Summary
Objective
Genetics contribute to variability in individual response to weight‐loss interventions. The objective of this study was to determine the efficacy of a commercially available exercise and weight‐loss program and whether alignment of diet to genotype related to lipid metabolism promotes greater success.
Design
Sedentary women with obesity (n = 63) had genotype (FABP2rs1799883, PPARG2rs1801282, ADRB3rs4994C3, ADRB2rs1042713, rs1042714) determined using a direct‐to‐consumer genetic screening kit purported to promote greater weight‐loss success through dietary recommendations based on these genes. Participants were randomly assigned to follow a moderate carbohydrate (MC) or lower carbohydrate (LC) hypo‐energetic diet that aligned (A) or did not align (NA) with genotype for 24 weeks while participating in a resistance training and walking program. Data were analysed by general linear model repeated measures adjusted for baseline variables and are presented as mean (95% confidence interval) changes from baseline.
Results
Participants in the LC group experienced greater improvements (p = 0.051, ηp 2 = 0.025) in per cent changes in body composition (weight: MC −3.32 [−1.4, −5.2], LC −5.82 [−4.1, −7.6]; fat mass: MC −7.25 [−3.2, −11.2], LC −10.93 [−7.3, −14.5]; fat‐free mass: MC −0.32 [1.4, −2.0], LC −1.48 [0.7, −3.0]; and body fat percentage: MC −4.19 [−1.6, −6.8], LC −5.60 [−3.3, −7.9] %). No significant differences were observed between genotype groups (weight: A −5.00 [−3.3, −6.7], NA −4.14 [−2.2, −6.1]; fat mass: A −10.15 [−7.0, −13.6], NA −8.02 [−4.0, −12.0]; fat‐free mass: A −1.23 [0.3, −2.8], NA −0.56 [1.12, −2.3]; and body fat: A −5.28 [−3.0, −7.6], NA −4.51 [−1.9, −7.1] %).
Conclusions
Adherence to this exercise and weight‐loss program promoted improvements in body composition and health outcomes. While individuals following the LC diet experienced greater benefits, alignment of these diets to this genetic profile did not promote greater health outcomes.
Keywords: Exercise, Nutrigenetic, Weight management
Introduction
Obesity, and its related comorbidities, increases risk of chronic disease and remains a significant public health problem 1, 2, 3, 4. For this reason, there is substantial interest in finding ways to promote weight loss and weight management 3, 4, 5. While a number of interventions have been studied, a considerable amount of variability has been reported in weight and/or fat loss among individuals, even when participating in the same intervention 5, 6, 7, 8. One factor that has been suggested to influence an individual's response to a weight management intervention is genetics 4, 9, 10, 11, 12, 13. For example, findings from the Genome Wide Association Studies have identified genetic polymorphisms associated with obesity and aspects related to obesity, such as energy expenditure, appetite control and lipid metabolism 10, 12, 14, 15, 16. Theoretically, identification of obesity‐related genes may be helpful in determining optimal behavioural, dietary and/or pharmacological interventions to promote weight loss and weight management.
In support of this contention, Dopler‐Nelson and colleagues 17 retrospectively evaluated the relationship of five candidate genes related to lipid metabolism (i.e. FABP2rs1799883, PPARG2rs1801282, ADRB3rs4994C3, ADRB2rs1042713 and rs1042714) to weight loss success of individuals participating in several commercially available diet interventions. The researchers reported a twofold to threefold greater reduction in body weight along with greater reductions in waist circumference and triglycerides and an increase in high‐density lipoprotein (HDL) cholesterol among participants following a diet aligned with genotype as opposed to individuals in which the diet intervention did not align with genotype 17. These findings prompted others to examine whether alignment of genotype to different macronutrient diets would promote greater weight and/or fat loss as well as development of direct‐to‐consumer genetic screening kits to help individuals select a diet that may optimize weight loss efforts 17, 18, 19, 20, 21.
Our group has conducted a number of studies evaluating the effects of various diet and exercise interventions on weight loss as part of a Women's Health & Fitness Initiative 22, 23, 24, 25, 26, 27, 28. These studies generally found that participating in a circuit‐style resistance training program while following a slightly hypo‐energetic, low fat and moderately higher protein diet promoted fat loss while maintaining fat free mass and resting energy expenditure (REE) 22, 23, 24, 25, 26, 27, 28. Additionally, adherence to the moderately higher protein versions of this diet while following the circuit‐style resistance training program promoted more favourable changes in markers of metabolic syndrome 22, 25. We also observed significant changes in exercise capacity 22, 23, 24, 25, 26, 28, which is relevant as cardiopulmonary fitness is a known to be associated with chronic disease risk and related mortality 29, 30, 31, 32, 33, 34, 35. The purpose of the present investigation was to (1) determine the efficacy of two moderately higher protein diets that varied in carbohydrate and fat content on changes in weight, body composition, exercise capacity and biomarkers of health in sedentary women with obesity participating in a circuit‐style resistance training program and (2) determine if these tested diets would result in greater health outcomes when aligned by the genetic profile within direct‐to‐consumer genetic screening kit previously tested by Dopler‐Nelson et al. 17. We hypothesized that the moderately higher protein diet with higher fat and lower carbohydrate content (as opposed to lower fat and higher carbohydrate) and alignment of diet to genotype would promote greater reductions in body weight and lead to more favourable changes in body composition, exercise capacity and biomarkers of health.
Methods
Study design
This study was conducted in a randomized, parallel arm, prospective manner at a university‐based research facility. This investigation was approved by the Texas A&M University Institutional Review Board (IRB2013‐0737FX), performed in accordance with the Declaration of Helsinki 36 and registered retrospectively (NCT03116711). Prior to initiation of the intervention, participants provided a buccal swab sample using a direct‐to‐consumer genetic screening kit. This kit was designed to align diet recommendation with several genes related to lipid metabolism in an effort to promote greater weight loss success. Participants were randomly assigned to hypo‐energetic, moderately higher protein diets that were moderate carbohydrate (MC) or lower carbohydrate (LC) and aligned (A) or did not align (NA) with genotype as per recommendations from the screening kit. For 24 weeks, participants followed the recommended diets while participating in a supervised circuit‐style resistance training (4 d week−1) and walking program. Assessment sessions for data collection were carried out at 0, 4, 12, 16, 20 and 24 weeks with the exception that fitness related data were collected at 0, 12 and 24 weeks. An overview of the study design is presented in Figure 1.
Figure 1.
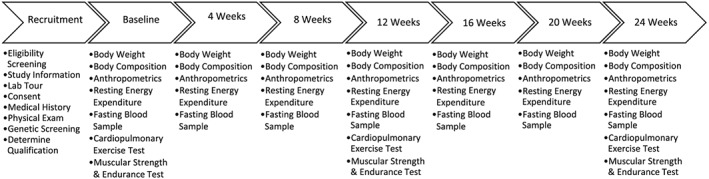
Overview of the study design.
Participant recruitment
Participants were recruited from university e‐mails and local advertisements. Inclusion criteria consisted of the following: female, apparently healthy, sedentary, between 18 and 60 years of age and a body mass index > 22 kg m−2 and/or body fat percentage >30%. Exclusion criteria consisted of uncontrolled metabolic disorder; participation in a structured exercise program within the past 3 months (>30 min d−1, 3 d week−1); recent weight change (+/−3.2 kg) within 12 weeks prior to initiation of the study; consumption of nutritional supplements that may affect muscle mass, anabolic/catabolic hormone levels, or weight loss; and pregnancy, nursing or intention to become pregnant during the subsequent 52 weeks.
A total of 255 individuals expressed interest in participating in the study and attended a recruitment session. Eligibility screening occurred at the recruitment session. A total of 64 individuals did not meet eligibility criteria. Thus, 191 eligible women signed the informed consent, underwent a standard medical exam and provided a buccal swab sample for genotyping. Of these, 43 participants were no longer eligible based on genotype results, and 51 declined participation due to scheduling conflicts (n = 9) or concerns with the time commitment required by the study (n = 42). A total of 97 participants were randomly assigned to diet based on results from the genetic screening kit and initiated participation in the study. Of these, 34 participants withdrew prior to or following the first assessment session, 63 participants completed at least 12 weeks of intervention and 51 women completed all 24 weeks. The primary reasons for attrition were non‐compliance (failure to follow diet, attend exercise or assessment sessions), concerns with the time commitment required by the study or personal reasons. Figure 2 presents a CONSORT diagram.
Figure 2.
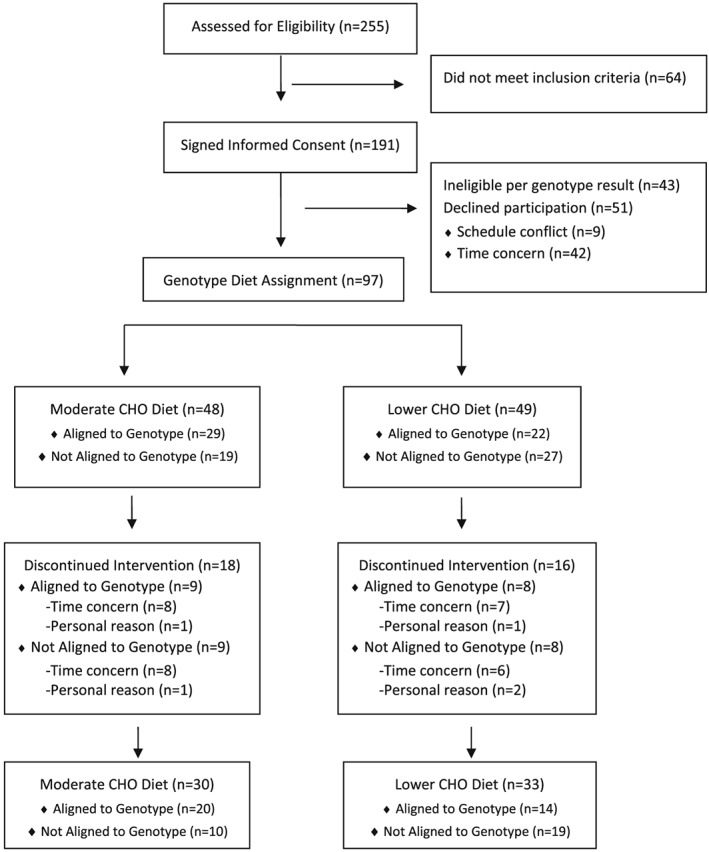
CONSORT diagram.
Genotyping
Buccal swab samples were collected in duplicate, and each swab was placed in an individual envelope with a barcode that corresponded with participant identification. Barcode and participant identification information were kept in a secured document accessible only to the study coordinator and primary investigator. Upon collection of the samples, one envelope per participant was placed in a container for shipment to Interleukin Genetics (Waltham, MA) for genotyping within a Clinical Laboratory Improvements Amendments certified molecular genetics laboratory. In this laboratory, DNA was extracted from buccal cheek swabs (Puritan, Gilford, ME) using standard procedures for determination of the five polymorphisms included within the profile (FABP2rs1799883, PPARG2rs1801282, ADRB3rs4994C3, ADRB2rs1042713 and rs1042714). Single nucleotide polymorphism (SNP) genotyping was performed by the single‐base extension method using a Biomek® FXP SNPstream® system (Beckman Coulter, Brea, CA) with SNPs multiplexed as needed to avoid interference. The multiplex polymerase chain reaction was treated with exonuclease I and shrimp alkaline phosphatase (USB). The single‐base extension reaction was performed per manufacturer's protocol, and ‘tagged’ products were hybridized to a microarray plate. The SNPstream instrument was used to read the plates, and the SNPstream software was used to determine initial allele calls, which were confirmed by a technician.
Dietary intervention
Dietary protocols
Participants followed either the MC or LC Curves Complete® (Curves International, Waco, TX) diet programs. The MC dietary prescription included a macronutrient breakdown of 30% kcal from carbohydrates, 25% kcal fat and 45% kcal protein. The LC diet prescription included 20% kcal carbohydrate, 35% kcal fat and 45% kcal protein. Participants on the MC program were provided instruction from a registered dietitian on how to use the Curves Complete online program to select, change and track daily meals and snacks. Participants assigned to the LC diet program were provided sample menus and instructions on how to make substitutions on each menu from a registered dietitian. Dietary instruction was provided to all participants at baseline and throughout the intervention as needed for each participant. Guidance provided by the registered dietitian to achieve the moderately higher dietary protein goals in both diet groups included increased intake of higher protein food sources and incorporation of protein shakes and bars. Both diets were designed to provide 1,400 kcal d−1 for the first week followed by 1,500 kcal d−1 for the remaining 23 weeks. Because the diets were hypo‐energetic, all participants were provided a daily multivitamin and mineral supplement containing calcium, vitamin D and omega‐3 fatty acids (Curves 2 Go, Curves International®).
Genotype alignment to diets
Results from the genotyping analysis were provided by the genetic testing company with diet types recommended based on the combination of allelic patterns among the polymorphisms within the tested genetic profile 9. The recommended diet types provided were LC diet, lower fat diet or balanced diet. In the present investigation, participants with a genetic profile suggesting the lower carbohydrate diet were assigned as aligned to the LC diet (20:35:45% kcal) or not aligned to the MC diet (30:25:45% kcal). Participants with a genetic profile suggesting lower fat diet were assigned as aligned to the MC diet (30:25:45% kcal) or not aligned to the LC diet (20:35:45% kcal). Individuals with a genetic profile suggesting a balanced diet were excluded from the present investigation and offered the opportunity to participate in another exercise and weight loss study.
Exercise intervention
Throughout the duration of the intervention, all participants completed four, 30‐min workouts per week in the lab, following the Curves International® exercise program. The circuit‐style resistance training program utilized the computerized Curves Smart system with software designed by MYTRAK® (version 4.2.1, copyright 2004–2011, MYTRAK Health System Inc., Mississauga, Ontario, Canada). This system provides feedback and records performance to ensure participants meet intensity expectations (i.e. 60% to 80% of one repetition maximum). The circuit included 13 bi‐directional hydraulic concentric only resistance exercise machines, which worked all major muscle groups (i.e. elbow flexion/extension, knee flexion/extension, shoulder press/lat pull, hip abductor/adductor, chest press/seated row, horizontal leg press, squat, abdominal crunch/back extension, chest flies, oblique, shoulder shrug/dip, hip extension and side bends). Before and after training sessions, participants performed whole body stretching. During training sessions, participants wore a Polar FT4 Heart Rate Monitor (Lake Success, NY) to monitor exercise intensity. Participants were coached to perform as many repetitions as possible while on each resistance machine. Three out of the four weekly workouts consisted of alternating between 30 s on the resistance machines and 30 s completing floor‐based aerobic callisthenic exercises following a video of a fitness instructor in order to maintain heart rate between 60% and 80% of age‐predicted maximal heart rate. Participants completed the entire circuit twice during these workouts. One of the four weekly workouts included Zumba dance between resistance machines. During these workouts, participants alternated between 1 min on the resistance machines and 1 min performing Zumba dance exercise as taught by a certified Zumba instructor. Participants complete the entire circuit once during these workouts. Participants were required to complete at least 90% of exercise training sessions in order to be considered compliant (i.e. 86 of 96 workouts).
We previously reported that women participating in this type of circuit‐style resistance training regimen elicit an average exercise heart rate of 126 ± 15 bpm (80% of maximal heart rate), an average exercise intensity of 65% ± 10% of peak oxygen uptake, resistance exercise intensities ranging between 61% and 82% of one repetition maximum (1RM) on the various exercise machines and an average energy expenditure of 314 ± 102 kcals per workout 37, 38, 39. Additionally, participants were provided with a standard pedometer and instructed to reach a goal of 10,000 steps per day on non‐circuit training days.
Procedures
Participants were instructed to refrain from exercising for 48 h and fast for 12 h prior to each assessment session. Participants reported to the lab at approximately the same time of day during each session. The following describes the methods of assessments performed.
Dietary intake
Participants were provided a detailed description of how to measure and record food and beverage intake on food logs by a registered dietitian. Participants recorded all food and energy containing fluids consumed for 4 d (three weekdays and one weekend day) prior to each testing session. Food logs were checked for accuracy when returning to the lab for each assessment session and analysed by a registered dietitian using dietary analysis software (ESHA Food Processor Version 8.6, Salem, OR).
Resting energy expenditure
Resting energy expenditure was measured using an open‐circuit method of indirect calorimetry with the ParvoMedics TrueMax 2400 Metabolic Measurement System (Parvomedics Inc., Sandy, UT) using previously described procedures 23, 25. Participants laid down motionless in a bed with legs resting on a padded box, while respiratory gases were collected from a metabolic hood. The five data points after at least 10 min of assessment in which oxygen uptake and carbon dioxide production varied by no more than 5% were used for REE measurements 40.
Body composition
Height and body weight were measured following standard procedures with the Health‐O‐Meter Professional 500KL scale (Pelstar LLC, Alsip, IL, USA) self‐calibrating digital scale with an accuracy of ±0.02 kg. Body composition was determined with a Hologic Discovery W Dual‐Energy X‐ray Absorptiometer (Hologic Inc., Waltham, MA, USA) equipped with APEX Software (APEX Corporation Software, Pittsburg, PA, USA) that estimated visceral adipose tissue by using procedures previously described 41. Prior to testing, quality control calibration procedures were performed following manufacturer's guidelines. The mean coefficient of variation for all scans throughout the study was between 0.31% and 0.33% with a mean intra‐class correlation of 0.935.
Exercise capacity
At 0, 12 and 24 weeks of intervention, participants performed muscular strength (1RM) and muscular endurance tests (maximum number of repetitions at 80% of 1RM) on the bench press and hip sled/leg press (Nebular Fitness, Versailles, OH) following standard procedures 42. Hand position on the bench press and foot and sled seat positions on the leg press were recorded and standardized among testing sessions. Participants were provided 2 min of recovery between 1RM attempts and 5 min of recovery between exercises. Participants also performed a symptom‐limited maximal cardiopulmonary exercise test on a treadmill following the Bruce protocol using standard procedures 42. Expired ventilation and respiratory gases were assessed using a calibrated ParvoMedics TrueMax 2400 Metabolic Measurement System (Parvomedics Inc.) metabolic measurement system. Heart rate and rhythm were assessed using a 12‐lead electrocardiogram (Nasiff Cardio Card Electrocardiograph, Central Square, NY). Resting and exercise blood pressure were measured by auscultation of the brachial artery using a mercurial sphygmomanometer using standard procedures 42.
Blood collection and biomarker analysis
Fasting whole blood was collected into two BD Vacutainer® SST™ Serum Separation Tubes (Becton, Dickinson and Company, Franklin Lakes, NJ) using standard phlebotomy procedures. The SST tubes sat at room temperature for 15 min and were then centrifuged at 3,500 rpm for 10 min using a Heraeus Megafuge 40R centrifuge (ThermoFisher Scientific, Inc., Watham, MA). Serum was transferred into micro‐centrifuge tubes and stored in a freezer at −80°C for later analyses. Serum glucose, triglycerides, total cholesterol, low‐density lipoprotein (LDL) and HDL levels were analysed using a Cobas C 111 (Roche Diagnostics, Indianapolis, IN) automated chemistry analyzer according to manufacturer's instructions. The tested intra‐assay coefficient of variation for these variables was approximately less than 3% along with an inter‐assay coefficient of variation of less than 2% 43. Serum insulin was determined using a commercially available enzyme linked immunoabsorbent assay kit (ALPCO Diagnostics, Salem, NH) using a BioTek ELX‐808 Ultramicoplate reader (BioTek Instruments Inc., Winooski, VT) set at an optical density of 450 nm with BioTek Gen5 Analysis software (BioTek Instruments Inc.). The intra‐assay coefficient of variation has been shown to range from 5.1% to 10.3%, and the inter‐assay coefficient of variation ranges from 6.7% to 16.6%. The homeostatic model of assessment for insulin resistance (HOMA‐IR) was calculated by dividing the product of glucose (mmol L−1) and insulin (μIU L−1) by 22.5 44.
Statistical analyses
A priori power calculation was set at >0.80 and was based on change in fat mass between diet groups from previous research in our lab utilizing similar diet and exercise interventions 22, 23, 25, 26, 27 as well as considering the change in body weight reported in a similar study 17. This analysis revealed that a sample size of 15–20 participants per group was sufficient to detect meaningful changes in fat mass. Given this, we employed an intent‐to‐treat procedure using expectation maximization 45 to replace missing values for participants who completed at least 12 weeks of the intervention (n = 63). A total of 51 participants completed the 24‐week intervention, and the following completed at least 12 weeks of the 24‐week intervention: three participants in NA‐MC, four in A‐MC, four in NA‐LC and one in A‐LC completed at least 12 weeks of the 24‐week intervention. This resulted in 628 data points that were imputed out of 10,458 total data points, which is 6% of the data. All data were analysed with IBM® SPSS® Version 25 software (IBM Corp., Armonk, NY, USA).
General linear model (GLM) multivariate analysis was used to determine differences between groups at baseline. Related variables were analysed using univariate, multivariate and repeated measures GLM. For body composition analysis, baseline variables were used as covariates. The overall multivariate Wilks' Lamda and Greenhouse–Geisser univariate p‐levels are reported. Data were considered significant when the probability of type I error was 0.05 or less (p < 0.05). Partial eta squared effect sizes (ηp 2) are reported as an indicator of effect size when the p‐level was close to significance (i.e. p < 0.1 but p > 0.05) 46. An eta squared around 0.02 was considered small, 0.13 medium and 0.26 large 46. Tukey's least significant differences post hoc analyses were performed to determine differences among groups. Mean changes from baseline as well as per cent changes from baseline were calculated and analysed using one‐way anova to determine mean changes with 95% confidence intervals (CIs). Mean changes with 95% CIs completely above or below baseline were considered significantly different 46. Data are presented as means ± standard deviations, mean change from baseline with 95% CIs or mean per cent change from baseline with 95% CIs.
Results
Baseline characteristics
Multivariate GLM analysis revealed no overall Wilks' Lamba diet (p = 0.22), genotype (p = 0.60) or diet × genotype (p = 0.15) effects. Table 1 presents univariate analysis of participant demographics. A statistically significant difference between genotype groups for height was observed (p = 0.01), suggesting that participants that were not aligned to genotype were taller. Additionally, the difference between fat‐free mass between genotype groups was close to significance (p = 0.06); therefore, we normalized data among groups by calculating delta values from baseline as well as per cent changes from baseline.
Table 1.
Participant demographics at baseline
| Variable | MC | LC | A | NA | A‐MC | NA‐MC | A‐LC | NA‐LC | Mean | Diet | Genotype | D × G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p‐level | p‐level | p‐level | ||||||||||
| Age (years) | 38.8 ± 12.5 | 41.9 ± 12.3 | 40.4 ± 11.7 | 40.5 ± 13.3 | 39.6 ± 12.6 | 37.4 ± 13.0 | 41.5 ± 10.8 | 42.2 ± 13.5 | 40.4 ± 12.4 | 0.31 | 0.82 | 0.67 |
| Height (cm) | 162.1 ± 5.9 | 163.7 ± 8.4 | 160.8 ± 6.0 | 165.5 ± 7.9 | 160.4 ± 4.7 | 165.5 ± 6.8 | 161.3 ± 7.6 | 165.6 ± 8.7 | 163.0 ± 7.3 | 0.78 | 0.01* | 0.82 |
| Weight (kg) | 93.3 ± 23.8 | 94.3 ± 24.5 | 89.8 ± 19.8 | 98.5 ± 24.4 | 88.2 ± 17.0 | 103.3 ± 22.4 | 92.0 ± 23.8 | 95.9 ± 25.5 | 93.8 ± 22.3 | 0.76 | 0.11 | 0.34 |
| BMI (kg m−2) | 35.5 ± 7.5 | 35.0 ± 8.2 | 34.6 ± 7.0 | 36.0 ± 8.7 | 34.3 ± 6.8 | 37.9 ± 8.8 | 35.0 ± 7.6 | 34.9 ± 8.7 | 35.2 ± 7.8 | 0.58 | 0.40 | 0.37 |
| Fat mass (kg) | 39.2 ± 11.7 | 39.7 ± 14.4 | 37.5 ± 41.8 | 41.8 ± 15.2 | 36.7 ± 10.7 | 44.1 ± 12.7 | 38.5 ± 14.2 | 40.6 ± 16.6 | 39.5 ± 13.6 | 0.81 | 0.19 | 0.47 |
| Fat‐free mass (kg) | 47.6 ± 8.3 | 47.6 ± 9.0 | 45.8 ± 7.9 | 49.7 ± 9.1 | 45.3 ± 6.9 | 52.1 ± 9.3 | 46.6 ± 9.4 | 48.4 ± 9.0 | 47.6 ± 8.6 | 0.59 | 0.06 | 0.27 |
| Body fat (%) | 44.3 ± 4.7 | 44.3 ± 6.0 | 44.1 ± 5.1 | 44.6 ± 5.3 | 43.9 ± 5.0 | 45.2 ± 4.0 | 44.3 ± 5.3 | 44.3 ± 6.6 | 44.3 ± 5.3 | 0.88 | 0.65 | 0.64 |
| Waist:Hip ratio | 0.79 ± 0.50 | 0.78 ± 0.05 | 0.79 ± 0.05 | 0.77 ± 0.05 | 0.80 ± 0.04 | 0.77 ± 0.06 | 0.78 ± 0.05 | 0.77 ± 0.05 | 0.78 ± 0.05 | 0.34 | 0.15 | 0.41 |
| Peak VO2 (mL/kg/min) | 21.4 ± 4.2 | 22.2 ± 5.4 | 22.2 ± 4.1 | 21.3 ± 5.6 | 22.7 ± 2.9 | 18.9 ± 3.9 | 21.6 ± 4.6 | 22.6 ± 6.0 | 21.8 ± 4.8 | 0.31 | 0.27 | 0.06 |
| Bench press 1RM (kg kg−1 body weight) | 0.39 ± 0.09 | 0.37 ± 0.11 | 0.40 ± 0.10 | 0.36 ± 0.10 | 0.42 ± 0.09 | 0.35 ± 0.05 | 0.39 ± 0.11 | 0.37 ± 0.12 | 0.38 ± 0.10 | 0.74 | 0.11 | 0.40 |
| Leg press 1RM (kg kg−1 body weight) | 2.50 ± 0.88 | 2.18 ± 0.73 | 2.44 ± 0.91 | 2.21 ± 0.67 | 2.63 ± 1.02 | 2.25 ± 0.45 | 2.18 ± 0.71 | 2.19 ± 0.77 | 2.34 ± 0.82 | 0.23 | 0.39 | 0.37 |
| Resting energy expenditure (kcal d−1) | 1,454 ± 293 | 1,415 ± 307 | 1,405 ± 299 | 1,468 ± 304 | 1,397 ± 274 | 1,568 ± 310 | 1,416 ± 342 | 1,415 ± 306 | 1,434 ± 304 | 0.40 | 0.29 | 0.29 |
| Resting energy expenditure (kcal/kg/d) | 19.20 ± 5.85 | 20.79 ± 8.57 | 20.51 ± 6.46 | 19.47 ± 8.42 | 19.89 ± 6.21 | 17.80 ± 5.05 | 21.38 ± 6.96 | 21.38 ± 8.75 | 20.03 ± 7.39 | 0.31 | 0.43 | 0.79 |
p < 0.05.
Data are presented as mean ± standard deviations. The p‐levels are from univariate analysis. A, diet aligned to genotype (n = 34); BMI, body mass index; D, diet; G, genotype; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19); VO2, oxygen uptake.
Dietary intake and resting energy expenditure
Tables 2 and 3 present univariate analysis of dietary intake data. Significant time effects were observed for all variables (p < 0.05). On average, all participants reduced energy intake by −449 ± 607 to −553 ± 505 kcals d−1. Additionally, the diet interventions were successful in decreasing carbohydrate (CHO) intake (−38.4 ± 28.9 to −42.0 ± 34.3% kcal CHO per day) and fat intake (−38.8 ± 6.2 to −34.0 ± 7.3% kcal fat per day) while increasing protein intake (31.2 ± 58.3 to 38.3 ± 56.3% kcal protein per day) with relative protein intake ranging from 1.01 ± 0.39 to 1.33 ± 0.79 g/kg/d among groups. Individuals in the LC group on average consumed approximately 28–31% kcals from CHO and 34–37% kcals from fat, which was slightly above the prescription for CHO (20% kcal CHO) but met the prescription for fat (35% kcal CHO). Individuals in the MC group on average consumed 33–36% kcal CHO and 32–34% kcal fat, which was very close to the prescription of CHO and slightly above the prescription for fat. However, those in the LC group consistently consumed less CHO (MC: −60.7 [−36.2, −85.2]; LC: −92.6 [−70.3, −114.8] g d−1, p = 0.059) throughout the study, and this observation was significantly different between groups when expressed as mean per cent change from baseline (MC: −30.2 [−20.4, −40.1], LC: −45.4 [−36.4, −54.3] %, p = 0.027).
Table 2.
Absolute energy intake and percentage of total kcals ingested per day from macronutrients
| Variable | Group | Baseline | 4 weeks | 8 weeks | 12 weeks | 16 weeks | 20 weeks | 24 weeks | p‐level | |
|---|---|---|---|---|---|---|---|---|---|---|
| Energy intake (kcal d−1) | MC | 1,743 ± 502 | 1,274 ± 239 | 1,305 ± 274 | 1,331 ± 312 | 1,340 ± 307 | 1,268 ± 277 | 1,280 ± 264 | Diet | 0.86 |
| LC | 1,838 ± 553 | 1,342 ± 369 | 1,279 ± 335 | 1,354 ± 430 | 1,246 ± 371 | 1,279 ± 352 | 1,203 ± 321 | Genotype | 0.97 | |
| A | 1,795 ± 553 | 1,355 ± 304 | 1,327 ± 307 | 1,342 ± 370 | 1,256 ± 298 | 1,251 ± 283 | 1,240 ± 221 | G × D | 0.08 | |
| NA | 1,790 ± 505 | 1,256 ± 320 | 1,250 ± 304 | 1,345 ± 388 | 1,331 ± 390 | 1,300 ± 354 | 1,239 ± 368 | T × D | 0.32 | |
| A‐MC | 1,738 ± 593 | 1,272 ± 266 | 1,299 ± 291 | 1,273 ± 298 | 1,283 ± 298 | 1,217 ± 242 | 1,195 ± 204 | T × G | 0.23 | |
| NA‐MC | 1,754 ± 260 | 1,277 ± 185 | 1,318 ± 252 | 1,446 ± 321 | 1,454 ± 308 | 1,369 ± 326 | 1,450 ± 296 | T × G × D | 0.42 | |
| A‐LC | 1,877 ± 498 | 1,474 ± 326 | 1,366 ± 336 | 1,440 ± 448 | 1,218 ± 305 | 1,299 ± 336 | 1,305 ± 237 | Time | 0.001 | |
| NA‐LC | 1,810 ± 602 | 1,244 ± 376 | 1,214 ± 328 | 1,291 ± 417 | 1,266 ± 420 | 1,264 ± 371 | 1,128 ± 358 | |||
| Mean | 1,793 ± 527 | 1,309* ± 313 | 1,291* ± 305 | 1,343* ± 376 | 1,291* ± 343 | 1,274* ± 316 | 1,240* ± 295 | |||
| Carbohydrate intake (% kcal d−1) | MC | 40.7 ± 8.8 | 35.8 ± 10.55*** | 33.6 ± 7.7 | 36.0 ± 9.0 | 32.7 ± 8.5 | 33.6 ± 7.8 | 32.4 ± 9.3 | Diet | 0.13 |
| LC | 41.7 ± 8.2 | 28.4 ± 11.0 | 30.0 ± 9.6 | 31.1 ± 13.8 | 29.3 ± 9.7 | 29.5 ± 10.8 | 29.3 ± 10.6 | Genotype | 0.27 | |
| A | 42.4 ± 8.5 | 33.4 ± 12.2 | 33.1 ± 8.4 | 35.8 ± 10.8 | 32.0 ± 8.5 | 32.8 ± 9.0 | 31.5 ± 10.3 | G × D | 0.95 | |
| NA | 39.8 ± 8.3 | 30.2 ± 10.2 | 30.1 ± 9.3 | 30.6 ± 12.7 | 29.6 ± 10.0 | 29.8 ± 10.3 | 30.0 ± 9.9 | T × D | 0.06 | |
| A‐MC | 42.3 ± 9.2 | 36.2 ± 11.8 | 34.4 ± 7.5 | 36.5 ± 8.0 | 32.8 ± 7.6 | 35.1 ± 6.8 | 33.0 ± 8.9 | T × G | 0.87 | |
| NA‐MC | 37.6 ± 7.2 | 35.0 ± 7.9 | 32.1 ± 8.3 | 35.0 ± 11.0 | 32.4 ± 10.4 | 30.4 ± 9.1 | 31.3 ± 10.4 | T × G × D | 0.52 | |
| A‐LC | 42.6 ± 7.5 | 29.4 ± 11.9 | 31.3 ± 9.5 | 34.8 ± 14.2 | 30.8 ± 9.7 | 29.5 ± 10.8 | 29.4 ± 11.9 | Time | <0.001 | |
| NA‐LC | 41.0 ± 8.7 | 27.6 ± 10.6 | 29.1 ± 9.9 | 28.3 ± 13.2 | 28.2 ± 9.8 | 29.4 ± 11.1 | 29.2 ± 9.9 | |||
| Mean | 41.2 ± 8.4 | 31.9* ± 11.3 | 31.7* ± 8.9 | 33.4* ± 11.9 | 30.9* , ** ± 9.2 | 31.4* , ** ± 9.7 | 30.8* , ** ± 10.1 | |||
| Protein intake (% kcal d−1) | MC | 19.5 ± 5.3 | 30.9 ± 7.8 | 31.4 ± 7.6 | 29.9 ± 10.2 | 32.0 ± 7.9 | 30.9 ± 8.0 | 33.0 ± 8.0 | Diet | 0.31 |
| LC | 18.4 ± 4.5 | 34.1 ± 9.1 | 33.9 ± 8.5 | 33.3 ± 9.4 | 34.5 ± 8.2 | 33.2 ± 8.5 | 34.6 ± 8.1 | Genotype | 0.24 | |
| A | 18.6 ± 4.6 | 31.8 ± 9.8 | 31.4 ± 8.1 | 29.6 ± 10.6 | 32.2 ± 7.1 | 31.8 ± 7.9 | 32.2 ± 8.1 | G × D | 0.87 | |
| NA | 19.4 ± 5.2 | 33.5 ± 6.9 | 34.2 ± 7.9 | 34.2 ± 8.4 | 34.6 ± 9.1 | 32.4 ± 8.8 | 35.8 ± 7.7 | T × D | 0.38 | |
| A‐MC | 18.6 ± 5.1 | 30.6 ± 8.2 | 31.0 ± 7.4 | 28.8 ± 10.8 | 32.1 ± 7.1 | 30.5 ± 6.6 | 32.2 ± 7.2 | T × G | 0.53 | |
| NA‐MC | 21.3 ± 5.3 | 31.5 ± 7.3 | 32.1 ± 8.1 | 32.3 ± 8.8 | 31.8 ± 9.7 | 31.7 ± 10.7 | 34.6 ± 9.7 | T × G × D | 0.63 | |
| A‐LC | 18.5 ± 4.0 | 33.6 ± 11.9 | 32.0 ± 9.3 | 30.9 ± 10.6 | 32.4 ± 7.5 | 33.8 ± 9.4 | 32.2 ± 9.5 | Time | <0.001 | |
| NA‐LC | 18.3 ± 4.9 | 34.5 ± 6.7 | 35.4 ± 7.8 | 35.1 ± 8.3 | 36.1 ± 8.6 | 32.8 ± 8.0 | 36.4 ± 6.7 | |||
| Mean | 18.9 ± 4.9 | 32.6* ± 8.6 | 32.7* ± 8.1 | 31.7* ± 9.9 | 33.3* ± 8.1 | 32.1* ± 8.3 | 33.8* , ** ± 8.1 | |||
| Fat intake (% kcal d−1) | MC | 38.6 ± 6.4 | 32.8 ± 7.2 | 34.1 ± 6.9 | 33.1 ± 5.3 | 34.4 ± 5.4 | 34.8 ± 8.2 | 33.9 ± 5.8 | Diet | 0.20 |
| LC | 39.0 ± 6.1 | 37.4 ± 9.7 | 34.7 ± 6.3 | 34.8 ± 8.7 | 36.2 ± 6.8 | 36.7 ± 8.3 | 35.8 ± 6.9 | Genotype | 0.51 | |
| A | 38.1 ± 6.4 | 34.4 ± 10.0 | 33.8 ± 6.0 | 33.3 ± 7.1 | 35.2 ± 5.4 | 34.7 ± 7.9 | 35.2 ± 6.5 | G × D | 0.71 | |
| NA | 39.6 ± 6.0 | 36.1 ± 7.4 | 35.2 ± 7.2 | 34.8 ± 7.5 | 35.5 ± 7.1 | 37.1 ± 8.6 | 34.5 ± 6.3 | T × D | 0.46 | |
| A‐MC | 38.3 ± 6.7 | 32.7 ± 8.0 | 33.8 ± 6.1 | 33.4 ± 5.6 | 34.0 ± 5.4 | 33.2 ± 6.7 | 33.3 ± 5.2 | T × G | 0.77 | |
| NA‐MC | 39.2 ± 6.1 | 32.9 ± 5.7 | 34.9 ± 8.7 | 32.5 ± 4.9 | 35.4 ± 5.5 | 38.0 ± 10.1 | 34.9 ± 6.9 | T × G × D | 0.28 | |
| A‐LC | 37.9 ± 6.2 | 36.8 ± 12.1 | 33.7 ± 6.0 | 33.1 ± 9.0 | 36.9 ± 5.1 | 36.9 ± 9.1 | 37.8 ± 7.5 | Time | 0.001 | |
| NA‐LC | 39.8 ± 6.1 | 37.8 ± 7.7 | 35.4 ± 6.6 | 36.0 ± 8.4 | 35.6 ± 8.0 | 36.6 ± 7.9 | 34.3 ± 6.2 | |||
| Mean | 38.8 ± 6.2 | 35.2* ± 8.8 | 34.4* ± 6.6 | 34.0* ± 7.3 | 35.3* ± 6.2 | 35.8* ± 8.2 | 34.9* ± 6.4 | |||
Different from baseline (p < 0.05).
Different from 12 weeks (p < 0.05).
Difference between diet groups (p < 0.05).
Data presented as mean ± standard deviations. The p‐levels are from univariate analysis. A, diet aligned to genotype (n = 34); D, diet effect; G, genotype match to diet effect; G × D, genotype match to diet × diet effect; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19); T, time effect; T × G, time × genotype match to diet effect.
Table 3.
Relative energy and macronutrient intake
| Variable | Group | Baseline | 4 weeks | 8 weeks | 12 weeks | 16 weeks | 20 weeks | 24 weeks | p‐level | |
|---|---|---|---|---|---|---|---|---|---|---|
| Energy intake (kcal/kg/d) | MC | 19.20 ± 5.85 | 14.35 ± 3.39 | 15.00 ± 4.36 | 15.36 ± 4.62 | 15.47 ± 4.26 | 14.76 ± 4.47 | 14.72 ± 3.66 | Diet | 0.32 |
| LC | 20.79 ± 8.57 | 15.59 ± 5.99 | 14.99 ± 5.20 | 15.91 ± 5.50 | 14.71 ± 5.53 | 15.18 ± 5.68 | 14.45 ± 5.30 | Genotype | 0.15 | |
| A | 20.51 ± 6.47 | 16.06 ± 4.96 | 15.77 ± 4.44 | 15.99 ± 4.34 | 15.26 ± 4.74 | 15.22 ± 4.75 | 14.97 ± 3.29 | G × D | 0.45 | |
| NA | 19.47 ± 8.42 | 13.75 ± 4.68 | 14.08 ± 5.07 | 15.25 ± 5.86 | 14.86 ± 5.25 | 14.70 ± 5.57 | 14.11 ± 5.72 | T × D | 0.34 | |
| A‐MC | 19.89 ± 6.21 | 14.93 ± 3.01 | 15.57 ± 4.39 | 15.45 ± 4.45 | 15.52 ± 3.74 | 14.90 ± 4.17 | 14.54 ± 3.42 | T × G | 0.26 | |
| NA‐MC | 17.80 ± 5.05 | 13.18 ± 3.95 | 13.84 ± 4.30 | 15.17 ± 5.17 | 15.39 ± 5.39 | 14.48 ± 5.26 | 15.07 ± 4.26 | T × G × D | 0.45 | |
| A‐LC | 21.38 ± 6.96 | 17.68 ± 6.66 | 16.04 ± 4.67 | 16.75 ± 4.21 | 14.88 ± 6.03 | 15.66 ± 5.61 | 15.58 ± 3.12 | Time | 0.005 | |
| NA‐LC | 20.35 ± 9.75 | 14.05 ± 5.09 | 14.21 ± 5.54 | 15.29 ± 6.33 | 14.58 ± 5.30 | 14.82 ± 5.85 | 13.61 ± 6.41 | |||
| Mean | 20.03 ± 7.39 | 15.00* ± 4.93 | 14.99* ± 5.19 | 15.65* ± 5.07 | 15.07* ± 4.94 | 14.98* ± 5.10 | 14.58* ± 4.56 | |||
| CHO intake (g/kg/d) | MC | 1.91 ± 0.72 | 1.27 ± 0.59 | 1.26 ± 0.50 | 1.35 ± 0.53 | 1.21 ± 0.50 | 1.22 ± 0.51 | 1.19 ± 0.48 | Diet | 0.61 |
| LC | 2.11 ± 1.07 | 1.07 ± 0.61 | 1.11 ± 0.56 | 1.17 ± 0.77 | 1.05 ± 0.56 | 1.09 ± 0.59 | 1.02 ± 0.57 | Genotype | 0.14 | |
| A | 2.10 ± 0.78 | 1.30 ± 0.61 | 1.30 ± 0.52 | 1.41 ± 0.69 | 1.17 ± 0.48 | 1.22 ± 0.53 | 1.18 ± 0.52 | G × D | 0.83 | |
| NA | 1.91 ± 1.07 | 1.01 ± 0.56 | 1.04 ± 0.53 | 1.07 ± 0.61 | 1.07 ± 0.60 | 1.07 ± 0.57 | 1.00 ± 0.54 | T × D | 0.10 | |
| A‐MC | 2.04 ± 0.72 | 1.35 ± 0.63 | 1.34 ± 0.51 | 1.39 ± 0.54 | 1.20 ± 0.44 | 1.29 ± 0.53 | 1.20 ± 0.49 | T × G | 0.62 | |
| NA‐MC | 1.65 ± 0.67 | 1.13 ± 0.49 | 1.11 ± 0.49 | 1.28 ± 0.53 | 1.21 ± 0.64 | 1.07 ± 0.46 | 1.15 ± 0.49 | T × G × D | 0.41 | |
| A‐LC | 2.19 ± 0.88 | 1.23 ± 0.60 | 1.26 ± 0.55 | 1.44 ± 0.89 | 1.13 ± 0.54 | 1.13 ± 0.52 | 1.16 ± 0.59 | Time | <0.001 | |
| NA‐LC | 2.06 ± 1.22 | 0.94 ± 0.60 | 1.00 ± 0.56 | 0.97 ± 0.63 | 0.99 ± 0.58 | 1.06 ± 0.64 | 0.92 ± 0.55 | |||
| Mean | 2.02 ± 0.92 | 1.16* ± 0.60 | 1.18* ± 0.54 | 1.26* ± 0.67 | 1.12* , ** ± 0.54 | 1.15* ± 0.55 | 1.10* , ** ± 0.53 | |||
| Protein intake (g/kg/d) | MC | 0.88 ± 0.31 | 1.45 ± 2.09 | 1.12 ± 0.30 | 1.12 ± 0.39 | 1.18 ± 0.38 | 1.10 ± 0.35 | 1.17 ± 0.33 | Diet | 0.62 |
| LC | 0.90 ± 0.35 | 1.29 ± 0.69 | 1.22 ± 0.54 | 1.27 ± 0.60 | 1.19 ± 0.61 | 1.22 ± 0.63 | 1.20 ± 0.54 | Genotype | 0.43 | |
| A | 0.89 ± 0.30 | 1.60 ± 2.01 | 1.18 ± 0.40 | 1.16 ± 0.47 | 1.20 ± 0.52 | 1.21 ± 0.55 | 1.17 ± 0.36 | G × D | 0.99 | |
| NA | 0.88 ± 0.36 | 1.09 ± 0.43 | 1.16 ± 0.49 | 1.25 ± 0.57 | 1.16 ± 0.51 | 1.11 ± 0.47 | 1.20 ± 0.54 | T × D | 0.75 | |
| A‐MC | 0.87 ± 0.29 | 1.67 ± 2.54 | 1.15 ± 0.26 | 1.10 ± 0.35 | 1.20 ± 0.36 | 1.13 ± 0.30 | 1.14 ± 0.28 | T × G | 0.23 | |
| NA‐MC | 0.91 ± 0.36 | 1.01 ± 0.39 | 1.07 ± 0.39 | 1.18 ± 0.47 | 1.12 ± 0.44 | 1.06 ± 0.45 | 1.23 ± 0.43 | T × G × D | 0.72 | |
| A‐LC | 0.93 ± 0.33 | 1.51 ± 0.90 | 1.24 ± 0.56 | 1.25 ± 0.60 | 1.20 ± 0.71 | 1.33 ± 0.79 | 1.22 ± 0.45 | Time | 0.09 | |
| NA‐LC | 0.87 ± 0.37 | 1.13 ± 0.46 | 1.21 ± 0.54 | 1.29 ± 0.62 | 1.18 ± 0.55 | 1.14 ± 0.49 | 1.18 ± 0.60 | |||
| Mean | 0.89 ± 0.33 | 1.37* ± 1.52 | 1.17* ± 0.44 | 1.20* ± 0.51 | 1.18* ± 0.51 | 1.17* ± 0.52 | 1.18* ± 0.45 | |||
| Fat intake (g/kg/d) | MC | 0.80 ± 0.27 | 0.51 ± 0.18 | 0.57 ± 0.23 | 0.56 ± 0.22 | 0.56 ± 0.18 | 0.56 ± 0.21 | 0.54 ± 0.16 | Diet | 0.37 |
| LC | 0.88 ± 0.43 | 0.62 ± 0.30 | 0.56 ± 0.22 | 0.58 ± 0.23 | 0.56 ± 0.21 | 0.60 ± 0.24 | 0.55 ± 0.24 | Genotype | 0.43 | |
| A | 0.85 ± 0.34 | 0.61 ± 0.28 | 0.58 ± 0.21 | 0.57 ± 0.20 | 0.57 ± 0.18 | 0.56 ± 0.19 | 0.57 ± 0.16 | G × D | 0.68 | |
| NA | 0.83 ± 0.40 | 0.52 ± 0.21 | 0.54 ± 0.23 | 0.57 ± 0.25 | 0.55 ± 0.21 | 0.59 ± 0.27 | 0.51 ± 0.25 | T × D | 0.38 | |
| A‐MC | 0.83 ± 0.31 | 0.53 ± 0.16 | 0.59 ± 0.24 | 0.57 ± 0.22 | 0.56 ± 0.16 | 0.53 ± 0.15 | 0.52 ± 0.15 | T × G | 0.51 | |
| NA‐MC | 0.72 ± 0.16 | 0.48 ± 0.21 | 0.52 ± 0.20 | 0.54 ± 0.25 | 0.58 ± 0.23 | 0.61 ± 0.30 | 0.56 ± 0.19 | T × G × D | 0.33 | |
| A‐LC | 0.88 ± 0.38 | 0.72 ± 0.38 | 0.57 ± 0.17 | 0.57 ± 0.17 | 0.58 ± 0.21 | 0.61 ± 0.23 | 0.63 ± 0.15 | Time | <0.001 | |
| NA‐LC | 0.88 ± 0.47 | 0.55 ± 0.22 | 0.55 ± 0.25 | 0.59 ± 0.26 | 0.54 ± 0.21 | 0.59 ± 0.26 | 0.48 ± 0.28 | |||
| Mean | 0.84 ± 0.36 | 0.57* ± 0.26 | 0.56* ± 0.22 | 0.57* ± 0.22 | 0.56* ± 0.20 | 0.58* ± 0.23 | 0.54* ± 0.21 | |||
Different from baseline (p < 0.05).
Different from 12 weeks (p < 0.05).
Data presented as mean ± standard deviations. The p‐levels are from univariate analysis. A, diet aligned to genotype (n = 34); D, diet effect; G, genotype match to diet effect; G × D, genotype match to diet × diet effect; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19); T, time effect; T × D, time × diet effect; T × G, time × genotype match to diet effect; T × G × D, time × diet × genotype match to diet effect.
Regarding REE, multivariate analysis revealed a significant overall Wilks' Lambda time effect (p < 0.001), but no significant interactions for time × diet (p = 0.993), time × genotype (p = 0.466) or time × diet × genotype (p = 0.54). Table 4 presents univariate analysis related to REE data. Significant group effects were observed for REE (p = 0.03) and respiratory exchange ratio (RER) (p = 0.02). Interestingly, participants aligned to genotype had significantly lower REE and RER values at several time points. Additionally, significant time effects were observed for REE and RER (p = 0.003 and p = 0.001, respectively) (Table 4). REE levels increased above baseline, while RER values declined in all groups towards the end of the study, which indicates greater fat oxidation.
Table 4.
Resting energy expenditure
| Variable | Group | Baseline | 4 weeks | 8 weeks | 12 weeks | 16 weeks | 20 weeks | 24 weeks | p‐level | |
|---|---|---|---|---|---|---|---|---|---|---|
| Resting energy expenditure (kcal d−1) | MC | 1,454 ± 293 | 1,410 ± 309 | 1,422 ± 320 | 1,476 ± 238 | 1,530 ± 258 | 1,516 ± 282 | 1,488 ± 229 | Diet | 0.32 |
| LC | 1,415 ± 317 | 1,415 ± 315 | 1,394 ± 295 | 1,466 ± 262 | 1,482 ± 230 | 1,469 ± 255 | 1,479 ± 257 | Genotype | 0.03 | |
| A | 1,405 ± 299 | 1,340 ± 287**** | 1,320 ± 302**** | 1,410 ± 248**** | 1,453 ± 232**** | 1,449 ± 261 | 1,452 ± 254 | G × D | 0.49 | |
| NA | 1,468 ± 311 | 1,497 ± 315 | 1,509 ± 280 | 1,542 ± 235 | 1,566 ± 246 | 1,541 ± 270 | 1,520 ± 226 | T × D | 0.91 | |
| A‐MC | 1,397 ± 274 | 1,315 ± 292 | 1,363 ± 324 | 1,419 ± 225 | 1,475 ± 239 | 1,451 ± 307 | 1,465 ± 259 | T × G | 0.24 | |
| NA‐MC | 1,568 ± 310 | 1,599 ± 260 | 1,539 ± 292 | 1,591 ± 232 | 1,641 ± 272 | 1,645 ± 174 | 1,533 ± 156 | T × G × D | 0.16 | |
| A‐LC | 1,416 ± 342 | 1,377 ± 287 | 1,259 ± 267 | 1,398 ± 286 | 1,422 ± 227 | 1,445 ± 188 | 1,433 ± 256 | Time | 0.003 | |
| NA‐LC | 1,415 ± 306 | 1,443 ± 335 | 1,493 ± 281 | 1,515 ± 238 | 1,526 ± 228 | 1,486 ± 299 | 1,513 ± 259 | |||
| Mean | 1,434 ± 304 | 1,412 ± 308 | 1,407 ± 305 | 1471*** ± 249 | 1,505**, *** ± 243 | 1,491** , *** ± 267 | 1,483*** ± 242 | |||
| Resting energy expenditure (kcal/kg/min) | MC | 1.01 ± 0.21 | 0.98 ± 0.2 | 0.99 ± 0.23 | 1.03 ± 0.16**** | 1.06 ± 0.2 | 1.05 ± 0.2 | 1.03 ± 0.2 | Diet | 0.87 |
| LC | 0.98 ± 0.22 | 1.00 ± 0.3 | 1.20 ± 1.42 | 1.01 ± 0.18 | 1.05 ± 0.2 | 1.02 ± 0.2 | 1.04 ± 0.2 | Genotype | 0.03 | |
| A | 0.97 ± 0.21 | 0.93 ± 0.2**** | 0.90 ± 0.26 | 0.98 ± 0.17**** | 1.01 ± 0.16**** | 1.00 ± 0.2 | 1.00 ± 0.2 | G × D | 0.87 | |
| NA | 1.02 ± 0.22 | 1.06 ± 0.3 | 1.33 ± 1.49 | 1.07 ± 0.16 | 1.1 ± 0.2 | 1.07 ± 0.2 | 1.07 ± 0.2 | T × D | 0.53 | |
| A‐MC | 0.97 ± 0.19 | 0.91 ± 0.2 | 0.95 ± 0.23 | 0.99 ± 0.15 | 1.02 ± 0.2 | 1.00 ± 0.2 | 1.01 ± 0.2 | T × G | 0.30 | |
| NA‐MC | 1.09 ± 0.22 | 1.11 ± 0.2 | 1.07 ± 0.20 | 1.10 ± 0.16 | 1.14 ± 0.5 | 1.14 ± 0.1 | 1.07 ± 0.1 | T × G × D | 0.30 | |
| A‐LC | 0.98 ± 0.24 | 0.96 ± 0.20 | 0.83 ± 0.29 | 0.96 ± 0.19 | 0.99 ± 0.2 | 1.00 ± 0.1 | 0.99 ± 0.2 | Time | 0.63 | |
| NA‐LC | 0.99 ± 0.21 | 1.03 ± 0.3 | 1.46 ± 1.84 | 1.05 ± 0.16 | 1.08 ± 0.2 | 1.04 ± 0.2 | 1.08 ± 0.2 | |||
| Mean | 1.00 ± 0.21 | 0.99 ± 0.2 | 1.10 ± 1.04 | 1.07 ± 0.16 | 1.05 ± 0.2 | 1.03 ± 0.2 | 1.03 ± 0.2 | |||
| Respiratory exchange ratio | MC | 0.86 ± 0.10 | 0.81 ± 0.1 | 0.82 ± 0.08 | 0.83 ± 0.07 | 0.80 ± 0.1 | 0.84 ± 0.1 | 0.83 ± 0.1 | Diet | 0.03 |
| LC | 0.84 ± 0.05 | 0.80 ± 0.1 | 0.82 ± 0.05 | 0.82 ± 0.07 | 0.81 ± 0.1 | 0.81 ± 0.1 | 0.82 ± 0.1 | Genotype | 0.02 | |
| A | 0.84 ± 0.07 | 0.80 ± 0.1 | 0.82 ± 0.08 | 0.81 ± 0.07 | 0.80 ± 0.05**** | 0.82 ± 0.1 | 0.81 ± 0.06**** | G × D | 0.12 | |
| NA | 0.86 ± 0.07 | 0.81 ± 0.1 | 0.82 ± 0.05 | 0.83 ± 0.07 | 0.82 ± 0.1 | 0.83 ± 0.1 | 0.84 ± 0.1 | T × D | 0.87 | |
| A‐MC | 0.85 ± 0.08 | 0.80 ± 0.1 | 0.82 ± 0.09 | 0.81 ± 0.07 | 0.80 ± 0 | 0.82 ± 0.1 | 0.81 ± 0.1 | T × G | 0.77 | |
| NA‐MC | 0.89 ± 0.09 | 0.82 ± 0.1 | 0.85 ± 0.04 | 0.85 ± 0.06 | 0.85 ± 0.1 | 0.86 ± 0 | 0.87 ± 0.1 | T × G × D | 0.85 | |
| A‐LC | 0.83 ± 0.05 | 0.80 ± 0.1 | 0.81 ± 0.06 | 0.81 ± 0.07 | 0.80 ± 0.05 | 0.81 ± 0.1 | 0.81 ± 0.1 | Time | 0.001 | |
| NA‐LC | 0.84 ± 0.05 | 0.81 ± 0.1 | 0.81 ± 0.05 | 0.82 ± 0.07 | 0.81 ± 0.1 | 0.81 ± 0.1 | 0.82 ± 0.1 | |||
| Mean | 0.85 ± 0.07 | 0.81* ± 0.1 | 0.82* ± 0.06 | 0.82* ± 0.07 | 0.81* ± 0.1 | 0.82* , ** ± 0.1 | 0.82* , ** ± 0.1 | |||
Different from baseline (p < 0.05).
Different from 4 weeks (p < 0.05).
Different from 8 weeks (p < 0.05).
Difference between genotype groups (p < 0.05).
Data presented as mean ± standard deviations. The p‐levels are from univariate analysis. A, diet aligned to genotype (n = 34); D, diet effect; G, genotype match to diet effect; G × D, genotype match to diet × diet effect; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19); T, time effect; T × D, time × diet effect; T × G, time × genotype match to diet effect; T × G × D, time × diet × genotype match to diet effect.
Body composition
Multivariate analysis revealed an overall Wilks' Lambda time effect (p < 0.001) with no significant time × diet (p = 0.278), time × genotype (p = 0.985) or time × diet × genotype (p = 0.914) interactions, group interactions or group effects for body composition variables. Table 5 presents univariate analysis of body composition data. Significant time effects were observed for all variables (p < 0.05). When adjusted for baseline values, multivariate GLM did not reveal significant time effects, group effects or interactions, or time interactions. Univariate analysis also did not reveal significant group effects or interactions, or time effects or interactions; however, a time × diet interaction for weight (p = 0.05, ηp 2 = 0.061) and fat mass (p = 0.064, ηp 2 = 0.048) that was close to statistical significance was observed. A significant diet interaction was not observed for fat‐free mass (p = 0.301) or body fat percentage (p = 0.40). Post hoc analysis revealed a significant difference between diet groups in body weight (p = 0.045) but not fat mass (p = 0.077, ηp 2 = 0.056) at week 24.
Table 5.
Body composition results
| Variable | Group | Baseline | 4 weeks | 8 weeks | 12 weeks | 16 weeks | 20 weeks | 24 weeks | p‐level | |
|---|---|---|---|---|---|---|---|---|---|---|
| Body weight (kg) | MC | 93.27 ± 19.93 | 91.63 ± 19.25 | 90.61 ± 19.15 | 89.87 ± 19.01 | 89.50 ± 18.99 | 89.39 ± 18.80 | 89.77 ± 19.17 | Diet | 0.61 |
| LC | 94.26 ± 24.50 | 91.98 ± 23.77 | 90.98 ± 23.41 | 90.04 ± 23.22 | 89.16 ± 22.86 | 88.89 ± 22.68 | 88.60 ± 22.97 | Genotype | 0.09 | |
| A | 89.80 ± 19.81 | 87.92 ± 19.20 | 87.11 ± 19.13 | 86.21 ± 18.97 | 85.48 ± 18.58 | 85.29 ± 18.54 | 85.24 ± 18.53 | G × D | 0.30 | |
| NA | 98.45 ± 24.36 | 96.38 ± 23.56 | 95.13 ± 23.22 | 94.34 ± 23.01 | 93.82 ± 22.92 | 93.63 ± 22.58 | 93.75 ± 23.22 | T × D | 0.14 | |
| A‐MC | 88.24 ± 16.99 | 86.59 ± 16.22 | 85.78 ± 16.21 | 84.80 ± 16.18 | 84.34 ± 15.89 | 84.23 ± 16.05 | 84.48 ± 16.50 | T × G | 0.65 | |
| NA‐MC | 103.34 ± 22.39 | 101.70 ± 21.66 | 100.28 ± 21.71 | 100.00 ± 20.98 | 99.81 ± 21.27 | 99.73 ± 20.43 | 100.33 ± 20.57 | T × G × D | 0.80 | |
| A‐LC | 92.05 ± 23.78 | 89.82 ± 23.34 | 89.01 ± 23.21 | 88.23 ± 22.88 | 87.12 ± 22.42 | 86.80 ± 22.17 | 86.33 ± 21.71 | Time | <0.001 | |
| NA‐LC | 95.89 ± 25.53 | 93.57 ± 24.59 | 92.43 ± 24.09 | 91.37 ± 24.01 | 90.66 ± 23.67 | 90.42 ± 23.52 | 90.28 ± 24.30 | |||
| Mean | 93.79 ± 22.27 | 91.81* ± 21.56 | 90.80* ± 21.32 | 89.96* ± 21.15 | 89.32* ± 20.94 | 89.13* ± 20.76 | 89.16* ± 21.08 | |||
| Fat mass (kg) | MC | 39.19 ± 11.70 | 37.80 ± 11.34 | 36.83 ± 10.96 | 36.31 ± 11.67 | 36.08 ± 11.15 | 35.84 ± 11.32 | 36.04 ± 11.33 | Diet | 0.71 |
| LC | 39.71 ± 15.39 | 38.40 ± 15.34 | 37.30 ± 15.31 | 36.97 ± 14.88 | 35.90 ± 14.76 | 35.68 ± 14.88 | 35.40 ± 14.62 | Genotype | 0.16 | |
| A | 37.45 ± 12.05 | 36.05 ± 11.91 | 35.03 ± 11.74 | 34.54 ± 12.01 | 34.01 ± 11.46 | 33.60 ± 11.54 | 33.70 ± 11.51 | G × D | 0.41 | |
| NA | 41.82 ± 15.19 | 40.54 ± 14.97 | 39.47 ± 14.79 | 39.13 ± 14.59 | 38.31 ± 14.58 | 38.28 ± 14.72 | 38.05 ± 14.51 | T × D | 0.25 | |
| A‐MC | 36.72 ± 10.67 | 35.18 ± 10.03 | 34.20 ± 9.77 | 33.60 ± 10.45 | 33.60 ± 10.19 | 33.04 ± 9.91 | 33.43 ± 10.29 | T × G | 0.83 | |
| NA‐MC | 44.11 ± 12.66 | 43.04 ± 12.51 | 42.10 ± 11.81 | 41.73 ± 12.64 | 41.05 ± 11.85 | 41.45 ± 12.37 | 41.27 ± 12.00 | T × G × D | 0.76 | |
| A‐LC | 38.49 ± 14.16 | 37.28 ± 14.49 | 36.22 ± 14.42 | 35.89 ± 14.26 | 34.60 ± 13.46 | 34.42 ± 13.89 | 34.10 ± 13.46 | Time | <0.001 | |
| NA‐LC | 40.61 ± 16.56 | 39.22 ± 16.28 | 38.09 ± 16.27 | 37.76 ± 15.67 | 36.87 ± 15.94 | 36.61 ± 15.87 | 36.36 ± 15.70 | |||
| Mean | 39.46 ± 13.65 | 38.11* ± 13.48 | 37.08* ± 13.31 | 36.65* ± 13.35 | 35.99* ± 13.06 | 35.76* ± 13.20 | 35.71* ± 13.05 | |||
| Fat‐free mass (kg) | MC | 47.59 ± 8.28 | 47.30 ± 8.15 | 47.01 ± 8.23 | 46.89 ± 7.60 | 46.80 ± 7.62 | 47.01 ± 7.57 | 47.16 ± 7.60 | Diet | 0.47 |
| LC | 47.64 ± 9.08 | 46.92 ± 8.71 | 47.04 ± 8.25 | 46.33 ± 8.17 | 46.73 ± 8.24 | 46.65 ± 7.88 | 46.68 ± 7.96 | Genotype | 0.05 | |
| A | 45.84 ± 7.90 | 45.43 ± 7.63 | 45.52 ± 7.71 | 45.09 ± 7.41 | 45.07 ± 7.25 | 45.26 ± 7.28 | 45.09 ± 6.89 | G × D | 0.20 | |
| NA | 49.70 ± 9.13 | 49.06 ± 8.93 | 48.79 ± 8.47 | 48.37 ± 8.09 | 48.74 ± 8.27 | 48.65 ± 7.84 | 49.05 ± 8.23 | T × D | 0.58 | |
| A‐MC | 45.32 ± 6.87 | 45.09 ± 6.79 | 45.02 ± 7.12 | 44.71 ± 6.73 | 44.37 ± 5.94 | 44.73 ± 6.59 | 44.66 ± 6.37 | T × G | 0.49 | |
| NA‐MC | 52.12 ± 9.34 | 51.71 ± 9.19 | 51.01 ± 9.19 | 51.25 ± 7.67 | 51.65 ± 8.57 | 51.57 ± 7.64 | 52.16 ± 7.66 | T × G × D | 0.57 | |
| A‐LC | 46.58 ± 9.39 | 45.91 ± 8.95 | 46.24 ± 8.71 | 45.63 ± 8.53 | 46.07 ± 8.94 | 46.02 ± 8.36 | 45.70 ± 7.77 | Time | 0.006 | |
| NA‐LC | 48.43 ± 9.01 | 47.66 ± 8.71 | 47.62 ± 8.08 | 46.85 ± 8.08 | 47.21 ± 7.90 | 47.11 ± 7.70 | 47.41 ± 8.24 | |||
| Mean | 47.62 ± 8.64 | 47.10* ± 8.39 | 47.03* ± 8.17 | 46.60* ± 7.84 | 46.76* ± 7.89 | 46.82* ± 7.67 | 46.91* ± 7.73 | |||
| Body fat (%) | MC | 44.33 ± 4.66 | 43.93 ± 4.85 | 43.24 ± 5.10 | 42.78 ± 5.65 | 42.76 ± 5.17 | 42.42 ± 5.36 | 42.42 ± 4.91 | Diet | 0.69 |
| LC | 44.33 ± 5.97 | 43.79 ± 5.94 | 42.93 ± 6.21 | 43.10 ± 6.02 | 42.15 ± 6.19 | 41.92 ± 6.47 | 41.81 ± 6.19 | Genotype | 0.60 | |
| A | 44.07 ± 5.06 | 43.40 ± 5.20 | 42.63 ± 5.49 | 42.41 ± 5.96 | 42.16 ± 5.44 | 41.69 ± 5.58 | 41.80 ± 5.41 | G × D | 0.98 | |
| NA | 44.63 ± 5.74 | 44.40 ± 5.67 | 43.60 ± 5.92 | 43.57 ± 5.64 | 42.77 ± 6.06 | 42.71 ± 6.36 | 42.44 ± 5.86 | T × D | 0.51 | |
| A‐MC | 43.89 ± 5.01 | 43.15 ± 5.31 | 42.53 ± 5.68 | 42.10 ± 6.28 | 42.33 ± 5.84 | 41.71 ± 5.69 | 41.95 ± 5.38 | T × G | 0.74 | |
| NA‐MC | 45.22 ± 3.96 | 45.51 ± 3.50 | 44.66 ± 3.48 | 44.13 ± 4.07 | 43.61 ± 3.60 | 43.82 ± 4.56 | 43.35 ± 3.90 | T × G × D | 0.74 | |
| A‐LC | 44.34 ± 5.31 | 43.76 ± 5.23 | 42.77 ± 5.40 | 42.86 ± 5.68 | 41.91 ± 5.00 | 41.65 ± 5.63 | 41.60 ± 5.65 | Time | <0.001 | |
| NA‐LC | 44.32 ± 6.56 | 43.82 ± 6.55 | 43.04 ± 6.89 | 43.28 ± 6.40 | 42.33 ± 7.07 | 42.12 ± 7.18 | 41.96 ± 6.71 | |||
| Mean | 44.33 ± 5.35 | 43.86 ± 5.40 | 43.08* ± 5.66 | 42.95* ± 5.80 | 42.44* ± 5.69 | 42.16* ± 5.93 | 42.10* ± 5.58 | |||
| VAT (cm2) | MC | 190 ± 86 | 171 ± 88 | 160 ± 69 | 158 ± 67 | 157 ± 70 | 159 ± 65 | 153 ± 60 | Diet | 0.78 |
| LC | 178 ± 92 | 181 ± 102 | 158 ± 83 | 157 ± 82 | 153 ± 81 | 154 ± 84 | 150 ± 71 | Genotype | 0.57 | |
| A | 180 ± 75 | 165 ± 77 | 152 ± 54 | 158 ± 65 | 154 ± 67 | 151 ± 61 | 148 ± 55 | G × D | 0.81 | |
| NA | 189 ± 103 | 189 ± 113 | 168 ± 95 | 156 ± 85 | 156 ± 86 | 163 ± 90 | 155 ± 77 | T × D | 0.66 | |
| A‐MC | 182 ± 74 | 161 ± 78 | 151 ± 54 | 160 ± 69 | 157 ± 70 | 153 ± 61 | 146 ± 52 | T × G | 0.28 | |
| NA‐MC | 206 ± 108 | 191 ± 107 | 179 ± 92 | 154 ± 64 | 159 ± 73 | 169 ± 74 | 165 ± 77 | T × G × D | 0.69 | |
| A‐LC | 176 ± 79 | 172 ± 78 | 152 ± 57 | 155 ± 61 | 150 ± 63 | 148 ± 63 | 151 ± 61 | Time | <0.001 | |
| NA‐LC | 180 ± 102 | 189 ± 119 | 163 ± 99 | 158 ± 96 | 154 ± 94 | 159 ± 99 | 150 ± 79 | |||
| Mean | 184 ± 89 | 176* ± 95 | 159* ± 76 | 157* ± 74 | 155* ± 75 | 156* ± 75 | 151* ± 66 | |||
Different from baseline (p < 0.05).
Data presented as mean ± standard deviations. The p‐levels are from univariate analysis. A, diet aligned to genotype (n = 34); D, diet effect; G, genotype match to diet effect; G × D, genotype match to diet × diet effect; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (NA, n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19); T, time effect; T × D, time × diet effect; T × G, time × genotype match to diet effect; T × G × D, time × diet × genotype match to diet effect; VAT, visceral adipose tissue.
When assessed as per cent change from baseline, significant time effects were observed for body composition variables (p < 0.001) (Figure 3). By 24 weeks, on average participants lost 4.75% body weight (−4.75 [−3.5, −6.0] %), 9.38% fat mass (−9.38 [−6.8, −11.9] %), 1.10% fat‐free mass (−1.10 [−0.002, −11.7] %) and 4.92% body fat (−4.93 [−3.3, −6.6] %). A significant diet effect was not observed (p = 0.051, ηp 2 = 0.025), but overall participants following LC exhibited greater improvements in body composition variables than those following the MC diet (weight: MC −3.32 [−1.4, −5.2], LC −5.82 [−4.1, −7.6]; fat mass: MC −7.25 [−3.2, −11.2], LC −10.93 [−7.3, −14.5]; fat‐free mass: MC −0.32 [1.4, −2.0], LC −1.48 [0.7, −3.0]; body fat: MC −4.19 [−1.6, −6.8], LC −5.60 [−3.3, −7.9] %). A significant genotype effect or time × genotype interaction was not observed (weight: A −5.00 [−3.3, −6.7], NA −4.14 [−2.2, −6.1]; fat mass: A −10.15 [−7.0, −13.6], NA −8.02 [−4.0, −12.0]; fat‐free mass: A −1.23 [0.3, −2.8], NA −0.56 [1.12, −2.3]; body fat: A −5.28 [−3.0, −7.6], NA −4.51 [−1.9, −7.1] %).
Figure 3.
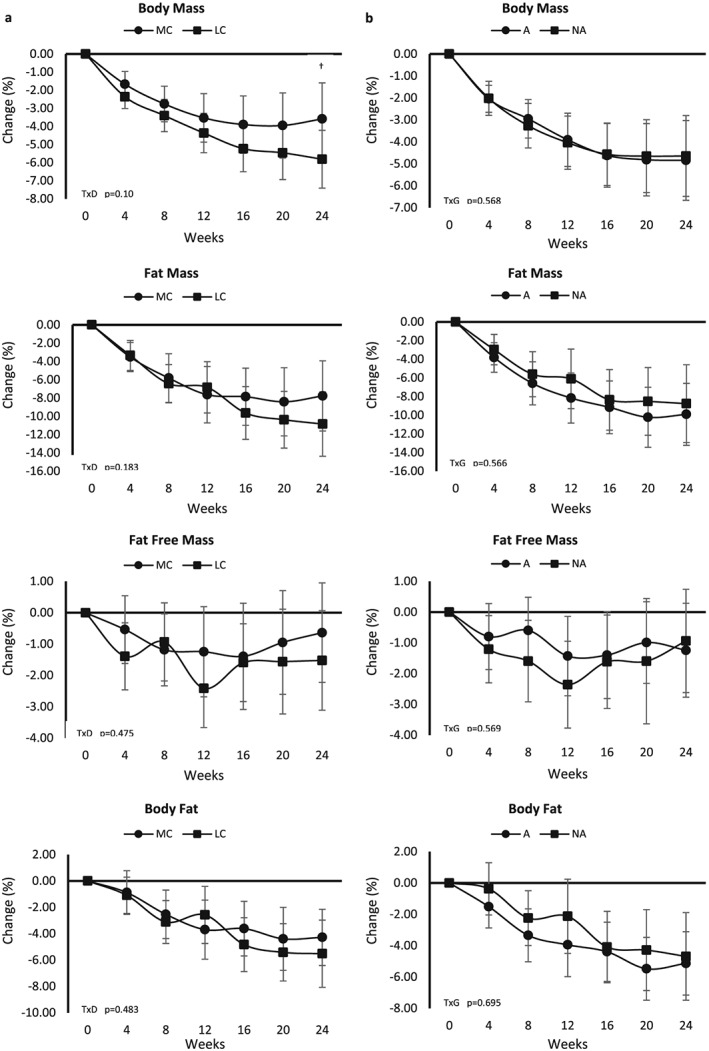
Per cent changes from baseline with 95% confidence intervals in body composition variables between diet groups (a) and genotype groups (b). A, diet aligned to genotype; D, diet; G, genotype; LC, lower carbohydrate; MC, moderate carbohydrate; NA, diet not aligned to genotype; T, time.
Exercise capacity
Table 6 presents univariate analysis of cardiopulmonary maximal exercise variables. Significant time effects were observed for absolute and relative peak oxygen uptake (p < 0.001), time to exhaustion (p < 0.001) and peak systolic blood pressure (p = 0.01) (Table 6). When assessed as per cent change from baseline, significant time effects were observed for absolute (9.8% ± 11%, p < 0.001) and relative (15.6% ± 13.3%, p < 0.001) peak oxygen uptake, time to reach maximal effort (14.9% ± 13.5%, p < 0.001), peak systolic blood pressure (5.4% ± 12.8%, p = 0.003) and peak rating of perceived exertion (4.4% ± 10.0%, p = 0.026) in all groups but were not observed for peak heart rate (3.1% ± 8.4%, p = 0.113) or peak diastolic blood pressure (−0.8% ± 13.2%, p = 0.319). Significant time × genotype × diet interactions were observed in per cent changes in peak oxygen uptake variables. Among individuals following the LC diet, post hoc analysis revealed greater improvements in absolute and relative peak oxygen uptake among individuals aligned to genotype diet (A‐LC) compared with individuals not aligned to genotype diet (NA‐LC) (absolute: A‐LC 12.9 ± 12.2, NA‐LC 4.4 ± 10.6 L min−1, p = 0.025; relative: A‐LC 14.7 ± 12.2, NA‐LC 9.6 ± 10.5 mL/kg/min, p = 0.047) after 24 weeks of training. No other significant interactions or group effects were observed.
Table 6.
Cardiopulmonary maximal exercise responses
| Variable | Group | Baseline | 12 weeks | 24 weeks | p‐level | |
|---|---|---|---|---|---|---|
| Absolute peak oxygen uptake (L min−1) | MC | 1.94 ± 0.31 | 2.14 ± 0.35 | 2.16 ± 0.34 | Diet | 0.98 |
| LC | 1.99 ± 0.34 | 2.12 ± 0.36 | 2.14 ± 0.34 | Genotype | 0.82 | |
| A | 1.95 ± 0.35 | 2.12 ± 0.38 | 2.15 ± 0.36 | G × D | 0.68 | |
| NA | 1.99 ± 0.29 | 2.14 ± 0.32 | 2.15 ± 0.32 | T × D | 0.08 | |
| A‐MC | 1.97 ± 0.36 | 2.14 ± 0.39 | 2.15 ± 0.37 | T × G | 0.90 | |
| NA‐MC | 1.89 ± 0.17 | 2.13 ± 0.26 | 2.19 ± 0.30 | T × G × D | 0.01 | |
| A‐LC | 1.92 ± 0.35 | 2.09 ± 0.38 | 2.14 ± 0.36 | Time | <0.001 | |
| NA‐LC | 2.05 ± 0.32 | 2.14 ± 0.36 | 2.13 ± 0.33 | |||
| Mean | 1.97 ± 0.32 | 2.13* ± 0.35 | 2.15* ± 0.34 | |||
| Relative peak oxygen uptake (mL/kg/min) | MC | 21.41 ± 4.20 | 24.48 ± 4.50 | 24.94 ± 4.65 | Diet | 0.41 |
| LC | 22.17 ± 5.40 | 24.65 ± 5.82 | 25.40 ± 6.04 | Genotype | 0.19 | |
| A | 22.24 ± 4.14 | 25.23 ± 4.68 | 25.90 ± 4.58 | G × D | 0.17 | |
| NA | 21.31 ± 5.59 | 23.80 ± 5.72 | 24.34 ± 6.16 | T × D | 0.65 | |
| A‐MC | 22.67 ± 3.88 | 25.62 ± 4.29 | 26.00 ± 4.22 | T × G | 0.43 | |
| NA‐MC | 18.89 ± 3.79 | 22.20 ± 4.20 | 22.80 ± 4.93 | T × G × D | 0.17 | |
| A‐LC | 21.62 ± 4.56 | 24.68 ± 5.29 | 25.76 ± 5.21 | Time | <0.001 | |
| NA‐LC | 22.58 ± 6.04 | 24.64 ± 6.32 | 25.14 ± 6.71 | |||
| Mean | 21.81 ± 4.85 | 24.57* ± 5.19 | 25.18* , ** ± 5.38 | |||
| Time to exhaustion (s) | MC | 463.2 ± 93.9 | 511.4 ± 82.5 | 528.4 ± 81.4 | Diet | 0.24 |
| LC | 488.7 ± 104.0 | 538.2 ± 115.5 | 550.3 ± 107.0 | Genotype | 0.58 | |
| A | 479.8 ± 92.0 | 525.9 ± 83.1 | 543.4 ± 79.4 | G × D | 0.39 | |
| NA | 472.7 ± 108.9 | 525.0 ± 120.7 | 535.7 ± 112.9 | T × D | 0.83 | |
| A‐MC | 476.3 ± 103.4 | 523.1 ± 85.5 | 539.8 ± 81.4 | T × G | 0.74 | |
| NA‐MC | 437.1 ± 68.7 | 488.2 ± 74.7 | 505.5 ± 80.5 | T × G × D | 0.77 | |
| A‐LC | 484.9 ± 76.4 | 529.9 ± 82.4 | 548.6 ± 79.2 | Time | <0.001 | |
| NA‐LC | 491.5 ± 122.5 | 544.3 ± 136.8 | 551.6 ± 125.8 | |||
| Mean | 476.5 ± 99.4 | 525.4* ± 101.2 | 539.9* , ** ± 95.6 | |||
| Peak heart rate (bpm) | MC | 167.6 ± 21.4 | 165.5 ± 17.4 | 171.7 ± 17.9 | Diet | 0.56 |
| LC | 169.0 ± 15.2 | 168.9 ± 13.5 | 173.7 ± 13.4 | Genotype | 0.85 | |
| A | 169.4 ± 18.4 | 165.2 ± 14.5 | 174.2 ± 15.1 | G × D | 0.31 | |
| NA | 167.0 ± 18.4 | 169.7 ± 16.3 | 171.0 ± 16.3 | T × D | 0.92 | |
| A‐MC | 166.8 ± 22.4 | 162.0 ± 16.7 | 173.0 ± 17.8 | T × G | 0.13 | |
| NA‐MC | 169.2 ± 20.4 | 172.5 ± 17.4 | 169.0 ± 18.9 | T × G × D | 0.33 | |
| A‐LC | 173.2 ± 10.3 | 169.9 ± 9.4 | 176.0 ± 10.7 | Time | 0.11 | |
| NA‐LC | 165.8 ± 17.6 | 168.2 ± 16.0 | 172.0 ± 15.1 | |||
| Mean | 168.3 ± 18.3 | 167.3 ± 15.4 | 172.7 ± 15.6 | |||
| Peak systolic blood pressure (mmHg) | MC | 156.8 ± 17.3 | 165.5 ± 17.4 | 160.6 ± 16.6 | Diet | 0.45 |
| LC | 151.6 ± 14.4 | 168.9 ± 13.5 | 161.3 ± 11.9 | Genotype | 0.86 | |
| A | 153.6 ± 16.7 | 165.2 ± 14.5 | 160.3 ± 16.1 | G × D | 0.61 | |
| NA | 154.7 ± 15.4 | 169.7 ± 16.3 | 161.7 ± 11.9 | T × D | 0.48 | |
| A‐MC | 157.0 ± 16.4 | 159.3 ± 15.5 | 159.7 ± 18.1 | T × G | 0.65 | |
| NA‐MC | 156.4 ± 20.2 | 154.2 ± 18.8 | 162.4 ± 13.8 | T × G × D | 0.60 | |
| A‐LC | 148.7 ± 16.5 | 155.1 ± 10.4 | 161.3 ± 13.3 | Time | 0.01 | |
| NA‐LC | 153.7 ± 12.6 | 156.0 ± 15.5 | 161.3 ± 11.2 | |||
| Mean | 154.1 ± 16.0 | 156.6 ± 14.9 | 161.0* ± 14.2 | |||
| Peak diastolic blood pressure (mmHg) | MC | 78.9 ± 9.8 | 77.5 ± 6.2 | 77.6 ± 6.0 | Diet | 0.14 |
| LC | 75.9 ± 7.9 | 78.2 ± 8.5 | 74.3 ± 6.8 | Genotype | 0.31 | |
| A | 76.2 ± 9.1 | 77.9 ± 7.9 | 75.6 ± 7.4 | G × D | 0.97 | |
| NA | 78.7 ± 8.6 | 77.8 ± 7.0 | 76.1 ± 5.6 | T × D | 0.13 | |
| A‐MC | 78.1 ± 9.9 | 77.3 ± 7.0 | 77.1 ± 6.6 | T × G | 0.35 | |
| NA‐MC | 80.4 ± 9.8 | 78.0 ± 4.6 | 78.5 ± 4.7 | T × G × D | 0.72 | |
| A‐LC | 73.4 ± 7.3 | 78.7 ± 9.3 | 43.5 ± 8.2 | Time | 0.24 | |
| NA‐LC | 77.8 ± 8.0 | 77.8 ± 8.1 | 74.8 ± 5.7 | |||
| Mean | 77.3 ± 8.9 | 77.8 ± 7.5 | 75.8 ± 6.6 | |||
| Peak rating of perceived exertion (20 = maximal exertion) | MC | 17.9 ± 1.2 | 18.2 ± 2.1 | 18.5 ± 1.0 | Diet | 0.24 |
| LC | 17.5 ± 1.7 | 17.8 ± 1.5 | 18.2 ± 1.3 | Genotype | 0.22 | |
| A | 17.4 ± 1.7 | 17.9 ± 2.0 | 18.4 ± 1.3 | G × D | 0.79 | |
| NA | 18.0 ± 1.1 | 17.2 ± 1.5 | 18.2 ± 1.1 | T × D | 0.74 | |
| A‐MC | 17.7 ± 1.3 | 18.0 ± 2.4 | 18.6 ± 0.7 | T × G | 0.23 | |
| NA‐MC | 18.3 ± 0.8 | 18.8 ± 1.4 | 18.3 ± 1.5 | T × G × D | 0.57 | |
| A‐LC | 17.1 ± 2.2 | 17.7 ± 1.4 | 18.2 ± 1.8 | Time | 0.05 | |
| NA‐LC | 17.8 ± 1.2 | 17.8 ± 1.5 | 18.2 ± 0.9 | |||
| Mean | 17.7 ± 1.5 | 18.0 ± 1.8 | 18.3* ± 1.2 | |||
Different from baseline (p < 0.05).
Different from 12 weeks (p < 0.05).
Data presented as mean ± standard deviations. The p‐levels are from univariate analysis. A, diet aligned to genotype (n = 34); D, diet effect; G, genotype match to diet effect; G × D, genotype match to diet × diet effect; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19); T, time effect; T × D, time × diet effect; T × G, time × genotype match to diet effect; T × G × D, time × diet × genotype match to diet effect.
Table 7 presents univariate analysis of muscular strength and endurance variables. Significant time effects were observed for bench press 1RM (p < 0.001) and leg press 1RM (p < 0.001). Participants improved bench press 1RM by 17.2% (17.2% ± 21.4%, p < 0.001) and leg press 1RM by 24% (24.0% ± 24.5%, p < 0.001) (Table 7). While a time effect was not observed for leg or bench press lifting volume, when assessed as per cent change from baseline, a significant change was observed in leg press lifting volume (35.0% ± 100%, p = 0.025) but not bench press lifting volume (22.9% ± 78.6%, p = 0.054).
Table 7.
Upper and lower extremity muscular strength and endurance
| Variable | Group | Baseline | 12 weeks | 24 weeks | p‐level | |
|---|---|---|---|---|---|---|
| Bench press 1RM (kg) | MC | 36.14 ± 8.13 | 38.94 ± 8.89 | 52.17 ± 51.70 | Diet | 0.07 |
| LC | 33.33 ± 7.00 | 35.48 ± 6.62 | 38.37 ± 15.91 | Genotype | 0.64 | |
| A | 35.16 ± 7.55 | 37.85 ± 8.12 | 49.79 ± 48.62 | G × D | 0.63 | |
| NA | 34.09 ± 7.80 | 36.29 ± 7.72 | 39.26 ± 17.78 | T × D | 0.35 | |
| A‐MC | 36.14 ± 8.47 | 38.98 ± 9.68 | 56.91 ± 62.77 | T × G | 0.56 | |
| NA‐MC | 36.14 ± 7.83 | 38.86 ± 7.53 | 42.67 ± 11.99 | T × G × D | 0.49 | |
| A‐LC | 33.77 ± 6.04 | 36.23 ± 5.05 | 39.61 ± 7.23 | Time | <0.001 | |
| NA‐LC | 33.01 ± 7.78 | 34.93 ± 7.66 | 37.47 ± 20.25 | |||
| Mean | 34.67 ± 7.63 | 37.13* ± 7.91 | 44.94* , ** ± 37.80 | |||
| Bench press lifting volume (kg) | MC | 239.85 ± 87.8 | 240.8 ± 97.0 | 232.2 ± 72.0 | Diet | 0.40 |
| LC | 213.02 ± 116.3 | 219.7 ± 94.3 | 231.0 ± 89.2 | Genotype | 0.60 | |
| A | 238.57 ± 94.4 | 235.4 ± 93.8 | 232.3 ± 87.2 | G × D | 0.48 | |
| NA | 210.82 ± 113.6 | 223.1 ± 98.5 | 230.7 ± 74.2 | T × D | 0.65 | |
| A‐MC | 238.52 ± 86.2 | 236.6 ± 100.7 | 234.5 ± 72.5 | T × G | 0.77 | |
| NA‐MC | 242.50 ± 95.6 | 249.1 ± 93.7 | 227.6 ± 74.7 | T × G × D | 0.56 | |
| A‐LC | 238.64 ± 108.6 | 233.7 ± 86.6 | 229.3 ± 107.7 | Time | 0.95 | |
| NA‐LC | 194.14 ± 121.1 | 209.4 ± 100.6 | 232.4 ± 75.9 | |||
| Mean | 225.79 ± 103.8 | 229.7 ± 95.4 | 231.6 ± 80.8 | |||
| Leg press 1RM (kg) | MC | 230.38 ± 84.1 | 269.8 ± 97.7 | 293.6 ± 111.9 | Diet | 0.03 |
| LC | 197.93 ± 59.7 | 221.6 ± 69.6 | 231.8 ± 75.0 | Genotype | 0.98 | |
| A | 214.57 ± 81.7 | 249.5 ± 92.8 | 271.5 ± 107.6 | G × D | 0.96 | |
| NA | 211.99 ± 64.2 | 238.6 ± 80.6 | 249.2 ± 87.2 | T × D | 0.06 | |
| A‐MC | 230.23 ± 96.5 | 270.6 ± 109.3 | 294.7 ± 127.4 | T × G | 0.48 | |
| NA‐MC | 230.68 ± 56.0 | 268.2 ± 74.3 | 291.6 ± 77.6 | T × G × D | 0.63 | |
| A‐LC | 192.21 ± 49.2 | 219.5 ± 52.7 | 238.4 ± 60.3 | Time | <0.001 | |
| NA‐LC | 202.15 ± 67.3 | 223.1 ± 81.3 | 226.9 ± 85.5 | |||
| Mean | 213.38 ± 73.6 | 244.5* ± 86.9 | 261.2* , ** ± 98.6 | |||
| Leg press lifting volume (kg) | MC | 2,828.03 ± 1,831 | 3,204 ± 2,868 | 3,445 ± 2,919 | Diet | 0.14 |
| LC | 2,237.67 ± 1,284 | 2,360 ± 1,292 | 2,388 ± 1,147 | Genotype | 0.62 | |
| A | 2,680.08 ± 1,784 | 2,997 ± 2,726 | 3,026 ± 2,714 | G × D | 0.30 | |
| NA | 2,329.70 ± 1,316 | 2,487 ± 1,386 | 2,733 ± 1,492 | T × D | 0.41 | |
| A‐MC | 3,057.50 ± 2,163 | 3,511 ± 3,388 | 3,652 ± 3,388 | T × G | 0.74 | |
| NA‐MC | 2,369.09 ± 760 | 2,590 ± 1,276 | 3,031 ± 1,712 | T × G × D | 0.94 | |
| A‐LC | 2,140.91 ± 842 | 2,263 ± 1,045 | 2,132 ± 677 | Time | 0.13 | |
| NA‐LC | 2,308.97 ± 1,551 | 2,432 ± 1,472 | 2,577 ± 1,385 | |||
| Mean | 2,518.80 ± 1,583 | 2,762 ± 2,211 | 2,891 ± 2,224 | |||
Different from baseline (p < 0.05).
Different from 12 weeks (p < 0.05).
Data presented as mean ± standard deviations. The p‐levels are from univariate analysis. 1RM, one repetition maximum; A, diet aligned to genotype (n = 34); D, diet effect; G, genotype match to diet effect; G × D, genotype match to diet × diet effect; T, time effect; T × D, time × diet effect; T × G, time × genotype match to diet effect; T × G × D, time × diet × genotype match to diet effect; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19).
Biomarkers of health
Table 8 presents univariate analysis of blood lipid variables. Significant time effects were observed for all blood lipid variables (p < 0.05), such that all participants experienced favourable changes in blood lipids by 24 weeks. Significant group effect for diet was observed for total cholesterol (p < 0.001), LDL cholesterol (p < 0.001) and triglycerides (p < 0.001) such that values were generally lower in the LC diet group. No other significant diet or genotype group effects were observed. Further, significant time × diet interactions were observed in total cholesterol (p < 0.001), HDL cholesterol (p = 0.02), LDL cholesterol (p = 0.001) and triglycerides (p < 0.001). No other significant interactions were observed. When assessed as per cent change from baseline, on average all participants experienced a 2.1% reduction in total cholesterol (−2.1% ± 16.4%, p < 0.001), 7.4% reduction in triglycerides (−7.4% ± 32%, p = 0.027), 9.1% reduction in total cholesterol to HDL cholesterol ratio (−9.1% ± 16.5%, p < 0.001) and a 10.8% improvement in HDL cholesterol (10.8% ± 26%, p < 0.001).
Table 8.
Lipid profile analysis
| Variable | Group | Baseline | 12 weeks | 24 weeks | p‐level | |
|---|---|---|---|---|---|---|
| Total cholesterol (mmol L−1) | MC | 6.30*** ± 1.38 | 7.02* , *** ± 1.38 | 5.66* , ** ± 0.99 | Diet | <0.001 |
| LC | 5.04 ± 0.99 | 5.30 ± 1.06 | 5.16 ± 1.05 | Genotype | 0.30 | |
| A | 5.87 ± 1.49 | 6.44 ± 1.55 | 5.55 ± 1.10 | G × D | 0.23 | |
| NA | 5.37 ± 1.13 | 5.75 ± 1.37 | 5.21 ± 0.95 | T × D | <0.001 | |
| A‐MC | 6.54 ± 1.34 | 7.21 ± 1.32 | 5.83 ± 0.99 | T × G | 0.95 | |
| NA‐MC | 5.85 ± 1.42 | 6.66 ± 1.50 | 5.31 ± 0.96 | T × G × D | 0.67 | |
| A‐LC | 4.93 ± 1.16 | 5.34 ± 1.14 | 5.16 ± 1.18 | Time | <0.001 | |
| NA‐LC | 5.12 ± 0.88 | 5.27 ± 1.04 | 5.16 ± 0.97 | |||
| Mean | 5.64 ± 1.35 | 6.12* ± 1.49 | 5.40* , ** ± 1.04 | |||
| High‐density lipoprotein (mmol L−1) | MC | 1.55 ± 0.53 | 1.80* ± 0.53 | 1.53* , ** ± 0.46 | Diet | 0.22 |
| LC | 1.36 ± 0.42 | 1.57 ± 0.42 | 1.54 ± 0.43 | Genotype | 0.63 | |
| A | 1.43 ± 0.50 | 1.69 ± 0.49 | 1.51 ± 0.40 | G × D | 0.58 | |
| NA | 1.46 ± 0.48 | 1.66 ± 0.50 | 1.57 ± 0.49 | T × D | 0.02 | |
| A‐MC | 1.53 ± 0.52 | 1.82 ± 0.54 | 1.54 ± 0.44 | T × G | 0.75 | |
| NA‐MC | 1.58 ± 0.59 | 1.75 ± 0.55 | 1.53 ± 0.53 | T × G × D | 0.71 | |
| A‐LC | 1.29 ± 0.45 | 1.50 ± 0.34 | 1.46 ± 0.33 | Time | <0.001 | |
| NA‐LC | 1.40 ± 0.41 | 1.62 ± 0.48 | 1.60 ± 0.49 | |||
| Mean | 1.45 ± 0.49 | 1.68* ± 0.49 | 1.54** ± 0.44 | |||
| Low‐density lipoprotein (mmol L−1) | MC | 3.94 ± 1.04 | 4.45 ± 1.11 | 3.58 ± 0.93 | Diet | <0.001 |
| LC | 2.97*** ± 0.77 | 3.21*** ± 0.77 | 3.09 ± 0.79 | Genotype | 0.68 | |
| A | 3.53 ± 1.17 | 3.99 ± 1.23 | 3.39 ± 0.92 | G × D | 0.03 | |
| NA | 3.31 ± 0.84 | 3.58 ± 0.96 | 3.25 ± 0.86 | T × D | 0.001 | |
| A‐MC | 4.14 ± 1.06 | 4.66 ± 1.11 | 3.71 ± 0.91 | T × G | 0.74 | |
| NA‐MC | 3.54 ± 0.95 | 4.03 ± 1.05 | 3.32 ± 0.98 | T × G × D | 0.45 | |
| A‐LC | 2.66 ± 0.69 | 3.04 ± 0.64 | 2.94 ± 0.76 | Time | <0.001 | |
| NA‐LC | 3.19 ± 0.77 | 3.34 ± 0.84 | 3.21 ± 0.82 | |||
| Mean | 3.43 ± 1.03 | 3.80 ± 1.13 | 3.32* , ** ± 0.89 | |||
| Triglycerides (mmol L−1) | MC | 1.90 ± 0.90 | 2.06* ± 0.99 | 1.51* , ** ± 0.56 | Diet | <0.001 |
| LC | 1.34*** ± 0.50 | 1.17* , *** ± 0.41 | 1.22* ± 0.50 | Genotype | 0.98 | |
| A | 1.64 ± 0.85 | 1.65 ± 0.85 | 1.46 ± 0.61 | G × D | 0.93 | |
| NA | 1.56 ± 0.67 | 1.53 ± 0.89 | 1.25 ± 0.45 | T × D | <0.001 | |
| A‐MC | 1.92 ± 0.94 | 1.99 ± 0.91 | 1.54 ± 0.57 | T × G | 0.18 | |
| NA‐MC | 1.85 ± 0.87 | 2.21 ± 1.16 | 1.44 ± 0.57 | T × G × D | 0.28 | |
| A‐LC | 1.25 ± 0.49 | 1.17 ± 0.45 | 1.34 ± 0.66 | Time | <0.001 | |
| NA‐LC | 1.41 ± 0.51 | 1.17 ± 0.39 | 1.14 ± 0.34 | |||
| Mean | 1.61 ± 0.77 | 1.60* ± 0.86 | 1.36** ± 0.55 | |||
| Cholesterol:HDL | MC | 4.41 ± 1.39 | 4.17 ± 1.31 | 3.96 ± 1.20 | Diet | 0.13 |
| LC | 3.95 ± 1.08 | 3.54 ± 0.95 | 3.50 ± 0.88 | Genotype | 0.28 | |
| A | 4.40 ± 1.39 | 4.00 ± 1.22 | 3.87 ± 1.11 | G × D | 0.72 | |
| NA | 3.89 ± 1.01 | 3.64 ± 1.10 | 3.53 ± 0.99 | T × D | 0.39 | |
| A‐MC | 4.64 ± 1.49 | 4.24 ± 1.34 | 4.06 ± 1.25 | T × G | 0.42 | |
| NA‐MC | 3.94 ± 1.10 | 4.02 ± 1.30 | 3.75 ± 1.13 | T × G × D | 0.40 | |
| A‐LC | 4.07 ± 1.22 | 3.67 ± 0.96 | 3.60 ± 0.86 | Time | <0.001 | |
| NA‐LC | 3.87 ± 1.00 | 3.45 ± 0.96 | 3.41 ± 0.91 | |||
| Mean | 4.17 ± 1.25 | 3.84* ± 1.17 | 3.72* ± 1.06 | |||
Different from baseline (p < 0.05).
Different from 12 weeks (p < 0.05).
Difference between diet groups (p < 0.05).
Data presented as mean ± standard deviations. The p‐levels are from univariate analysis. A, diet aligned to genotype (n = 34); D, diet effect; G, genotype match to diet effect; G × D, genotype match to diet × diet effect; HDL, high‐density lipoprotein; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19); T, time effect; T × D, time × diet effect; T × G, time × genotype match to diet effect; T × G × D, time × diet × genotype match to diet effect.
Table 9 presents univariate analysis for variables related to glucose homeostasis. A significant time effect was observed for insulin (p = 0.001) and HOMA‐IR (p = 0.001). A significant time effect was not observed for glucose (p = 0.18); however, a significant diet effect was observed for glucose (p < 0.001), such that individuals in MC generally had lower fasting levels of glucose. No other significant diet effects were observed, but a significant genotype effect was observed for insulin (p = 0.03) and HOMA‐IR (p = 0.04), where levels were generally higher in individuals not aligned to genotype diet. No significant interactions were observed. When assessed as per cent change from baseline, insulin increased by 31.5% (31.5% ± 52.8%, p < 0.001) and HOMA‐IR increased by 37.7% (37.7% ± 63.6%, p < 0.001).
Table 9.
Glucose homeostasis‐related variables
| Variable | Group | Baseline | 24 weeks | p‐level | |
|---|---|---|---|---|---|
| Glucose (mmol L−1) | MC | 4.66 ± 0.41 | 4.93 ± 0.45 | Diet | <0.001 |
| LC | 5.25 ± 0.50 | 5.24 ± 0.53 | Genotype | 0.57 | |
| A | 4.91 ± 0.57 | 5.10 ± 0.51 | G × D | 0.52 | |
| NA | 5.04 ± 0.51 | 5.08 ± 0.54 | T × D | 0.15 | |
| A‐MC | 4.61 ± 0.45 | 4.97 ± 0.47 | T × G | 0.48 | |
| NA‐MC | 4.76 ± 0.32 | 4.83 ± 0.41 | T × G × D | 0.24 | |
| A‐LC | 5.34 ± 0.45 | 5.29 ± 0.52 | Time | 0.18 | |
| NA‐LC | 5.18 ± 0.53 | 5.21 ± 0.56 | |||
| Mean | 4.97 ± 0.54 | 5.09 ± 0.52 | |||
| Insulin (microIU mL−1) | MC | 12.22 ± 7.56 | 14.35 ± 5.30 | Diet | 0.93 |
| LC | 13.67 ± 6.99 | 15.09 ± 5.50 | Genotype | 0.03 | |
| A | 11.18 ± 6.30 | 13.35 ± 4.81 | G × D | 0.45 | |
| NA | 15.08 ± 7.81 | 16.36 ± 5.63 | T × D | 0.53 | |
| A‐MC | 10.71 ± 7.24 | 12.74 ± 4.63 | T × G | 0.52 | |
| NA‐MC | 15.24 ± 7.64 | 17.59 ± 5.28 | T × G × D | 0.35 | |
| A‐LC | 11.86 ± 4.82 | 14.22 ± 5.10 | Time | 0.001 | |
| NA‐LC | 15.00 ± 8.10 | 15.72 ± 5.83 | |||
| Mean | 12.98 ± 7.24 | 14.74* ± 5.38 | |||
| HOMA‐IR | MC | 2.52 ± 1.53 | 3.09 ± 1.02 | Diet | 0.27 |
| LC | 3.24 ± 1.77 | 3.56 ± 1.43 | Genotype | 0.04 | |
| A | 2.44 ± 1.34 | 3.04 ± 1.20 | G × D | 0.50 | |
| NA | 3.43 ± 1.91 | 3.69 ± 1.27 | T × D | 0.45 | |
| A‐MC | 2.18 ± 1.44 | 2.78 ± 0.94 | T × G | 0.31 | |
| NA‐MC | 3.20 ± 1.53 | 3.72 ± 0.92 | T × G × D | 0.49 | |
| A‐LC | 2.82 ± 1.13 | 3.41 ± 1.46 | Time | 0.001 | |
| NA‐LC | 3.55 ± 2.10 | 3.67 ± 1.44 | |||
| Mean | 2.90 ± 1.69 | 3.34* ± 1.27 |
Different from baseline (p < 0.05).
Data presented as mean ± standard deviations. The p‐levels are from univariate analysis. A, diet aligned to genotype (n = 34); D, diet effect; G, genotype match to diet effect; G × D, genotype match to diet × diet effect; LC, low carbohydrate diet (n = 33); MC, moderate carbohydrate diet (n = 30); NA, diet not aligned to genotype (n = 29), A‐MC (n = 20), NA‐MC (n = 10), A‐LC (n = 14), NA‐LC (n = 19); T, time effect; T × D, time × diet effect; T × G, time × genotype match to diet effect; T × G × D, time × diet × genotype match to diet effect.
Discussion
The purpose of this study was to (1) determine the efficacy of two moderately higher protein diets that varied in carbohydrate and fat content on changes in weight, body composition, exercise capacity and biomarkers of health in sedentary women with obesity participating in a circuit‐style resistance training program and (2) determine if alignment of genotype related to lipid metabolism using a direct‐to‐consumer genetic screening kit would promote greater benefits. Results indicated that at 24 weeks, all participants experienced significant reductions in weight, fat mass, per cent body fat and several biomarkers of health, with some evidence that participants in the LC group experienced greater improvements. However, alignment of these diets to the genotype profile used in the current investigation did not promote greater changes in weight, body composition, exercise capacity or biomarkers of health. These findings do not support use of the tested direct‐to‐consumer genetic screening kit to optimize weight loss or other health outcomes when participating in a circuit‐style resistance training program while following moderately higher protein diet programs.
Our findings contrast results from Dopler‐Nelson and colleagues 17 retrospective study, who reported a twofold to threefold greater weight loss in individuals following diets that were retrospectively aligned to this genetic profile yet support results from a larger study this research group recently reported 19. In this regard, Gardner and colleagues 19 randomly assigned 609 participants to a lower fat (48–53% kcal carbohydrate; 24–29% kcal fat; 21–22% kcal protein) or lower carbohydrate (23–30% kcal carbohydrate; 45–49% kcal fat; 23–26% kcal protein) diet for 52 weeks. Participants were instructed to continue normal physical activity habits. Insulin response to an oral glucose challenge as well as three of the five candidate genes included in the profile of the present investigation (PPARGrs1801282, ADRB2rs1042714 and FABP2rs1799883) were assessed. At 52 weeks, weight change in participants following the lower fat diet was −5.3 kg, while individuals following the lower carbohydrate diet were −6.0 kg (mean difference 0.7 kg [95% CI, −0.2 to 1.6 kg]). There were no significant interactions observed for diet to genotype or diet to insulin response to an oral glucose challenge. The researchers concluded that aligning these diets to the tested genotype patterns did not promote greater weight loss success.
Accordingly, in Frankwich and colleagues 47 24‐week diet intervention, 51 participants were randomized to a standard balanced diet or a diet that aligned with dietary recommendations related to carbohydrate and fat intake based on candidate genes in the tested profile (APOA2rs5082, ADIPOQrs17300539, FTOrs9939609, KCTD10rs10850219, LIPCrs1800588, MMABrs2241201 and PPARFrs1801280). At 24 weeks, statistically significant differences between diet groups were not observed for per cent weight loss, glucose homeostasis or fasting blood lipids. Investigators concluded that aligning these diets to the tested genotype patterns also did not optimize weight loss.
With that said, some investigations have observed statistically significant changes in health outcomes when aligning a diet based on one genetic polymorphism as opposed to a profile of multiple genes. For example, Grau et al. 48 prospective trial evaluated the interaction between allele patterns of transcription factor 7‐like 2 (TCF7L2 rs7903146) and dietary fat and carbohydrate intake among 739 obese individuals. Participants followed a high‐fat (40–45% kcal carbohydrate, 40–45% kcal fat, 15% kcal protein) or lower fat/higher carbohydrate (60–65% kcal carbohydrate, 20–25% kcal fat, 15% kcal protein) hypo‐energetic diet (2,600 kcal d−1) for 10 weeks. Overall individuals who were homozygous for the T‐allele and followed the high‐fat diet experienced less weight loss and changes in fat‐free mass, waist circumference and homeostatic model insulin resistance than those in the low‐fat diet groups. Additionally, for those following the high‐fat diet, each additional T‐allele was associated with less fat loss. The investigators concluded that individuals with obesity who were homozygous for T allele in TCF7L2 rs7903146 were more sensitive to lower fat/higher carbohydrate diet versus high‐fat diets.
Furthermore, Qi and associates 49 examined whether adherence to a higher carbohydrate/low fat (55–65% kcal carbohydrate, 20% kcal fat, 15% kcal protein) or lower carbohydrate/higher fat diet (35–45% kcal carbohydrate, 40% kcal fat, 15% kcal protein) were modified by the effects of allele patterns of glucose‐dependent insulinotropic polypeptide receptor (GIPRrs2287019) on body weight and glucose and insulin resistance among 737 individuals with obesity. After 24 weeks of intervention, the T allele of GIPRrs2287019 was associated with greater weight loss and reductions in fasting glucose and insulin in participants who followed the higher carbohydrate/low‐fat diet. Consequently, while much more research needs to be done to evaluate the relationship of specific genes to favourable changes in body weight, composition and other health outcomes related to chronic disease risk, results of these studies offer some support to the potential role of aligning diet with genotype in an effort to optimize health outcomes.
Strengths of the present investigation include the prospective study design (a randomized controlled trial), length of study and use of both a direct‐to‐consumer genetic screening kit for weight management and a commercially available exercise and weight loss program. Limitations include a relatively small sample size, use of a prior power calculation based on two‐way interactions, a wide age range that may result in the possibility of age‐related anabolic resistance to the moderately higher protein diet and resistance training program and the possibility of inter‐individual variations of epigenetic factors that may have impacted gene expression of the candidate genes in the genetic profile. Future research should continue to test potential candidate genes prospectively and consider testing genes individually as opposed to in panels, to identify alternative methods to optimize weight loss and promote favourable changes in health outcomes associated with chronic disease risk.
Overall, our findings and findings from previous work do not support use of the genetic polymorphisms tested here to select diet and optimize weight loss success and changes in body composition, exercise capacity and biomarkers of health; however, available evidence suggests use of other genes for diet selection to improve health outcomes. From a public health perspective, this work is important, as once we identify the correct genetic polymorphism or combination of polymorphisms for diet selection to optimize health outcomes, this knowledge may contribute significantly to disease prevention.
Conflict of Interest Statement
Curves International (Waco, TX, USA) provided funding for this research through unrestricted grants to Texas A&M University and Richard Kreider who served as Principal Investigator for this research. No other authors declare a conflict of interest.
Author Contributions
R. B. K designed the research (project conception, development of overall research plan, obtained external funding and provided study oversight). A. M. C. served as study coordinator. A. M. C., E. G., C. R., K. L., B. L., S. Y. S., Y. P. J., M. K., M. M., J. O., R. D., M. B., D. K., B. S. and A. J. conducted research (hands‐on conduct of the experiments and data collection). A. M. C. and R. B. K. wrote the paper with R. B. K. having primary responsibility for final content. All contributors reviewed and approved submission of the paper.
Funding
We would like to thank Curves International (Waco, TX) for funding the study. We would also like to thank the National Cancer Institute of the National Institutes of Health under award number R25CA057730 (PI: Shine Chang, PhD) for their recent support for AMC.
Acknowledgements
We would like to thank all the individuals who participated in this study. We would also like to thank Dr. J. Timothy Lightfoot for his contributions to data interpretation and Sunday Simbo, Minye Cho, Aimee Reyes and Ryan Sowinski who assisted with data collection.
Coletta, A. M. , Sanchez, B. , O'Connor, A. , Dalton, R. , Springer, S. , Koozehchian, M. S. , Murano, P. S. , Woodman, C. R. , Rasmussen, C. , and Kreider, R. B. (2018) Alignment of diet prescription to genotype does not promote greater weight loss success in women with obesity participating in an exercise and weight loss program. Obesity Science & Practice, 4: 554–574. 10.1002/osp4.305.
References
- 1. Byrd AS, Toth AT, Stanford FC. Racial disparities in obesity treatment. Curr Obes Rep 2018; 7: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ogden CLCM, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA 2014; 311: 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daniels J. Obesity: America's epidemic. Am J Nurs 2006; 106: 40–49. quiz 49‐50. [DOI] [PubMed] [Google Scholar]
- 4. Wilborn C, Beckham J, Campbell B, et al. Obesity: prevalence, theories, medical consequences, management, and research directions. J Int Soc Sports Nutr 2005; 2: 4–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kouvelioti R, Vagenas G, Langley‐Evans S. Effects of exercise and diet on weight loss maintenance in overweight and obese adults: a systematic review. J Sports Med Phys Fitness 2014; 54: 456–474. [PubMed] [Google Scholar]
- 6. Matsuo T, Okura T, Nakata Y, et al. The influence of physical activity‐induced energy expenditure on the variance in body weight change among individuals during a diet intervention. Obes Res Clin Pract 2007; 1: I–II. [DOI] [PubMed] [Google Scholar]
- 7. Teixeira PJ, Palmeira AL, Branco TL, et al. Who will lose weight? A reexamination of predictors of weight loss in women. Int J Behav Nutr Phys Act 2004; 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zibellini J, Seimon RV, Lee CM, Gibson AA, Hsu MS, Sainsbury A. Effect of diet‐induced weight loss on muscle strength in adults with overweight or obesity – a systematic review and meta‐analysis of clinical trials. Obes Rev 2016; 17: 647–663. [DOI] [PubMed] [Google Scholar]
- 9. Coletta A, Kreider RB. Genetic profiling for weight loss: potential candidate genes. Bioenergetics 2015; 4. [Google Scholar]
- 10. Fox CS, Liu Y, White CC, et al. Genome‐wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet 2012; 8: e1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mutch DM, Temanni MR, Henegar C, et al. Adipose gene expression prior to weight loss can differentiate and weakly predict dietary responders. PLoS One 2007; 2: e1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome‐wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009; 41: 18–24. [DOI] [PubMed] [Google Scholar]
- 13. Heianza Y, Qi L. Gene‐diet interaction and precision nutrition in obesity. Int J Mol Sci 2017; 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Willer C, Speliotes E, Loos R, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009; 21: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010; 42: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goni L, Cuervo M, Milagro FI, Martinez JA. Future perspectives of personalized weight loss interventions based on nutrigenetic, epigenetic, and metagenomic data. J Nutr 2016. [DOI] [PubMed] [Google Scholar]
- 17. Dopler‐Nelson M, Prahakar P, Kornman K, Gardner C. Genetic phenotypes predict weight loss success: the right diet does matter. AHA Abstracts 2010: 79–80. [Google Scholar]
- 18. Gardner CD, Kiazand A, Alhassan S, et al. Comparison of the Atkins, Zone, Ornish, and LEARN diets for change in weight and related risk factors among overweight premenopausal women: the A TO Z Weight Loss Study: a randomized trial. JAMA 2007; 297: 969–977. [DOI] [PubMed] [Google Scholar]
- 19. Gardner CD, Trepanowski JF, Del Gobbo LC, et al. Effect of low‐fat vs low‐carbohydrate diet on 12‐month weight loss in overweight adults and the association with genotype pattern or insulin secretion: the DIETFITS randomized clinical trial. JAMA 2018; 319: 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heianza Y, Ma W, Huang T, et al. Macronutrient intake‐associated FGF21 genotype modifies effects of weight‐loss diets on 2‐year changes of central adiposity and body composition: The POUNDS Lost trial. Diabetes Care 2016; 39: 1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abete IN‐CS, Marti A, Alfredo MJ. Nutrigenetics and nutrigenomics of caloric restriction In: Elsevier I. (ed.). Progress in Molecular Biology and Translational Science, Vol. 108, 2012, pp. 323–346. [DOI] [PubMed] [Google Scholar]
- 22. Baetge C, Earnest CP, Lockard B, et al. Efficacy of a randomized trial examining commercial weight loss programs and exercise on metabolic syndrome in overweight and obese women. Appl Physiol Nutr Metab 2017; 42: 216–227. [DOI] [PubMed] [Google Scholar]
- 23. Kerksick C, Thomas A, Campbell B, et al. Effects of a popular exercise and weight loss program on weight loss, body composition, energy expenditure and health in obese women. Nutr Metab (Lond) 2009; 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerksick CM, Wismann‐Bunn J, Fogt D, et al. Changes in weight loss, body composition and cardiovascular disease risk after altering macronutrient distributions during a regular exercise program in obese women. Nutr J 2010; 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kreider R, Rasmussen C, Kerksick C, et al. A carbohydrate‐restricted diet during resistance training promotes more favorable changes in body composition and markers of health in obese women with and without insulin resistance. Phys Sportsmed 2011; 39: 27–40. [DOI] [PubMed] [Google Scholar]
- 26. Kreider RB, Serra M, Beavers KM, et al. A structured diet and exercise program promotes favorable changes in weight loss, body composition, and weight maintenance. J Am Diet Assoc 2011; 111: 828–843. [DOI] [PubMed] [Google Scholar]
- 27. Lockard B, Earnest CP, Oliver J, et al. Retrospective analysis of protein‐ and carbohydrate‐focused diets combined with exercise on metabolic syndrome prevalence in overweight and obese women. Metab Syndr Relat Disord 2016; 14: 228–237. [DOI] [PubMed] [Google Scholar]
- 28. Magrans‐Courtney T, Wilborn C, Rasmussen C, et al. Effects of diet type and supplementation of glucosamine, chondroitin, and MSM on body composition, functional status, and markers of health in women with knee osteoarthritis initiating a resistance‐based exercise and weight loss program. J Int Soc Sports Nutr 2011; 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blair S, Kampert J, Kohl H, et al. Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all‐cause mortality in men and women. JAMA 1996; 276: 205–210. [PubMed] [Google Scholar]
- 30. DeFina L, Barlow C, Radford N, Leonard D, Willis B. The association between midlife cardiorespiratory fitness and later life chronic kidney disease: the Cooper Center Longitudinal Study. Prev Med 2016; 89: 178–183. [DOI] [PubMed] [Google Scholar]
- 31. Heber S, Assinger A, Pokan R, Volf I. Correlation between cardiorespiratory fitness and platelet function in healthy women. Med Sci Sports Exerc 2016; 48: 1101–1110. [DOI] [PubMed] [Google Scholar]
- 32. Lakoski S, Willis B, Barlow C, et al. Midlife cardiorespiratory fitness, incident cancer, and survival after cancer in men: the Cooper Center Longitudinal Study. JAMA Oncol 2015; 1: 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lavie C, Schutter A, Archer E, McAuley P, Blair S. Obesity and prognosis in chronic diseases – impact of cardiorespiratory fitness in the obesity paradox. Curr Sports Med Rep 2014; 13: 240–245. [DOI] [PubMed] [Google Scholar]
- 34. Sieverdes J, Sui X, Lee D, et al. Physical activity, cardiorespiratory fitness and the incidence of type 2 diabetes in a prospective study of men. Br J Sports Med 2010; 44: 238–244. [DOI] [PubMed] [Google Scholar]
- 35. Stathokostas L, Dogra S, Paterson D. The independent roles of cardiorespiratory fitness and sedentary time on chronic conditions and body mass index in older adults. J Sports Med Phys Fitness 2015; 55: 1200–1206. [PubMed] [Google Scholar]
- 36. World Medical A . World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 2013; 310: 2191–2194. [DOI] [PubMed] [Google Scholar]
- 37. Farris G, Wismann J, Farris R, et al. Exercise intensity and energy expenditure analysis of women participating in the Curves® exercise program. FASEB J 2006; LB: 93–94. [Google Scholar]
- 38. Henderson S, Magu B, Rasmussen C, et al. Effects of coleus forskohlii supplementation on body composition and hematological profiles in mildly overweight women. J Int Soc Sports Nutr 2005; 2: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. La Bounty P, Wilborn C, Marcello B, et al. Analysis of exercise intensities of women using the Curves® hydraulic training equipment. FASEB J 2006; LB: 93. [Google Scholar]
- 40. Sanchez‐Delgado G, Alcantara JMA, Ortiz‐Alvarez L, et al. Reliability of resting metabolic rate measurements in young adults: impact of methods for data analysis. Clin Nutr 2017. [DOI] [PubMed] [Google Scholar]
- 41. Almada A, Kreider R, Ransom J, Rasmussen C, Tutko R, Milnor P. Comparison of the reliability of repeated whole body DEXA scans to repeated spine and hip scans. J Bone Miner Res 1999; 14: S369. [Google Scholar]
- 42. Pescatello LS, American College of Sports M . ACSM's Guidelines for Exercise Testing and Prescription. Wolters Kluwer/Lippincott Williams & Wilkins Health: Philadelphia, 2014. [Google Scholar]
- 43. Bowling JL, Katayev A. An evaluation of the Roche Cobas c 111. Laboratory Medicine 2010; 41: 398–402. [Google Scholar]
- 44. Song Y, Manson JE, Tinker L, et al. Insulin sensitivity and insulin secretion determined by homeostasis model assessment and risk of diabetes in a multiethnic cohort of women: the Women's Health Initiative Observational Study. Diabetes Care 2007; 30: 1747–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Little R, Kang S. Intention‐to‐treat analysis with treatment discontinuation and missing data in clinical trials. Stat Med 2015; 34: 2381–2390. [DOI] [PubMed] [Google Scholar]
- 46. Page P. Beyond statistical significance: clinical interpretation of rehabilitation research literature. Int J Sports Phys Ther 2014; 9: 726–736. [PMC free article] [PubMed] [Google Scholar]
- 47. Frankwich K, Egnatios J, Kenyon M, et al. Differences in weight loss between persons on standard balanced vs nutrigenetic diets in a randomized controlled trial. Clin Gastroenterol Hepatol 2015; 13: 1625–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grau K, Cauchi S, Holst C, et al. TCF7L2 rs7903146‐macronutrient interaction in obese individuals' responses to a 10‐wk randomized hypoenergetic diet. Am J Clin Nutr 2010; 91: 472–479. [DOI] [PubMed] [Google Scholar]
- 49. Qi Q, Bray GA, Hu FB, Sacks FM, Qi L. Weight‐loss diets modify glucose‐dependent insulinotropic polypeptide receptor rs2287019 genotype effects on changes in body weight, fasting glucose, and insulin resistance: the Preventing Overweight Using Novel Dietary Strategies trial. Am J Clin Nutr 2012; 95: 506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]


