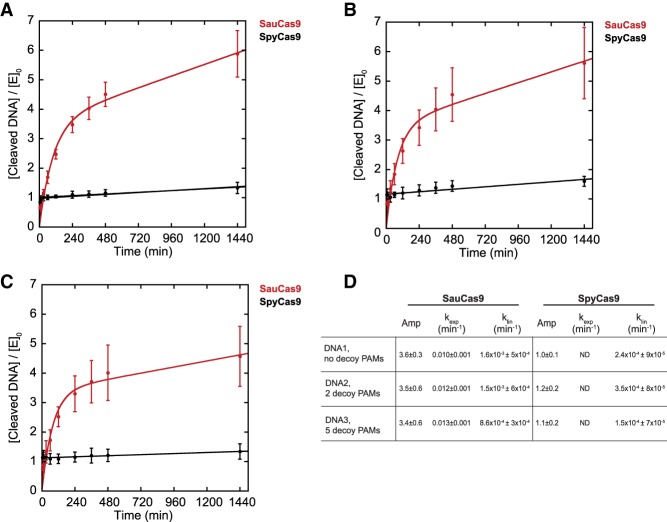
FIGURE 2.
S. aureus Cas9 is a multiple turnover enzyme. A total of 25 nM SpyCas9 (black) or SauCas9 (red) was preincubated with 100 nM sgRNA for 15 min prior to the addition of 250 nM DNA. The amount of HNH-cleaved non-PAM strand of the DNA, divided by the enzyme concentration, was plotted versus time. (A) DNA1, no decoy PAMs. SauCas9: amplitude = 3.6 ± 0.3; kexp = 0.010 ± 0.001 min−1; klin = 1.6 × 10−3 ± 6 × 10−4 min−1 (R2 = 0.98). SpyCas9: amplitude = 1.0 ± 0.1; klin = 2.4 × 10−4 ± 9 × 10−5 min−1 (R2 = 0.95). (B) DNA2, 2 decoy PAMs. SauCas9: amplitude = 3.5 ± 0.60; kexp = 0.012 ± 0.001 min−1; klin = 1.5 × 10−3 ± 6 × 10−4 min−1 (R2 = 0.96). SpyCas9: amplitude = 1.2 ± 0.2; klin = 3.5 × 10−4 ± 8 × 10−5 min−1 (R2 = 0.83). (C) DNA3, 5 decoy PAMs. SauCas9: amplitude = 3.4 ± 0.6; kexp = 0.013 ± 0.001 min−1; klin = 8.6 × 10−4 ± 3 × 10−4 min−1 (R2 = 0.96). SpyCas9: amplitude = 1.1 ± 0.2; klin = 1.5 × 10−4 ± 7 × 10−5 min−1 (R2 = 0.83). (D) Summary of the data in A–C for convenience. ND, the rate of the burst could not be resolved by manual quenching. All of the values are the result of at least three independent experiments and are reported as mean ± average deviation.