Abstract
Purpose
MicroRNAs play a prominent role in a variety of physiological and pathological biological processes, including cancer. For rectal cancers, only limited data are available on microRNA expression profiles, while the underlying genomic and transcriptomic aberrations have been firmly established. We therefore aimed to comprehensively map the microRNA expression patterns of this disease.
Experimental design
Tumor biopsies and corresponding matched mucosa samples were prospectively collected from 57 patients with locally advanced rectal cancers. Total RNA was extracted, and tumor and mucosa microRNA expression profiles were subsequently established for all patients. The expression of selected microRNAs was validated using semi-quantitative real-time PCR.
Results
Forty-nine microRNAs were significantly differentially expressed (log2-fold difference >0.5 and p<0.001) between rectal cancer and normal rectal mucosa. The predicted targets for these microRNAs were enriched for the following KEGG pathways: Wnt, TGF-beta, mTOR, insulin, MAPK, and ErbB signaling. Thirteen of these 49 microRNAs seem to be rectal cancer-specific, and have not been previously reported for colon cancers: miR-492, miR-542-5p, miR-584, miR-483-5p, miR-144, miR-2110, miR-652*, miR-375, miR-147b, miR-148a, miR-190, miR-26a/b, and miR-338-3p. Of clinical impact, miR-135b expression correlated significantly with disease-free and cancer-specific survival in an independent multicenter cohort of 116 patients.
Conclusion
This comprehensive analysis of the rectal cancer microRNAome uncovered novel microRNAs and pathways associated with rectal cancer. This information contributes to a detailed view of this disease. Moreover, the identification and validation of miR-135b may help to identify novel molecular targets and pathways for therapeutic exploitation.
Introduction
Cancer cells exhibit complex alterations of their genome, transcriptome, and epigenome. More recently, evidence has accumulated that a class of small non-coding RNAs, termed microRNAs (miRNAs), also contributes to tumor initiation and progression, in addition to their pivotal role in essential biological processes such as development, cellular differentiation, proliferation and apoptosis. The number of verified human miRNAs is constantly growing.
Colorectal cancer is the third most common cause of cancer and the second leading cause of cancer death in Europe and the United States (1). From a clinical point of view, cancers of the colon and the rectum are two distinct entities that require different treatment strategies. Accordingly, when analyzing the genetics and biology of these tumor entities, they should be separated from each other. With respect to colon cancer, extensive catalogues of deregulated microRNAs have been identified in the past few years. For rectal cancer, however, only very limited data are available (2), particularly because previous contributions failed to separate these entities.
For this reason, we aimed to establish comprehensive microRNA expression patterns of this common tumor. We therefore prospectively collected tumor biopsies from 57 patients with locally advanced rectal cancers enrolled in phase-III clinical trials to ensure standardized acquisition of biomaterial and follow-up. To enable a direct comparison of rectal cancer to matched normal rectal mucosa, corresponding non-tumor biopsies from each individual patient were ascertained as well. Accordingly, we present here the largest and most comprehensive comparative analysis of locally advanced rectal cancers, which serves as the basis for the rectal cancer microRNAome.
Materials and Methods
Patients and sample collection
For this study, we prospectively collected pre-therapeutic biopsies from 57 patients with locally advanced rectal cancer who were treated at the Department of General and Visceral Surgery, University Medical Center, Göttingen, Germany. To further validate miRNA findings, a completely independent set of 116 patients was recruited from our and four additional surgical departments in Germany (Goettingen, Erlangen, Moenchengladbach, Oldenburg, and Kassel). All patients were enrolled or treated according to the CAO/ARO/AIO-94 (3) and CAO/ARO/AIO-04 trial of the German Rectal Cancer Study Group. Written informed consent was obtained from all patients according to the guidelines approved by the local ethic committee, and the clinical data are summarized in Supplementary Table S1.
All patients received a total radiation dose of 50.4 Gy (single dose of 1.8 Gy) accompanied by either a 120-hour continuous intravenous application of 5-FU (1000 mg/m2/day on days 1–5 and days 29–33), or a combination of an intravenous infusion of oxaliplatin (50 mg/m2 on days 1, 8, 22 und 29 over 2-hour) and a continuous infusion of 5-FU (250 mg/m2/day on days 1–14 and 22–35). Six weeks after preoperative radiochemotherapy (RCT), surgery with quality controlled total mesorectal excision (TME) was performed. Four to six weeks after surgery, multimodal therapy was completed by either four cycles of 5-FU (500 mg/m2/d) bolus infusion (days 1 to 5) or with eight cycles of 5-FU (2400 mg/m2) continuous infusion with folinic acid (400 mg/m2) and oxaliplatin (100 mg/m2) infusion.
Biopsies were acquired before any treatment was administered. Along with the tumor (T) adjacent non-tumorous mucosa (N) was taken that had to be at least 2 cm distant from the tumor. Biopsies were then immediately transferred into RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany), and stored over night at 4°C to allow saturation of the entire biopsy. Long-term storage was performed at −20°C.
RNA isolation and microRNA expression profiling
RNA was extracted using TRIZOL (Invitrogen, Carlsbad, CA) as previously described (4). Nucleic acid quantity, quality and purity were determined using a spectrophotometer (Nanodrop, Rockland, DE) and a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Subsequently, microRNA expression profiling was carried out on LNA™ (Locked Nucleic Acid) enhanced miRCURY™ microarrays (Exiqon, Vedbaek, Denmark). These arrays contain Tm-normalized capture probes for 2,090 microRNAs. All hybridizations were performed against a common reference pool consisting of an equimolar mixture of total RNA from all samples. This enables both one- and two-channel data analysis, as described in detail by Søkilde et al. (5).
1 μg of total RNA from each sample was labeled using the miRCURY™ LNA microRNA Power Labeling Kit (Exiqon) following a two-step protocol: First, Calf Intestinal Alkaline Phosphatase (CIAP) was applied to remove terminal 5′phosphates. Second, fluorescent labels were attached enzymatically to the 3′-end of the microRNAs: Sample-specific RNA was labeled with Hy3 (green channel) fluorophore, while the common reference RNA pool was labeled with Hy5 (red channel). The labeled RNA was hybridized to miRCURY arrays for 16 hours at 65°C in a Tecan HS4800 hybridization station. After washing and drying, the microarrays were scanned in an Agilent G2565BA Microarray Scanner System (Agilent Technologies, Santa Clara, CA) and the images were quantified using ImaGene® v. 8.0 (BioDiscovery, El Segundo, CA).
Data analysis
Low-level analysis was performed in the R statistical computing environment, including importing and pre-processing of the data using the LIMMA package (available at http://bioconductor.org). After excluding flagged spots from the analysis, the “normexp” background correction method, plus “offset=50” was applied. For single channel analysis, the intensities were log2 transformed and quantile normalized as implemented in LIMMA. Both log2 intensities (single channel analysis) and log2 ratios (dual channel analysis) of four intra-slide replicates were averaged.
To identify microRNAs that were differentially expressed, we applied the empirical Bayes, moderated t-statistics implemented in LIMMA. Linear models were fitted to the data, and comparisons of interest were extracted as contrasts. Unsupervised hierarchical clustering with complete linkage, using both Euclidean and 1-Pearson correlation as distance metrics, was applied to cluster the samples according to their microRNA expression levels. Both group comparisons (tumor versus normal) as well as within-patient comparisons (individual T/N ratios) were applied to identify the most reliable subset of microRNAs specific for rectal cancer. Only those miRNAs that were significant in both analyses based on (|log2(T/N)|>0.5 tumor versus mucosa and p<0.001) were used for validation and are discussed in the results. All p-values were adjusted for multiple testing using the function “p.adjust” and the False Discovery Rate (FDR) method described by Benjamini and Hochberg (6). Data visualization, including dynamic principle component analysis (PCA; based on variance filtering, significance level and FDR), heat maps and clustering was performed in Qlucore Omics Explorer v.2.2 (Qlucore AB, Lund, Sweden).
For disease-free (DFS) and cancer-specific survival (CSS) analysis, Kaplan-Meier plots and the Cox proportional hazards model were applied using the R package “survival”. A step-forward ANOVA model was used for pair-wise comparison of biomarkers, the expression of which was log2 transformed and treated as continuous variables. DFS was defined as time from resection of the tumor (patients considered as tumor-free) until the development of distant or local recurrence, and CSS as time until tumor-related death.
Bioinformatic analysis and target prediction
Differentially expressed microRNAs (see criteria above) were analyzed by using the commercial software IPA v.8.8 (Ingenuity® Systems, www.ingenuity.com, Ingenuity Systems, Redwood City, CA), and by DIANA-miRPath, a free web-based tool that performs enrichment analysis of microRNA target genes to all known KEGG pathways (7). Messenger RNA expression values of genes indicated in the IPA network were overlaid accordingly and were retrieved from a previous publication (8) comparing 65 matched rectal cancer and mucosa samples (GSE20842). This was possible because the samples used within the present study for miRNA expression profiling have been previously analyzed using whole genome expression arrays. Potential microRNA targets were assessed by several target prediction algorithms to identify overlapping target genes: MicroCosm Targets (http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/search.pl), Diana Lab microT (http://diana.cslab.ece.ntua.gr/microT/), PicTar (http://pictar.mdc-berlin.de/), and TargetScan 5 (http://www.targetscan.org/).
As an additional approach, miRNA target predictions were retrieved from miRecords V.3 (http://mirecords.biolead.org/) and compared to the mRNA expression levels from our previous analysis (8) (GSE20842). Target genes were matched to the array probes via ENSEMBL ID or Gene Symbol. In case of hits to multiple probes, only the probe with the most significant differential expression between tumor and mucosa was taken. A cutoff of an FDR < 0.05 for differential gene expression between tumor and mucosa was used to determine significantly differentially up or down-regulated genes. Fisher’s Exact Test was used to determine statistical overrepresentation in each target gene set, in the case of up-regulated miRNAs a one sided test was used on the down-regulated genes, in case of down-regulated miRNAs a one sided test on up-regulated genes; False Discovery Rate was used to adjust the p-values for multiple testing (6).
Semi-quantitative real-time PCR
Semi-quantitative real-time PCR was performed using Taqman microRNA assays (Applied Biosystems, Foster City, California) and a BioMark 48.48 Dynamic Array System (Fluidigm, South San Francisco, CA) according to the manufacturer’s instructions. After reverse transcription, pre-amplified cDNA of 24 rectal cancers and 24 matched mucosa samples was loaded into the Dynamic Array along with 2x Master Mix (Applied Biosystems) and Loading Agent (Fluidigm). Based on the microarray data, 10 microRNAs differentially expressed between cancerous and normal tissues as well as three housekeeping microRNAs were selected for PCR validation. Based on their stable expression over all patients and representing different levels of expression intensity (high, medium and low), three small nucleolar RNAs (snoRNA), i.e., U66, U44, and U48, were chosen as endogenous normalization controls. U44 and U48 have previously been used to normalize miRNA experiments (9, 10). A potential bias due to its localization within the growth arrest specific 5 (GAS5) transcript has been reported for U44 (11). These data, however, were retrieved from a mixed cohort of breast as well as head and neck cancers and did not hold true in our data set.
All assays were performed in triplicate. The target sequences for the PCR reactions are listed in Supplementary Table S2. Replicates with a Ct standard deviation greater than 1 were omitted from further analysis. MicroRNA expression was quantified as ΔCt values, where Ct = threshold cycle, ΔCt = (Ct target microRNA − average Ct of RNU66, U44, and U48), and ΔΔCt values, ΔΔCt = (ΔCt target microRNA tumor tissue − ΔCt target microRNA matched normal tissue) were used to quantify microRNA expression of tumor compared to matched mucosa. ΔCt was calculated using Fluidigm Real-Time PCR Analysis software (v2.1).
In addition to this technical validation of the miRNA array data, semi-quantitative real-time PCR was used to measure the expression levels of miR-135b in a completely independent set of 116 rectal cancer patients. These PCR experiments were performed using the ABI 7900HT system (Applied Biosystem). Specific primers for miR-135b as well as for U66, U44 and U48 were obtained from Qiagen (Hilden, Germany). According to the manufacturer’s instructions, cDNA containing universal tag was generated from total RNA using the miScript Reverse Transcription Kit (Qiagen). Quantification was carried out using QuantiTect SBYR Green® PCR Master Mix (Qiagen).
Results
The rectal cancer miRNAome – differentially expressed microRNAs
In contrast to colon cancer, only very limited data on microRNA expression profiles are available for rectal cancer (2, 12, 13). Towards the establishment of comprehensive microRNA expression patterns, we profiled pre-therapeutic tumor biopsies that were prospectively collected from 57 patients with locally advanced rectal cancers. All patients were enrolled in phase-III clinical trials to ensure standardized acquisition of biomaterial. To enable a direct comparison of rectal cancer to matched normal rectal mucosa, corresponding non-tumor biopsies from each individual patient were hybridized as well. An updated version of a recently described microarray platform (14) containing LNA-enhanced capture probes for quantitation of 2,090 mature microRNAs, including 238 proprietary microRNAs identified by 454 sequencing (15), 10 snoRNA, and 55 viral microRNAs relevant for human was used for this analysis.
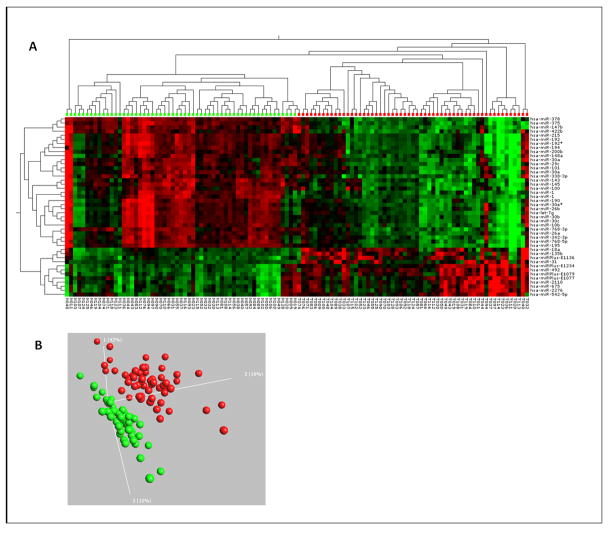
Following variance filtering (SD > 0.2) and inclusion of only signals that were higher than twice the background, 732 microRNAs were left for further analysis and data were uploaded to GEO Database GSE38389. Of these, 49 differed with high significance (|log2(T/N)|>0.5 and p<0.001) between normal and rectal cancer tissue. This miRNA signature was able to separate the tumor from the normal group in the 2-way hierarchical cluster shown in Figure 1A. Twenty of these differentially expressed microRNAs were up-regulated, and 29 were down-regulated in tumor versus mucosa (Table 1), suggesting a general repression of microRNA expression in cancer. This would be in line with the original observations by Lu et al. (16) for a variety of human cancers. However, using a looser filter, e.g. |log2(T/N)|>0.2 and p<0.01, we identified 266 microRNAs as differentially expressed, and of these, 107 are down-regulated in rectal cancer compared to normal mucosa, while 159 are up-regulated (Supplementary Table S3). Therefore, the general perception that microRNAs are down-regulated in human cancer might not apply to all tumor entities.
Figure 1.
A) Heat-map and two-way hierarchical clustering based on 49 miRNAs that were differentially expressed between tumor and normal samples. Normal (green label) and tumor (red label) samples fall in separate clusters. B) PCA plot based on the 100 most variable miRNAs (SD>0.2) showing complete, unsupervised separation of the 114 array samples into 57 tumor (red) and 57 matched normal (green) samples.
Table 1.
miRNAs that are up- or down-regulated in rectal cancer tissue compared to normal mucosa
| miRNA | log2(T/N) | p | q | miRNA cluster | CC |
|---|---|---|---|---|---|
| Up-regulated (20) | |||||
| hsa-miR-492 | 1.44 | 4.45E-20 | 2.92E-19 | N | |
| hsa-miR-223 | 0.99 | 9.75E-07 | 1.09E-06 | Y | |
| hsa-miRPlus-E1234 | 0.95 | 1.84E-14 | 6.06E-14 | N | |
| hsa-miR-135b | 0.93 | 2.83E-11 | 5.02E-11 | Y | |
| hsa-miR-542-5p | 0.90 | 6.05E-09 | 8.19E-09 | 424~503~542~450a-2/1~450b | N |
| hsa-miRPlus-E1245 | 0.86 | 1.01E-06 | 1.10E-06 | N | |
| hsa-miRPlus-E1079 | 0.76 | 3.32E-15 | 1.53E-14 | N | |
| SNORD12B | 0.69 | 1.39E-11 | 2.66E-11 | N | |
| hsa-miR-144 | 0.69 | 5.28E-05 | 1.06E-04 | 144~451 | N |
| hsa-miRPlus-E1077 | 0.68 | 4.03E-10 | 6.18E-10 | N | |
| hsa-miR-2110 | 0.65 | 1.95E-11 | 3.59E-11 | N | |
| hsa-miR-18a | 0.65 | 5.80E-08 | 7.21E-08 | 17~18a~19a~20a~19b1~92a1 | Y |
| hsa-miR-17 | 0.54 | 1.90E-04 | 3.66E-04 | 17~18a~19a~20a~19b1~92a1 | Y |
| hsa-miR-675 | 0.59 | 6.95E-09 | 9.14E-09 | Y | |
| hsa-miR-584 | 0.59 | 1.11E-06 | 1.18E-06 | N | |
| hsa-miRPlus-A1027 | 0.57 | 5.59E-07 | 6.43E-07 | N | |
| hsa-miR-31 | 0.56 | 4.79E-09 | 6.67E-09 | Y | |
| hsa-miR-21 | 0.56 | 2.94E-06 | 3.08E-06 | Y | |
| hsa-miR-483-5p | 0.56 | 3.06E-07 | 3.70E-07 | N | |
| hsa-miR-652* | 0.52 | 1.63E-03 | 2.85E-03 | new | N |
|
| |||||
| Down-regulated (29) | |||||
| hsa-miR-375 | −1.47 | 8.88E-30 | 4.09E-28 | N | |
| hsa-miR-195 | −1.30 | 3.47E-22 | 3.99E-21 | 497~195 | Y |
| hsa-miR-378 | −1.21 | 5.68E-29 | 1.31E-27 | Y | |
| hsa-miR-215 | −1.08 | 1.72E-14 | 6.06E-14 | 194-1~215 | Y |
| hsa-miR-194 | −0.79 | 5.39E-13 | 1.24E-12 | 194-1~215/194-2~192 | Y |
| hsa-miR-192 | −1.06 | 3.84E-15 | 1.61E-14 | 194-2~192 | Y |
| hsa-miR-192* | −0.78 | 4.68E-14 | 1.35E-13 | 194-2~192 | Y |
| hsa-miR-143 | −0.98 | 2.41E-15 | 1.23E-14 | 143~145 | Y |
| hsa-miR-145 | −0.89 | 3.19E-21 | 2.45E-20 | 143~145 | Y |
| hsa-miR-29c | −0.98 | 3.11E-21 | 2.45E-20 | 29b-2~29c | N |
| hsa-miR-147b | −0.92 | 1.50E-23 | 2.29E-22 | N | |
| hsa-miR-190 | −0.88 | 3.89E-13 | 9.42E-13 | N | |
| hsa-miR-1 | −0.87 | 1.81E-18 | 1.04E-17 | 1-2~133a-1 | Y |
| hsa-miR-30c | −0.86 | 4.06E-12 | 8.89E-12 | 30e~30c-1 | Y |
| hsa-miR-30e | −0.72 | 6.80E-12 | 1.42E-11 | 30e~30c-1 | Y |
| hsa-miR-30b | −0.85 | 8.74E-12 | 1.75E-11 | 30d~30b | Y |
| hsa-miR-26b | −0.81 | 1.78E-08 | 2.28E-08 | N | |
| hsa-miR-342-3p | −0.80 | 2.84E-13 | 7.26E-13 | Y | |
| hsa-miR-26a | −0.77 | 6.19E-15 | 2.37E-14 | N | |
| hsa-miR-10b | −0.72 | 7.72E-11 | 1.23E-10 | Y | |
| hsa-miR-148a | −0.70 | 8.81E-10 | 1.27E-09 | N | |
| hsa-miR-100 | −0.70 | 4.88E-11 | 8.02E-11 | 100~let-7a-2 | Y |
| hsa-let-7a | −0.57 | 3.59E-06 | 3.67E-06 | 100~let-7a-2 | Y |
| hsa-let-7f | −0.63 | 1.39E-05 | 3.09E-05 | let-7a-1~let-7f-1~let-7d | Y |
| hsa-miR-200b | −0.69 | 4.77E-11 | 8.02E-11 | 200b~200a~429 | Y** |
| hsa-miR-429 | −0.52 | 5.18E-06 | 5.18E-06 | 200b~200a~429 | Y** |
| hsa-miR-338-3p | −0.68 | 2.59E-13 | 7.00E-13 | 1250~338~3065~657 | N |
| hsa-miR-101 | −0.61 | 7.70E-10 | 1.14E-09 | 101-1~3671 | Y |
| hsa-miRPlus-E1141 | −0.60 | 1.64E-05 | 3.52E-05 | N | |
The 20 most up- and the 29 most down-regulated microRNAs (and one snoRNA) in rectal cancer (T) compared to normal adjacent tissue (N) are listed. Inclusion criteria: Var>0.2, |log2(T/N)|>0.5, p<0.001. The log-fold change, p-value of the two-sided t-test, and the q-value (FDR) for each microRNA are listed, as well as the microRNA cluster, where relevant. miR-652* is a new microRNA, not yet registered in miRBase. The CC column to the right indicates if the microRNA has been identified as differentially expressed in colon cancer (Y, yes; N, no; Y**, yes, but in opposite direction). Bold miRNAs have been validated by qRT-PCR.
We observed a larger variation in microRNA expression levels among rectal cancer samples (around 50% higher SD between all tumor samples than between all adjacent mucosa samples) than among the corresponding normal tissue, which is illustrated by the more scattered distribution of tumor samples (red) compared to normal samples (green) in the PCA plot, Figure 1B. This suggests that the tumor population is more heterogeneous than the corresponding non-tumor tissue.
Comparison of microRNA expression profiles from rectal and colon cancer
Cancers of the colon and the rectum represent two distinct clinical entities that require different treatment strategies. We therefore compared our rectal cancer specific microRNA profiles to previously published microRNA data on colon cancer. In Table 1, we have indicated that out of the 49 significantly differentially expressed microRNAs, 28 have already been identified in colon cancer, including miR-21, miR-31, miR-135b, miR-223 (all up-regulated), miR-195, miR-378, miR-192~194~215, miR-143~145, miR-1, and members of the miR-30 and let-7 family (all down-regulated). Therefore, the remaining 21 microRNAs potentially define rectal cancer as a disease molecularly distinct from colon tumors.
It should be kept in mind, however, that most data on colon cancer rely on older microarrays that do not contain many of the “novel” microRNAs detected in this study: besides the six proprietary miRPlus sequences, the following 15 microRNA appear to be rectal cancer-specific: miR-492, miR-542-5p, miR-584, miR-483-5p, miR-144, miR-2110, miR-652*, and the C/D box snoRNA, SNORD12B, was detected as up-regulated in rectal cancer. MiR-375, miR-147b, miR-148a, miR-190, miR-26a/b, miR-29c and miR-338-3p were found to be down-regulated. However, the comparison of colon and rectal cancer miRNA expression patterns should be performed with caution as these results have been retrieved from several different studies.
Of great interest miR-652* was identified to be differentially regulated. As, in compliance with the latest release 17 of miRBase(17), it has not been entered yet. For future reference, this microRNA (which has the sequence ACAACCCUAGGAGAGGGUGCCA) should be denoted miR-652-5p (Supplementary Figure S1).
Technical validation of microRNA expression levels by qPCR
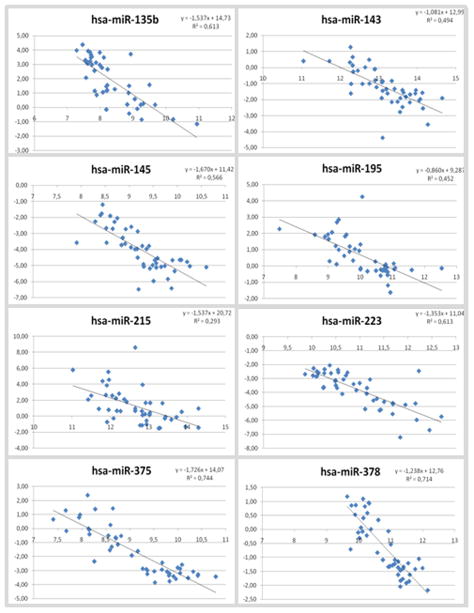
The expression of 10 microRNAs, which were selected based on a combination of fold-change and p-value (indicated in Table 1), was validated using semi-quantitative real-time PCR in 48 samples (24 matched tumor-mucosa pairs). Except for miR-492 and miR-29c (Supplementary Figure S2), the microRNA expression levels correlated very well between the two methods (Figure 2), with a range of coefficient of correlations (R2) from 0.293 to 0.744.
Figure 2.
Validation of selected miRNAs using semi-quantitative real-time PCR. Overall, there is a good correlation between the microarray data (X-axis, log2 transformed values) and the qRT-PCR data (Y-axis, ΔCt values).
Predicted targets are enriched for cancer-related pathways
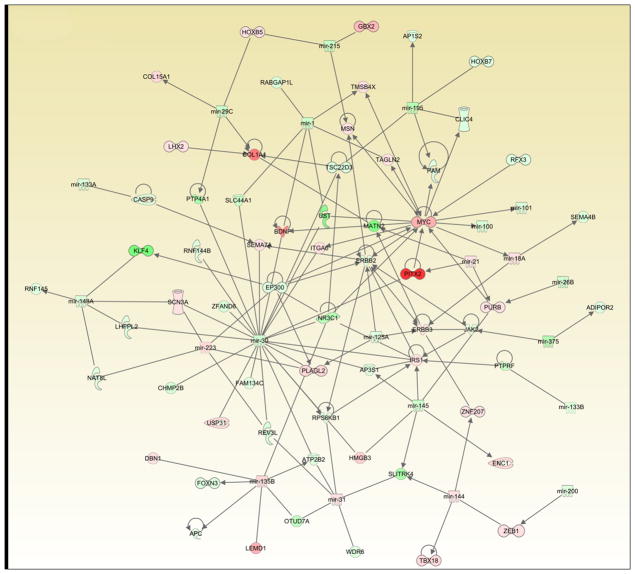
We applied both IPA and DIANA-miRpath (7) to visualize potential interaction networks based on target predictions for the 49 microRNAs that were differentially expressed between normal and rectal cancer tissue (Table 1); mRNA expression levels for the 57 patients were overlaid on the IPA network. One such interaction map is illustrated in Figure 3, which represents a composite of merging the three top networks with higher significant scores according to the IPA. The genes included to generate each individual network were identified as predicted target genes of our differentially expressed set of miRNAs according to the Diana microT4 databases. Ultimately, this network aims at representing the set of predicted target genes whose gene expression might change as a consequence of differentially expressed miRNAs in the tumor compared to matched normal mucosa. The mRNA levels of these genes were retrieved from a previous analysis (8). Genes whose expression did not change in the tumor versus matched normal mucosa comparison were excluded from the network, as we believe that their biological significance in this particular context was unclear and could be misleading. Within this figure, some of the notable miRNA-regulated targets are MYC, APC, ERBB2-ERBB3, ZEB1, and FOXD3. All of these genes have previously been implicated in tumorigenesis. Based on target enrichment for the top-regulated microRNAs, the predicted KEGG pathway analysis revealed Wnt, TGF-beta, mTOR, insulin, MAPK, and ErbB signaling.
Figure 3.
Network interaction map based on combined miRNA expression (Table 1), target prediction and mRNA expression data for all tumor and normal samples. “Red” indicates up-regulation, “green” indicates down-regulation in the tumors compared to the corresponding normal tissue. Precise description of shapes, lines and arrows can be found in Supplementary Figure legend 4-1.
Comparison of different databases revealed highly discrepant results. For example, miR-135b was found to have 428 predicted targets by PicTar, 510 targets by TargetScan, and 663 targets by micro-T3, but only seven targets were overlapping among the top-100 hits (Supplementary Figure S3). So far, only one miR-135b target, namely APC, has been experimentally verified (18); this gene was ranked 117 by TargetScan, 181 by micro-T3, and was not identified by PicTar.
Difficulties in miRNA target prediction became obvious again when we used IPA to generate the interaction network shown in Figure 3. The miR-135b-APC association was not detected, nor was there any reference to it in the IPA knowledge database, which is the basis for generating the pathways. Only after manual curation, the interactions for both LEMD1 and APC with miR-135b could be included into this figure. Therefore, the number of possible pathways increases, and we present other probable interactions that are based on the most up- and down-regulated microRNAs and their associated predicted mRNA targets in Supplementary Figure S4. However, the biological significance of our observations is strengthened through the identification of many microRNAs that appear to participate in regulatory networks involved in tumorigenesis, such as the Wnt/β-catenin, MYC, and EGFR/KRAS pathways (19).
As an additional approach to increase the number of reliable miRNA target genes, we used an mRNA expression data set and compared these results to the predicted targets retrieved from a database for validated miRNA target prediction. This was possible because we recently published the mRNA expression signatures for the tumor and mucosa samples used in this analysis (8). Twenty-six miRNAs could be retrieved from the database predicting 203 mRNA targets. We based this analysis on the principle assumption that, in general, the up-regulation of a miRNA should result in a down-regulation of the “corresponding” mRNA, and vice versa. Although target enrichment analysis was only significant for miRNA-21 (p=0.003), a total of 104 target genes were found to be regulated as expected, 74 were regulated the opposite direction (Supplementary Table S4).
Expression of miR-135b correlates with prognosis
Finally, we aimed to test whether the expression of any of the differentially expressed microRNAs was associated with patients’ disease-free survival (DFS) or cancer-specific survival (CCS). Towards this goal, we performed a rank regression analysis using tumor, normal and tumor/normal ratios for all microRNAs. Interestingly, in tumors, expression of miR-135b significantly correlated with CCS. It appears that patients with above median expression levels of miR-135b have a more favorable prognosis (no deaths) than patients with below median miR-135b expression (p=0.0015; data not shown).
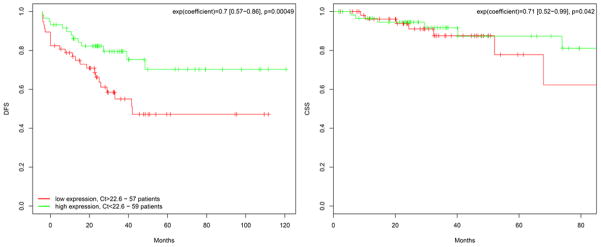
To validate these findings, semi-quantitative real-time PCR was performed to measure the expression levels of miR-135b in a completely independent set of 116 rectal cancer patients that have been treated at five different hospitals in Germany (Goettingen, Erlangen, Moenchengladbach, Oldenburg, and Kassel). Based on the median expression, both DFS (p=0.042) and CCS (p=0.00049) were significantly better in patients with high expression levels of miR-135b (Figure 4).
Figure 4.
Kaplan-Meier plots showing estimates of DFS (left panel) and CCS (right panel) probabilities grouped by their miR-135b expression levels in a completely independent set of 116 rectal cancer patients from five different institutions. The green curve represents samples with high (above median) miR-135b expression, while the red curve corresponds to samples with low (below median) miR-135b levels. Displayed is the hazard ratio as well as the 95% confidence interval. Abbreviations: DFS, disease free survival; CCS, cancer-specific survival.
No significant correlation was found between miR-135b expression levels and age (cor=−0.0009 [95%CI: −0.16–0.16], p=0.991), clinical UICC stage (cor=−0.018 [95%CI: 0.25, 0.07], Pearson correlation, p=0.26) or ypN stage (cor=0.11 [−0.27, 0.05], Pearson correlation, p=0.16). However, ypUICC stage (cor=0.21 [95%CI: 0.36, −0.05], Pearson correlation, p=0.01) as well as tumor regression grading (cor=0.2 [95%CI: 0.04, 0.36], Pearson correlation, p=0.01) turned out to be significantly correlated.
Discussion
Several groups have published microRNA expression signatures for colorectal cancer based on RT-PCR (20), microarray (21–23), bead array (24), and sequencing (25) approaches. However, colon and rectal cancer should be studied separately based on their molecular and clinical differences. We therefore focused our analysis on a well-defined group of locally advanced rectal cancer. To screen for differential microRNA expression, both unpaired (tumor versus normal group) as well as paired (individual tumor/normal ratios) analyses were performed. Both analyses gave very similar results in terms of significance. Since the group-wise comparison is more conservative, we focused on these results.
microRNAs upregulated in cancer
Based on our filter criteria (|log2 tumor/mucosa| >0.5 and p<0.001), 49 miRNAs were differentially expressed between cancer and mucosa. As expected, the up-regulation of several miRNAs could be confirmed such as miR-21, miR-223, miR-31 as well as miR-675 (20, 23, 26). The latter was demonstrated to down-regulate the expression of the retinoblastoma tumor suppressor gene, RB1 (27). MiR-18a (p<5.8E-8) and miR-17 (p<1.9–4) were found to be up-regulated in the tumor samples as well. Both miRNAs belong to the miR17-92 cluster (oncomir-1) located on chromosome 13, which is frequently increased in copy number in colorectal cancer (28).
Of the remaining miRNAs that were identified to be up-regulated in the tumors, none has been reported to occur in comparison of colon or colorectal cancer to normal tissue. Therefore, these miRNAs are likely to be rectal cancer specific. However, because previous studies are limited due to the lack of stratification into colon and rectal cancers, herein identified miRNAs were potentially not included in the prior analyses. To our knowledge, only one recent paper by Slattery et al. (2) has examined the microRNA profiles of rectal and colon cancer in a systematic manner. However, two distinct differences are evident. First, FFPE tissue was used by Slattery and colleagues. Second, only subgroups like CpG island methylator phenotype positive (CIMP), KRAS mutated or TP53 mutated colon or rectal cancer tumors were compared to normal tissue. Nevertheless, eight out of 23 potentially rectal cancer specific microRNAs have not been described by Slattery et al. (2).
One of the newly identified microRNAs was miR-492. In our dataset, miR-492 was the single most up-regulated microRNA (up to 16-fold higher). Its pre-microRNA sequence overlaps a pseudo-gene of Keratin-19 (KRT19) that is highly expressed in tumors of the lower gastrointestinal tract (29). Their co-expression has recently been shown for hepatoblastoma (30). However, our attempts to validate the miR-492 expression with qRT-PCR data failed. As a potential explanation we hypothesized that this could be due to cross-reaction on the array, for example between the complementary mRNA or the pre-miRNA and the probe for miR-492. An alternative explanation might be that the PCR assay did not detect post-transcriptionally modified microRNAs (31). Another hypothesis is that although we found miR-492 to be highly expressed in rectal cancer by array analysis, this microRNA has escaped detection in all deep sequencing studies conducted so far according to deepBase (http://deepbase.sysu.edu.cn/browseAllRNA.php) (32). Therefore, it is likely, that miR-492 is either modified, for example by post-transcriptional processing (33) or not a genuine microRNA after all. However, because the signal-to-noise ratio for miR-492 detection is very high, we do not consider this finding as a technical artifact.
Among the remaining microRNAs, a cancer related expression was reported for miR-483-5p (34), miR-542-5p (35), miR-584 (36) as well as for miR-144 (37) and miR-2110 (38). Of interest was the identification of miR-652*. While miR-652 has previously been identified to be differentially regulated in colorectal cancer (25), miR-652* has not been entered to miRBase (17) yet, and can therefore be considered as a newly identified microRNA and should be denoted miR-652-5p.
microRNAs downregulated in cancer
Just as for the up-regulated miRNAs, many of the down-regulated microRNAs have previously been described in studies analyzing colorectal cancer (39), such as miR-378 (40), miR-195 (41) or the miR-143~145 cluster (39). Additionally, let-7a, let -7f and representative miRNAs from the miR 200 family confirmed previous findings. One miRNA that has not been associated with colon cancer was miR-375. miR-375 was the single most down-regulated microRNA in rectal cancer (3-fold change). Along with miR-148a that targets BCL2 (42), we also identified miR-190, which plays a role in pancreatic cancer tissues and cell lines (43).
Another miRNA, miR-29c, has been very recently shown to be down-regulated in the mesenchymal part of endometrial carcinosarcoma (44). For miR-29c, however, we only observed a poor correlation between microarray and qRT-PCR data. This discrepancy could be due to either cross-reaction on the array, for example between the highly similar microRNAs, miR-29a/miR-29b/miR-29c, or because the PCR assay may not detect post-transcriptionally modified microRNAs (31).
Interestingly, miR-26 that was described to be associated with inhibition of tumorigenesis (45, 46) was found to be down-regulated as well. However, in previous studies, it has been found to be stably expressed in colorectal cancer. Therefore, together with let-7a, it was suggested as a reference gene in colorectal miRNA studies (10). Finding both miR-26 and let-7a to be down-regulated in rectal cancer compared to adjacent mucosa, their application as “housekeeping” miRNAs for rectal cancer analyses should be considered with caution.
Pathway and target predictions – caveats
Depending on the target prediction tools applied, very different results were obtained, and there was only little overlap between the various algorithms. This may be due to both methodological differences, including diverse criteria for defining miRNA binding regions and differences in the databases used for the target prediction (47, 48).
Therefore, in order to decrease the number of potential miRNA targets, we only applied validated targets, and compared this list of mRNA targets to the true mRNA expression values that we previously established for these samples (8). For miR-21, the predicted targets from the databank and the regulated mRNAs correlated significantly (p=0.003). Furthermore, the identification of miR-135b and APC (18), miR-200b and ZEB1 (49) or miR-195 and CCND1 (50) validates the principle approach to screen for potential miRNA targets.
Difficulties in miRNA target prediction not only affect the target prediction itself but also the generation of interaction networks. In this particular analysis, manual curation was necessary for both LEMD1 and APC with miR-135b to identify regulatory networks involved in tumorigenesis, such as the Wnt/ß-catenin, MYC, and EGFR/KRAS pathways (19).
Conclusively, any study on pathway analyses and/or the prediction of miRNA targets that does not follow-up on the in silico predictions with functional validation of the putative targets or at least applies mRNA data from corresponding samples should be regarded with great caution. Additionally, this underlines the importance of an analytical, knowledge-based approach, rather than on relying solely on “streamlined” statistical visualization tools.
Clinical perspective
From a clinical point of view, the identification of miR-135b (p<2.8E-11) was of particular interest. In the microRNA array analysis, miR-135b was the only microRNA that correlated with survival. We therefore performed a completely independent PCR-based validation on a large and multi-center cohort of 116 patients. Based on the mean expression we were able to confirm that both disease-free and cancer-specific survival were significantly better for patients high expression levels of miR-135b in the tumor. The clear separation between patients with a good and a bad prognosis based on miR-135b expression is promising for the development of microRNA-based biomarkers for rectal cancer. However, the application of this marker needs further validation in an even larger patient cohort to assess the feasibility of setting-up a cut-off as this cut-off might vary between different patients groups.
The finding that miR-135b is differentially regulated between tumor and mucosa as well as being associated with prognosis may be explained by its possible association with rectal tumors in terms of being a marker rather than having a “true” biological function within the tumorigenic process itself. As such, it would be useful as a prognostic marker, but not as a therapeutic target. In addition, miRNAs are known to have a myriad of potential target genes, likely with different binding affinities. One could therefore imagine that while increased expression of miR-135b could lead to tumor formation (i.e., through inhibition of a tumor suppressor gene), expression at higher levels could additionally result in suppression of genes required for a more aggressive phenotype (i.e., invasion or metastasis) or resistance to chemotherapy, resulting in a better prognosis.
In summary, by applying global microRNA expression profiling to screen 57 locally advanced rectal carcinomas and 57 corresponding matched normal mucosa biopsies, we identified 49 differentially expressed miRNAs. The robustness of our results was demonstrated through rigorous real-time PCR validation. A comprehensive analysis revealed miR-492, miR-542-5p, miR-584, miR-483-5p, miR-144, miR-2110, miR-652*, miR-375, miR-147b, miR-148a, miR-190, miR-26a/b, and miR-338-3p as rectal cancer-specific. Target prediction analyses suggested that these microRNAs regulate prominent KEGG pathways such as Wnt, TGF-beta, mTOR, insulin, MAPK, and ErbB signaling, which adds weight to the biological significance of this study. Importantly, the expression levels of miR-135b correlated significantly with DFS and CCS.
Supplementary Material
Translational Relevance.
The response of locally advanced rectal cancers to preoperative radiochemotherapy varies tremendously. Although a good response to preoperative treatment is associated with favorable outcome, molecular markers that allow therapy stratification are still lacking. The identification of differentially regulated miRNAs could therefore contribute to a better understanding of the underlying mechanisms of rectal cancer and its response to preoperative treatment. Furthermore, the detection of miRNAs that correlate with prognosis prior to preoperative therapy would open up the possibility for treatment individualization in rectal cancer.
Acknowledgments
Grant Support
This work was supported by the Deutsche Forschungsgemeinschaft (KFO 179) and by the Intramural Research Program of the NIH, National Cancer Institute. This work was part of a CRADA between the NCI and Exiqon A/S (CRADA #2254).
We thank Dara Wangsa and Chang-Rong Lai for their assistance with the PCR assays, and Yue Hu for help with the microRNA target correlation analysis. We are very grateful to our collaborators in the Departments of Surgery in Erlangen (Dr. Hohenberger, Dr. Matzel), Moenchengladbach (Dr. Kania, Dr. Grünewald), Oldenburg (Dr. Weyhe, Dr. Burkowski) and Kassel (Dr. Hesterberg, Dr. Schrader) who helped us to realize this multicenter validation study.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed for J.G., M.G., J.C., A.J.S., M.J.D., C.C.H., B.M.G., T.B., and T.R.
R.S., B.K., S.M., and T.L. are former employees of Exiqon A/S.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011;50:196–206. doi: 10.1002/gcc.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauer R, Becker H, Hohenberger W, Rodel C, Wittekind C, Fietkau R, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351:1731–40. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 4.Grade M, Hormann P, Becker S, Hummon AB, Wangsa D, Varma S, et al. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res. 2007;67:41–56. doi: 10.1158/0008-5472.CAN-06-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sokilde R, Kaczkowski B, Podolska A, Cirera S, Gorodkin J, Moller S, et al. Global microRNA analysis of the NCI-60 cancer cell panel. Mol Cancer Ther. 2011;10:375–84. doi: 10.1158/1535-7163.MCT-10-0605. [DOI] [PubMed] [Google Scholar]
- 6.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 7.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–3. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 8.Gaedcke J, Grade M, Jung K, Camps J, Jo P, Emons G, et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 2010;49:1024–34. doi: 10.1002/gcc.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wotschofsky Z, Meyer HA, Jung M, Fendler A, Wagner I, Stephan C, et al. Reference genes for the relative quantification of microRNAs in renal cell carcinomas and their metastases. Anal Biochem. 417:233–41. doi: 10.1016/j.ab.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer. 2010;10:173. doi: 10.1186/1471-2407-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 104:1168–77. doi: 10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi S, Kopetz S, Davuluri R, Hamilton SR, Calin GA. MicroRNAs, ultraconserved genes and colorectal cancers. Int J Biochem Cell Biol. 2009;42:1291–7. doi: 10.1016/j.biocel.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Faber C, Kirchner T, Hlubek F. The impact of microRNAs on colorectal cancer. Virchows Arch. 2009;454:359–67. doi: 10.1007/s00428-009-0751-9. [DOI] [PubMed] [Google Scholar]
- 14.Castoldi M, Benes V, Hentze MW, Muckenthaler MU. miChip: a microarray platform for expression profiling of microRNAs based on locked nucleic acid (LNA) oligonucleotide capture probes. Methods. 2007;43:146–52. doi: 10.1016/j.ymeth.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Nygaard S, Jacobsen A, Lindow M, Eriksen J, Balslev E, Flyger H, et al. Identification and analysis of miRNAs in human breast cancer and teratoma samples using deep sequencing. BMC Med Genomics. 2009;2:35. doi: 10.1186/1755-8794-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 19.Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanza G, Ferracin M, Gafa R, Veronese A, Spizzo R, Pichiorri F, et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–75. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 23.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA, et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. doi: 10.1186/1471-2407-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Earle JS, Luthra R, Romans A, Abraham R, Ensor J, Yao H, et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12:433–40. doi: 10.2353/jmoldx.2010.090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–8. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 28.Camps J, Grade M, Nguyen QT, Hormann P, Becker S, Hummon AB, et al. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008;68:1284–95. doi: 10.1158/0008-5472.CAN-07-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devor EJ. Primate microRNAs miR-220 and miR-492 lie within processed pseudogenes. J Hered. 2006;97:186–90. doi: 10.1093/jhered/esj022. [DOI] [PubMed] [Google Scholar]
- 30.von Frowein J, Pagel P, Kappler R, von Schweinitz D, Roscher A, Schmid I. MicroRNA-492 is processed from the keratin 19 gene and up-regulated in metastatic hepatoblastoma. Hepatology. 2011;53:833–42. doi: 10.1002/hep.24125. [DOI] [PubMed] [Google Scholar]
- 31.Findeiss S, Langenberger D, Stadler PF, Hoffmann S. Traces of post-transcriptional RNA modifications in deep sequencing data. Biol Chem. 2011;392:305–13. doi: 10.1515/BC.2011.043. [DOI] [PubMed] [Google Scholar]
- 32.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–30. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson EE, Holloway AK, Weng J, Fojo T, Kebebew E. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer. 2011 doi: 10.1002/cncr.25724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bray I, Tivnan A, Bryan K, Foley NH, Watters KM, Tracey L, et al. MicroRNA-542-5p as a novel tumor suppressor in neuroblastoma. Cancer Lett. 2011;303:56–64. doi: 10.1016/j.canlet.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueno K, Hirata H, Shahryari V, Chen Y, Zaman MS, Singh K, et al. Tumour suppressor microRNA-584 directly targets oncogene Rock-1 and decreases invasion ability in human clear cell renal cell carcinoma. Br J Cancer. 2011;104:308–15. doi: 10.1038/sj.bjc.6606028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Peng B, Wang D, Ma X, Jiang D, Zhao J, et al. Human tumor MicroRNA signatures derived from large-scale oligonucleotide microarray datasets. Int J Cancer. 2010 doi: 10.1002/ijc.25818. [DOI] [PubMed] [Google Scholar]
- 38.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, et al. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–41. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael MZ, SMOC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 40.Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50–4. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 41.Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–40. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng H, et al. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castilla MA, Moreno-Bueno G, Romero-Perez L, Van De Vijver K, Biscuola M, Lopez-Garcia MA, et al. Micro-RNA signature of the epithelial-mesenchymal transition in endometrial carcinosarcoma. J Pathol. 2011;223:72–80. doi: 10.1002/path.2802. [DOI] [PubMed] [Google Scholar]
- 45.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–33. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 46.Zhang B, Liu XX, He JR, Zhou CX, Guo M, He M, et al. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis. 2011;32:2–9. doi: 10.1093/carcin/bgq209. [DOI] [PubMed] [Google Scholar]
- 47.Ritchie W, Flamant S, Rasko JE. Predicting microRNA targets and functions: traps for the unwary. Nat Methods. 2009;6:397–8. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
- 48.Sioud M, Cekaite L. Profiling of miRNA expression and prediction of target genes. Methods Mol Biol. 2010;629:257–71. doi: 10.1007/978-1-60761-657-3_16. [DOI] [PubMed] [Google Scholar]
- 49.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–54. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 50.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–21. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.