Abstract
Purpose
Centromere protein U (CENPU) abnormally exhibits high expression in various types of human tumor tissues and participates in tumor progression; however, its expression pattern and biological function in lung cancer have not yet been elucidated. In the present study, we explored the clinical significance and biological function of CENPU in lung cancer.
Materials and methods
The Cancer Genome Atlas (TCGA) data analyses, quantitative real-time PCR (RT-PCR), and Western blotting were performed to quantify CENPU and FOXM1 expression in non-small-cell lung cancer (NSCLC) samples. Survival data were obtained from Kaplan–Meier plotter or PROGgene V2 prognostic database. The function of CENPU in lung cancer cell proliferation was determined using 5-ethynyl-2′-deoxyuridine (EdU), Cell Counting Kit-8 (CCK-8), and cell cycle assays, and the underlying mechanism was determined through bioinformatic analyses and validated by in vitro siRNA or plasmid transfection experiments.
Results
CENPU was abnormally overexpressed in NSCLC samples compared with matched paired normal tissues. Higher expression of CENPU predicted worse overall survival (OS) and relapse-free survival (RFS) in NSCLC patients. Knockdown of CENPU expression by siRNA significantly inhibited proliferation and delayed cell cycle progression of lung cancer cells. To figure out the mechanism, bioinformatic analyses were performed and the results showed that the transcription factor, FOXM1, positively correlated with CENPU. Further in vitro experiments indicated that FOXM1 was the possible downstream transcription factor of CENPU as the knockdown of CENPU led to lower expression of FOXM1 and the overexpression of FOXM1 significantly reversed the inhibition of proliferation caused by CENPU knockdown. Furthermore, FOXM1 was highly expressed in NSCLC. The knockdown of FOXM1 also attenuated proliferation and induced G1 arrest in lung cancer cells.
Conclusion
CENPU was highly expressed in NSCLC tissues, wherein it promoted lung cancer cell proliferation via the transcription factor, FOXM1, which could be a potential target for therapeutic strategies.
Keywords: CENPU, NSCLC, proliferation, FOXM1
Introduction
Lung cancer remains the leading cause of cancer-related death in the world currently.1,2 Non-small-cell lung cancer (NSCLC) accounts for ~85% of lung cancers, and >50% of patients with NSCLC are initially diagnosed in advanced stages of the disease.3 Over the past decade, major progress has been made in the application of advanced medical technologies such as targeted therapies and immunotherapy to improve survival in patients with advanced NSCLC.4–6 Although the overall 5-year survival rate for lung cancer has increased slightly, it remains at <18%.1,2 Moreover, even for lung cancer patients diagnosed at an early stage, a higher recurrence rate cannot be ignored.7 Therefore, further elucidation of the molecular mechanism of lung cancer progression is important for exploring new effective therapeutic targets.
Centromere protein U (CENPU), also known as myelodysplasia/myeloid leukemia factor 1 interacting protein (MLF1IP),8 KHSV LNA-interacting protein 1 (KLIP1),9 or Cenp-5010 is a constitutive component of the centromere that is required for recovery from spindle damage. It also plays an important role in orchestrating kinetochore–microtubule attachment by interacting with Hec1.10-11 CENPU has been reported to repress herpes simplex virus thymidine kinase promoter activity, implying that it is also a transcriptional repressor.9 More recently, the upregulation of CENPU was found in several types of cancer including hereditary non-polyposis colorectal cancer (Lynch syndrome), glioblastoma (GBM), prostate cancer, breast cancer, bladder cancer, and ovarian cancer.12–17 Moreover, CENPU overexpression predicts poor prognosis in breast cancer, bladder cancer, and ovarian cancer.15–17 However, the role and mechanism of CENPU in the development of NSCLC have not yet been explored.
In the present study, we found that CENPU was signifi-cantly upregulated in human NSCLC tumor samples, which was consistent with the results of the Cancer Genome Atlas (TCGA) data analyses. Overexpression of CENPU was associated with shorter overall survival (OS) in NSCLC patients. Knockdown of CENPU in A549 and SPC-A1 NSCLC cell lines significantly inhibited cell proliferation and induced G1 phase arrest. Further research demonstrated that CENPU promoted proliferation in NSCLC cell lines through the transcription factor, FOXM1. In summary, it is suggested that CENPU promoted lung cancer progression partially through FOXM1, highlighting its potential as a new therapeutic target for lung cancer.
Materials and methods
Human NSCLC samples
Twenty human NSCLC tumor samples and paired normal lung tissues were obtained from consenting patients at the Southwest Hospital (Chongqing, China) between 2017 and 2018. None of the patients received anticancer treatment before operation. NSCLC samples included squamous cell carcinoma (eight cases) and adenocarcinoma (12 cases). All samples were frozen in liquid nitrogen. The study was performed after being approved by the research ethics boards at the Southwest Hospital, and we confirm that all patients included in this study provided written informed consent.
Cell culture
Human NSCLC cell lines, SPC-A1 and A549 (Chinese Academy of Sciences Shanghai Cell Bank, Shanghai, China), were cultured in Roswell Park Memorial Institute (RPMI) 1640 (Thermo Fisher Scientific, Waltham, MA, USA) and DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific), 100 IU/mL penicillin, and 100 mg/mL streptomycin (Thermo Fisher Scientific), respectively. All cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C.
RNA isolation and quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from tissues or cells using RNAiso Plus (TaKaRa Bio, Otsu, Japan) according to the manufacturer’s instructions. For cDNA synthesis, 1 µg of total RNA was reverse transcribed using the PrimeScript™ RT Reagent Kit (TaKaRa Bio) according to the manufacturer’s instructions. qRT-PCR was performed with SYBR Green Premix Ex Taq kit (Takara Bio) and the ABI 7500 Prism Sequence Detection System (Thermo Fisher Scientific). The amplification conditions were as follows: 95°C for 30 seconds, followed by 40 cycles of 95°C for 5 seconds and 60°C for 34 seconds. For normalization of all qRT-PCR data, β-actin expression was used as a reference gene. Primers used for qRT-PCR are summarized in Table 1.
Table 1.
Primers and siRNA sequences
| Targets | Sequences | |
|---|---|---|
| β-Actin | Forward | 5′-GAGCTACGAGCTGCCTGACG-3′ |
| Reverse | 5′-GTAGTTTCGTGGATGCCACAG-3′ | |
| CENPU | Forward | 5′-ATGAACTGCTTCGGTTAGAGC-3′ |
| Reverse | 5′-TATTTCGCAGATGGCTTTCGG-3′ | |
| FOXM1 | Forward | 5′-CGTCGGCCACTGATTCTCAAA-3′ |
| Reverse | 5′-GGCAGGGGATCTCTTAGGTTC-3′ | |
| siCENPU | Forward | 5′-CCACCUAGAGCAUCAACAATT-3′ |
| Reverse | 5′-UUGUUGAUGCUCUAGGUGGGT-3′ | |
| siFOXM1 | Forward | 5′-CACUAUCAACAAUAGCCUATT-3′ |
| Reverse | 5′-UAGGCUAUUGUUGAUAGUGTT-3′ | |
| NC | Forward | 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Reverse | 5′-ACGUGACACGUUCGGAGAATT-3′ | |
Abbreviations: CENPU, centromere protein U; FOXM1, Forkhead box protein M1; NC, negative control; siCENPU, CENPU siRNA; siFOXM1, FOXM1 siRNA.
Western blot analysis
Tissues and cells were lysed on ice for 10 minutes in RIPA lysis buffer (Beyotime, Jiangsu, China) including 1% pro tease inhibitor and phenylmethanesulfonyl fluoride (PMSF; Beyotime). The concentration of proteins was quantified using the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific). Next, 40 µg of total protein from each sample was separated using 10% polyacrylamide gels (Beyotime), which was followed by transfer to polyvinylidene fluoride (PVDF) membranes (Hoffman-La Roche Ltd., Basel, Switzerland). Membranes were subsequently blocked for 1 hour at room temperature with 5% BSA (Boster Biological Technology Co. Ltd., Wuhan, China). The PVDF membranes were, respectively, incubated overnight with mouse monoclonal anti-β-actin (dilution 1:1,000; BOSTER, BM0627), rabbit polyclonal anti-CENPU (dilution 1:500; LS-C158151; LifeSpan BioSciences, Seattle, WA, USA), and rabbit monoclonal anti-FOXM1 (1:500; 20459; Cell Signaling Technology, Danvers, MA, USA). After washing with tris buffered saline tween, membranes were probed with goat anti-rabbit IgG (dilution 1:5,000) or goat anti-mouse IgG (dilution 1:5,000) conjugated with horseradish peroxidase for 1 hour at room temperature. Bands were detected using BeyoECL Plus (Beyotime). Results are expressed relative to β-actin band density, which was used as a loading control.
Knockdown and overexpression treatment
Small interfering RNAs specific for CENPU and FOXM1, as well as negative control (NC) siRNA (siCENPU, siFOXM1, and NC), were chemically synthesized by GenePharma (Shanghai, China). CENPU expression plasmid and control plasmid were purchased from Genechem (Shanghai, China). FOXM1 expression plasmid and control plasmid were purchased from Vigene Biosciences (Shandong, China). The sequences of siCENPU, siFOXM1, and NC are summa-rized in Table 1. Cells were seeded into 24-well plates, and siRNA transfection was performed using the ribo FECT™ CP Transfection Kit (Ribobio, Shanghai, China) according to the manufacturer’s instructions. CENPU or FOXM1 expression plasmid transfection was performed using the Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cells were harvested and subjected to subsequent analysis 48 hours after transfection.
Cell Counting Kit-8 (CCK-8) assays
Cell proliferation was assessed using the CCK-8 (Dojindo, Kumamoto, Japan). SPC-A1 or A549 cells were seeded in 96-well plates at 3,000 cells/well. siCENPU, siFOXM1, or siNC was used for transfection. In addition, after 24 hours, 10 µL of CCK-8 solution was added. Cells were continuously incubated for 2 hours. The absorbance was measured using the iMARK (Bio-Rad Laboratories Inc., Hercules, CA, USA) microplate reader at 450 nm. Experiments were repeated three times under the same conditions.
5-Ethynyl-2′-deoxyuridine (EdU) assays
Differences in siCENPU- or siFOXM1-treated lung cancer cells were studied using the iClick™ EdU Andy Fluor™ 647 Flow Cytometry Assay Kit (GeneCopoeia, Rockville, MD, USA). Cells were seeded in 12-well plates at 1.5×105 per well. After 24 hours, cells were transfected with siCENPU, siFOXM1, siNC, CENPU, or FOXM1 expression plasmid. After 48 hours, all cells were treated with 40 µM EdU, and after 2 hours the cells were detected according to the recommended staining protocol. Then, the cells were collected and flow cytometry was performed following the manufacturer’s instructions.
Cell cycle assays
Cells were digested with trypsin, collected in a 1.5 mL centrifuge tube, and washed twice with cold saline; 1 mL of precooled 70% ethanol was added, and samples were incubated at 4°C overnight. The next day, the fixed cells were stained with 500 µL of propidium iodide (PI) staining buffer (containing 2 g/L RNase A and 50 µg/mL PI) at room temperature for 30 minutes in the dark. Cell cycle analysis was then performed by flow cytometry (BD Biosciences, San Jose, CA, USA). Cells in G0–G1, S, and G2–M phases were measured by ModFit LT software.
TCGA data analyses
To analyze the differential CENPU/FOXM1 expression between normal lung tissues and lung cancer tissues, we generated a CENPU differential plot using UALCAN (http://ualcan.path.uab.EdU./analysis.html).18
Survival data of NSCLC patients were obtained from Kaplan–Meier plotter (http://kmplot.com/analysis/)19,20 or PROGgeneV2 prognostic database (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php).21 These databases integrate gene expression and clinical data. Currently, Kaplan–Meier plotter can assess the effect of 54,675 genes on survival based on 10,461 cancer samples. The PROGgene V2 contains more than 19,000 samples from 20 cancer types. The cohort was split based on high and low CENPU/FOXM1 expression. OS was analyzed with an auto-select best cutoff. Relapse-free survival (RFS) was analyzed using PROGgene V2.
The genes that were co-expressed with CENPU were analyzed using cBioPortal (http://www.cbioportal.org/index.do). The expression data of CENPU, FOXM1, PCNA, and Ki-67 in the same patient cohort were obtained from University of California Santa Cruz (UCSC) Xena (http://xena.ucsc.EdU/).
Statistical analyses
Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical significance was assessed by performing a Student’s t-test. Pearson’s correlation coefficients were calculated to compare the expression levels of CENPU and FOXM1, FOXM1and PCNA, and FOXM1 and Ki-67. All statistical data are presented as mean ± standard error of the mean (SEM). Statistical significance was indicated as *P<0.05; **P<0.01; ***P<0.001; and ****P<0.0001. All experiments were independently repeated at least three times.
Results
CENPU is overexpressed in NSCLC and predicts poor survival for NSCLC patients
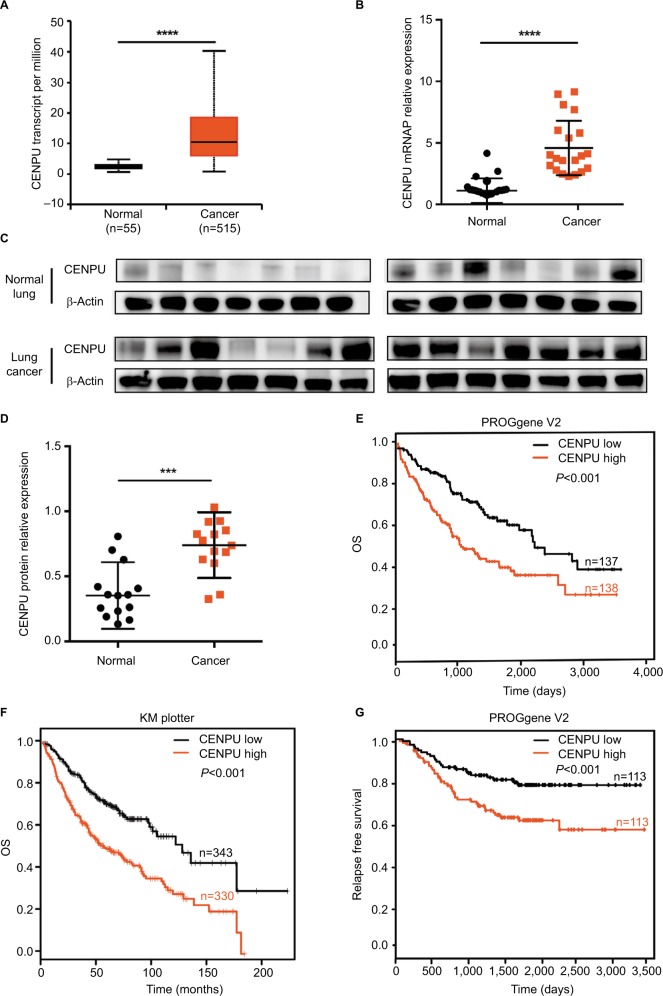
To determine whether CENPU is overexpressed in NSCLC, we first analyzed the CENPU mRNA levels using UALCAN, a portal for facilitating tumor subgroup gene expression analysis. Results showed that the expression of CENPU was significantly upregulated in cancer tissues compared with normal lung tissues (Figure 1A). We next examined CENPU mRNA level in 20 paired NSCLC tissues and normal lung tissues. As shown in Figure 1B, the CENPU mRNA level in cancer tissues was higher than that in normal lung tissues, which was consistent with the online analysis using UALCAN. In addition, Western blot analysis revealed that CENPU protein level was also elevated in lung cancer tissues (Figure 1C and D).
Figure 1.
CENPU is overexpressed in NSCLC and predicts poor prognosis.
Notes: (A) The mRNA expression data for CENPU in 515 human lung cancer tissues and 55 adjacent normal tissues were obtained from the UALCAN website. CENPU expression in lung cancer tissues was higher than that in adjacent normal tissues. (B) Quantitative reverse transcription PCR was performed to analyze the expression of CENPU in human NSCLC cancer tissues and paired adjacent normal tissues (n=20). (C) Western blotting was performed to analyze the expression level of CENPU in 14 paired human NSCLC cancer tissues and adjacent normal tissues. (D) The relative protein expression level was normalized to that of β-actin. (E, F) KM survival curve available on PROGene V2 and KM plotter website demonstrating decreased OS in the CENPU high expression group. (G) KM survival curve available on PROGene V2 website demonstrating decreased RFS in the CENPU high expression group. ****P<0.0001; ***P<0.001.
Abbreviations: CENPU, centromere protein U; KM, Kaplan–Meier; NSCLC, non-small-cell lung cancer; OS, overall survival; RFS, relapse-free survival.
To further explore the prognostic value of CENPU overexpression in NSCLC, we performed survival analysis using PROGgene V2 and Kaplan–Meier plotter. High expression of CENPU in lung cancer tissues was strongly associated with poor OS in lung cancer patients (PROGgene V2: HR =1.35, 95% CI: 1.17–1.55, P<0.001; Kaplan–Meier plotter: HR =1.98, 95% CI: 1.55–2.54, P<0.001; Figure 1E and F). Online PROGgene V2 analysis also revealed that the CENPU overexpression was negatively associated with the RFS of these patients (HR =1.47, 95% CI: 1.17–1.84, P<0.001; Figure 1G).
These findings indicated the involvement of CENPU in the progression of NSCLC as a tumor-promoting factor.
CENPU silencing inhibits proliferation and induces G1 arrest in lung cancer cells
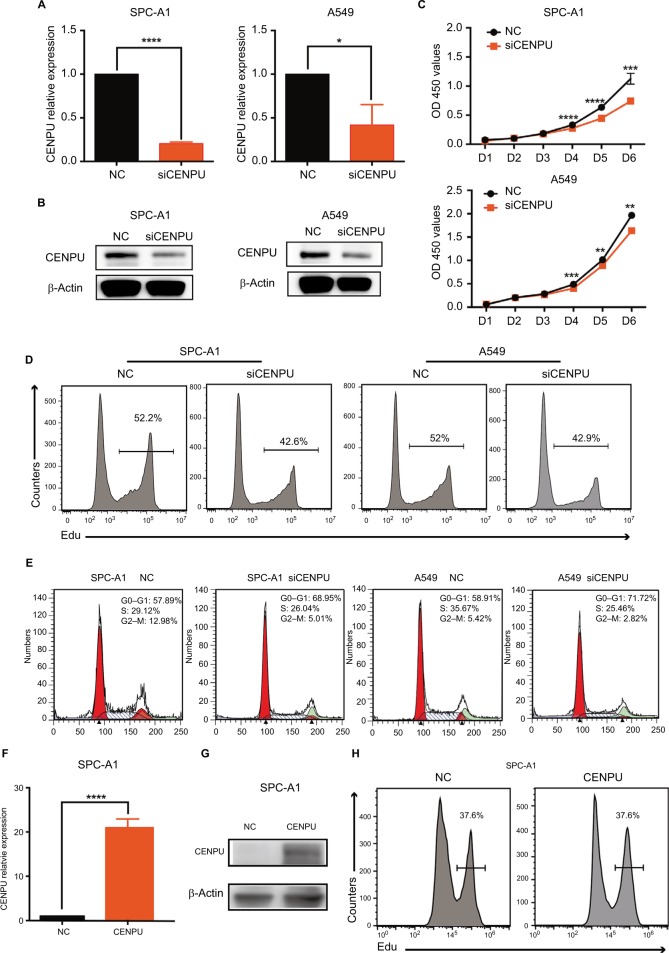
We next explored the biological function of CENPU by knocking down its expression using siRNA in SPC-A1 and A549 lung cancer cell lines. The efficacy of siCENPU transfection was confirmed by qRT-PCR and Western blotting. As shown in Figure 2A and B, CENPU mRNA and protein levels were significantly reduced compared with those in the NC group. These results indicated that the specific siRNA was able to effectively knock down CENPU expression at both the mRNA and protein levels in lung cancer cells.
Figure 2.
Knockdown of CENPU by RNA interference inhibits proliferation and induces G1 arrest in lung cancer cells.
Notes: (A) Quantitative reverse transcription PCR results showed that the mRNA expression level of CENPU was decreased in the siCENPU group compared with that in the NC group in SPC-A1 and A549 lung cancer cells 48 hours after transfection. (B) Western blotting was performed to analyze the expression level of CENPU 48 hours after siRNA transfection. (C) CCK-8 assays were performed to evaluate the effect of CENPU knockdown on the growth of SPC-A1 and A549 cells. (D, E) Cell proliferation rates were determined by EdU assays in SPC-A1 and A549 cells between the siCENPU and NC groups. (E) Cell cycle analysis of SPC-A1 and A549 cells transfected with siCENPU or NC siRNA. ****P<0.0001; ***P<0.001; **P<0.01 and *P<0.05 (mean ± SEM). (F) Quantitative reverse transcription results showed that the mRNA expression level of CENPU was increased in the CENPU group compared with that in the NC group in SPC-A1 cells 48 hours after transfection. (G) Western blotting showed that the expression of CENPU protein increased 48 hours after CENPU plasmid transfection in SPC-A1 cells. (H) Cell cycle analysis of SPC-A1 cells transfected with CENPU plasmid or NC plasmid.
Abbreviations: CCK-8, Cell Counting Kit-8; CENPU, centromere protein U; EdU, 5-ethynyl-2’-deoxyuridine; NC, negative control; SEM, standard error of the mean; siCENPU, CENPU siRNA.
We then investigated the effects of CENPU on the proliferative capacity of lung cancer cells by CCK-8 and EdU assays. CCK-8 assay results revealed that CENPU knockdown dramatically reduced the absorbance (corresponding to viability) of SPC-A1 and A549 cell cultures after 4, 5, and 6 days compared with that of the NC group (Figure 2C). Further in the two cell lines transfected with siCENPU, the EdU-positive cell rates were remarkably decreased compared with NC cells (Figure 2D). These results indicated that RNAi-mediated CENPU knockdown inhibited the proliferation of SPC-A1 and A549 lung cancer cells.
The cell cycle is closely related to cell proliferation. Thus, we next examined the effect of CENPU knockdown on the cell cycle progression of lung cancer cells. Compared with that in the NC group, the siCENPU group exhibited a significant increase in G0–G1 phase cells in both SPC-A1 (68.95% vs 57.89%) and A549 (71.72% vs 58.91%) cell lines (Figure 2E). At the same time, a significant decrease in G2–M phase cells was observed in the siCENPU group compared with the NC group (5.01% vs 12.98% in SPC-A1; 2.82% vs 5.42% in A549). These data demonstrated that CENPU knockdown induced G1 phase arrest and inhibited lung cancer cell mitosis to affect proliferation.
To confirm the promoter role of CENPU in lung cancer cell, we transfected the CENPU plasmid into SPC-A1 cell lines to achieve CENPU overexpression (Figure 2F and G). Consistent with the knockdown experiment results, the overexpression of CENPU significantly accelerated the proliferation of SPC-A1 cells compared with the NC group (Figure 2H).
CENPU expression is positively correlated with the expression of the transcription factor, FOXM1
To address the molecular mechanism responsible for CENPU-medicated lung cancer cell proliferation, we analyzed genes that are co-expressed with CENPU using cBioPortal (http://www.cbioportal.org/index.do); we found 287 genes that were positively correlated and 32 genes that were negatively correlated with CENPU expression. Table 2 summarizes some of the positively correlated genes. Among these, we noticed that Forkhead box protein M1 (FOXM1) showed a relatively high correlation. It is well known that FOXM1 was essential for cell proliferation and cell cycle progression.22 Recent studies showed that FOXM1 was overexpressed in lung cancer tissues and predicted poor prognosis in lung cancer, which also played an important role in the proliferation and mitosis of lung cancer cells.23–26 Therefore, FOXM1 was our primary choice for further study. But this did not rule out the possibility that CENPU might work through other proliferation-related genes, which need more research to verify.
Table 2.
A proportion of genes co-expressed with CENPU
| Correlated gene | Cytoband | Spearman’s correlation | Correlated gene | Cytoband | Spearman’s correlation |
|---|---|---|---|---|---|
| TACC3 | 4p16.3 | 0.70 | DIAPH3 | 13q21.2 | 0.67 |
| KIF188 | 17q21.31 | 0.70 | RANBP1 | 22q11.21 | 0.67 |
| RFC4 | 3q27.3 | 0.70 | MND1 | 4q31.3 | 0.67 |
| CENPM | 22q13.2 | 0.69 | FEN1 | 11q12.2 | 0.67 |
| CDCA3 | 12p13.31 | 0.69 | FOXM1 | 12p13.33 | 0.67 |
| ANLN | 7p14.2 | 0.69 | CDCA2 | 8p21.2 | 0.67 |
| CEP55 | 10q23.33 | 0.69 | KIF11 | 10q23.33 | 0.67 |
| ORC1 | 1p32.3 | 0.69 | CENPE | 4q24 | 0.66 |
| H2AFZ | 4q23 | 0.69 | KPNA2 | 17q24.2 | 0.66 |
| STIL | 1p33 | 0.69 | HMGB2 | 4q34.1 | 0.66 |
| SGO2 | 2q33.1 | 0.69 | PARPBP | 12q23.2 | 0.66 |
| OIP5 | 15q15.1 | 0.69 | WDR76 | 15q15.3 | 0.66 |
| CCNE1 | 19q12 | 0.69 | POLQ | 3q13.33 | 0.66 |
| MCM2 | 3q21.3 | 0.69 | DTL | 1q32.3 | 0.66 |
| TK1 | 17q25.3 | 0.68 | TRAIP | 3p21.31 | 0.66 |
| SPAG5 | 17q11.2 | 0.68 | CDT1 | 16q24.3 | 0.66 |
| KIF20A | 5q31.2 | 0.68 | DBF4 | 7q21.12 | 0.66 |
| GINS1 | 20p11.21 | 0.68 | CHAF1B | 21q22.12-q22.13 | 0.66 |
| POC1A | 3p21.2 | 0.68 | TUBA1B | 12q13.12 | 0.66 |
| FAM72B | 1p11.2 | 0.68 | BRCA1 | 17q21.31 | 0.65 |
| PBK | 8p21.1 | 0.68 | ASPM | 1q31.3 | 0.65 |
| CDC25A | 3p21.31 | 0.68 | GINS3 | 16q21 | 0.65 |
| UBE2C | 20q13.12 | 0.67 | TICRR | 15q26.1 | 0.65 |
| UBE2T | 1q32.1 | 0.67 | SHCBP1 | 16q11.2 | 0.65 |
| SPC24 | 19p13.2 | 0.67 | HASPIN | 17p13.2 | 0.65 |
Note: A proportion of genes co-expressed with CENPU was obtained from cBioPortal (http://www.cbioportal.org/index.do).
Abbreviations: CENPU, centromere protein U; FOXM1, Forkhead box protein M1.
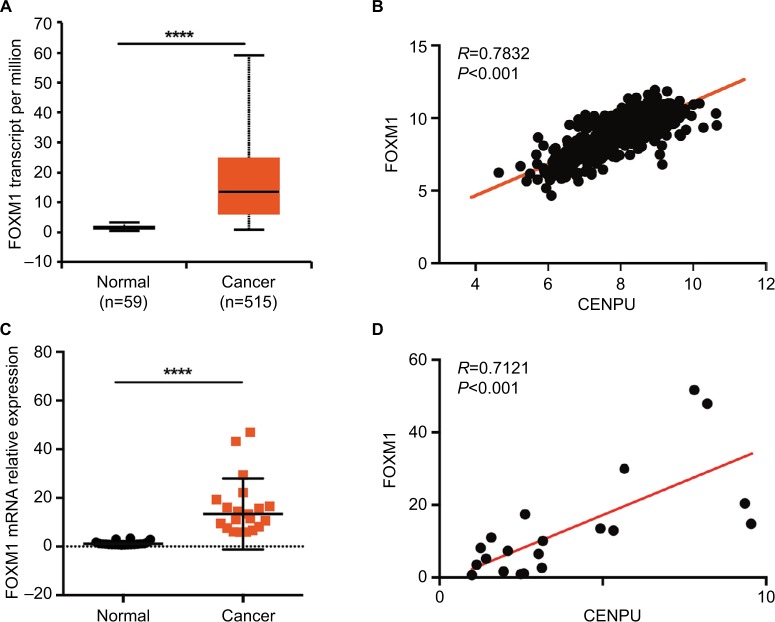
We first analyzed the mRNA expression levels of FOXM1 in NSCLC. Online analysis results showed that FOXM1 was significantly upregulated in cancer tissues compared with expression in normal tissues (Figure 3A). Then, we assessed the correlation between CENPU and FOXM1 expression in human NSCLC. Using mRNA expression data (n=576 NSCLC patients) from UCSC Xena, we found that the expression of CENPU was positively correlated with FOXM1 (R=0.7832, P<0.001; Figure 3B); these results were consistent with those of cBioPortal analysis.
Figure 3.
CENPU expression is positively correlated with FOXM1 expression.
Notes: (A) The mRNA expression data for FOXM1 was downloaded from the UALCAN website. FOXM1 was highly expressed in lung cancer tissues compared with expression in adjacent normal tissues; ****P<0.0001. (B) Pearson’s correlation was employed to analyze the correlation between CENPU and FOXM1 mRNA expression in 576 lung cancer samples in the available RNA-seq database downloaded from UCSC Xena (http://xena.ucsc.edu/; R=0.7832, P<0.001). (C) Quantitative reverse transcription was performed to analyze the expression of FOXM1 in human NSCLC tissues and paired adjacent normal tissues (n=20); ****P<0.0001. (D) The correlation between CENPU and FOXM1 mRNA expression in 20 lung cancer samples was measured by Pearson’s correlation (R=0.7121, P<0.001).
Abbreviations: CENPU, centromere protein U; FOXM1, Forkhead box protein M1; NSCLC, non-small-cell lung cancer.
After analyzing the data from the online database, we used qRT-PCR to detect FOXM1 expression in 20 paired lung cancer and normal lung tissues and performed correlation analysis with respect to CENPU expression as shown in Figure 1B. In accordance with the online analysis results, our own data also indicated that FOXM1 was upregulated in lung cancer tissues. The expression of CENPU was positively correlated with FOXM1 in lung cancer tissues (Figure 3C and D).
These results suggested that in lung cancer tissues the CENPU expression was positively correlated with FOXM1 levels and that CENPU might exert its biological function through FOXM1.
FOXM1 is a downstream mediator of CENPU in lung cancer cells
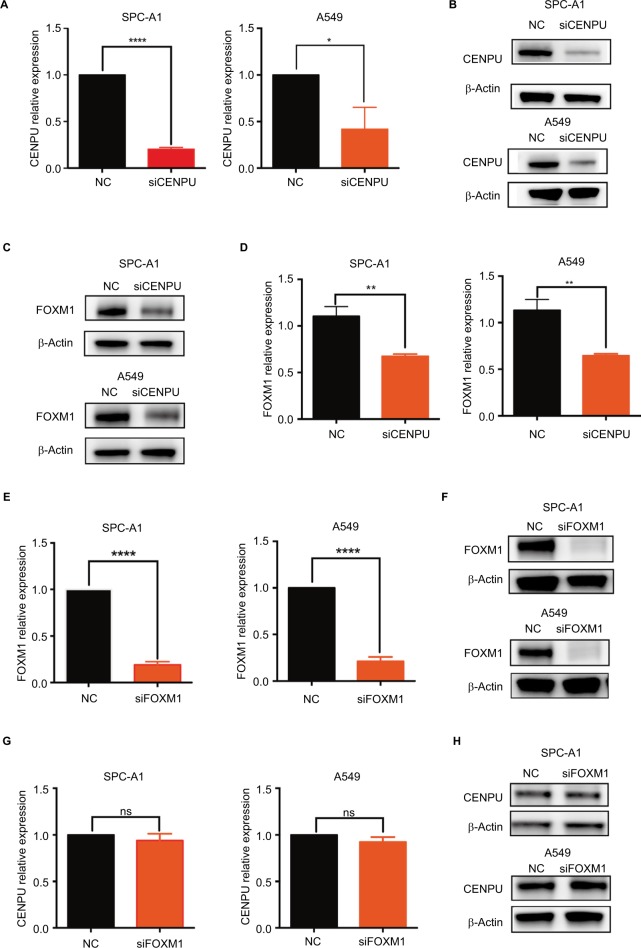
To determine whether FOXM1 is a downstream mediator of CENPU, we used siRNA to knock down the mRNA and protein expression of CENPU (Figure 4A and B). The Western blot results showed that FOXM1 protein was significantly downregulated after CENPU knockdown, about 40% lower than that of the NC group in both SPC-A1 and A549 cells (Figure 4C and D). These data suggested that CENPU knockdown could decrease the expression of FOXM1 in lung cancer cells.
Figure 4.
Knockdown of CENPU inhibits FOXM1 expression in lung cancer cells.
Notes: (A) Quantitative reverse transcription results showed that the mRNA expression of CENPU was downregulated in the siCENPU group compared with that expression in the NC group in SPC-A1 and A549 lung cancer cells. (B) Western blotting showed that the expression of CENPU declined 48 hours after siCENPU transfection in SPC-A1 and A549 cells. (C) Western blotting showed that the protein expression of FOXM1 was significantly downregulated in the siCENPU group. (D) Relative protein levels of FOXM1 were determined by Western blotting. The protein levels of FOXM1 were normalized to those of β-actin. (E) Quantitative reverse transcription PCR results showed that the mRNA expression of FOXM1 was downregulated in the siFOXM1 group compared with that expression in the NC group. (F) Western blotting showed that the expression of FOXM1 protein declined 48 hours after siFOXM1 transfection. (G) Knockdown of FOXM1 did not affect the mRNA expression level of CENPU, which was determined by RT-qPCR. (H) Western blotting showed that the expression of CENPU protein was not changed in the siFOXM1 group in both SPC-A1 and A549 cells; ***P<0.001; **P<0.01; and *P<0.05 (mean ± SEM).
Abbreviations: CENPU, centromere protein U; FOXM1, Forkhead box protein M1; NC, negative control; NS, no significance; RT-qPCR, real-time quantitative PCR; SEM, standard error of the mean; siCENPU, CENPU siRNA; siFOXM1, FOXM1 siRNA.
Next, we used siRNA to suppress FOXM1 expression in SPC-A1 and A549 lung cancer cells and investigated the effect on CENPU expression. Figure 4E and F shows that the siFOXM1 effectively knocked down the mRNA and protein expression level of FOXM1 in SPC-A1 and A549 cells. qRT-PCR and Western blotting results showed that the mRNA and protein expression levels of CENPU were not affected by FOXM1 knockdown in both SPC-A1 and A549 cell lines (Figure 4G and H). Overall, these results indicated that FOXM1 did not impact the expression level of CENPU.
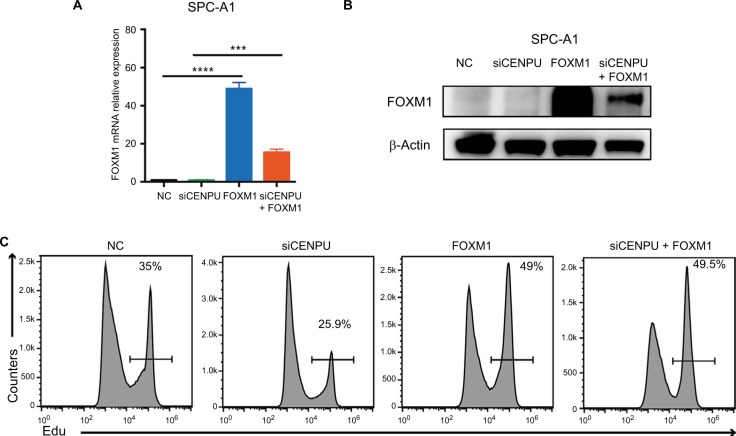
To further confirm that FOXM1 is a downstream molecule of CENPU, we transfected FOXM1 plasmid into CENPU knockdown SPC-A1 cell lines, and the EdU assay was performed. The results showed that the mRNA and protein levels of FOXM1 were significantly upregulated (Figure 5A and B) in both the FOXM1 group and cotransfected group. As shown in Figure 5C, the overexpression of FOXM1 in CENPU knocked down cells increased the EdU-positive cell rate from 25.9% to 49.5%, suggesting that the overexpression of FOXM1 significantly reversed the inhibition of proliferation caused by CENPU knockdown. These results further confirmed that CENPU affected the proliferation of lung cancer cells through FOXM1.
Figure 5.
Overexpression of FOXM1 significantly reverses the inhibition of proliferation caused by CENPU knockdown.
Notes: (A) The mRNA expression level of FOXM1 was significantly upregulated 48 hours after FOXM1 plasmid transfection. (B) Western blotting showed that the protein expression of FOXM1 was significantly upregulated 48 hours after FOXM1 plasmid transfection. (C) Cell proliferation rates were determined by EdU assays in SPC-A1 and A549 cells between NC, siCENPU, FOXM1, and siCENPU plus FOXM1 group. ***P<0.001; **P<0.01; and *P<0.05 (mean ± SEM).
Abbreviations: CENPU, centromere protein U; EdU, 5-ethynyl-2’-deoxyuridine; FOXM1, Forkhead box protein M1; NC, negative control; SEM, standard error of the mean; siCENPU, CENPU siRNA.
FOXM1 promotes proliferation of lung cancer cells
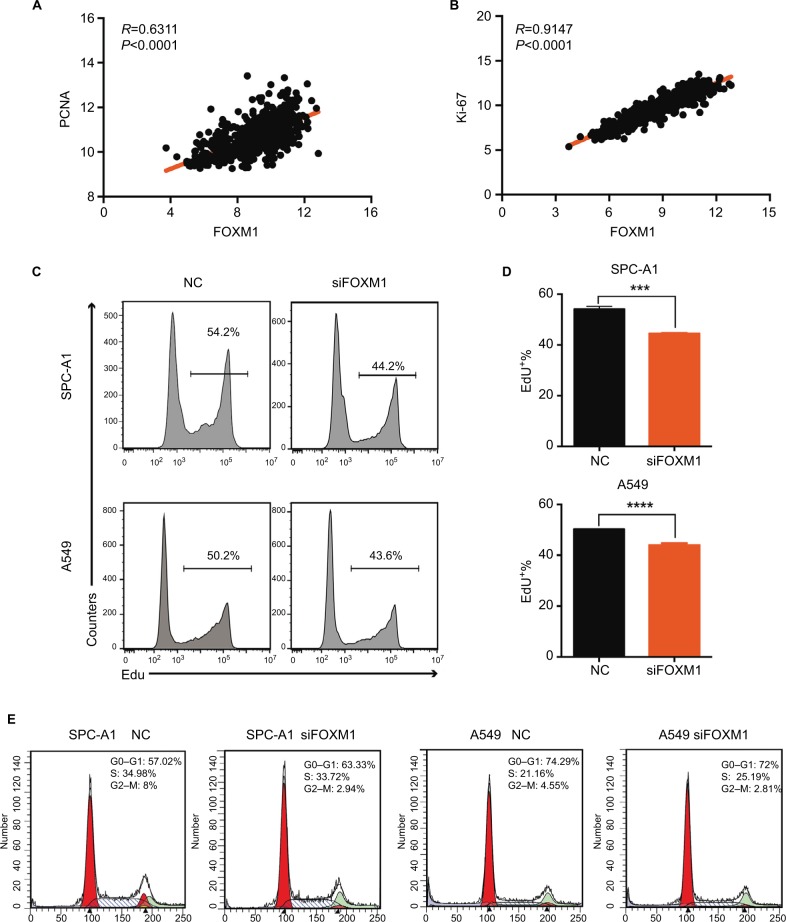
To reveal the role of FOXM1 in lung cancer cell proliferation, we screened paired expression data of FOXM1 and the proliferation markers, PCNA and Ki-67, using the UCSC Xena website. Correlation analysis showed that the expression of FOXM1 was significantly positively correlated with PCNA and Ki-67 proliferation indices (PCNA, R=0.6311, P<0.0001; Ki-67, R=0.9147, P<0.0001; Figure 6A and B).
Figure 6.
FOXM1 promotes proliferation in lung cancer cells.
Notes: (A) Pearson’s correlation was employed to analyze the correlation between PCNA and FOXM1 mRNA expression in 576 lung cancer samples in the available RNA-seq database downloaded from UCSC Xena (http://xena.ucsc.edu/; R=0.6311, P<0.0001). (B) The correlation between Ki-67 and FOXM1 mRNA expression in 576 lung cancer samples in the available RNA-seq database downloaded from UCSC Xena was measured by Pearson’s correlation (R=0.9147, P<0.001). (C, D) EdU assays were performed to evaluate the effect of CENPU knockdown on the growth of SPC-A1 and A549 cells. (E) Cell cycle analysis of SPC-A1 and A549 cells transfected with siFOXM1 or NC siRNA. ****P<0.0001; ***P<0.001 (mean ± SEM).
Abbreviations: EdU, 5-ethynyl-2’-deoxyuridine; FOXM1, Forkhead box protein M1; NC, negative control; PCNA, proliferating cell nuclear antigen; SEM, standard error of the mean; siFOXM1, FOXM1 siRNA.
Next, we performed an EdU assay to detect the effects of FOXM1 knockdown on the proliferation of lung cancer cells. We found that the proportion of EdU-positive cells in the FOXM1 knockdown group was significantly lower than that in the NC group for both SPC-A1 and A549 cells (Figure 6C and D).
Cell cycle assays also revealed that FOXM1 knockdown induced an increase in G0–G1 phase in SPC-A1 cells (63.33% vs 57.02%), but not in A549 cells (72% vs 72.49%; Figure 6E). In addition, FOXM1 knockdown also led to a decrease in G2–M phase cells in both cell lines, as compared with that in the respective NC groups (2.94% vs 8%) in SPC-A1 and (2.81% vs 4.55%) in A549 cells. Moreover, there were no significant changes in S phase cells between these two groups. These data demonstrated that FOXM1 is closely related to lung cancer cell proliferation and that its knockdown could induce G1 phase arrest in SPC-A1 cells and G2–M phase inhibition in both SPC-A1 and A549 cells, which ultimately inhibited lung cancer cell proliferation. In conclusion, FOXM1 might promote proliferation in lung cancer.
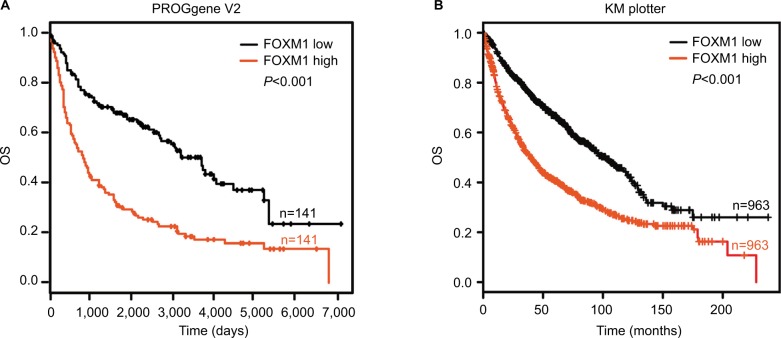
FOXM1 expression predicts poor prognosis for NSCLC patients
Kaplan–Meier analysis, based on the TCGA database from the PROGgene V2 and Kaplan–Meier plotter website, revealed that increased FOXM1 expression was also associated with poorer patient outcome (P<0.001; Figure 7A and B).
Figure 7.
FOXM1 expression predicts poor prognosis in NSCLC patients.
Notes: (A) Kaplan–Meier curves showed OS according to FOXM1 mRNA expression in 141 lung cancer samples from the PROGgene V2 website. (B) Kaplan–Meier curves showed OS according to the FOXM1 mRNA expression level in 963 lung cancer samples from the Kaplan–Meier plotter website.
Abbreviations: FOXM1, Forkhead box protein M1; NSCLC, non-small-cell lung cancer; OS, overall survival.
Discussion
As the most lethal malignancy,1,2 the diagnosis and treatment of lung cancer continue to be the focus of medical attention. However, due to the lack of specific early symptoms, most patients present with advanced-stage disease. Although breakthroughs have been made in targeted therapy and immunotherapy for lung cancer in recent years, the 5-year survival rate of lung cancer is still low.4,6 The high proliferation rate of tumor cells is an important cause of rapid disease progression. Therefore, it is important to explore the molecular mechanisms that promote the proliferation of lung cancer cells.
CENPU encoded a protein that contains two nuclear receptor-binding motifs and several potential phosphorylation sites, which was first identified in 2004 by Hanissian et al.8 It is found to be an important factor for centromere assembly27 and is responsible for the initial recruitment and maintenance of the kinetochore PLK1, which ensures proper chromosome segregation.28,29 CENPU has been implicated in the pathogenesis of polycythemia vera.30 Subsequently, some research groups found that CENPU was abnormally overexpressed in some types of tumors and is closely associated with poor prognosis in breast, bladder, and ovarian cancers.12–17 Nevertheless, the expression and function of CENPU in lung cancer have not yet been reported. In this study, we found that this marker was abnormally overexpressed in lung cancer tissues, compared with expression in normal lung tissues, as determined by qRT-PCR and Western blotting. Moreover, online TCGA data analyses found that CENPU overexpression was negatively correlated with OS and RFS in lung cancer patients. These findings revealed that this protein might play an important role in the pathological process of lung cancer. Therefore, we next explored the effect of CENPU knockdown on the proliferation of lung cancer cells. CCK-8 and EdU assays showed that the proliferation of lung cancer cells obviously declined after CENPU knockdown. As this protein is a centromere component that participates in mitosis, we wondered whether it was involved in lung cancer cell cycle progression. The results confirmed that the knockdown of CENPU could induce G1 arrest in lung cancer cells. Overall, the expression of CENPU is abnormally elevated in lung cancer and its suppression can inhibit cell proliferation and cell cycle progression.
We then explored the mechanism through which CENPU promotes the proliferation of lung cancer cells. Our qRT-PCR results confirmed a positive correlation between CENPU and FOXM1 expression in lung cancer tissues. FOXM1 is an important member of the Forkhead box (FOX) family, which participates in several physiological and pathological processes including the regulation of cell proliferation, differentiation, organ development, and carcinogenesis.31–34 Many studies have demonstrated that FOXM1 expression is elevated in several solid human carcinomas including gastric cancer, breast cancer, hepatocellular carcinoma, and lung cancer.23,26,35–37 Furthermore that FOXM1 overexpression has been shown to promote lung cancer cell proliferation and inhibit apoptosis.22 Moreover, it has been shown that FOXM1 is associated with poor prognosis in NSCLC.38–39 Therefore, considering that CENPU was found to be closely related to the expression of FOXM1 and that the latter plays an important role in the development of lung cancer, we speculated that CENPU might promote in lung cancer progression by regulating FOXM1. The current results showed that the knockdown of CENPU expression in lung cancer cells can downregulate the expression of FOXM1, whereas the knockdown of FOXM1 did not affect the expression of CENPU. This suggested that FOXM1 was downstream of CENPU. TCGA data analyses showed that FOXM1 is positively correlated with the expression of proliferation markers, PCNA and Ki-67, in lung cancer. Knockdown of FOXM1 in lung cancer cells restrained cell proliferation and induced G1 arrest, similar to the phenomenon observed with CENPU knockdown. Furthermore, the higher expression of FOXM1 could also be an indicator of poor prognosis in lung cancer.
Overall, our results confirmed that CENPU is remarkably overexpressed in lung cancer and that the knockdown of CENPU inhibits proliferation and induces G1 arrest in lung cancer cells by modulating FOXM1. It is thus suggested that CENPU could represent a novel therapeutic target and indictor of prognosis in lung cancer.
Conclusion
We found that CENPU was elevated in NSCLC tissues, and the higher expression of CENPU predicted poorer OS and RFS. Silencing of CENPU could inhibit lung cancer cell proliferation through the transcription factor, FOXM1. These findings indicated that CENPU could be a new therapeutic target for lung cancer therapy.
Acknowledgments
This study was supported by grants from the National Nature Science Foundation of China (no. 31700681; no. 81302025).
Footnotes
Author contributions
WW and BZ designed the study. XW, DC, JG, and HL performed the experiments and prepared the manuscript. CS, LZ, and FY collected the lung cancer samples and adjacent normal lung tissues. HZ and AZ processed the data. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Witt C. European respiratory society/american thoracic society/international association for the study of lung cancer international multidisciplinary classification of lung adenocarcinoma: state of the art. J Thorac Oncol. 2011;6(8):1451. doi: 10.1097/JTO.0b013e318224643b. [DOI] [PubMed] [Google Scholar]
- 4.Parums DV. Current status of targeted therapy in non-small cell lung cancer. Drugs Today (Barc) 2014;50(7):503–525. doi: 10.1358/dot.2014.50.7.2185913. [DOI] [PubMed] [Google Scholar]
- 5.Deslypere G, Gullentops D, Wauters E, Vansteenkiste J. Immunotherapy in non-metastatic non-small cell lung cancer: can the benefits of stage IV therapy be translated into earlier stages? Ther Adv Med Oncol. 2018;10:1758835918772810. doi: 10.1177/1758835918772810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumarakulasinghe NB, van Zanwijk N, Soo RA. Molecular targeted therapy in the treatment of advanced stage non-small cell lung cancer (NSCLC) Respirology. 2015;20(3):370–378. doi: 10.1111/resp.12490. [DOI] [PubMed] [Google Scholar]
- 7.Chen YY, Huang TW, Tsai WC, et al. Risk factors of postoperative recurrences in patients with clinical stage I NSCLC. World J Surg Oncol. 2014;12:10. doi: 10.1186/1477-7819-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanissian SH, Akbar U, Teng B, et al. cDNA cloning and characterization of a novel gene encoding the MLF1-interacting protein MLF1IP. Oncogene. 2004;23(20):3700–3707. doi: 10.1038/sj.onc.1207448. [DOI] [PubMed] [Google Scholar]
- 9.Pan HY, Zhang YJ, Wang XP, Deng JH, Zhou FC, Gao SJ. Identification of a novel cellular transcriptional repressor interacting with the latent nuclear antigen of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2003;77(18):9758–9768. doi: 10.1128/JVI.77.18.9758-9768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minoshima Y, Hori T, Okada M, et al. The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol Cell Biol. 2005;25(23):10315–10328. doi: 10.1128/MCB.25.23.10315-10328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua S, Wang Z, Jiang K, et al. CENP-U cooperates with Hec1 to orchestrate kinetochore-microtubule attachment. J Biol Chem. 2011;286(2):1627–1638. doi: 10.1074/jbc.M110.174946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez-Valentin M, Therkildsen C, Veerla S, et al. Distinct gene expression signatures in lynch syndrome and familial colorectal cancer type x. PLoS One. 2013;8(8):e71755. doi: 10.1371/journal.pone.0071755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanissian SH, Teng B, Akbar U, et al. Regulation of myeloid leukemia factor-1 interacting protein (MLF1IP) expression in glioblastoma. Brain Res. 2005;1047(1):56–64. doi: 10.1016/j.brainres.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Ji G, Shao Y, et al. MLF1 interacting protein: a potential gene therapy target for human prostate cancer? Med Oncol. 2015;32(2):454. doi: 10.1007/s12032-014-0454-1. [DOI] [PubMed] [Google Scholar]
- 15.Huang DP, Luo RC. MLF1IP is correlated with progression and prognosis in luminal breast cancer. Biochem Biophys Res Commun. 2016;477(4):923–926. doi: 10.1016/j.bbrc.2016.06.159. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Liu B, Zhang J, et al. Centromere protein U is a potential target for gene therapy of human bladder cancer. Oncol Rep. 2017;38(2):735–744. doi: 10.3892/or.2017.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Zhang H, Wang Y. Centromere protein U facilitates metastasis of ovarian cancer cells by targeting high mobility group box 2 expression. Am J Cancer Res. 2018;8(5):835–851. [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gyo˝rffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 21.Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280(23):22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- 23.Kim IM, Ackerson T, Ramakrishna S, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66(4):2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 24.Kong FF, Qu ZQ, Yuan HH, et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep. 2014;31(6):2660–2668. doi: 10.3892/or.2014.3129. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Zhang J, Cui X, et al. FoxM1: a novel tumor biomarker of lung cancer. Int J Clin Exp Med. 2015;8(3):3136–3140. [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Sun L, Li H, et al. DT-13 synergistically enhanced vinorelbine-mediated mitotic arrest through inhibition of FOXM1-BICD2 axis in non-small-cell lung cancer cells. Cell Death Dis. 2017;8(5):e2810. doi: 10.1038/cddis.2017.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8(5):458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 28.Kang YH, Park JE, Yu LR, et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol Cell. 2006;24(3):409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Kang YH, Park CH, Kim TS, et al. Mammalian polo-like kinase 1-dependent regulation of the PBIP1-CENP-Q complex at kinetochores. J Biol Chem. 2011;286(22):19744–19757. doi: 10.1074/jbc.M111.224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng G, Zhang T, Liu J, et al. MLF1IP promotes normal erythroid proliferation and is involved in the pathogenesis of polycythemia vera. FEBS Lett. 2017;591(5):760–773. doi: 10.1002/1873-3468.12587. [DOI] [PubMed] [Google Scholar]
- 31.Katoh M, Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25(5):1495–1500. [PubMed] [Google Scholar]
- 32.Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016;144:194–201. doi: 10.1016/j.lfs.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Gartel AL. FOXM1 in Cancer: interactions and vulnerabilities. Cancer Res. 2017;77(12):3135–3139. doi: 10.1158/0008-5472.CAN-16-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res. 2013;119:191–419. doi: 10.1016/B978-0-12-407190-2.00016-2. [DOI] [PubMed] [Google Scholar]
- 35.Li Q, Zhang N, Jia Z, et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69(8):3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song X, Fiati Kenston SS, Zhao J, Yang D, Gu Y. Roles of FoxM1 in cell regulation and breast cancer targeting therapy. Med Oncol. 2017;34(3):41. doi: 10.1007/s12032-017-0888-3. [DOI] [PubMed] [Google Scholar]
- 37.Meng FD, Wei JC, Qu K, et al. FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma. World J Gastroenterol. 2015;21(1):196–213. doi: 10.3748/wjg.v21.i1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Wen L, Zhao SH, Ai ZH, Guo JZ, Liu WC. FoxM1 expression is significantly associated with cisplatin-based chemotherapy resistance and poor prognosis in advanced non-small cell lung cancer patients. Lung Cancer. 2013;79(2):173–179. doi: 10.1016/j.lungcan.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 39.Kong FF, Qu ZQ, Yuan HH, et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep. 2014;31(6):2660–2668. doi: 10.3892/or.2014.3129. [DOI] [PubMed] [Google Scholar]