Abstract
Vibrio parahaemolyticus is a Gram-negative, halophilic bacterium isolated from marine environments globally. After the consumption of contaminated seafood, V. parahaemolyticus causes acute gastroenteritis. To initiate infection, a wide range of virulence factors are required. A complex group of genes is known to participate in the pathogenicity of V. parahaemolyticus; however, to understand the full mechanism of infection, extensive research is yet required. V. parahaemolyticus has become the leading cause of seafood-related gastroenteritis in Japan, the United States and several other parts of the world. In addition, outbreaks caused by the pandemic clone of this organism are escalating and spreading universally. To minimize the risk of V. parahaemolyticus infection and warrant the safety of seafood, collaboration between governments and scientists is required. We herein provide an updated review of the pathogenicity determinants and distribution of V. parahaemolyticus to deliver a better understanding of the significance of V. parahaemolyticus and its host–pathogen interactions.
Keywords: Pandemic clone, pathogenesis, Vibrio Parahaemolyticus, virulence determinants
Abstract
ملخص البحث: تؤدي بكتيريا ضمة الدم والتي يمكن عزلها من البيئات البحرية إلى التهاب حاد في الجهاز الهضمي عند تناول أطعمة بحرية ملوثة ولكي تسبب هذه البكتيريا عدوى فأنها تحتاج إلى تواجد عوامل خطورة، والتي لازالت تحتاج إلى مزيد من البحوث. أصبحت هذه البكتيريا المسبب الرئيسي لالتهابات الجهاز الهضمي الناتجة عن تناول الأطعمة البحرية في اليابان، وأمريكا ودول أخرى. يقدم الباحثون تحديثاً عن هذه البكتيريا ومحدداتها المرضية، ولتقليل العدوى بهده البكتيريا ينصح الباحثون بالتعاون بين العلماء والحكومات على مستوى العالم لمعرفة المزيد عن هذه الجرثومة.
INTRODUCTION
The constant interaction of human populations with their surroundings continues to transform the spectrum of infectious diseases. Therefore, the search of emerging pathogens must not come to a pause. An emerging infectious disease is one for which the rate of incidence has increased within the past two decades or one which threatens to spread rapidly.[1] Examples of these are infections caused by Vibrio parahaemolyticus, which is progressing as a significant human pathogen.[2] V. parahaemolyticus is a Gram-negative halophilic bacterium that produces a capsule with different somatic (O) and capsular (K) antigens.[3] V. parahaemolyticus is isolated from coastal and estuarine environments universally.[4] In addition, it has been recovered from a wide variety of marine organisms.[5] The consumption of raw or undercooked seafood contaminated by virulent strains of V. parahaemolyticus leads to acute gastroenteritis.[5] The symptoms of the disease include diarrhea, nausea, abdominal pain, vomiting and low-grade fever.[6] In most cases, the disease is self-resolving. However, incidences where V. parahaemolyticus caused a more debilitating and dysenteric form of gastroenteritis have been reported.[7] In addition, when open wounds come in contact with contaminated seawater, wound infections occur.[8] Uncommonly, in immunocompromised patients, it may progress into a life-threatening fulminant necrotizing fasciitis characterized by the rapid necrosis of subcutaneous tissue.[9,10] In rare cases, V. parahaemolyticus causes septicemia, which is associated with a high mortality rate.[11] It has been mostly reported in individuals who are immunocompromised due to underlying medical conditions such as liver diseases.[12] To initiate infection, a wide range of virulence factors are used by V. parahaemolyticus including adhesins, toxins and secretion systems.[11] V. parahaemolyticus has become the leading cause of seafood-related gastroenteritis in Japan, the United States and several other parts of the world.[13] Further, outbreaks caused by the pandemic clone of this organism are escalating and spreading universally.[13] To minimize the risk of V. parahaemolyticus infection and warrant the safety of seafood, collaboration between governments and scientists is required.[14] Therefore, the objective of this study was to provide an updated review of the pathogenicity determinants and distribution of V. parahaemolyticus and use this information to deliver a better understanding of the V. parahaemolyticus significance and its host–pathogen interactions.
QUORUM SENSING
Quorum sensing (QS) is a term that defines the cell–cell communication process by which bacteria respond to released signaling molecules (known as autoinducers), on the basis of cell density fluctuations, to regulate gene expression.[11] As the density of QS bacteria increases, the concentration of the autoinducers increases until it reaches a critical threshold, at which point bacteria forms a response.[15] In the process of QS, individual cells' function in unison to coordinately alter their gene expression and control their synchrony-requiring activities such as virulence factor secretion.[16] At high cell densities, V. parahaemolyticus bacteria produce their major transcriptional regulator OpaR in response to the QS system.[11] To summarize the role of OpaR in controlling the phenotypic output of V. parahaemolyticus, Kernell Burke et al.[16] demonstrated 11 transcription factors under the downstream regulation of OpaR. They fall into the following four broad themes: first, genes related to cell surface and adhesion; second, virulence factors and cell–cell interactions including type III and type VI secretion systems (T3SS and T6SS); third, surface-specific regulon including the lateral flagellar system, chemotaxis and the swarm-specific sRNA and finally, other functions such as competency.[16]
ADHESION TO HOST CELLS
Multivalent adhesion molecules (MAMs) are present in a wide range of Gram-negative pathogens.[17] It enables high-affinity binding to the host cells during preliminary stages of infection, which is essential for delivery of virulence factors.[17] MAM7 is the adhesin expressed constitutionally by V. parahaemolyticus. With its aid, V. parahaemolyticus is able to attach different types of host cells including macrophages, fibroblasts and epithelial cells.[18] Correct localization and outer membrane anchoring of the protein are achieved by the hydrophobic stretch of 44 amino acids in MAM7 N-terminus.[5] MAM7 has two host surface receptors: host membrane phosphatidic acid lipids (PA), to which MAM7 has a high affinity of binding, and extracellular matrix protein fibronectin, which acts as a co-receptor.[19] MAM7 is constituted of seven mammalian cell entry domains, and each of them is capable of PA binding.[19] The binding of MAM7 to PA in the host membrane causes downstream activation of small GTPase RhoA, which eventually leads to redistribution of epithelial tight junction proteins.[18] The consequences of this pathway are the free migration of bacteria across epithelial layers and the depolarization of the barrier, leaving apical and basolateral surfaces with no particular markers.[18]
IRON ACQUISITION
Iron is essential for the survival of organisms. Therefore, bacteria develop different methods to acquire iron from their hosts.[8] Intracellularly, iron is involved in many processes ranging from signaling to metabolism.[20] Furthermore, many organisms use the intracellular “low-iron conditions” to stimulate the expression of virulence genes.[21] In humans, iron is present as part of multiple molecular complexes such as transferrin and hemoglobin.[8] During infection, V. parahaemolyticus utilizes at least two methods of iron acquisition: production of siderophores and use of heme as a direct source of iron.[22] Siderophores are compounds with a low molecular weight and a high iron affinity. They can scavenge extracellular iron, remove transferrin and lactoferrin-bound iron and facilitate its uptake by the bacteria.[23] V. parahaemolyticus produces a siderophore known as vibrioferrin, which is synthesized by proteins from the pvsABCDE operon.[8] An outer membrane receptor, composed of PvuA1 and PvuA2 proteins, recognizes the ferric-charged vibrioferrin.[21] Since this receptor is coupled to the inner membrane ATP-binding cassette, it imports the ferric-charged vibrioferrin to the inner side of the cell.[21] As it acquires the energy required for iron-siderophore transportation across the outer membrane, V. parahaemolyticus contains three sets of the TonB energy transduction system.[21] The process of iron uptake should be highly regulated because the concentration of iron inside the cell is critical; too little prevents the completion of certain cellular processes and too much causes the accumulation of free radicals, leading to cell death.[20] In V. parahaemolyticus, this vital process is controlled by the ferric uptake regulation protein “Fur.”[23] By the downstream regulation of several genes, it represses the iron absorption.[21]
TOXINS
The late 1980s witnessed the study of the first V. parahaemolyticus virulence factors: thermostable direct hemolysin (TDH)[24] and the thermostable direct-related hemolysin (TRH).[25] Today, TDH and TRH remain the most distinct virulence factors, as almost all clinically isolated strains of V. parahaemolyticus possess hemolytic activity attributed to these two genes.[3] TDH is composed of four soluble monomers, in which a central pore is formed to allow the diffusion of small molecules.[26] In a process termed as Kanagawa phenomenon (KP), tdh + strains of V. parahaemolyticus exhibit β-hemolytic activity when plated on special blood media known as Wagatsuma agar.[6] Purified TDH is heat stable after being exposed to 100°C for 10 min.[27] Hemolysis, enterotoxicity, cytotoxicity and cardiotoxicity represent a group of biological activities caused by TDH.[28] TDH is a pore-forming toxin. The fairly large pores it creates on erythrocytes allow both water and ions to flow through membranes.[29] Eventually, this results in colloidal osmotic lysis.[30] The primary targets for TDH activity are the epithelial and intestinal cells. The effect of TDH on these cells is very crucial for biological functions, such as diarrhea, during infection.[27] TDH binds to phospholipid bilayers, in which single channel events occur.[31] The mechanisms of cell binding and calcium ions influx result in the increase of intracellular Ca+ and release of chloride ions in human colonic epithelial cells.[27] When the osmotic pressure of the cell exceeds its limit of self-regulation, pathological changes follow, leading to cell expansion and even death.[32] Homologs of V. parahaemolyticus TDH are expressed in other vibrios causing diarrhea, such as non-O1 Vibrio cholerae and Vibrio mimicus. They have all been included in the tdh family because the coding sequences of these genes share >93% homology.[33] Clinical samples from an outbreak of gastroenteritis in the Republic of Maldives presented KP-negative strains of V. parahaemolyticus. However, these strains were found to express a new hemolysin, termed as TRH.[25] TRH is immunologically similar to TDH and their genes share approximately 70% homology.[29] TRH demonstrates hemolytic activity similar to that of TDH on blood cells.[6] Moreover, TRH activates Cl− channels and causes altered ion influx, in a manner analogous to TDH.[29] However, TRH is heat-labile when exposed to 60°C for 10 min.[27] Besides, when compared to TDH, evidence of a lesser role of TRH in the pathogenicity of V. parahaemolyticus was proposed.[34] Although the association between the expression of tdh and trh and the pathogenicity of V. parahaemolyticus is well acknowledged, it should be perceived that not all clinical strains possess these genes.[3] Some of the isolated clinical strains do not contain tdh and/or trh although these main hemolysins are absent, V. parahaemolyticus remains pathogenic, indicating the expression of other virulence activities.[29] It should not escape our notice that V. parahaemolyticus bacteria express an additional toxin, known as thermolabile hemolysin (TLH).[33] It is encoded by the tlh gene, which is targeted during the genetic detection of V. parahaemolyticus because it is a species-specific marker.[13] TLH exhibits phospholipase activity and the ability to lyse human erythrocytes.[8] Furthermore, the expression of tlh was strongly upregulated under conditions simulating the human–host intestinal environment.[35] Therefore, TLH is assumed to have a role in V. parahaemolyticus similar to TDH.[32] However, its direct contribution to the pathogenicity of V. parahaemolyticus is yet to be elucidated.[33]
TYPE III SECRETION SYSTEMS
The T3SS bacterial machinery is a needle-like apparatus that injects bacterial proteins (termed effectors) directly into the cytoplasm of eukaryotic cells, without encountering the extracellular environment.[11] Those effectors disrupt the regular cell signaling processes to modify the biological activities of the host cell.[36] The basic structure of the T3SS apparatus is conserved among different species of bacteria. However, their effectors' functions and targets differ.[11] According to the needs of pathogens, the production of effectors may be up- or downregulated.[8] The secretion apparatus of T3SS consists of first, the basal body spanning the bacterial inner and outer membrane; second, a needle that connects between the bacteria and the host cells and finally, a translocon pore that penetrates the eukaryotic cell membrane.[5] The whole genome sequencing of V. parahaemolyticus RIMD 22106633 (O3:K6) revealed the acquisition of two sets of T3SS gene clusters, with one on each of its two circular chromosomes, and thus they were entitled T3SS1 and T3SS2.[37]
Type three III secretion system 1
The T3SS1 gene cluster is encoded in the first pathogenicity island on chromosome 1. Nearly all clinical and environmental strains of V. parahaemolyticus encode T3SS1.[32] The sequence homology of T3SS1 gene cluster with Yersinia spp. and other vibrios suggests that it was ancestrally acquired and has been evolutionarily conserved.[3] A dual regulatory system consisting of ExsACDE regulatory cascade and heat-stable nucleoid-structuring protein (H-NS) orchestrates the transcription of T3SS1.[38] ExsA, a member of AraC family of transcription regulators, is the master transcriptional regulator of T3SS1 expression.[39] Under noninducing conditions, ExsD (an anti-activator) directly binds to ExcA and renders it inactive. Meanwhile, the anti-anti-activator ExsC binds to ExsE. In inducing conditions, ExsE is secreted to release ExsC. ExsC then sequesters ExsD and liberates ExsA. Free ExsA binds to the promoter genes and activates T3SS1.[39] H-NS is a major component of the bacterial nucleoid.[38] In V. parahaemolyticus, H-NS was found to repress the T3SS1-related genes' expression by suppressing exsA gene.[38] During V. parahaemolyticus infection, the activation of T3SS1 initiates a reproducible series of events. Their outcome includes the induction of rapid autophagy followed by cell rounding, eventually leading to cell lysis.[11] To date, four T3SS1 effectors have been recognized: Vibrio outer protein (Vop) Q, VopS, VPA0450 and VopR (VP1638) [Table 1].[32] The effector VopQ contributes to T3SS1 cytotoxicity.[40] Through membrane permeations, VopQ manipulates lysosomal homeostasis and autophagic flux, leading to rapid induction of autophagy.[41] The mediated autophagy inhibits recruitment of phagocytosis-related cellular machinery, leading to debilitated phagocytes engulfment of V. parahaemolyticus bacteria during infection.[40] Moreover, Vop-Q triggers mitogen-activated protein kinases (MAPKs) pathway that regulates transcription of inflammatory cytokines. Through MAPK, VopQ induces production of interleukin-8, which is known for its leukocyte chemotactic properties and role in inflammatory disease.[42] VopS (VP1686) modifies a conserved threonine residue on Rho, Rac and Cdc42, with adenosine-5′-monophosphate (AMP).[43] The AMPylation of Rho GTPases prevents subsequent interaction with downstream effectors, by which actin assembly is inhibited.[43] The upshots of VopS activities in V. parahaemolyticus infection are the interference with the assembly of specks in infected macrophages, the hindrance of inflammasome activation and the assistance in the bacterial evasion from inflammatory responses.[49] VPA0450 is another T3SS1 effector protein contributing to host cell death.[8] In the human host, VPA0450 disrupts the cytoskeletal binding sites on the inner surface of the membrane. The subsequent plasma membrane blebbing compromises the membrane integrity, facilitates cell lysis and participates in cytotoxicity.[8] Finally, by binding to the phosphoinositide on the host cell, VopR localizes to the cellular membrane. It may also play a role in the infection of V. parahaemolyticus by promoting the refolding of T3SS effector proteins after their delivery into host cells.[32]
Table 1.
List of Vibrio parahaemolyticus type III secretion systems effectors
| T3SS | T3SS effectors | Activity | References |
|---|---|---|---|
| T3SS1 | VopQ | Rapid induction of autophagy leading to debilitated phagocytes engulfment of V. parahaemolyticus during infection | [40,41,42] |
| VopS | Actin assembly inhibition | [43] | |
| VPA0450 | Compromises membranes integrity, facilitates cell lysis and participates in cytotoxicity | [8] | |
| VopR | May play a role in infection by promoting refolding of T3SS effector proteins after their delivery into host cell | [32] | |
| T3SS2 | VopA/P (VPA1346) | Suppresses the host innate immune response | [44,45] |
| VopL | Actin nucleation, induction of stress fibers and contribute to bacterial uptake into the host cells | [6] | |
| VopC | Changes in actin cytoskeleton and facilitates bacterial invasion | [46] | |
| VopT | Cytotoxicity | [47] | |
| VopV | Enterotoxicity | [36] | |
| VopZ | Mediates pathological phenotypes during V. parahaemolyticus infection | [48] | |
| VPA1380 | Unknown | [32] |
T3SS – Type III secretion systems; Vop – Vibrio outer protein; V. parahaemolyticus – Vibrio parahaemolyticus
Type three III secretion system 2
The second cluster of genes encoding T3SS2 is found on the pathogenicity island Vp-PAI (VPaI-7) on chromosome 2. Along with T3SS2 and its known effectors, VPaI-7 typically encodes two tdh genes.[37] Accordingly, it is primarily expressed in clinical isolates of V. parahaemolyticus and associated with large outbreaks of the disease.[8] The G + C content of VPaI-7 is lower than the genomic average and has a higher number of genes unique to each Vibrio spp. These annotations indicate that it was recently acquired by V. parahaemolyticus through lateral transfer.[6] The genes of T3SS2 are mainly regulated by two transcriptional regulators, VtrA and VtrB.[38] Gotoh et al.[35] demonstrated that V. parahaemolyticus recognizes the intestinal environment through detecting bile acids, which induce VtrA-mediated VtrB transcription and subsequent production of T3SS2 effector proteins. Noteworthy, VtrA and VtrB do not only regulate the expression of T3SS2 but also regulate the expression of both tdh genes on Vp-PAI. Thus, they play a major role in V. parahaemolyticus cytotoxicity.[6] The delivery of effector proteins to host cells requires the presence of a translocon complex. During V. parahaemolyticus infection, VopB2, VopD2 and the recently identified VopW are necessary for the T3SS2 effector translocation through permeation of the host cell membrane.[6] While the cytotoxicity of T3SS2 is limited and associated with the function of TDH, T3SS2 appears to be the major contributor to the enterotoxicity occurring in rabbit ileal loop model.[50] In addition, the T3SS2 effectors are capable of adhering to human cells, leading to cytoskeletal disruption and loss of membrane integrity.[51] To date, seven T3SS2 effector proteins have been characterized: first, Vop A/P (VPA1346), which shares around 55% homology with the YopJ-like proteins of Yersinia and Salmonella.[44] VopA is an acetyltransferase that prevents the activation of MAPK kinases, inhibits MAPK signaling pathway and ultimately suppresses the host's innate immune response.[45] Second, VopL contains three N-terminal Wiskott–Aldrich homology 2 motifs and a unique VopL C-terminal domain. Its main function is actin nucleation and induction of stress fibers.[52] Ham and Orth[6] suggested that the role of VopL in actin manipulation may contribute to the bacterial uptake into the host cells. Third, VopC, which displays homology to the catalytic domain of cytotoxic necrotizing factor toxins.[46] VopC exhibits transglutaminase activity, by which it modifies Rho family GTPases. Once Rho GTPase is activated, it triggers changes in the actin cytoskeleton of infected cells.[46] In the meantime, the activation of CDC42 facilitates the invasion of V. parahaemolyticus into host cells.[46] Fourth, VopT is a cytotoxin and ADP-ribosyltransferase effector that targets Ras GTPase in V. parahaemolyticus-infected cells.[47] Fifth, VopV is the homolog of VopM protein that is involved in non-O1/non-O139 V. cholerae enterotoxicity. Parallel to VopM, VopP possesses multiple F-actin-binding domains and an enterotoxic activity during infection of V. parahaemolyticus.[36] Sixth, VopZ is a multifunctional effector crucial for the pathological phenotypes induced by V. parahaemolyticus. Strains lacking vopZ genes fail to colonize the intestine and cause diarrhea.[48] VopZ inhibits the activation of transforming growth factor β-activated kinase-1, TAK1, which is essential for MAPK and NF-κB signaling pathway activation. TAK1 has a profound influence on the preservation of intestinal integrity, and its absence leads to different consequences including inflammation, apoptosis and reduced transepithelial resistance.[48] Finally, VPA1380 is the most recently identified T3SS2 effector protein. It is a typical cysteine protease that catalyzes its targeted substrates.[53] VPA1380 possibly contributes to the invasion of the host cell by V. parahaemolyticus. However, its direct role in the pathogenicity of V. parahaemolyticus has not been fully recognized.[32]
TYPE VI SECRETION SYSTEM
Of Gram-negative bacterial secretion systems, the latest to be described is the T6SS. It is a complex molecular machine that utilizes a bacteriophage-like cell-puncturing device to inject effector proteins into target cells.[54] Homologs of V. parahaemolyticus T6SS are found in a range of Gram-negative bacteria. During the infection of those cells, T6SS is predicted to take part in actin cross-linking, intracellular trafficking, secretion and vesicular transport.[55] V. parahaemolyticus holds two sets of putative T6SSs, one on each chromosome.[56] VPT6SS1 is mainly associated with clinical isolates, while all strains of V. parahaemolyticus encode VPT6SS2. Both systems have different aspects of adherence to Caco-2 and/or Hela cells.[56] Furthermore, there is recent evidence of a significant role of T6SS of V. parahaemolyticus in inducing autophagy in macrophages.[57]
Spread of Vibrio parahaemolyticus and emergence of pandemic clone
During 1950, Japan witnessed an outbreak of acute gastroenteritis, in which 272 people were infected and 20 died. An investigation of the leading cause, held by Fujino et al., resulted in the first isolation of the Gram-negative rods presently known as V. parahaemolyticus.[58] Some of the earliest outbreaks caused by V. parahaemolyticus were reported in the United States and Europe [Figure 1].[59,60] The occurrence of V. parahaemolyticus cases had a typical sporadic manner, with no clear association between distinct serotypes of V. parahaemolyticus and gastroenteritis incidence.[61] This situation remained until 1996, during which a surveillance of V. parahaemolyticus gastroenteritis incidence in Kolkata, India, witnessed a sudden surge due to the new unique serotype, O3:K6.[62] Since then, cases of gastroenteritis caused by O3:K6 V. parahaemolyticus have been reported worldwide: in Africa,[63] Asia,[64] Europe[65] and the American continent [Figure 1].[66] The rapid spread of O3:K6 marked the first pandemic of V. parahaemolyticus and placed this pathogen on the global public health agenda.[3] Matsumoto et al.[67] exploited the intraspecies variation of the toxRS sequence to develop a group-specific polymerase chain reaction method that permits the confirmation of the clonality of the new O3:K6 strains. During this period, emerging strains that belong to different serovars were found to be almost indistinguishable from the new O3:K6 clone.[67,68] By 2007, 21 different serotypes of V. parahaemolyticus appeared to have identical genotypes and molecular profiles to those of O3:K6 and were collectively entitled as “serovariants” of O3:K6 isolates, in which the most common serotypes were O4:K68, O1:K25 and O1:KUT (untypeable).[67,69] The understanding of the genetic diversity of V. parahaemolyticus was enhanced by the establishment of a multilocus sequence typing (MLST) scheme.[70] It is the elected method for determining the global epidemiology of bacterial pathogens based on sequence analysis of chosen housekeeping genes.[70] After gathering data from the pubMLST database and different studies, Han et al.[71] generated a comprehensive overview of the spread of clinical and environmental pandemic V. parahaemolyticus strains [Table 2]. They concluded that 49 serotypes, widely distributed in 22 countries, represent the pandemic isolates. The comparison between the genetic organizations of O3:K6 and other serotypes of V. parahaemolyticus disclose the complexity of the pandemic clones.[3] One possible explanation for the widespread of O3:K6 is the acquisition of open reading frame 8 (orf8) through phage f237 infection.[110] It was suggested that O3:K6 strains have a higher epidemic potency because the protein product of orf8 caused them to be more adhesive to host intestine cells.[110] Although orf8 appears to be a suitable genetic marker for the identification of pandemic clones, its reliability became debatable after findings of O3:K6 strains lacking f237 were reported.[111,112] Soon after that, the gene sequence VP2905 was proposed as an alternative genetic marker exclusive to the pandemic clones.[113] The gene is located in a 16-kb region inserted in the open reading frame of the histone-like DNA-binding protein HU-α, causing a frameshift in the amino acid sequence. However, its role in the pathogenicity of V. parahaemolyticus is yet undecided.[113,114]
Figure 1.
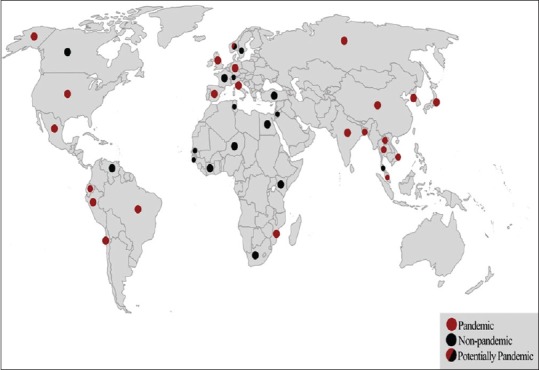
World map presenting the global dissemination of V. parahaemolyticus (illustrative representation of Table 2)
Table 2.
Overview of the global clinical and environmental distribution of Vibrio parahaemolyticus
| Continent | Country | Source(s) | Pandemic serotypes | Reference(s) |
|---|---|---|---|---|
| Asia | Japan | Clinical and environmental | Yes | [72] |
| Korea | Clinical and environmental | Yes | [71,73] | |
| China | Clinical and environmental | Yes | [64] | |
| Vietnam | Clinical and environmental | Yes | [71,74] | |
| Laos | Clinical and environmental | Yes | [71,72] | |
| Indonesia | Clinical and environmental | Yes | [69] | |
| Malaysia | Clinical and environmental | No | [75] | |
| Singapore | Clinical and environmental | Yes | [3] | |
| Thailand | Clinical and environmental | Yes | [76,77] | |
| India | Clinical and environmental | Yes | [78,79] | |
| Bangladesh | Clinical and environmental | Yes | [80] | |
| Jordan | Environmental | No | [81] | |
| Europe | Russia | Clinical and environmental | Yes | [82] |
| Sweden | Environmental | No | [83] | |
| Norway | Environmental | Potentially yes | [84] | |
| United Kingdom | Clinical and environmental | Yes | [85,86] | |
| Spain | Clinical and environmental | Yes | [87,88] | |
| France | Clinical and environmental | Yes | [89,90] | |
| Italy | Clinical and environmental | Yes | [46,91] | |
| Switzerland | Environmental | No | [92] | |
| Germany | Environmental | No | [93] | |
| Turkey (Black Sea) | Environmental | No | [94] | |
| Africa | Egypt | Environmental | No | [95] |
| Tunisia | Environmental | No | [96] | |
| Nigeria | Environmental | No | [97] | |
| Kenya | Environmental | No | [98] | |
| Mozambique | Clinical and environmental | Yes | [63] | |
| Côte d’Ivoire | Environmental | No | [99] | |
| Senegal | Environmental | No | [100] | |
| South Africa | Environmental | No | [101] | |
| Guinea-Bissau | Environmental | No | [102] | |
| America | Canada | Clinical and environmental | No | [103] |
| United States | Clinical and environmental | Yes | [104,105] | |
| Mexico | Clinical and environmental | Yes | [66] | |
| Ecuador | Clinical and environmental | Yes | [71,106] | |
| Brazil | Clinical and environmental | Yes | [107] | |
| Peru | Clinical and environmental | Yes | [108] | |
| Chile | Clinical and environmental | Yes | [109] |
CONCLUSIONS AND RECOMMENDATIONS
Studying the virulence determinants used by V. parahaemolyticus reveals a complex combination of genes orchestrating the host–pathogen interactions. However, the exact mechanism by which the infection is initiated is yet unknown. Therefore, further research is required to fill gaps in the literature. This is especially emphasized because V. parahaemolyticus may act as a reservoir from which genes may be transferred to other bacteria. In addition, the transient nature of the gastroenteritis infection caused by V. parahaemolyticus masks the true burden of this pathogen. However, reviewing the emergence of the pandemic clone and its ability to cause large outbreaks highlights the significance of V. parahaemolyticus and its impact on the population health as well as calls for the systematic monitoring of its existence and potential pathogenicity in the region.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.van Doorn HR. Emerging infectious diseases. Medicine. 2014;42:60–3. doi: 10.1016/j.mpmed.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Igbinosa EO, Okoh AI. Emerging Vibrio species: An unending threat to public health in developing countries. Res Microbiol. 2008;159:495–506. doi: 10.1016/j.resmic.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Ceccarelli D, Hasan NA, Huq A, Colwell RR. Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front Cell Infect Microbiol. 2013;3:97. doi: 10.3389/fcimb.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabbro C, Cataletto B, Del Negro P. Detection of pathogenic Vibrio parahaemolyticus through biochemical and molecular-based methodologies in coastal waters of the Gulf of Trieste (North Adriatic Sea) FEMS Microbiol Lett. 2010;307:158–64. doi: 10.1111/j.1574-6968.2010.01969.x. [DOI] [PubMed] [Google Scholar]
- 5.Letchumanan V, Chan KG, Lee LH. Vibrio parahaemolyticus: A review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol. 2014;5:705. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ham H, Orth K. The role of type III secretion system 2 in Vibrio parahaemolyticus pathogenicity. J Microbiol. 2012;50:719–25. doi: 10.1007/s12275-012-2550-2. [DOI] [PubMed] [Google Scholar]
- 7.Levin RE. Vibrio Parahaemolyticus, a notably lethal human pathogen derived from seafood: A review of its pathogenicity, characteristics, subspecies characterization, and molecular methods of detection. Food Biotechnol. 2006;20:93–128. [Google Scholar]
- 8.Broberg CA, Calder TJ, Orth K. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 2011;13:992–1001. doi: 10.1016/j.micinf.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tena D, Arias M, Alvarez BT, Mauleón C, Jiménez MP, Bisquert J. Fulminant necrotizing fasciitis due to Vibrio Parahaemolyticus. J Med Microbiol. 2010;59(Pt 2):235–8. doi: 10.1099/jmm.0.014654-0. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad A, Brumble L, Maniaci M. Vibrio Parahaemolyticus induced necrotizing fasciitis: An atypical organism causing an unusual presentation. Case Rep Infect Dis. 2013;2013:216854. doi: 10.1155/2013/216854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Orth K. Virulence determinants for Vibrio parahaemolyticus infection. Curr Opin Microbiol. 2013;16:70–7. doi: 10.1016/j.mib.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Nelapati S, Nelapati K, Chinnam BK. Vibrio parahaemolyticu s – An emerging foodborne pathogen – A review. Vet World. 2012;5:48. [Google Scholar]
- 13.Klein SL, Gutierrez West CK, Mejia DM, Lovell CR. Genes similar to the Vibrio parahaemolyticus virulence-related genes tdh, tlh, and vscC2 occur in other vibrionaceae species isolated from a pristine estuary. Appl Environ Microbiol. 2014;80:595–602. doi: 10.1128/AEM.02895-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canizalez-Roman A, Flores-Villaseñor H, Zazueta-Beltran J, Muro-Amador S, León-Sicairos N. Comparative evaluation of a chromogenic agar medium-PCR protocol with a conventional method for isolation of Vibrio parahaemolyticus strains from environmental and clinical samples. Can J Microbiol. 2011;57:136–42. doi: 10.1139/w10-108. [DOI] [PubMed] [Google Scholar]
- 15.Henke JM, Bassler BL. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol. 2004;186:3794–805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kernell Burke A, Guthrie LT, Modise T, Cormier G, Jensen RV, McCarter LL, et al. OpaR controls a network of downstream transcription factors in Vibrio parahaemolyticus BB22OP. PLoS One. 2015;10:e0121863. doi: 10.1371/journal.pone.0121863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krachler AM, Ham H, Orth K. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. Proc Natl Acad Sci U S A. 2011;108:11614–9. doi: 10.1073/pnas.1102360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stones DH, Krachler AM. Dual function of a bacterial protein as an adhesin and extracellular effector of host GTPase signaling. Small GTPases. 2015;6:153–6. doi: 10.1080/21541248.2015.1028609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim J, Stones DH, Hawley CA, Watson CA, Krachler AM. Multivalent adhesion molecule 7 clusters act as signaling platform for host cellular GTPase activation and facilitate epithelial barrier dysfunction. PLoS Pathog. 2014;10:e1004421. doi: 10.1371/journal.ppat.1004421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kustusch RJ, Kuehl CJ, Crosa JH. Power plays: Iron transport and energy transduction in pathogenic vibrios. Biometals. 2011;24:559–66. doi: 10.1007/s10534-011-9437-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.León-Sicairos N, Angulo-Zamudio UA, de la Garza M, Velázquez-Román J, Flores-Villaseñor HM, Canizalez-Román A. Strategies of Vibrio parahaemolyticus to acquire nutritional iron during host colonization. Front Microbiol. 2015;6:702. doi: 10.3389/fmicb.2015.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Malley SM, Mouton SL, Occhino DA, Deanda MT, Rashidi JR, Fuson KL, et al. Comparison of the heme iron utilization systems of pathogenic Vibrios. J Bacteriol. 1999;181:3594–8. doi: 10.1128/jb.181.11.3594-3598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanabe T, Funahashi T, Nakao H, Miyoshi S, Shinoda S, Yamamoto S. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J Bacteriol. 2003;185:6938–49. doi: 10.1128/JB.185.23.6938-6949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishibuchi M, Fasano A, Russell RG, Kaper JB. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun. 1992;60:3539–45. doi: 10.1128/iai.60.9.3539-3545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honda T, Ni YX, Miwatani T. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect Immun. 1988;56:961–5. doi: 10.1128/iai.56.4.961-965.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bechlars S, Wüstenhagen DA, Drägert K, Dieckmann R, Strauch E, Kubick S. Cell-free synthesis of functional thermostable direct hemolysins of Vibrio parahaemolyticus. Toxicon. 2013;76:132–42. doi: 10.1016/j.toxicon.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Shimohata T, Takahashi A. Diarrhea induced by infection of Vibrio parahaemolyticus. J Med Invest. 2010;57:179–82. doi: 10.2152/jmi.57.179. [DOI] [PubMed] [Google Scholar]
- 28.Kodama T, Hiyoshi H, Okada R, Matsuda S, Gotoh K, Iida T. Regulation of Vibrio parahaemolyticus T3SS2 gene expression and function of T3SS2 effectors that modulate actin cytoskeleton. Cell Microbiol. 2015;17:183–90. doi: 10.1111/cmi.12408. [DOI] [PubMed] [Google Scholar]
- 29.Raghunath P. Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front Microbiol. 2014;5:805. doi: 10.3389/fmicb.2014.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi A, Yamamoto C, Kodama T, Yamashita K, Harada N, Nakano M, et al. Pore formation of thermostable direct hemolysin secreted from Vibrio parahaemolyticus in lipid bilayers. Int J Toxicol. 2006;25:409–18. doi: 10.1080/10915810600868181. [DOI] [PubMed] [Google Scholar]
- 31.Hardy SP, Nakano M, Iida T. Single channel evidence for innate pore-formation by Vibrio parahaemolyticus thermostable direct haemolysin (TDH) in phospholipid bilayers. FEMS Microbiol Lett. 2004;240:81–5. doi: 10.1016/j.femsle.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 32.Wang R, Zhong Y, Gu X, Yuan J, Saeed AF, Wang S. Corrigendum: The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front Microbiol. 2015;6:437. doi: 10.3389/fmicb.2015.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Tang X, Zahan W. Cloning, expressing, and hemolysis of tdh, trh and tlh genes of Vibrio parahaemolyticus. J Ocean Univ China. 2011;10:275–9. [Google Scholar]
- 34.Saito S, Iwade Y, Tokuoka E, Nishio T, Otomo Y, Araki E, et al. Epidemiological evidence of lesser role of thermostable direct hemolysin (TDH)-related hemolysin (TRH) than TDH on Vibrio parahaemolyticus pathogenicity. Foodborne Pathog Dis. 2015;12:131–8. doi: 10.1089/fpd.2014.1810. [DOI] [PubMed] [Google Scholar]
- 35.Gotoh K, Kodama T, Hiyoshi H, Izutsu K, Park KS, Dryselius R, et al. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS One. 2010;5:e13365. doi: 10.1371/journal.pone.0013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiyoshi H, Kodama T, Saito K, Gotoh K, Matsuda S, Akeda Y, et al. VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe. 2011;10:401–9. doi: 10.1016/j.chom.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 37.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, et al. Genome sequence of Vibrio parahaemolyticus: A pathogenic mechanism distinct from that of V.cholerae. Lancet. 2003;361:743–9. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 38.Kodama T, Yamazaki C, Park KS, Akeda Y, Iida T, Honda T. Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiol Lett. 2010;311:10–7. doi: 10.1111/j.1574-6968.2010.02066.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhou X, Konkel ME, Call DR. Regulation of type III secretion system 1 gene expression in Vibrio parahaemolyticus is dependent on interactions between ExsA, ExsC, and ExsD. Virulence. 2010;1:260–72. doi: 10.4161/viru.1.4.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burdette DL, Seemann J, Orth K. Vibrio VopQ induces PI3-kinase-independent autophagy and antagonizes phagocytosis. Mol Microbiol. 2009;73:639–49. doi: 10.1111/j.1365-2958.2009.06798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sreelatha A, Bennett TL, Zheng H, Jiang QX, Orth K, Starai VJ. Vibrio effector protein, VopQ, forms a lysosomal gated channel that disrupts host ion homeostasis and autophagic flux. Proc Natl Acad Sci U S A. 2013;110:11559–64. doi: 10.1073/pnas.1307032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimohata T, Nakano M, Lian X, Shigeyama T, Iba H, Hamamoto A, et al. Vibrio parahaemolyticus infection induces modulation of IL-8 secretion through dual pathway via VP1680 in Caco-2 cells. J Infect Dis. 2011;203:537–44. doi: 10.1093/infdis/jiq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–72. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- 44.Trosky JE, Mukherjee S, Burdette DL, Roberts M, McCarter L, Siegel RM, et al. Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J Biol Chem. 2004;279:51953–7. doi: 10.1074/jbc.M407001200. [DOI] [PubMed] [Google Scholar]
- 45.Trosky JE, Li Y, Mukherjee S, Keitany G, Ball H, Orth K. VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J Biol Chem. 2007;282:34299–305. doi: 10.1074/jbc.M706970200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Krachler AM, Broberg CA, Li Y, Mirzaei H, Gilpin CJ, et al. Type III effector VopC mediates invasion for Vibrio species. Cell Rep. 2012;1:453–60. doi: 10.1016/j.celrep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kodama T, Rokuda M, Park KS, Cantarelli VV, Matsuda S, Iida T, et al. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol. 2007;9:2598–609. doi: 10.1111/j.1462-5822.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhou X, Gewurz BE, Ritchie JM, Takasaki K, Greenfeld H, Kieff E, et al. A Vibrio parahaemolyticus T3SS effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting TAK1 activation. Cell Rep. 2013;3:1690–702. doi: 10.1016/j.celrep.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higa N, Toma C, Koizumi Y, Nakasone N, Nohara T, Masumoto J, et al. Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLoS Pathog. 2013;9:e1003142. doi: 10.1371/journal.ppat.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiyoshi H, Kodama T, Iida T, Honda T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect Immun. 2010;78:1772–80. doi: 10.1128/IAI.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caburlotto G, Haley BJ, Lleò MM, Huq A, Colwell RR. Serodiversity and ecological distribution of Vibrio parahaemolyticus in the Venetian Lagoon, Northeast Italy. Environ Microbiol Rep. 2010;2:151–7. doi: 10.1111/j.1758-2229.2009.00123.x. [DOI] [PubMed] [Google Scholar]
- 52.Yu B, Cheng HC, Brautigam CA, Tomchick DR, Rosen MK. Mechanism of actin filament nucleation by the bacterial effector VopL. Nat Struct Mol Biol. 2011;18:1068–74. doi: 10.1038/nsmb.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calder T, Kinch LN, Fernandez J, Salomon D, Grishin NV, Orth K. Vibrio type III effector VPA1380 is related to the cysteine protease domain of large bacterial toxins. PLoS One. 2014;9:e104387. doi: 10.1371/journal.pone.0104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coulthurst SJ. The Type VI secretion system – A widespread and versatile cell targeting system. Res Microbiol. 2013;164:640–54. doi: 10.1016/j.resmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 55.Boyd EF, Cohen AL, Naughton LM, Ussery DW, Binnewies TT, Stine OC, et al. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 2008;8:110. doi: 10.1186/1471-2180-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Y, Yang H, Li J, Zhang P, Wu B, Zhu B, et al. Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch Microbiol. 2012;194:827–35. doi: 10.1007/s00203-012-0816-z. [DOI] [PubMed] [Google Scholar]
- 57.Yu Y, Fang L, Zhang Y, Sheng H, Fang W. VgrG2 of type VI secretion system 2 of Vibrio parahaemolyticus induces autophagy in macrophages. Front Microbiol. 2015;6:168. doi: 10.3389/fmicb.2015.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujino T, Okuno Y, Nakada D, Aoyama A, Mukai T, Ueho T. On the bacteriological examination of shirasu food poisoning. Med J Osaka Univ. 1953;4:299–304. [Google Scholar]
- 59.Molenda JR, Johnson WG, Fishbein M, Wentz B, Mehlman IJ, Dadisman TA., Jr Vibrio parahaemolyticus gastroenteritis in Maryland: Laboratory aspects. Appl Microbiol. 1972;24:444–8. doi: 10.1128/am.24.3.444-448.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hooper WL, Barrow GI, McNab DJ. Vibrio parahaemolyticus food-poisoning in Britain. Lancet. 1974;1:1100–2. doi: 10.1016/s0140-6736(74)90570-4. [DOI] [PubMed] [Google Scholar]
- 61.de Jesús Hernández-Díaz L, Leon-Sicairos N, Velazquez-Roman J, Flores-Villaseñor H, Guadron-Llanos AM, Martinez-Garcia JJ, et al. A pandemic Vibrio parahaemolyticus O3:K6 clone causing most associated diarrhea cases in the Pacific Northwest coast of Mexico. Front Microbiol. 2015;6:221. doi: 10.3389/fmicb.2015.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay AK, et al. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–5. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ansaruzzaman M, Chowdhury A, Bhuiyan NA, Sultana M, Safa A, Lucas M, et al. Characteristics of a pandemic clone of O3: K6 and O4: K68 Vibrio parahaemolyticus isolated in Beira, Mozambique. J Med Microbiol. 2008;57(Pt 12):1502–7. doi: 10.1099/jmm.0.2008/004275-0. [DOI] [PubMed] [Google Scholar]
- 64.Li J, Xue F, Yang Z, Zhang X, Zeng D, Chao G, et al. Vibrio parahaemolyticus strains of pandemic serotypes identified from clinical and environmental samples from Jiangsu, China. Front Microbiol. 2016;7:787. doi: 10.3389/fmicb.2016.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez-Urtaza J, Simental L, Velasco D, DePaola A, Ishibashi M, Nakaguchi Y, et al. Pandemic Vibrio parahaemolyticus O3:K6, Europe. Emerg Infect Dis. 2005;11:1319–20. doi: 10.3201/eid1108.050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Velazquez-Roman J, León-Sicairos N, de Jesus Hernández-Díaz L, Canizalez-Roman A. Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front Cell Infect Microbiol. 2014;3:110. doi: 10.3389/fcimb.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, et al. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol. 2000;38:578–85. doi: 10.1128/jcm.38.2.578-585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chowdhury NR, Chakraborty S, Ramamurthy T, Nishibuchi M, Yamasaki S, Takeda Y, et al. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg Infect Dis. 2000;6:631–6. doi: 10.3201/eid0606.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20:39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.González-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, DePaola A. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol. 2008;190:2831–40. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han C, Tang H, Ren C, Zhu X, Han D. Sero-prevalence and genetic diversity of pandemic V. parahaemolyticus strains occurring at a global scale. Front Microbiol. 2016;7:567. doi: 10.3389/fmicb.2016.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hara-Kudo Y, Kumagai S. Impact of seafood regulations for Vibrio parahaemolyticus infection and verification by analyses of seafood contamination and infection. Epidemiol Infect. 2014;142:2237–47. doi: 10.1017/S0950268814001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang CH, Shin Y, Kim W, Kim Y, Song K, Oh EG, et al. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from oysters in Korea. Environ Sci Pollut Res Int. 2016;23:918–26. doi: 10.1007/s11356-015-5650-9. [DOI] [PubMed] [Google Scholar]
- 74.Tai DT, Vinh Thuy A, Nhi NT, Ngoc NT, Lan NT. Virulence and antimicrobial resistance characteristics of Vibrio parahaemolyticus isolated from environment, food and clinical samples in the south of Vietnam, 2010. BMC Proc. 2011;5:P94. [Google Scholar]
- 75.Letchumanan V, Yin WF, Lee LH, Chan KG. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front Microbiol. 2015;6:33. doi: 10.3389/fmicb.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thongjun J, Mittraparp-Arthorn P, Yingkajorn M, Kongreung J, Nishibuchi M, Vuddhakul V. The trend of Vibrio parahaemolyticus infections in Southern Thailand from 2006 to 2010. Trop Med Health. 2013;41:151–6. doi: 10.2149/tmh.2013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kongrueng J, Yingkajorn M, Bunpa S, Sermwittayawong N, Singkhamanan K, Vuddhakul V. Characterization of Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease in Southern Thailand. J Fish Dis. 2015;38:957–66. doi: 10.1111/jfd.12308. [DOI] [PubMed] [Google Scholar]
- 78.Alagappan KM, Deivasigamani B, Somasundaram ST, Kumaran S. Occurrence of Vibrio parahaemolyticus and its specific phages from shrimp ponds in east coast of India. Curr Microbiol. 2010;61:235–40. doi: 10.1007/s00284-010-9599-0. [DOI] [PubMed] [Google Scholar]
- 79.Pazhani GP, Bhowmik SK, Ghosh S, Guin S, Dutta S, Rajendran K, et al. Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl Trop Dis. 2014;8:e2815. doi: 10.1371/journal.pntd.0002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akther F, Neogi SB, Chowdhury WB, Sadique A, Islam A, Akhter MZ, et al. Major tdh(+) Vibrio parahaemolyticus serotype changes temporally in the Bay of Bengal estuary of Bangladesh. Infect Genet Evol. 2016;41:153–9. doi: 10.1016/j.meegid.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 81.Alaboudi AR, Ababneh M, Osaili TM, Al Shloul K. Detection, identification, and prevalence of pathogenic Vibrio parahaemolyticus in fish and coastal environment in Jordan. J Food Sci. 2016;81:M130–4. doi: 10.1111/1750-3841.13151. [DOI] [PubMed] [Google Scholar]
- 82.Rykovskaia OA, Mazrukho AV, Smolikova LM, Monakhova EV, Chemisova OS, Podoinitsyna OA, et al. O3:K6 serogroup Vibrio parahaemolyticus – The causative agent of food toxic infection outbreaks in Primorsky region of Russian federation. Zh Mikrobiol Epidemiol Immunobiol. 2014;4:57–61. [PubMed] [Google Scholar]
- 83.Collin B, Rehnstam-Holm AS. Occurrence and potential pathogenesis of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus on the South Coast of Sweden. FEMS Microbiol Ecol. 2011;78:306–13. doi: 10.1111/j.1574-6941.2011.01157.x. [DOI] [PubMed] [Google Scholar]
- 84.Ellingsen AB, Olsen JS, Granum PE, Rørvik LM, González-Escalona N. Genetic characterization of trh positive Vibrio spp. isolated from Norway. Front Cell Infect Microbiol. 2013;3:107. doi: 10.3389/fcimb.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wagley S, Koofhethile K, Wing JB, Rangdale R. Comparison of V. parahaemolyticus isolated from seafoods and cases of gastrointestinal disease in the UK. Int J Environ Health Res. 2008;18:283–93. doi: 10.1080/09603120801911064. [DOI] [PubMed] [Google Scholar]
- 86.Powell A, Baker-Austin C, Wagley S, Bayley A, Hartnell R. Isolation of pandemic Vibrio parahaemolyticus from UK water and shellfish produce. Microb Ecol. 2013;65:924–7. doi: 10.1007/s00248-013-0201-8. [DOI] [PubMed] [Google Scholar]
- 87.Rodriguez-Castro A, Ansede-Bermejo J, Blanco-Abad V, Varela-Pet J, Garcia-Martin O, Martinez-Urtaza J. Prevalence and genetic diversity of pathogenic populations of Vibrio parahaemolyticus in coastal waters of Galicia, Spain. Environ Microbiol Rep. 2010;2:58–66. doi: 10.1111/j.1758-2229.2009.00064.x. [DOI] [PubMed] [Google Scholar]
- 88.Martinez-Urtaza J, Powell A, Jansa J, Rey JL, Montero OP, Campello MG, et al. Epidemiological investigation of a foodborne outbreak in Spain associated with US West Coast genotypes of Vibrio parahaemolyticus. Springerplus. 2016;5:87. doi: 10.1186/s40064-016-1728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quilici ML, Robert-Pillot A, Picart J, Fournier JM. Pandemic Vibrio parahaemolyticus O3:K6 spread, France. Emerg Infect Dis. 2005;11:1148–9. doi: 10.3201/eid1107.041008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robert-Pillot A, Copin S, Himber C, Gay M, Quilici ML. Occurrence of the three major Vibrio species pathogenic for human in seafood products consumed in France using real-time PCR. Int J Food Microbiol. 2014;189:75–81. doi: 10.1016/j.ijfoodmicro.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 91.Ottaviani D, Leoni F, Rocchegiani E, Santarelli S, Canonico C, Masini L, et al. First clinical report of pandemic Vibrio parahaemolyticus O3:K6 infection in Italy. J Clin Microbiol. 2008;46:2144–5. doi: 10.1128/JCM.00683-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schärer K, Savioz S, Cernela N, Saegesser G, Stephan R. Occurrence of Vibrio spp. in fish and shellfish collected from the Swiss market. J Food Prot. 2011;74:1345–7. doi: 10.4315/0362-028X.JFP-11-001. [DOI] [PubMed] [Google Scholar]
- 93.Huehn S, Eichhorn C, Urmersbach S, Breidenbach J, Bechlars S, Bier N, et al. Pathogenic vibrios in environmental, seafood and clinical sources in Germany. Int J Med Microbiol. 2014;304:843–50. doi: 10.1016/j.ijmm.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 94.Terzi Gulel G, Martinez-Urtaza J. Molecular characterizations of Vibrio parahaemolyticus in seafood from the Black Sea, Turkey. Lett Appl Microbiol. 2016;62:494–500. doi: 10.1111/lam.12579. [DOI] [PubMed] [Google Scholar]
- 95.Abd-Elghany SM, Sallam KI. Occurrence and molecular identification of Vibrio parahaemolyticus in retail shellfish in Mansoura, Egypt. Food Control. 2013;33:399–405. [Google Scholar]
- 96.Khouadja S, Suffredini E, Spagnoletti M, Croci L, Colombo MM, Amina B. Presence of pathogenic Vibrio parahaemolyticus in waters and seafood from the Tunisian Sea. World J Microbiol Biotechnol. 2013;29:1341–8. doi: 10.1007/s11274-013-1297-1. [DOI] [PubMed] [Google Scholar]
- 97.Eja ME, Abriba C, Etok CA, Ikpeme EM, Arikpo GE, Enyi-Idoh KH, et al. Seasonal occurrence of vibrios in water and shellfish obtained from the Great Kwa River estuary, Calabar, Nigeria. Bull Environ Contam Toxicol. 2008;81:245–8. doi: 10.1007/s00128-008-9482-x. [DOI] [PubMed] [Google Scholar]
- 98.Kagiko MM, Damiano WA, Kayihura MM. Characterisation of Vibrio parahaemolyticus isolated from fish in Kenya. East Afr Med J. 2001;78:124–7. doi: 10.4314/eamj.v78i3.9076. [DOI] [PubMed] [Google Scholar]
- 99.Traoré SG, Bonfoh B, Krabi R, Odermatt P, Utzinger J, Rose KN, et al. Risk of Vibrio transmission linked to the consumption of crustaceans in coastal towns of Côte d'Ivoire. J Food Prot. 2012;75:1004–11. doi: 10.4315/0362-028X.JFP-11-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coly I, Sow AG, Seydi M, Martinez-Urtaza J. Vibrio cholerae and Vibrio parahaemolyticus detected in seafood products from Senegal. Foodborne Pathog Dis. 2013;10:1050–8. doi: 10.1089/fpd.2013.1523. [DOI] [PubMed] [Google Scholar]
- 101.Okoh AI, Sibanda T, Nongogo V, Adefisoye M, Olayemi OO, Nontongana N. Prevalence and characterisation of non-cholerae Vibrio spp. in final effluents of wastewater treatment facilities in two districts of the Eastern Cape Province of South Africa: Implications for public health. Environ Sci Pollut Res Int. 2015;22:2008–17. doi: 10.1007/s11356-014-3461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Machado A, Bordalo AA. Detection and quantification of Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus in Coastal Waters of Guinea-Bissau (West Africa) Ecohealth. 2016;13:339–49. doi: 10.1007/s10393-016-1104-1. [DOI] [PubMed] [Google Scholar]
- 103.Khaira G, Galanis E. Descriptive epidemiology of Vibrio parahaemolyticus and other Vibrio species infections in British Columbia: 2001-2006. Can Commun Dis Rep. 2007;33:12–22. [PubMed] [Google Scholar]
- 104.Gutierrez West CK, Klein SL, Lovell CR. High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl Environ Microbiol. 2013;79:2247–52. doi: 10.1128/AEM.03792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haendiges J, Timme R, Allard MW, Myers RA, Brown EW, Gonzalez-Escalona N. Characterization of Vibrio parahaemolyticus clinical strains from Maryland (2012-2013) and comparisons to a locally and globally diverse V. parahaemolyticus strains by whole-genome sequence analysis. Front Microbiol. 2015;6:125. doi: 10.3389/fmicb.2015.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sperling L, Alter T, Huehn S. Prevalence and antimicrobial resistance of Vibrio spp. in retail and farm shrimps in Ecuador. J Food Prot. 2015;78:2089–92. doi: 10.4315/0362-028X.JFP-15-160. [DOI] [PubMed] [Google Scholar]
- 107.Raszl SM, Froelich BA, Vieira CR, Blackwood AD, Noble RT. Vibrio parahaemolyticus and Vibrio vulnificus in South America: Water, seafood and human infections. J Appl Microbiol. 2016;121:1201–22. doi: 10.1111/jam.13246. [DOI] [PubMed] [Google Scholar]
- 108.Gil AI, Miranda H, Lanata CF, Prada A, Hall ER, Barreno CM, et al. O3:K6 serotype of Vibrio parahaemolyticus identical to the global pandemic clone associated with diarrhea in Peru. Int J Infect Dis. 2007;11:324–8. doi: 10.1016/j.ijid.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 109.Fuenzalida L, Hernández C, Toro J, Rioseco ML, Romero J, Espejo RT. Vibrio parahaemolyticus in shellfish and clinical samples during two large epidemics of diarrhoea in southern Chile. Environ Microbiol. 2006;8:675–83. doi: 10.1111/j.1462-2920.2005.00946.x. [DOI] [PubMed] [Google Scholar]
- 110.Nasu H, Iida T, Sugahara T, Yamaichi Y, Park KS, Yokoyama K, et al. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J Clin Microbiol. 2000;38:2156–61. doi: 10.1128/jcm.38.6.2156-2161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bhuiyan NA, Ansaruzzaman M, Kamruzzaman M, Alam K, Chowdhury NR, Nishibuchi M, et al. Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J Clin Microbiol. 2002;40:284–6. doi: 10.1128/JCM.40.1.284-286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Osawa R, Iguchi A, Arakawa E, Watanabe H. Genotyping of pandemic Vibrio parahaemolyticus O3:K6 still open to question. J Clin Microbiol. 2002;40:2708–9. doi: 10.1128/JCM.40.7.2708-2709.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Williams TL, Musser SM, Nordstrom JL, DePaola A, Monday SR. Identification of a protein biomarker unique to the pandemic O3:K6 clone of Vibrio parahaemolyticus. J Clin Microbiol. 2004;42:1657–65. doi: 10.1128/JCM.42.4.1657-1665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Okura M, Osawa R, Arakawa E, Terajima J, Watanabe H. Identification of Vibrio parahaemolyticus pandemic group-specific DNA sequence by genomic subtraction. J Clin Microbiol. 2005;43:3533–6. doi: 10.1128/JCM.43.7.3533-3536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


