was then reimported back to the ArcturusXT, a microdissection map was created with AutoScan, and the annotated PSA+ glands were microdissected [Figure 4d].
Figure 4.
Probabilistic pairwise Markov model-SIVQ- laser capture microdissection (a) Probabilistic pairwise Markov model identified region on a pseudo-coverslipped prostate tumor that was immunostained. (b) High magnification of the same region in (a). (c) SIVQ applied to the high-power image. (d) Dissected cells on the laser capture microdissection cap. (e) Probabilistic pairwise Markov model of pseudo-coverslipped region. (f) High magnification of the same region in (e). (g) SIVQ applied to the image. (h) Laser capture microdissection cap showing the dissected targets
To demonstrate additional utility, an H&E stained prostate tissue recut section from use case #1 on a glass slide was imaged instead of an immunostained slide [Figure 4e]. An area within the PPMM annotation was selected [Figure 4f] and analyzed using SIVQ. A ring vector predicate image was captured from the epithelium and used to identify additional epithelium [Figure 4g].
The annotated region was microdissected via the AutoScan software and the ArcturusXT instrument [Figure 4h]. Due to the low resolution of the image and morphologic heterogeneity of the tumor epithelium, a small proportion of tumor epithelium was not annotated for dissection.
Use Case #4: Integration of SIVQ with the Leica LMD7000
The next two use cases illustrate the integration of third-party pattern matching and segmentation software with the Leica LMD7000 system. This approach and implementation differed from the ArcturusXT in that the Leica AutoShape software subsystem was modified to accommodate the setup and running of third-party tools. Furthermore, the specification of a generalized communications strategy between third-party software and the Leica laser dissection and tissue positioning subsystems was required. Thus, an interface file was designed as the system integration method that enabled the necessary communication between the findings of a third-party pattern matching software system (e.g., SIVQ) and then passing these tissue-specific coordinates back to the Leica LMD7000 tissue positioning and laser cutting subsystems. Finally, the operator reviewed, and edited candidate tissue sites as needed for the laser dissection process.
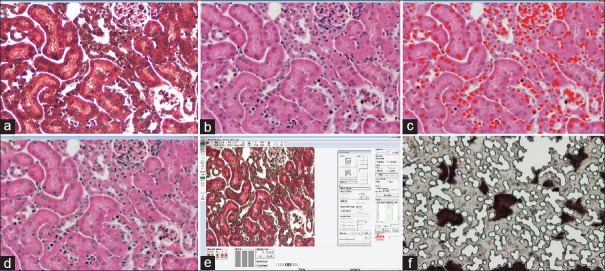
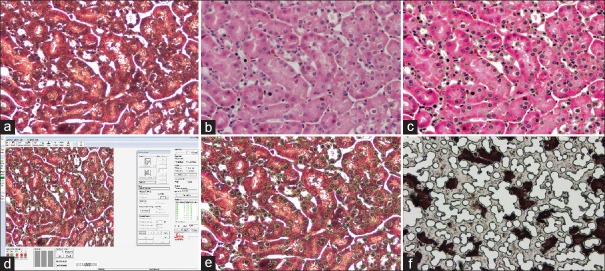
A FFPE mouse kidney block was cut at the 5-μ thickness, deparaffinized, mounted on a Leica PET metal frame slide, and stained with H&E [Figure 5a]. The image was enhanced using xylenes as a pseudocoverslip [Figure 5b] and imported into SIVQ where a nuclei-specific vector was selected for pattern matching (red color) [Figure 5c]. The SIVQ mask was imported into the Leica display console, and the nuclei were dissected [Figure 5d–f].
Figure 5.
SIVQ with the Leica LMD7000 (a) Haemotoxylin and Eosin image of a formalin-fixed, paraffin-embedded mouse kidney tissue uncoverslipped. (b) Same image as (a) with xylenes pseudocoverslip. (c) SIVQ selected the nuclear targets (red dots). (d) SIVQ output imported into Leica and green contours are shown. (e) Leica software with imported SIVQ data. (f) Leica ultraviolet dissection of the target nuclei
Use Case #5: Integration of eSeg with the Leica LMD7000
Finally, using the Leica LMD7000, we evaluated the integration of a novel segmentation algorithm called Extensible Segmentor (eSeq). The same mouse kidney FFPE tissue block and staining process as Use Case #4 were again utilized, including the use of xylenes for a pseudocoverslip [Figure 6a and b]. The results of eSeg pattern matching (green contours) for nuclei were identified [Figure 6c]. The data from eSeg were mported into the Leica LMD7000 instrument, and the nuclear targets were dissected [Figure 6d–f].
Figure 6.
eSeg with the Leica LMD7000 (a) Deparaffinized, Haemotoxylin and Eosin stained tissue on a PET membrane metal frame slide. (b) Same image as in (a), but with the xylenes pseudocoverslip. (c) eSeg applied to the image and the green contours around the target nuclei are shown. (d) The Leica display following transfer of pattern matching contours from eSeg integrated into the Leica LMD7000 workflow. (e) The Leica display console screen following transfer of pattern matching contours from eSeg. (f) The Leica dissection following eSeg-based computer-aided laser dissection on the LMD7000
DISCUSSION
In this study, we presented the computer-aided laser dissection (CALD) workflow with five use cases demonstrating the integration of third-party software (individually or in tandem) onto two different laser microdissection commercial instruments (ThermoFisher Scientific ArcturusXT and Leica LMD7000). Recent studies have revealed the use of microdissection to obtain specific cell populations for downstream molecular analysis, such as gene expression arrays and proteomic studies.[2,4,5,8,22,32,33] However, there is an unmet need to improve the speed, and overall usability of laser microdissection to facilitate these studies as current microdissection technologies limit the ability to dissect specific cells in a rapid and semi-automated manner. The use of algorithms, such as PPMM, ImageJ, eSeg, and SIVQ streamline this process and reduce the amount of user hands-on operator time. Here, we sought to show the diversity in the types of algorithms and LCM platforms that can be integrated and utilized. One novelty of this work is the successful application of algorithms that had originally been designed for WSI to the field of LCM (PPMM, SIVQ). Moreover, we have demonstrated a modular workflow where not only those WSI algorithms but also custom software (eSeg, ImageJ plugins) can be applied to annotate tissue for microdissection. This modularity will enable investigators to identify the right algorithm for the LCM platform appropriate for their specific application.
CALD-based dissection could become, especially important in the burgeoning age of precision medicine. As molecular assays become more sophisticated and potentially provide clinicians with greater information, there likely will be a need to more quickly obtain pure populations of the target tissue and thus improve specificity. However, today, there are few clinical laboratories that employ LCM due to the time and effort required to identify the cells of interest and the fact that there has not been a strong clinical need for dissected cells. This may be changing now in the era of precision medicine and the use of image analysis algorithms, such as those presented here, may be able to facilitate the implementation of microdissection into the clinical workflow in the future.
The use of algorithm cascades (PPMM-SIVQ), as opposed to single analytic image analysis methods, can be particularly beneficial for improving the microdissection workflow and providing a means to rapidly assess regions of tumor heterogeneity. For example, PPMM can identify the tumor area in a WSI of a prostate tissue section very rapidly and then SIVQ can be used to more finely identify and tag the epithelial cells for microdissection. The use of an algorithm that can quickly identify an area of interest (PPMM) can greatly improve the speed of the cellular identification step in the microdissection dissection workflow.
In the current practice, a pathologist highlights the tumor area on a glass slide, and then a technician scrapes that area on a serial unstained tissue section. Although this method is quick and inexpensive, procurement of a mixed cell population may negatively impact the molecular results and potentially patient care. In addition, for cases that have tumor cells dispersed throughout the specimen, the sample may either be rejected or a repeat clinical biopsy procedure for the patient may be required to obtain enough material for molecular analysis, putting the patient at increased risk and increasing financial costs associated with obtaining the proper diagnosis.
In addition to PPMM, there are some CAD algorithms that have been developed to aid pathologists.[16,34,35,36,37] As we proceed forward, it will be interesting to explore the use of these novel algorithms to drive and guide the microdissection process. In the future, it may be possible to employ new algorithms in a “plug and play” fashion with the microdissection devices. Steps towards this goal were illustrated with the Leica LMD7000 where two different third-party pattern matching and segmentation tools were integrated into the workflow in a fairly seamless manner.
Another microdissection tool, Expression Microdissection (xMD), is not image driven and performs high-throughput microdissection based on IHC staining.[25,38,39,40] Image analysis tools, such as those described here, could enhance xMD by providing an upfront image analysis step to identify specific tumor regions (ex. PPMM) or limit the laser to only the immunostained regions (ex. SIVQ). The combination of these tools could further improve the microdissection workflow and move the field toward user-independent dissections.
Despite these advancements, there are several limitations that inhibit the implementation of algorithms into the microdissection workflow. One of the major limitations is that the current commercial technologies are not easily amenable to integration. For example, there were some steps that had to be leveraged to work around the limitations of the ArcturusXT and LMD7000 software and instrumentation. For the ArcturusXT, the computing limit and the ability to only export, analyze, and import low-resolution jpeg images hampered our ability to advance the technology. When integrating SIVQ with the LMD7000 device, it was necessary to work directly with Leica representatives to obtain access to their proprietary software, which took additional time and effort. For the field to move forward, the microdissection instrument companies will need to appreciate the need for high-end computing and embrace standards for open integration of third-party software.
Future directions
The algorithms for the region of interest detection and segmentation used here are based on image processing and traditional machine learning, engineering of morphological or cytological features known to pathologists, and manual adjustment of thresholds and parameters to optimize the result. The advantages of these approaches are that they are readily available without much training data, but they have several limitations: a large amount of manual intervention is required, the precision of the dissection boundaries is relatively low, and moreover, they can only capture ROI that a human can identify. Recent advances in applying deep learning to histopathological images[10,41,42,43,44,45,46] provide an opportunity to overcome some of these limitations for LCM. For instance, deep learning-based segmentation of the tissue sample will allow for better scalability through reduced manual intervention, enabling potentially a much larger number of experiments to be performed. Even more importantly, it will likely result in a higher precision of the dissection boundaries and higher purity within each sample, which translates to an increased signal-to-noise ratio of the molecular assay. Finally, since the features detected by convolutional neural networks are not predefined by the user but instead are learned by the model itself, applying deep learning to LCM has the potential of yielding ROI for sequencing that would not be detected by a human.
CONCLUSION
In summary, we present five CALD use cases that highlight the integration of new computer algorithms (PPMM and ImageJ) and combinatorial approaches (PPMM-SIVQ) with microdissection on the Arcturus XT, to facilitate the procurement of specific cell populations. In addition, we demonstrate the integration of both SIVQ and a new image segmenter named eSeg, onto the Leica LMD7000 microdissection device. This methodology highlights the potential importance of applying image analysis tools to improve the microdissection workflow for both clinical and life science applications.
Financial support and sponsorship
Dr. Emmert-Buck is an inventor on all NIH held patents covering laser capture microdissection technology and receives royalty-based payments through the NIH Technology Transfer Program.
Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. He is also a scientific advisory consultant for Inspirata Inc. In addition, he currently serves as a scientific advisory board member for Inspirata Inc. He also has sponsored research agreements with Philips and Inspirata Inc. His technology has been licensed to Elucid Bioimaging and Inspirata Inc. He is also involved in a NIH U24 grant with PathCore Inc, and 3 different R01 grants with Inspirata Inc.
This work was supported in part by the NCI's Center for Cancer Research in the Intramural Program at NIH, and by funding from the Burroughs Wellcome fund travel grant.
Dr. Madabhushi's research is supported in part by the National Cancer Institute of the National Institutes of Health under award numbers:
1U24CA199374-01,
R01CA202752-01A1
R01CA208236-01A1
R01 CA216579-01A1
R01 CA220581-01A1
National Center for Research Resources under award number
1 C06 RR12463-01
The DOD Prostate Cancer Idea Development Award;
The DOD Lung Cancer Idea Development Award;
The DOD Peer Reviewed Cancer Research Program W81XWH-16-1-0329
The Ohio Third Frontier Technology Validation Fund
The Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering and the Clinical and Translational Science Award Program (CTSA) at Case Western Reserve University.
Dr. Tangrea's research is supported by the Kahlert Foundation, Mr. Curtis Fichter and the Eddie Dopkin Family.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2018/9/1/45/247296
REFERENCES
- 1.Frost AR, Eltoum IE, Siegal GP, Emmert-Buck MR, Tangrea MA. Laser microdissection. Curr Protoc Mol Biol. 2015;112:25A11–30. doi: 10.1002/0471142727.mb25a01s112. [DOI] [PubMed] [Google Scholar]
- 2.Silvestri A, Colombatti A, Calvert VS, Deng J, Mammano E, Belluco C, et al. Protein pathway biomarker analysis of human cancer reveals requirement for upfront cellular-enrichment processing. Lab Invest. 2010;90:787–96. doi: 10.1038/labinvest.2010.47. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Canales J, Hanson JC, Hipp JD, Balis UJ, Tangrea MA, Emmert-Buck MR, et al. Optimal molecular profiling of tissue and tissue components: Defining the best processing and microdissection methods for biomedical applications. Methods Mol Biol. 2013;980:61–120. doi: 10.1007/978-1-62703-287-2_5. [DOI] [PubMed] [Google Scholar]
- 4.Klee EW, Erdogan S, Tillmans L, Kosari F, Sun Z, Wigle DA, et al. Impact of sample acquisition and linear amplification on gene expression profiling of lung adenocarcinoma: Laser capture micro-dissection cell-sampling versus bulk tissue-sampling. BMC Med Genomics. 2009;2:13. doi: 10.1186/1755-8794-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrell JC, Dye WW, Harvell DM, Sartorius CA, Horwitz KB. Contaminating cells alter gene signatures in whole organ versus laser capture microdissected tumors: A comparison of experimental breast cancers and their lymph node metastases. Clin Exp Metastasis. 2008;25:81–8. doi: 10.1007/s10585-007-9105-7. [DOI] [PubMed] [Google Scholar]
- 6.Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, et al. Laser-capture microdissection. Nat Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 7.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 8.El-Serag HB, Nurgalieva ZZ, Mistretta TA, Finegold MJ, Souza R, Hilsenbeck S, et al. Gene expression in Barrett's esophagus: Laser capture versus whole tissue. Scand J Gastroenterol. 2009;44:787–95. doi: 10.1080/00365520902898127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martel AL, Hosseinzadeh D, Senaras C, Zhou Y, Yazdanpanah A, Shojaii R, et al. An image analysis resource for cancer research: PIIP-pathology image informatics platform for visualization, analysis, and management. Cancer Res. 2017;77:e83–6. doi: 10.1158/0008-5472.CAN-17-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhargava R, Madabhushi A. Emerging themes in image informatics and molecular analysis for digital pathology. Annu Rev Biomed Eng. 2016;18:387–412. doi: 10.1146/annurev-bioeng-112415-114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal. 2016;33:170–5. doi: 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellors RC, Glassman A, Papanicolaou GN. A microfluorometric scanning method for the detection of cancer cells in smears of exfoliated cells. Cancer. 1952;5:458–68. doi: 10.1002/1097-0142(195205)5:3<458::aid-cncr2820050306>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 14.Hipp J, Flotte T, Monaco J, Cheng J, Madabhushi A, Yagi Y, et al. Computer aided diagnostic tools aim to empower rather than replace pathologists: Lessons learned from computational chess. J Pathol Inform. 2011;2:25. doi: 10.4103/2153-3539.82050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, et al. Computerized image-based detection and grading of lymphocytic infiltration in HER2+ breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:642–53. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 16.Doyle S, Feldman M, Tomaszewski J, Madabhushi A. A boosted bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans Biomed Eng. 2012;59:1205–18. doi: 10.1109/TBME.2010.2053540. [DOI] [PubMed] [Google Scholar]
- 17.Fatakdawala H, Xu J, Basavanhally A, Bhanot G, Ganesan S, Feldman M, et al. Expectation-maximization-driven geodesic active contour with overlap resolution (EMaGACOR): Application to lymphocyte segmentation on breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:1676–89. doi: 10.1109/TBME.2010.2041232. [DOI] [PubMed] [Google Scholar]
- 18.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B, et al. Histopathological image analysis: A review. IEEE Rev Biomed Eng. 2009;2:147–71. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lexe G, Monaco J, Doyle S, Basavanhally A, Reddy A, Seiler M, et al. Towards improved cancer diagnosis and prognosis using analysis of gene expression data and computer aided imaging. Exp Biol Med (Maywood) 2009;234:860–79. doi: 10.3181/0902-MR-89. [DOI] [PubMed] [Google Scholar]
- 20.Madabhushi A. Digital pathology image analysis: Opportunities and challenges. Imaging Med. 2009;1:7–10. doi: 10.2217/IIM.09.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson JC, Tangrea MA, Kim S, Armani MD, Pohida TJ, Bonner RF, et al. Expression microdissection adapted to commercial laser dissection instruments. Nat Protoc. 2011;6:457–67. doi: 10.1038/nprot.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberle FC, Hanson JC, Killian JK, Wei L, Ylaya K, Hewitt SM, et al. Immunoguided laser assisted microdissection techniques for DNA methylation analysis of archival tissue specimens. J Mol Diagn. 2010;12:394–401. doi: 10.2353/jmoldx.2010.090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hipp J, Cheng J, Hanson JC, Yan W, Taylor P, Hu N, et al. SIVQ-aided laser capture microdissection: A tool for high-throughput expression profiling. J Pathol Inform. 2011;2:19. doi: 10.4103/2153-3539.78500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monaco JP, Tomaszewski JE, Feldman MD, Hagemann I, Moradi M, Mousavi P, et al. High-throughput detection of prostate cancer in histological sections using probabilistic pairwise Markov models. Med Image Anal. 2010;14:617–29. doi: 10.1016/j.media.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tangrea MA, Hanson JC, Bonner RF, Pohida TJ, Rodriguez-Canales J, Emmert-Buck MR, et al. Immunoguided microdissection techniques. Methods Mol Biol. 2011;755:57–66. doi: 10.1007/978-1-61779-163-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hipp JD, Cheng JY, Toner M, Tompkins RG, Balis UJ. Spatially invariant vector quantization: A pattern matching algorithm for multiple classes of image subject matter including pathology. J Pathol Inform. 2011;2:13. doi: 10.4103/2153-3539.77175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hipp J, Smith SC, Cheng J, Tomlins SA, Monaco J, Madabhushi A, et al. Optimization of complex cancer morphology detection using the SIVQ pattern recognition algorithm. Anal Cell Pathol (Amst) 2012;35:41–50. doi: 10.3233/ACP-2011-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hipp J, Cheng J, Daignault S, Sica J, Dugan MC, Lucas D, et al. Automated area calculation of histopathologic features using SIVQ. Anal Cell Pathol (Amst) 2011;34:265–75. doi: 10.3233/ACP-2011-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng J, Hipp J, Monaco J, Lucas DR, Madabhushi A, Balis UJ, et al. Automated vector selection of SIVQ and parallel computing integration MATLAB™: Innovations supporting large-scale and high-throughput image analysis studies. J Pathol Inform. 2011;2:37. doi: 10.4103/2153-3539.83752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hipp J, Monaco J, Kunju LP, Cheng J, Yagi Y, Rodriguez-Canales J, et al. Integration of architectural and cytologic driven image algorithms for prostate adenocarcinoma identification. Anal Cell Pathol (Amst) 2012;35:251–65. doi: 10.3233/ACP-2012-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy Chowdhuri S, Hanson J, Cheng J, Rodriguez-Canales J, Fetsch P, Balis U, et al. Semiautomated laser capture microdissection of lung adenocarcinoma cytology samples. Acta Cytol. 2012;56:622–31. doi: 10.1159/000342984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HK, Kim J, Korolevich S, Choi IJ, Kim CH, Munroe DJ, et al. Distinctions in gastric cancer gene expression signatures derived from laser capture microdissection versus histologic macrodissection. BMC Med Genomics. 2011;4:48. doi: 10.1186/1755-8794-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J, Garg S, Wang J, Loose DS, Hauer-Jensen M. Laser capture microdissected mucosa versus whole tissue specimens for assessment of radiation-induced dynamic molecular and pathway changes in the small intestine. PLoS One. 2013;8:e53711. doi: 10.1371/journal.pone.0053711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A. Selective invocation of shape priors for deformable segmentation and morphologic classification of prostate cancer tissue microarrays. Comput Med Imaging Graph. 2015;41:3–13. doi: 10.1016/j.compmedimag.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leo P, Lee G, Shih NN, Elliott R, Feldman MD, Madabhushi A, et al. Evaluating stability of histomorphometric features across scanner and staining variations: Prostate cancer diagnosis from whole slide images. J Med Imaging (Bellingham) 2016;3:047502. doi: 10.1117/1.JMI.3.4.047502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sridhar A, Doyle S, Madabhushi A. Content-based image retrieval of digitized histopathology in boosted spectrally embedded spaces. J Pathol Inform. 2015;6:41. doi: 10.4103/2153-3539.159441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee G, Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A, et al. Cell orientation entropy (COrE): Predicting biochemical recurrence from prostate cancer tissue microarrays. Med Image Comput Comput Assist Interv. 2013;16:396–403. doi: 10.1007/978-3-642-40760-4_50. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg AZ, Armani MD, Fetsch PA, Xi L, Pham TT, Raffeld M, et al. High-throughput microdissection for next-generation sequencing. PLoS One. 2016;11:e0151775. doi: 10.1371/journal.pone.0151775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tangrea MA, Chuaqui RF, Gillespie JW, Ahram M, Gannot G, Wallis BS, et al. Expression microdissection: Operator-independent retrieval of cells for molecular profiling. Diagn Mol Pathol. 2004;13:207–12. doi: 10.1097/01.pdm.0000135964.31459.bb. [DOI] [PubMed] [Google Scholar]
- 40.Blackler AR, Morgan NY, Gao B, Olano LR, Armani MD, Romantseva E, et al. Proteomic analysis of nuclei dissected from fixed rat brain tissue using expression microdissection. Anal Chem. 2013;85:7139–45. doi: 10.1021/ac400691k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Gadepalli K, Norouzi M, Dahl GE, Kohlberger T, Boyko A, et al. Detecting cancer metastases on gigapixel pathology images. arXiv: 170302442. 2017 [Google Scholar]
- 43.Cruz-Roa AA, Arevalo Ovalle JE, Madabhushi A, González Osorio FA. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. Med Image Comput Comput Assist Interv. 2013;16:403–10. doi: 10.1007/978-3-642-40763-5_50. [DOI] [PubMed] [Google Scholar]
- 44.Cruz-Roa A, Gilmore H, Basavanhally A, Feldman M, Ganesan S, Shih NN, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep learning approach for quantifying tumor extent. Sci Rep. 2017;7:46450. doi: 10.1038/srep46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing. 2016;191:214–23. doi: 10.1016/j.neucom.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Xiang L, Liu Q, Gilmore H, Wu J, Tang J, et al. Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Trans Med Imaging. 2016;35:119–30. doi: 10.1109/TMI.2015.2458702. [DOI] [PMC free article] [PubMed] [Google Scholar]