Abstract
The gold-standard of preclinical micro-computed tomography (μCT) data processing is still manual delineation of complete organs or regions by specialists. However, this method is time-consuming, error-prone, has limited reproducibility, and therefore is not suitable for large-scale data analysis. Unfortunately, robust and accurate automated whole body segmentation algorithms are still missing. In this publication, we introduce a database containing 225 murine 3D whole body μCT scans along with manual organ segmentation of most important organs including heart, liver, lung, trachea, spleen, kidneys, stomach, intestine, bladder, thigh muscle, bone, as well as subcutaneous tumors. The database includes native and contrast-enhanced, regarding spleen and liver, μCT data. All scans along with organ segmentation are freely accessible at the online repository Figshare. We encourage researchers to reuse the provided data to evaluate and improve methods and algorithms for accurate automated organ segmentation which may reduce manual segmentation effort, increase reproducibility, and even reduce the number of required laboratory animals by reducing a source of variability and having access to a reliable reference group.
Subject terms: 3-D reconstruction, X-ray tomography, Image processing
Background & Summary
Micro-computed tomography (μCT) is one of the most commonly used imaging technologies in preclinical research. It provides detailed information about the volume, textures, and abnormal alterations of internal structures in high-resolution1–4. Because of its high reliability and reproducibility, μCT is often used as a single imaging modality. It offers many advantages including homogenous resolution, fast acquisition, and well-calibrated voxel intensities5–9. In addition, other imaging modalities such as nuclear or optical imaging technologies are often combined with μCT due to the need of an anatomical reference7,10–12. Thus, μCT provides accurate anatomical information on the basis of its good contrast recognition especially of dense tissues such as bones or calcified structures13–18. The main drawback of μCT imaging is a low soft tissue contrast, which can be improved by the utilization of radiopaque contrast agents19,20. Nowadays, a wide range of clinical and preclinical CT contrast-enhancing agents are available. Preclinical contrast agents often show a longer blood half-life time or a more specific uptake than their clinical counterparts. Examples of them are contrast-producing lipids, iodine-containing aqueous colloids, or alkaline earth metal-based nanoparticulate contrast agents21,22.
When μCT scans are acquired at a low dose of X-ray, longitudinal measurements in the same animal can be performed10,23,24. Hence, more information per animal can be acquired and disease or treatment progression within the same animal can be determined. This leads to a reduction in the required animal number, which is in accordance with the 3 R aims (Refinement, Replacement, Reduction)25 for animal protection.
Nevertheless, most preclinical μCT imaging studies result in a huge amount of data that needs to be processed. Currently, the gold-standard of μCT image processing is still manual delineation of regions of interest or complete organs, although this method is laborious and limited in its reproducibility due to high user-dependence9,26–28. Especially in preclinical imaging studies5,26,29–31, the sophisticated analysis of the immense amount of μCT data is more time-consuming than the scanning procedure alone, because of the high manual effort to generate whole body organ segmentations32. Consequently, there is a significant need for automated segmentation tools for preclinical imaging studies.
Automated segmentation (AS) or machine-learning algorithms could address the aforementioned problems by introducing consistency, reliability, and reproducibility to the process9,26,33–37. Although the development of AS algorithms has gained much interest among researchers, no universal algorithm has been established yet. Multi-atlas segmentation (MAS) is one promising candidate for a new gold-standard in image annotation26. MAS has been successful used in both multi- and single-organ segmentations, despite the general shortcomings of abdominal imaging, i.e. shifting of organs inside the abdominal cavity. Wang et al. presented a MAS atlas dedicated to preclinical image analysis including multiple training subjects29. This atlas consists of 103 μCT whole body mouse images and reflects more realistically the deformation of internal organs following the changes of pose and weight due to interspecies variations and within one individual along longitudinal studies.
Nevertheless, to our knowledge no atlas or database of preclinical μCT data including organ segmentations exists, because, so far, most CT databases only include reconstructed scans or segmented bone structures17,38. Therefore, the aim of our study is to provide the first preclinical μCT database including whole body mouse images and their organ segmentations. Our open-access database includes 225 native and contrast-enhanced whole-animal μCT volumes along with manual organ segmentations acquired from mice scanned longitudinally in different positions. Organ parameters such as volume, surface, and distances in one individual remain stable over time. Furthermore, we calculated the Sørensen-DICE coefficient to compare the similarity between segmentations of two independent experts. This coefficient may help to compare the achieved accuracy of automated methods with the inter-user variability of manual segmentation. We highly encourage researchers to use these 3D datasets, e.g. for further comparative analysis of organ morphology or to determine relevant μCT features such as intensity or variations between voxels. Ideally, this introduced database will be used to validate segmentation and machine-learning approaches and thus, facilitate the development of reliable, simplified, and user-independent analysis tools for whole body organ segmentation. In addition, the anatomical 3D data of the whole mouse body including the main organs will serve as a visual and education resource to train researchers for segmentation of tumors and organs.
Methods
Datasets
For generating this database, two μCT datasets from other studies were reused: one native dataset without using a contrast agent and one dataset with contrast-enhanced μCT scans, where the contrast agent ExiTron™ nano 6000 (Viscover, Berlin, Germany) was injected, see Fig. 1. The native μCT dataset is part of an already published study23. Publishing the contrast-enhanced μCT data is currently in progress. In both studies, all animal experiments were approved by the Governmental Review Committee on Animal Care. Thus, for generating this database no additional mice were required.
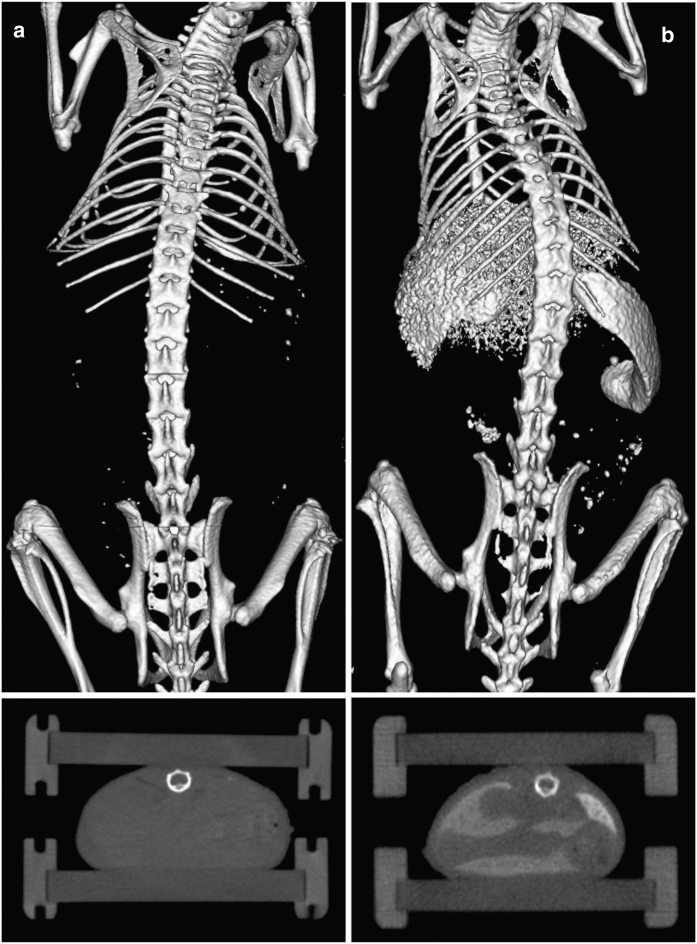
Figure 1. 3D visualization of native and contrast-enhanced μCT data.
Each μCT scan consists of a set of isotropic voxels, whereby all voxel intensities are calibrated in Hounsfield units allowing a direct comparison between native and contrast-enhanced μCT data. Using a gray scale, structures with high attenuation of X-rays appear brighter, e.g. the bones, whereas structures with low attenuation appear dark such as lung and soft tissue. (a) 3D rendering (upper panel) and 2D axial slice view (lower panel) for a native (#M01-0.25 h) and a contrast-enhanced scan (#M03-008h) are depicted. (b) Spleen and liver appear brighter after the injection of the contrast agent.
The native dataset includes 140 3D whole body scans acquired from 20 female BALB/c nu/nu mice (Charles River Laboratory, Sulzfeld, Germany) measured at seven time points by a preclinical μCT (Tomoscope Duo, CT Imaging GmbH, Erlangen, Germany), see Table 1. For the μCT scanning process, the mice were anesthetised using 2.5% isoflurane vaporised in 1.0 l/min of oxygen gas using a dedicated vaporiser. Afterwards, they were placed in an animal cassette as described before10,39. While acquiring μCT data, mice were constantly under anesthesia. For each time point; 0.25 h, 002 h, 004 h, 008 h, 024 h, 048 h, and 072 h; mice were newly anesthetised, positioned in the mouse bed, and scanned. A dual energy μCT scan (HQD-6565-360-90) was used, where tubes were operated with a voltage of 65 kV and a current of 1 mA acquiring 720 projections with 1032 × 1012 pixels during one full rotation, respectively as it was described in detail before10,39,40. Per scan a time of 90 s was required, whereby two scans per mouse were needed at each time point to entirely cover the mouse body. The acquired voxel sizes were 0.28 mm × 0.28 mm × 0.28 mm and the field of view was 40.32 mm × 28.84 mm × 55.44 mm. The spatial resolution of the system is in the order of 80 μm with a fixed geometry.
Table 1. Characterization of the native dataset.
| Source | Mouse ID | Temporal range (bold: 2 organ segmentations are available) |
|---|---|---|
| The table shows the details of the native dataset, mouse IDs, measured time points, and the time points where 2 organ segmentations are available. | ||
| native | M01 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M02 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M03 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M04 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M05 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M06 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M07 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M08 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M09 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M10 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M11 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M12 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M13 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M14 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M15 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M16 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M17 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M18 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M19 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
| native | M20 | 0.25 h; 002 h; 004 h; 008 h; 024 h; 048 h; 072 h |
The contrast-enhanced dataset consists of 85 3D whole body scans from ten female A431-tumor bearing BALB/cAnNRj-Foxn1nu mice (Janvier, Le Genest-Saint-Isle, France), see Table 2. They were scanned with the InSyTe μCT scanner (BMIF TriFoil Imaging, Dijon, France). One hour before the first scan, the preclinical μCT contrast agent ExiTron™ nano 6000 (100 μl, 640 mg iodine/kg body weight) was intravenously injected. This non-toxic, commercially available, alkaline earth metal-based nanoparticulate contrast agent circulates in the blood stream and is taken up by the Kupffer cells. It significantly enhances the CT-contrast in spleen and liver21,41 as clearly shown in Fig. 1. A single dose of ExiTron™ nano 6000 results in longstanding enhancement of liver and spleen tissue for longer than 3 weeks peaking for the liver at approximately 4 h and for spleen contrast at 48 h post injection41. For scanning procedure, the mice were anaesthetised in the same way and placed in the same animal cassette as described in the case of the native dataset. A special adapter was designed and built for this μCT. Hence, the same mouse bed from the previous study was used among the different μCT systems in order to increase the consistency of μCT analysis. Similar to the protocol of the native μCT scans, the mice were repeatedly anesthetised, positioned in the mouse bed, and scanned at the different time points; pre (−001h), 0.25 h, 002 h, 004 h, 006 h, 008 h, 024 h, 048 h, 072 h, 144 h, 168 h, 192 h, and 240 h. For a full-rotation μCT scan, 207 views with a frame rate of 1 frame per view, an X-ray tube voltage of 75 kV, and an exposure time of 230 ms were acquired. The acquired voxel sizes were 0.28 mm × 0.28 mm × 0.28 mm and the field of view was 43.12 mm × 33.88 mm × 67.76 mm.
Table 2. Characterization of the contrast-enhanced dataset.
| Source | Mouse ID | Temporal range (bold: 2 organ segmentations are available) |
|---|---|---|
| The table shows the details of the contrast-enhanced dataset, mouse IDs, measured time points, and the time points where 2 organ segmentations are available. | ||
| Contrast-enhanced | M01 | −001h; 002 h; 004 h; 006 h; 024 h; 048 h; 072 h; 120 h; 168 h; 240 h |
| Contrast-enhanced | M02 | 0.25 h; 002 h; 004 h; 006 h; 008 h; 024 h |
| Contrast-enhanced | M03 | −001h; 0.25 h; 002 h; 004 h; 006 h; 008 h; 024 h; 048 h; 072 h |
| Contrast-enhanced | M04 | −001h; 0.25 h; 002 h; 004 h; 006 h; 008 h; 024 h; 048 h; 072 h; 120 h; 144 h; 192 h; 240 h |
| Contrast-enhanced | M05 | −001h; 0.25 h; 002 h; 004 h; 006 h; 008 h; 024 h; 048 h; 072 h |
| Contrast-enhanced | M06 | −001h; 0.25 h; 002 h; 004 h; 006 h; 008 h; 024 h; 048 h; 072 h; 120 h; 144 h; 192 h; 240 h |
| Contrast-enhanced | M07 | −001h; 0.25 h; 002 h; 004 h; 006 h; 008 h; 024 h; 048 h; 072 h; 120 h |
| Contrast-enhanced | M08 | −001h; 0.25 h; 002 h; 004 h; 006 h; 008 h; 024 h; 048 h; 072 h; 120 h; 144 h; 168 h; 192 h |
| Contrast-enhanced | M09 | 024 h |
| Contrast-enhanced | M10 | 024 h |
Image reconstruction and analysis - 3D whole body organ segmentation
All acquired 3D μCT images were reconstructed at an isotropic voxel size of 28 μm using a Feldkamp type algorithm and a smooth kernel as previously described10,23,39. 3D organ segmentations based on the μCT data were performed for all mice at the different time points. The standardised segmentation protocol, used for both datasets, was developed in our group and has been previously described42. Briefly, bone structures and lung were semi-automatically segmented using threshold functions above a certain value, for bone >1000 HU, or below a certain value, for lung <300 HU, and selecting a seed point for region growing. Organs with defined and clearly visible boundaries such as the heart, bladder, and kidneys were segmented by manual delineation. Scribbles were drawn around the organ boundaries, see Fig. 2d. Other organs such as the stomach and intestine were segmented approximated by a few convex regions and manual delineation of them. Liver segmentation was performed slab wise due to the complex shape of the lobes. As an example of muscle, a part of the thigh was segmented. Despite their polymorphic shape subcutaneous tumors displayed clearly distinguishable boundaries and were segmented by manual delineation.
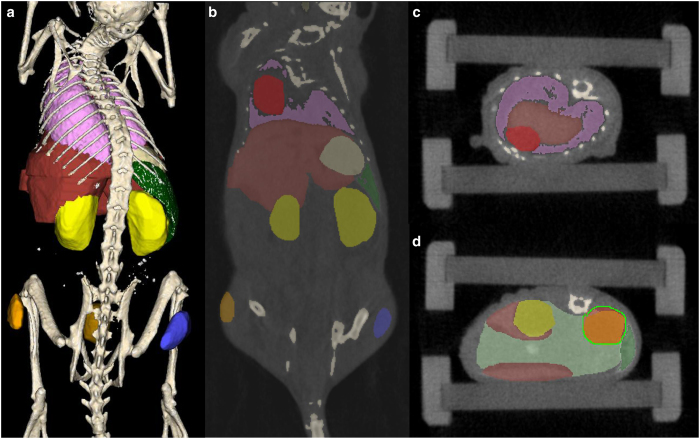
Figure 2. Interactive organ segmentation based on whole body μCT data.
(a–d) A μCT-based 3D whole body organ segmentation of a mouse is performed semi-automatically for bones (beige), lung (pink), and spleen (dark green). Other organs are segmented by manual delineation: liver (brown), stomach (light tan), kidneys (yellow), intestine (light green, only depicted in d), tumor (orange), part of the thigh muscle (blue), and bladder (gold). For the segmentation process, organs need to be encircled in several slices such as (b) coronal and (c) axial from which a program can interpolate the remaining slices. (d) Example of the manual delineation procedure by drawing scribbles (green line) around the right kidney. Mouse #M03-004h of the contrast-enhanced dataset was used in this example.
Statistics and calculation of the Sørensen–Dice coefficient
The quality of the whole body organ segmentations by manual delineation between two trained scientists was compared by calculating the Sørensen–Dice coefficient (Sørensen index, Dice’s coefficient). This similarity coefficient is widely used in image analysis, for example, to evaluate the reproducibility of manual segmentations and the overlap accuracy of automated probabilistic fractional segmentation of MR images28,43. Here in particular, it is used to investigate the similarity between the same organ analysed independently by two experts. The Sørensen-Dice similarity coefficient for image segmentation is calculated using this formula:
For each particular organ, X and Y represent the set of segmented voxels of user 1 and 2, respectively. The Sørensen–Dice coefficient computes the ratio of segmentation overlap to the segmentation size. A higher Sørensen–Dice coefficient represents a higher degree of similarity. A score of 1.0 denotes a perfect overlap and a score of 0.0 represents no overlap. Thus, the Sørensen–Dice coefficient can be used to determine the accuracy of automated segmentation methods by comparison with manual segmentations.
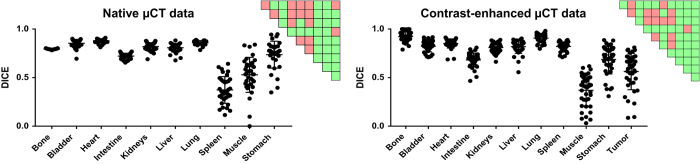
The Sørensen–Dice coefficient was computed for both datasets and all segmented organs to assess inter-user segmentation variability, see Table 3. For the native dataset, 35 whole body μCT-based organ segmentations were performed by a second evaluator. All mice that received the fluorescent probe OsteoSense 750 EX (PerkinElmer, USA) at all seven time points were chosen for this analysis23, see Table 1. This probe has no decreasing or enhancing effect on CT-contrast. For the contrast-enhanced μCT dataset, 39 organ segmentations were used for calculating the Sørensen-Dice coefficient. All eight mice, but only the time points 0.25 h, 002 h, 004 h, 006 h, and 008 h were chosen for this analysis, see Table 2. Time point 008 h of #M01 is missing due to some technical problems during the scanning process. Statistical analysis was performed using GraphPad Prism version 7.0. For the comparison between organs, a multi-comparison one-way ANOVA was performed in combination with a Tukey posttest. A p-value below 0.05 was considered to represent statistical significance. Statistical significances are shown as pair-wise significance matrices (P < 0.05 in green) in Fig. 3, detailed explanation has been previously described23.
Table 3. Comparison of the Sørensen–Dice coefficient.
| Native μCT data |
Contrast-enhanced μCT data |
|||||||
|---|---|---|---|---|---|---|---|---|
| Organ | Dice | Std dev | Minimum | Maximum | Dice | Std dev | Minimum | Maximum |
| The coefficients for all organs of the native and the contrast-enhanced datasets are depicted to assess the quality of two organ segmentations by manual delineation. Furthermore, the calculated standard deviation (Std dev) and the minimum and maximum values of the Sørensen–Dice coefficient are shown. The main difference between the native and the contrast-enhanced μCT data is the increase in Sørensen–Dice coefficient showing the higher similarity in segmentation of the spleen (increase from 0.373 to 0.820, as highlighted with *). The data are also graphically depicted in Fig. 3. | ||||||||
| Bone | 0.793 | 0.005 | 0.782 | 0.803 | 0.923 | 0.050 | 0.789 | 0.999 |
| Bladder | 0.854 | 0.039 | 0.694 | 0.900 | 0.822 | 0.057 | 0.710 | 0.915 |
| Heart | 0.879 | 0.021 | 0.812 | 0.910 | 0.851 | 0.047 | 0.687 | 0.913 |
| Intestine | 0.722 | 0.029 | 0.654 | 0.768 | 0.686 | 0.149 | 0.308 | 0.886 |
| Kidneys | 0.819 | 0.040 | 0.689 | 0.878 | 0.809 | 0.051 | 0.662 | 0.888 |
| Liver | 0.808 | 0.044 | 0.677 | 0.883 | 0.818 | 0.068 | 0.555 | 0.903 |
| Lung | 0.859 | 0.021 | 0.784 | 0.888 | 0.907 | 0.048 | 0.797 | 0.980 |
| Spleen | *0.373 | 0.137 | 0.115 | 0.642 | *0.820 | 0.046 | 0.710 | 0.888 |
| Muscle | 0.528 | 0.179 | 0 | 0.839 | 0.369 | 0.162 | 0.031 | 0.618 |
| Stomach | 0.736 | 0.138 | 0.348 | 0.947 | 0.682 | 0.070 | 0.466 | 0.809 |
| Tumor | - | 0.562 | 0.187 | 0.087 | 0.810 | |||
Figure 3. Analysis of the Sørensen–Dice coefficient including pair-wise significance matrices.
The highest variability occurs for the spleen in the native dataset due to the low soft tissue contrast of μCT images (DICE of 0.373) as well as for muscle (native: 0.528 and contrast-enhanced: 0.369), intestine (0.722 and 0.682), and stomach (0.736 and 0.686) for both datasets. All other organs, especially those with clear organ boundaries such as bladder and heart, depict a good Sørensen-Dice coefficient, nearly reaching the optimum 1.0 (=perfect overlap). Statistical significances are shown as pair-wise significance matrices (p < 0.05 in green). The matrices demonstrate that analysing the native μCT data, the highest user-dependent errors occur for spleen, muscle, and stomach. For the contrast-enhanced μCT data, the highest user-dependent errors occur in segmenting muscle, stomach, and tumor.
Data Records
The μCT database published in this article consists of native and contrast-enhanced μCT scans. The native dataset comprises 140 murine 3D whole body scans and organ segmentations, where 35 scans include organ segmentations from two different evaluators. The contrast-enhanced dataset includes 85 murine 3D whole body scans with enhanced contrast in spleen, liver, and other organs, where 39 scans include two organ segmentations. Both datasets have been deposited in an online Figshare repository (Data Citation 1). For each scan, there is a subfolder labeled with mouse ID (M01, M02, etc.) and time point of measurement (0.25 h, 002 h, etc.) which contains a pyramid of μCT data with different resolutions (CT140, CT280) in the Analyze file format (consisting of pairs of .HDR and .IMG files). CT280 is generated by averaging eight neighboring voxels of CT140 to one average voxel, which results in a lower resolution. For the organ segmentations of the native data, the CT280 scan was used. The CT140 scans were initially used for the segmentations of the contrast-enhanced data, but, additionally, the organ segmentations were saved using CT280, clearly marked in the file names (Organ_140 or Organ_280). All 3D organ segmentations are saved as Analyze files with 8-bit voxels containing different indices for each segmented organ. Every voxel belongs exactly to one class index, either to an organ class or to class 0 (unclassified). The folder also includes a text file ending with .CLS, describing the assignment of the class indices to the respective organ and class color, for example: ClassColors = 0 0 0 255|201 238 255 255|255 170 255 255, ClassIndices = 0|1|2, ClassNames = unclassified|Bone|Lung. Additionally, a segmentation file named *Bed including the mouse bed, the whole body of the mouse, and fiducial markers, is included in every folder.
Technical Validation
The intensity values of μCT images are usually provided in Hounsfield units, which are calibrated in such a way that air generates intensities of −1000 and water 0. Therefore all CT images acquired by different scanners can be compared with each other due to the general calibration. Both preclinical μCT scanners were regularly maintained including calibration and quality control under the responsibility of qualified service personnel from the respective companies. However, occurring image artifacts, ring or beam-hardening artifacts, or motion artifacts due to breathing or cardiac movements can result in discrepancies between reconstructed values and true attenuation coefficients. In our study, these artifacts are negligible, because the manual segmentation is not influenced by any kind of artifacts, because when organ segmentation by manual delineation is performed, most organ boundaries can be seen by eye even if they are blurred. Nevertheless, the artifacts might interfere with some automated organ segmentation algorithms under certain conditions and should be considered in detail. Furthermore, the used multimodal mouse bed places the animal in a fixed position which leads to a reduction of breathing and motion artefacts. This mouse cassette is routinely used in many research institutes and companies, for several applications such as FMT-CT, PET-CT.
Usage Notes
Researchers are highly encouraged to download the 3D μCT scans of the native and/or contrast-enhanced μCT datasets from Figshare (Data Citation 1). The μCT data including organ segmentations could be used for the development of automated organ segmentation algorithms. By computing the Sørensen-DICE coefficient, the accuracy of existing or newly developed approaches can be compared. Usage of the well-known Analyze file format ensures that the μCT data can be loaded by many 3D analysis software packages. For all analysis, we used the software “Imalytics Preclinical”42, which was developed in our group.
Additional information
How to cite this article: Rosenhain, S. et al. A preclinical micro-computed tomography database including 3D whole body organ segmentations. Sci. Data. 5:180294 doi: 10.1038/sdata.2018.294 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the German Higher Education Ministry (BMBF) (Biophotonics program/ 13N13355), the European Union (FP7 program), the German Research Foundation (DFG; GR 5027/2-1, and FOR 2591), and by the excellence initiative of the German federal and state governments (I3TM seed fund program).
Footnotes
The authors declare no competing interests.
Data Citations
- Rosenhain S., et al. . 2018. figshare. https://doi.org/10.6084/m9.figshare.c.4224377
References
- Burghardt A. J., Link T. M. & Majumdar S. High-resolution computed tomography for clinical imaging of bone microarchitecture. Clin. Orthop. 469, 2179–2193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahkri H. et al. Checkpoint kinase Chk2 controls renal Cyp27b1 expression, calcitriol formation, and calcium-phosphate metabolism. Pflugers Arch. 467, 1871–1880 (2015). [DOI] [PubMed] [Google Scholar]
- ElAyouti A. et al. Apical constriction: location and dimensions in molars-a micro-computed tomography study. J. Endod. 40, 1095–1099 (2014). [DOI] [PubMed] [Google Scholar]
- Schambach S. J., Bag S., Schilling L., Groden C. & Brockmann M. A. Application of micro-CT in small animal imaging. Methods 50, 2–13 (2010). [DOI] [PubMed] [Google Scholar]
- Wang H., Stout D. B. & Chatziioannou A. F. Estimation of mouse organ locations through registration of a statistical mouse atlas with micro-CT images. IEEE Trans. Med. Imaging 31, 88–102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Transgenic mice as probes into complex systems. Science 246, 1265–1275 (1989). [DOI] [PubMed] [Google Scholar]
- Deroose C. M. et al. Multimodality Imaging of Tumor Xenografts and Metastases in Mice with Combined Small-Animal PET, Small-Animal CT, and Bioluminescence Imaging. J. Nucl. Med. 48, 295–303 (2007). [PMC free article] [PubMed] [Google Scholar]
- Tuveson D. & Hanahan D. Translational medicine: Cancer lessons from mice to humans. Nature 471, 316–317 (2011). [DOI] [PubMed] [Google Scholar]
- Zhou X., Takayama R., Wang S., Hara T. & Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med. Phys. 44, 5221–5233 (2017). [DOI] [PubMed] [Google Scholar]
- Gremse F. et al. Hybrid μCT-FMT imaging and image analysis. J. Vis. Exp. JoVE 100, e52770 ; DOI: 10.3791/52770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremse F. et al. Absorption reconstruction improves biodistribution assessment of fluorescent nanoprobes using hybrid fluorescence-mediated tomography. Theranostics 4, 960–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriese J. et al. Preclinical validation of automated dual-energy X-ray absorptiometry and computed tomography-based body composition measurements. Q. J. Nucl. Med. Mol. Imaging Off. Publ. Ital. Assoc. Nucl. Med. AIMN Int. Assoc. Radiopharmacol. IAR Sect. Soc. Of 60, 40–47 (2016). [PubMed] [Google Scholar]
- Bonnet N. et al. Assessment of trabecular bone microarchitecture by two different x-ray microcomputed tomographs: a comparative study of the rat distal tibia using Skyscan and Scanco devices. Med. Phys. 36, 1286–1297 (2009). [DOI] [PubMed] [Google Scholar]
- Donnelly E. Methods for assessing bone quality: a review. Clin. Orthop. 469, 2128–2138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R. et al. Morphometric analysis of human bone biopsies: a quantitative structural comparison of histological sections and micro-computed tomography. Bone 23, 59–66 (1998). [DOI] [PubMed] [Google Scholar]
- Ranjanomennahary P. et al. Comparison of radiograph-based texture analysis and bone mineral density with three-dimensional microarchitecture of trabecular bone. Med. Phys. 38, 420–428 (2011). [DOI] [PubMed] [Google Scholar]
- Ranzoni A. M., Corcelli M., Arnett T. R. & Guillot P. V. Micro-computed tomography reconstructions of tibiae of stem cell transplanted osteogenesis imperfecta mice. Sci. Data 5, 180100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Neves Borges P., Vincent T. L. & Marenzana M. Automated assessment of bone changes in cross-sectional micro-CT studies of murine experimental osteoarthritis. PloS One 12, e0174294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disselhorst J. A., Bezrukov I., Kolb A., Parl C. & Pichler B. J. Principles of PET/MR Imaging. J. Nucl. Med. Off. Publ. Soc. Nucl. Med 55, 2S–10S (2014). [DOI] [PubMed] [Google Scholar]
- Hallouard F., Anton N., Choquet P., Constantinesco A. & Vandamme T. Iodinated blood pool contrast media for preclinical X-ray imaging applications–a review. Biomaterials 31, 6249–6268 (2010). [DOI] [PubMed] [Google Scholar]
- Mannheim J. G. et al. Comparison of small animal CT contrast agents. Contrast Media Mol. Imaging 11, 272–284 (2016). [DOI] [PubMed] [Google Scholar]
- Li X., Anton N., Zuber G. & Vandamme T. Contrast agents for preclinical targeted X-ray imaging. Adv. Drug Deliv. Rev. 76, 116–133 (2014). [DOI] [PubMed] [Google Scholar]
- Al Rawashdeh W. et al. Noninvasive Assessment of Elimination and Retention using CT-FMT and Kinetic Whole-body Modeling. Theranostics 7, 1499–1510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R. D., Rudmann C., Wood R. W., Schwarz E. M. & Rahimi H. Longitudinal micro-CT as an outcome measure of interstitial lung disease in TNF-transgenic mice. PloS One 13, e0190678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W. M. S. & Burch R. L. The principles of humane experimental technique. Methuen (1959). [Google Scholar]
- Iglesias J. E. & Sabuncu M. R. Multi-atlas segmentation of biomedical images: A survey. Med. Image Anal. 24, 205–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiker M. et al. Atlas-based whole-body segmentation of mice from low-contrast Micro-CT data. Med. Image Anal. 14, 723–737 (2010). [DOI] [PubMed] [Google Scholar]
- Zou K. H. et al. Statistical Validation of Image Segmentation Quality Based on a Spatial Overlap Index. Acad. Radiol. 11, 178–189 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Stout D. B. & Chatziioannou A. F. A deformable atlas of the laboratory mouse. Mol. Imaging Biol. MIB Off. Publ. Acad. Mol. Imaging 17, 18–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogdas B., Stout D., Chatziioannou A. F. & Leahy R. M. Digimouse: a 3D whole body mouse atlas from CT and cryosection data. Phys. Med. Biol. 52, 577 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskó L., Bekes G. & Fidrich M. Automatic segmentation of the liver from multi- and single-phase contrast-enhanced CT images. Med. Image Anal. 13, 871–882 (2009). [DOI] [PubMed] [Google Scholar]
- Akselrod-Ballin A. et al. Multimodal Correlative Preclinical Whole Body Imaging and Segmentation. Sci. Rep 6, 27940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue R. T. H. M., Defraene G., De Ruysscher D., Lambin P. & van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br. J. Radiol. 90, 20160665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling F. The changing face of cancer diagnosis: From computational image analysis to systems biology. Eur. Radiol 28(8), 3160–3164, ; DOI: 10.1007/s00330-018-5347-9 (2018). [DOI] [PubMed] [Google Scholar]
- Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc 31, 198–211 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giger M. L., Karssemeijer N. & Schnabel J. A. Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annu. Rev. Biomed. Eng. 15, 327–357 (2013). [DOI] [PubMed] [Google Scholar]
- Aguilar C. et al. Automated CT-based segmentation and quantification of total intracranial volume. Eur. Radiol 25, 3151–3160 (2015). [DOI] [PubMed] [Google Scholar]
- Messmer P. et al. A CT database for research, development and education: concept and potential. J. Digit. Imaging 20, 17–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjachan S. et al. Noninvasive optical imaging of nanomedicine biodistribution. ACS Nano 7, 252–262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremse F. et al. Virtual Elastic Sphere Processing Enables Reproducible Quantification of Vessel Stenosis at CT and MR Angiography. Radiology 260, 709–717 (2011). [DOI] [PubMed] [Google Scholar]
- Boll H. et al. Comparison of Fenestra LC, ExiTron nano 6000, and ExiTron nano 12000 for Micro-CT Imaging of Liver and Spleen in Mice. Acad. Radiol. 20, 1137–1143 (2013). [DOI] [PubMed] [Google Scholar]
- Gremse F. et al. Imalytics Preclinical: Interactive Analysis of Biomedical Volume Data. Theranostics 6, 328–341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullo S. et al. Warping an atlas derived from serial histology to 5 high-resolution MRIs. Sci. Data 5, 180107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Rosenhain S., et al. . 2018. figshare. https://doi.org/10.6084/m9.figshare.c.4224377