Abstract
Many bacteria use flavin mononucleotide (FMN) riboswitches to control the expression of genes responsible for the biosynthesis and transport of this enzyme cofactor or its precursor, riboflavin. Rare variants of FMN riboswitches found in strains of Clostridium difficile and some other bacteria typically control the expression of proteins annotated as transporters, including multidrug efflux pumps. These RNAs no longer recognize FMN, and differ from the original riboswitch consensus sequence at nucleotide positions normally involved in binding of the ribityl and phosphate moieties of the cofactor. Representatives of one of the two variant subtypes were found to bind the FMN precursor riboflavin and the FMN degradation products lumiflavin and lumichrome. Although the biologically relevant ligand sensed by these variant FMN riboswitches remains uncertain, our findings suggest that many strains of C. difficile might use rare riboswitches to sense flavin degradation products and activate transporters for their detoxification.
Keywords: aptamer, flavin mononucleotide, lumichrome, lumiflavin, noncoding RNA, riboflavin biosynthesis
INTRODUCTION
Riboswitches are noncoding RNAs (ncRNAs) typically found in the 5′ untranslated regions of bacterial mRNAs, where they regulate gene expression in response to the binding of specific small molecule or ion ligands (Henkin 2008; Breaker 2011, 2012; Serganov and Nudler 2013). To date, more than 40 riboswitch classes have been discovered (McCown et al. 2017), many of which control fundamental metabolic processes such as the biosynthesis and transport of amino acids, enzyme cofactors, and nucleotides. In addition, several riboswitch classes have been recently discovered that respond to ligands whose roles in biology were poorly known until recently. These include riboswitches for fluoride (Baker et al. 2012; Ren et al. 2012), guanidine (Nelson et al. 2017; Sherlock and Breaker 2017; Sherlock et al. 2017), c-di-AMP (Nelson et al. 2013), and c-AMP-GMP (Kellenberger et al. 2015; Nelson et al. 2015a). Thus, the continued identification of novel riboswitch classes provides opportunities to discover hidden or otherwise underappreciated aspects of biology.
Current computational search methods based on comparative sequence analysis (Barrick et al. 2004; Weinberg et al. 2007, 2010, 2017a) have been used to identify numerous widespread riboswitch classes. Unfortunately, the vast majority of the thousands of additional riboswitch classes that likely remain undiscovered (McCown et al. 2017) are probably exceedingly rare, and therefore will be difficult to uncover by using current bioinformatic search techniques. Many of the rare riboswitch classes remaining to be discovered could represent ancient ncRNAs on the brink of evolutionary extinction or they might be niche-specific riboswitches that have more recently emerged within organisms. For example, a newly found SAM-sensing riboswitch, called SAM-VI, was identified exclusively in Bifidobacterium species (Mirihana Arachchilage et al. 2018). Discoveries such as this support the hypothesis that numerous additional rare riboswitch classes likely exist, and that some of these might have emerged through evolution very recently. If the rarest riboswitch classes are largely distinct in their conserved sequence and architectural features, computational approaches that rely on comparative sequence and structure analysis algorithms simply have too few representatives to evaluate.
However, some exceedingly rare riboswitch classes might remain hidden due to their mistaken classification as members of known riboswitch classes. These “variant” riboswitches might exhibit RNA sequence and structural features that are shared with previously validated riboswitch classes, but carry key mutations that have altered their ligand specificities (Weinberg et al. 2017b). Examples of rare riboswitch classes include variants of guanine riboswitches that sense 2′-deoxyguanosine (2′-dG). To date, the 2′-dG-I riboswitch class (McCown et al. 2017) has only been identified in Mesoplasma florum (Kim et al. 2007). Similarly, the recently discovered 2′-dG-II riboswitch class has only been found within metagenomic sequence data sets (Weinberg et al. 2017b). Additional examples include riboswitches for the bacterial alarmone ppGpp (Sherlock et al. 2018a) and the purine biosynthetic precursor phosphoribosyl pyrophosphate (PRPP) (Sherlock et al. 2018b), which were both identified as rare variants of the far more abundant guanidine-I riboswitch class (Nelson et al. 2017).
Riboswitches that bind to flavin mononucleotide (FMN) represent the seventh most abundant riboswitch class that has been experimentally validated to date (McCown et al. 2017). Members of this common riboswitch class usually control the expression of genes required for the biosynthesis and transport of the coenzyme precursor riboflavin (Gelfand et al. 1999; Mironov et al. 2002; Winkler et al. 2002). FMN and flavin adenine dinucleotide (FAD) are ordinarily used as redox cofactors for flavoenzymes involved in various aspects of cellular metabolism (Fischer and Bacher 2005). However, due to the high photoreactivity of these compounds, they can degrade into various toxic byproducts such as lumiflavin and lumichrome (Fall and Petering 1956; West and Owen 1973; Choe et al. 2005). Some bacterial and archaeal cells protect themselves from these phototoxic products with riboflavin-binding proteins called dodecins that act to both store riboflavin and prevent its undesirable breakdown (Grininger et al. 2006, 2009). Presumably, some bacterial species also have detection and remediation systems in the event that substantial amounts of FMN breakdown products accumulate.
Herein, we describe our analysis of two classes of variant FMN riboswitches whose initial members were previously discovered in various strains of Clostridium difficile (Blount 2013; Blount et al. 2012; Weinberg et al. 2017b) and originally called CD3299 RNA. Furthermore, bioinformatic analysis has yielded additional representatives of these variant FMN motif RNAs in strains of Atopobium and in bacterial metagenomic DNA sequences. We describe two highly similar variant subtypes, and demonstrate that all representatives tested completely reject FMN. Intriguingly, subtype 2 representatives strongly bind riboflavin and certain light-mediated FMN degradation products. We hypothesize that some variant FMN RNAs might have evolved to recognize flavin derivatives to activate transporters involved in their disposal. These findings reinforce the hypothesis that many riboswitch classes remaining to be discovered will be exceptionally rare and challenging to match with their corresponding ligands.
RESULTS AND DISCUSSION
Variant RNAs have key characteristics that differ from typical FMN riboswitches
Examples of FMN riboswitch variants were previously reported (Blount 2013; Blount et al. 2012) to exist only in several strains of C. difficile. Additional examples were subsequently uncovered in a bioinformatics study that purposefully examined known riboswitch classes for the existence of variant riboswitch aptamers that have possibly altered their ligand binding specificities (Weinberg et al. 2017b). Given the ongoing rapid expansion of bacterial genomic sequence data sets, we searched (see Materials and Methods) for additional representatives of the FMN variants and identified a total of 57 unique-sequence examples (Supplemental Data File) present in certain Clostridium and Atopobium species, as well as in bacterial metagenomic DNA sequence data (RefSeq version 76 and additional environmental DNA sequence databases).
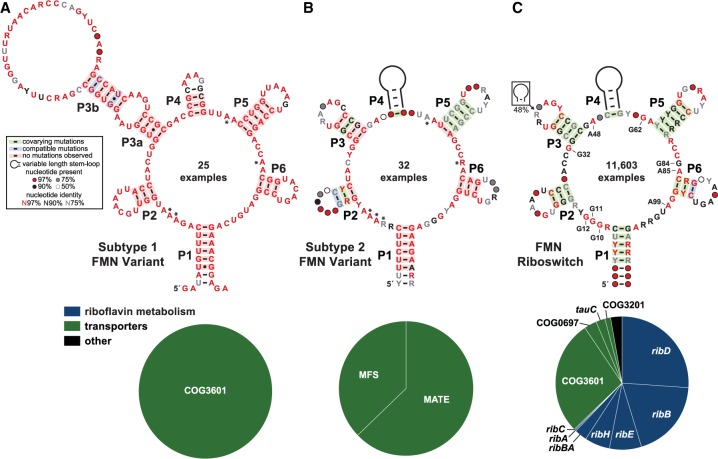
These findings allowed us to create revised consensus sequence models for the variants, and to increase the list of genes known to be associated with these RNAs. Specifically, the variants have been organized into two groups, called subtype 1 (25 examples) (Fig. 1A) and subtype 2 (32 examples) (Fig. 1B). In addition, we updated the collection of FMN riboswitch representatives from the same genomic and metagenomic sequence databases to establish a current consensus model for this well-established riboswitch class (Fig. 1C). The variants exhibit many similarities, both with regard to their conserved nucleotide sequences and their major secondary structure elements, to the FMN riboswitch consensus model. Due to these abundant similarities, it is not surprising that the rare variants previously had been grouped with the much larger collection of FMN riboswitch representatives (11,603 examples), including in the Rfam Database (Nawrocki et al. 2015). This lumping of multiple riboswitch classes into a single collection can occur when variants do not substantially differ from the established aptamer consensus, or when too few representatives exist for their discovery by careful comparison of nucleotide sequences.
FIGURE 1.
Variant FMN motif RNAs differ from the FMN riboswitch consensus and its typical genetic associations. (A) Consensus sequence and secondary structure model of 25 unique examples of subtype 1 variant FMN RNAs. Asterisks identify key nucleotides that differ from FMN riboswitch consensus. The key (box) describes the annotations in the consensus models. (Bottom) Pie chart of the genes located immediately downstream from subtype 1 RNAs. (B) Consensus model and gene associations for 32 unique examples of subtype 2 RNAs. Additional annotations are as described for A. (C) Consensus model and gene associations for 11,603 unique examples of FMN riboswitch aptamers. Certain nucleotides that are most directly involved in forming the ligand binding pocket for FMN are numbered according to the crystal structure model (PDB ID code 3F2Q) published previously (Serganov et al. 2009).
Despite these commonalities, the differences between authentic FMN riboswitches and the two variant subtypes are striking. The distinct features of the variants can be best appreciated by comparison with the gene associations, conserved sequences, and structural features of FMN-sensing riboswitches. FMN riboswitches usually reside upstream of genes involved in riboflavin biosynthesis (Fig. 1C, bottom; Gelfand et al. 1999; Mironov et al. 2002; Winkler et al. 2002). Genes encoding COG3601, COG0697, COG3201, and TauC family proteins are also commonly associated with FMN riboswitches. These proteins have been characterized as riboflavin transporters in various organisms (Vogl et al. 2007; García Angulo et al. 2013; Gutiérrez-Preciado et al. 2015).
In stark contrast to FMN riboswitch regulation of genes involved in the biosynthesis and import of riboflavin, subtype 1 variant FMN riboswitches are never associated with FMN biosynthetic genes, but are currently exclusively observed in association with genes for members of the COG3601 transporter family (Fig. 1A, bottom). Curiously, genes annotated as COG3601 are also commonly regulated by true FMN riboswitches and, as noted above, in those instances are predicted to be riboflavin transporters (Gutiérrez-Preciado et al. 2015). However, there are many examples of COG3601 genes that are linked to other metabolic processes, and the specificities of those transporters are proposed to be different (Gutiérrez-Preciado et al. 2015). Thus, it seems possible that the COG3601 genes whose expression is regulated by subtype 1 RNAs might have a distinct substrate compared to those regulated by true FMN riboswitches.
Likewise, the subtype 2 variant FMN riboswitches are exclusively found upstream of genes encoding putative multidrug efflux transporters (Fig. 1B, bottom). In various strains of C. difficile, subtype 2 variant FMN riboswitches are commonly associated with genes coding for proteins annotated as putative EmrB and NorM multidrug transporters from the major facilitator superfamily (MFS) and multidrug and toxic compound extrusion (MATE) superfamily, respectively (Fig. 1B, bottom). Certain members of the MATE superfamily, such as the Vibrio parahaemolyticus protein NorM, have been demonstrated to transport a wide variety of cationic dyes, quaternary ammonium compounds, and quinolones (Morita et al. 1998). Moreover, transporters associated with variant FMN RNAs have been previously shown to contain MFS 1 domains (Gutiérrez-Preciado et al. 2015). Proteins that contain these domains exhibit homology with the Escherichia coli protein EmrB. Members of this protein class are part of the EmrAB-TolC tripartite system, which has been shown to confer resistance to hydrophobic toxins (Lomovskaya and Lewis 1992; Lewis 2000).
Not a single example of a norM or emrB gene is associated with the many thousands of FMN riboswitches that have been identified by using bioinformatics search methods. Again, the patterns of gene associations between true FMN riboswitches and these rarer variants strongly indicate that the ligand for the variant riboswitches has been changed from FMN to another, but perhaps structurally related, compound. Presumably, the natural substrate for these specific transporters will match the ligand for the variant riboswitches. Unfortunately, the precise ligands transported by the proteins whose genes are regulated by the variant riboswitches are not yet known. Indeed, as described in greater detail later in this report, we used ligand-binding assays to demonstrate that these variants do not recognize the cofactor FMN. These findings are consistent with our hypothesis that the variants do not function as FMN-responsive riboswitches, but carry nucleotide differences that have altered their ligand specificities.
Most importantly, the nucleotide sequence differences between true FMN riboswitches and the rare variants almost exclusively reside in otherwise highly conserved nucleotides that are known to form direct contacts with the FMN ligand (Fig. 2). Previously, the FMN riboswitch aptamer from the Bacillus subtilis ribD mRNA was found to exhibit a dissociation constant (KD) for FMN of 5 nM (Winkler et al. 2002). X-ray crystallography was subsequently performed with another member of the FMN riboswitch class from Fusobacterium nucleatum (Serganov et al. 2009). FMN is recognized by the RNA predominantly through interactions with the isoalloxazine ring and through contacts with the phosphate on the ribityl moiety (Fig. 2A). The isoalloxazine ring system intercalates between the bases of two adenosine nucleotides (A48 and A85) (Fig. 1C) of the aptamer, and also forms two hydrogen bonds with another highly conserved adenosine (A99) (Fig. 2A). The ribityl and phosphate moieties are selectively recognized by the formation of numerous hydrogen bonds with several conserved guanosine nucleotides distributed throughout the riboswitch aptamer (G10, G11, G32, G62, G84) (Fig. 2A). The absence of the phosphate moiety, such as with riboflavin, has been found to reduce ligand affinity by almost three orders of magnitude (Winkler et al. 2002), which demonstrates the importance of this set of highly conserved guanosine nucleotides.
FIGURE 2.
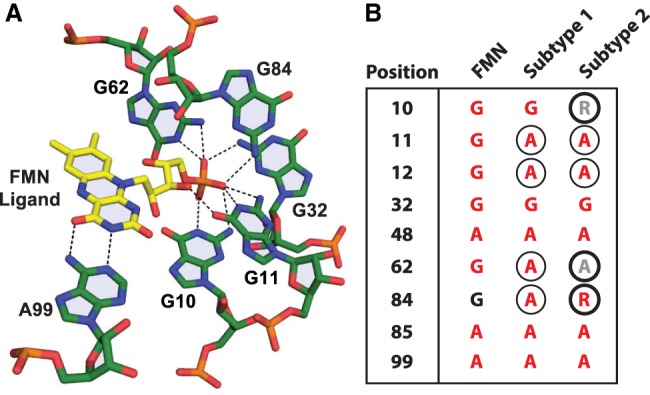
Atomic-resolution structural model for the binding site of an FMN riboswitch from F. nucleatum (PDB ID code 3F2Q) (Serganov et al. 2009). Dashed lines represent hydrogen-bonding contacts between aptamer nucleotides (numbered as depicted in Fig. 1C) and the ligand, FMN. (B) Comparison of key equivalent nucleotide positions among FMN aptamer, subtype 1, and subtype 2 RNAs. Nucleotides are depicted according to their level of sequence conservation as described in the key to Figure 1A. Circled nucleotides are distinct from the consensus model for FMN riboswitches. Thick-lined circles identify nucleotides in the consensus for subtype 2 RNAs that differ from the consensus for subtype 1 RNAs. Nucleotide positions are depicted in A, except for positions 12, 48, and 85, which were omitted for image clarity.
The two variant FMN RNA subtypes (Fig. 1A,B) have many sequence and structural features resembling those of the FMN riboswitch consensus (Fig. 1C), but importantly they deviate from the consensus sequence at certain nucleotide positions that are directly involved in recognizing FMN. These changes to the ligand binding pocket typically convert the highly conserved (>97%) guanosine nucleotides that are known to contact the phosphate and ribityl moieties of FMN (Fig. 2A) to adenosine nucleotides (Fig. 2B). Presumably, the variant RNAs no longer have the ability to create productive binding interactions with the phosphate moiety of FMN, which explains why the variant RNAs examined for FMN ligand binding completely reject this enzyme cofactor (see below). However, the conservation of nucleotide identities at positions equivalent to 48, 85, and 99, which contact the flavin moiety, suggest that this part of the natural ligand for subtypes 1 and 2 RNAs might remain unchanged.
There are some notable sequence and structural features that also differ between the two subtypes. Most obvious is the presence of an extended P3 stem (called P3a and P3b) in the consensus model for subtype 1 RNAs (Fig. 1B) that is lacking in subtype 2 RNAs (Fig. 1B). However, nearly half of all true FMN riboswitches carry an extended P3 stem, and so it is not certain that this distinction affects the ligand specificity of subtype 1 RNAs. There are modest differences in the key nucleotides that form the original FMN binding pocket between subtype 1 and 2 RNAs (Fig. 2B). Given the differences in gene associations between these two RNAs, it seems possible that these nucleotide changes also result in different ligand specificities.
Subtype 2 variant FMN RNAs recognize photolytic products of FMN and FAD
Frequently, the ligand that is sensed by a riboswitch class is identified by examining the genes that reside immediately downstream. Unfortunately, as noted above, the selectivities of the transporter proteins encoded by the genes downstream from the variant FMN RNAs remain unknown, and thus provide limited clues regarding the identity of the cognate ligand. Because many C. difficile strains already carry a gene for a putative riboflavin importer whose expression is controlled by an FMN riboswitch, we hypothesized that some bacteria might use variant FMN RNAs to control the expression of efflux pumps to alleviate the deleterious buildup of toxic FMN breakdown products (Fall and Petering 1956; West and Owen 1973). This would be analogous to the observed use of guanidine riboswitches for controlling the production of multidrug efflux pumps (Nelson et al. 2017; Kermani et al. 2018). Both FMN and riboflavin are highly photoreactive and have been shown to generate various light-promoted breakdown products such as lumiflavin, lumichrome, and carboxymethylflavin (Choe et al. 2005). Small riboflavin-binding proteins called dodecins are used by archaeal and bacterial cells to prevent flavin derivatives from damaging key molecules in the cell by stabilizing riboflavin against photochemical breakdown or by sequestering toxic byproducts, such as lumichrome (Grininger et al. 2006, 2009).
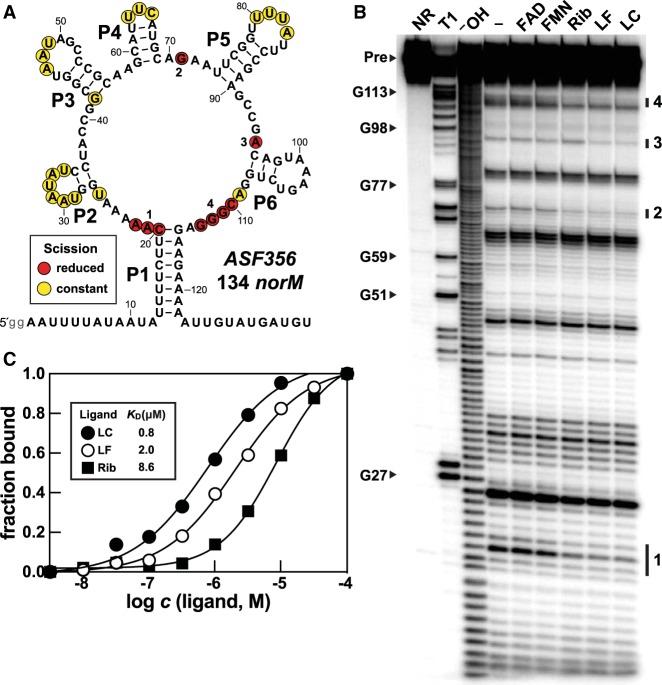
To assess the ability of variant FMN RNAs to directly bind photochemical breakdown products of FMN, in-line probing assays were performed with a 134 nt RNA, named 134 norM, which is derived from the 5′-untranslated region of the norM gene of Clostridium sp. ASF356 (Fig. 3A). In-line probing reveals ligand-mediated shape changes in RNA structures by monitoring the products of spontaneous RNA phosphodiester cleavage (Soukup and Breaker 1999). We observed that both UV-treated FMN and UV-treated FAD cause decreased levels of spontaneous RNA strand scission at various locations of the 134 norM RNA chain (Supplemental Fig. S1A), suggesting that various breakdown products of FMN (Supplemental Fig. S1B) such as lumichrome and lumiflavin (Supplemental Fig. S1C) are bound by this putative aptamer.
FIGURE 3.
A representative of the subtype 2 variant FMN motif RNAs recognize FMN degradation products. (A) Sequence and secondary structure model of the 134 norM RNA construct. Lowercase letters on the 5′ terminus represent guanosine nucleotides added to the DNA template to promote transcription by T7 RNA polymerase. Regions of constant and reduced spontaneous RNA cleavage upon addition of ligand are indicated by yellow and red circles, respectively. These data are derived from the in-line probing analysis depicted in B. (B) PAGE analysis of in-line probing assays with 5′ 32P-radiolabeled 134 norM RNA in the absence (−) or presence of FMN, riboflavin (Rib), lumiflavin (LF), or lumichrome (LC) at 100 µM. NR, T1, and −OH designate RNAs that have undergone no reaction, that have been partially digested with RNase T1, or that have been partially digested with alkali, respectively. Bands corresponding to certain RNase T1 digestion products (scission after G residues) are labeled according to the numbering system in A. Groups of bands denoted 1 through 4 identify regions of the RNA that undergo substantial change in response to ligand addition. (C) Plot of the fraction of RNA bound to the ligand versus the logarithm of the molar concentration (c) of the ligand. Fraction bound values were estimated based on the extent of band intensity changes at region 1 from data presented in Supplemental Figure S4.
Indeed, reproducible changes in the banding pattern produced by in-line probing incubations are also exhibited by the 134 norM RNA in the presence of riboflavin, lumiflavin, and lumichrome, when each was individually tested at 100 µM (Fig. 3B). Similar ligand-induced structural modulation of subtype 2 RNA constructs from other organisms was also observed (Supplemental Figs. S2, S3). These changes to the RNA structure as inferred from differences in the banding patterns produced by the in-line probing assays are occurring at nucleotide positions that are at or immediately adjacent to those corresponding to the binding site nucleotides for FMN riboswitches (Figs. 1, 2A). In contrast, FMN is rejected by the 134 norM construct and the other subtype 2 RNAs examined, presumably because most of the original nucleotides of the ribityl-phosphate binding pocket are different in subtype 2 RNAs (Fig. 2B). Similarly, the 134 norM RNA binds lumichrome and lumiflavin more tightly than it does riboflavin (Fig. 3C; Supplemental Fig. S4), suggesting that the ribityl moiety of riboflavin (which is lacking in lumichrome and lumiflavin) also is discriminated against by the variant binding pocket. This is consistent with our prediction that the extensive changes in the identities of nucleotides forming the original ribityl-phosphate binding pocket that occur with subtypes 1 and 2 RNAs would affect recognition of that portion of the ligand, but would not necessarily affect binding of the flavin moiety.
The in-line probing data for 134 norM also indicate that the various ligands bind with a 1:1 interaction (Fig. 3C; Supplemental Fig. S4). Similar results are exhibited by another subtype 2 RNA construct called 136 emrB RNA derived from C. difficile CD196 (Supplemental Fig. S5). These findings are consistent with the data observed previously for FMN riboswitches (Winkler et al. 2002; Serganov et al. 2009), suggesting that the nucleotide differences in the binding pocket of FMN riboswitches compared to subtype 2 RNAs cause a change in ligand specificity, but not stoichiometry.
Unlike subtype 2 RNA constructs, we have not yet observed ligand binding by subtype 1 RNA constructs. In-line probing assays were used to examine the function of a 186-nt construct associated with the COG3601 gene from the bacterium Atopobium sp. BS2, called 186 COG3601 (Supplemental Fig. S6). The construct fails to respond to the addition of 100 μM FMN, which suggests that these RNAs have also altered their ligand specificity. However, both UV-treated samples and known photochemical breakdown products of FMN fail to induce any substantial structural modulation. Also tested was the FMN analog 10-(3-(4-fluorophenyl)propyl)-7,8-dimethylbenzo[g]pteridine-2,4(3H,10H)-dione (called 10FDPD) (Blount 2013; Blount et al. 2012), which contains an aryl-alkyl moiety in place of the ribityl phosphate moiety (Supplemental Fig. S1B). Compounds like 10FDPD were developed as FMN analogs that function as antibacterial agents that target FMN riboswitches (Blount et al. 2015), and 10FDPD modulates the structure of a subtype 2 RNA construct (Supplemental Fig. S1A). Again, unlike subtype 2 RNAs, the subtype 1 RNA construct 186 COG3601 fails to bind this FMN analog. It is not known whether subtype 1 RNAs recognize a natural ligand that is different from that of the other variants, or whether the construct tested in vitro fails to respond as they do within their native biological context.
Subtype 2 RNAs from Clostridium species are genetic “ON” riboswitches
The B. subtilis ribD FMN riboswitch controls the expression of the entire riboflavin biosynthesis operon through transcription termination regulation (Winkler et al. 2002). When FMN is bound by the riboswitch, an intrinsic transcription terminator stem (Gusarov and Nudler 1999; Yarnell and Roberts 1999) forms that halts extension of the nascent mRNA transcript. In the absence of FMN, an anti-terminator structure can form to prevent formation of the terminator. Thus, the competing terminator and anti-terminator structures constitute the “expression platform” (Breaker 2012) of the riboswitch. A second FMN riboswitch representative also found in B. subtilis controls expression of the ribU gene (previously called ypaA), which encodes a riboflavin transporter of the COG3601 protein family (Vogl et al. 2007). This riboswitch is predicted to regulate translation initiation by preventing ribosome access to the ribosome binding site (Shine-Dalgarno sequence) upon binding to FMN (Winkler et al. 2002). These examples provided by FMN riboswitches from the same organism showcase two of the most common expression platform mechanisms by which riboswitches control gene expression in response to the accumulation of their target metabolites (Breaker 2012).
Regardless of the mechanism used, riboswitches almost always incorporate an expression platform downstream from the aptamer to regulate the level of gene expression upon ligand binding. Therefore, to assess how variant FMN riboswitches control gene expression, we manually inspected sequences downstream from several variant FMN RNA aptamers for evidence of expression platform structures. Subtype 2 RNAs, which associate with genes annotated as multidrug transporters, typically precede a putative intrinsic terminator stem. These subtype 2 aptamers therefore most probably regulate gene expression via transcription termination, and should activate the expression of genes for efflux pumps if they indeed recognize toxic FMN breakdown products.
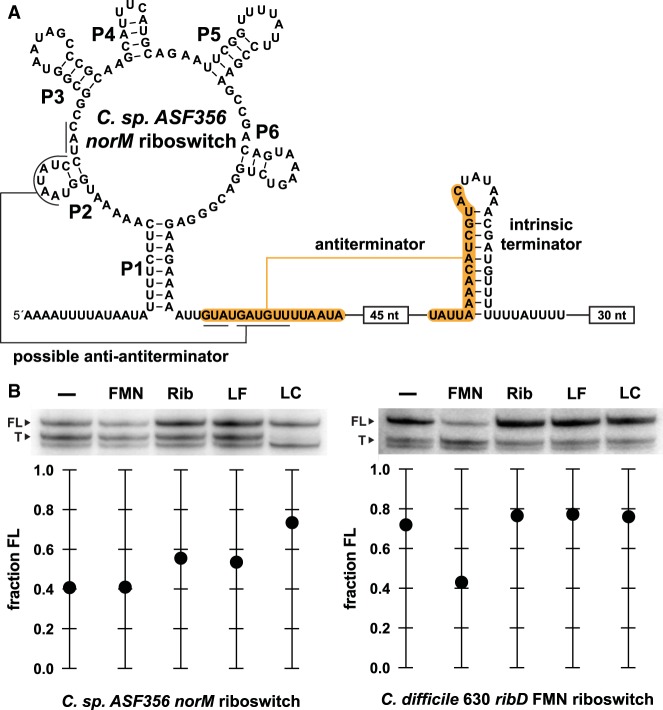
To experimentally assess the mechanism of variant 2 RNAs most similar to those found in C. difficile, we conducted single-round transcription termination assays (Landick et al. 1996) using a DNA template derived from the region upstream of the norM gene from C. sp. ASF356. The RNA derived from this norM construct has the potential to form a strong antiterminator stem within the expression platform (orange shading, Fig. 4A). Folding of the aptamer structure in the presence of ligand should permit formation of this antiterminator stem and thereby promote transcription of the full-length mRNA. In the absence of ligand, alternative folding appears possible because nucleotides in the right shoulder of P2 are complementary to nucleotides required to form the antiterminator. Formation of this putative “anti-antiterminator” stem in the absence of ligand should halt transcription by allowing the formation of the intrinsic terminator stem.
FIGURE 4.
Subtype 2 and FMN riboswitch RNAs regulate transcription termination in response to different ligands. (A) Sequence and secondary structure model of the subtype 2 variant FMN riboswitch construct derived from the norM gene of C. sp. ASF356 used for transcription termination assays. Terminated transcripts end within the U-rich tract immediately following the intrinsic terminator stem, whereas full-length transcripts from the DNA template used for this assay include the 30 nt following the U-rich tract. (B, top) PAGE analysis of single-round transcription termination assays conducted in the absence of ligand (−), or in the presence of 1 mM FMN, riboflavin (Rib), lumiflavin (LF), or lumichrome (LC). FL and T denote full-length and terminated RNA transcripts, respectively. (Bottom) Values for the fraction of FL RNA generated are derived from the PAGE data presented.
The amounts of terminated and full-length RNA transcripts generated in the presence of FMN and several analogs were measured with the C. sp. ASF356 norM DNA template, and with a template carrying an FMN riboswitch derived from the ribD gene from C. difficile 630 (Fig. 4B). We observed modest changes in the fraction of full-length transcripts when riboflavin, lumiflavin, and particularly lumichrome are added to single-round in vitro transcription reactions (Fig. 4B, left). Lumichrome also has the best observed KD value determined by in-line probing assays for these variant RNAs (Fig. 3C). In contrast, no changes in the fraction of elongated transcripts are seen for the ribD FMN riboswitch when these same compounds are introduced (Fig. 4B, right). However, as expected, FMN does alter the relative transcription yields for this natural FMN riboswitch, whereas FMN has no effect on the distribution of norM RNA transcripts.
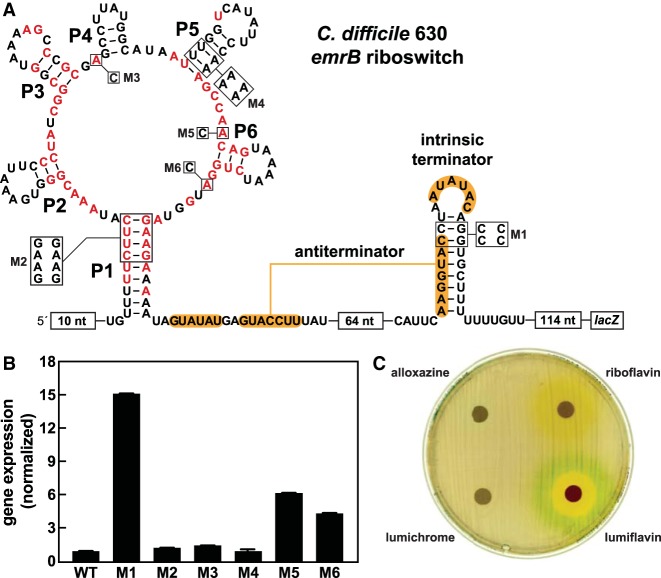
Another subtype 2 riboswitch representative, this one derived from the emrB gene of C. difficile 630, was also examined for in vivo gene control activity by fusing the construct or several mutant versions of this construct to a lacZ β-galactosidase reporter gene (Fig. 5A). When B. subtilis cells carrying the wild-type (WT) emrB variant construct are cultured in rich (lysogeny broth, LB) medium liquid, reporter gene expression is negligible (Fig. 5B). In contrast, when mutations (construct M1) are introduced to disrupt the predicted intrinsic terminator stem, robust reporter gene expression occurs. Furthermore, disruption of key conserved RNA sequence or structural features that form the aptamer, such as in mutant reporter constructs M2, M3, and M4, fails to change reporter gene expression. These results suggest that, under these culture conditions, the WT riboswitch is in its “OFF” state and that ligand is not present at concentrations sufficient to be bound to the aptamer. Reporter construct M5 and M6 do exhibit modest gene expression increases relative to the WT construct, which is not entirely consistent with our hypothesis that subtype 2 RNAs function as “ON” switches. However, we speculate that these mutations might cause folding problems that interfere somewhat with terminator stem formation, rather than serve as evidence for “OFF” switch function by this riboswitch representative.
FIGURE 5.
In vivo assessment of gene control by a subtype 2 variant RNA from C. difficile. (A) Sequence and secondary structure of the regulatory region of the riboswitch-reporter construct made by joining the variant FMN riboswitch RNA from the emrB gene of C. difficile 630 to a lacZ gene. Red nucleotides are >97% conserved. Constructs carrying mutations to conserved nucleotides and secondary structures are designated M1 through M6. (B) Plot of the normalized β-galactosidase reporter gene expression levels of B. subtilis cells containing WT and various mutant reporter constructs in rich medium. Error bars indicate the standard deviation from three independent experiments. (C) Agar diffusion assays were conducted with LB agar plates supplemented with 100 µg mL−1 X-gal and inoculated with B. subtilis cells carrying the WT reporter construct as described in A. Filter disks were infused with 10 µL of alloxazine (50 mM), riboflavin (35 mM), lumichrome (10 mM), or lumiflavin (80 mM). Note that the yellow-orange color surrounding the riboflavin-applied disk is due to the color of this compound, which is diffusing outward.
Similarly, the C. difficile 630 subtype 2 riboswitch reporter construct in B. subtilis cells grown on LB medium agar plates exhibits no apparent signs of gene expression (Fig. 5C). Agar-diffusion assays revealed that riboflavin, lumichrome, and alloxazine (Supplemental Fig. S1D) also do not trigger expression of the reporter construct in B. subtilis. However, all three compounds lack zones of cell growth inhibition, meaning that they do not accumulate to toxic levels inside B. subtilis cells. Thus, we cannot be certain that concentrations necessary to trigger riboswitch function can be achieved by supplying these molecules in the culture medium. Although reporter gene expression (blue color) is seen with the addition of lumiflavin to a filter disk, reporter constructs M2 through M4 failed to disrupt reporter gene expression (data not shown). This result is most likely caused by nonspecific activation of our reporter gene, as lumiflavin was also seen to modestly activate reporter gene expression when fused to FMN riboswitches (data not shown). This false positive signal might occur when toxic compounds affect overall mRNA transcription or translation levels.
We did not conduct parallel studies on a representative of subtype 1 variant riboswitches, in part due to the fact that we have not observed binding for any of the candidate ligands tested in vitro. In contrast to subtype 2 RNAs, all subtype 1 riboswitches located upstream of genes encoding COG3601 family proteins in Atopobium species are predicted to translationally control gene expression via sequestration of the ribosome binding site. Formation of the P1 stem of the variant aptamer in the presence of ligand would preclude access of the ribosome binding site and repress gene expression. These RNAs thus appear to repress the expression of their associated transporters in response to binding by their natural ligand, which is opposite of that predicted for subtype 2 variants. Either subtype 1 RNAs have yet another ligand specificity, or cells that carry these RNAs benefit by repressing the expression of their associated transporters, whereas subtype 2 variants activate the expression of their associated transporters.
Variant subtype 2 FMN riboswitches hint at an unknown flavin-like detoxification pathway
Toxic compounds often build up from the spontaneous chemical breakdown of coenzymes and cofactors, or from enzymatic reactions that yield unwanted side products (Linster et al. 2013). Flavins, such as FMN and FAD, which are commonly utilized by flavoproteins to catalyze a wide range of redox reactions, are no exception from this problem particularly due to their high photoreactivity (Fischer and Bacher 2005). Various types of damage control systems are utilized by bacteria and other species to prevent the harmful buildup of toxic agents. For example, many bacteria utilize riboswitches to sense toxic ligands, such as S-adenosylhomocysteine (Wang et al. 2008), fluoride (Baker et al. 2012), or guanidine (Nelson et al. 2017), and to activate genes important for toxicity resistance.
It also seems possible that subtype 2 FMN RNAs activate the production of efflux proteins to alleviate the harmful buildup of their cognate ligand. Consistent with this hypothesis is the observation that the subtype 2 norM riboswitch includes an expression platform with terminator, antiterminator, and possible anti-antiterminator base-pairing potential (Fig. 4A). Indeed, lumichrome, which was found to bind most tightly to representative subtype 2 RNAs (Fig. 3C), promotes elongation of the norM riboswitch construct in single-round transcription termination assays (Fig. 4B, left).
Unfortunately, due to the limited genetic contexts and limited analysis of ligand analogs, we cannot yet conclude that any of these molecules are the natural ligand for the subtype 2 riboswitch class. As noted above, many of the genes predicted to be controlled by subtype 2 RNAs are annotated as putative multidrug transporters. However, one of these genes from C. difficile has been recently proposed to code for a riboflavin import protein (Gutiérrez-Preciado et al. 2015). It is important to note that multidrug resistance transporters have also been observed to function inefficiently in the opposite direction by bringing molecules into the cell (Robinson et al. 2017), and many of the C. difficile strains that carry variant FMN RNAs often already carry another gene regulated by an FMN riboswitch that codes for a riboflavin importer. Therefore, the natural function of the transporters associated with the variant riboswitches might involve the efflux of toxic flavin compounds, rather than the import of riboflavin.
Consistent with this hypothesis is the observation that ligand binding activates production of the full-length RNA transcript, which is opposite of what cells would need if riboflavin were the ligand, and the associated transporter were riboflavin importers. Transporters for fluoride (Baker et al. 2012; Stockbridge et al. 2013) and guanidine (Nelson et al. 2017; Kermani et al. 2018) are two examples of proteins whose annotations have been changed in recent years as orphan riboswitch classes have been solved. Thus, the identification of the true natural ligand for the FMN riboswitch variants described herein would also help establish the substrates targeted by the transporters.
Due to the lack of compound availability, we were unable to generate sufficient data to formulate a structural model for the cognate ligand for variant FMN RNAs using structure–activity relationship (SAR) assays. Many of the predicted photochemical breakdown products of FMN and FAD contain a variation of the ribityl tail (Supplemental Fig. S1C). Although subtype 2 variant riboswitch RNAs bind lumichrome with the tightest binding affinity measured in this study (Fig. 3C), drug-like FMN analogs similar to 10FDPD have been previously identified to bind FMN riboswitches just as tightly as the cognate ligand (Blount et al. 2015). Thus, we cannot be certain that tight-binding ligand analogs are perfect guides to the chemical structure of the true ligand. Also, lumichrome reduces the amount of terminated product during in vitro transcription assays, but fails to inhibit B. subtilis cell growth or trigger reporter gene expression. Perhaps the cognate ligand for variant FMN RNAs is a distinct compound endogenous to C. difficile. Or, perhaps the B. subtilis surrogate organism used in our study to evaluate reporter gene regulation already carries an efficient detoxification pathway to deal with lumichrome toxicity, which prevents our in vivo gene regulation assays from revealing gene regulation by this compound.
Conclusions
In the current study, we provide data on variant riboswitches that carry mutations in the ligand-binding aptamer domain of the FMN class. Most notably, these variant RNAs carry mutations of nucleotide positions that directly recognize the ribityl and phosphate moieties, and so the mechanism for the loss of FMN recognition is readily apparent. We demonstrate that degradation products of FMN, such as riboflavin, lumiflavin, and lumichrome, are recognized by certain variant FMN RNAs of the subtype 2 group (Fig. 3). However, due to obscure gene associations, it is not immediately evident what cognate ligand this variant riboswitch class senses. Previous work has determined that it is possible to design synthetic analogs that trick riboswitch classes into altering gene expression (Kim et al. 2009; Blount et al. 2015; Howe et al. 2015). Thus, the isoalloxazine derivatives might only be close mimics of the cognate ligand, and the true cognate ligand might await discovery.
The expression platform for subtype 2 variant RNAs is predicted to activate gene expression in the presence of ligand (Fig. 4). This logic is similar to other riboswitch classes that sense toxic ligands and activate the expression of transporter proteins (Baker et al. 2012; Nelson et al. 2017; Sherlock and Breaker 2017; Sherlock et al. 2017). Consistent with this hypothesis is the fact that these variant RNAs are commonly found upstream of genes annotated as multidrug transporters. These transporters have similarity to members of the EmrB/QacA family of MFS exporters whose typical substrates are polyaromatic cations. In other Gram-positive bacteria, QacA transporters are regulated by the QacR protein, which functions as a transcriptional repressor of the qacA gene (Grkovic et al. 1998). When substrates for QacA accumulate in cells, they bind to QacR and prevent it from binding DNA, thereby increasing expression levels of QacA transporters. If the sensory and regulatory functions of subtype 2 variant FMN RNAs are analogous to those of QacR, then these RNAs most likely function to turn on gene expression in response to toxic flavin metabolites.
Many flavin compounds, such as riboflavin, are highly photoreactive, and can become harmful for cells when they accumulate or degrade into various phototoxic byproducts (Choe et al. 2005). However, in the riboflavin overproducing strain, Ashbya gossypii, riboflavin overproduction during sporulation has been shown to protect spores from ultraviolet (UV) light (Stahmann et al. 2001). Therefore, anaerobic spore-forming bacteria, such as C. difficile, might utilize variant FMN riboswitches to activate detoxification pathways necessary to efflux toxic flavin compounds that accumulated as a spore. Since C. difficile is a major cause of nosocomial infections, understanding the mechanism by which C. difficile spores are highly resistant to environmental stresses such as UV could aid in the development of new therapeutic or decontamination methods.
MATERIALS AND METHODS
Chemicals, DNA oligonucleotides, and bacterial strains
Chemical compounds were purchased from Sigma-Aldrich with the exception of 10FDPD, which was acquired from the former assets of BioRelix. [γ-32P]-ATP and [α-32P]-ATP were purchased from PerkinElmer. DNA oligonucleotides (Supplemental Table S1) were purchased from Sigma-Aldrich or Integrated DNA Technologies. All enzymes were purchased from New England BioLabs unless otherwise specified. Plasmid pDG1661 and B. subtilis strain 168 (BGSC 1A747) cells were obtained from the Bacillus Genetic Stock Center (BGSC) at The Ohio State University.
Bioinformatic analyses
Additional homologs of the variant FMN motif RNAs were identified using Infernal 1.1 (Nawrocki and Eddy 2013). Searches were conducted with RefSeq (O'Leary et al. 2016) version 76 and various metagenomic databases as previously described (Weinberg et al. 2017b). Sequence and secondary structure consensus model of the variant FMN motif RNAs (57 sequences) were constructed using R2R (Weinberg and Breaker 2011).
Preparation of RNA oligonucleotides
RNAs were prepared by in vitro transcription using DNA oligonucleotides containing a T7 RNA polymerase promoter sequence upstream of the desired template sequence (Supplemental Table S1). Specifically, in vitro transcription reactions were performed using laboratory-prepared bacteriophage T7 RNA polymerase (2 units µL−1) in 80 mM HEPES (pH 7.5 at 23°C), 40 mM DTT, 24 mM MgCl2, 2 mM spermidine, and 2 mM of each NTP. RNA was purified using denaturing (8 M urea) 10% polyacrylamide gel electrophoresis (PAGE). RNA bands were visualized by UV shadowing, excised, and then eluted from the gel slice by the crush-soak method overnight in 10 mM Tris-HCl (pH 7.5 at 23°C), 200 mM NaCl, and 1 mM EDTA. The RNA was precipitated by adding two volumes of 100% ethanol and incubating at −20°C for 30 min, and then pelleted by centrifugation.
To generate RNAs 32P-radiolabeled at the 5′-terminus, RNA was subjected to dephosphorylation using rAPid Alkaline Phosphatase (Roche Diagnostics), and then radiolabeled with [γ-32P]-ATP at the 5′-terminus using T4 polynucleotide kinase. Radiolabeled RNAs were purified by denaturing 10% PAGE and recovered as described above.
In-line probing of RNAs
In-line probing assays were performed as previously described (Soukup and Breaker 1999; Regulski and Breaker 2008). Briefly, 5′ 32P-labeled RNAs were incubated in the presence or absence of different concentrations of desired ligand at 23°C for 42–48 h in the presence of 20 mM MgCl2, 100 mM KCl, and 50 mM Tris-HCl (pH 8.3 at 23°C). The spontaneous RNA cleavage products were separated by denaturing 10% PAGE. The gels were dried and imaged with a Typhoon phosphorImager (GE Healthcare). Dissociation constants were established by varying the concentration of ligand added and quantifying the changes in band intensities via ImageQuaNT at nucleotide positions that exhibited ligand-induced structural modulation. Values for band intensities were normalized to a nonmodulating band, scaled between 0 and 1, and then plotted as a function of the logarithm of the ligand concentration. Values for apparent dissociation constants were determined using a sigmoidal dose-response equation and GraphPad Prism 7 as described previously (Sherlock and Breaker 2017).
In vitro transcription termination assays
The protocol for single-round in vitro transcription was adapted from one previous description (Landick et al. 1996). DNA constructs were designed to include the promoter sequence of the lysC gene from B. subtilis, the riboswitch aptamer, and the expression platform of the ribD gene or emrB gene from C. difficile to 57 or 42 nt, respectively, following the terminator stem. Approximately 2 pmol of the purified, PCR-amplified DNA template was added to a transcription initiation mixture (20 mM HEPES [pH 8.0 at 23°C], 5 mM magnesium acetate, 4 mM DTT, 1 mM EDTA, 10 µg mL−1 bovine serum albumin [BSA], 130 µM ApA dinucleotide, 1% glycerol, 0.04 U µL−1 E. coli RNA polymerase holoenzyme, 2.5 µM GTP, 2.5 µM UTP, and 1.5 µM ATP). Approximately 8 µCi [α-32P]-ATP was added to the 90 µL transcription reaction and transcription was allowed to proceed at 37°C for 10 min, leading to formation of a stalled polymerase complex at the first cytidine nucleotide of each transcript. The reaction mixture was then distributed in 8 µL aliquots into separate tubes, which contained 1 µL of a 10× solution of the ligand of interest and 1 µL of 10× elongation buffer (20 mM HEPES [pH 8.0 at 23°C], 5 mM magnesium acetate, 4 mM DTT, 1 mM EDTA, 1 mg mL−1 heparin, 75 µM each of ATP, GTP, and CTP, and 25 µM UTP). Products were separated by denaturing 10% PAGE then imaged and quantified using a Typhoon phosphorImager and ImageQuaNT software. The band intensities for both the full-length (FL) and terminated (T) RNA transcripts were measured, and the fraction FL values were calculated by using the equation Fraction FL = (FL intensity)/(FL intensity + T intensity). This simple equation can be used because the differences in specific activities between the FL and T products due to [α-32P]-ATP incorporation are negligible.
Design of reporter gene constructs
The nucleotide region −299 to +1 relative to the emrB translation start site of the downstream gene was amplified by PCR from C. difficile 630 genomic DNA. Subsequently, the B. subtilis lysC promoter was introduced via PCR amplification. This fragment was cloned into the vector pDG1661 as a transcriptional fusion with a lacZ reporter gene. The resulting plasmid was transformed and integrated into the amyE locus of B. subtilis 1A1.
Agar diffusion assays
B. subtilis strains carrying riboswitch reporter constructs were grown on LB agar plates containing X-gal (100 µg mL−1) and appropriate antibiotic. Autoclaved 6 mm diameter paper discs prepared from 0.35 mm thick pure cellulose chromatography paper (Fisher Scientific) were soaked with 10 µL of compound at specific concentrations and transferred to prepared agar plates. The plates were incubated overnight at 37°C prior to analysis.
Liquid-based β-galactosidase assays
Liquid-based β-galactosidase assays were performed as previously described (Vidal-Aroca et al. 2006; Nelson et al. 2015b). Bacterial cell cultures were grown overnight in 3 mL of LB medium with appropriate antibiotics at 37°C with shaking. The next day, cells were diluted 1:10 in LB and 80 µL of the resulting cultures were transferred to Costar black 96-well clear-bottom plates. The resulting samples were incubated for 4 h at 37°C with shaking. The absorbance at 595 nm was measured using a Tecan Infinite M200 PRO microplate reader. Subsequently, 80 µL of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) and 40 µL of 1 mg mL−1 4-methylumbelliferyl-β-D-galactopyranoside (4-MUG) were added to each well. The mixture was allowed to incubate at room temperature for 15 min, and 40 µL of 1 M Na2CO3 was used to quench the reaction. Excitation and emission values were measured at 360 nm and 460 nm, respectively, using a Tecan Infinite M200 PRO microplate reader. Fluorescence units were calculated as previously described (Vidal-Aroca et al. 2006).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adam Roth, Narasimhan Sudarsan, Etienne Greenlee, and other members of the Breaker laboratory for helpful conversations. R.M.A. was supported by the National Science Foundation Graduate Research Fellowship Program (DGE1122492). K.R.P was supported in part by the Medical Scientist Training Program (NIH/NIGMS T32 GM007205). This work was also supported by a National Institutes of Health grant to R.R.B. (GM022778), and R.R.B. is supported by the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067975.118.
REFERENCES
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. 2012. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335: 233–235. 10.1126/science.1215063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. 2004. New riboswitch motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426. 10.1073/pnas.0308014101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF. 2013. Methods for treating or inhibiting infection by Clostridium difficile. U.S. patent appl. no. 13/576,989.
- Blount KF, Coish PDG, Dixon BR, Myung J, Osterman D, Wickens P, Avola S, Baboulas N, Bello A, Berman J, et al. 2012. Flavin derivatives. U.S. patent appl. no. 13/381,809.
- Blount KF, Megyola C, Plummer M, Osterman D, O'Connell T, Aristoff P, Quinn C, Chrusciel RA, Poel TJ, Schostarez HJ, et al. 2015. Novel riboswitch-binding flavin analog that protects mice against Clostridium difficile infection without inhibiting cecal flora. Antimicrob Agents Chemother 59: 5736–5746. 10.1128/AAC.01282-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol Cell 43: 867–879. 10.1016/j.molcel.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2012. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 4: a003566 10.1101/cshperspect.a003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe E, Huang R, Min DB. 2005. Chemical reactions and stability of riboflavin in foods. J Food Sci 70: R28–R36. 10.1111/j.1365-2621.2005.tb09055.x [DOI] [Google Scholar]
- Fall HH, Petering HG. 1956. Metabolite inhibitors. I. 6, 7-dimethyl-9-formylmethyl-isoalloxazine, 6, 7-dimethyl-9-(2′-hydroxyethyl)-isoalloxazine and derivatives. J Am Chem Soc 78: 377–380. 10.1021/ja01583a035 [DOI] [Google Scholar]
- Fischer M, Bacher A. 2005. Biosynthesis of flavocoenzymes. Nat Prod Rep 22: 324–350. 10.1039/b210142b [DOI] [PubMed] [Google Scholar]
- García Angulo VA, Bonomi HR, Posadas DM, Serer MI, Torres AG, Zorreguieta Á, Golbaum FA. 2013. Identification and characterization of RibN, a novel family of riboflavin transporters from Rhizobium leguminosarum and other proteobacteria. J Bacteriol 195: 4611–4619. 10.1128/JB.00644-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet 15: 439–442. 10.1016/S0168-9525(99)01856-9 [DOI] [PubMed] [Google Scholar]
- Grininger M, Zeth K, Oesterhelt D. 2006. Dodecins: a family of lumichrome binding proteins. J Mol Biol 357: 842–857. 10.1016/j.jmb.2005.12.072 [DOI] [PubMed] [Google Scholar]
- Grininger M, Staudt H, Johansson P, Wachtveitl J, Oesterhelt D. 2009. Dodecin is the key player in flavin homeostasis of Archaea. J Biol Chem 284: 13068–13076. 10.1074/jbc.M808063200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grkovic S, Brown MH, Roberts NJ, Paulsen IT, Skurray RA. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem 273: 18665–18673. 10.1074/jbc.273.29.18665 [DOI] [PubMed] [Google Scholar]
- Gusarov I, Nudler E. 1999. The mechanism of intrinsic transcription termination. Mol Cell 3: 495–504. 10.1016/S1097-2765(00)80477-3 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Preciado A, Torres AG, Merino E, Bonomi HR, Goldbaum FA, García-angulo VA. 2015. Extensive identification of bacterial riboflavin transporters and their distribution across bacterial species. PLoS One 10: e0126124 10.1371/journal.pone.0126124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM. 2008. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 22: 3383–3390. 10.1101/gad.1747308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Todd M, Villafania A, Nahvi A, et al. 2015. Selective small-molecule inhibition of an RNA structural element. Nature 526: 672–677. 10.1038/nature15542 [DOI] [PubMed] [Google Scholar]
- Kellenberger CA, Wilson SC, Hickey SF, Gonzalex TL, Su Y, Halberg ZF, Brewer TF, Lavarnoe AT, Carolson HK, Hsieh YF, et al. 2015. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc Natl Acad Sci 112: 5383–5388. 10.1073/pnas.1419328112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermani AA, Macdonald CB, Gundepudi R, Stockbridge RB. 2018. Guanidinium export is the primal function of SMR family transporters. Proc Natl Acad Sci 115: 3060–3065. 10.1073/pnas.1719187115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Roth A, Breaker RR. 2007. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci 104: 16092–16097. 10.1073/pnas.0705884104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. 2009. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem Biol 4: 915–927. 10.1021/cb900146k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R, Wang D, Chan CL. 1996. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol 274: 334–353. 10.1016/S0076-6879(96)74029-6 [DOI] [PubMed] [Google Scholar]
- Lewis K. 2000. Translocases: a bacterial tunnel for drugs and proteins. Curr Biol 10: R678–R681. 10.1016/S0960-9822(00)00682-5 [DOI] [PubMed] [Google Scholar]
- Linster CL, Van schaftingen E, Hanson AD. 2013. Metabolite damage and its repair or pre-emption. Nat Chem Biol 9: 72–80. 10.1038/nchembio.1141 [DOI] [PubMed] [Google Scholar]
- Lomovskaya O, Lewis K. 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci 89: 8938–8942. 10.1073/pnas.89.19.8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. 2017. Riboswitch diversity and distribution. RNA 23: 995–1011. 10.1261/rna.061234.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirihana Arachchilage G, Sherlock ME, Weinberg Z, Breaker RR. 2018. SAM-VI RNAs selectively bind S-adenosylmethionine and exhibit similarities to SAM-III riboswitches. RNA Biol 15: 371–378. 10.1080/15476286.2017.1399232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111: 747–756. 10.1016/S0092-8674(02)01134-0 [DOI] [PubMed] [Google Scholar]
- Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 42: 1778–1782. 10.1128/AAC.42.7.1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43: D130–D137. 10.1093/nar/gku1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. 2013. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol 9: 834–839. 10.1038/nchembio.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Phillips GE, Stav S, Lünse CE, McCown PJ, Breaker RR. 2015a. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc Natl Acad Sci 112: 5389–5394. 10.1073/pnas.1419264112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Plummer MS, Blount KF, Ames TD, Breaker RR. 2015b. Small molecule fluoride toxicity agonists. Chem Biol 22: 527–534. 10.1016/j.chembiol.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Atilho RM, Sherlock ME, Stockbridge RB, Breaker RR. 2017. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol Cell 65: 220–230. 10.1016/j.molcel.2016.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR. 2008. In-line probing analysis of riboswitches. Methods Mol Biol 419: 53–67. [DOI] [PubMed] [Google Scholar]
- Ren A, Rajashankar KR, Patel DJ. 2012. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 486: 85–89. 10.1038/nature11152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AE, Thomas NE, Morrison EA, Balthazor BM, Henzler-Wildman KA. 2017. New free-exchange model of EmrE transport. Proc Natl Acad Sci 114: E10083–E10091. 10.1073/pnas.1708671114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152: 17–24. 10.1016/j.cell.2012.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. 2009. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458: 233–237. 10.1038/nature07642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock ME, Breaker RR. 2017. Biochemical validation of a third guanidine riboswitch class in bacteria. Biochemistry 56: 359–363. 10.1021/acs.biochem.6b01271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock ME, Malkowski SN, Breaker RR. 2017. Biochemical validation of a second guanidine riboswitch class in bacteria. Biochemistry 56: 352–358. 10.1021/acs.biochem.6b01270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock ME, Sudarsan N, Breaker RR. 2018a. Riboswitches for the alarmone ppGpp expand the collection of RNA-based signaling systems. Proc Natl Acad Sci 115: 6052–6057. 10.1073/pnas.1720406115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock ME, Sudarsan N, Stav S, Breaker RR. 2018b. Tandem riboswitches form a natural Boolean logic gate to control purine metabolism in bacteria. Elife 7: e33908 10.7554/eLife.33908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308–1325. 10.1017/S1355838299990891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahmann KP, Arst HN, Althöfer H, Revuelta JL, Monschau N, Schlüpen C, Gätgens C, Wiesenburg A, Schlösser T. 2001. Riboflavin, overproduced during sporulation of Ashbya gossypii, protects its hyaline spores against ultraviolet light. Environ Microbiol 3: 545–550. 10.1046/j.1462-2920.2001.00225.x [DOI] [PubMed] [Google Scholar]
- Stockbridge RB, Robertson JL, Kolmakova-Partensky L, Miller C. 2013. A family of fluoride-specific ion channels with dual-topology architecture. Elife 2: e01084 10.7554/eLife.01084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Aroca F, Giannattasio M, Brunelli E, Vezzoli A, Plevani P, Muzi-Falconi M, Bertoni G. 2006. One-step high-throughput assay for quantitative detection of β-galactosidase activity in intact Gram-negative bacteria, yeast, and mammalian cells. Biotechniques 40: 433–440. 10.2144/000112145 [DOI] [PubMed] [Google Scholar]
- Vogl C, Grill S, Schilling O, Stülke J, Mack M, Stolz J. 2007. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J Bacteriol 189: 7367–7375. 10.1128/JB.00590-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. 2008. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell 29: 691–702. 10.1016/j.molcel.2008.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Breaker RR. 2011. R2R—software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12: 3 10.1186/1471-2105-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35: 4809–4819. 10.1093/nar/gkm487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11: R31 10.1186/gb-2010-11-3-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR. 2017a. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res 18: 10811–10823. 10.1093/nar/gkx699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Nelson JW, Lünse CE, Sherlock ME, Breaker RR. 2017b. Bioinformatic analysis of riboswitch structures uncovers variant classes with altered ligand specificity. Proc Natl Acad Sci 114: E2077–E2085. 10.1073/pnas.1619581114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DW, Owen EC. 1973. Degradation of riboflavin by alimentary bacteria of the ruminant and man: production of 7,8-dimethyl-10-carboxymethylisoalloxazine. Br J Nutr 29: 33–41. 10.1079/BJN19730074 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR. 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci 99: 15908–15913. 10.1073/pnas.212628899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284: 611–615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.