Abstract
Brown adipose tissue (BAT) dissipates chemical energy as heat via thermogenesis and protects against obesity by increasing energy expenditure. However, regulation of BAT by dietary factors remains largely unexplored at the mechanistic level. We investigated the effect of eicosapentaenoic acid (EPA) on BAT metabolism. Male C57BL/6J (B6) mice fed either a high-fat diet (HF, 45% kcal fat) or HF diet supplemented with EPA (HF-EPA, 6.75% kcal EPA) were used for 11 weeks. RNA sequencing (RNA-Seq) and microRNA (miRNA) profiling were performed on RNA from BAT using Illumina HiSeq and miSeq respectively. We conducted pathway analyses using ingenuity pathway analysis software (IPA®) and validated some genes and miRNAs using qPCR. We identified 479 genes that were differentially expressed (2-fold change, n=3, p ≤ 0.05) in BAT from HF compared to HF-EPA. Genes negatively correlated with thermogenesis such as hypoxia Inducible factor 1 alpha subunit inhibitor (Hif1αn), was downregulated by EPA. Pathways related to thermogenesis such as peroxisome proliferator-activated receptor (PPAR) were upregulated by EPA while pathways associated with obesity and inflammation such as nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) were downregulated by EPA. MiRNA profiling identified nine and six miRNAs that were upregulated and downregulated by EPA, respectively (log2 fold change > 1.25, n=3, P ≤ 0.05). Key regulatory miRNAs were involved in thermogenesis, such as miR-455–3p and miR-129–5p were validated using qPCR. In conclusion, the depth of transcriptomic and miRNA profiling revealed novel mRNA-miRNA interaction networks in BAT which are involved in thermogenesis which regulated by EPA.
Keywords: Brown adipose tissue, microRNA, obesity, omega 3 fatty acids, RNA sequencing
1. Introduction
Obesity is a major metabolic disease that has reached global epidemic proportions. It increases the risk of obesity-associated comorbidities including type 2 diabetes, cardiovascular diseases, and cancer [1]. White adipose tissue (WAT), includes both subcutaneous adipose tissue (SAT) and intra-abdominal visceral adipose tissue (VAT), which are depots for major fat storage in obesity [2]. On the other hand, brown adipose tissue (BAT) which is another adipose depot, acts as an anti-obesity target due to its thermogenic function [3]. BAT is further classified into classical, (uncoupling protein 1 positive; (UCP-1+); found in the interscapular and perirenal depots and non-classical, “beige” or “brite” fat which are also UCP-1+, but found in WAT depots [4].
BAT induces non-shivering thermogenesis by producing heat which is activated by cold, exercise, therapeutic and pharmacological approaches [5]. Cold temperature stimulates sympathetic nervous system (SNS) to activate β-adrenergic function which in turn increases lipolysis of triglycerides (TG) leading to increased release of fatty acids and fuel substrates for thermogenesis [6]. BAT also consumes circulating glucose and lipids for its thermogenic function making it a valuable anti-obesity target [7]. Recent research indicates that activation of thermogenesis leads to a higher weight loss in individuals with obesity, which has spiked interest in identifying potential pharmacological and dietary approaches to activate BAT [3].
While energy restriction is the main dietary approach to prevent and treat obesity, bioactive compounds such as fish oil which contains long-chain ω−3 polyunsaturated fatty acids (PUFA) such as docosahexaenoic acid (22:6n-3, DHA) and eicosapentaenoic acid (20:5n-3, EPA) have potential anti-obesity effects [8]. These fatty acids have established roles as anti-inflammatory, cardioprotective and triglyceride-lowering agents [9]. Moreover, our laboratory has reported that feeding a high-fat (HF) diet enriched with EPA to mice significantly reduced body weight, fat mass and glucose intolerance while improving insulin sensitivity and inflammation (to levels comparable to low-fat fed mice) [10]. Additionally, we recently reported that EPA increased levels of several classical thermogenic biomarkers at both gene and protein levels in BAT of mice fed EPA and in clonal brown adipocytes [11].
Recent studies also demonstrate the role of microRNAs (miRNAs) to regulate BAT [12]. However, to our knowledge, no studies have conducted a global search for mRNAs and miRNAs pairs which coordinately mediate dietary regulation of BAT. We hypothesize that EPA regulates novel genes and miRNAs to mediate BAT regulation. For these purposes, we employed RNA sequencing (RNA-Seq), which is an unbiased and integrated approach to identify differential regulation of BAT genes, along with small RNA-Seq to detect miRNA profiles. We also identified integrated pathways and validated few targets to gain further insight into mechanisms mediating metabolic effects of EPA.
2. Materia and Methods:
2.1. Animals and Experimental Design:
The experimental groups used in this study have been previously described in detail [10]. Briefly, male B6 mice aged 5–6 weeks were fed a HF (45% kcal from fat) or a HF diet supplemented with 6.57% kcal EPA for 11 weeks (HF-EPA) [10]. BAT from interscapular depot was carefully dissected and weighed for subsequent analyses [10]. These protocols were all approved by the Institutional Animal Care and Use Committee at the University of Tennessee, Knoxville and Texas Tech University.
2.2. RNA Extraction and cDNA Library Preparation
Approximately 70 mg of frozen BAT was homogenized in QIAzol® (QIAGEN, Redwood City, CA, USA). RNA was isolated according to manufacturer’s protocol using RNeasy Mini Kit (QIAGEN, Redwood City, CA, USA) and quantified using Nanodrop, (Thermo Scientific, Waltham, MA, USA). Quality of the RNA samples was assessed using Agilent 2200 Tape station (Agilent Technologies, Santa Clara, CA, USA). RNA integrity numbers (RIN) for the samples ranged from 7.2 to 8.1. cDNA libraries were made using Illumina’s TruSeq RNA sample preparation kit v2 (Illumina Inc., San Diego, CA, USA) for 2–4 μg of RNA per sample. Briefly, the poly-A containing mRNA was purified, fragmented from total RNA and double stranded cDNA was made. cDNA was recovered using Agencourt, AMPure XP beads (Beckman Coulter, IN, USA). The sample identifier adapters were ligated to the ends of cDNA using 3’ adenylation and adapters ligation. We used specific index for each sample to facilitate multiplexing then adaptor ligated fragments were purified by using AMPure XP beads and amplified with polymerase chain reaction (PCR). Validation and identification of mid insert size of cDNA libraries was performed using Agilent 2200 Tape station. Libraries were quantified using Qubit 2.0 fluorimeter (Invitrogen, Life Technologies Waltham, MA, USA). Prior to loading into HiSeq rapid flow cell, the cDNA libraries were diluted, and denatured.
2.3. mRNA and miRNA Sequencing Strategy
For transcriptomic studies, paired end sequencing was performed using Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) with a 108 bp read length at the Center for Biotechnology and Genomics Core Facility at Texas Tech University to study gene expression. For miRNA profiling, Illumina small RNA protocol (Illumina, Inc., San Diego, CA, USA) was used for libraries preparation and sequencing, then the Illumina Genome Analyzer NextSeq 500 (Illumina, Inc., San Diego, CA, USA) was used to perform sequencing at Department of Molecular and Cell Biology, Baylor College of Medicine, Houston.
2.4. Quality Control (QC) and reads alignment
For gene expression, FastQC v0.10.1 High Throughput Sequence QC report (Version 0.11.2) [13] was used to check the quality of raw sequence reads. The average base call quality Q score based on fastqc data was 38 (Supplementary Figure S1). The reads were mapped to the Mus musculus genome (GRCm38.p4) using Qseq® software Version 12 (DNASTAR Madison, WI). For miRNA expression, the Gunaratne Next Generation pipeline was used to identify miRNA expression profiles [14]. Fastqc data on quality control for small RNA sequencing are provided in supplementary figure S2. In the normalization process, counts of each unique read of miRNA were normalized to total usable reads and then 40 counts were added. Normalized expression values of genes and miRNAs were presented as reads per kilo base per million mapped reads (RPKM).
2.5. Data availability
RNA sequencing data for HF and HF-EPA were submitted in the BioProject at NCBI under PRJNA353387 individual accession numbers for HF Sequence Read Archive (SRA) submission: BAT1 (SAMN05717642), BAT2 (SAMN05717643), and BAT3 (SAMN05717644). For HF-EPA SRA submission: BAT-EPA1 (SAMN05991740), BAT-EPA2 (SAMN05991741), and BAT-EPA3 (SAMN05991742). The small RNA-Seq data were submitted to Gene Expression Omnibus (GEO) data repository (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE85101 and GSE99506 for BAT HF and BAT HF-EPA respectively.
2.6. Quantitative real-time polymerase chain reaction (q-PCR) validation
RNA was isolated from BAT using RNeasy kit (QIAGEN, Redwood City, CA, USA) followed by cDNA synthesis using the iScript kit (Bio-Rad Laboratories, Inc. CA.USA) and TaqMan® Advanced miRNA cDNA Synthesis Kit (Life Technologies Corporation, Pleasanton, CA, USA) for genes and miRNAs respectively. Quantitative polymerase chain reaction (qPCR) was performed using BioRad CFX-96 real time PCR detection system (Bio-Rad Laboratories, Inc. CA.USA). 18S and miR-191–5p was used as housekeeping control for genes and microRNAs respectively.
2.7. Statistical analyses:
Differentially expressed (DE) genes between HF (control) and HF-EPA groups were identified as those with 2 or more-fold change at 95% confidence using Moderated t-test with false discovery rate (FDR) of 5% (using Benjamini & Hochberg test) with QSeq software. P ≤ 0.05 was considered significantly different based on Student’s t-test. The Ingenuity Pathways Analysis (IPA®, QIAGEN Redwood City, CA, USA; www.qiagen.com/ingenuity) software was used for analysis of target pathways, genes and networks. We used RPKM 0.3 or more in both HF and HF-EPA as the threshold for mRNA expression [15]. In the canonical pathway analyses, Fischer’s exact test p values indicates the significance of enrichment of pathways by DE genes and the Z score to determine whether the canonical pathway is activated or inhibited based on the DE genes in the dataset. Z score ≤ −2 is inhibited and ≥ 2 is activated. For miRNAs, a cut off of log2 fold change > 1.25 was used to determine differentially regulated ones that were DE between HF vs. HF-EPA, RPKM ≥ 3.76. Also, we submitted differentially expressed miRNAs with log2 fold change > 1.25 to canonical pathways using the grow function in IPA®. For gene and miRNA validation, results are presented as means ± SEM and the ΔΔCt method was used to determine mRNA expression and fold changes. Three to five replicates were used for each dietary or treatment groups.
3. Results
We have previously reported significant reductions in body weight, insulin resistance, and body fat in HF-EPA and low fat (LF) group compared to HF fed mice [10]. The mean final body weights of the HF, HF-EPA, and LF fed mice were 40.4 ± 1.2 (g), 35.9 ± 0.9 (g), and 31.7 ± 1.0 (g) respectively (Supplementary Figure S3A), (P ≤ 0.05). Mean BAT weights of the HF, HF-EPA, and LF groups were 0.28 ± 0.02 (g), 0.18 ± 0.02 (g), and 0.2± 0.02 (g) respectively (Supplementary Figure S3B), (P ≤ 0.05). No significant differences were observed in food intake between the HF, HF-EPA, and LF groups (Supplementary Figure S3C) [10]. Furthermore, hematoxylin and eosin stained histological sections of BAT (HF, HF-EPA, and LF) showed reduced lipid content in HF-EPA compared to HF; while HF-EPA was comparable with LF (supplementary Figure 4).
3.1. RNA sequencing data Quality
BAT from HF and HF-EPA groups were used for Illumina paired-end sequencing for whole transcriptome mRNA sequencing. Total sequences per sample were about 24 million paired-end reads of 108 bp length. For small RNA sequencing, an average of 13.8 million sequences reads per sample was mapped to the Mus musculus genome (build mm 10) and a total of 307 miRNAs were identified in BAT.
3.2. Analyses of gene and miRNA differences between two diets
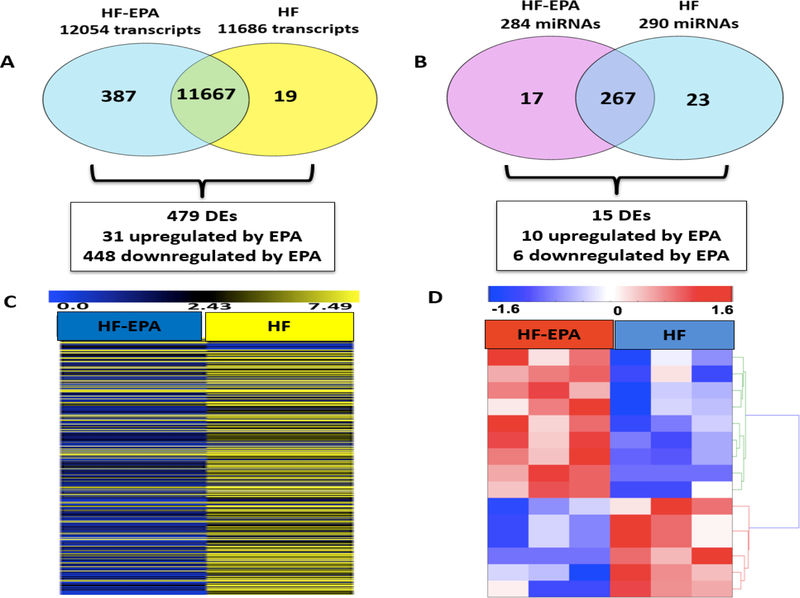
Using Qseq software analysis, genes with RPKM cut off ≥0.3 was filtered to avoid poorly expressed genes. HF and HF-EPA groups expressed a total of 11686 and 12054 mRNAs genes respectively; among these 11667 were common genes that existed between HF and HF-EPA as shown by venn diagram (Figure 1.A). Hence, only 19 and 387 genes, were found to be exclusively expressed in HF or HF-EPA groups respectively. Out of 11667 common genes identified, 479 of them were differentially expressed (DE) between HF and HF-EPA with a criterion of 2-fold change and significance of p ≤0.05 (Supplementary Table 1). Out of the 479 DE genes, 31 were upregulated while 448 were downregulated with EPA respectively (Figure 1.A).
Figure 1. Gene and miRNA profiling and Hierarchical clustering process in B6 mice.
(A) Gene filtering process using venn diagram following RNA sequencing for HF vs. HF-EPA in BAT. Four hundred and seventy nine DE genes were identified using QSeq software of which 31 and 448 genes were upregulated and downregulated by EPA respectively (P ≤ 0.05, fold change ≥2, RPKM ≥0.3). (B) MiRNA filtering process using venn diagram following miRNA profiling for HF vs. HF-EPA in BAT. Fifteen DE miRNAs were identified of which 9 and 6 miRNAs were upregulated and downregulated by EPA respectively (P ≤ 0.05, log 2 fold change > 1.25, RPKM ≥ 3.76). (C) Four hundred and seventy nine DE genes in BAT, which were differentially regulated by EPA. The vertical line represents the different diets (HF vs. HF-EPA) and the horizontal line represents the list of genes. Lower values are represented with blue tones while higher values have more intense yellow tones (P ≤ 0.05, fold change ≥ 2, RPKM ≥ 0.3). (D) Fifteen DE miRNAs in BAT, which were differentially regulated by EPA. The vertical line represents the different diets (HF vs. HF-EPA) and the horizontal line represents the list of miRNAs. Lower values are represented with blue tones while higher values have hotter red tones (P ≤ 0.05, log 2 fold change > 1.25, RPKM ≥ 3.76). HF, high fat; HF-EPA, HF diet supplemented with EPA; DE, differentially expressed.
In the miRNA analyses, a total of 307 miRNAs were expressed in both groups with 267 common miRNAs, out of which, 290 and 284 miRNAs were expressed in HF and HF-EPA respectively. Using an RPKM cut off ≥ 3.76, we identified 23 and 17 miRNAs that were expressed only in HF or HF-EPA respectively. Next, using the Gunaratne Next Generation pipeline, we identified 15 miRNAs that were DE with a log2 fold change > 1.25 (P ≤ 0.05). Out of the 15 miRNAs DE, 9 were upregulated and 6 were downregulated with EPA respectively (Figure 1.B). Based on the DE expression of genes and miRNAs, we used hierarchical clustering (HC) to determine the similarity in expression level in different treatment groups (HF and HF-EPA), which is represented by specific colors (Figure 1.C and 1.D).
3.3. Top up and downregulated mRNAs and miRNAs by EPA
In this study, we identified several DE genes by EPA using Qseq software in BAT (Table 1). Among these, Serine/threonine-protein kinase (Sgk2) gene which is associated with thermogenesis [16], was significantly upregulated (2.11 fold change) by EPA compared to HF diet. Also, significant increases of Paraoxonase 1 (Pon1) gene (2.11 fold change), which has antioxidant properties [17] was observed with EPA. Top downregulated genes by EPA (Table 1) included genes negatively associated with thermogenesis such as translocation-associated Notch protein TAN-1(Notch1) [18], hypoxia Inducible factor 1 alpha subunit inhibitor (Hif1αn) [19], adenosine A1 receptor (Adora1) [20], nuclear receptor corepressor 2 (Ncor2) [21], early growth response 1 (Egr1) [22], insulin like growth factor 2 (Igf2) [23], transforming growth factor receptor 3 (Tgfβr3), and smad family member 3 (Smad3) [24]. Genes associated with obesity, and type 2 diabetes such as CREB binding protein (Crebbp), inhibin beta B (Inhbb), Sp1 transcription factor (Sp1), caspase 7 (Casp7), and forkhead box O1 (Foxo1) were also downregulated [25–27]. Additionally, genes associated with inflammation, Jun proto-oncogene (Jun) [28], and lipid metabolism, Glycerol-3-phosphate dehydrogenase 1 (Gpd1) were downregulated by EPA (P ≤ 0.05, fold change ≥ 2, RPKM ≥0.3) [29].
Table 1.
Top upregulated and downregulated mRNA transcripts in BAT from B6 mice (HF vs. HF-EPA), which were differentially regulated by EPA using QSeq software (P ≤ 0.05, fold change ≥2, RPKM ≥0.3). HF, high fat; HF-EPA, HF diet supplemented with EPA; RPKM, reads per kilo base per million mapped reads.
| Gene Name | RPKM HF | RPKM HF-EPA | Fold Change | P value | |
|---|---|---|---|---|---|
| Top upregulated genes | |||||
| Klhl6 | Kelch-Like Family Member 6 | 0.65 | 1.39 | 2.14 | 0.01 |
| Sgk2 | Serine/threonine-protein kinase | 0.5 | 1.05 | 2.11 | 0 |
| Pon1 | Paraoxonase 1 | 8.43 | 17.8 | 2.11 | 0 |
| Rrl41 | Catalyze Protein Synthesis, Ribosomal Protein L41 | 5.48 | 11.49 | 2.1 | 0.01 |
| Top downregulated genes | |||||
| Notch1 | translocation-associated Notch protein TAN-1 | 9.43 | 4.66 | 0.49 | 0.01 |
| Casp7 | Caspase 7 | 3.8 | 1.79 | 0.47 | 0.05 |
| Hif1an | Hypoxia Inducible Factor 1 Alpha Subunit Inhibitor | 2.44 | 1.13 | 0.46 | 0 |
| Gpd1 | Glycerol-3-Phosphate Dehydrogenase 1 | 3.53 | 1.63 | 0.46 | 0.00 |
| Crebbp | CREB Binding Protein | 4.57 | 2.04 | 0.45 | 0 |
| Adora1 | Adenosine A1 Receptor | 19 | 8.25 | 0.43 | 0 |
| Foxo1 | Forkhead Box O1 | 4.65 | 1.95 | 0.42 | 0 |
| Smad3 | Smad Family Member 3 | 2.51 | 0.9 | 0.36 | 0 |
| Igf2 | Insulin Like Growth Factor 2 | 2.67 | 0.97 | 0.36 | 0.02 |
| JUN | Jun Proto-Oncogene | 28.37 | 9.56 | 0.34 | 0 |
| Inhbb | Inhibin Beta B | 1.57 | 0.52 | 0.33 | 0.01 |
| Sp1 | Sp1 Transcription Factor | 2.96 | 0.95 | 0.32 | 0.01 |
| Egr1 | Early Growth Response 1 | 5.41 | 1.75 | 0.32 | 0 |
| Tgfbr3 | Transforming Growth Factor Receptor 3 | 5.52 | 1.64 | 0.3 | 0 |
| Ncor2 | Nuclear Receptor Corepressor 2 | 14.14 | 5.58 | 0.2 | 0.01 |
We identified 15 miRNAs which were differentially expressed between HF and HF-EPA diets (Table 2). Candidates with high differential expression were further validated; these include miR-455–3p, miR-129–5p, miR-129–2-3p, miR-129–1-3p, miR-181c-3p, miR-199a-3p, and miR-31023p. Of these, miR-455 was previously reported to play a critical role in BAT adipogenesis [19]. Also, miR-129–5p which was identified in this study, was also shown previously to regulate thermogenesis and energy expenditure [30]. However, to our knowledge, no studies have identified regulation of these miRNAs in response to EPA. The rest of the miRNAs we identified are novel and not previously known to be involved in BAT regulation, specifically in response to EPA.
Table 2. List of miRNAs regulated by EPA.
Fifteen miRNAs were differentially expressed between HF and HF-EPA diets in B6 mice. Nine and 6 miRNAs were upregulated and downregulated by EPA respectively. HF, high fat; HF-EPA, HF diet supplemented with EPA; RPKM, reads per kilo base per million mapped reads.
| miRNA Name | RPKM HF | RPKM HF-EPA | Fold Change | P value |
|---|---|---|---|---|
| Upregulated miRNA | ||||
| miRNA-17–3p | 3.74 | 5.33 | 1.59 | 0.03 |
| miRNA-129–2-3p | 9.75 | 11.30 | 1.55 | 0.01 |
| miRNA-129–5p | 8.70 | 10.27 | 1.57 | 0.01 |
| miRNA-129–1-3p | 3.32 | 4.94 | 1.61 | 0.01 |
| miRNA-148a-5p | 6.56 | 7.22 | 0.67 | 0.02 |
| miRNA-150–5p | 14.52 | 15.26 | 0.74 | 0.02 |
| miRNA-181c-3p | 3.85 | 5.33 | 1.7 | 0.04 |
| miRNA-2137 | 12.48 | 13.27 | 0.79 | 0.03 |
| miRNA-455–3p | 12.15 | 13.52 | 1.38 | 0.05 |
| Downregulated miRNA | ||||
| miRNA-125b-1–3p | 7.66 | 6.44 | −1.22 | 0.03 |
| miRNA-199a-3p | 10.3 | 9 | −1.03 | 0.03 |
| miRNA-199b-3p | 10.3 | 9 | −1.03 | 0.03 |
| miRNA-3102–3p | 7.50 | 4.26 | −3.24 | 0.03 |
| miRNA-503–3p | 4.89 | 3.32 | −1.57 | 0.00 |
| miRNA-674–5p | 7.22 | 5.52 | −1.7 | 0.02 |
3.4. Validation of gene and miRNA profiling results
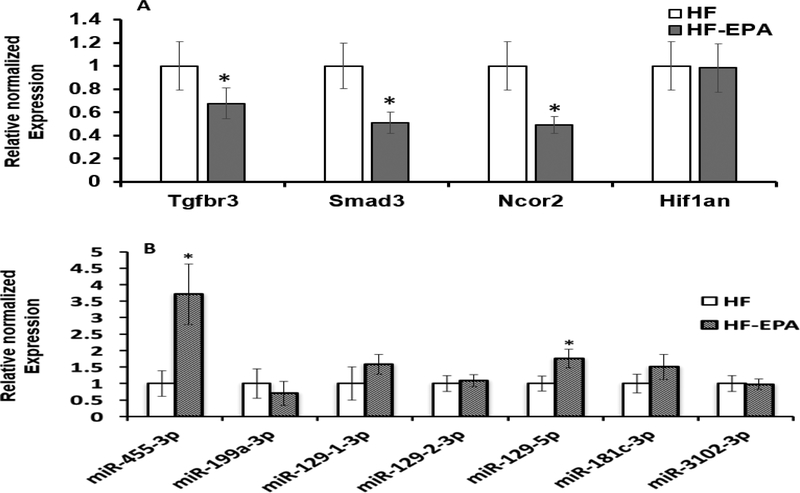
qPCR was performed for validation of target genes and miRNAs using BAT from HF vs. HF-EPA groups. As previously reported, we observed significantly higher expression of key thermogenesis genes, peroxisome proliferator-activated receptor gamma coactivator 1 alpha (Pgc1α), PR domain containing 16 (Prdm16), Ppar-γ, and Ucp1 in HF-EPA Vs. HF diet [11]. Additionally, we observed significantly lower expression of genes known to have a negative correlation with thermogenesis, such as Tgfbr3, Smad3, and Ncor2 (P≤ 0.05, Figure 2.A). We further tested 5 miRNAs (miR-455–3p, miR-129–1-3p, miR-1292–3p, miR-129–5p, and miR-181c-3p), which were significantly upregulated by EPA and 2 miRNAs (miR-3102–3p and miR-199a-3p), significantly downregulated by EPA in our miRNA profiling. qPCR validation showed that the expression levels of miR-455–3p and miR-129–5p were significantly higher (P≤ 0.05) in HF-EPA compared to HF groups. Other miRNAs validated showed trends towards significance (P<0.1) (Figure 2.B). The validation of gene and miRNA (HF vs. HF-EPA) compared to low LF diet is included in supplementary figure 5.
Figure 2. Validation of genes and miRNAs in B6 mice:
Genes and miRNAs were validated using RT-qPCR (A) Gene expression of thermogenesis markers Tgfbr3, Smad3, Ncor2, and Hif1an. Data are expressed as mean ± SEM, P ≤ 0.05, n=5. (B) MiR-455–3p, miR-199a3p, miR-129–1-3p, miR-129–2-3p, miR-129–5p, miR-181c-3p, and miR-3102–3p were validated. Data are expressed as mean ± SEM, P ≤ 0.05, n=5. HF, high fat; HF-EPA, HF diet supplemented with EPA.
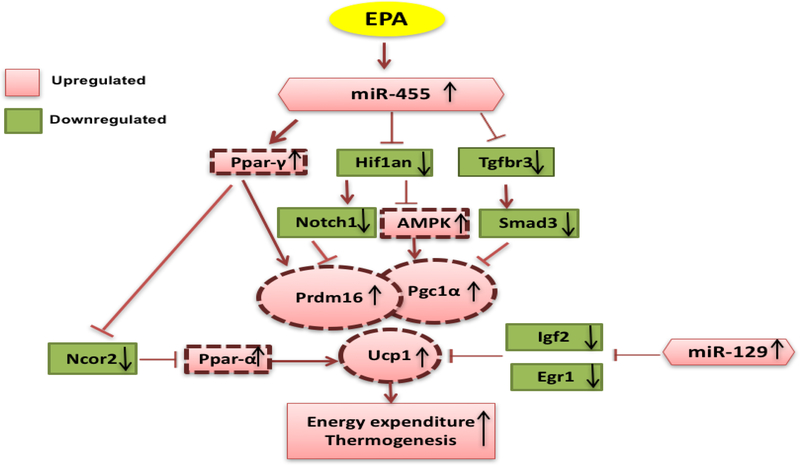
We conducted further analysis of the DE miRNAs in canonical pathways to create a proposed mRNA/ miRNA network model in IPA (Figure 3). We previously reported that mRNAs such as Ppar-γ, Ppar-α, Prdm16, Pgc1α, and Ucp1 mainly involved in energy expenditure and fatty acid oxidation were upregulated by EPA [11], while some genes negatively associated with thermogenesis such as Hif1αn [19], Tgfβr3 [24], Notch1 [18], Smad3 [24], Ncor2 [21], Igf2 [23], and Egr1 [22], were all downregulated by EPA. Also, in our main network analysis, miR-455–3p and miR-129–5p, were upregulated by EPA, consistent with their role in BAT thermogenic function, and were therefore incorporated from our miRNA profiling analysis [19, 30].
Fig 3. mRNAs and miRNA networks leading to activation of BAT in B6 mice.
Solid lines around boxes represent the genes, which were identified by our study (mean ± SEM, P ≤ 0.05, n=5), while dashed lines represent genes from our previously published work or published literature.
3.5. IPA analysis including canonical pathways and networks
Using IPA, we identified that EPA upregulated pathways such as peroxisome proliferator-activated receptor (PPAR), which is involved in lipid metabolism and fatty acid oxidation and pathways involved in energy balance such as PPAR and retinoid X receptors (PPAR/RXR) [31]. Additionally, energy expenditure pathways such as phosphatase and tensin homolog (PTEN) [32], Rho GDP-dissociation inhibitor (Rho-GDI) signaling pathway which is involved in multi-molecular functions such as shuttling capability for the particular membrane microenvironment were also upregulated by EPA (Table 3) [33]. As expected, inflammatory pathways were downregulated by EPA. They include transforming growth factor beta 1 (TGFβ1) [34], signal transducer and activator of transcription 3 (STAT3) [35], nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) [36], high mobility group box 1 (HMGB1) [37], integrin linked kinase (ILK) [38], interleukin 6 (IL6) [39], tumor necrosis factor receptor 1 (TNFR1) [40], and toll like receptor signaling (TLR) [41] (Table 3).
Table 3.
Top upregulated and downregulated ingenuity canonical pathways, which were differentially regulated by EPA in BAT from B6 (HF vs. HF-EPA) using IPA. HF, high fat; HFEPA, HF diet supplemented with EPA.
| Ingenuity Canonical Pathway | Gene Name | -log (p-value) | Ratio |
|---|---|---|---|
| Top upregulated | |||
| PPAR Signaling | Crebbp, Fos, Insr, Jun, Ncor2, Ppard, Rxra | 2.47E | 7.53E-02 |
| PTEN Signaling | Bmpr2, Cbl, Foxo1, Foxo3, Foxo4, Igf1r, Insr, Pdpk1, Tgfbr3 | 3.07E | 7.63E-02 |
| PPAR/RXR Activation | Bmpr2, Crebbp, Gpd1, Insr, Jun, Map2k6, Ncor2, Rxra, Smad3, Tgfbr3 | 2.88E | 6.21E-02 |
| RHO-GDI Signaling | Arhgap35, Arhgef17, Crebbp, Esr1, Fnbp1, Gnao1, Pip4k2b, Rhob, Wasf2 | 1.96E | 5.20E-02 |
| Top downregulated | |||
| TGFB1 Signaling | Bmpr2, Crebbp, Fos, Inhbb, \Jun, Map2k6, Pmepa1, Smad3 | 3.32E | 9.20E-02 |
| STAT3 Pathway | Bmpr2, Cish, Igf1r, Insr, Tgfbr3 | 1.73E | 6.85E-02 |
| NF-κB Signaling | Bmpr2, Crebbp, Igf1r, Insr, Map2k6, Map3k1, Pik3c2b, Tgfbr3, Tnfaip3 | 1.98E | 5.23E-02 |
| HMGB1 Signaling | Fnbp1, Fos, Jun, Kat6a, Map2k6, Pik3c2b, Rhob, Sp1 | 2.42E | 6.67E-02 |
| IL-k Signaling | Crebbp, Fnbp1, Fos, Irs1, Irs2, Jun, Map2k6, Myh9, Pdpk1, Pik3c2b, Rhob | 2.73E | 5.95E-02 |
| IL-6 Signaling | Fos, IL-6r, Jun, Map2k6, Pik3c2b, Ptpn11 | 1.45E | 5.17E-02 |
| CD 40 Signaling | Fos, Jon, Map2k6, Pik3c2b, Tnfaip3 | 1.93E | 7.69E-02 |
| TNFR1 Signaling | Fos, Jun, Map3k1, Tnfaip3, Casp7 | 2.45E | 1.02E-01 |
| Toll-like Receptor Signaling | Map2k6, Fos, Jun, Map3k1, Tnfaip3 | 1.71E | 6.76E-02 |
4. Discussion
Our study is the first report of transcriptomic and miRNA profiling in BAT from mice fed a HF compared to HF diet supplemented with EPA. The depth of transcriptomic and miRNA profiling performed in this study uncovered many genes and miRNAs involved in pathways for thermogenesis, energy expenditure, brown fat development, which may link to lipid homeostasis, and fatty acid oxidation. Our findings suggest that EPA increased markers of thermogenesis, similar to cold exposure or diet induced thermogenesis.
In the previous report [11], we investigated the effects of both EPA and DHA on cultured brown adipocytes. We observed more mitochondrial content with EPA than DHA. Therefore, for this study we only used EPA to detect its effect on BAT metabolism related to thermogenesis. While, DHA is a key component of membranes specifically in the central nervous system and contribute to brain development [42], studies have shown that EPA suppresses inflammation and major coronary events better than DHA [43].
Out of four canonical pathways activated by EPA, we focused on PTEN and PPAR pathways, which are related to thermogenesis. PTEN is a tumor suppressor protein which inhibits the activity of phosphatidylinositol 3-kinase (PI3K). Moreover, transgenic mice with higher copies of Pten displayed increased energy expenditure and higher levels of Ucp1 in BAT [32]. Corroborating with this, deficiency of Pten in mice is linked to obesity and insulin resistance suggesting Pten activation is beneficial [44]. These reports and our transcriptomic profiling together suggests that EPA could activate PTEN in BAT, and this in turn may be involved in increasing thermogenesis and energy expenditure.
Another pathway that is modulated by EPA in BAT includes PPAR. This is consistent with previous studies which show that omega-3 fatty acids are ligands for PPARs, namely Ppar-α and Ppar-γ, leading to activation of adipogenesis, thermogenesis, fat catabolism, mitochondrial fatty acid oxidation rates and energy expenditure in BAT and WAT [45]. Ppar-α is highly expressed in BAT and increases levels of thermogenic marker including Pgc1α and Ucp1 [46], all of which are suitable targets for BAT activation and metabolic disorders. Moreover, some studies have already reported that n-3 PUFAs induce thermogenesis in BAT by increasing the level of Ppar-α and Pparγ [47–49]. Beside the activation of BAT, omega 3 fatty acids act as natural ligands and activators of Ppar-α, which blocks NF-KB activity in WAT to prevent inflammation [50]. Furthermore, the free fatty acid receptor 4 (Ffar4), also known as G-protein-coupled receptor 120 (Gpr120), was also significantly upregulated by EPA in our study. However, it was not listed in the differentially expressed genes in the tables above, due to the higher cut off used in our data analyses. These findings are consistent with a recent report demonstrating that Ffar4 was a functional receptor for omega 3 fatty acids in mouse BAT [51].
Our results show that EPA significantly increased miR-455 expression (3). This miRNA targets key brown adipogenic signaling molecules including Hif1αn, Ppar-γ, and Tgfβr3 [19]. Mir-455 inhibits Hif1an, which in turn induces Ucp1. Indeed, higher BAT Ucp1 levels were reported in Hif1αn whole-body knockout mice [52]. Furthermore, Hif1αn is upstream of the AMPK-PGC1α regulatory pathway [19], and miR-455 also activates protein kinase, AMP-activated, alpha1 (AMPKa1), an enzyme which catalytic subunit of AMP-activated protein kinase (AMPK) by targeting Hif1an [19]. AMPK1 activation in turn induces Pgc1α and Ucp1 expression to induce mitochondrial biogenesis and thermogenesis [53]. Thus, our findings are consistent with these reports and validate the importance of this pathway in BAT regulation.
Moreover, the main network pathway in figure 3 shows that Hif1αn serves as an intermediate target between miR-455 and another gene, Notch1. Mir-455 reduces the expression of Notch1, which in turn promotes browning of white adipose tissue to prevent obesity [54, 55]. Additionally, mRNA levels of beige cell markers such as CD137 and T-Box1 (Tbx1), and mitochondria markers such as carnitine palmitoyltransferase 1A (Cpt1a) and Cpt2, were also higher, suggesting beneficial effects of Notch depletion [54]. This has been also confirmed in mouse studies, where Notch specific deletion in inguinal WAT induced Ucp1, cell death-inducing DFFA-like effector A (Cidea), Pgc1α and Prdm16 expression [56]. Consistent with these reports, our data predict an inhibitory effect of EPA on Hif1αn and Notch1 genes, which may increase thermogenesis and energy expenditure by increasing levels of Prdm16, Pgc1α, and Ucp1 as the key regulators of thermogenesis.
Our results further demonstrated that miR-455 also downregulated Tgfβr3 level and its downstream member, Smad2/3. TGF-β signals through dual serine/threonine kinase receptors and its transcription factors named Smads, with Smad3 serving as the major facilitator of TGF-β signal [57]. Blockade of TGF-β/Smad3 signaling leads to enhanced metabolic profile and energy expenditure [24]. Interestingly, Smad3 global knockout mouse is resistant to diet induced obesity and has elevated thermogenic markers and increased basal rate of oxygen consumption in the WAT compared to controls [24]. These results confirm that loss of Tgfβ/Smad3 triggers browning of WAT. In agreement with these studies, our study validates these findings and identified a link between miR-455 and its Tgfβ/Smad3 targets.
PPAR-γ is a nuclear receptor highly expressed in BAT and WAT, which serves as a master transcriptional regulator of brown adipocyte differentiation and it is crucial for tissue development, function, and survival [58]. In our study, EPA induced miR-455 and PPAR-γ expression, inhibiting Ncor expression, known for its role in skeletal muscle and BAT energy homeostasis [59]. Ncor1 is a direct corepressor of PPAR-γ. Hence, clearance of Ncor from the PPAR-γ complex is necessary for recruitment of the brown cofactor Prdm16 [21]. Furthermore, inhibition of Ncor leads to higher levels of PPAR-α, which also stimulates mitochondrial activity, fatty acid β-oxidation via Ucp1 [60]. Thus, our results demonstrate that EPA increased thermogenic markers, in part via regulation of miR-455, a novel EPA target, upstream of PPAR-γ and PPAR-α, which inhibit Ncor to induce key thermogenic biomarkers. However, other studies have also reported potential roles of miR-455 in cancer, specifically as a tumor suppressor [61–63]. Taken together these studies suggest potential dual benefits for miR-455 in both cancer and energy balance.
Another miRNA, miR-129–5p was also significantly upregulated by EPA. Its reported targets are Igf2 and Egr1 [30, 64]. Interestingly, mouse mutant for Igf2 gene demonstrated massive BAT hypertrophy, less lean mass and higher levels of Ucp1 and Prdm16 compared to wildtype mice [23]. Additionally, inactivation of Igf2 in preadipocytes increased Ucp1 and Prdm16 expression consistent with in vivo studies. The other target for miR-129–5p, Egr1 when knocked down in the WAT induced expression of BAT Ucp1 and Pgc1α [22]. These data indicate that miR-129–5p regulates important genes involved in thermogenesis. Although the fold differences in the expression of miRNAs were small in the current study, previous studies have shown that even a small (2-fold) change will have a significant impact on target protein levels [65].
In the present study, we used a diet containing 36 g/kg EPA, which is equivalent to 6.75% of energy intake in diet. In comparison, the intakes of EPA and DHA in the United States are low for omega 3s ~0.1–0.2 g/d [66] and the current recommendations vary from 1 g/d to 4 g/d [67] with upper levels necessary to reduce hypertriglyceridemia. Higher doses have been used in human studies from 4.2 g/d [68] to doses up to 15 g/d described in a recent meta-analysis [69, 70]. However, in human studies, the effect of fish oil in improving fat oxidation and whole-body metabolism is inconsistent. Some factors responsible for theses inconclusive results include usage of lower doses in human studies compared to animal studies, genetic variabilities in humans compared to selected animals for research, and different ratio of EPA/DHA which were used for clinical studies [71, 72]
5. Conclusion
We report several previously identified as well as new genes, miRNAs, and pathways involved in thermogenesis, energy expenditure, and brown fat development. Some of the identified targets of EPA in BAT include miRNAs 455, 129 and PPAR/NCOR, and SMAD3/TGF pathways. These all may lead to improved metabolic homeostasis, suggesting that omega-3 fatty acids, and potentially other nutritional interventions are viable therapies for preventing and/or treating obesity and related metabolic disorders, in part via regulation of BAT. While our study has many strengths and novel findings, it has some limitations. We did not measure the energy expenditure directly in our mice, thus we did not validate whether increased expression of thermogenic biomarkers by EPA translated into increased energy expenditure in the EPA fed mice. Moreover, additional molecular and physiological studies are necessary to further validate the mRNA-miRNA regulatory pairs that we identified in our unbiased screens, their direct role in thermogenesis and mechanism of their regulation by EPA both in vitro and in vivo. Such studies include using miRNA mimics and inhibitors in cells and animals, to validate target genes which are beyond the scope of the current report. Lastly, it will be worthwhile in future to determine if lower doses of EPA compared to DHA or whole fish oil recapitulate the responses reported here for EPA in animal models; and whether these findings can be translated in clinical studies.
Supplementary Material
Highlights:
EPA upregulated thermogenic markers similar to cold exposure or exercise.
MiR-455 and −129 are novel miRNAs involved in regulation of thermogenesis by EPA.
Smad3, Tgfbr3, and Notch are novel RNAs down-regulated by EPA for thermogenesis.
Modulation of BAT function using EPA is a potential novel target for obesity
Acknowledgements
We thank members of the Dr. Moustaid-Moussa’s lab for their assistance with this project, especially Shane Scoggin for his assistance with various technical aspects of this research. We also would like to thank the center of Biotechnology and Genomics at Texas Tech University, the department of Biology and Biochemistry, University of Houston and Baylor College of Medicine, for assistance from their facilities used in this study. The authors’ responsibilities were as follows N.M.M. and N.S.K. designed the study; M. P.; N. N. W; N. S. K conducted research; M. P. drafted the first version of this paper; M. P; N.M.M. L. R; P. G., C. C; and K. R. were involved in the data analyses; P. K; P. H. G; C.C. and N. M. M. provided facilities for research; N. M. M; provided facilities and materials for this research, and is the primary responsibility for this research and final content of the manuscript. Funding for this research was supported in part by NIH/NCCIH grant #R15AT008879-01A1 (NMM), USDA NIFA Exploratory award (2015-67030-23452), startup funds from Texas Tech University, and The Obesity Research Cluster.
Abbreviations used:
- Adra1
adenosine A1 receptor
- AMPK
AMP-activated protein kinase
- Arhgap35
Rho GTPase activating protein 35
- Arhgef17
Rho guanine nucleotide exchange factor 17
- Atg1
autophagy related 1
- BAT
brown adipose tissue
- BMPR
bone morphogenetic protein receptor
- Casp7
caspase 7
- Cbl
Cbl proto-oncogene
- Cidea
cell death-inducing DFFA-like effector A
- Cish
cytokine inducible SH2 containing protein
- Cpt-1
carnitine palmitoyl transferase I
- Crebbp
CREB binding protein
- DE
differentially expressed
- DHA
docosahexaenoic acid
- DMEM
dulbecco’s modified eagle medium
- Egr1
early growth response 1
- EPA
eicosapentaenoic Acid
- Esr1
estrogen receptor 1
- FDR
false discovery rate
- Ffar4
Free fatty acid receptor 4
- Fnbp1
formin binding protein 1
- Fos
fos proto-oncogene
- Foxo1
forkhead box O1
- Gapdh
glyceraldehyde 3phosphate dehydrogenase
- GEO
gene expression omnibus
- Gnao1
G protein subunit alpha O1
- Gpd1
glycerol-3-phosphate dehydrogenase 1
- (Gpr120)
G-protein-coupled receptor 120
- HC
hierarchical clustering
- HF
high-fat diet
- HF-EPA
HF diet supplemented with EPA
- Hif1an
hypoxia inducible factor 1 alpha subunit inhibitor
- Hi-Seq
high sequencing
- HMGB
high mobility group box
- HS
hematopoietic lineage cell-specific protein
- Igf2
insulin like growth factor 2
- IL
interleukin
- ILK
integrin linked kinase
- Inhbb
inhibin beta B
- Insr
insulin receptor
- IPA
ingenuity pathway analysis
- Jun
Jun proto-oncogene
- Kat6a
lysine acetyltransferase 6A
- Klhl6
kelch-like family member 6
- LF
low fat
- miRNA and miR
microRNA
- mRNA
messenger RNA
- Myh9
myosin heavy chain 9
- Ncor
nuclear receptor corepressor
- NF-κB
nuclear factor kappa-light-chainenhancer of activated B cells
- Notch1
Notch homolog 1, translocation-associated (Drosophila)
- Pdpk1
3-phosphoinositide dependent protein kinase 1
- Pgc1α
peroxisome proliferator-activated receptor gamma coactivator 1 alpha
- Pik3c2b
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta
- Pmepa1
prostate transmembrane protein, androgen induced 1
- Pon
Paraoxonase
- Ppar
peroxisome proliferator-activated receptor
- Prdm16
PR domain containing 16
- Prl41
catalyze protein synthesis, ribosomal protein L41
- PTEN
phosphatase and tensin homolog
- Ptpn11
protein tyrosine phosphatase, Non-Receptor Type 11
- PUFA
polyunsaturated fatty acid
- Rhob
ras homolog family member B
- RHO-GDI
Rho GDP-dissociation inhibitor
- PI3K
phosphatidylinositol 3-kinase
- RIN
RNA integrity numbers
- RNA-Seq
RNA sequencing
- RPKM
reads per kilo base per million mapped reads
- RT-qPCR
quantitative reverse transcription
- RXR
retinoid X receptor
- SAT
subcutaneous adipose tissue
- SEM
standard error of mean
- Sgk
serine/threonine-protein kinase
- Smad
SMAD family member
- SNS
sympathetic nervous system
- Sp1
Sp1 transcription factor
- SRA
sequence read archive
- STAT
signal transcription and activator of transcription
- Tbx
T-box transcription factor
- TG
triglycerides
- TGFβ
transforming growth factor –β
- TGFβR
transforming growth factor beta receptor
- TLR
toll-like receptor
- Tnfaip3
TNF Alpha Induced Protein 3
- TNFR
tumor necrosis factor receptor
- UCP
uncoupling protein
- VAT
visceral adipose tissue
- Wasf2
WAS protein family member 2
- WAT
white adipose tissue
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Ogden CL, Carroll MD, Flegal KM, Prevalence of obesity in the United States, Jama 312(2) (2014) 189–90. [DOI] [PubMed] [Google Scholar]
- [2].Cummins TD, Holden CR, Sansbury BE, Gibb AA, Shah J, Zafar N, Tang Y, Hellmann J, Rai SN, Spite M, Bhatnagar A, Hill BG, Metabolic remodeling of white adipose tissue in obesity, American journal of physiology. Endocrinology and metabolism 307(3) (2014) E262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chechi K, Nedergaard J, Richard D, Brown adipose tissue as an anti-obesity tissue in humans, Obesity reviews : an official journal of the International Association for the Study of Obesity 15(2) (2014) 92–106. [DOI] [PubMed] [Google Scholar]
- [4].Symonds ME, Brown Adipose Tissue Growth and Development, Scientifica 2013 (2013) doi: 10.1155/2013/305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sidossis L, Kajimura S, Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis, The Journal of clinical investigation 125(2) (2015) 478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pfeifer A, Hoffmann LS, Brown, beige, and white: the new color code of fat and its pharmacological implications, Annual review of pharmacology and toxicology 55 (2015) 207–27. [DOI] [PubMed] [Google Scholar]
- [7].Poher AL, Altirriba J, Veyrat-Durebex C, Rohner-Jeanrenaud F, Brown adipose tissue activity as a target for the treatment of obesity/insulin resistance, Frontiers in physiology 6 (2015) doi: 10.3389/fphys.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wortman P, Miyazaki Y, Kalupahana NS, Kim S, Hansen-Petrik M, Saxton AM, Claycombe KJ, Voy BH, Whelan J, Moustaid-Moussa N, n3 and n6 polyunsaturated fatty acids differentially modulate prostaglandin E secretion but not markers of lipogenesis in adipocytes, Nutrition & metabolism 6 (2009) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Siriwardhana N, Kalupahana NS, Moustaid-Moussa N, Health benefits of n-3 polyunsaturated fatty acids: eicosapentaenoic acid and docosahexaenoic acid, Advances in food and nutrition research 65 (2012) 211–22. [DOI] [PubMed] [Google Scholar]
- [10].Kalupahana NS, Claycombe K, Newman SJ, Stewart T, Siriwardhana N, Matthan N, Lichtenstein AH, Moustaid-Moussa N, Eicosapentaenoic acid prevents and reverses insulin resistance in high-fat diet-induced obese mice via modulation of adipose tissue inflammation, The Journal of nutrition 140(11) (2010) 1915–22. [DOI] [PubMed] [Google Scholar]
- [11].Pahlavani M, Razafimanjato F, Ramalingam L, Kalupahana NS, Moussa H, Scoggin S, Moustaid-Moussa N, Eicosapentaenoic Acid Regulates Brown Adipose Tissue Metabolism in High Fat Fed Mice and in Clonal Brown Adipocytes, The Journal of Nutritional Biochemistry 39 (2016) 101–109. [DOI] [PubMed] [Google Scholar]
- [12].Trajkovski M, Lodish H, MicroRNA networks regulate development of brown adipocytes, Trends in Endocrinology & Metabolism 24(9) (2013) 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Andrews S, FastQC A Quality Control tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (Accessed April 2016).
- [14].Creighton CJ, Reid JG, Gunaratne PH, Expression profiling of microRNAs by deep sequencing, Brief Bioinform 10(5) (2009) 490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ramsköld D, Wang ET, Burge CB, Sandberg R, An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data, PLoS Comput Biol 5(12) (2009) e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rosell M, Kaforou M, Frontini A, Okolo A, Chan Y-W, Nikolopoulou E, Millership S, Fenech ME, MacIntyre D, Turner JO, Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice, American Journal of Physiology-Endocrinology and Metabolism 306(8) (2014) E945–E964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rupérez AI, Gil A, Aguilera CM, Genetics of Oxidative Stress in Obesity, International Journal of Molecular Sciences 15(2) (2014) 3118–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bi P, Shan T, Liu W, Yue F, Yang X, Liang X-R, Wang J, Li J, Carlesso N, Liu X, Kuang S, Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity, Nat Med 20(8) (2014) 911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang H, Guan M, Townsend KL, Huang TL, An D, Yan X, Xue R, Schulz TJ, Winnay J, Mori M, Hirshman MF, Kristiansen K, Tsang JS, White AP, Cypess AM, Goodyear LJ, Tseng YH, MicroRNA-455 regulates brown adipogenesis via a novel HIF1anAMPK-PGC1alpha signaling network, EMBO reports 16(10) (2015) 1378–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hampton M, Melvin RG, Andrews MT, Transcriptomic Analysis of Brown Adipose Tissue across the Physiological Extremes of Natural Hibernation, PloS one 8(12) (2013) e85157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li P, Fan W, Xu J, Lu M, Yamamoto H, Auwerx J, Sears DD, Talukdar S, Oh D, Chen A, Bandyopadhyay G, Scadeng M, Ofrecio JM, Nalbandian S, Olefsky JM, Adipocyte NCoR knockout decreases PPARgamma phosphorylation and enhances PPARgamma activity and insulin sensitivity, Cell 147(4) (2011) 815–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang J, Zhang Y, Sun T, Guo F, Huang S, Chandalia M, Abate N, Fan D, Xin HB, Chen YE, Fu M, Dietary obesity-induced Egr-1 in adipocytes facilitates energy storage via suppression of FOXC2, Scientific reports 3 (2013) 1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Borensztein M, Viengchareun S, Montarras D, Journot L, Binart N, Lombes M, Dandolo L, Double Myod and Igf2 inactivation promotes brown adipose tissue development by increasing Prdm16 expression, FASEB journal : official publication of the Federation of American Societies for Experimental Biology 26(11) (2012) 4584–91. [DOI] [PubMed] [Google Scholar]
- [24].Yadav H, Quijano C, Kamaraju AK, Gavrilova O, Malek R, Chen W, Zerfas P, Zhigang D, Wright EC, Stuelten C, Sun P, Lonning S, Skarulis M, Sumner AE, Finkel T, Rane SG, Protection from obesity and diabetes by blockade of TGF-beta/Smad3 signaling, Cell metabolism 14(1) (2011) 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu J, Li T, Yang D, Ma R, Moran TH, Smith WW, Synphilin-1 alters metabolic homeostasis in a novel Drosophila obesity model, International journal of obesity (2005) 36(12) (2012) 1529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tinahones FJ, Coin Araguez L, Murri M, Oliva Olivera W, Mayas Torres MD, Barbarroja N, Gomez Huelgas R, Malagon MM, El Bekay R, Caspase induction and BCL2 inhibition in human adipose tissue: a potential relationship with insulin signaling alteration, Diabetes care 36(3) (2013) 513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim HJ, Kobayashi M, Sasaki T, Kikuchi O, Amano K, Kitazumi T, Lee YS, YokotaHashimoto H, Susanti VY, Kitamura YI, Nakae J, Kitamura T, Overexpression of FoxO1 in the hypothalamus and pancreas causes obesity and glucose intolerance, Endocrinology 153(2) (2012) 659–71. [DOI] [PubMed] [Google Scholar]
- [28].Schonthaler HB, Guinea-Viniegra J, Wagner EF, Targeting inflammation by modulating the Jun/AP-1 pathway, Annals of the rheumatic diseases 70 Suppl 1 (2011) i109–12. [DOI] [PubMed] [Google Scholar]
- [29].Gao YZ, Jiang Y, Wu X, Bai CY, Pan YC, Sun YZ, Molecular characteristics and expression profiles of glycerol-3-phosphate dehydrogenase 1 (GPD1) gene in pig, Molecular Biology Reports 38(3) (2011) 1875–1881. [DOI] [PubMed] [Google Scholar]
- [30].Døssing KBV, Binderup T, Kaczkowski B, Jacobsen A, Rossing M, Winther O, Federspiel B, Knigge U, Kjær A, Friis-Hansen L, Down-regulation of miR-129–5p and the let7 family in neuroendocrine tumors and metastases leads to up-regulation of their targets Egr1, G3bp1, Hmga2 and Bach1, Genes 6(1) (2014) 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Plutzky J, The PPAR-RXR transcriptional complex in the vasculature: energy in the balance, Circulation research 108(8) (2011) 1002–16. [DOI] [PubMed] [Google Scholar]
- [32].Ortega-Molina A, Efeyan A, Lopez-Guadamillas E, Munoz-Martin M, Gomez-Lopez G, Canamero M, Mulero F, Pastor J, Martinez S, Romanos E, Mar Gonzalez-Barroso M, Rial E, Valverde AM, Bischoff JR, Serrano M, Pten positively regulates brown adipose function, energy expenditure, and longevity, Cell metabolism 15(3) (2012) 382–94. [DOI] [PubMed] [Google Scholar]
- [33].Dovas A, John R Couchman, RhoGDI: multiple functions in the regulation of Rho family GTPase activities, Biochemical Journal 390(Pt 1) (2005) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Craft CS, Pietka TA, Schappe T, Coleman T, Combs MD, Klein S, Abumrad NA, Mecham RP, The extracellular matrix protein MAGP1 supports thermogenesis and protects against obesity and diabetes through regulation of TGF-beta, Diabetes 63(6) (2014) 192032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Priceman SJ, Kujawski M, Shen S, Cherryholmes GA, Lee H, Zhang C, Kruper L, Mortimer J, Jove R, Riggs AD, Yu H, Regulation of adipose tissue T cell subsets by Stat3 is crucial for diet-induced obesity and insulin resistance, Proceedings of the National Academy of Sciences of the United States of America 110(32) (2013) 13079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baker RG, Hayden MS, Ghosh S, NF-κB, inflammation and metabolic disease, Cell metabolism 13(1) (2011) 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gunasekaran MK, Viranaicken W, Girard AC, Festy F, Cesari M, Roche R, Hoareau L, Inflammation triggers high mobility group box 1 (HMGB1) secretion in adipose tissue, a potential link to obesity, Cytokine 64(1) (2013) 103–11. [DOI] [PubMed] [Google Scholar]
- [38].Ahmed AU, Sarvestani ST, Gantier MP, Williams BR, Hannigan GE, Integrin-linked kinase modulates lipopolysaccharide- and Helicobacter pylori-induced nuclear factor kappaB-activated tumor necrosis factor-alpha production via regulation of p65 serine 536 phosphorylation, The Journal of biological chemistry 289(40) (2014) 27776–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sindhu S, Thomas R, Shihab P, Sriraman D, Behbehani K, Ahmad R, Obesity Is a Positive Modulator of IL-6R and IL-6 Expression in the Subcutaneous Adipose Tissue: Significance for Metabolic Inflammation, PloS one 10(7) (2015) e0133494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Romanatto T, Roman EA, Arruda AP, Denis RG, Solon C, Milanski M, Moraes JC, Bonfleur ML, Degasperi GR, Picardi PK, Hirabara S, Boschero AC, Curi R, Velloso LA, Deletion of tumor necrosis factor-alpha receptor 1 (TNFR1) protects against diet-induced obesity by means of increased thermogenesis, The Journal of biological chemistry 284(52) (2009) 36213–22. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [41].Fresno M, Alvarez R, Cuesta N, Toll-like receptors, inflammation, metabolism and obesity, Archives of physiology and biochemistry 117(3) (2011) 151–64. [DOI] [PubMed] [Google Scholar]
- [42].Lauritzen L, Brambilla P, Mazzocchi A, Harsløf LBS, Ciappolino V, Agostoni C, DHA Effects in Brain Development and Function, Nutrients 8(1) (2016) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Swanson D, Block R, Mousa SA, Omega-3 Fatty Acids EPA and DHA: Health Benefits Throughout Life, Advances in Nutrition 3(1) (2012) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim YJ, Sherwin R, Devaskar S, Lesche R, Magnuson MA, Wu H, Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected], Proceedings of the National Academy of Sciences of the United States of America 101(7) (2004) 2082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Marion-Letellier R, Savoye G, Ghosh S, Fatty acids, eicosanoids and PPAR gamma, European journal of pharmacology (2015). [DOI] [PubMed] [Google Scholar]
- [46].Contreras AV, Torres N, Tovar AR, PPAR-α as a Key Nutritional and Environmental Sensor for Metabolic Adaptation, Advances in Nutrition 4(4) (2013) 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Takahashi Y, Ide T, Dietary n-3 fatty acids affect mRNA level of brown adipose tissue uncoupling protein 1, and white adipose tissue leptin and glucose transporter 4 in the rat, The British journal of nutrition 84(2) (2000) 175–84. [PubMed] [Google Scholar]
- [48].Bargut TC, Silva-e-Silva AC, Souza-Mello V, Mandarim-de-Lacerda CA, Aguila MB, Mice fed fish oil diet and upregulation of brown adipose tissue thermogenic markers, European journal of nutrition 55(1) (2016) 159–69. [DOI] [PubMed] [Google Scholar]
- [49].Pahlavani M, Razafimanjato F, Ramalingam L, Kalupahana NS, Moussa H, Scoggin S, Moustaid-Moussa N, Eicosapentaenoic Acid Regulates Brown Adipose Tissue Metabolism in High Fat Fed Mice and in Clonal Brown Adipocytes, The Journal of Nutritional Biochemistry (2016). [DOI] [PubMed] [Google Scholar]
- [50].Grygiel-Górniak B, Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications – a review, Nutrition journal 13 (2014) 17–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kim J, Okla M, Erickson A, Carr T, Natarajan SK, Chung S, Eicosapentaenoic Acid Potentiates Brown Thermogenesis through FFAR4-dependent Up-regulation of miR-30b and miR-378, The Journal of biological chemistry 291(39) (2016) 20551–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhang N, Fu Z, Linke S, Chicher J, Gorman JJ, Visk D, Haddad GG, Poellinger L, Peet DJ, Powell F, Johnson RS, The asparaginyl hydroxylase factor inhibiting HIF-1alpha is an essential regulator of metabolism, Cell metabolism 11(5) (2010) 364–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ahmadian M, Abbott MJ, Tang T, Hudak CS, Kim Y, Bruss M, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, Wang Y, Duncan RE, Kang C, Sul HS, Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype, Cell metabolism 13(6) (2011) 739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bi P, Shan T, Liu W, Yue F, Yang X, Liang XR, Wang J, Li J, Carlesso N, Liu X, Kuang S, Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity, Nat Med 20(8) (2014) 911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zheng X, Linke S, Dias JM, Zheng X, Gradin K, Wallis TP, Hamilton BR, Gustafsson M, Ruas JL, Wilkins S, Bilton RL, Brismar K, Whitelaw ML, Pereira T, Gorman JJ, Ericson J, Peet DJ, Lendahl U, Poellinger L, Interaction with factor inhibiting HIF1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways, Proceedings of the National Academy of Sciences of the United States of America 105(9) (2008) 3368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Di Zazzo E, De Rosa C, Abbondanza C, Moncharmont B, PRDM Proteins: Molecular Mechanisms in Signal Transduction and Transcriptional Regulation, Biology 2(1) (2013) 107–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Xu P, Liu J, Derynck R, Post-translational regulation of TGF-beta receptor and Smad signaling, FEBS Lett 586(14) (2012) 1871–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Petrovic N, Shabalina IG, Timmons JA, Cannon B, Nedergaard J, Thermogenically competent nonadrenergic recruitment in brown preadipocytes by a PPARgamma agonist, American journal of physiology. Endocrinology and metabolism 295(2) (2008) E287–96. [DOI] [PubMed] [Google Scholar]
- [59].Yamamoto H, Evan G Williams L Mouchiroud C Cantó W Fan M Downes C Héligon, Grant D Barish B Desvergne, Ronald M Evans K Schoonjans J Auwerx, NCoR1 Is a Conserved Physiological Modulator of Muscle Mass and Oxidative Function, Cell 147(4) (2011) 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hondares E, Rosell M, Diaz-Delfin J, Olmos Y, Monsalve M, Iglesias R, Villarroya F, Giralt M, Peroxisome proliferator-activated receptor alpha (PPARalpha) induces PPARgamma coactivator 1alpha (PGC-1alpha) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16, The Journal of biological chemistry 286(50) (2011) 43112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhao Y, Yan M, Yun Y, Zhang J, Zhang R, Li Y, Wu X, Liu Q, Miao W, Jiang H, MicroRNA-455–3p functions as a tumor suppressor by targeting eIF4E in prostate cancer, Oncology reports 37(4) (2017) 2449–2458. [DOI] [PubMed] [Google Scholar]
- [62].Zhan T, Huang X, Tian X, Chen X, Ding Y, Luo H, Zhang Y, Downregulation of MicroRNA-455–3p Links to Proliferation and Drug Resistance of Pancreatic Cancer Cells via Targeting TAZ, Molecular Therapy - Nucleic Acids 10 (2018) 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mao QD, Zhang W, Zhao K, Cao B, Yuan H, Wei LZ, Song MQ, Liu XS, MicroRNA455 suppresses the oncogenic function of HDAC2 in human colorectal cancer, Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas 50(6) (2017) e6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xu H-L, Feng Y-Y, Jia Z-K, Zhang Y-G, Li J, Xia M-L, MicroRNA-129–5p inhibits the proliferation and migration in renal cell carcinoma via targeting IGF2BP1, INTERNATIONAL JOURNAL OF CLINICAL AND EXPERIMENTAL PATHOLOGY 9(8) (2016) 8254–8260. [Google Scholar]
- [65].Chugh P, Dittmer DP, Potential Pitfalls in microRNA Profiling, Wiley interdisciplinary reviews. RNA 3(5) (2012) 601–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Papanikolaou Y, Brooks J, Reider C, Fulgoni VL, adults US are not meeting recommended levels for fish and omega-3 fatty acid intake: results of an analysis using observational data from NHANES 2003–2008, Nutrition journal 13(1) (2014) 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jain AP, Aggarwal KK, Zhang PY, Omega-3 fatty acids and cardiovascular disease, European review for medical and pharmacological sciences 19(3) (2015) 441–5. [PubMed] [Google Scholar]
- [68].Krebs JD, Browning LM, McLean NK, Rothwell JL, Mishra GD, Moore CS, Jebb SA, Additive benefits of long-chain n-3 polyunsaturated fatty acids and weight-loss in the management of cardiovascular disease risk in overweight hyperinsulinaemic women, International journal of obesity (2005) 30(10) (2006) 1535–44. [DOI] [PubMed] [Google Scholar]
- [69].Du S, Jin J, Fang W, Su Q, Does Fish Oil Have an Anti-Obesity Effect in Overweight/Obese Adults? A Meta-Analysis of Randomized Controlled Trials, PLoS One 10(11) (2015) e0142652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Miller PE, Van Elswyk M, Alexander DD, Long-chain omega-3 fatty acids eicosapentaenoic acid and docosahexaenoic acid and blood pressure: a meta-analysis of randomized controlled trials, American journal of hypertension 27(7) (2014) 885–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pedersen MH, Mølgaard C, Hellgren LI, Matthiessen J, Holst JJ, Lauritzen L, The Effect of Dietary Fish Oil in addition to Lifestyle Counselling on Lipid Oxidation and Body Composition in Slightly Overweight Teenage Boys, Journal of Nutrition and Metabolism 2011 (2011) 348368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].DeFina LF, Marcoux LG, Devers SM, Cleaver JP, Willis BL, Effects of omega-3 supplementation in combination with diet and exercise on weight loss and body composition, The American journal of clinical nutrition 93(2) (2011) 455–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data for HF and HF-EPA were submitted in the BioProject at NCBI under PRJNA353387 individual accession numbers for HF Sequence Read Archive (SRA) submission: BAT1 (SAMN05717642), BAT2 (SAMN05717643), and BAT3 (SAMN05717644). For HF-EPA SRA submission: BAT-EPA1 (SAMN05991740), BAT-EPA2 (SAMN05991741), and BAT-EPA3 (SAMN05991742). The small RNA-Seq data were submitted to Gene Expression Omnibus (GEO) data repository (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE85101 and GSE99506 for BAT HF and BAT HF-EPA respectively.