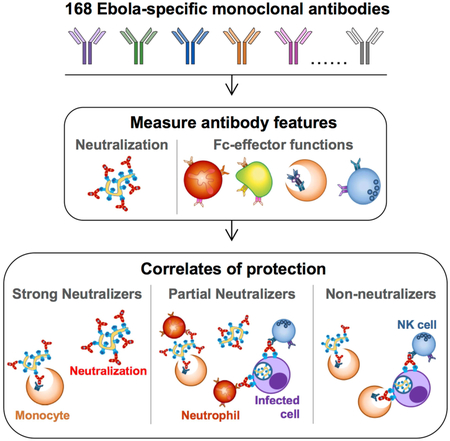
SUMMARY.
The recent Ebola virus (EBOV) epidemic highlighted the need for effective vaccines and therapeutics to limit/prevent outbreaks. Host antibodies against EBOV are critical for controlling disease, and recombinant monoclonal antibodies (mAbs) can protect from infection. However, antibodies mediate an array of antiviral functions including neutralization as well as engagement of Fc-domain receptors on immune cells, resulting in phagocytosis or NK cell-mediated killing of infected cells. Thus, to understand the antibody features mediating EBOV protection, we examined specific Fc-features associated with protection using a library of EBOV-specific mAbs. Neutralization was strongly associated with therapeutic protection against EBOV. However, several neutralizing mAbs failed to protect, while several non-neutralizing or weakly neutralizing mAbs could protect. Antibody-mediated effector functions, including phagocytosis and NK cell activation were associated with protection, particularly for antibodies with moderate neutralizing activity. This framework identifies functional correlates that can inform therapeutic and vaccine design strategies against EBOV and other pathogens.
GRAPHICAL ABSTRACT
eTOC Blurb:
While antibodies provide protection against Ebola virus, the mechanism is unclear. Gunn et al. dissect the contribution of Fc-functions to protection using a library of Ebola virus-specific antibodies. Fc-function was a critical predictor of protection across neutralizing and non-neutralizing antibodies, pointing to synergy between Fc- and Fab-mediated antibody-functions against Ebola.
INTRODUCTION.
The high mortality rate and rapid spread of the recent 2013–2016 Ebola virus disease (EVD) epidemic in West Africa underscores the urgency for the development of interventions that can prevent and limit the spread of the disease. Currently, a licensed vaccine or an effective treatment against Ebola virus (EBOV) does not exist, although swift progress has been made in the wake of the recent EVD epidemic (Henao-Restrepo et al., 2015; Kanapathipillai et al., 2014). Beyond vaccines, monoclonal antibody (mAb) therapeutics, including the mAb cocktail, ZMapp™, have shown protection in post-exposure animal models (Qiu et al., 2014). While ZMapp did not meet the pre-specified cutoff for efficacy in humans, treatment reduced the length of stay in Ebola treatment units (PREVAIL II Writing Group et al., 2016). Yet, ZMapp was constructed from the limited pool of mAbs available prior to 2013, rather than rationally designed through the selection of mAbs with the highest potential antiviral efficacy. Novel EBOV mAbs have rapidly emerged over the course of the recent EVD epidemic (Bornholdt et al., 2016; Corti et al., 2016; Flyak et al., 2016; Wec et al., 2017; Zhao et al., 2017). As many have been tested in vivo, these new mAbs provide an opportunity to not only define the correlates of mAb-mediated protection against EBOV, but to also guide the rational design of next-generation mAb cocktails for use in future epidemics.
While neutralizing activity has generally been regarded as a critical mechanism underlying Ab-mediated protection from EBOV infection (Crowe, 2017), early studies demonstrated that both neutralizing and non-neutralizing mAbs could confer protection from infection in animal models (Wilson et al., 2000). Along these lines, mAb cocktails composed of both neutralizing and non-neutralizing mAbs, such as ZMapp (Qiu et al., 2014), or composed of non-neutralizing mAbs alone, such as the cocktail MB-003 (Olinger et al., 2012; Pettitt et al., 2013), both showed protection in animal models following EBOV challenge. In contrast, no protection from infection was observed in non-human primate (NHP) models following the administration of one of the first identified potently neutralizing mAbs, KZ52 (Oswald et al., 2007; Parren et al., 2002), suggesting that neutralizing activity alone may not always be sufficient for protection. An in-depth dissection of the humoral correlates of protective immunity has been lacking but could guide both the generation of more effective mAb cocktails as well as correlates of immunity to support licensure of a safe and effective EBOV vaccine.
Beyond neutralization, Abs mediate an array of additional antiviral functions via their ability to interact with Fc-domain receptors (Fc receptors, FcR) found on all innate immune cells (Bruhns, 2012). These innate immune effector functions include phagocytosis of virions and infected cells by monocytes, macrophage, and neutrophils, as well as direct killing of infected cells by natural killer (NK) cells and/or complement activation that contribute to protective immunity by clearing infected cells and limiting viral dissemination (Pincetic et al., 2014). These functions are governed by two variables in the Ab Fc-domain which govern the affinity of the Fc for different FcRs: 1) the selection of particular isotype/subclasses and 2) alteration of Fc N-glycosylation (Gunn and Alter, 2016), both of which have been exploited in the mAb therapeutics field to enhance mAb efficacy (Goede et al., 2014; Umana et al., 1999; Weiner et al., 2009). Accordingly, the protective efficacy of the non-neutralizing EBOV-specific mAb, 13F6 (Zeitlin et al., 2011), and of the MB-003 cocktail (Olinger et al., 2012) have both been linked to the production in glyco-engineered Nicotiana benthamiana plants, which generate mAbs with highly functional agalactosylated, afucosylated, bisected glycan structures that enhance Ab dependent cellular cytotoxicity (ADCC) (Shields et al., 2002; Strasser et al., 2008). These data suggest that Fc-mediated effector functions are key to the protection conferred by these non-neutralizing mAbs. Moreover, recent data argue that the Fc-domain can augment neutralizing mAb-mediated protection from infection in the context of HIV and influenza (Bournazos et al., 2014; DiLillo et al., 2016), indicating a synergy between the antigen-binding domain (Fab)- and Fc-mediated functions to maximize protective efficacy. However, whether a specific set of humoral functions are key to protection for EBOV-specific mAbs, whether specific functions are more critical for neutralizing or non-neutralizing Abs, and if these functions may be defined to aid in the design of a highly efficacious mAb cocktail or vaccine is not understood.
Defining the specific protective features of Abs against EBOV thus far has been difficult as most available Abs have been studied in isolation by individual labs, making comparisons between Abs challenging. To overcome this problem, the Viral Hemorrhagic Fever Immunotherapeutics Consortium (VIC) collected 168 EBOV glycoprotein (GP)-specific mAbs from research laboratories around the globe with the goal of comparing all 168 mAbs in an array of assays and define their relation to protective efficacy in a lethal EBOV murine infection model (Saphire et al., 2018). Neutralization was clearly linked to protection; however analysis of Ab effector function suggested a role for additional Ab functions in protection against EBOV.
With the hypothesis that Fc-effector function plays a critical role in Ab-mediated protection against EBOV, here we systematically evaluated the intersection between neutralization and Fc-effector function. We present the in-depth functional profiling of this panel of EBOV-specific Abs used to begin to define the Fc-mediated functional correlates of protection against EBOV. We observed significant functional heterogeneity among the Abs with respect to both neutralizing activity and non-neutralizing Fc-effector functions, and while protection was largely observed among potently neutralizing Abs, Fc-effector function was associated with protection for mAbs with moderate neutralizing activity, some of which provided robust protection in vivo. Machine learning analyses on this unprecedented dataset illuminated the minimal required functional correlates of protection, which if integrated into mAb screening, could accelerate development of rationally designed and functionally enhanced mAb-based therapies. Together, our data form a critical framework to both define and exploit immune correlates of protection to rationally guide the development of effective therapeutics against EBOV and other human pathogens.
RESULTS.
Heterogeneous Fc-effector functions induced by mAbs in the ZMapp and MB-003 cocktails.
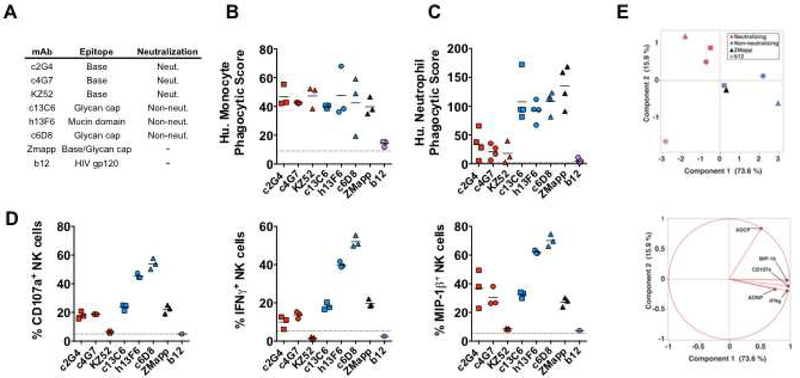
ZMapp was formed by combining the best mAbs of the protective mAb cocktails MB-003, composed of three non-neutralizing mAbs (c13C6, h13F6 and c6D8), and ZMab, composed of two neutralizing (c2G4, c4G7) and one non-neutralizing (1H3) mAbs, resulting in the final ZMapp cocktail containing c2G4, c4G7, and c13C6 (Qiu et al., 2014). The essential inclusion of c13C6 in the formulation of both the three mAb and two mAb ZMapp cocktails (Qiu et al., 2016; Qiu et al., 2014) strongly suggested that functions beyond neutralization are critical for protection against lethal EBOV infection. Thus, we hypothesized that specific functional characteristics of these mAbs could account for alternate modes of protection within the ZMapp and MB-003 cocktails (Figure 1A). The ability of individual mAbs to recruit NK cell, monocyte, and neutrophil activation were interrogated, and compared to the ZMapp cocktail, the neutralizing mAb KZ52, and the HIV-specific mAb b12, which served as a negative control. We found that all of the EBOV mAbs induced similar levels of Ab-dependent monocyte phagocytosis (ADCP) (Figure 1B). In contrast, we observed significant differences in the capacity of non-neutralizing mAbs (c13C6, h13F6, and c6D8) to recruit Ab-dependent neutrophil phagocytosis (ADNP), as compared to the neutralizing mAbs (c2G4, c4G7, KZ52) (Figure 1C). Monocytes express high levels of the Fc receptor FcγRIIA, an FcR implicated in phagocytosis (Ackerman et al., 2013; Indik et al., 1995), while neutrophils express an alternate Fc-receptor, FcγRIIIB (Bruhns, 2012); thus, these differences between neutralizing and non-neutralizing mAbs may reflect different abilities to bind to FcγRIIA vs. FcγRIIIB.
Figure 1. Functionality of Ebola virus GP-specific monoclonal antibodies.
The mAbs within the Zmapp and MB-003 cocktails (A) were evaluated for the induction the following innate immune effector functions: Ab-dependent phagocytosis of GP-coated beads by human monocytes (B); Ab-dependent phagocytosis of GP-coated beads by human neutrophils (C); Ab-dependent activation of NK cells as measured by degranulation (CD107a), and secretion of cytokines (IFNγ) and chemokines (MIP-1β) (C). Red and blue symbols indicate neutralizing and non-neutralizing mAbs, respectively. Each dot represent an independent replicate, the solid line represents the mean response across replicates, and the dashed line indicates the level of the no Ab control. Unsupervised principal component analysis of functional responses separates neutralizing and non-neutralizing mAbs (D), and the scoring plot of component 1 and 2 is shown on the top and the loadings plot is shown on the bottom.
Beyond phagocytosis, Abs can also induce NK cell activity through the FcγRIIIA receptor (Bruhns et al., 2009), resulting in release of cytotoxic granules (degranulation) that kill infected cells/pathogens, and/or production of cytokines and chemokines (Alter et al., 2004). We observed greater heterogeneity in the ability of the different mAbs to induce NK cell activation measured by three NK cell functions (degranulation, IFNγ cytokine and MIP-1β chemokine secretion) (Figure 1D). The mucin domain-specific mAbs (h13F6 and c6D8) drove the best responses in NK cells, whereas the neutralizing mAb KZ52, which binds to the GP1/GP2 interface at the base of the cleaved GP (Lee et al., 2008), exhibited the lowest NK cell activating capacity and induced negligible NK cell activation (Figure 1D). In contrast, the ZMapp components (c2G4 and c4G7, which both bind the base and neutralize, and c13C6, which binds the glycan cap and does not neutralize), each induced intermediate levels of NK activity. When all the functional data were analyzed together in an unsupervised principal component analysis, the functional profiles of neutralizing and non-neutralizing mAbs appeared distinct, driven predominantly by higher NK cell and neutrophil responses by non-neutralizing mAbs, as compared to the neutralizing mAbs (Figure 1E; Table S1), pointing to different Fc-effector profiles potentially associated with their distinct protective mechanisms of action in vivo (Olinger et al., 2012; Qiu et al., 2014; Zeitlin et al., 2011).
Functional heterogeneity across a large pool of Ebola virus-specific monoclonal Abs.
Given the differences in functionality between the protective neutralizing and non-neutralizing mAbs within the MB-003 and ZMapp cocktails, we next hypothesized that the precise Fc-effector correlates of protection, linked to neutralization, could be defined using a larger panel of monoclonal Abs collected by the VIC (n=168). The neutralizing activity of each mAb was evaluated in three separate neutralization assays, and the EBOV GP epitope targeted by each mAb was defined (Saphire et al., 2018). Importantly, the protective activity of each mAb was evaluated in a mouse postexposure challenge model, where ten BALB/c mice were infected with 100 plaque forming units of mouse-adapted EBOV followed by passive transfer of 100μg of a given mAb two days post-infection. The mAb panel included both human and mouse Abs with varying neutralizing activity ranging from no activity to potent neutralizing activity, spanning a variety of epitopes across the EBOV GP, and showed variables levels of protection (Saphire et al., 2018). Of note, within the panel of mAbs, VIC 1-136 were generated prior to the 2013-2016 EVD epidemic, and included both mouse and human neutralizing and non-neutralizing mAbs, whereas VIC 137-171 were donated after the recent EVD epidemic, and many were selected specifically for their strong neutralizing activity and generated as human IgG1. Thus, analysis of the ability of each mAb to induce effector functions offered a unique opportunity to dissect the relationships between functional activity, neutralization, epitope, and protection.
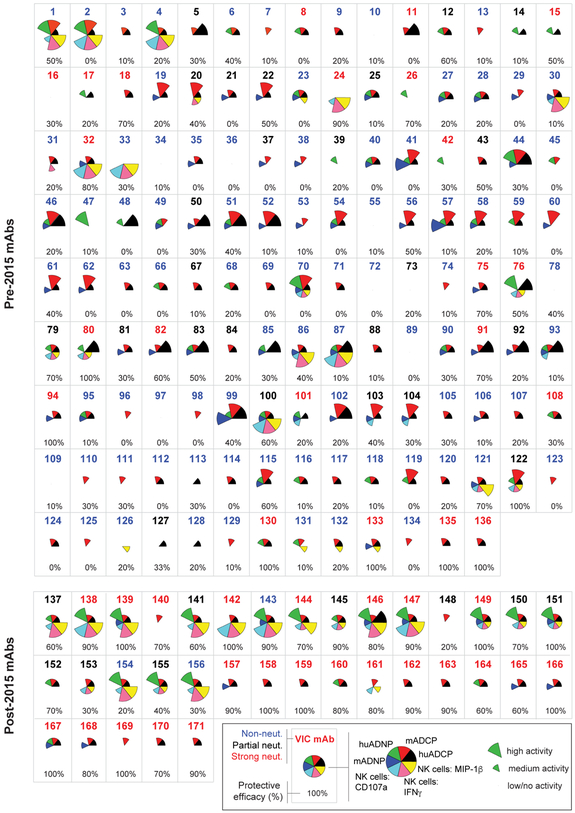
Protective mAbs were defined as those that protected ≥60% of mice from death following administration of 100μg of the mAb two days after a lethal EBOV challenge, and the 60% threshold represents 3X the level of protection observed in mice that received PBS alone (0-20%). Nearly all of the protective mAbs exhibited some level of neutralizing activity (Saphire et al., 2018), and neutralizing activity was significantly correlated with protection (Figure S1). Yet, several protective mAbs had no or low neutralizing activity, similar to the MB-003 mAbs, highlighting the possibility that additional functions beyond neutralization may contribute to protection. As ADCC in mice is thought to be primarily mediated by monocytes/macrophages rather than NK cells (DiLillo and Ravetch, 2015), we evaluated the ability of mAbs to recruit both human and mouse innate immune cells to objectively define the functional characteristics that were associated with protection (Figure S2A). We observed significant heterogeneity among the panel of mAbs in their functional profiles (Figure 2). Despite differences in FcRs between mouse and human innate immune cells (Bruhns, 2012), strong concordance was observed in mAb-mediated recruitment of corresponding murine and human innate effector cells (Figure S2B), consistent with the cross-talk observed between human IgG and mouse FcRs (Overdijk et al., 2012). Remarkably, some mAbs induced robust activity in nearly all functional assays (e.g. VIC 2, 87, 146) whereas others exhibited nearly no detectable activity (e.g. VIC 10, 16, 34). Given the broad heterogeneity of functional profiles among both the neutralizing and non-neutralizing Abs, the data presented here suggest that particular combinations, rather than any single discrete function, may be more relevant for protection following infection.
Figure 2. Functionality of Ebola virus GP-specific mAbs.
The functional profile of each mAb within the panel is graphed by characterizing the magnitude of the functional response for each mAb into high, medium, and low/negligible for each function based on cutoffs defined in Figure S2. Each wedge is color-coded by effector function, with the size of the wedge corresponding to the magnitude of the response in the respective functional assay. The specific VIC Ab number is located at the top of each Ab box, and color-coded according to neutralization group (strongly neutralizing mAbs are indicated in red text, partially neutralizing mAbs in black text, and non-neutralizing mAbs in blue text). The percent protection provided by the indicated Ab is located at the bottom of the Ab box, with protection ranging from 0% (no animals protected) to 100% (all animals protected). The protection data is derived from (Saphire et al., 2018).
Both neutralization and Fc effector function are associated with mAb-mediated protection.
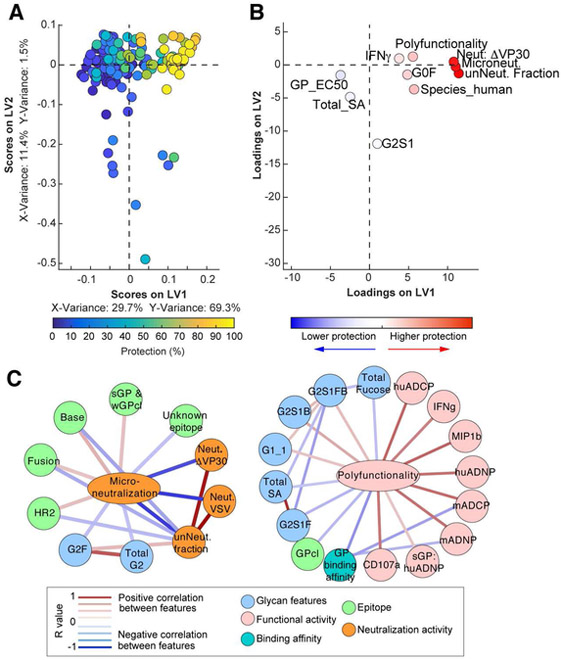
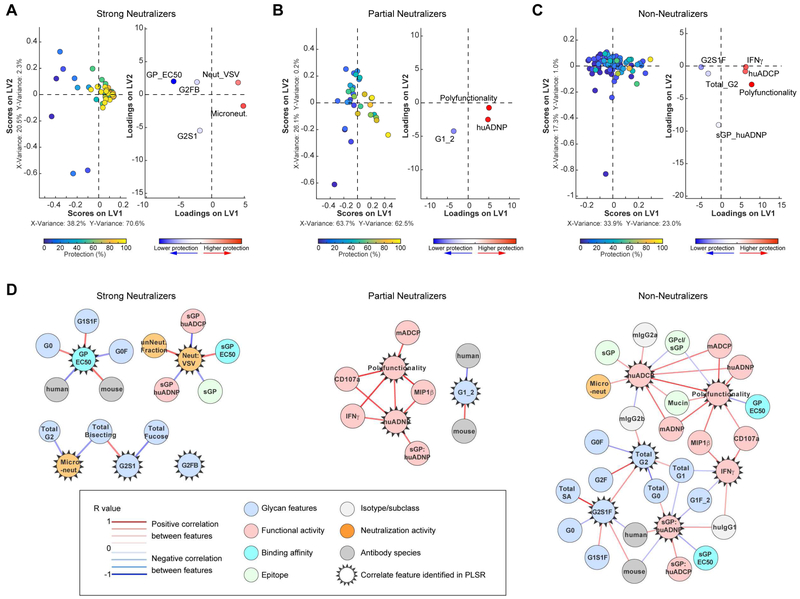
Given the breadth of functional profiles, we next used a multivariate analysis approach that integrateds all functional and biophysical data to provide enhanced resolution of the precise Ab features that track with protective immunity. Specifically, we used an unbiased multivariate machine learning approach that eliminates co-correlated variables to explore the minimal Ab features associated with protection. While 40 Ab features were measured (Table S2), including Fab-associated features such as neutralization and epitope specificity (Saphire et al., 2018), and Fc-associated features such as function (Figure 2, Figure S2), and Fc-glycan content (Figure S3), many of the features correlated with one another. Thus, to hone in on the minimal Ab profiles that were essential in predicting protection, we used an Elastic Net algorithm to down select the features to the fewest features that captured the overall variation in the Ab dataset followed by partial least square regression (PLSR) analysis to effectively separate protective from non-protective Abs (Figure 3A), The mirror loadings plot (Figure 3B) shows the 10 Ab features that drove separation of the protective and non-protective Abs, where the relative location of an individual feature in the loadings plot is associated with the geographical position of the corresponding Ab on the dot plot (Figure 3A). Thus, features in the top right quadrant on the loadings plot were selectively enriched among the most protective Abs. Importantly, two key features were located in the same region as the protective Abs, neutralization and polyfunctionality (i.e., the number of effector functions the mAb can induce on a scale of 0-7 functions), pointing to a critical role for both Fab- and Fc-mediated functions with protection.
Figure 3. Antibody features that predict protection.
Elastic Net/PSLR were used to identify the minimal mAb features that best predicted level of protection across all mAbs (A-B). Each mAb is color coded by level of protection (blue = 0%; yellow = 100%) in the scoring plot (A), and features are color coded by importance to positively (red) or negatively (blue) predicting protection in the mirrored loadings plot as determined by variable influence on projection (B). All models were tested using two permutation tests (size matched random selection of features and outcome shuffling), and the predictive power of the true model was assessed using 10-fold cross-validation, providing a test for robustness of each true model. The cross validation MSE, cross validation correlation R2 value, and prediction correlation R2 values are shown in Table S5. Correlation network analysis (C) was performed to identify features associated with microneutralization (left) or polyfunctionality (right). Positive correlations between features above a threshold adjusted p-value<0.05 after Benjamini-Hochberg correction for multiple comparisons are indicated by a red connecting line, and negative correlations (adjusted p-value <0.05) are indicated by a blue connecting line. Strength of correlation is indicated by weight of the connecting line, and the background color of the features indicates the category of feature (e.g. neutralization, functional activity, etc.), as indicated in the boxed legend.
As the Elastic Net feature reduction algorithm aims to minimize correlated features to the minimal set of features that capture the overall variation of the dataset, individual features often are linked to other Ab metrics that may provide additional biological insights. Thus, to define the potentially biologically relevant features linked to the minimal Ab features selected by Elastic Net that were enriched among the more protective Abs, networks were constructed based on covariation between features (Figure 3C). Neutralizing activity was linked to Fab targeting of specific epitopes on EBOV GP (HR2, base, and fusion) that are known sites of vulnerability (Wec et al., 2017; Zhao et al., 2017). Conversely, polyfunctionality was positively associated with multiple innate immune cell effector functions as well as afucosylated and bisected glycosylation structures (G1, G2S1B, G2S1FB). Importantly, the finding that these glycan structures were linked to protection via polyfunctionality is consistent with the role of glycans in enhancing the protective efficacy of MB-003 (Olinger et al., 2012). Moreover, PLSR analysis using neutralizing features alone was insufficient to separate protective and non-protective mAbs (Figure S4A), and full separation required the inclusion of polyfunctionality (Figure S4B). Together, these data argue that both Fab-mediated neutralizing activity and Fc-mediated functions may synergize to drive protection following EBOV infection.
Fc-effector function augments the protective activity of partial or non-neutralizing mAbs.
While we found that neutralization of EBOV was significantly associated with protection within the VIC mAbs (Figure S1A, Figure 3A), we observed two anomalies. First, a subset of highly protective mAbs only exhibited moderate neutralizing activity (Figure S1), suggesting that strong neutralizing activity is not always necessary for protection. Second, a subset of potently neutralizing mAbs showed no protection, suggesting that for these Abs, neutralization was not sufficient for protection, and additional functions may be needed for these mAbs to protect.
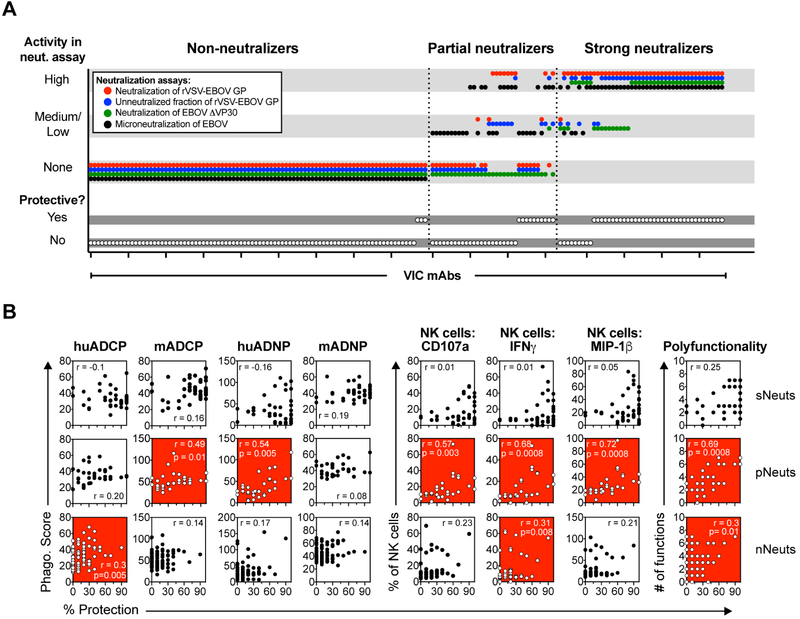
Three distinct neutralization assays with four readouts were performed to define the neutralizing activity of each of the mAbs: neutralization of a rVSV-EBOV GFP pseudotype virus (both IC50 and the fraction of cells left unneutralized at maximal Ab concentration were measured); neutralization of EBOVΔVP30-Luciferase (% Luciferase+ cells measured); and neutralization of live EBOV performed in BSL-4 (% uninfected cells measured) (Table S3). While all the neutralization assays correlated with one another (Figure S1B), a subset of the mAbs showed medium/high neutralizing activity in all assays while another subset displayed no neutralizing activity in any assay, and the remaining mAbs showed variation in neutralizing activity across the different assays, with high neutralizing activity in some assays, yet low or no activity in others (Figure 4A). We performed K-means clustering to identify three groups of neutralizing mAb profiles within the mAb library (Saphire et al., 2018): 1) mAbs with consistent medium/high neutralizing activity in all assays (strong neutralizers; sNeuts); 2) mAbs with more heterogeneous neutralizing activity (partial neutralizers; pNeuts) that have high neutralizing activity in at least one assay; and 3) mAbs with no activity in any of the assays (non-neutralizers; nNeuts). Cognizant of the functional differences between neutralizing and non-neutralizing mAbs among the handful of mAbs in the ZMapp and MB-003 cocktails (Figure 1), we used this classification scheme as a framework to further probe the role of Fc-effector function within the much more extensive mAb panel.
Figure 4. Protective efficacy of partially neutralizing mAbs is associated with induction of effector functions.
The neutralizing activity of the mAbs was evaluated in four neutralization assays, and mAbs were grouped into strong neutralizers (sNeuts; n=44), partial neutralizers (pNeuts; n=33), and non-neutralizers (nNeuts; n=90) by K-means clustering as described in (Saphire et al., 2018). For representation (A), each mAb was categorized into high (neut. activity value = 6-8), medium/low (neut. activity value = 1-5), and no activity (neut. activity value = 0) according to defined cutoff values (Table S3). The neutralizing activity and protection ≥60% (yes/no) of each mAb is plotted, with individual mAbs on the X-axis, and the neutralizing activity and protection on the Y-axis. The colored dots indicate the different neutralization readouts, and white dots indicate protection. The VIC number of the mAbs within each category are listed in Table S4. Correlation analyses was performed between the response in the indicated functional assay and protection (% of mice protected from death) within the sNeut (B; top row), pNeut (B; middle row), or nNeut groups (B; bottom row,). Spearman rho was used to determine significance of association, and an adjusted p<0.05 was considered significant after Benjamini-Hochberg correction for multiple comparisons. Red, blue, and white background of graph indicates a significant positive, negative (none register), or no significant association, respectively.
We first evaluated the association between protection and individual Fc-effector functions within each neutralization category (Figure 4B). Among the sNeuts, we did not observe any association between specific functions and protection (Figure 4B, top). Conversely, among the pNeut and nNeut mAbs, we found that several Fc-effector functions were associated with protection (Figure 4B, middle and bottom). Specifically, the protective efficacy of pNeuts was associated with Ab-dependent human neutrophil phagocytosis (huADNP), activation of NK cells (degranulation, IFNγ and MIP1β secretion), and with the ability to induce polyfunctional mAb effector profiles (Figure 4B, middle). Similarly, the protective efficacy of nNeuts was significantly, but weakly associated with recruitment of human monocyte-mediated ADCP (huADCP), NK cell-mediated secretion of IFNγ, and polyfunctionality (Figure 4B, bottom). Together, these data suggest that recruitment of innate immune effector functions may be critical for mAbs that cannot completely block viral entry via mechanical means.
Given that both Fab- and Fc-domain functions were critical for predicting protection following EBOV infection (Figure 3, (Saphire et al., 2018)), we performed Elastic Net/PLSR analyses for each of the neutralizing mAb categories to define the features associated with protection within each category. Despite the importance of neutralization as a key predictor of protection across the entire mAb panel (Figure 3), different features predicted protection in the sNeut and pNeut models (Figure 5A-B). Only 5 features were needed to predict protection within the sNeut group of mAbs, with neutralization activity as the key positive predictor of protection whereas low affinity Fabs and surprisingly, digalactosylated (G2) glycans were negative predictors of protective activity, implicating both Fab and Fc characteristics in protection even in the context of potent neutralizing activity (Figure 5A). Within the pNeut group, Elastic Net/PLSR required only 3 Fc features to predict protection, with polyfunctionality, huADNP and the afucosylated monogalactosylated (G1) Fc glycan structure as key predictors of protection (Figure 5B). Although protection was low on average among the nNeut mAbs, several effector functions were key to separating the few protective nNeut mAbs from those that had no in vivo activity (Figure 5C). Co-correlate network analyses of the Elastic Net-selected features provided enhanced resolution of the overall Ab profiles that tracked with protective immunity in each category (Figure 5D). Most notably, neutralizing activity in the sNeuts was inversely associated with the sGP epitope, consistent with the limited number of sGP-binding Abs with neutralizing activity. Additionally, several glycan structures were associated with binding affinity to EBOV GP and neutralizing activity, possibly highlighting a bias in glycosylation in the production of high affinity, strongly neutralizing monoclonal Abs. Polyfunctionality within the pNeuts and nNeuts was associated with both phagocytic and NK cell functions, further highlighting the importance of Ab function for protection by mAbs with reduced neutralizing activity.
Figure 5. Antibody functionality predicts protection mediated by partially and non-neutralizing mAbs.
The minimal features of mAbs within the sNeuts, pNeuts, and nNeuts that predict mAb-mediated protection from lethal EBOV challenge were determined using machine learning analyses. Elastic Net/PLSR analysis was used to identify the minimal mAb features that best predicted level of protection within the sNeuts (A), pNeuts (B), and nNeuts Abs (C). Plots are color coded similar to Figure 3A. Correlation network analyses were performed to identify features associated with Elastic Net-selected features of sNeuts (D; left), pNeuts (D; middle), or nNeuts (D, right). Positive or negative correlations between features above a threshold adjusted p-value<0.05 after Benjamini-Hochberg correction are indicated by a red or blue connecting line, respectively. Strength of correlation and the category of feature are indicated in the boxed legend.
Even strongly neutralizing Abs may depend on Fc-effector function to mediate protection.
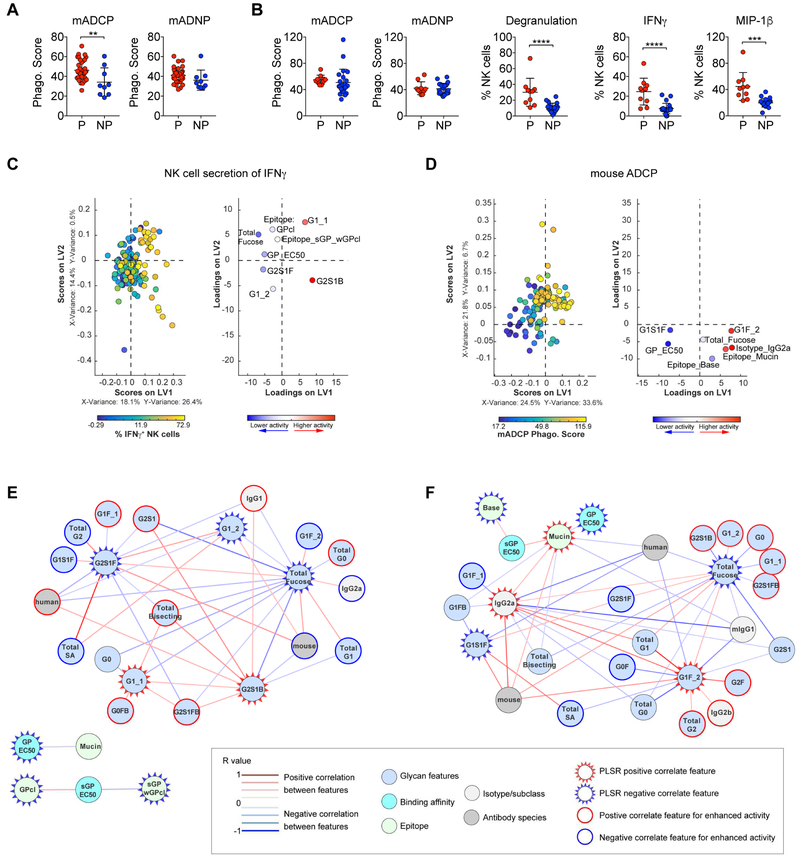
While a majority of the sNeut mAbs provided protection, a subset of sNeut mAbs did not provide protection, begging the question of why these mAbs did not protect. Several glycan structures (G2FB and G2S1) were negatively associated with protection in the sNeut group (Figure 5A), while others were positively associated with GP binding affinity, which was enriched among the protective sNeuts (Figure 5D). Thus, it is plausible that specific Fc-glycan modifications may augment functional activity to synergize with neutralization and provide protection following infection. Indeed, comparison of the phagocytic scores induced by protective and non-protective sNeut mAbs showed that the non-protective sNeuts induced lower phagocytic scores in mouse monocytes than protective sNeuts (Figure 6A, left). However, mouse neutrophil phagocytosis was similar (Figure 6A, right), suggesting that the failure of sNeut mAbs to leverage specific host innate immune cells may contribute to failure of these neutralizing mAbs to achieve in vivo protection.
Figure 6. Non-protective neutralizing mAbs are impaired in recruitment of certain effector functions.
The functional responses (mADCP, mADNP) of protective (≥60% protection; n=35) and non-protective (≤50% protection; n=9) within the sNeut group (A), and the functional responses (mADCP, mADNP, NK cell secretion of IFNγ and MIP1β) of protective (≥60% protection; n=10) and non-protective (≤50% protection; n=23) within pNeut group (B) was compared. Unpaired t-test was used to determine significance between protective and non-protective groups. ****p<0.0005 ***p<0.001 **p<0.005 after correction for multiple comparisons by Benjamini-Hochberg correction. The horizonal line represents the mean functional response within groups, and the error bars represent the standard deviation. The minimal Ab features needed to predict higher NK cell mediated IFNγ secretion (C) or mADCP activity (D) were determined by PLSR analysis using Elastic Net-selected features. All mAbs within the VIC were used in the analysis, and each mAb is color coded in the scoring plot (left graph) by activity response in each assay (blue to yellow = lowest to highest response in assay), and features are color coded by importance to positively (red) or negatively (blue) predicting activity as determined by variable influence on projection in the mirrored loadings plot (right graph). Correlation network analysis was performed to identify features associated with Elastic Net-selected features of NK cell mediated IFNγ secretion (E) or mADCP activity (F). Positive or negative correlations between features above a threshold adjusted p-value<0.05 after Benjamini-Hochberg correction are indicated by a red or blue connecting line, respectively. Strength of correlation and the category of feature are indicated in the boxed legend.
In the context of the pNeuts, huADNP and NK cell activation were associated with protection (Figure 4B). Moreover, univariate comparison of induction of effector functions by protective and non-protective pNeuts demonstrated that the protective pNeuts induced higher NK cell activity than non-protective pNeuts but did not differ in the ability to induce host (mouse) monocyte or neutrophil phagocytosis (Figure 6B). These data argue that the ability to leverage NK cell activity, not captured by the mouse monocyte and neutrophil assays, predicts higher protection by Abs that can only partially block viral entry. Thus, these data support a role for distinct functions in protection mediated by Fab domains with different neutralizing activities, with monocyte phagocytic activity enriched among protective sNeuts and NK cell activity enriched among protective pNeuts.
Given the importance of specific functions in mAb-mediated protection, we next aimed to define the biophysical features associated with these functions within the whole mAb library. PLSR/Elastic Net analysis defined the minimal mAb features associated with NK cell activation indicated by higher IFNγ secretion (Figure 6C) or higher mADCP activity (Figure 6D). Higher NK cell IFNγ secretion was associated with several known glycan structures, including afucosylated glycans (G2S1B, G1) and reduced overall fucose levels, consistent with previous reports of the negative role of fucose in activation of NK cells through FcγRIIIA (Shields et al., 2002). Consistent with other studies (Chung et al., 2014; Herter et al., 2014), network analysis of Elastic Net-selected features highlighted an enrichment of bisecting GlcNAc and particular monogalactosylated (G1) glycans with enhanced NK cell activation (Figure 6E). Only 7 features were needed to predict the most phagocytic Abs, including both Fc and Fab features such as binding affinity to EBOV GP, the glycans G1S1F and G1F, and particular isotypes (Figure 6C). In contrast to the known glycan structures associated with NK cell activity, several additional glycan structures were connected to Elastic Net-selected features for phagocytosis, pointing to structures that may be exploited to modulate phagocytic activity (Figure 6F). Thus collectively, the systems level analyses of mAb panels associated with protection provide a unique opportunity to dissect and define the linked Fab/Fc combinations that provide the greatest level of protection, providing insights for the rational engineering of next generation protective therapeutics.
DISCUSSION.
Viral neutralization is often regarded as the primary mechanism of Ab-mediated protection in the context of vaccines and therapeutics, and is commonly used as a strategy to select mAbs for in vivo protection studies, with little attention to Fc-functionality. However, the failure of some neutralizing mAbs and protective activity of several non-neutralizing mAbs has raised questions as to the specific mechanistic correlates that underlie Ab protective efficacy (Oswald et al., 2007; Wilson et al., 2000). Here, we hypothesized that Fc-effector function contributes to mAb-mediated protection against EBOV in the context of both non-neutralizing and neutralizing Abs. We analyzed a large panel of EBOV GP-specific mAbs to begin to dissect the intersection of functional and neutralizing correlates of Ab-mediated protection against EBOV, and found that the ability to leverage specific innate immune effector functions, in combination with Fab-mediated neutralizing activity, was associated with protection from EBOV. Phagocytosis and NK cell activation were particularly important for protection mediated by mAbs that were not able to strongly neutralize. However, even for those mAbs that strongly neutralized, an inability to induce phagocytosis correlated with death from EBOV challenge, arguing for a critical collaborative role for the Fc in supporting Fab protective activity. Similar findings have been reported in the HIV and influenza fields, where complete in vivo protection mediated by broadly neutralizing mAbs required recruitment of innate immune cells by the mAb Fc domain (Bournazos et al., 2014; DiLillo et al., 2016).
Our data highlight the critical importance of Fc-effector function in the setting of incomplete neutralization, and suggest that the additional capacity of pNeuts to recruit multiple functions may compensate for incomplete neutralization (Figure 7A). In addition, although only a few nNeuts included in the mAb panel provided protection, these mAbs were all highly functional, indicating that immune effector function alone can be sufficient for protection. Along these lines, the nNeut mAb in ZMapp, c13C6, is a potent inducer of all of Fc-mediated innate immune effector functions evaluated (Figure 1), whereas the neutralizing mAbs in ZMapp, c2G4 and c4G7, demonstrate reduced effector functionality. Thus, the critical inclusion of c13C6 in ZMapp suggests that the addition of effector functionality may complement the direct neutralizing activity provided by c2G4 and c4G7 to confer complete protection after disease onset. Importantly, this combination of neutralization and Fc functionality may be key to protection in the NHP challenge model, which has shown some discrepancy with small animal models in the past, particularly in the context of the potently neutralizing Ab, KZ52. (Oswald et al., 2007). While KZ52 provided protection in guinea pigs (Parren et al., 2002), it did not protect NHP (Oswald et al., 2007), thus calling into question the relevance of the small animal models for mAb down selection for EBOV. However, we show here that KZ52 exhibited negligible innate immune effector function, and specifically failed to induce activation of human NK cells (Figure 1). Thus, while neutralization alone may be sufficient to provide protection in the mouse model, the data presented here strongly argue that the mouse model may not always capture the added need for neutralizing Abs to recruit innate immune effector function, which may be critical for protection in NHP. Thus, a complementary set of Ab functional assays may be necessary to support the down selection of neutralizing mAbs to ensure that they fully leverage the protective antiviral immune response in higher order species.
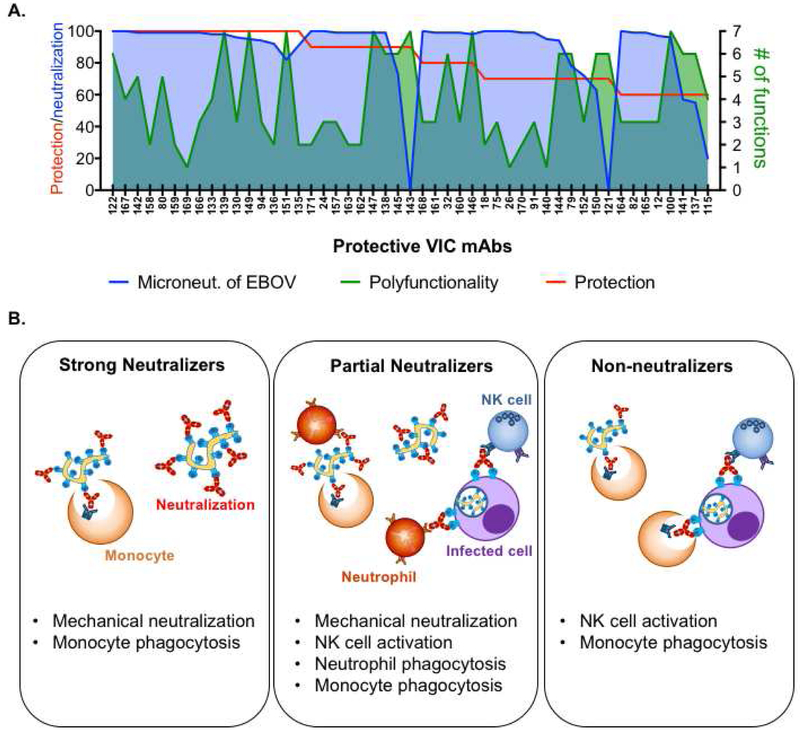
Figure 7. Exploiting both ends of the antibody to fight Ebola.
A. The overlay of neutralization of live Ebola virus (microneutralization; blue shade), number of functions (polyfunctionality; green shade) and protection of protective Abs is depicted.
B. Summary of collaboration between the Fc- and Fab- mediated Ab functions in protective Abs against EBOV.
The high level of protective efficacy observed for the mAbs isolated after 2015 highlights a potential bias towards neutralizing activity in the Ab panel, as many of these mAbs were selected from large pools by neutralizing activity alone. However, our analysis also finds a few non-neutralizing, but highly functional, and many partially neutralizing and highly functional Abs that were also protective, indicating that different combinations of neutralizing activity and function can be protective (Figure 7B). Using this dataset, we developed an unbiased computational analysis framework to define the Ab features and effector functions that are best associated increased protective efficacy in the setting of differing levels of neutralizing activity. This information may serve to help guide the design of next generation mAbs against EBOV to optimize functional activity. Our data points to glycan structures that are associated with enhanced phagocytic activity and NK cell activation (Figure 6). Currently, only a limited number of engineered glycans can be produced on monoclonal Abs (Dekkers et al., 2016). However, efforts to produce Abs with any one of the ~36 naturally occurring Ab glycan substructures are underway and will allow the dissection of the role of specific structures in modulating functional activity (Mimura et al., 2018). In addition, engineered Fc-point mutations that selectively enhance binding to different FcRs may also provide a means to enhance specific functions in the development of future mAbs. Thus, there are multiple avenues by which mAbs may be rationally designed in the future to optimize both ends of the Ab, resulting in the development of potentially highly effective and potent therapies for use in future epidemics.
Beyond the implications for immunotherapeutic design, our findings also provide unique insights for evaluating both natural and vaccine-induced immunity, where total Ab titers remain steady for years, yet neutralizing Ab titers wane over time (Regules et al., 2017; Wauquier et al., 2009). Thus, polyclonal pools of Abs may contribute to long-lived immunity through similar mechanisms as those observed for the non-neutralizing mAbs profiled in this study. Thus, our results point to the potential need to evaluate specific and additional innate immune-recruiting functions in the context of polyclonal EBOV Ab responses to fully elucidate correlates of immunity to the virus.
Ultimately, the systematic and comprehensive profiling approach used here to identify protective Ab features against EBOV provides potential avenues to modulate efficacy of existing therapeutics through enhancement of functional activity and points to future opportunities to fully explore, down select, and engineer next generation Ab therapeutics. Together, these approaches may serve as a template to rapidly develop therapeutic mAbs against other hemorrhagic fever viruses, such as Marburg virus and Lassa virus, that may re-emerge as epidemic threats in the future.
STAR Methods.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact, Dr. Galit Alter (galter@mgh.harvard.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS.
Ethics Statement for primary human innate immune cells.
Innate immune effector cells were isolated from fresh peripheral blood samples collected by the Ragon Institute or the MGH Blood bank from healthy human volunteers. All subjects signed informed consent and the study was approved by the MGH Institutional Review Board. Samples were collected from human adults older than 18 years of age, and were completely deidentified prior to use and thus, researchers were blinded to gender and age of donors.
Culture of primary human innate immune cells.
Primary human neutrophils and NK cells from deidentified donors were cultured and maintained in RPMI1640 supplemented with 10% fetal bovine serum, L-Glutamine, and penicillin/streptomycin at 37°C in a humidified incubator at 5% CO2 for the duration of the assays.
Cell lines.
The human monocyte cell line, THP-1, originally isolated from a human male infant, and mouse monocyte cell line, RAW264.7, originally isolated from an adult male BALB/c mouse, were purchased and authenticated through ATCC. Cells were maintained in RPMI1640 supplemented with 10% fetal bovine serum, L-Glutamine, penicillin/streptomycin, and 55μM beta-mercaptoethanol (THP-1 only) at 37°C in a humidified incubator at 5% CO2.
Animals.
Naïve 6-12 week old female BALB/c mice used for immune effector assays were purchased from The Jackson Laboratories and were housed in specific pathogen free conditions at the Ragon Institute of MGH, MIT, and Harvard. naïve 6-8 week old female BALB/c mice used for challenge studies were purchased from Charles River Laboratory, and were housed in microisolator cages at no more than 5 mice/cage at the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) and the Public Health Agency of Canada (PHAC). Animals had access to water and food ad libitum. All animal work involving infectious EBOV was performed in the BSL-4 containment at USAMRIID or PHAC. All USAMRIID animal studies were conducted in accordance with the Animal Welfare Act and other Federal statutes and regulations relating to animals and experiments involving animals and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 1996 edition. All procedures were reviewed and approved by the Institutional Animal Care and Use Committee. Animal work performed at PHAC was approved by the Canadian Science Centre for Human and Animal Health Animal Care Committee following the guidelines of the Canadian Council on Animal Care.
Antibodies.
All monoclonal antibodies used in this study were supplied through the Viral Hemorrhagic Immunotherapeutics Consortium (VIC) and distributed through The Scripps Research Institute, with the exception of the following: h13F6 and c6D8 (IBT Biotherapeutics); c2G4, c4G7, c13C6, and ZMapp (Mapp Biopharmaceuticals), and b12 (Polymun). All mAbs were assayed at 5μg/ml in all functional assays, and were performed in duplicate. The average of the duplicate wells were used for subsequent analysis.
METHOD DETAILS.
Protection studies.
Evaluation of monoclonal antibody protective efficacy was performed in a post-exposure therapeutic mouse model of Ebola virus infection as previously described (Bray et al., 1998; Gibb et al., 2001). Ten BALB/c female mice were infected with 100 plaque forming units (pfu) of mouse adapted Ebola virus (Mayinga isolate, Yambuku variant) two days prior to intraperitoneal administration of 100μg of a given monoclonal antibody. Ten animals per antibody were used, and ten antibodies were evaluated at a time alongside a control group of ten animals that received PBS alone, for a total of 110 animals used per round. A total of 16 rounds were performed in two BSL-4 facilities (United States Army Research Institute of Infectious Diseases and Public Health Agency of Canada). Between 0-20% of PBS-administered control animals survived infection (Saphire et al., 2018), thus establishing a threshold of >20% survival for some level of protection. A higher threshold of ≥60% was used to classify a given antibody as protective, and thus antibodies that conferred 50% protection or less were classified as non-protective for the analysis.
Measurement of antibody-mediated innate immune effector functions.
Antibody concentrations.
All mAbs were assayed at 5μg/ml in all functional assays, and were performed in duplicate. The average of the duplicate wells were used for subsequent analysis.
Antibody-mediated cellular phagocytosis by human monocytes.
ZEBOV GPΔTM (IBT Biotherapeutics) was biotinylated and conjugated to streptavidin-coated Alexa488 beads (Life Technologies). Antibodies (5μg/ml) were incubated with beads for 2 hours at 37°C. Human monocytic cells (THP-1) were added at a concentration of 2.5 × 104 cells/well and incubated for approximately 18 hours at 37°C. Cells were fixed and analyzed by flow cytometry on a BD LSRII flow cytometer and a phagocytic score was determined as follows: (percentage of FITC+ cells) * (the geometric mean fluorescent intensity (MFI) of the FITC+ cells)/10,000.
Antibody-mediated cellular phagocytosis by mouse monocytes.
ZEBOV GPΔTM-coated beads were generated as described above. Antibodies (5μg/ml) were incubated with beads for 2 hours at 37°C. Mouse monocytic cells (RAW264.7) were added at a concentration of 1.0 × 105 cells/well and incubated for 1 hour at 37°C in low adherence 96-well plates. Cells were fixed and analyzed by flow cytometry and a phagocytic score was determined as described above.
Antibody-mediated human neutrophil phagocytosis.
ZEBOV GPΔTM-coated beads were generated as described above. Antibodies (5μg/ml) were incubated with beads for 2 hours at 37°C. Human neutrophils freshly isolated from peripheral blood were added at a concentration of 5.0 × 104 cells/well and incubated for 1 hour at 37°C. Cells were stained for neutrophil markers (neutrophils were defined as high granularity SSC-A high CD66b+, CD14−, CD3−) were analyzed by flow cytometry. A phagocytic score was determined as described above.
Antibody-mediated mouse neutrophil phagocytosis.
ZEBOV GPΔTM-coated beads were generated as described above. Antibodies (5μg/ml) were incubated with beads for 2 hours at 37°C. Mouse neutrophils were freshly isolated from bone marrow of BALB/c mice, and were added at a concentration of 1.0 × 105 cells/well and incubated for 1 hour at 37°C. Cells were stained for neutrophil markers (neutrophils were defined as high granularity SSC-A high Gr-1+, CD11b+, CD3−) were analyzed by flow cytometry. A phagocytic score was determined as described above.
NK cell activation and degranulation.
3μg/ml of ZEBOV-GPΔTM was coated on a Maxisorp ELISA plate. Plates were blocked with 5% BSA prior to addition of Abs for 2 hours at 37C. Human NK cells were enriched from peripheral blood by negative selection using RosetteSep (Stem Cell Technologies) followed by Ficoll separation. The Abs were removed and the wells washed prior to addition of NK cells. The NK cells were added at 5 × 104 cells/well in the presence of brefeldin A (Sigma Aldrich), GolgiStop (BD), and anti-CD107a and incubated for 5 hours at 37C. NK cells were stained for surface markers of NK cells (CD3, CD56, CD16, BD Biosciences), followed by intracellular staining to detect the production of cytokines and chemokines (IFNγ and MIP-β, BD Biosciences). Cells were analyzed by flow cytometry on a BD LSR2 flow cytometer and data was analyzed using FlowJo software.
Analysis of glycan content of antibodies.
The relative abundance of antibody glycan structures were quantified by capillary electrophoresis as previously described (Mahan et al., 2015). Briefly, antibodies were digested with IDES (FabRICATOR, Genovis) to separate Fc and F(ab)2, and N-linked glycans were removed using PNGase F (New England Biolabs). Free glycans were labeled with 50mM APTS in 1.2M citric acid and 1M sodium cyanoborohydride in THF, and excess dye was removed using a size exclusion resin. Labeled glycans were loaded onto a 3130 XL ABI DNA Sequencer using a POP7 polymer in a 36cm capillary. Peaks of 19 substructures were identified, and the relative abundance of a given structure was determined by calculating the area under the curve of each peak divided by the total area of all peaks.
Normalization of data.
The mAbs c13C6 and b12 were used as positive and negative controls, respectively, on each plate for every assay. Data for each individual antibody is normalized to c13C6 within a given experiment, and data within each assay was batch corrected to account for experimental batch effects.
Determination of polyfunctionality.
Functional responses were divided into “high”, “medium”, and “low/none” tritiles based on the responses of c13C6 and b12. Responses equal or greater to c13C6 were considered “high”, and responses equal or less than b12 were considered “low”, and responses between b12 and c13C6 were considered “medium”. mAbs that scored medium/high for a given assay were considered functional for that assay. The number of functions each antibody induced was summed to generate a polyfunctionality score ranging from a minimum of zero functions to a maximum of 7 functions.
Calculation of Neutralizing Activity Score.
The neutralizing activity of the VIC mAbs was evaluated in four neutralization assays, and each mAbs was categorized into high (neut. activity value = 2), medium/low (neut. activity value = 1), and no activity (neut. activity value = 0) within each assay according to defined cutoff values (Table S3). A composite neutralization score was calculated based on the activity in each assay, where a neutralization score of 8 represents high activity in all four assays (4 assays * neut. activity score of 2), and a neutralization score of 0 represent no activity in any neutralization assay (4 assays * neut. activity score of 0). The mAbs were then further categorized into “strong neutralizers” (neut. activity score 6-8), “partial neutralizers” (neut. activity score of 1-5), and “non-neutralizers” (neut. activity score of 0).
QUANTIFICATION AND STATISTICAL ANALYSIS.
Association analyses.
Spearman correlation coefficients were used to determine significance of association between antibody features (functional activity, glycan abundance, etc.) and protection. P-values are corrected for multiple comparisons using the Benjamini-Hochberg correction for false discovery rate using MATLAB statistical software. The number of antibodies used in each analysis is indicated in the relevant figure legends.
Univariate statistical analyses.
Mann-Whitney test, Kruskal-Wallis test with Dunn’s multiple comparison test, unpaired t-tests, and Benjamini-Hochberg test for false discovery rate correction were used to determine statistical differences between protective and non-protective groups within sNeuts, pNeuts, and nNeuts. Analysis was performed using GraphPad Prism 7 and MATLAB. The number of antibodies used in each analysis is indicated in the relevant figure legends.
Machine learning and computational analyses:
Identification of protection-specific antibody correlates using Elastic Net/PLSR.
The minimal set of antibody features and functional parameters that collectively predicted antibody-mediated protection was defined by using a two-step model: Elastic-Net regularization followed by partial least squares regression analysis (PLSR). The Elastic-Net method was used to reduce the number of the features and to select the features most relevant to the outcome, and the PLSR was used to model the covariance relationship between the feature variables (X) and the outcome variables (Y) in the way that PLSR decomposes both X and Y into a hyperplane and maximizes covariance between the hyperplanes. To perform feature reduction, the Elastic Net method which integrates the penalty functions of least absolute shrinkage and selection operator (LASSO) and ridge regression, was used to remove irrelevant features in order to improve the robustness of high-dimensional modeling (Zou, 2005). The alpha (α) in Elastic Net, controlling the weight of L1 norm and L2 norm, was determined by 10-fold cross validation. Features with spearman r ranging between −0.05 and 0.05 were excluded from the analysis._After feature selection, PLSR was used to define the relationship between the input as a linear combination of the selected features and the outcome (protection [0-100%] or antibody function profiles). Specifically, PLSR seeks the latent variables, which linearly combine the features, which explain the maximum variance of the outcome. As a result, this approach produced a model that generates the greatest separation among the antibodies from low to high outcome (protection/antibody functions) with the lowest possible mean squared error (MSE). In order to generate a robust PLSR model less sensitive to the outliers, a bootstrap framework for Elastic Net/PLSR analysis was applied, which contained 5000 repetitions of 10-fold cross validation (Cawley, 2010). In each repetition, the dataset was randomly divided into 10 folds, where 9-fold of the dataset was used for Elastic-net/PLSR model optimization (inner cross-validation) and the remaining holdout set was used to test the model (outer cross-validation). Ultimately, each feature was weighted by the frequency of being selected among 5000 cycles and was ranked by the weights. To determine the minimal set of the correlates, we applied a sequential step-forward approach where the algorithm started with the best feature and, for each step, the next best feature was added into the PLSR model. The model prediction performance in each step was evaluated by MSE calculated from 10-fold cross-validation, and the set of the features showing the lowest MSE was defined as the minimal set of the correlates. Finally, variable importance in projection (VIP) along with the final model was calculated, a weighted sum of squares of the PLS weights, which summarized the importance of the features in a PLSR model (Galindo-Prieto, 2014)
To estimate statistical significance of the correlation (R2), the correlation of the predicted and the observed outcome, two types of random permutation tests were computed, 1) outcome shuffling, shuffling the order of the outcome labels. 2) size-matched random feature selection, randomly selecting the features in the same size as the actual minimal set of the correlates. In total 10,000 random models from each of both tests were collected to estimate the null distributions for the R2. Eventually an empirical, nominal p-value for the R2 in each test was calculated relative to this null distribution, which represents the statistical significance of the true model.
Network interactions.
The Elastic Net/PLSR model selects the features the most relevant to the outcome, those of which can be viewed as the biomarkers for predicting antibody protection. The correlation network, which interrogates correlations among PLSR-selected correlates and other features, may provide further mechanistic insight of antibody-mediated immunity. To do this, the network was constructed based on the correlation coefficients between antibody features. Edges between nodes was weighted based on the correlation coefficient, between the features represented by the two nodes. Using significant correlation coefficients after correcting for multiple comparisons (Benjamini-Hochberg q-value <0.05) testing the null hypothesis of zero correlation) an adjacency matrix was constructed using soft thresholding to define edge weights. The adjacency network was built by using Cytoscape.
DATA AND SOFTWARE AVAILABILITY.
The dataset generated and used in this study is available through http://apps.vhfimmunotherapy.org/vic-data-explorer/ and is derived from (Saphire et al., 2018).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| c2G4 | Mapp Bio | n/a |
| c4G7 | Mapp Bio | n/a |
| c13C6 | Mapp Bio | n/a |
| ZMapp | Mapp Bio | n/a |
| h13F6 | IBT Bioservices | Cat#: 0201-022 |
| c6D8 | IBT Bioservices | Cat#: 0201-021 |
| KZ52 | IBT Bioservices | Cat#: 0260-001 |
| b12 | Polymun | Cat#: AB011 |
| Mouse anti-human CD66b (clone G10F5) | BioLegend | Cat#: 305112 |
| Mouse anti-human CD3 (clone UCHT1) | BD Biosciences | Cat#: 557943 |
| Mouse anti-human CD107a (clone H4A3) | BD Biosciences | Cat#: 555802 |
| Mouse anti-human IFNγ (clone B27) | BD Biosciences | Cat#: 554702 |
| Mouse anti-human MIP-1β (clone D21-1351) | BD Biosciences | Cat#: 550078 |
| Mouse anti-human CD56 (clone B159) | BD Biosciences | Cat#: 557747 |
| Mouse anti-human CD14 (clone MφP9) | BD Biosciences | Cat#: 557943 |
| Mouse anti-human CD16 (clone 3G8) | BD Biosciences | Cat#: 557758 |
| Rat anti-mouse Ly-6G/Ly-6C (GR-1) (clone RB6-8C5) | BioLegend | Cat#: 108412 |
| Rat anti-CD11b (clone M1/70) | BD Biosciences | Cat#: 560455 |
| Hamster anti-mouse CD3 (clone 145-2C11) | BioLegend | Cat#: 100334 |
| VIC Antibody Panel | Saphire et al. 2018 | n/a |
| VIC Antibody set 10 | Wec et al., 2017; Bornholdt et al., 2016 | n/a |
| VIC Antibody set 2 | Takada et al., 2003 | n/a |
| VIC Antibody set 3 | Qiu et al., 2011; Qiu et al., 2012 | n/a |
| VIC Antibody set 4 | Flyak et al., 2016 | n/a |
| VIC Antibody set 5 | Ewer et al., 2016; Huang et al., 2015 | n/a |
| VIC Antibody set 6 | Keck et al., 2015 | n/a |
| VIC Antibody set 7 | Goh et al., 2014 | n/a |
| VIC Antibody set 8 | EMORY | |
| VIC Antibody set 9 | Murphy et al., 2014; Macdonald et al., 2014 | n/a |
| Bacterial and Virus Strains | ||
| Mouse-adapted EBOV/Mayinga (EBOV/M.mustc/COD/76/Yambuku-Mayinga | Bray et al., 1998 | n/a |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 9-aminopyrene-1,4,6-trisulfonic acid (APTS) | Life Technologies | Cat#: A6257 |
| Brefeldin A | Sigma Aldrich | Cat#: B7651 |
| GolgiStop | BD Biosciences | Cat#: 554724 |
| EBOV GP∆TM | IBT Bioservices | Cat#: 0501-015 |
| FabRICATOR IdeS | Genovis | Cat#: A0-FR1-096 |
| PNGase F | New England Biolabs | Cat#: P0704S |
| FluoSpheres® NeutrAvidin®-Labeled Microspheres, 1.0 μm, yellow-green fluorescent (505/515), 1% solids | Life Technologies | Cat#: F-8776 |
| Critical Commercial Assays | ||
| RosetteSep NK cell enrichment kit | Stem Cell Technologies | Cat#: 15025 |
| Experimental Models: Cell Lines and Primary Cells | ||
| Human: THP-1 monocytes | ATCC | Cat#: TIB-202 |
| Mouse: RAW264.7 monocytes | ATCC | Cat#: TIB-71 |
| Primary human immune cells | MGH Blood Bank; Ragon Institute Clinical Core | n/a |
| Experimental Models: Organisms/Strains | ||
| Mouse: Female BALB/c | Charles River; The Jackson Laboratories | Strain Code 028 (CR); Stock number 000651 (JAX) |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad Software, Inc. | RRID:SCR_002798 |
| MATLAB | MathWorks | RRID:SCR_001622 |
| R Studio | R Project for Statistical Computing | RRID:SCR_000432 |
| Cytoscape | RRID:SCR_003032 | |
| Other | ||
| 3130 XL ABI DNA Sequencer with 36 cm capillary and POP7 polymer | Applied Biosystems | n/a |
| BD LSR2 flow cytometer | BD Biosciences | n/a |
| Deposited Data | This manuscript, Saphire et al., 2018 | https://data.mendeley.com/datasets/5ckb2jrk7t/1 |
HIGHLIGHTS.
The Fc-effector profile of 168 Ebola-specific monoclonal antibodies was defined
Effector function correlates with protection for partially neutralizing antibodies
Non-protective, strongly neutralizing antibodies fail to induce phagocytosis
Profiles of protective antibodies may aid in design of future immunotherapeutics
ACKNOWLEDGEMENTS.
We would like to thank all those who contributed their monoclonal antibodies to the VIC for this study, the members of the VIC, and members of the Alter Lab for valuable discussion and insights. This work was supported by a supplement to the NIH U19 AI109762, the Ragon Institute, and the MGH Scholars Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS.
L.Z. is an employee, shareholder, and owner of Mapp Biopharmaceutical.
REFERENCES
- Ackerman ME, Dugast AS, McAndrew EG, Tsoukas S, Licht AF, Irvine DJ, and Alter G (2013). Enhanced phagocytic activity of HIV-specific antibodies correlates with natural production of immunoglobulins with skewed affinity for FcgammaR2a and FcgammaR2b. Journal of virology. 87(10), 5468–5476. DOI: 10.1128/JVI.03403-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G, Malenfant JM, and Altfeld M (2004). CD107a as a functional marker for the identification of natural killer cell activity. Journal of immunological methods. 294(1-2), 15–22. DOI: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Turner HL, Murin CD, Li W, Sok D, Souders CA, Piper AE, Goff A, Shamblin JD, Wollen SE, et al. (2016). Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science. 351(6277), 1078–1083. Published online 2016/02/26 DOI: 10.1126/science.aad5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, and Ravetch JV (2014). Broadly Neutralizing Anti-HIV-1 Antibodies Require Fc Effector Functions for In Vivo Activity. Cell. 158(6), 1243–1253. DOI: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Davis K, Geisbert T, Schmaljohn C, and Huggins J (1998). A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis 178(3), 651–661. Published online 1998/09/05. [DOI] [PubMed] [Google Scholar]
- Bruhns P (2012). Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 119(24), 5640–5649. DOI: 10.1182/blood-2012-01-380121. [DOI] [PubMed] [Google Scholar]
- Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, and Daeron M (2009). Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 113(16), 3716–3725. DOI: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- Cawley GC, Talbot NLC (2010). On Over-fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation. Journal of Machine Learning Research. 11, 2079–2107. [Google Scholar]
- Chung AW, Crispin M, Pritchard L, Robinson H, Gorny MK, Yu X, Bailey-Kellogg C, Ackerman ME, Scanlan C, Zolla-Pazner S, et al. (2014). Identification of antibody glycosylation structures that predict monoclonal antibody Fc-effector function. Aids. 28(17), 2523–2530. DOI: 10.1097/QAD.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Misasi J, Mulangu S, Stanley DA, Kanekiyo M, Wollen S, Ploquin A, Doria-Rose NA, Staupe RP, Bailey M, et al. (2016). Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 351(6279), 1339–1342. DOI: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- Crowe JE Jr. (2017). Principles of Broad and Potent Antiviral Human Antibodies: Insights for Vaccine Design. Cell host & microbe. 22(2), 193–206. Published online 2017/08/12 DOI: 10.1016/j.chom.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers G, Plomp R, Koeleman CA, Visser R, von Horsten HH, Sandig V, Rispens T, Wuhrer M, and Vidarsson G (2016). Multi-level glyco-engineering techniques to generate IgG with defined Fc-glycans. Scientific reports. 6, 36964 Published online 2016/11/23 DOI: 10.1038/srep36964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, Palese P, Wilson PC, and Ravetch JV (2016). Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. The Journal of clinical investigation. 126(2), 605–610. DOI: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, and Ravetch JV (2015). Differential Fc-Receptor Engagement Drives an Anti-tumor Vaccinal Effect. Cell. 161(5), 1035–1045. DOI: 10.1016/j.cell.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, et al. (2016). Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection. Cell. DOI: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-Prieto B, Eriksson L, Trygg J (2014). Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). Journal of Chemometrics. 28(8), 623–632. DOI: doi: 10.1002/cem.2627. [DOI] [Google Scholar]
- Gibb TR, Bray M, Geisbert TW, Steele KE, Kell WM, Davis KJ, and Jaax NK (2001). Pathogenesis of experimental Ebola Zaire virus infection in BALB/c mice. J Comp Pathol. 125(4), 233–242. Published online 2002/01/19 DOI: 10.1053/jcpa.2001.0502. [DOI] [PubMed] [Google Scholar]
- Goede V, Fischer K, Busch R, Engelke A, Eichhorst B, Wendtner CM, Chagorova T, de la Serna J, Dilhuydy MS, Illmer T, et al. (2014). Obinutuzumab plus chlorambucil in patients with CLL and coexisting conditions. N Engl J Med 370(12), 1101–1110. Published online 2014/01/10 DOI: 10.1056/NEJMoa1313984. [DOI] [PubMed] [Google Scholar]
- Gunn BM, and Alter G (2016). Modulating Antibody Functionality in Infectious Disease and Vaccination. Trends in molecular medicine. Published online 2016/10/21 DOI: 10.1016/j.molmed.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, et al. (2015). Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet. 386(9996), 857–866. DOI: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- Herter S, Birk MC, Klein C, Gerdes C, Umana P, and Bacac M (2014). Glycoengineering of therapeutic antibodies enhances monocyte/macrophage-mediated phagocytosis and cytotoxicity. Journal of immunology. 192(5), 2252–2260. Published online 2014/02/04 DOI: 10.4049/jimmunol.1301249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indik ZK, Park JG, Hunter S, and Schreiber AD (1995). The molecular dissection of Fc gamma receptor mediated phagocytosis. Blood. 86(12), 4389–4399. Published online 1995/12/15. [PubMed] [Google Scholar]
- Kanapathipillai R, Henao Restrepo AM, Fast P, Wood D, Dye C, Kieny MP, and Moorthy V (2014). Ebola vaccine--an urgent international priority. The New England journal of medicine. 371(24), 2249–2251. DOI: 10.1056/NEJMp1412166. [DOI] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, and Saphire EO (2008). Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 454(7201), 177–182. DOI: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan AE, Tedesco J, Dionne K, Baruah K, Cheng HD, De Jager PL, Barouch DH, Suscovich T, Ackerman M, Crispin M, et al. (2015). A method for high-throughput, sensitive analysis of IgG Fc and Fab glycosylation by capillary electrophoresis. Journal of immunological methods. 417, 34–44. DOI: 10.1016/j.jim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura Y, Katoh T, Saldova R, O'Flaherty R, Izumi T, Mimura-Kimura Y, Utsunomiya T, Mizukami Y, Yamamoto K, Matsumoto T, et al. (2018). Glycosylation engineering of therapeutic IgG antibodies: challenges for the safety, functionality and efficacy. Protein Cell. 9(1), 47–62. Published online 2017/06/10 DOI: 10.1007/s13238-017-0433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinger GG Jr., Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, et al. (2012). Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proceedings of the National Academy of Sciences of the United States of America. 109(44), 18030–18035. DOI: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald WB, Geisbert TW, Davis KJ, Geisbert JB, Sullivan NJ, Jahrling PB, Parren PW, and Burton DR (2007). Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 3(1), e9 Published online 2007/01/24 DOI: 10.1371/journal.ppat.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overdijk MB, Verploegen S, Ortiz Buijsse A, Vink T, Leusen JH, Bleeker WK, and Parren PW (2012). Crosstalk between human IgG isotypes and murine effector cells. J Immunol. 189(7), 3430–3438. Published online 2012/09/08 DOI: 10.4049/jimmunol.1200356. [DOI] [PubMed] [Google Scholar]
- Parren PW, Geisbert TW, Maruyama T, Jahrling PB, and Burton DR (2002). Pre- and postexposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. Journal of virology. 76(12), 6408–6412. Published online 2002/05/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt J, Zeitlin L, Kim do H, Working C, Johnson JC, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, et al. (2013). Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Science translational medicine. 5(199), 199ra113 DOI: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- Pincetic A, Bournazos S, DiLillo DJ, Maamary J, Wang TT, Dahan R, Fiebiger BM, and Ravetch JV (2014). Type I and type II Fc receptors regulate innate and adaptive immunity. Nat Immunol. 15(8), 707–716. Published online 2014/07/22 DOI: 10.1038/ni.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PREVAIL II Writing Group, Multi-National PREVAIL II Study Team, Davey RTJ, Dodd L, Proschan MA, Neaton J, Neuhaus Nordwall J, Koopmeiners JS, Beigel J, Tierney J, et al. (2016). A Randomized, Controlled Trial of ZMapp for Ebola Virus Infection. The New England journal of medicine. 375(15), 1448–1456. Published online 2016/10/13 DOI: 10.1056/NEJMoa1604330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Audet J, Lv M, He S, Wong G, Wei H, Luo L, Fernando L, Kroeker A, Bovendo HF, et al. (2016). Two-mAb cocktail protects macaques against the Makona variant of Ebola virus. Science translational medicine. 8(329), 329ra333 DOI: 10.1126/scitranslmed.aad9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, et al. (2014). Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 514(7520), 47–53. Published online 2014/08/30 DOI: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regules JA, Beigel JH, Paolino KM, Voell J, Castellano AR, Hu Z, Munoz P, Moon JE, Ruck RC, Bennett JW, et al. (2017). A Recombinant Vesicular Stomatitis Virus Ebola Vaccine. The New England journal of medicine. 376(4), 330–341. DOI: 10.1056/NEJMoa1414216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire EO, Schendel SL, Fusco ML, Gangavarapu K, Gunn BM, Wec AZ, Halfmann PJ, Brannan JM, Herbert AS, Qiu X, et al. (2018). Systematic analysis of monoclonal antibodies against Ebola virus GP defines features contributing to protection. In Press at Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, and Presta LG (2002). Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. The Journal of biological chemistry. 277(30), 26733–26740. DOI: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- Strasser R, Stadlmann J, Schahs M, Stiegler G, Quendler H, Mach L, Glossl J, Weterings K, Pabst M, and Steinkellner H (2008). Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant biotechnology journal. 6(4), 392–402. DOI: 10.1111/j.1467-7652.2008.00330.x. [DOI] [PubMed] [Google Scholar]
- Umana P, Jean-Mairet J, Moudry R, Amstutz H, and Bailey JE (1999). Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol. 17(2), 176–180. Published online 1999/03/03 DOI: 10.1038/6179. [DOI] [PubMed] [Google Scholar]
- Wauquier N, Becquart P, Gasquet C, and Leroy EM (2009). Immunoglobulin G in Ebola outbreak survivors, Gabon. Emerging infectious diseases. 15(7), 1136–1137. DOI: 10.3201/eid1507.090402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec AZ, Herbert AS, Murin CD, Nyakatura EK, Abelson DM, Fels JM, He S, James RM, de La Vega MA, Zhu W, et al. (2017). Antibodies from a Human Survivor Define Sites of Vulnerability for Broad Protection against Ebolaviruses. Cell. 169(5), 878–890 e815. Published online 2017/05/20 DOI: 10.1016/j.cell.2017.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner LM, Dhodapkar MV, and Ferrone S (2009). Monoclonal antibodies for cancer immunotherapy. Lancet. 373(9668), 1033–1040. Published online 2009/03/24 DOI: 10.1016/S0140-6736(09)60251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, and Hart MK (2000). Epitopes involved in antibody-mediated protection from Ebola virus. Science. 287(5458), 1664–1666. [DOI] [PubMed] [Google Scholar]
- Zeitlin L, Pettitt J, Scully C, Bohorova N, Kim D, Pauly M, Hiatt A, Ngo L, Steinkellner H, Whaley KJ, et al. (2011). Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proc Natl Acad Sci U S A 108(51), 20690–20694. Published online 2011/12/07 DOI: 10.1073/pnas.1108360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Howell KA, He S, Brannan JM, Wec AZ, Davidson E, Turner HL, Chiang CI, Lei L, Fels JM, et al. (2017). Immunization-Elicited Broadly Protective Antibody Reveals Ebolavirus Fusion Loop as a Site of Vulnerability. Cell. 169(5), 891–904 e815. DOI: 10.1016/j.cell.2017.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T (2005). Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology). 67(2), 301–320. DOI: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.