Abstract
The mitochondrial genome encodes several protein components of the oxidative phosphorylation (OXPHOS) pathway and is critical for aerobic respiration. These proteins have evolved adaptively in many taxa, but linking molecular-level patterns with higher-level attributes (e.g., morphology, physiology) remains a challenge. Turtles are a promising system for exploring mitochondrial genome evolution as different species face distinct respiratory challenges and employ multiple strategies for ensuring efficient respiration. One prominent adaptation to a highly aquatic lifestyle in turtles is the secondary loss of keratenized shell scutes (i.e., soft-shells), which is associated with enhanced swimming ability and, in some species, cutaneous respiration. We used codon models to examine patterns of selection on mitochondrial protein-coding genes along the three turtle lineages that independently evolved soft-shells. We found strong evidence for positive selection along the branches leading to the pig-nosed turtle (Carettochelys insculpta) and the softshells clade (Trionychidae), but only weak evidence for the leatherback (Dermochelys coriacea) branch. Positively selected sites were found to be particularly prevalent in OXPHOS Complex I proteins, especially subunit ND2, along both positively selected lineages, consistent with convergent adaptive evolution. Structural analysis showed that many of the identified sites are within key regions or near residues involved in proton transport, indicating that positive selection may have precipitated substantial changes in mitochondrial function. Overall, our study provides evidence that physiological challenges associated with adaptation to a highly aquatic lifestyle have shaped the evolution of the turtle mitochondrial genome in a lineage-specific manner.
Keywords: molecular evolution, respiratory physiology, ecomorphology, mitochondria, Cryptodira
Introduction
The organisms on Earth are faced with a wide variety of ecological and physiological challenges, and exhibit a corresponding diversity of ecophysiological adaptations. Uncovering the evolutionary mechanisms by which this phenotypic variation arose has long been a major goal for evolutionary biologists (Parsons and Albertson 2013). In particular, considerable efforts have been directed towards understanding how and why closely related organisms differ at the molecular level in otherwise highly conserved genes with critical organismal functions (Vitti etal. 2013). This approach is biologically meaningful considering its potential for revealing the evolutionary history of phenotypic traits at multiple biological scales (i.e., molecular, cellular, physiological, organismal), and for the insights it can provide into the adaptability of organisms to the environment (Kaessmann 2010).
In eukaryotic cells, the mitochondrial genome provides a powerful system for examining adaptation at the molecular level. This maternally inherited, nonrecombining organellar genome encodes 13 peptide subunits that contribute to four of the five complexes of the oxidative phosphorylation (OXPHOS) system: seven NADH dehydrogenases (ND1–ND6 and ND4L; Complex CI), Cytochrome b (CYTB; CIII), three cytochrome c oxidases (COX1–COX3; CIV), and two ATPases (ATP6 and ATP8; CV). These respiratory complexes organize into various supercomplexes (SCs), and one such SC, involving complexes CI, CIII, and CIV, has recently been subject to detailed structural analysis (Gu etal. 2016; Wu etal. 2016). Notably, this SC forms an interlinked structure by means of accessory subunits that contribute to oligomerization and both stabilize and regulate the catalytic core (which is composed of the mitochondrially encoded subunits as well as several nuclear-encoded subunits) (Fiedorczuk etal. 2016; Gu etal. 2016). The OXPHOS biochemical pathway produces 95% of the cell’s “energy currency” (i.e., adenosine triphosphate or ATP); the OXPHOS process is performed by means of electron flow in the inner mitochondrial membrane, with CI working in concert with CII, CIII, and CIV to create a proton energy gradient that is used by CV to synthesize ATP (Sazanov 2015). Amino acid changes in the constituent subunits can have large functional effects and several mutations have been linked to human diseases (Bridges etal. 2011). Accordingly, a wealth of information exists on the structure and function of the mitochondrially encoded proteins (Xia etal. 2013; Sazanov 2015; Zhou etal. 2015).
It had classically been believed that most polymorphisms and substitutions in mitochondrial DNA (mtDNA) were selectively neutral (Avise etal. 1987), but two key lines of research called this notion into question (Dowling etal. 2008). First, it was shown that mitochondrial amino acid replacements can be beneficial, underlying adaptive improvements in metabolic performance (Garvin etal. 2011). Second, protein-coding mtDNA variation has been associated with differences in thermal or metabolic needs (e.g., heat production, oxygen consumption, physical activity) across species that either experience distinct environmental conditions (da Fonseca etal. 2008; Garvin, Thorgaard, etal. 2015) or employ different locomotive strategies (Shen etal. 2010; Zhang and Broughton 2015). In light of these findings, the evolution of mitochondrial function has become the subject of numerous studies testing for Darwinian selection in a wide range of nonmodel organisms (reviewed in Garvin, Bielwaski, etal. 2015), particularly those living in challenging habitats (Welch etal. 2014) and those distributed across geographic clines or ecological gradients (Morales etal. 2015).
Despite the growing recognition of nonneutral genetic variation in mtDNA, most work supporting this relationship in vertebrates comes from mammals and birds, while reptiles are under-represented. Unlike mammals and birds, reptiles are ectothermic, making them an interesting group to further explore the mode of evolution of the mtDNA: the body temperatures of ectotherms are similar to that of their surroundings, resulting in lower metabolic rates and lower energy requirements relative to endotherms. This fundamental difference is expected to result in ectotherms having unique patterns of mitochondrial evolution (Rand 1993). Consistent with this hypothesis, environmental factors (i.e., latitude and habitat) appear to be major determinants of mitochondrial and nuclear mutation rates in ectotherms but not in endotherms (LourençO etal. 2012). Furthermore, some reptiles have distinct metabolic strategies (e.g., metabolic down-regulation in anoxia-tolerant turtles—Galli etal. 2013, or increased metabolic rates in the largest marine turtle Dermochelys coriacea—Bostrom etal. 2010) or accelerated rates of mitochondrial genome evolution (e.g., snakes; Castoe etal. 2008) that differ substantially from other vertebrates.
Among reptiles, turtles (order Chelonia) possess remarkable adaptations to pronounced environmental stresses that are unique among amniotes. Turtles originated at least 240 Ma (Schoch and Hans-Dieter 2015) and have diversified and invaded a number of habitats including marine, freshwater, and terrestrial ecosystems, and ranging from tropical to temperate latitudes (Ernst and Barbour 1989). Numerous morphological and physiological specializations have been described in turtles, the protective shell being the most familiar example (Wyneken etal. 2008). However, a unique feature of some highly aquatic turtles is a shell devoid of protective keratinized scutes. This low degree of shell ossification represents a secondary loss that evolved independently along the lineages leading to the following: 1) the softshells clade (family Trionychidae), 2) the pig-nosed turtle (Carettochelys insculpta), and 3) the leatherback sea turtle (Dermochelys coriacea) (Joyce 2007, 2014). This loss likely occurred in the Late Cretaceous to early Eocene for both Carettochelys and Dermochelys (Joyce 2014), and in the Early Cretaceous for Trionychidae (Georgalis and Joyce 2017). The lack of shell scutes in all three lineages is accompanied by a reduction of the plastron, which increases body flexibility and aquatic maneuverability, allowing for greater swimming speed (Scheyer etal. 2007; Davenport etal. 2011) and enhanced hydrodynamic performance (Alibardi and Toni 2006; Bang etal. 2016). Furthermore, in some species the loss of shell scutes has been linked to cutaneous respiration, in which gas exchange (O2 and CO2) occurs across the skin surface (Ultsch etal. 1984). This has proven to be important in Trionychidae, as it enables the members of this group to remain aerobic for long periods while submerged (Scheyer etal. 2007). For the pig-nosed turtle, cutaneous respiration has not yet been confirmed, but the remarkable similarities of its skin surface features with those of Trionychidae (i.e., thin, permeable, and vascularized) suggest that it is likewise capable of employing cutaneous respiration (Joyce etal. 2012). Both the softshells and the pig-nosed turtle also employ buccopharyngeal respiration (Winokur 1988). By contrast, the skin of leatherbacks does not have respiratory function (John Davenport, personal communication); instead, leatherbacks rely on their capacity to store oxygen in blood and tissues to remain aerobic during deep dives (Lutcavage etal. 1990).
These suites of adaptations relate to energetic requirements and physiological processes that impact respiration capacity (Storey etal. 2008), and thus may depend on or influence the OXPHOS system. As such, it seems reasonable to hypothesize that turtle mtDNA encoded proteins may have undergone adaptive evolutionary changes associated with the loss of shell scutes and the invasion of highly aquatic eco-physiological niches. Here, we investigated patterns of evolution in mtDNA protein coding genes across 53 Cryptodiran turtle species (representing a total of 10 families), testing for adaptive or divergent patterns of mtDNA evolution associated with the evolution of soft-shells. To do so, we employed a variety of maximum likelihood (ML) codon-based models of molecular evolution that estimate the nonsynonymous-to-synonymous substitution rate ratio (ω) (Goldman and Yang 1994). This parameter measures the form and strength of selective pressure on the protein-coding gene: when 0 < ω < 1 (dN < dS) indicates purifying selection; when ω = 1 (dN = dS) is consistent with mutations that are neutral, and ω > 1 (dN > dS) is indicative of fixation of advantageous mutations or positive selection.
Past studies that used similar approaches successfully uncovered links between mtDNA evolution and the origin of flight in bats (Shen etal. 2010), swimming performance in fishes (Zhang and Broughton 2015), and adaptation of polar bears to life in the artic (Welch etal. 2014). Our analyses revealed signatures of positive and divergent selection among turtle mtDNA protein-coding genes, supporting the notion that the adoption of highly aquatic lifestyles in soft-shelled turtles was associated with altered patterns of selection on mitochondrial function. We discuss our findings in the context of mitochondrial protein biochemistry and turtle ecology.
Results
Phylogenetic Tree Estimates
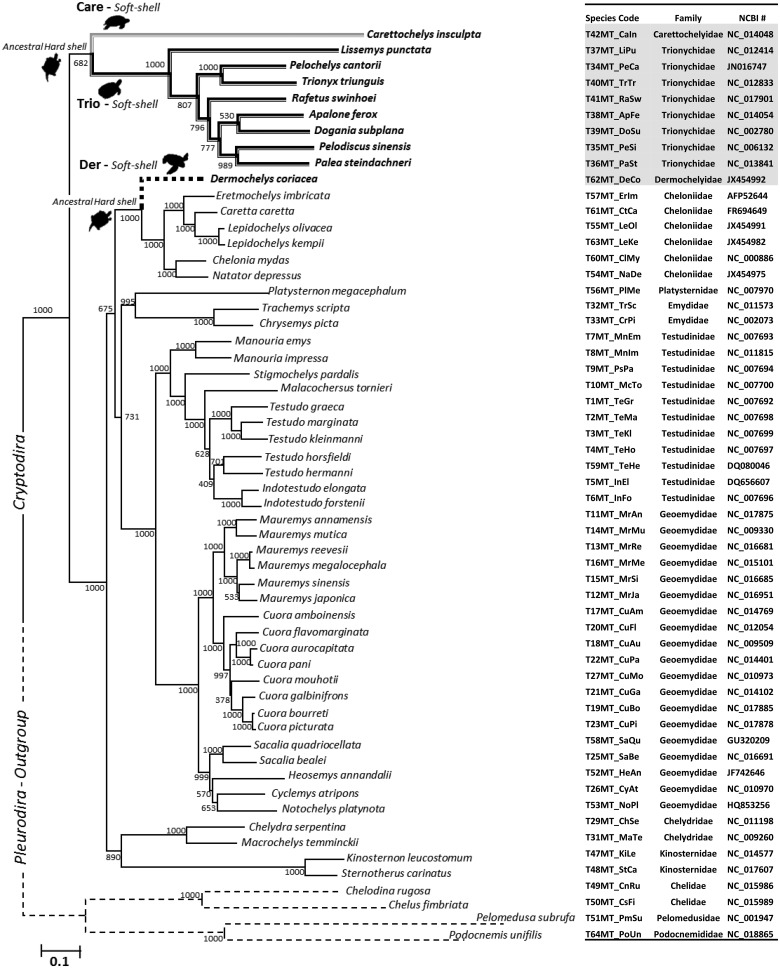
ML and Bayesian (BY) approaches were used to reconstruct the turtle phylogeny given a concatenated nucleotide alignment of all 12 mitochondrial heavy strand protein-coding genes. The two approaches recovered highly similar and well resolved topologies; the only differences involved the positioning of closely related taxa. Most nodes had relatively high ML bootstrap proportions (BP > 50%) and BY posterior probabilities (PP > 90%) (fig. 1 andsupplementary fig. S1, Supplementary Material online). Most importantly, both were generally congruent with accepted turtle relationships based on comprehensive phylogenetic analyses of both nuclear and mtDNA genes (Shaffer etal. 1997; Guillon etal. 2012). Given the close match between the two trees, only the ML tree was used in subsequent analyses.
Fig. 1.
Maximum Likelihood (ML) phylogenetic tree for Cryptodiran turtles. The tree was estimated using the concatenated data set of 12 mtDNA protein-coding genes (with Pleurodira as the outgroup; thin dashed branches). Species codes and NCBI Genbank accession numbers are provided alongside the phylogeny. Paleontological evidence indicates that soft-shells evolved 3 times independently, as denoted by the thicker branches (Trionychidae (Trio)-black, Carettochelyidae (Care)-gray, Dermochelyidae (Der)-dashed). Branch lengths and the scale bar indicate the number of nucleotide substitutions per site. Numbers adjacent to nodes represent ML bootstrap values (1,000 replicates).
Signatures of Positive and Divergent Selection in the OXPHOS System in Soft-Shelled Turtles
Random-Sites Analyses
We used random-sites codon models to gain insights into longstanding patterns of molecular evolution of mitochondrial genes in Cryptodiran turtles, analyzing both individual alignments for each of the 12 heavy strand protein-coding genes and a single concatenated alignment of all 12 genes (Tconc). Fitting the M0 model, which assumes that ω is invariant across both sites and branches (Goldman and Yang 1994), resulted in an overall ω estimate of 0.07 for the concatenated data set. Gene-specific estimates of ω were <0.10 for all genes except for ATP8 (ω = 0.18), with COX1, 2, and 3 having the lowest gene-specific estimates (ω ≤ 0.03) (table 1). Standard errors for the ω estimates were always ≤0.001. The elevated ω estimate for ATP8 suggests that selective constraint is not equally strong across all genes.
Table 1.
Data Set Properties.
| Genes | ω | κ | lnL | Branch-Specific dS Estimates |
Alignment Length (Codons) |
||
|---|---|---|---|---|---|---|---|
| Range | Median | With Gaps | Without Gaps | ||||
| ATP6 | 0.08 | 7.43 | −11571.913 | 0.00–1.59 | 0.23 | 232 | 224 |
| ATP8 | 0.18 | 9.31 | −2877.671 | 0.00–2.01 | 0.18 | 67 | 50 |
| COX1 | 0.01 | 8.53 | −19307.359 | 0.00–3.45 | 0.24 | 516 | 502 |
| COX2 | 0.03 | 8.33 | −8744.865 | 0.00–2.24 | 0.22 | 228 | 224 |
| COX3 | 0.03 | 8.36 | −10351.057 | 0.00–2.49 | 0.24 | 261 | 260 |
| CYTB | 0.04 | 7.38 | −16859.833 | 0.00–1.51 | 0.26 | 381 | 368 |
| ND1 | 0.04 | 6.64 | −14905.527 | 0.00–2.56 | 0.29 | 325 | 315 |
| ND2 | 0.08 | 7.27 | −17925.309 | 0.00–3.21 | 0.26 | 347 | 342 |
| ND3 | 0.09 | 5.96 | −6169.866 | 0.00–2.51 | 0.24 | 116 | 109 |
| ND4 | 0.06 | 7.96 | −22801.071 | 0.00–2.47 | 0.29 | 465 | 448 |
| ND4L | 0.08 | 7.22 | −4889.322 | 0.00–1.74 | 0.23 | 99 | 92 |
| ND5 | 0.07 | 6.30 | −30245.036 | 0.00–1.96 | 0.30 | 627 | 568 |
| Tconc | 0.07 | 6.55 | −171582.577 | 0.00–1.18 | 0.23 | 3664 | 3502 |
Note.—The M0 codon model was fit to each gene-specific alignment and to the concatenated (Tconc) alignment to obtain estimates of omega (ω), transition/transversion rate ratio (κ), and, for each branch, the number of synonymous substitutions per synonymous site (dS). The log-likelihood (lnL) for each fit is provided. Also shown are the lengths of each alignment (in codons).
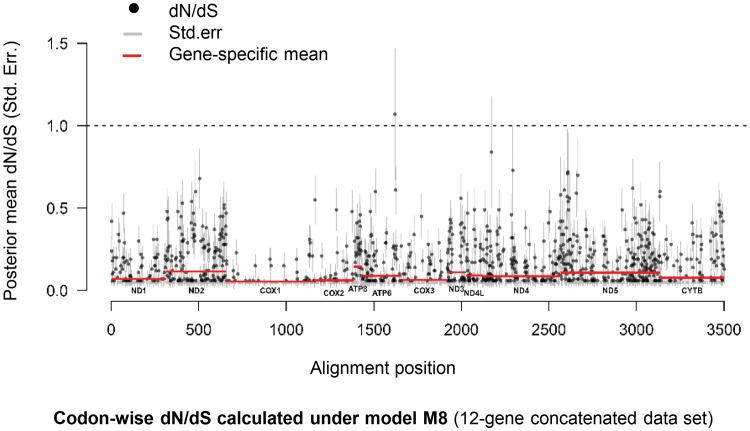
We applied the M8a-M8 random-sites likelihood ratio test (LRT) to test for site-specific positive selection. In no case did the test reveal evidence for positive selection (P = 1 for all tests; supplementary table S2, Supplementary Material online), indicating that negative selection is the dominant evolutionary force shaping turtle mitochondrial genes. That said, in all cases the log-likelihood score for the M8 model was substantially greater than that of the simple M0 model, indicating considerable variation in dN/dS among sites. A graphical examination of site-specific posterior mean estimates of dN/dS across the concatenated data set showed that, overall, dN/dS was lowest for COX1–COX3, and highest for ATP8, consistent with the M0 model results (fig. 2). However, many sites appeared to be evolving with weak-to-no selective constraint. For example, under the concatenated data set, 27 sites distributed across seven genes (ATP6, ND2, ND3, ND4, ND5, COX2, CYTB) had dN/dS estimates of at least 0.5 (fig. 2). This is consistent with very weak purifying selection operating at these sites, or possibly even the action of episodic (branch-specific) positive-selection.
Fig. 2.
Codon-wise dN/dS estimates for the concatenated data set of 12 mtDNA protein-coding genes. Codon-wise dN/dS estimates were calculated using the M8 random-site model. The y-axis shows the BEB posterior mean estimate of dN/dS for each site (±standard error). Horizontal solid lines show the mean estimate for each gene.
Branch-Site and Clade Model Analyses
Branch-site (BrS) and clade (CmC) models were used to determine whether patterns of mtDNA evolution and the evolution of shell type are linked in Cryptodiran turtles. Among the 53 Cryptodiran species in our data set are 10 species with soft-shells. Based on fossil evidence (Joyce 2007, 2014) this trait evolved independently at least 3 times: once along the lineage leading to Dermochelys coriacea, once along the lineage leading to Carettochelys insculpta, and once along the lineage leading to the Trionychidae clade (fig. 1). Our BrS and CmC models found evidence for positive and divergent selection associated with soft-shelled lineages, albeit in a manner that varied across branches and genes.
Branch-Site Model
BrS analysis of the concatenated data set identified the signature of positive selection along both the Trionychidae branch and the Care branch (table 2). For Trionychidae, the positive selection site-class was estimated to comprise 2.8% of the data set, with a ω2 estimate of 9.7. The corresponding values for Carettochelyidae were roughly similar (4.0%, with ω2 = 17.6). In both cases, the LRTs were highly significant (P < 0.001). For Dermochelyidae, ω2 was estimated at 3.6 but here the positively selected site class was of negligible size and the LRT did not indicate significance (P = 0.488). BEB analysis of the Tconc data set identified well-supported positively selected sites (PP ≥ 0.90) in nine genes when Trionychidae was the foreground (ATP6, ATP8, COX3, CYTB, ND1, ND2, ND4, ND4L, ND5: total of 21 sites) and six genes when Carettochelyidae was the foreground (ATP6, ATP8, COX3, CYTB, ND2, ND4: total of 19 sites). In no cases were the same sites identified on both the Carettochelyidae branch and the Trionychidae branch, though there was limited overlap in sites when considering all positively selected sites (i.e., with PP ≥ 0.50) (table 2). However, for both branches, the majority of the well-supported positively selected sites were in CI proteins: positively selected sites were over-represented in CI (and, to a lesser degree, CV), even when taking into account the number of codons analyzed for each complex (table 3). No sites were likewise identified as positively selected when the Dermochelyidae branch was set as the foreground, though a single site in ATP8 was identified with PP = 0.87; notably, this site was also identified along the Trionychidae branch (table 2 andsupplementary table S3, Supplementary Material online). All told, positive selection appears to have targeted the protein-coding components of the mitochondria in a function (OXPHOS Complex) and lineage specific manner. Gene-specific analyses were conducted to explore this variation in greater detail.
Table 2.
Results of Branch-Site (BrS) Model Analyses of the Concatenated Data Set of 12 mtDNA Protein-Coding Genes (Tconc).
| Branch | BrS Model (n.p.) | Site Class 2 |
lnL | P | Positively Selected Sites Gene: site | |
|---|---|---|---|---|---|---|
| ω2 | p2 | |||||
| Trionychidae | A (108) | 9.746 | 0.028 | −168594.01 | <0.001 | ATP6: 31 |
| N (107) | 1 | 0.044 | −168615.34 | ATP8: 15,23,36 | ||
| COX3: 41 | ||||||
| CYTB: 39,58 | ||||||
| ND1: 2 | ||||||
| ND2: 83,117,327,332 | ||||||
| ND4: 354 | ||||||
| ND4L: 10,20,25,46,80 | ||||||
| ND5: 63,418,523 | ||||||
| Carettochelyidae | A (108) | 17.57 | 0.040 | −168579.48 | <0.001 | ATP6: 11 |
| N (107) | 1 | 0.046 | −168598.43 | ATP8: 50 | ||
| COX3: 224 | ||||||
| CYTB: 225 | ||||||
| ND2: 2,46,87,143,146,150,205,268,274,307,320 | ||||||
| ND4: 130,193,374,378 | ||||||
| Dermochelyidae | A (108) | 3.555 | 0.000 | −168641.74 | 0.448 | ATP8: 23 (PP = 0.87) |
| N (107) | 1 | 0.000 | −168641.75 | |||
Note.—ω estimates for site class 2, obtained under either the BrS alternative (A) or null (N) model, are shown alongside the proportional size estimates for site class 2 (p2). Sites identified as positively selected by Bayes empirical Bayes (BEB) analysis with posterior probability (PP) ≥ 0.90 are listed, along with selected sites with lower PP values. Other abbreviations: number of estimated parameters (n.p.); log likelihood score (lnL). Parameter estimates are shown only for the positive-selection site class (Site Class 2). Full results (including all site classes and support values for the positively selected sites) are provided in supplementary table S3, Supplementary Material online.
Table 3.
The Distribution of Positively Selected Sites (Branch-Site Model) Among Genes and OXPHOS Complexes.
| OXPHOS Complex | Number of Analyzed Codons | Number of Positively Selected Sites: Concatenated Analysis | Number of Positively Selected Sites: Gene Specific Analyses |
|---|---|---|---|
| CI | 1874 (54% of total)
|
29 (72% of total)
|
47 (73% of total)
|
| CIII | 368 (11%)
|
3 (8%)
|
0 (0%)
|
| CIV | 986 (28%)
|
2 (5%)
|
5 (8%)
|
| CV | 274 (8%)
|
6 (15%)
|
12 (19%)
|
Note.—Only well-supported positively selected sites were considered (PP ≥ 0.90). Values represent the total number of positively selected sites summed across the branch-site analyses of the Trionychidae and Carettochelyidae branches; results for each individual branch are provided in parentheses next to the relevant gene (number for Trionychidae, number for Carettochelyidae). Positively selected sites were nonrandomly distributed among complexes (G-tests of independence: G = 16.64, df = 3, P = 0.001 for the concatenated data analysis; G = 38.45, df = 3, P < 0.001 for the gene-specific analyses).
Gene-specific LRTs identified the signature of positive selection in seven genes along the Trionychidae branch (ATP6, ATP8, COX3, ND1, ND2, ND4L, ND5), five genes along the Carettochelyidae branch (ATP6, ATP8, COX3, ND2, ND4), and two genes along the Dermochelyidae branch (ATP8, ND1) (supplementary table S4, Supplementary Material online). Positive selection was not detected along any of the tested lineages for COX1, COX2, CYTB, or ND3 (supplementary table S4, Supplementary Material online). The fact that BrS tests were not significant for CYTB is odd considering that CYTB sites were identified as positively selected along the Trionychidae and Carettochelyidae branches when considering the concatenated data set. Conversely, ATP8 and ND1 were identified as positively selected along the Dermochelyidae branch despite there being minimal evidence for positive selection along this lineage in the concatenated analysis. Presumably these discrepancies reflect a trade-off between the benefit of increased data and the cost of shared parameter estimates across genes in the larger concatenated data set compared with the smaller gene-specific data sets. These few points notwithstanding, the gene-specific analyses largely confirmed the results of the concatenated analyses: positive selection was strong and affected many genes for the Trionychidae and Carettochelyidae branches, but not for the Dermochelyidae branch.
The size of the positively selected site class was generally ≤10% in the gene-specific analyses. The exceptions were ATP8, where it ranged from 16% to 23% depending the foreground branch; ND2, where it reached 15% when Carettochelyidae was set as the foreground, and ND4L, where it reached 13% with Trionychidae as the foreground. Thus, in most cases, the target of positive selection was estimated to be a relatively small portion of the gene. The ω2 estimates reached high values (ω2 > 10) for several gene/branch combinations, even sometimes for those with large positively selected site classes (e.g., ATP8), indicating cases where positive selection was particularly strong. Gene-specific BEB analyses identified more sites as positively selected than had been found through analysis of the concatenated data set, suggesting greater power for the gene-specific analyses. Reassuringly, almost all of the sites identified using the concatenated data set were also identified through gene-specific analyses. Some sites were identified multiple times in different lineages, particularly when considering all identified sites (i.e., PP ≥ 0.50, rather than the more strict cutoff of PP ≥ 0.90). For example, three nearby sites identified as positively selected along the Trionychidae branch in ATP8 (sites 14, 15, and 23; each with PP ≥ 0.98) were also identified along the Carettochelyidae branch (each with PP ≥ 0.75). Ancestral reconstruction of the amino acids at these three sites showed that different residues fixed along the Trionychidae (S14L, S15I, L23Y) and Carettochelyidae (S14Y, S15L, L23I) lineages (supplementary table S4, Supplementary Material online). Thus, even when the same sites were targeted by positive selection, the molecular-level outcomes differed.
For the softshells (Trionychidae), the focus of our BrS analysis was the ancestral branch at the base of the clade, not the branches within the clade. We therefore examined patterns of AA variation at the 43 sites identified as positively selected along this ancestral branch (supplementary fig. S2, Supplementary Material online). Thirteen of the sites were fixed for a particular AA within the Trionychidae clade and another eight had singleton AA differences. As most of these sites were highly conserved across the rest of the phylogeny, this pattern is consistent with a restricted period of positive selection at the base of the Trionychidae clade, before and after which stabilizing selection acted. At the other extreme, three of the positively selected sites were highly variable within the Trionychidae clade, with four or five different residues across the eight species. These sites may represent cases where positive selection began operating along the ancestral branch and then continued to act throughout the history of the clade. However, applying a M8a-M8 LRT to test for positive selection within the softshells clade (using a concatenated alignment consisting of only the eight Trionychidae species and using a pruned version of the tree in fig. 1) did not reveal convincing evidence for positive selection (LRT P = 0.225). Moreover, posterior mean estimates of dN/dS were <1 for all 43 focal sites. These observations suggest that adaptive evolution of mitochondrially encoded OXPHOS subunits in the Trionychidae was restricted to the group’s early history, and that subsequent evolution at the positively selected sites was typically characterized by moderate-to-strong constraint.
Clade Mode C
CmC was used to test for variation in site-specific selective constraint associated with shell evolution. Considering first the concatenated data set, and the case where the various soft-shelled lineages were combined into a single foreground branch set, our analyses identified a significant decrease in ω on lineages associated with soft-shells: ω estimates for the divergent site class, which comprised 31% of the total data set, were 0.10 for soft-shelled lineages and 0.17 for hard-shelled lineages (supplementary table S5, Supplementary Material online). Gene-specific analyses uncovered similar patterns for most genes: the M2a_rel null model was strongly rejected for all data sets except for ATP8 (where M2a_rel was the best fitting AIC model) and ND4L (where AICΔ for M2a_rel was <2) (supplementary table S5, Supplementary Material online). Comparing models via LRTs did not change this general conclusion. In no case did the ω estimates for the divergent class approach or exceed 1, indicating that the shifts in constraint detected by CmC analyses were always in the relative strength of purifying selection. The size of the divergent site class varied among genes with significant signatures of divergent selection, ranging from 12% (COX1) to 44% (ATP6). COX3 was an outlier, with a divergent site class size of 82%, and here the large size seems to have resulted in a statistically significant test result despite the ω difference being small and presumably biologically insignificant (0.01 vs. 0.00; background vs. foreground); this outcome has been observed for other data sets analyzed using CmC (Chang etal. 2012). In all other cases, the divergent site-classes were smaller and the ω estimates more separated (supplementary table S5, Supplementary Material online). While we fit models multiple times from varying starting points, we cannot rule out the possibility that this unusual result for COX3 is due to suboptimal model fitting. Regardless, the evidence from the Tconc data set and the bulk of the gene-specific data sets indicate that, for many sites, selective constraint increased along the set of lineages where soft-shells evolved.
Comparing the fit of models that focused only on one of the three soft-shelled lineages at a time (either Trionychidae, Carettochelyidae, or Dermochelyidae) indicated that the signature of divergent selection varied both among lineages and among genes (supplementary table S5, Supplementary Material online). There was no obvious relationship between whether divergent selection was identified for a given gene/branch and whether positive selection was identified for that same gene/branch. For example, a matching pattern was found for ND1 (positive selection and divergent selection were both detected for Trionychidae and for Dermochelyidae, but not for Carettochelyidae), but contrasting patterns were found for ATP8 (positive selection detected for all branches, but divergent selection detected for none) and CYTB (divergent selection detected for all branches, but positive selection detected for none). The CmC codon model is not constrained to strictly focus on qualitative shifts in the selection regime (i.e., from either purifying selection or neutrality to positive selection), so it is not surprising that these two sets of analyses identified different evolutionary patterns. This lack of correspondence indicates that functional divergence can occur in multiple ways (positive selection vs. strengthened or relaxed constraint) and that these alternatives are evolutionarily uncoupled (Bielawski and Yang 2004; Zhang etal. 2005).
Structural and Functional Analysis
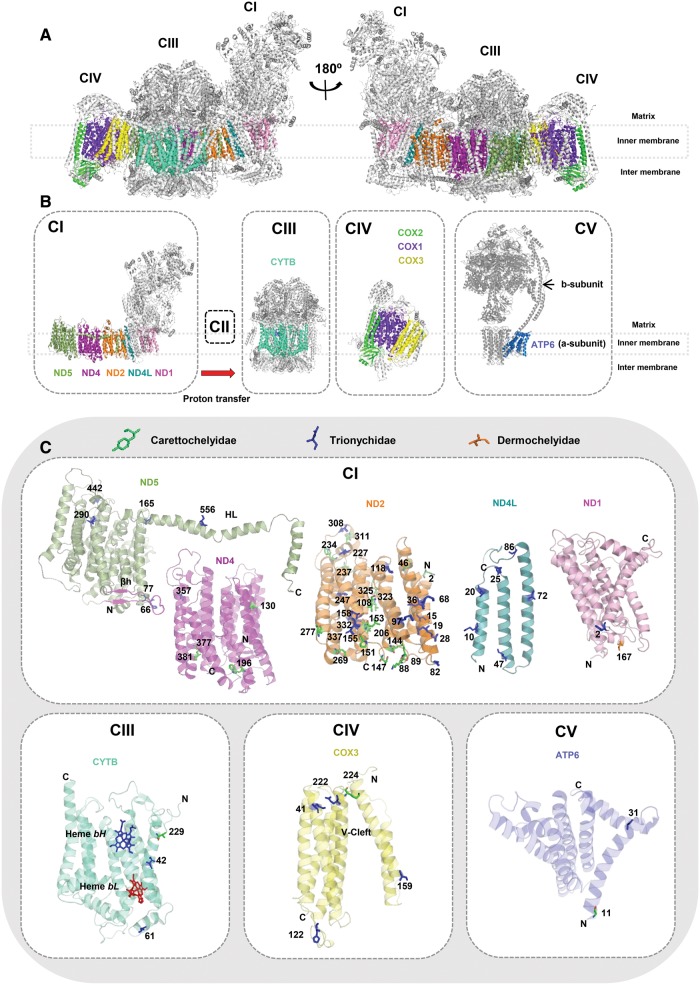
Positively selected sites identified through our BrS analyses were mapped onto the mitochondrial protein three-dimensional (3D) crystal structures and each site was evaluated in light of past and recent structural or biomedical studies of the OXPHOS system. Combining the results of the Tconc and gene-specific analyses, and considering only sites with PP ≥ 0.90, our analyses identified a total of 71 positively selected sites (Trionychidae 43; Carettochelyidae 27; Dermochelyidae 1). Several of these sites were located in or very near key residues and critical protein regions, supporting the notion that many of the identified sites are functionally relevant, and that substitutions at these sites were adaptive. In particular, we note close proximity between many of Complex I’s positively selected sites and both the proton translocation pathway and the coupling elements that help coordinate conformational changes among subunits, as can be seen in figures 3 and 4. Moreover, we found that some of the positively selected sites, particularly in CI subunits ND2 (TM4, 6, 9, 10), ND4 (TM7) and ND5 (α-helix HL), are near accessory subunits or the lipid molecules that pack between complexes, suggesting that selection may have shaped higher-order interactions among subunits and complexes (supplementary fig. S3, Supplementary Material online). Detailed results for each OXPHOS complex are provided as Supplementary Material online (supplementary table S6 and text S1).
Fig. 3.
Spatial distribution of the positively selected sites identified along soft-shelled turtle lineages. Sites identified by branch-site analysis of the 12 heavy chain mitochondrially encoded proteins of the OXPHOS pathway were mapped onto homologous solved protein structures from Sus scrofa (Complexes CI, CIII, CIV; PDB 5GUP) and Bos taurus (Complexes CV; PDB 5ARA). Grey structures represent nuclear-encoded subunits, and no structure currently exists for CV subunit ATP8. Only positively selected sites with PP ≥ 0.90 are considered; no sites met this threshold for the ND3, COX1, and COX2 subunits. (a) The CI–CIII–CIV supercomplex. The supercomplex is shown in two differently rotated views along the membrane. CIII is present twice as a homodimer. Mitochondrially encoded subunits are represented in different colors, as indicated in (b). (b) Individual OXPHOS complexes, with mitochondrially encoded subunits colored as follows (from L to R): CI-ND5 in green: CI-ND4 in purple; CI-ND2 in orange; CI-ND4L in cyan; CI-ND1 in light magenta; CIII (homodimer)–CYTB in aquamarine; CIV-COX2 in green; CIV-COX1 in purple, CIV-COX3 in yellow; CV-ATP6 in blue. (c) Individual core subunits with positively selected sites shown as stick models, with the color indicating the branch along which the site was identified: the softshells (Trionychidae) branch in blue; the pig-nose turtle branch (Carettochelyidae) branch in green; the leatherback branch (Dermochelyidae) in orange. Abbreviations: OXPHOS Complex I (CI), Complex III (CIII), Complex IV (CIV), Complex V (CV); Long horizontal α-helix coupling element (HL); β hairpin coupling element (βh); Quinone-binding site (QN); C-terminus (C); N-terminus (N); V-shaped cleft (V-Cleft); Bound cofactors HemebH and HemebL; Complex V subunits (a and b-subunit). Site numbering follows that of the relevant crystal structure.
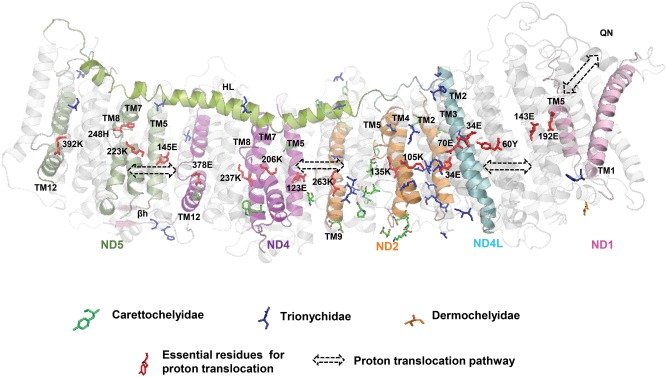
Fig. 4.
Positively selected sites and the proton translocation pathway in OXPHOS Complex I. The figure illustrates the position of positively selected sites in relation to the proposed proton translocation pathway that runs throughout Complex I. Subunits with positively selected sites (with PP ≥0.90) are colored (from left-to-right: ND5 in green; ND4 in purple; ND2 in orange; ND4L in blue; ND1 in magenta); for clarity, only relevant structural elements are colored. Positively selected sites are shown as stick models, with the color indicating the branch along which the site was identified: blue represents the softshell clade (Trionychidae), green the pig-nose turtle branch (Carettochelyidae), and orange the leatherback sea turtle branch (Dermochelyidae). The proposed proton translocation pathway is indicated by double-headed arrows, and essential residues that line the channel are shown in red (Sazanov 2015; Fiedorczuk etal. 2016). Also noted on the structure: 1) relevant transmembrane (TM) helices; 2) the coupling elements HL and βh; 3) the quinone-binding cavity (QN). The structure is derived from Sus scrofa (PDB 5GUP) and residue and TM numbering follows that of the original structure.
Discussion
We tested for adaptive and divergent selection in the mtDNA protein coding genes of turtles in relation to the evolution of soft-shells, reasoning that the highly aquatic lifestyles of soft-shelled turtles may have precipitated adaptive modification of the mitochondrially encoded OXPHOS proteins. Random-site models, which ignore among-branch variation in selection, returned results indicative of strong negative selection (ω < 1), consistent with the notion that mtDNA protein-coding genes are conserved to maintain OXPHOS function in Cryptodiran turtles, as they are broadly in eukaryotes (Castellana etal. 2011). That said, the degree of selective constraint varied considerably among and within OXPHOS proteins, indicating that positive selection may have operated intermittently. This notion was confirmed through branch-site (BrS) analyses, which uncovered strong signatures of positive selection in a branch and gene specific manner: when all genes were concatenated, positive selection was detected along the branch leading to the softshells clade (branch ID: Trionychidae) and the branch leading to the pig-nosed turtle (Carettochelyidae), but not along the branch leading to the leatherback (Dermochelyidae). A similar pattern was identified through gene-specific analyses: positive selection was readily detected along the branch leading to the Trionychidae clade (7 genes) and the Carettochelyidae branch (5 genes), but was relatively limited along the Dermochelyidae branch (2 genes). Moreover, only a single site was deemed positively selected along the Dermochelyidae branch through empirical Bayesian identification of positively selected sites, whereas dozens were identified as such along the Trionychidae and Carettochelyidae branches. Our analyses thus revealed that positive selection strongly affected mtDNA evolution along two of the three lineages associated with the evolution of soft-shells, and that positive selection targeted multiple mtDNA genes in both cases.
Molecular Evolution of Soft-Shelled Turtle mtDNA: Biochemistry
Our results suggest that the convergent morphological evolution of soft-shells in the Carettochelyidae and Trionychidae lineages may have been coupled with convergent molecular evolution of the mitochondrially encoded OXPHOS components. Of the five positively selected genes along the Carettochelyidae lineage, four were also positively selected along the Trionychidae lineage (ATP6, ATP8, COX3, ND2). Furthermore, for both of these lineages, the greatest number of positively selected sites was found in ND2, followed by ATP8 (according to gene-specific analyses). Although we did not find overlap between the lists of positively selected sites for the two lineages, we did observe concordance at the gene and complex scales. For both the Trionychidae branch and the Carettochelyidae branch we observed that the majority of the identified sites, regardless of data set (i.e., all genes concatenated or gene-specific), were located in CI, followed by CV, CIV, and finally CIII. Taken together, this indicates that convergence may apply at higher functional levels (i.e., at the OXPHOS complex level). This observation is consistent with the findings of a recent meta-analysis of natural selection in mitochondria, which identified CI as the most frequent target of positive selection in metazoans (Garvin, Bielwaski, etal. 2015). This general pattern appears to be due to the fact that CI is both the first and the largest domain of the OXPHOS pathway, responsible for an estimated 40% of the proton current that drives ATP synthase (Sasanov etal. 2013). Furthermore, among the complexes, CI is the target of the most mutations that cause mitochondrial disorders in humans (Bridges etal. 2011; Mimaki etal. 2012). These observations, combined with ours, highlight the vital role of CI in initiating the proton-pumping process and strengthen our view that CI is an essential domain in shaping the evolution of bioenergetics in soft-shelled turtles and in metazoans in general.
Within CI, the majority of positively selected sites were found in ND2 for both the Trionychidae branch and the Carettochelyidae branch, followed by ND5 (for the Trionychidae branch) or ND4 (for the Carettochelyidae branch). These three subunits are homologous to one another and are referred to as antiporter-like subunits because they are related to the Mrp Na+/H+ antiporter protein family (Sazanov 2015). These subunits have been proposed to serve as key parts of the actual proton-pumping mechanism of CI, thereby playing a crucial role for energy production (Wu etal. 2016). Remarkably, most of the positively selected sites identified in these three subunits are in the most conserved and functionally relevant transmembrane (TM) regions. For ND2 and ND4, this corresponds to the regions stretching from TM1 to TM10 and TM4 to TM13, respectively, which are critical to ion transport (Sazanov 2015; Fiedorczuk etal. 2016), whereas for ND5 this corresponds to TM9 as well as the coupling elements, HL and βh, which coordinate conformational cycling via interactions with TM domains within and with neighboring subunits ND4 and ND2 (Sazanov 2015). By considering the position of positively selected sites in light of recently solved structures of the CI-CIII-CIV supercomplex, we observed that some of the sites from ND2, ND4, and ND5 (α-helix HL) are in close proximity to CI accessory subunits (e.g., NDUFA11, NDUFB4, NDUFB8, NDUFB9) and/or lipid molecules that pack among the subunits; these accessory subunits and lipid molecules are proposed to help stabilize the supercomplex and play an important role in biological function (Wu etal. 2016). Also in CI, we detected signatures of adaptive evolution in ND1 (for the Trionychidae and Dermochelyidae branches) and ND4L (for the Trionychidae branch), both of which are known to interact with ND2; these subunits play an important structural role in linking the membrane-embedded hydrophobic domain and the peripheral hydrophilic arm of CI, and substitutions within these domains can impact the overall efficiency of proton-pumping dynamics (Bridges etal. 2011). Interestingly, the Trionychidae ND1 site is in TM1, which frames the entrance to the quinone-binding cavity and which is presumed to be involved in the coupling mechanism that links electron transfer and proton translocation (Sazanov 2015; Wu etal. 2016). Notably, several of the Trionychidae ND4L sites are close to conserved residues that form part of the proton translocation E (Glu)-channel (Sazanov 2015). Our results therefore indicate that even though the mitocondrially encoded subunits of CI are generally subject to stringent constraints, they seem to have been targeted by positive selection in a manner that likely affects proton flux.
Positively selected sites were also identified outside of CI and they too may affect critical aspects of the OXPHOS system. Positive selection was detected within the ATP6, ATP8, and COX3 genes along both the Carettochelyidae branch and the Trionychidae branch (as were sites within CYTB, though only when considering the analysis of the concatenated data set). ATP6 and ATP8 form part of the regulatory system of CV and contribute to the proton translocation path that drives ATP production (Castellana etal. 2011; Zhou etal. 2015). CYTB and COX3 also participate in proton translocation: CYTB has an essential catalytic activity (cytochrome c reduction) and uses direct coupling through important cofactors (haems, metal centers) for electron transfer and proton translocation (Sazanov 2015), as does COX3, albeit indirectly via its interaction with neighboring COX1 (Sharma etal. 2015). Several of the identified CYTB and COX3 sites are situated in TM domains that interact with the cofactors or molecules that influence proton uptake and translocation, such as the bound cofactors HemebL or HemebH in CIII (Xia etal. 2013) and the bound charged lipid molecules of the V-shaped cleft of COX3 in CIV (Sharma etal. 2015). Taken together, we infer that proton flow efficiency was targeted by positive selection along the Trionychidae and Carettochelyidae soft-shelled turtle lineages, primarily (but not exclusively) through adaptive evolution of the CI subunit ND2.
Molecular Evolution of Soft-Shelled Turtle mtDNA: Ecology and Physiology
Our branch-site analyses provided strong and consistent evidence for positive selection along the branches leading to the softshells (Trionychidae) and the branch leading to the pig-nosed turtle, but not along the branch leading to the leatherback sea turtle. Why do we see such different patterns on these different lineages given that they all represent highly aquatic lineages that evolved soft-shells? One possibility is that adaptation of OXPHOS system did in fact occur along the leatherback branch, but that it involved nuclear-encoded subunits (Rand etal. 2004; Welch etal. 2014) or that it involved other factors that increase OXPHOS efficiency, such as increased mitochondrial content and/or cristae surface density (Dalziel etal. 2006; Zhang and Broughton 2015). Another possibility is that morphological, physiological, or behavioral adaptations occurred along the Dermochelyidae lineage and that these were sufficient to ensure oxygen needs were met without adaptive modification of the mitochondrially encoded OXPHOS subunits. For instance, the leatherback can store a high concentration of dissolved oxygen in its blood and muscle tissue and has multiple anatomical adaptations associated with efficient swimming performance (e.g., a ridged, tear-dropped shaped carapace) (Davenport etal. 2011; Bang etal. 2016) and deep and prolonged diving (e.g., collapsible lungs, a pulmonary sphincter) (Spotila and Tomillo 2015).
Furthermore, given that the leatherback is a long distance migratory species that travels from cold to warm waters, the OXPHOS system of leatherbacks may be subject to thermoregulatory constraints not found in other soft-shelled lineages. Most notably, leatherbacks are endotherms (Bostrom etal. 2010) and endothermy has been linked to OXPHOS proton leakage (Clarke and Pötner 2010). Consistent with this notion, a recent study of marine fish mitochondrial genomes reported strong negative selection along the branches leading to endothermic tunas and billfishes (Zhang and Broughton 2015). Finally, it may be that the primary driver of positive selection in soft-shelled turtles is cutaneous respiration, which is unlikely to be employed by leatherbacks as their skin is thick and lined with insulating blubber (Davenport etal. 2011), making it poorly suited for cutaneous respiration. Moreover, the potential benefit of cutaneous respiration to leatherbacks (assuming dermal permeability) would presumably be limited, owing both to their large size and to the lower dissolved oxygen content of marine water relative to freshwater. That said, it would be interesting to explore differences between marine and freshwater turtles in greater detail, as past work with fishes suggests that osmoregulatory strategies associated with the two environments differ in terms of metabolic costs and oxygen requirements (Boeuf and Payan 2001). Since there was only a single invasion of the marine environment in turtles, this work would be particularly informative if combined with comparable studies of other taxonomic groups that have invaded both aquatic habitats (e.g., fishes, sharks, cetaceans, snakes).
While we found evidence for strong positive selection along the branch leading to the pig-nosed turtle (the Carettochelyidae branch), the signature of positive selection was even stronger along the branch leading to the clade of softshells (the Trionychidae branch). This may reflect the fact that shell loss, and the adoption of a highly aquatic lifestyle, is more “complete” in softshells than it is in the pig-nosed turtle. Both taxa employ oropharyngeal respiration (Winokur 1988) and presumably also cutaneous respiration (Ultsch etal. 1984; Joyce etal. 2012), yet the shells of softshells are much more highly vascularized and flattened (Scheyer etal. 2007). This increases the surface area-to-volume ratio available for cutaneous uptake of oxygen from water (Stone etal. 1992), allowing for greater dermal gas exchange and enabling the turtles to remain submerged for prolonged amounts of time (Ultsch etal. 1984). For instance, cutaneous respiration in the softshell Trionyx triunguis accounts for ∼70% of total respiration, with the remaining 30% attributed to pharyngeal breathing (Girgis 1961). For the pig-nosed turtle, dermal gas exchange diffusion capacity has not yet been studied. However, having a domed shell and overall a greater degree of mineralization presumably reduces surface area-to-volume ratio and slows the diffusion of respiratory gases.
Conclusions
Our study provides evidence of strong positive Darwinian selection operating on the mitochondrial genome of two lineages of soft-shelled turtles, suggesting a link between shell morphology, respiratory physiology, and OXPHOS molecular biology in turtles. Combined with the results of several recent studies of mtDNA evolution (da Fonseca etal. 2008; Garvin etal. 2011; Caballero etal. 2015; Consuegra etal. 2015; Zhang and Broughton 2015), our work highlights the importance of the aquatic environment as a driver of adaptive evolutionary change in the OXPHOS system. Moreover, because we identified positively selected sites in functionally important regions of the OXPHOS system, our work provides a foundation for future experiments on the biochemical and physiological impact of mtDNA variation in turtles. However, our findings also indicate a complex pattern of molecular evolution, with the intensity of positive selection differing both among genes and lineages. Moreover, it is plausible that adaptive evolution of mtDNA has occurred along other lineages within the turtle phylogeny due to the evolution of other ecophysiological adaptations for efficient respiration. Finally, we note that the mitochondrial genome is not functionally independent of the nuclear genome, and it would be beneficial to compare and contrast patterns of molecular evolution between mtDNA genes and the nuclear genome-encoded components of the OXPHOS system; these two sets of genes are presumably coevolving due to structural and functional interactions.
Materials and Methods
Data Sets
Whole mitochondrial genome sequences from 57 turtle species (53 Cryptodiran species plus four outgroup Pleurodiran species) were downloaded from the NCBI database. This data set covers 10 of 11 Cryptodiran families (Trionychidae (8 species); Carettochelyidae (1); Dermochelyidae (1); Cheloniidae (6); Platysternidae (1); Emydidae (2); Testudinididae (11); Geomydidae (19); Kinosternidae (2); Chelydridae (2)) and all Pleurodiran families (Pelomedusidae (1); Podocnemidae (1); Chelidae (2) (fig. 1). We restricted our phylogenetic and molecular evolutionary analyses to the 12 heavy or H-strand protein-coding genes (i.e., ATP6 and ATP8, COX1–3, CYTB, ND1–5 and ND4L, but excluding ND6) because the light or L-strand has a significantly different base composition from the heavy-chain (Bradshaw etal. 2005). Two data set types were examined: 1) each of the 12 genes individually, and 2) all 12 coding genes concatenated into a single sequence. Species names and accession numbers are provided in figure 1.
Alignments
We generated multiple sequence alignments using three different methods: ClustalW (Thompson etal. 1994), which was implemented using the program MEGA 6 (Tamura etal. 2013), and MUSCLE (Edgar 2004) and PAGAN (Löytynoja etal. 2012), which were both implemented using the GUIDANCE web-server (Penn etal. 2010). For all methods, nucleotide sequences were first translated into amino acids, aligned, then back-translated into nucleotides and manually inspected. Stop codons were removed from the sequences prior to alignment. For the concatenated data set, we aligned each gene separately and then manually combined them into a single data set. Position-wise confidence scores estimated using GUIDANCE (Penn etal. 2010) were consistently high (range = 0.92–1.00), suggesting that the alignments were reliable and robust. Alignment properties are summarized in table 1.
A preliminary examination of the translated sequences revealed a few species with frameshifting insertions and/or deletions (indels) of 1 or 2 bp. These indels may simply represent PCR/sequencing errors or they may be true indels that are corrected during protein production via translational frameshifting (as has been previously reported for turtle mtDNA sequences; Russell and Beckenbach 2008). We manually edited these sequences to preserve the expected reading frame (0-frame) prior to alignment, following Russell and Beckenbach (2008) (supplementary table S1, Supplementary Material online).
Saturation in Codon Positions
We tested for codon position-specific patterns of saturation in the concatenated alignment using the method of Xia etal. 2003 in DAMBE 5.3.17 (Xia 2013). Estimates of the substitution saturation index were significantly lower than the critical value when considering all sites (1st + 2nd + 3rd codon positions) and when excluding 3rd positions, indicating minimal saturation (P < 0.001). This was true regardless of assumptions about the underlying tree topology (completely symmetrical vs. fully asymmetrical). A similar result was obtained when considering only 3rd positions and assuming a symmetric tree topology (P < 0.001), though we could not rule out some degree of saturation for 3rd positions when assuming a fully asymmetrical topology (P = 0.877). However, given that the phylogenetic trees we derived are largely congruent with previous reports of turtle relationships (see below), we concluded that saturation is likely minimal in our data.
Phylogenetic Tree Estimates
The individual nucleotide alignments for the 12 heavy strand protein-coding genes were concatenated and used for phylogeny estimation. Phylogenetic trees were inferred by first testing for the best fitting model of evolution using JModelTest (Darriba etal. 2012), comparing models via Akaike Information Criterion (AIC) scores. GTR + I + Γ was deemed the best-fit nucleotide substitution model and used to generate maximum likelihood and Bayesian estimates of tree topology. Alignment gaps were retained during model testing and tree building.
The Bayesian tree was estimated using MrBayes 3.2 (Ronquist etal. 2012). To create the Bayesian tree, the nucleotide model parameter was set to 4×4, the MCMC chain was run with four chains (three heated and one cold) for 10 million generations, and every 100 samples were taken to estimate the posterior distribution (discounting the first 25% of the samples). Convergence was confirmed by ensuring that the standard deviation of split frequencies was <0.01 and was further checked visually with Tracer v1.5 (http://beast.bio.ed.ac.uk/Tracer). The 50% majority rule consensus tree was taken as our Bayesian estimate of the phylogeny. PhyML 3 was used to infer the ML tree (Guindon etal. 2010). We used BIONJ as the starting tree and SPR (subtree-pruning and regrafting) as the search algorithm. Node support was evaluated using nonparametric bootstrapping (1,000 replicates).
Phylogenetic analyses of the concatenated alignments built using different alignment algorithms produced similar topologies with strong node support (data not shown). As such, alignments built using the MUSCLE approach were used in all subsequent analyses. Moreover, the ML and Bayesian approaches yielded highly similar topologies (supplementary fig. S1, Supplementary Material online); subsequent analyses were therefore conducted solely using a pruned (Cryptodira clade) version of the ML tree (fig. 1).
Adaptive Evolution Analyses
We used the CODEML program in the PAML4 software package (Yang 2007) to explore patterns of natural selection and identify sites targeted by positive selection in each of the 12 heavy-strand mtDNA protein coding genes, as well as in the concatenated data set. This was performed using codon substitution models and ML to estimate the ratio of nonsynonymous-to-synonymous substitution rates, that is dN/dS (or ω) (Goldman and Yang 1994; Muse and Gaut 1994). Three different classes of codon-based likelihood models of evolution were employed: random-site, branch-site, and clade models.
The random-site models assume that selection pressure varies among sites in the alignment but not among branches in the phylogeny. We used nested model comparisons (likelihood ratio tests; LRTs) to test for positive selection, comparing the fit of alternative models that allow for site-specific positive selection to the fit of similarly constructed null models that do not (null M8a vs. M8) (Yang etal. 2000; Swanson etal. 2003). LRT statistics (twice the difference in log-likelihood scores) were compared with a χ2 null distribution with the degrees of freedom set to the number of additional parameters estimated in the alternative model.
The branch-site (BrS) models of Zhang etal. (2005) allow the selection process to vary across lineages and sites, and were used to test for positive selection on pre-specified lineages (“foreground branches”). The BrS alternative model (BrS-A) assumes four categories of sites: the first two assume either purifying selection (0 ≤ ω0 ≤ 1) or neutrality (ω1 = 1), respectively, on all branches, whereas the remaining two assume switches from either purifying selection or neutrality along the background to positive selection along the foreground (ω2 > 1). The null model (BrS-N) is similarly built, but is constrained such that ω2 = 1. BrS model comparisons were conducted using LRTs; here, because the null is formed by constraining a parameter in the alternative model to a boundary, the test statistic was evaluated using a 50:50 mixture distribution of 0 and χ21 (see Goldman and Whelan 2000). Because BrS models do not allow for multiple foreground branches, we conducted separate tests for each branch of interest.
Clade models are similar to BrS models in that they jointly consider among-site and among-branch variation, but are designed for detecting shifts in selective constraint rather than positive selection. We employed clade model C (CmC) (Bielawski and Yang 2004), which assumes three classes of sites: those subject to constant purifying selection (0 < ω0 < 1), constant neutrality (ω1 = 1), or divergent selection (ω2 > 0 and ω3 > 0, assuming two pre-specified sets of foreground branches). As a null model, we used M2a_rel, which allows for among-site but not among-branch variation (Weadick and Chang 2012). Because the alternative model contains one extra parameter relative to the null, the LRT was evaluated using a χ21 distribution. We also used AIC comparisons to help evaluate relative model fit.
Foreground Branch Assignments
For BrS and CmC models, the phylogeny was partitioned into background and foreground branches in multiple ways (fig. 1). First, we considered the branch leading to the softshell turtles (Trionychidae clade) as the foreground. For BrS models this branch alone comprised the foreground, whereas for CmC models this branch and all descendent branches were collectively set as the foreground. Second, the branch leading to the pig-nosed turtle Carettochelys insculpta was set as the foreground. Third, the branch leading to the leatherback sea turtle Dermochelys coriacea was set as the foreground. Fourth, for CmC only, we combined the above three sets into a single foreground group. These branch labelling decisions were based on our a priori knowledge of the evolution of the “soft-shelled trait” in turtles, as indicated by fossil evidence (Joyce 2007, 2014).
Codon Model Fitting
Codon models were run with initial branch length estimates derived from a preliminary fit of the M0 model (which assumes no site-wise or branch-wise dN/dS variation). Alignment columns with gaps and uncertainties were dropped during analyses to avoid false positives (Fletcher and Yang 2010). Codon frequencies were approximated using the F3×4 calculation. To avoid potential local optima, each model was fit multiple times, varying initial starting points for the transition-to-transversion rate ratio parameter (κ) or ω, depending on the model tested. Positively selected sites were identified for the M8 and BrS-A models using the Bayes empirical Bayes (BEB) approach (Yang etal. 2005) with a posterior probability (PP) cutoff of 0.90. The distribution of positively selected sites among OXPHOS complexes was examined using a G-test of independence, as implemented in R (R Core Team 2014). Sequence logos were generated using WebLogo (under default settings, except with no small sample correction) in order to visualize patterns of AA variability at selected sites (Crooks etal. 2004). Branch-specific estimates of the rate of synonymous substitutions per synonymous site (dS) calculated under the M0 model were <1 for nearly all branches, indicating that we have sufficient phylogenetic resolution to accurately estimate dN/dS across the phylogeny (table 1).
Structural Analysis
To gain insights into how substitutions at positively selected sites may influence the structure and function of the mitochondrial proteins, we aligned our data sets against homologous proteins for which X-ray crystal structures are available in the Protein Data Bank (http://www.rcsb.org/pdb). CI, CIII, and CIV subunits were aligned against homologs from the recently released Sus scrofa CI-CIII-CIV supercomplex structure (PDB 5GUP), and the CV subunit ATP6 was aligned with its Bos taurus homolog (PDB 5ARA). (No structure is available for ATP8). Finally, we also aligned each gene with its homologous human protein to relate our findings to biomedically important sites as reported in the literature, and used the infer ancestral sequences ML tool in MEGA 6 (Tamura etal. 2013) to infer specific amino acids changes at key sites and along key branches. We used PyMOL version 1.8.6.0 (https://www.pymol.org/) for protein visualization.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
The work presented here was conceived and designed by T.E. T.E. conducted the study in close collaboration with C.J.W. Data were analyzed and interpreted by T.E. with assistance from C.J.W. The manuscript was written by T.E. C.J.W. contributed to the writing and refining of the manuscript. A.A. participated in the initial design and coordination of the project, contributed materials and computational resources, and assisted in editing. All authors read and approved the final version.
Supplementary Material
Acknowledgments
This work was supported by a PostDoctoral Fellowship (SFRH/BPD/66448/2009) from Fundação para a Ciência e a Tecnologia (FCT) to T.E. A.A. was partially supported by the Strategic Funding UID/Multi/04423/2013 through FCT and the European Regional Development Fund (ERDF) program PT2020; by the European Structural and Investment Funds (ESIF) under the Competitiveness and Internationalization Operational Program—COMPETE 2020 through FCT under PTDC/AAG-GLO/6887/2014 (POCI-01-0124-FEDER-016845), and by the Structure Programs R&D&I INNOVMAR (NORTE-01-0145-FEDER-000035-NOVELMAR) and CORAL NORTE (NORTE-01-0145-FEDER-000036), and the Northern Regional Operational Program (NORTE2020) through ERDF. We owe special thanks to John Davenport (University College Cork) and Walter Joyce (University of Fribourg) for valuable comments on Dermochelys coriacea and the origin of the soft-shelled trait in turtles, respectively. We are also grateful to Romain Studer for comments and advice on analyses, Aldo Barreiro for R scripting advice, Daniela Almeida for figure designing advice, and two anonymous reviewers for their thoughtful comments provided on this manuscript.
References
- Alibardi L, Toni M.. 2006. Skin structure and cornification proteins in the soft-shelled turtle Trionyx spiniferus. Zoology 109:182–195. [DOI] [PubMed] [Google Scholar]
- Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC.. 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 18:489–522. [Google Scholar]
- Bang K, Kim J, Lee S-I, Choi H.. 2016. Hydrodynamic role of longitudinal dorsal ridges in a leatherback turtle swimming. Sci Rep. 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski JP, Yang ZH.. 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol. 59:121–132. [DOI] [PubMed] [Google Scholar]
- Boeuf G, Payan P.. 2001. How should salinity influence fish growth? Comp Biochem Physio Part C 130:411–423. [DOI] [PubMed] [Google Scholar]
- Bostrom BL, Jones TT, Hastings M, Jones DR.. 2010. Behaviour and physiology: the thermal strategy of leatherback turtles. PLoS One 5:e13925.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw PC, Rathi A, Samuels DC.. 2005. Mitochondrial-encoded membrane protein transcripts are pyrimidine-rich while soluble protein transcripts and ribosomal RNA are purine-rich. BioMed Central Genomics 6:136.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges RT, Birrell JA, Hirst J.. 2011. The mitochondrial-encoded subunits of respiratory complex I NADH: ubiquinone oxidoreductase): identifying residues important in mechanism and disease. Biochem Soc Trans. 39:799–806. [DOI] [PubMed] [Google Scholar]
- Caballero S, Duchêne S, Garavito MF, Slikas B, Baker CS.. 2015. Initial evidence for adaptive selection on the NADH subunit two of freshwater dolphins by analyses of mitochondrial genomes. PLoS One 10:e0123543.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana S, Vicario S, Saccone C.. 2011. Evolutionary patterns of the mitochondrial genome in metazoa: exploring the role of mutation and selection in mitochondrial protein–coding genes. Genome Biol Evol. 3:1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, Jiang ZJ, Gu W, Wang ZO, Pollock DD.. 2008. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS One 3:e2201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BSW, Du J, Weadick CJ, Morrow JM.. 2012. The future of codon models in studies of molecular function: ancestral reconstruction and clade models of functional divergence In: Cannarozzi GM, Schneider A, editors. Codon evolution: mechanisms and models. Oxford (NY: ): Oxford University Press; p. 145–164. [Google Scholar]
- Clarke A, Pötner H-O.. 2010. Temperature, metabolic power and the evolution of endotermy. Biol Rev. 85:703–727. [DOI] [PubMed] [Google Scholar]
- Consuegra S, John E, Verspoor E, de Leaniz CG.. 2015. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet Select Evol. 47:58.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca RR, Johnson WE, O’Brien SJ, Ramos MJ, Antunes A.. 2008. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics 9:119.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel AC, Moyes CD, Fredriksson E, Lougheed SC.. 2006. Molecular evolution of cytochrome c oxidase in high-performance fish (Teleostei: Scombroidei). J Mol Evol. 62:319–331. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and high-performance computing. Nature Methods 9:772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport J, Plot V, Georges J-Y, Doyle TK, James MC.. 2011. Pleated turtle escapes the box – shape changes in Dermochelys coriacea. J Exp Biol. 214:3474–3479. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Lindell J.. 2008. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol Evol. 23:546–554. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst CH, Barbour RW.. 1989. Turtles of the world. Washington, D.C.: Smithsonian Institution Press; p. 313. [Google Scholar]
- Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA.. 2016. Atomic structure of the entire mammalian mitochondrial complex I. Nat Lett. Accelerated Article Preview. Nature538:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher W, Yang Z.. 2010. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 27:2257–2267. [DOI] [PubMed] [Google Scholar]
- Galli GLJ, Lau GY, Richards JG.. 2013. Beating oxygen: chronic anoxia exposure reduces mitochondrial F1FO-ATPase activity in turtle (Trachemys Scripta) heart. J Exp Biol. 216:3283–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin MR, Bielawski JP, Gharrett AJ.. 2011. Positive Darwinian selection in the piston that powers proton pumps in Complex I of the mitochondria of Pacific salmon. PLoS One 6:e24127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin MR, Bielwaski JP, Sazanov LA, Gharrett AJ.. 2015. Review and meta-analysis of natural selection in mitochondrial complex I in metazoans. J Zool Syst Evol Res. 53:1–17. [Google Scholar]
- Garvin MR, Thorgaard GH, Narum SR.. 2015. Differential expression of genes that control respiration contribute to thermal adaptation in redband trout (Oncorhynchus mykiss gairdneri). Genome Biol Evol. 7:1404–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgalis G, Joyce WG.. 2017. A review of the fossil record of Old World turtles of the clade Pan-Trionychidae. Bull Peabody Mus Nat Hist. 58:115–208. [Google Scholar]
- Girgis S. 1961. Aquatic respiration in the common Nile turtle, Trionyx triunguis (Forskal). Comp Biochem Physiol. 3:206–217. [DOI] [PubMed] [Google Scholar]
- Goldman N, Whelan S.. 2000. Statistical tests of gamma-distributed rate heterogeneity in models of sequence evolution in phylogenetics. Mol Biol Evol. 17:975–978. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z.. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Gu J, Guo R, Yan K, Lei J, Gao N, Yang M.. 2016. The arquitecture of the mammalian respirasome. Nature 537:639–644. [DOI] [PubMed] [Google Scholar]
- Guillon J-M, Guéry L, Hulin V, Girondot M.. 2012. A large phylogeny of turtles (Testudines) using molecular data. Contrib Zool. 81:147–158. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Joyce WG. 2007. Phylogenetic relationships of Mesozoic turtles. Bull Peabody Mus Nat Hist. 48:3–102. [Google Scholar]
- Joyce WG. 2014. A review of the fossil record of turtles of the clade Pan-Carettochelys. Bull Peabody Mus Nat Hist. 55:3–33. [Google Scholar]
- Joyce WG, Micklich N, Schaal SFK, Scheyer TM.. 2012. Caught in the act: the first record of copulating fossil vertebrates. Biol Lett. 8:846–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LourençO JM, Glémin S, Chiari Y, Galtier N.. 2012. The determinants of the molecular substitution process in turtles. J Evol Biol. 26:38–50. [DOI] [PubMed] [Google Scholar]
- Löytynoja A, Vilella AJ, Goldman N.. 2012. Accurate extension of multiple sequence alignments using a phylogeny-aware graph algorithm. Bioinformatics 28:1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutcavage ME, Bushnell PG, Jones DR.. 1990. Oxygen transport in the leatherback sea turtle Dermochelys coriacea. Physiol Zool. 63:1012–1024. [Google Scholar]
- Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT.. 2012. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta 1817:851–862. [DOI] [PubMed] [Google Scholar]
- Morales HE, Pavlova A, Joseph L, Sunnucks P.. 2015. Positive and purifying selection in mitochondrial genomes of a bird with mitonuclear discordance. Mol Ecol. 24:2820–2837. [DOI] [PubMed] [Google Scholar]
- Muse SV, Gaut BS.. 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to chloroplast genome. Mol Biol Evol. 11:715–724. [DOI] [PubMed] [Google Scholar]
- Parsons KJ, Albertson RC.. 2013. Unifying and generalizing the two strands of evo-devo. Trends Ecol Evol. 28:584–591. [DOI] [PubMed] [Google Scholar]
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T.. 2010. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38 (Web Server issue):W23–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; http://www.R-project.org/. [Google Scholar]
- Rand DM. 1993. Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol. 37:281–295. [DOI] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ.. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 19:645–653. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RD, Beckenbach AT.. 2008. Recoding of translation in turtle mitochondrial genomes: programmed frameshift mutations and evidence of a modified genetic code. J Mol Evol. 67:682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanov LA, Baradaran R, Efremov RG, Berrisford JM, Minhas G.. 2013. A long road towards the structure of respiratory complex I, a giant molecular proton pump. Biochem SocTrans. 41:1265–1271. [DOI] [PubMed] [Google Scholar]
- Sazanov LA. 2015. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol. 16:375–388. [DOI] [PubMed] [Google Scholar]
- Scheyer TM, Sander PM, Joyce WG, Böhmec W, Witze U.. 2007. A plywood structure in the shell of fossil and living soft-shelled turtles (Trionychidae) and its evolutionary implications. Org Divers Evol. 7:136–144. [Google Scholar]
- Schoch RR, Hans-Dieter S.. 2015. A middle Triassic stem-turtle and the evolution of the turtle body plan. Nature 523:584–587. [DOI] [PubMed] [Google Scholar]
- Shaffer HB, Meylan P, McKnight ML.. 1997. Tests of turtle phylogeny: molecular, morphological, and paleontological approaches. Syst Biol. 46:235–268. [DOI] [PubMed] [Google Scholar]
- Sharma V, Ala-Vannesluoma P, Vattulainen I, Wikströmc M, Róga T.. 2015. Role of subunit III and its lipids in the molecular mechanism of cytochrome c oxidase. Biochim Biophys Acta 1847:690–697. [DOI] [PubMed] [Google Scholar]
- Shen Y-Y, Liang L, Zhu Z-H, Zhoua W-P, Irwine DM, Zhang Y-P.. 2010. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc Nat Acad Sci. 107:8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spotila JR, Tomillo PS.. 2015. The leatherback turtle: biology and conservation. Baltimore (MD: ): Johns Hopkins University Press; p. 228. [Google Scholar]
- Stone PA, Dobie JL, Raymond PH.. 1992. Cutaneous surface area and bimodal respiration in soft-shelled (Trionyx spiniferus), stinkpot (Sternotherus odoratus), and mud turtles (Kinosternon subrubrum). Physiol Biochem Zool. 65:311–330. [Google Scholar]
- Storey EM, Kayes SM, De Vries I, Franklin CE.. 2008. Effect of water depth, velocity and temperature on the surfacing frequency of the bimodally respiring turtle Elseya albagula. Funct Ecol. 22:840–846. [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q.. 2003. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 20:18–20. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kuma S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ.. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch GR, Herbert CV, Jackson DC.. 1984. The comparative physiology of diving in North American freshwater turtles. I. Submergence tolerance, gas exchange and acid–base balance. Physiol Zool. 57:620–631. [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC.. 2013. Detecting natural selection in genomic data. Ann Rev Genet. 47:97–120. [DOI] [PubMed] [Google Scholar]
- Weadick CJ, Chang BS.. 2012. An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol Biol Evol. 29:1297–1300. [DOI] [PubMed] [Google Scholar]
- Welch AJ, Bedoya-Reina OC, Carretero-Paulet L, Miller W, Rode KD, Charlotte L.. 2014. Polar bears exhibit genome-wide signatures of bioenergetic adaptation to life in the arctic environment. Genome Biol Evol. 6:433–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winokur RM. 1988. The buccopharyngeal mucosa of the turtles (Testudines). J Morphol. 196:33–52. [DOI] [PubMed] [Google Scholar]
- Wu M, Gu J, Guo R, Huang Y, Yang M.. 2016. Structure of mammalian respiratory supercomplex I1III2IV1. Cell 167:1598–1609. [DOI] [PubMed] [Google Scholar]
- Wyneken J, Godfrey MH, Bels V.. 2008. Biology of turtles: from structures to strategies of life. Boca Raton, FL: Taylor and Francis Group, CRC Press; p. 486. [Google Scholar]
- Xia D, Esser L, Tang W-K, Zhou F, Zhou Y, Yu L, Yu CA.. 2013. Structural analysis of cytochrome bc1 complexes: implications to the mechanism of function. Biochim Biophys Acta 1827:1278–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XH, Xie Z, Salemi M, Chend L, Wang Y.. 2003. An index of substitution saturation and its application. Mol Phylogenet Evol. 26:1–7. [DOI] [PubMed] [Google Scholar]
- Xia XH. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 30:1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM.. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R.. 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22:1107–1118. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang F, Broughton RE.. 2015. Heterogeneous natural selection on oxidative phosphorylation genes among fishes with extreme high and low aerobic performance. BMC Evol Biol. 15:173.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhou A, Rohou A, Schep DG, Bason JV, Montgomery MG, Walker JE, Grigorieff K, Rubinstein JL.. 2015. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife 4:e10180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.