Abstract
α-Ketoglutarate (αKG) is a key node in many important metabolic pathways. The αKG analogue N-oxalylglycine (NOG) and its cell-permeable pro-drug dimethyloxalylglycine (DMOG) are extensively used to inhibit αKG-dependent dioxygenases. However, whether NOG interference with other αKG-dependent processes contributes to its mode of action remains poorly understood. Here we show that, in aqueous solutions, DMOG is rapidly hydrolysed to yield methyloxalylglycine (MOG). MOG elicits cytotoxicity in a manner that depends on its transport by monocarboxylate transporter 2 (MCT2) and is associated with decreased glutamine-derived TCA-cycle flux, suppressed mitochondrial respiration and decreased ATP production. MCT2-facilitated entry of MOG into cells leads to sufficiently high concentrations of NOG to inhibit multiple enzymes in glutamine metabolism, including glutamate dehydrogenase (GDH). These findings reveal that MCT2 dictates the mode of action of NOG by determining its intracellular concentration, and have important implications for the use of (D)MOG in studying αKG-dependent signalling and metabolism.
Introduction
Many tumours have increased reliance on glutamine utilisation, a process known as glutaminolysis1, which is promoted by major oncogenic pathways2,3. Glutamine can contribute to biosynthetic processes directly or after conversion to glutamate by glutaminases (GLSs)1. Glutamate can then be deaminated to α-ketoglutarate (αKG, 1), which has pleiotropic roles in cell physiology4 from TCA cycle metabolism to epigenetic regulation. Glutamate deamination is catalysed by either glutamate dehydrogenase (GDH), producing αKG and ammonia, or by transaminases (TAs) that transfer the amino group of glutamate onto a recipient α-ketoacid, generating a non-essential amino acid. In addition to GLS, both GDH and TAs have been implicated in cancer3. αKG can also be metabolised via isocitrate dehydrogenase (IDH) in a process known as reductive carboxylation (RC). RC mediates synthesis of fatty acids from glutamine5 and is important for cancer cell survival upon matrix detachment by supporting the detoxification of reactive oxygen species (ROS)6.
αKG is also a co-substrate, together with oxygen and the co-factor iron, for αKG-dependent dioxygenases (αKGDDs), a family of approximately 60 enzymes with broad-ranging substrates and functions7. Prolyl hydroxylases (PHDs) are amongst the most studied dioxygenases, particularly because of their role in regulating hypoxia signalling. PHD-catalysed hydroxylation of hypoxia-inducible transcription factors (HIFs) tags these proteins for proteasome-mediated degradation8. Since PHD activity is oxygen-dependent, hypoxia inhibits the hydroxylation of HIF and leads to its stabilisation.
Supporting the functional importance of αKG, oncogenic mutations in various TCA cycle enzymes generate oncometabolites that inhibit αKGDD activity by competing with αKG for binding to the catalytic pocket8,9. Similarly, exogenous αKG can alone, or in combination with other metabolites, complement inhibition of glutaminolysis10–12 suggesting that, although glutamine may have diverse roles in cells, αKG is a major mediator of glutamine-supported cancer metabolism.
Genetic ablation studies in mice have suggested that inhibition of PHD function can be beneficial in various pathological settings13–15, prompting the development of small molecule inhibitors of PHD activity9,16, which commonly function by competing with αKG for the PHD catalytic pocket17. N-oxalylglycine (NOG, 2, Fig. 1a) is an αKG analogue where the central methylene group of αKG is replaced with an NH moiety. NOG inhibits collagen prolyl hydroxylases18 as well as several αKGDDs19–21 but displays minimal plasma membrane permeability, therefore for cellular and animal studies it is administered as the prodrug dimethyl-oxalylglycine (DMOG, 3, Fig. 1a), which is de-esterified within cells to form NOG22. Importantly, DMOG administration to mice phenocopies the effects of genetic PHD inactivation23,24, which instigated the development of αKGDDs inhibitors for various disease conditions9.
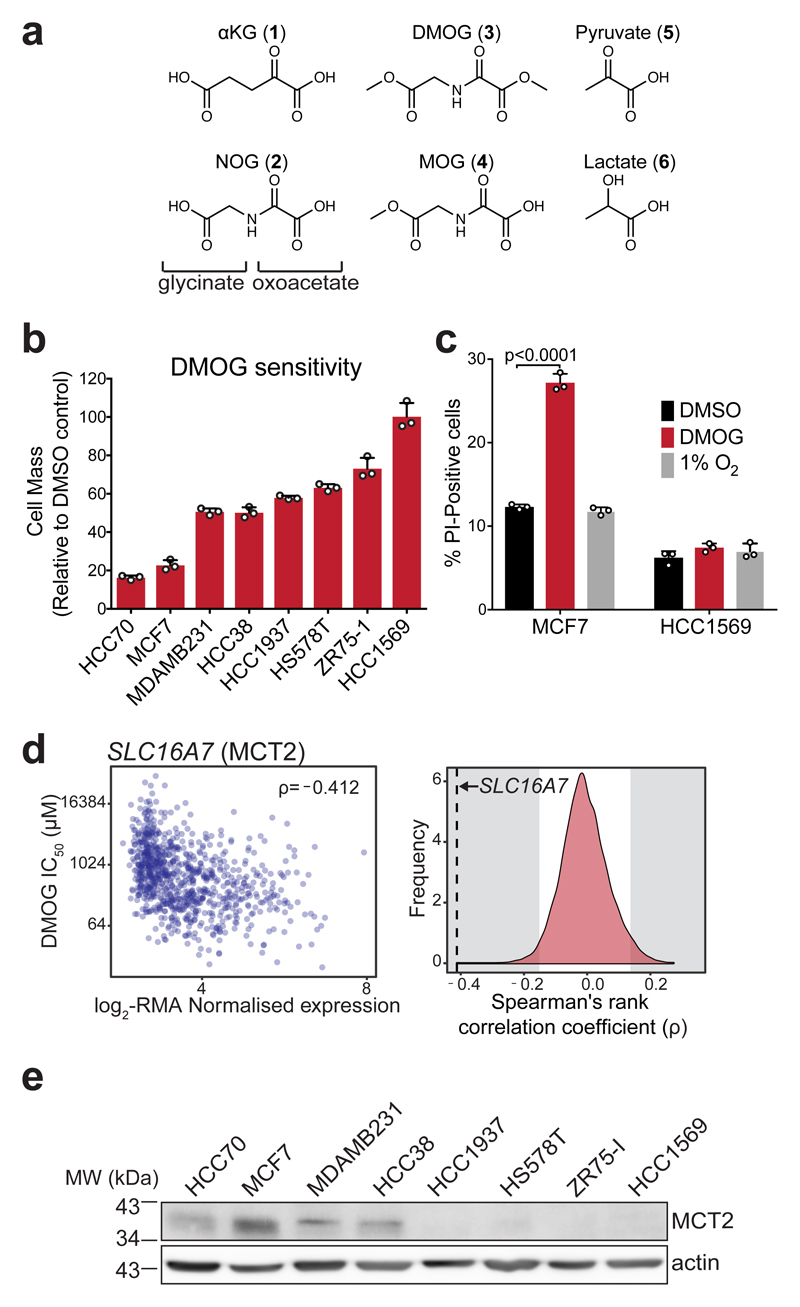
Figure 1. DMOG induces cytotoxicity that correlates with MCT2 expression and is not explained by differential inhibition of oxygen-sensitive dioxygenases.
a) Structures of αKG (1), NOG (2), DMOG (3) and MOG (4) (formed by de-esterification of DMOG in cells) shown alongside and the monocarboxylates pyruvate (5) and lactate (6) to illustrate structural similarities.
b) Cell mass accumulation of human breast cancer cell lines after 48 h treatment with 1 mM DMOG, relative to their respective vehicle (0.1% DMSO)-treated controls. Data shown as mean ± SD (n = 3 experimental replicates).
c) Measurement of propidium iodide (PI) uptake by flow cytometry to quantify cell death in MCF7 and HCC1569 cells treated for 48 h with vehicle (0.1% DMSO), 1 mM DMOG, or cultured at 1% O2 for 48 h (to inhibit dioxygenases). DMOG- and DMSO-treated cells were cultured at 21% O2. Data shown as mean ± SD (n = 3 experimental replicates), significance tested by 2-way ANOVA with Tukey’s multiple comparison correction.
d) Left: Correlation of robust multi-array average (RMA)-normalised SLC16A7 (encoding MCT2) mRNA expression and IC50DMOG across 850 different cancer cell lines. Data obtained from the Genomics of Drug Sensitivity in Cancer project (http://www.cancerrxgene.org). Spearman’s rank correlation coefficient is shown in the top right corner. Right: Spearman’s rank correlation coefficient of SLC16A7 (black dashed line) with respect to those of all other transcripts. Grey shaded region on either side indicates ±2-standard deviations cut-off used to define sensitivity-associated genes.
e) Western blot to assess MCT2 protein expression in lysates from breast cancer cell lines used in (b). Experiment performed once. Uncropped blot available in Supplementary Fig. 13a.
Beyond interfering with dioxygenase function, αKG analogues such as NOG will likely affect other αKG-dependent metabolic and signalling processes. Some of the inferred roles for PHDs, primarily through stabilisation of HIF1α, have been linked to metabolism and, in some cases, DMOG has been used as a means to study these processes14,25–27. However, little is known about the direct effects of NOG on αKG metabolism, therefore understanding its mode of action is important for interpreting its functional effects.
Here we show that DMOG is selectively toxic to cells that express monocarboxylate transporter 2, which we identify as a transporter of methyloxalylglycine (MOG, 4, Fig. 1a), a previously undescribed product of DMOG hydrolysis. MCT2 facilitates MOG entry into cells, leading to concentrations of NOG that are sufficiently high to inhibit multiple metabolic pathways, as exemplified by GDH, which binds NOG with low affinity, thereby attenuating glutamine metabolism through the TCA cycle.
Results
Selective DMOG toxicity independently of αKGDD inhibition
DMOG is widely used to study hypoxia signalling in cells because its hydrolysis product NOG inhibits PHDs leading to stabilisation of HIF1α19,20. We observed that treatment of different human breast cancer (BrCa) cell lines with DMOG inhibited cell mass accumulation to varying degrees (Fig. 1b) and, in sensitive cells, the morphological changes were consistent with cell death (Supplementary Fig. 1a). Using MCF7 and HCC1569 cells as model “sensitive” and “resistant” lines, respectively, we observed increased propidium iodide (PI) staining only in MCF7 cells (Fig. 1c and Supplementary Fig. 1b), suggesting that DMOG-induced inhibition of cell mass accumulation was due to cytotoxicity.
To test whether inhibition of dioxygenases accounted for differential sensitivity to DMOG, we cultured MCF7 and HCC1569 cells in 1% oxygen, to inhibit dioxygenases8. Consistent with dioxygenase inhibition, we observed an increase in HIF1α protein levels in both MCF7 and HCC1569 cells (Supplementary Fig. 1c). Hypoxia did not affect viability (Fig. 1c) but decreased cell mass accumulation similarly for both cell lines compared to normoxia (Supplementary Fig. 1d). Nevertheless, MCF7 cells had identical IC50DMOG in both conditions (Supplementary Fig. 1e) and the kinetics of HIF1α stabilisation by DMOG were similar between sensitive and resistant cells (Supplementary Fig. 1c). These data suggested that the selective cytotoxicity of DMOG cannot be explained by differential sensitivity to αKGDD inhibition.
DMOG toxicity correlates with MCT2 expression
To identify factors that contribute to selective DMOG cytotoxicity, we probed data from the Genomics of Drug Sensitivity in Cancer project (http://www.cancerrxgene.org)28, in which the DMOG IC50 (IC50DMOG) was determined for 850 cell lines with available gene expression data. Similarly to our BrCa cell lines, DMOG inhibited cell viability with a broad range of IC50s (0.010-58 mM) across all tested cancer types (Supplementary Fig. 2a). We defined (see Methods) a set of 341 gene transcripts that negatively correlated (high expression-low IC50DMOG) and 557 gene transcripts that positively correlated with IC50DMOG (Supplementary Dataset 1). SLC16A7, a transcript that encodes for monocarboxylate transporter 2 (MCT2)29, showed the highest correlation (ρ=-0.412) with sensitivity (Fig. 1d). MCT2 belongs to the solute carrier 16 (Slc16) family of transporters that comprises 14 members, including MCT1 (encoded by SLC16A1) and MCT4 (SLC16A3), which are the best studied MCTs and have both been implicated in cancer30. Neither SLC16A1 or SLC16A3 were present in the list of IC50DMOG-correlating transcripts (Supplementary Fig. 2b, c) but, interestingly, EMB, which encodes for Embigin, an accessory protein involved in trafficking of MCT2 to the plasma membrane29, was the 4th transcript most highly correlated with DMOG sensitivity (Supplementary Fig. 2d, Supplementary Dataset 1).
Consistent with IC50DMOG correlation at the transcript level, MCT2 protein expression was higher in DMOG-sensitive BrCa cell lines (Fig. 1e). We also investigated whether MCT2 expression correlated with DMOG sensitivity in cancer cell lines from other tissues of origin (Supplementary Fig. 2e). To this end, we selected high sensitivity cell lines within different tissue of origin groups and confirmed that they had low IC50DMOGs (comparable to MCF7). Interestingly, MCT2 expression was variable indicating that even low amounts of MCT2 are sufficient to confer sensitivity (Supplementary Fig. 2e, f).
These data suggested that expression of MCT2 is linked to DMOG-induced cytotoxicity.
The methyl oxoacetate ester of DMOG is rapidly hydrolysed
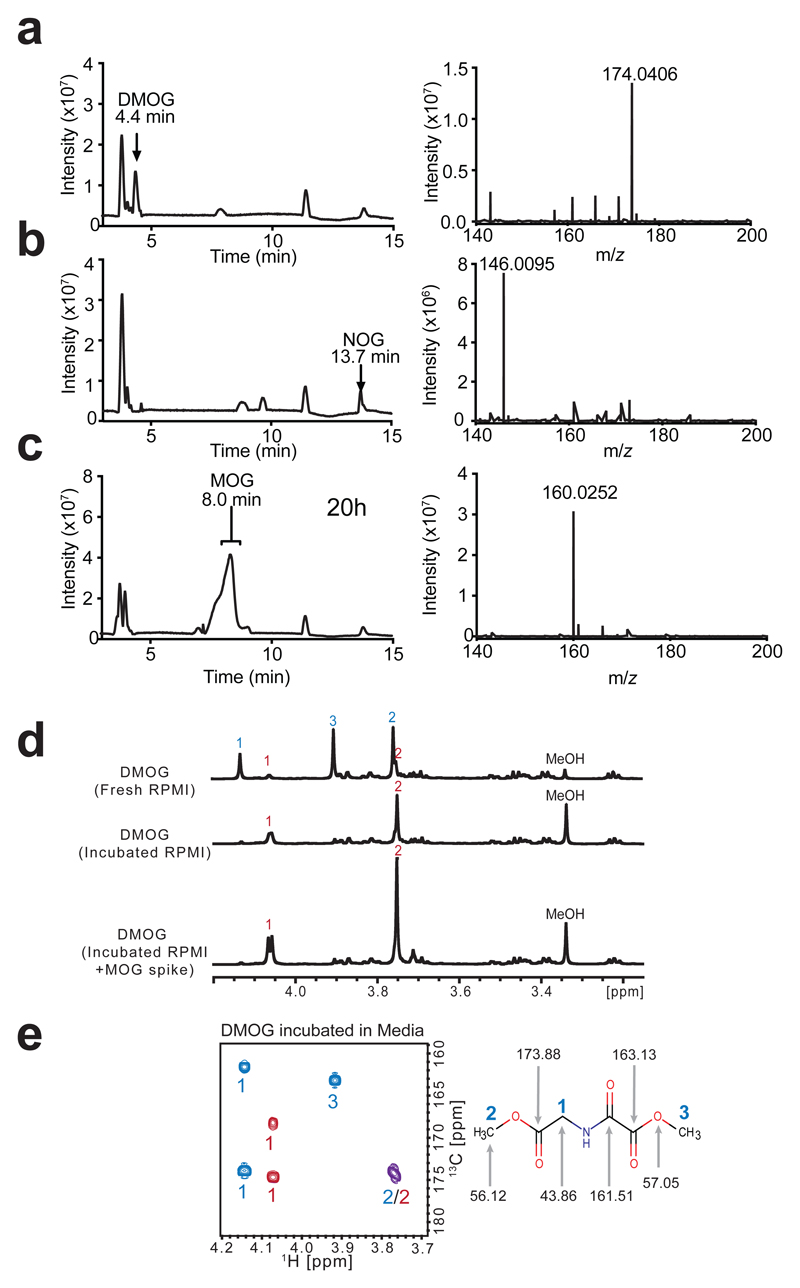
We hypothesised that, as a transporter, MCT2 could mediate DMOG-induced cytotoxicity by facilitating DMOG entry into cells. We developed a liquid chromatography-mass spectrometry (LC-MS) assay for the detection of DMOG (Fig. 2a) and its predicted end-product NOG (Fig. 2b). In control experiments, we observed loss of DMOG signal from culture media even in the absence of cells (Supplementary Fig. 3a). However, we did not detect NOG, suggesting that DMOG degraded to an unknown product. Inspection of the chromatograms of DMOG standard after 20 h incubation in water revealed an unknown peak, dominated by an ion with m/z 160.0252 (Fig. 2c). The mass difference to DMOG (m/z 174.0406) Δm/z = 14, indicated loss of a single methyl group, so we tentatively designated this ion species as methyl-oxalylglycine (MOG).
Figure 2. The methyl oxoacetate ester of DMOG is rapidly hydrolysed in cell culture media to yield MOG.
a) LC-MS base-peak chromatogram and corresponding mass spectrum of 10 µM DMOG in water, with peak and ion annotated.
b) LC-MS base-peak chromatogram and corresponding mass spectrum of 10 µM NOG in water, with peak and ion annotated.
c) LC-MS base-peak chromatogram demonstrating the MOG peak formed after incubation in water for 20 h at room temperature. Right: mass spectrum of MOG peak, with ion corresponding to MOG annotated.
d) 1D-1H-NMR spectra of DMOG freshly resuspended in RPMI medium, or after incubation in RPMI medium overnight, with and without the addition of a synthesised MOG standard. Signals annotated according to the labelled structure of DMOG in (e), DMOG peaks with blue numbers and MOG peaks with red numbers.
e) 2D-1H,13C-HMBC-NMR spectrum of DMOG incubated in RPMI media, DMOG peaks with blue numbers and MOG peaks with red numbers, overlapping cross-peak shown in purple. Right: DMOG structure annotated with the relevant 13C signal shifts.
Data are representative of more than 3 independent experiments each with similar results.
We next used nuclear magnetic resonance (NMR) spectroscopy to determine which of the two methyl groups of DMOG is hydrolysed. 1H NMR of DMOG incubated for 20 h in RPMI showed that the major DMOG peaks at 4.14, 3.77 and 3.92 ppm were lost, whereas methanol and two low abundance peaks (3.76 and 4.07 ppm) increased (Fig. 2d). These data indicated that the 3.76 and 4.07 peaks correspond to products that arise from ester hydrolysis of DMOG to release methanol. Further analyses assigned the 1H NMR resonances at 3.92 ppm to the protons of the methyl oxoacetate moiety (see Methods, Fig. 2e). In line with this interpretation, MOG authentic standard had identical retention time and m/z (Supplementary Fig. 3b) to the ion we observed by LC-MS in Fig. 2c, and a 1H NMR spectrum that precisely superimposed those signals corresponding to the DMOG degradation species (Fig. 2d). These results conclusively showed that the methyl oxoacetate ester within DMOG is hydrolysed to form the oxoacetic acid derivative MOG (Fig. 1a).
DMOG conversion to MOG in RPMI media was complete in < 1 h (t½ = 7.5 min, Supplementary Fig. 3c, d), and occurred also in water, albeit at a slower rate (t½ = 6.5 h, Supplementary Fig. 3e) suggesting that the selective hydrolysis of the methyl oxoacetate ester is non-enzymatic and likely due to the α-effect31. Additional analyses revealed that proton release during hydrolysis acidifies media and attenuates further hydrolysis, the rate of which is therefore influenced by the buffering capacity of the media (Supplementary Fig. 4).
Collectively, these data showed that, in buffered aqueous solutions, DMOG is rapidly converted to MOG.
MCT2-mediated MOG transport correlates with cytotoxicity
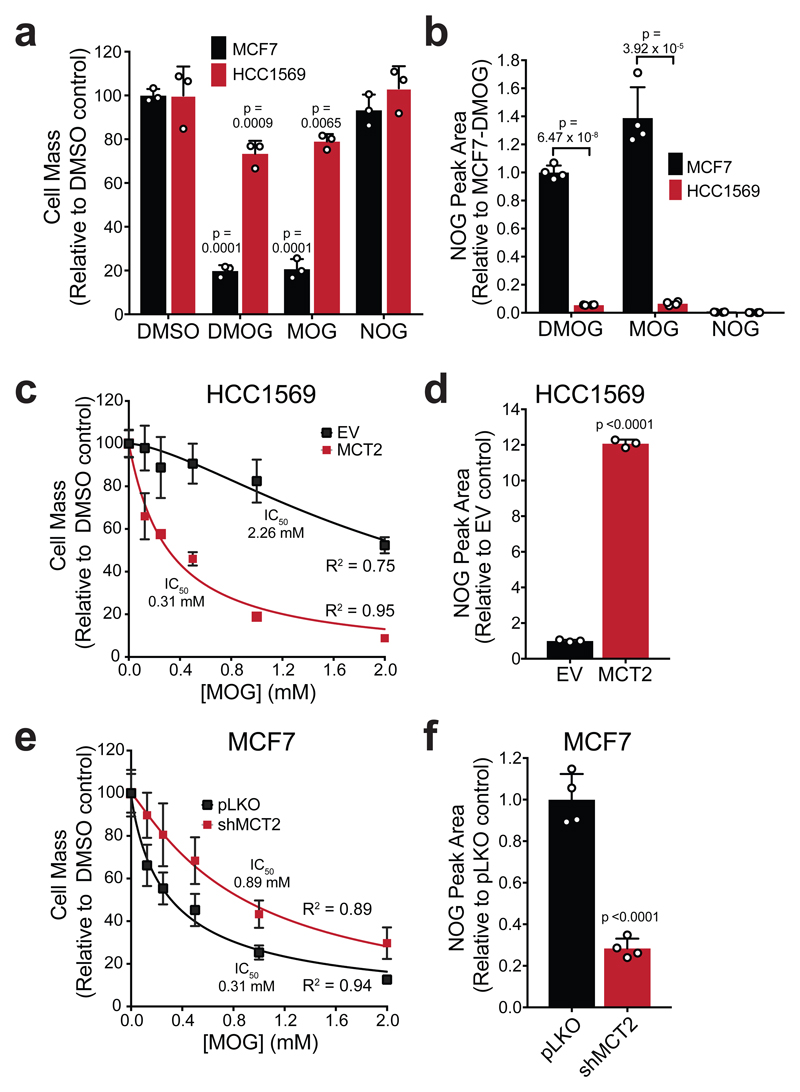
The rapid DMOG-to-MOG conversion raised the possibility that MOG suffices to elicit cytotoxicity. Consistent with this idea, DMOG and MOG inhibited cell mass accumulation equally in MCF7 cells, whereas NOG had no effect (Fig. 3a). Furthermore, intracellular NOG concentration ([NOG]ic) was similarly high in MCF7 cells treated with either DMOG or MOG (Fig. 3b). Intracellular MOG was not detected, suggesting rapid hydrolysis of the methyl-glycinate moiety in cells. Under these experimental conditions, HCC1569 cells had >10-fold lower [NOG]ic when incubated with either DMOG or MOG (Fig. 3b). Lower [NOG]ic correlated with lower cytotoxicity (Fig. 3a). We observed minimal [NOG]ic when either cell line was treated with NOG itself showing that NOG is not transported into these cells. These results suggested that MOG-induced toxicity is due to higher [NOG]ic in sensitive, relative to resistant, cells. Furthermore, since the conversion of DMOG to MOG in media occurs in <1 h and both compounds induce comparable cytotoxicity, these data indicated that MOG is the active cytotoxic compound in media. Henceforth, we used MOG to further explore the mechanism of cytotoxicity.
Figure 3. MOG is sufficient to cause cytotoxicity in an MCT2-dependent manner.
a) Cell mass accumulation of MCF7 and HCC1569 cells after treatment with 1 mM DMOG, MOG or NOG for 48 h relative to the respective 0.1% DMSO controls. Data shown as mean ± SD (n = 3 experimental replicates), significance was tested by 1-way ANOVA and corrected for multiple comparisons to the DMSO control using Dunnet’s post-hoc test.
b) Intracellular NOG concentrations ([NOG]ic) in MCF7 and HCC1569 cells after 4 h incubation with 1 mM of either DMOG, MOG or NOG. Reported concentrations are normalized to cell number. Data shown as mean ± SD (n = 4 experimental replicates), and significance was tested using 2-sided multiple t-tests with Holm-Sidak multiple comparison correction.
c) IC50 curve of cell mass accumulation for HCC1569 cells expressing either empty vector (EV) or MCT2, after incubation with increasing concentrations of MOG for 48 h, relative to vehicle-only control (0.2% DMSO). Data shown as the mean ± SD of n = 3 experimental replicates and are representative of three independent experiments. Curve was fitted using the [inhibitor] vs normalised response (variable slope) algorithm in GraphPad Prism.
d) Relative [NOG]ic in HCC1569 cells described in (c), after 4 h of incubation with 1 mM MOG. Data shown as mean ± SD (n = 3 experimental replicates) and significance was tested using a 2-sided, unpaired t-test.
e) IC50 curve of cell mass accumulation for MCF7 cells expressing either empty vector (pLKO) or shMCT2, after incubation with increasing concentrations of MOG for 48 h, relative to vehicle-only control (0.2% DMSO). Data shown as the mean ± SD of n = 3 experimental replicates and are representative of three independent experiments. Curve was fitted as in (c).
f) Relative [NOG]ic in MCF7 cells described in (e), after 4 hours of incubation with 1 mM MOG. Data shown as mean ± SD (n = 4 experimental replicates), and significance was tested using a 2-sided unpaired t-test.
Our gene expression correlation analysis implicated MCT2 in cell sensitivity to DMOG, so, we next tested whether exogenous MCT2 expression can sensitise cells to MOG and whether this is related to [NOG]ic. The IC50MOG of MOG-resistant HCC1569 cells (HCC1569-EV) was 2.26 ± 0.35 mM (Fig. 3c, Supplementary Fig. 5a). Stable expression of MCT2 in these cells (HCC1569-MCT2) decreased the IC50MOG to 0.31 ± 0.03 mM with a concomitant 12-fold increase in [NOG]ic after 4 h incubation with MOG (Fig. 3d). Similarly to Fig. 3b, we detected minimal NOG in cells incubated with NOG in the media, in either HCC1569-EV or HCC1569-MCT2 cells (Supplementary Fig. 5b), confirming that NOG cannot enter cells either via the plasma membrane or MCT2. These results suggested that expression of MCT2 leads to increased [NOG]ic and therefore increased sensitivity to MOG. To test whether MCT2 is necessary for MOG uptake, we stably knocked down MCT2 in MCF7 cells (MCF7-shMCT2) (Supplementary Fig. 5a). IC50MOG of MCF7-shMCT2 was 65% lower than control MCF7 cells (MCF7-pLKO) (Fig. 3e) while [NOG]ic decreased by 72% (Fig. 3f). Collectively, these data showed that MCT2 expression determines [NOG]ic and thereby modulates MOG-induced cytotoxicity.
Given the varying MCT2 levels among sensitive lines (Supplementary Fig. 2e, f), we measured [NOG]ic and toxicity in cells engineered to express doxycycline-inducible MCT2 (Supplementary Fig. 5c). Supplementary Fig. 5d shows that 12% of maximal MCT2 expression suffices to attain half-maximal [NOG]ic, while half-maximal toxicity occurred at just 3% of MCT2 maximal expression (Supplementary Fig. 5e) and correlated well with [NOG]ic (Supplementary Fig. 5f). These data indicated that even low levels of MCT2 can lead to sufficiently high [NOG]ic to elicit toxicity.
Since MOG is a monocarboxylate, we asked whether other MCTs could also mediate its transport into cells, focusing on MCT1 and MCT4, which share similar endogenous substrates with MCT2. We expressed MCT1, MCT2 or MCT4 in INS1 cells, a rat pancreatic β-cell line with very low endogenous MCT activity32 (Supplementary Fig. 6a). MCT2 expression led to the highest [NOG]ic after MOG treatment, followed by MCT1 and MCT4 (Supplementary Fig. 6b), which was reflected by the corresponding IC50MOG values (Supplementary Fig. 6c). These results indicated that MCT1 and MCT4 can transport MOG, albeit less efficiently than MCT2. MCT2 has the highest affinity for all tested substrates followed by MCT1 and MCT4, with a 10-fold selectivity for pyruvate (5) over lactate (6) 33. MOG treatment of MCF7 cells in the presence of super-stoichiometric pyruvate levels resulted in an 83 ± 3.5% decrease in [NOG]ic compared to MOG alone (Supplementary Fig. 6d), whereas super-stoichiometric lactate only minimally decreased the [NOG]ic. Accordingly, pyruvate but not lactate, prevents MOG-induced toxicity (Supplementary Fig. 6e). Together, these data showed that although other members of the MCT family can transport MOG, MCT2 is the primary MOG transporter, reflecting its higher affinity for endogenous substrates.
High [NOG]ic inhibits glutamine catabolism and respiration
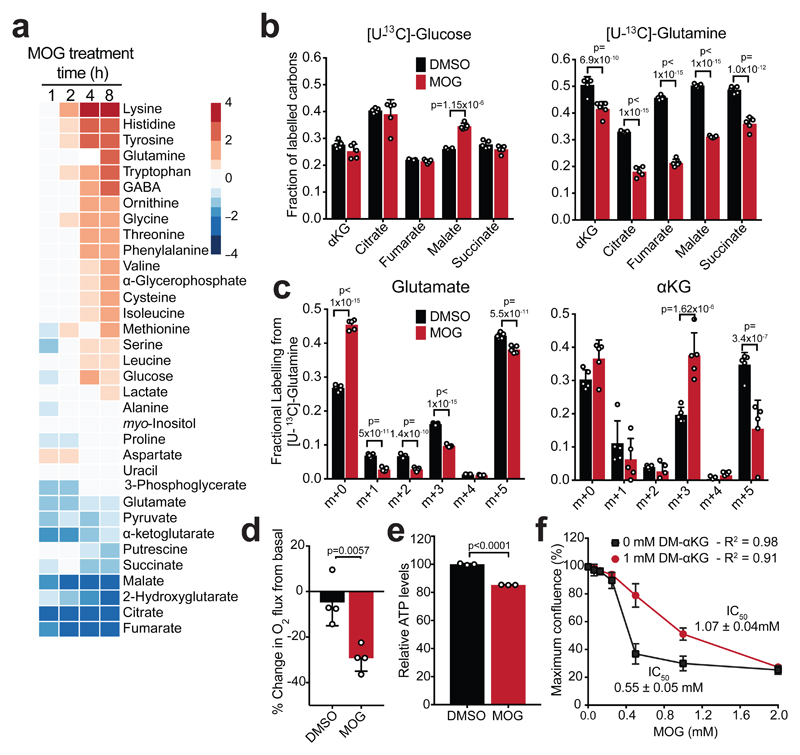
MOG cytotoxicity is associated with increased [NOG]ic but not with differential engagement of its known substrates, dioxygenases, in sensitive cells (Fig. 1). As an αKG analogue, high [NOG]ic could influence metabolism. To test this possibility, we treated MCF7 cells with MOG and analysed metabolite concentrations over time. Within an hour of treatment, several TCA intermediates (citrate, αKG, fumarate, malate) were depleted, followed by an increase in amino acid levels at 2-4 hours (Fig. 4a, Supplementary Fig. 7a). Increased amino acids were not due to attenuated translation (Supplementary Fig. 7b) as further supported by the observation that the non-proteinogenic amino acids ornithine and GABA also increased. We observed similar metabolic changes in HCC1569-MCT2 but not HCC1569-EV cells (Supplementary Fig. 7c), while, conversely, these metabolic effects were less pronounced in MCF7-shMCT2 cells (Supplementary Fig. 7d), indicating that they occur in an MCT2-dependent manner. MOG induced similar metabolic changes across other sensitive cell lines (Supplementary Fig. 7e). Importantly, the metabolic effects of MOG were not due to HIF1α stabilisation, because they were preserved in MCF7 cells defective for HIF1α function (Supplementary Fig. 8a, b), which also retained sensitivity to MOG (Supplementary Fig. 8c).
Figure 4. MOG inhibits glutamine catabolism in an MCT2-dependent manner.
a) Heatmap showing log2 fold-changes in the abundance of indicated metabolites in MCF7 cells treated with 1 mM MOG, relative to the 0 h control treatment (n = 4 experimental replicates for each condition and time-point). Metabolites are ordered from highest to lowest fold-change value using the 8 h time point.
b) Fraction of labelled carbons in TCA cycle metabolite pools in MCF7 cells after 4 h of labelling with [U-13C]-glucose or [U-13C]-glutamine in the presence or absence of 1 mM MOG. Data are shown as mean ± SD (n = 5 experimental replicates for each label). Significance was tested using 2-sided multiple t-tests with Holm-Sidak multiple comparisons correction.
c) Isotopologue distribution of glutamate and αKG in MCF7 cells after 4 h of labelling with [U-13C]-glutamine in the presence or absence of 1 mM MOG. Data shown as mean ± SD (n = 5 experimental replicates). Significance was tested using 2-sided multiple t-tests with Holm-Sidak multiple comparisons correction.
d) Change in respiration of MCF7 cells from basal after incubation with 0.1% DMSO or 1 mM MOG in RPMI medium. Data shown as mean ± SD (n = 3 experimental replicates). Significance was tested using a 2-sided, unpaired t-test.
e) ATP levels in MCF7 cells treated with 0.1% DMSO or 1 mM MOG in RPMI medium for 4 h. Data are shown as mean ± SD of n = 3 experimental replicates and are representative of 3 independent experiments. Significance was tested using a 2-sided, unpaired t-test.
f) IC50 curves of cell mass accumulation in MCF7 cells after incubation with increasing concentrations of MOG for 48 h in the absence or presence of 1 mM DM-αKG. Data are shown relative to vehicle-only control (0.2% DMSO) and represent mean ± SD (n = 3 experimental replicates). IC50 calculated using the [inhibitor] vs normalised response (variable slope) algorithm in GraphPad Prism.
To investigate the cause of TCA cycle intermediate depletion, we labelled MCF7 cells with [U-13C]-glucose or [U-13C]-glutamine in the presence or absence of MOG. MOG treatment decreased labelling from [U-13C]-glutamine, but not from [U-13C]-glucose, into TCA intermediates (Fig. 4b). This was associated with a 56 ± 24% decrease in fully-labelled (m+5) αKG that was produced from [U-13C]-glutamine although the proportion of fully-labelled (m+5) glutamate remained largely unchanged (Fig. 4c). Together, these data showed that treatment of cells with MOG inhibits entry of glutamine-derived carbons into the TCA cycle at the point of glutamate-to-αKG conversion.
A major function of glutamine-fuelled TCA metabolism is ATP production through respiration34. MOG-treated MCF7 cells showed a 29 ± 5.7% decrease in oxygen consumption compared to basal respiration, similar to H1299 cells expressing inducible MCT2 (Fig. 4d, Supplementary Fig. 9a), and consistent with a previous report35. Decreased respiration was associated with a significant decrease in ATP levels within 4 h in MOG-treated MCF7 cells (Fig. 4e), and in a number of cell lines (Supplementary Fig. 9b). Similarly, ATP levels decreased in MOG-treated HCC1569-MCT2, but not in the control HCC1569-EV cells, while MOG failed to decrease ATP levels in MCF7-shMCT2 cells (Supplementary Fig. 9c). These observations showed that decreased TCA cycle flux in MOG-treated cells is accompanied by impaired respiration and decreased ATP levels.
αKG replenishes TCA and partly rescues MOG-cytotoxicity
To explore whether MOG-induced inhibition of TCA metabolism is related to cytotoxicity, we treated MCF7 cells with MOG in the presence of dimethyl-glutamate (DM-Glu) or dimethyl-αKG (DM-αKG), cell permeable analogues of glutamate and αKG, respectively. DM-Glu restored 50% of the decrease in glutamate levels but failed to prevent MOG-induced cytotoxicity or the decrease in TCA intermediate abundances (Supplementary Fig. 10a-c). DM-αKG increased αKG levels in a concentration-dependent manner and, although the magnitude of the MOG-induced decrease was preserved regardless of the total αKG pool size (Supplementary Fig. 10d), DM-αKG restored the levels of most TCA intermediates and suppressed the MOG-induced increase in amino acid levels (Supplementary Fig. 10e). Furthermore, addition of αKG fully prevented the inhibition of glutamine-driven respiration by MOG in MCF7 cells (Supplementary Fig. 10f). Supplementation of media with DM-αKG doubled the IC50MOG of MCF7 cells (Fig. 4f), suggesting that replenishment of TCA intermediates with an exogenous source of αKG can alleviate MOG-induced toxicity. Together, these results showed that, in contrast to exogenous glutamate, exogenous αKG can restore TCA intermediates and respiration and partly rescues MOG-induced death.
NOG inhibits multiple targets in glutamine metabolism
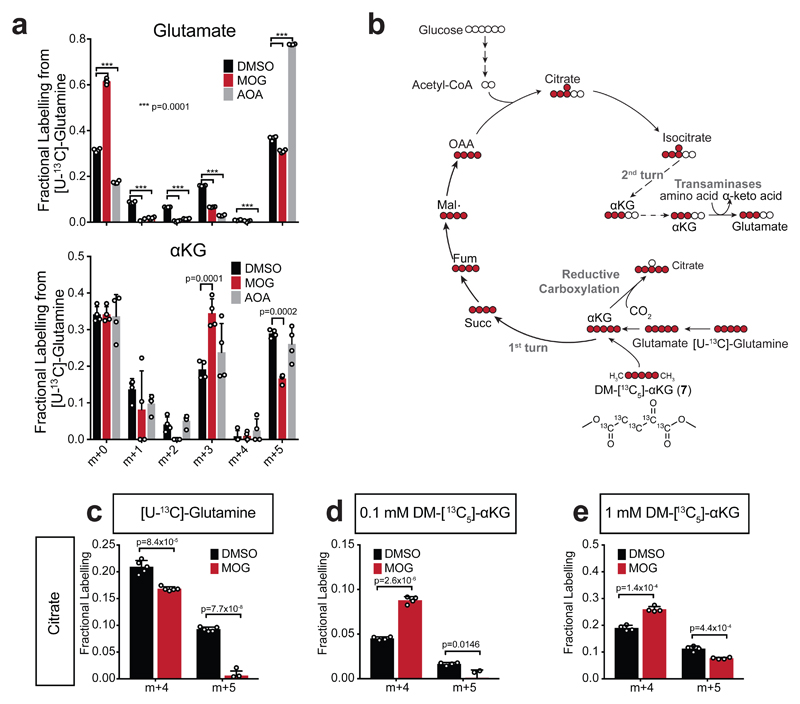
To gain insight into the NOG target(s) that mediate the observed metabolic changes, we examined the [U-13C]-glutamine labelling experiments further. Glutamate can be converted to αKG by either TAs or GDH, so to test involvement of the former, we compared the labelling pattern of metabolites from [U-13C]-glutamine in cells treated with MOG or aminooxyacetate (AOA), a pan-transaminase inhibitor. AOA caused an increase in the glutamate m+5 isotopologue but, unlike MOG, did not change αKG m+5 (Fig. 5a) nor labelling of TCA intermediates (Supplementary Fig. 11a) suggesting that, similarly to other cell lines11, TAs do not significantly contribute substrates to the TCA cycle in MCF7 cells. Therefore, inhibition of TAs by MOG cannot account for the decreased labelling of TCA metabolites from [U-13C]-glutamine, implicating GDH as the relevant NOG target.
Figure 5. Evidence that inhibition of GDH-mediated glutamine carbon flux accounts for MOG-induced metabolic changes associated with cytotoxicity.
a) Isotopologue distribution of glutamate and αKG in MCF7 cells after 4 h of labelling with [U-13C]-glutamine in the presence or absence of 1 mM MOG or 1 mM aminooxyacetate (AOA) compared to 0.1% DMSO control. Data shown as mean ± SD (n = 4 experimental replicates) and significance was tested with 2-way ANOVA with Dunnett’s multiple comparisons correction.
b) Scheme illustrating theoretical labelling pattern in the indicated metabolites, generated by incubation of cells with either [U-13C]-glutamine or DM-[13C5]-αKG (7). 13C-carbons are shown in red circles and 12C shown in white.
c) Quantification citrate m+4 isotopologue (generated by TCA in the oxidative direction), or citrate m+5 isotopologues [generated by reductive carboxylation (RC) of αKG] in MCF7 cells incubated with [U-13C]-glutamine for 4 h in the presence of 0.1% DMSO or 1 mM MOG. Data shown as mean ± SD (n = 5 experimental replicates). Significance was tested with 2-sided multiple t-tests using Holm-Sidak’s correction for multiple comparisons.
d) As in (c) but labelling was with tracer amounts (0.1 mM) of DM-[13C5]-αKG (n = 4 experimental replicates).
e) As in (c) but labelling was with rescue amounts (1 mM) of DM-[13C5]-αKG (n = 4 experimental replicates).
Intriguingly, AOA also caused a decrease in glutamate m+3 indicating that, in MCF7 cells, transaminases use αKG m+3 produced from the first turn of the TCA cycle as a nitrogen acceptor in order to degrade amino acids (Fig. 5b). MOG caused a decrease in glutamate m+3, similarly to AOA, but also an increase in αKG m+3, suggesting that in MOG-treated cells TAs are inhibited in the glutamate-producing direction. This interpretation is supported by the fact that MOG-treated cells contain a lower αKG/glutamate ratio (Supplementary Fig. 11b), which would shift the equilibrium of TA reactions against glutamate production. Consistent with this model, MOG treatment leads to increased amino acid concentrations (Fig. 4a). Together, these findings suggest that inhibition of GDH by NOG contributes to attenuated TCA cycle and a lower αKG/glutamate ratio that indirectly inhibits amino acid degradation by transaminases.
In MOG-treated MCF7 cells labelled with [U-13C]-glutamine, we also observed decreased labelling in citrate m+5, fumarate m+3 and aspartate m+3 (Fig. 5c, Supplementary Fig. 11c), which are produced via RC of αKG6. To assess whether decreased reductive TCA flux was also due to GDH inhibition, we labelled cells with DM-[13C5]-αKG (7) used at 0.1 mM, a concentration that minimally perturbs metabolism (Supplementary Fig. 11d). MOG inhibited labelling from DM-[13C5]-αKG in citrate m+5, but not citrate m+4 (Fig. 5d, Supplementary Fig. 11c) the levels of which actually increased, likely as a result of decreased unlabelled carbon contribution from glutamine into this isotopologue during MOG treatment. These data indicated that inhibition of RC by MOG occurs downstream of αKG, consistent with the target being IDH, which has been previously shown to be inhibited by NOG in vitro36. Importantly, 13C incorporation into RC-produced isotopologues of TCA intermediates decreased even when cells were labelled with 1 mM DM-[13C5]-αKG, demonstrating that IDH inhibition is not fully rescued by DM-αKG (Fig. 5e, Supplementary Fig. 11c), which may explain the only partial rescue of toxicity by DM-αKG (Fig. 4f).
In summary, these results provided evidence that MOG treatment independently inhibits both oxidative and reductive TCA cycle, as well as amino acid degradation, indicating that NOG simultaneously engages multiple targets in glutamine metabolism.
NOG binds to and inhibits GDH with low affinity
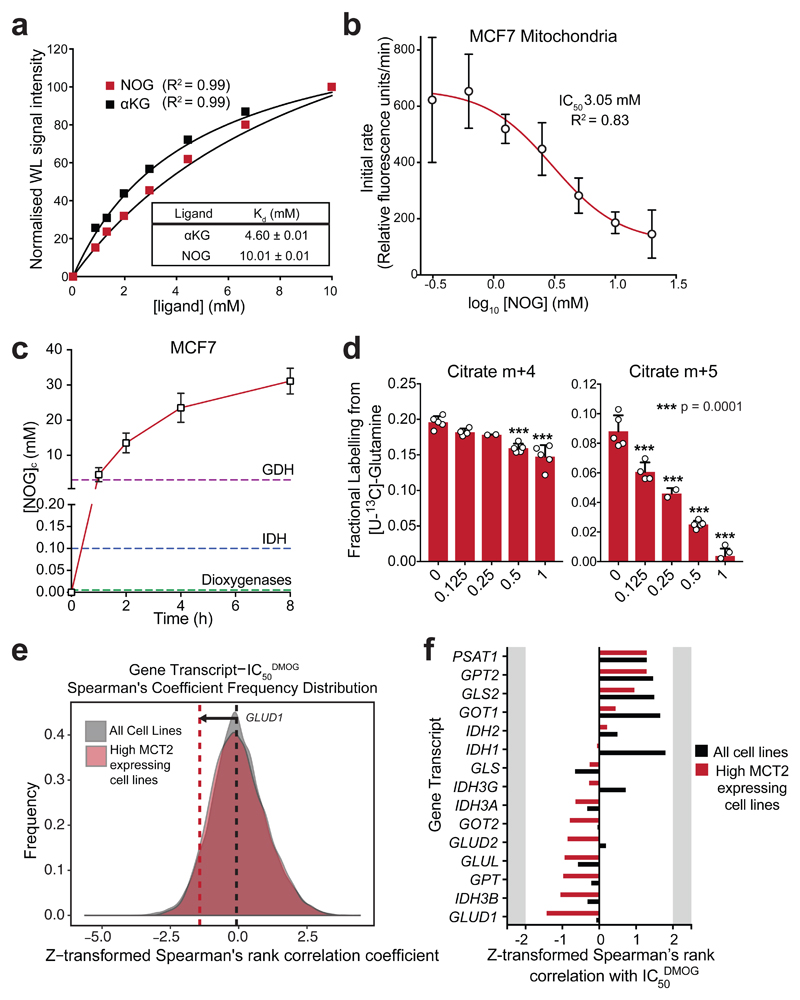
DM-αKG attenuates MOG-induced toxicity associated with inhibition of oxidative TCA cycle flux, which is mediated through GDH. We therefore focused on this enzyme to explore the basis of high [NOG]ic-dependent metabolic changes. WaterLOGSY NMR showed that NOG binds to bovine GDH directly with KdNOG = 10 mM, which is comparable to that of αKG (4.6 mM) (Fig. 6a, Supplementary Fig. 12a). Accordingly, we observed a dose-dependent decrease in GDH activity in MCF7 mitochondrial lysates (Fig. 6b) and purified GDH (Supplementary Fig. 12b) with IC50’s of 3.05 ± 1.39 mM and 6.09 ± 2.20 mM, respectively, confirming that NOG inhibits GDH activity.
Figure 6. NOG binds to GDH and inhibits its enzymatic activity.
a) Normalised waterLOGSY signal intensities in the presence of increasing concentrations of either αKG or NOG. Kd values were determined by fitting a one-site specific binding curve in GraphPad Prism. Single replicates were taken at each ligand concentration; experiments were performed 3 times with similar results.
b) GDH activity in MCF7 cell mitochondrial lysates pre-incubated for 15 min in the presence of increasing concentrations of NOG. Data shown as mean ± SD (n = 3 experimental replicates). IC50 determined by fitting a log[inhibitor] vs response (variable slope, 4 parameters) curve in GraphPad Prism.
c) [NOG]ic in MCF7 cells incubated with 1 mM MOG for increasing durations. Data shown as mean ± SD (n = 4 experimental replicates). Dashed lines indicate the measured IC50NOG values of GDH from (a), and those reported for IDH36 and dioxygenases17.
d) Quantification of the citrate m+4 isotopologue (generated by TCA in the oxidative direction), and citrate m+5 isotopologue (generated by RC) in MCF7 cells incubated with [U-13C]-glutamine for 4 h in the presence of different concentrations of MOG. Data shown as mean ± SD (5 experimental replicates). Statistical significance tested by one-way ANOVA, and multiple comparisons to the 1 mM MOG control were corrected using Dunnet’s method.
e) Frequency distribution graphs of Spearman’s rank correlation coefficient values from all genes vs. IC50DMOG, using all cells lines (850 different cell lines, grey) or only the top quartile of SLC16A7-expressing cell lines (213 cell lines, red) as in (f). The dashed lines represent the correlation coefficient for GDH (GLUD1) and DMOG IC50 in the analysis with all cell lines (850, black), and with only the top quartile of SLC16A7-expressing cell lines (red).
f) Correlation coefficients from Spearman’s rank analysis of transcripts encoding proteins involved in glutamine metabolism and IC50DMOG. Black bars represent correlation coefficients when all cell lines are included in the analysis (850 different cell lines, as for Fig. 1d), while red bars represent coefficients when only the cell lines that express the highest levels of SLC16A7 (213 cell lines) are included.
Considering the high KdNOG for GDH, we asked whether NOG can accumulate at sufficient concentrations to inhibit GDH in MOG-treated intact MCF7 cells. We found a time-dependent increase in the absolute (Supplementary Fig. 12c and Methods) [NOG]ic that, after 8h, reached 29 mM, thereby exceeding the levels required to substantially inhibit GDH (Fig. 6c).
Intriguingly, KdNOG of GDH is 10-fold higher than that reported for IDH36 suggesting that, at a given MOG concentration, glutamine metabolism would be more susceptible to inhibition in the reductive than in the oxidative direction. Consistent with this, 125 µM MOG caused a significant decrease in the production of citrate m+5 from [U-13C]-glutamine, but did not significantly affect the citrate m+4 species at concentrations below 0.5 mM (Fig. 6d). These data suggested that the metabolic effects of MOG depend on the [NOG]ic in a manner that reflects the relative potency of NOG towards its intracellular targets.
Finally, the finding that GDH is a target of MOG raised the possibility that expression levels of low affinity targets could determine sensitivity to MOG. However, neither GDH nor IDH transcripts correlated with IC50DMOG in our initial analysis (Supplementary Dataset 1). Given that MCT2 is required for NOG to reach sufficiently high concentrations to inhibit low affinity targets, we asked whether expression of genes involved in glutamine metabolism correlated with IC50DMOG in a subset of cell lines that expressed high levels of MCT2 mRNA (Supplementary Dataset 2). Within this subset, expression of several transcripts associated with glutamine metabolism exhibited increased correlation with IC50DMOG compared to the entire cell line panel (Fig. 6e, f). However, none of these targets alone can predict sensitivity, consistent with the idea that interference with multiple proteins by NOG contributes to toxicity. Notably, GDH mRNA expression showed the highest correlation with IC50DMOG among all genes involved in glutamine metabolism (Fig. 6e, f).
Together, these data show that NOG directly binds to and inhibits GDH with a high Kd. Although it is likely that other low affinity targets will be inhibited at this [NOG]ic, this finding rationalises the selective cytotoxicity observed only in cells that accumulate high [NOG]ic.
Discussion
In this study, we showed that the methyl oxoacetate ester of DMOG is rapidly hydrolysed in cell culture media to produce MOG. MCT2 facilitates the transport of MOG into cells and thereby determines [NOG]ic. At high concentrations, NOG inhibits glutamine metabolism, leading to ATP depletion and cytotoxicity. These new insights into the mode of action of DMOG provide important considerations for its use in mechanistic studies of αKG-dependent metabolism and signalling.
Despite the notion that relative lipophilicity determines drug entry into cells, increasing evidence suggests that most drugs enter cells via transporters37. Accordingly, transporter expression or polymorphisms can influence drug pharmacokinetics and pharmacodynamics, e.g. as demonstrated for metformin and the transporter OCT1 (SLC22A1)38. In addition to MCT2 expression, which is increased in some human cancers30,39,40, additional factors may influence MOG uptake, also in normal tissues, including the relative expression of other MCTs. MCT1 expression in INS-1 cells also led to increased MOG entry, albeit to a lower level than MCT2, indicating that unless expressed at high levels, MCT1 is unlikely to significantly contribute to MOG uptake. Moreover, as MCT function requires co-transport of protons, MOG pharmacodynamics may be influenced by an acidic tumour microenvironment29. Availability of endogenous MCT2 substrates may also be dictated by the tissue microenvironment41, and we showed that pyruvate can outcompete MOG for cell entry.
The observation that high [NOG]ic persists in cells suggests minimal turnover, making MOG a suitable candidate to image42 MCT2-positive tissues. Beyond its potential applications in cancer, the identification of MOG as an MCT2 substrate will help provide mechanistic insights into the in vivo function of MCT2, a poorly understood transporter, as well as MOG pharmacodynamics.
Furthermore, our results indicate that high [NOG]ic is cytotoxic due to simultaneous targeting of multiple enzymes in glutamine metabolism. DMOG has been previously reported to decrease mitochondrial respiration and ATP levels in HCT116 cells35 and to cause various metabolic effects, including depletion of TCA cycle intermediates, in primary peripheral mononuclear cells (PMCs)43. However, the metabolic targets of NOG remained, until now, unknown. Notably, we showed that HCT116 cells express sufficient MCT2 to confer sensitivity to MOG, and PMCs43 have also been reported to express MCT244.
We found that depletion of TCA cycle intermediates following MOG treatment was attributable to attenuated glutamine carbon entry into the TCA cycle, which can be mediated by two major pathways, TA’s and GDH. The pan-transaminase inhibitor AOA did not recapitulate the effects of MOG on TCA labelling from glutamine, pointing to GDH as the relevant NOG target. MOG also led to an increase in amino acid levels, which occurred after TCA cycle inhibition and reflected a decreased αKG/Glu ratio, indicating attenuated amino acid degradation through TAs. Finally, we also observed inhibition of RC, likely through IDH, which has been previously shown to be inhibited by NOG in vitro36. DM-αKG, which provides the product of GDH, restored TCA intermediates and respiration in MOG-treated cells. This was associated with a partial rescue of MOG-induced cytotoxicity further supporting the involvement of GDH-mediated metabolic effects in cytotoxicity. We therefore used GDH to understand why toxicity only occurs in cells with high [NOG]ic. Comparison of the absolute [NOG]ic to the in vitro KdNOG of GDH supports a model where GDH inhibition occurs only when [NOG]ic exceeds its KdNOG thereby explaining why expression of MCT2, which drives high [NOG]ic, correlates with toxicity.
Beyond GDH, our findings suggest that the ability of NOG to interfere with multiple αKG-mediated processes (“polypharmacology”45) underlies its effectiveness in disrupting cellular metabolism, thereby causing toxicity. Polypharmacology is emerging as a desirable trait in new drug development37,46, as best exemplified by kinase inhibitors47, so further systematic studies are warranted to define the spectrum of intracellular NOG targets.
Our findings could also be relevant for the targeting of dioxygenases, where competitive inhibition of the αKG binding pocket is a common pharmacological strategy9,17. Where known, the IC50 of NOG for dioxygenases is in the low μM range17. We showed that even when MCT function is low, [NOG]ic can reach mM concentrations, suggesting that many dioxygenases are likely inhibited, as also corroborated by indistinguishable kinetics of HIF1α stabilisation between MOG-sensitive and MOG-resistant cells. However, as more dioxygenases become better characterised, it would be of interest to compare their relative sensitivity to NOG. The availability of isoform-specific PHD inhibitors will help elucidate to which extent metabolic effects contributed to the action of DMOG in previous studies that explored the therapeutic potential of PHD inhibition. Finally, PHDs modulate metabolism in both a HIF-dependent and -independent48 manner. In particular, HIF-dependent gene expression leads to suppression of mitochondrial respiration14,25–27,35,49, similarly to the direct effects of MOG. Use of DMOG to probe metabolic functions of dioxygenases should therefore be evaluated in the light of its instability, MCT2 expression and the actual concentration that NOG reaches in cells.
Online Methods
Correlation of gene expression with DMOG IC50
Correlation of gene expression (Robust multi-array averaging (RMA)-normalised basal expression profiles for all cell lines) with DMOG sensitivity (IC50 values for all cell lines) was performed using publically available data from the Genomics of Drug Sensitivity in Cancer dataset (http://www.cancerrxgene.org), using RStudio version 1.0.136 (RStudio Team (2016). RStudio: Integrated Development for R. RStudio, Inc., Boston, MA URL http://www.rstudio.com/). Spearman’s rank correlation coefficient (ρ) was calculated for DMOG sensitivity against all genes. Since the data were normally distributed, we assumed a false-discovery rate of 5% and therefore used a cut-off of ± 2 standard deviations from the mean to define transcripts that were positively or negatively associated with DMOG IC50 (Supplementary Dataset 1).
For Fig. 6e and 6f, to investigate how intracellular target gene transcripts correlated with sensitivity to DMOG, Spearman’s rank correlation analysis was performed using only the top quartile of MCT2-expressing cell lines. This approach was based on the assumption that all high-MCT2-expressing cell lines accumulate [NOG]ic at sufficiently high levels to engage low affinity intracellular targets, thereby alleviating the bias imposed by the dependence of DMOG sensitivity on MCT2 expression status (Supplementary Dataset 2).
Data and Statistical analyses
For metabolomics analyses, data exported from Mass Hunter Workstation (see GC-MS section of Methods) were analysed using an in-house generated R script. To generate heatmaps, data were expressed as a log2-fold change relative to the appropriate control condition, averaged across all replicates and subsequently used to generate heatmaps using the ‘pheatmap’ package, version 1.0.8 (https://CRAN.R-project.org/package=pheatmap). Metabolites were ordered from those with the largest positive fold-change to those with the largest negative fold-change, based on one of the conditions in each experiment as indicated in the figure legend for the respective heatmaps. All metabolomics data were corrected for natural isotope abundance using a script provided by Sean O'Callaghan (Bio21 institute, The University of Melbourne).
Statistical analyses throughout this work were performed using GraphPad Prism® 7.0b. Comparisons were made using either 2-sided unpaired t-tests, 2-sided multiple t-tests with the Holm-Sidak method for multiple comparison testing, one-way ANOVA with Dunnett’s correction for multiple comparisons or two-way ANOVA with Tukey’s test for multiple comparisons, as indicated in the respective figure legends.
To determine IC50 values, an ‘[inhibitor] vs normalised response – variable slope’ curve was fitted. To determine degradation rates, curves were fitted using the one phase-decay model. To determine Kd values, a one-site specific binding curve model was used. All curve-fitting was performed using GraphPad Prism® 7.0b using standard parameters.
Chemicals
DMOG (Cayman Chemicals, 71210), MOG (synthesised as described in Supplementary Note – Synthetic Procedures, or generated from DMOG by incubating DMOG at 20 mM in RPMI medium for 16 h) or NOG (Santa Cruz, sc202720A) were used at 1 mM unless otherwise stated. 13C-labelled tracers were obtained from Cambridge Isotope Laboratories, except for DM-13C-αKG that was synthesised in-house (see Supplementary Note – Synthetic Procedures). All other chemicals were obtained from Sigma unless otherwise stated. Bovine GDH was obtained from Sigma Aldrich (G2626).
Cell lines, cell culture and viral transduction
All cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Human breast cancer cell lines (Fig. 1b) were obtained as NCI-ICBP45 kit. All cell lines were cultured in RPMI 1640 medium (Gibco, 31840) supplemented with 10 % foetal calf serum (FCS), 2 mM glutamine, 100 U/mL penicillin/streptomycin in a humidified incubator at 37 °C, 5 % CO2. In the case of INS-1 cells, medium was also supplemented with 10 mM HEPES, and 0.05 mM β-mercaptoethanol. All cell lines were tested mycoplasma-free and cell identity was confirmed by short tandem repeat (STR) profiling by The Francis Crick Institute Cell Services Science Technology Platform. Hypoxia treatment was carried out using an InVivo2 400 humidified workstation (Ruskinn, Pencoed, UK), set to 1% O2, 5% CO2 and 70% humidity.
Retroviruses were produced in 293T cells by co-transfecting pBabePuro-based vectors containing the cDNA of interest, and a plasmid containing the amphotropic receptor gene. 48h after transfection, viral supernatants were harvested and supplemented with 4 µg/mL polybrene, before adding to target cells for 6-8h. Cells were allowed to recover for 24h prior to selection with 1 µg/mL puromycin (in all cells except for INS1, for which 0.25 µg/mL puromycin was used) for at least 3 days.
Lentiviral transduction was performed as with retroviruses, but co-transfection of the pLVX-TightPuro-based vectors into 293T cells was done with pMD2.G (VSV-G), pMDLg/pRRE (GAG/POL) and pRSV-Rev.
Cloning of expression constructs
Plasmids containing cDNAs of human MCTs were as follows: MCT2 was obtained from OriGene (Clone ID: SC108858, NM_004731) and cDNAs for MCT1 (Dharmacon MHS6278-202806621, BC026317.1), and MCT4 (MHS6278-211688899, BC112267.1). cDNAs were amplified using the primers below and then cloned into pBabePuro or pLVX-TightPuro to enable retrovirus production. Wherever these constructs were used, results were always compared to a control cell line transduced with virus encoding empty pBabePuro or pLVX-TightPuro vector. Primers used can be found in Supplementary Table 1.
For knockdown of MCT2, pLKO vector based shRNAs were obtained from Dharmacon (TRCN0000038504, sequence: GCAGGTAAATTGGTGGATTTA).
Generation of HIF1α knockout cell line by CRISPR
CRISPR constructs were designed and cloned as previously described50 with minor modifications detailed below. CRISPR guide sequences (sgRNAs) were designed using the MIT CRISPR Design Tool (crispr.mit.edu) (forward oligo: caccgTTCTTTACTTCGCCGAGATC, reverse oligo: aaacGATCTCGGCGAAGTAAAGAAc). Guide oligonucleotides were phosphorylated using T4 Polynucleotide Kinase (New England Biolabs) and annealed in a thermocycler using the following parameters: 37°C for 30 minutes, 95°C for 5 minutes, decrease temperature to 25°C at 0.1°C/min. The empty Cas9 expression plasmid (pSpCas9(BB)-2A-Puro (PX459) V2.0) was linearised using BbsI and annealed oligonucleotides were ligated in before transforming into DH5a E. coli. Colonies were tested for successful insertion by colony PCR, with an expected band at 150bp.
To generate knockout clones, MCF7 cells were seeded 24 h prior to transfection with the expression plasmid encoding for Cas9 from S. pyogenes, the CRISPR target sequence and the puromycin resistance marker. Transfection was performed at 70-90% confluency using FuGENE HD Transfection Reagent (Promega), according to the manufacturer’s instructions. The day after transfection, puromycin selection (1 μg/mL, #P7255) was added to the medium for 72 h. The following day, selection was removed and cells were seeded at limiting dilutions to obtain monoclonal colonies (500-1000 cells per 15 cm cell culture dish). Monoclonal populations were grown for 2 weeks, during which the medium was replaced every 2-3 days. Colonies (>100 cells) were isolated and expanded until they could be tested for loss of the target protein by western blot.
Continuous cell proliferation and apoptosis measurements
To measure cell proliferation and death in real-time, MCF7 cells were seeded in 96-well plates at 9000 cells per well. DMOG and where appropriate DM-αKG were added 16-20 h after seeding. Cells were imaged once every three hours using an IncuCyteZoom (Essen Bioscience) and automated analysis of phase images was used to determine confluence.
Cell mass accumulation assay
To measure cell mass, cells were plated in 24-well plates at 50,000 cells per well and allowed to settle overnight. The following day, DMOG (and where applicable 10 mM pyruvate, 10 mM Lactate or 1 mM DM-αKG, unless otherwise stated) was added at the indicated concentration. After 48h, cells were washed with PBS, fixed with 10 % formalin at room temperature, and then washed once more with PBS before staining with 0.1% w/v crystal violet in 20% methanol (with shaking) at room temperature for 15 min. Plates were then washed twice more with water for 10 min, before being air-dried. Staining was quantified by solubilising in 250µl 10% v/v acetic acid and measuring absorbance at 595 nm in a Tecan Infinite M1000 plate reader.
Cell volume determination by measuring cell diameter
Cell volume determination was also calculated using median diameter measurements obtained from the Nexcelcom Bioscience Cellometer Auto T4. Since this measures diameter of trypsinised cells, we assumed a spherical shape, and therefore used cell diameter to calculate volume. Volumes determined by imaging cell diameter agreed well with TFA uptake experiments therefore this method was used subsequently.
Evaluation of cell death by propidium iodide incorporation and flow cytometry
Cells were seeded in 6-well plates 24 h prior to the experiment. Following treatment with DMOG for 48 h, cells were trypsinised, pelleted by centrifugation, re-suspended in 500 µL FACS solution [Phenol Red-free RPMI + 2% FCS, 50 μM propidium iodide (PI)], and filtered through a cell strainer to remove cell aggregates. Samples were analysed using a CS&T-calibrated LSR Fortessa (BD Biosciences). The laser configuration of the instrument was 488 nm (50 mW), 635 nm (40 mW), 406 nm (50 mW), and 561 nm (50 mW). PI fluorescence was detected using the 561nm laser for excitation and a 610/20 emission filter. Acquisition gates were set using FACS DIVA software (version 8.0.1) and post-acquisition analysis performed using FlowJo software (Treestar). Briefly, single cells were identified based on FSC-A and FSC-H as shown in Supplementary Fig. 1b. Then, live and dead cells were gated based on FSC-A and PI-A. Small debris was excluded from the analysis. The threshold was set to 5,000 events on FSC signal. A minimum of 20,000 single cells were acquired for analysis.
Cell lysis and Western Blotting
Cells on cell culture dishes were washed twice with PBS, snap-frozen in liquid nitrogen and stored at -80°C. Cells were scraped from the cell culture plate in TNN lysis buffer [50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 0.5% NP40] supplemented freshly with 1 mM dithiothreitol (DTT), and protease inhibitors [1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 4 μg/mL aprotinin, 4 μg/mL leupeptin, and 4 μg/mL pepstatin (pH 7.4)] and lysed for 20 min on ice. Lysates were centrifuged at 20,000 × g for 10 min at 4°C, supernatants were boiled in SDS sample buffer for 5 min, resolved by SDS-PAGE and proteins were transferred to PVDF membranes by electroblotting. Membranes were blocked with 5% milk in Tris-buffered saline (50 mM Tris-HCl pH 7.5, 150 mM NaCl) containing 0.05% Tween 20 (TBS-T) and subsequently incubated with the primary antibody overnight at 4°C. Membranes were washed with TBS-T and incubated with the secondary antibody conjugated to horseradish peroxidase for 1 h at RT in 5% milk TBS-T. Antibodies were visualised by chemiluminescence and imaged using the Amersham Imagequant 600 RGB, according to manufacturer’s instructions.
Primary antibodies used: mouse anti-α-tubulin (Clone DM1A, Sigma, T9026), 1:2000 in 5% BSA/TBS-T; rabbit anti-MCT1 (Millipore, AB3538P), 1:500 in 5% milk/TBS-T; rabbit anti-MCT2 (L-11, Santa Cruz, SC-22034-R), 1:500 in 5% milk/TBS-T; rabbit anti-MCT4 (H-90, Santa Cruz, SC-20329), 1:500 in 5% milk/TBS-T; mouse anti-β-actin antibody (Sigma, A2228), 1:1000 in 5% BSA/TBS-T. Secondary antibodies: Goat anti-rabbit IgG antibody conjugated to HRP, goat anti-mouse IgG antibody conjugated to HRP.
Puromycin incorporation to assess translation
Puromycin incorporation (Supplementary Fig. 7b) was carried out as in [Starck et al. Chem Biol 11: 999-1008 (2004)] with modifications as indicated in the figure legend and using an anti-puromycin antibody (clone 12D10, Merck, MABE343) at 1:25,000 in 5% BSA/TBS-T.
ATP quantification assay
15,000 cells per well (in all cases except HCT116, where 20,000 cells per well were used) were plated in 96-well plates, and the following day after treatment with MOG, ATP-dependent luciferase luminescence was measured in cells incubated with 0.1% DMSO or 1 mM MOG in RPMI medium using the CellTiterGlo kit (G7570, Promega) according to manufacturer’s instructions.
Respirometry
Cell respiration was determined in 1.2-1.5×106 non-permeabilised cells by measuring oxygen flux in an Oroboros Oxygraph-2K oxygen electrode system. For each measurement, one confluent plate of cells was used. For Supplementary Fig. 9a, cells were treated with 75 ng/ml doxycycline for 24 h prior to the experiment. Trypsinised cells were resuspended in RPMI medium and a basal oxygen flux reading was taken before 0.1% DMSO was added to one chamber or 1 mM MOG was added to the other. Oxygen flux was taken once more after respiration had plateaued (approximately 30-40 mins).
For the experiments in Supplementary Fig. 10f, cell respiration was measured as above except that cells were permeabilised and the following modifications: trypsinised cells were pre-incubated at 37 °C with stirring in working buffer (130 mM sucrose, 50 mM KCl, 5 mM KH2PO4, 5 mM MgCl2, 5 mM HEPES, 50 μM EDTA, pH 7.2) containing either 1 mM MOG or 0.1% DMSO for 30 mins. 1 nM xF was used to permeabilise cells. Glutamine and α-ketoglutarate were titrated to 10 mM final concentration, ADP was titrated to 3 mM final concentration.
Glutamate dehydrogenase (GDH) activity assays
Steady-state GDH activity was measured as previously described51. Briefly, initial velocities of oxidative deamination of glutamate were measured in a Tecan Infinite M1000 plate reader by monitoring the direct reduction of NAD to NADH at 37°C in a buffer containing 100 mM Sodium Phosphate buffer (pH 8.0), 25 mM glutamate and 0.2 mM NAD.
Bovine GDH was used at 400 ng/reaction and human mitochondria were isolated from MCF7 cells as previously described52. Mitochondria were lysed in TNN lysis buffer [50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 0.5% NP40] supplemented freshly with 1 mM dithiothreitol (DTT), and protease inhibitors [1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 4 μg/mL aprotinin, 4 μg/mL leupeptin, and 4 μg/mL pepstatin (pH 7.4)]. Mitochondrial protein lysate was used at 20 µg/reaction. All reactions were performed in a total volume of 200 µl. IC50 values were determined by fitting a ‘log(inhibitor) vs. response - Variable slope (four parameters)’ curve in GraphPad Prism.
Stable isotope labelling and metabolite extraction for metabolomics
1-2 days prior to the experiment, cells were seeded in 6 cm dishes in RPMI media (as described above), containing FCS that had been dialysed against PBS (3500 Da MWCO). At t = -1 h, medium was replaced with fresh medium, and then at t = 0, medium was changed again to medium containing the 13C-labelled nutrient and either 1 mM MOG or 0.1% DMSO (vehicle control). Tracers were used at the following concentrations: [U-13C]-Glucose, 11 mM; [U-13C]-Glutamine, 2 mM; Dimethyl [13C1,2,3,4,5]-2-ketoglutarate, 0.1 mM. Labelling was carried out for 4 h, unless otherwise stated. For each condition, 4-5 technical replicate plates were used. 2-3 plates of each cell line used in the experiment were counted and cell numbers were used to normalise metabolite measurements. Where applicable, cell diameter was also recorded in order to determine cell volumes and therefore the calculation of intracellular concentrations. Both cell number and diameter were measured using a Nexcelcom Bioscience Cellometer Auto T4.
At the end of the experiment, each plate was washed twice with ice-cold PBS, and the cells were quenched with the addition of 725 µl dry-ice-cold methanol to the plate. Plates were scraped on ice and contents were transferred to an Eppendorf tube containing 180 µl H2O (with 2 nmol of scyllo-inositol added as an internal standard) and 160 µl CHCl3. Plates were re-scraped with an additional 725 µl of cold MeOH, and this was added to the same corresponding Eppendorf tube. Samples were sonicated in a waterbath for 3 x 8 mins and metabolites were extracted at 4°C overnight. After removing precipitated material by centrifugation, samples were dried and resuspended in 3:3:1 v/v/v MeOH/H2O/CHCl3 (350 µl total), to separate polar metabolites into an upper aqueous phase and apolar metabolites in the lower organic phase.
Gas chromatography-mass spectrometry (GC-MS)
For GC-MS analysis, 150 µl of the aqueous phase were dried down in a vial insert, washed twice with 40 µl MeOH and dried again. Samples were then derivatised by methoximation (20 µl of 20 mg/mL methoxyamine in pyridine, RT overnight) before addition of 20 µl of N,O-bis(trimetylsilyl)trifluoroacetamide (BSTFA) + 1% trimethylchlorosilane (TMCS) (Sigma, 33148) for ≥1 h. Metabolite analysis was performed by GC-MS using an Agilent 7890B-5977A system. Splitless injection (injection temperature 270°C) onto a 30 m + 10 m × 0.25 mm DB-5MS+DG column (Agilent J&W) was used, with a helium carrier gas, using electron impact ionization (EI) mode. Oven temperature was initially 70 °C (2 min), followed by a temperature increase to 295 °C at 12.5 °C/min and subsequently to 320 °C at 25 °C/min (held for 3 min). MassHunter Workstation software (B.06.00 SP01, Agilent Technologies) was used for metabolite identification and quantification by comparison to the retention times, mass spectra, and responses of known amounts of authentic standards. Fractional labelling of individual metabolites was calculated as the fraction of carbons in the metabolite pool that were 13C atoms after correction for natural abundance.
Liquid chromatography-mass spectrometry (LC-MS)
The LC-MS method was adapted from Zhang et al. 201253. Samples were injected into a Dionex UltiMate LC system (Thermo Scientific) with a ZIC-pHILIC (150 mm x 4.6 mm, 5 μm particle) column (Merck Sequant). A 15 min elution gradient of 80% Solvent A to 20% Solvent B was used, followed by a 5 min wash of 95:5 Solvent A to Solvent B and 5 min re-equilibration, where Solvent B was acetonitrile (Optima HPLC grade, Sigma Aldrich) and Solvent A was 20 mM ammonium carbonate in water (Optima HPLC grade, Sigma Aldrich). Other parameters were as follows: flow rate 300 µL/min; column temperature 25°C; injection volume 10 µL; autosampler temperature 4°C. MS was performed with positive/negative polarity switching using an Q Exactive Orbitrap (Thermo Scientific) with a HESI II (Heated electrospray ionization) probe. MS parameters were as follows: spray voltage 3.5 kV and 3.2 kV for positive and negative modes, respectively; probe temperature 320°C; sheath and auxiliary gases were 30 and 5 arbitrary units, respectively; full scan range: 70 to 1050 m/z with settings of AGC target and resolution as Balanced and High (3 x 106 and 70,000), respectively. Data were recorded using Xcalibur 3.0.63 software (Thermo Scientific). Mass calibration was performed for both ESI polarities before analysis using the standard Thermo Scientific Calmix solution. To enhance calibration stability, lock-mass correction was also applied to each analytical run using ubiquitous low-mass contaminants. Parallel reaction monitoring (PRM) acquisition parameters: resolution 17,500, auto gain control target 2 × 105, maximum isolation time 100 ms, isolation window m/z 0.4; collision energies were set individually in HCD (high-energy collisional dissociation) mode. Quality control samples were prepared by pooling equal volumes of each sample and analysed throughout the run to provide a measurement of the stability and performance of the system. Qualitative and quantitative analysis was performed using Xcalibur Qual Browser and Tracefinder 4.1 software (Thermo Scientific) according to the manufacturer’s workflows. Details of compound detection are shown in Supplementary Table 2.
Intracellular NOG accumulation
Cells were incubated with DMOG or MOG for the indicated time period. Cells were then washed and metabolites were extracted as described in “GC-MS” above. After polar and apolar phases had been separated, a sample of the polar phase was diluted 50-fold in 1:1 v/v MeOH/H2O containing 5 µM [U-13C,15N]-Valine as an internal standard and analysed by LC-MS as described above.
DMOG degradation in water
DMOG was freshly resuspended at 10 µM in water containing 5 µM [U-13C,15N]-Valine as an internal standard, and 10 µL aliquots were removed every 25 minutes and analysed by LC-MS to monitor conversion to MOG and NOG. Sample temperature was maintained at 25 °C throughout the run in a thermostated autosampler.
DMOG degradation in media
DMOG was freshly resuspended at 1 mM in standard RPMI growth medium (containing 10 % FCS, penicillin/streptomycin and 2 mM glutamine), and incubated at 37°C. At the indicated times, 5 µl were removed (in triplicate), and added to 495 µl of ice-cold methanol to extract proteins. Samples were vortexed, snap frozen in liquid nitrogen and stored at -80°C. At the end of the experiment, samples were spun to remove any precipitate and then samples were analysed by LC-MS.
NMR spectroscopy
General
NMR samples were prepared in 3mm NMR tubes, except for the 19F-based cell volume measurement (see below). Data were acquired using a 600 MHz Bruker Avance III NMR spectrometer.
HSQC and HMBC measurements
2D 13C,1H heteronuclear single quantum coherence (HSQC) and heteronuclear multiple bond correlation (HMBC) experiments were performed on a 600 MHz Bruker Avance III spectrometer with 5mm TCI cryoprobe at 25 °C, using the standardhsqcetgpsisp2.2 and hmbcgplpndqf pulse sequences (Bruker TopSpin 3.5). HSQC: non-uniform sampling (35%); 1H sweepwidth 14 ppm, acquisition time 160 ms (2690 complex points); 13C sweepwidth 165 ppm, acquisition time 16 ms (800 complex points). HMBC: conventional sampling, and default parameters (1H sweepwidth 13 ppm, acquisition time 200 ms (1561 complex points); 13C sweepwidth 220 ppm, acquisition time 3.85 ms (256 complex points); long range coupling evolution time 50 ms). 4 transients were recorded for each t1 increment, with a relaxation delay of 1.8 s, yielding a total measurement time of 36 mins. 2D Fourier transformation was applied after apodization with unshifted sinebell window functions, and zerofilling to a 4K*1K data matrix. Chemical shifts were referenced to the methyl group signals of 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) at 0 ppm in each dimension.
Assignment of DMOG peaks
1D 1H-NMR analysis of DMOG in water revealed three major singlet resonances with chemical shifts at δ = 4.14, 3.92 and 3.77 ppm, integrating for 2, 3 and 3 protons, respectively (Fig. 2D, peaks numbered in blue 1, 3 and 2, respectively). On the basis of the relative peak intensities, the two upfield resonances (3.92 and 3.77 ppm) are attributable to the two methyl groups, and the peak at 4.14 ppm to the central methylene moiety. We also observed two minor peaks at 4.07 ppm and 3.76 ppm (peaks in crimson 1 and 2, respectively), as well as a peak at 3.34 ppm from methanol. To assign the NMR resonances, we analysed DMOG, either freshly prepared or incubated for 20 h in media, by 2D heteronuclear single quantum coherence (HSQC)54 and heteronuclear multiple bond correlation (HMBC)55 spectroscopy, the latter of which reports long-range (2-3 bonds) connectivity between 1H and 13C atoms. The assignment of the DMOG resonances obtained is shown in Fig. 2E. We detected long-range coupling of the methylene protons (δ = 4.14 ppm) with two distinct 13C signals (δ = 161.5 and 173.9 ppm). The methyl protons at 3.76 ppm were also coupled to the carbon resonance at 173.9 ppm. As the HMBC experiment does not detect coupling between atoms more than three bonds apart, we concluded that the 13C signal at 173.9 ppm corresponds to the ester carboxyl carbon 3J-coupled to the methyl group at 1H 3.76 ppm and that the 1H signal at 3.92 ppm corresponds to the protons of the methyl oxoacetate moiety.
1D-1H and waterLOGSY experiments
Bovine GDH was dialysed against 50 mM Sodium Phosphate buffer, containing 100 mM NaCl, pH7.4 before use. Measurements were conducted in the same buffer, using 10 µM GDH with increasing concentration of ligand, in the presence of 200 µM NAD. One-dimensional 1H NMR experiments were conducted at either 600 (Avance III) or 700 MHz with (Avance IIIHD) spectrometers equipped with 5mm TCI or QCI cryoprobes at 25 °C. Standard excitation sculpting56 (zgesgp) and WaterLOGSY57 (ephogsygpno) pulse sequences were employed with typical acquisition parameters: sweepwidth 16 ppm; relaxation delay 3 s; acquisition time 2 s (22320 complex data points); mixing time 1.5 s; 4 dummy scans; 256 transients; total measurement time 29 mins. Free induction decays were apodized with 4 Hz line broadening, and zerofilled to 128K complex points prior to Fourier transformation. WaterLOGSY peak intensities were fit in a non-linear least squares paradigm to the function described in Dalvit et al.10 using an in-house Python script with Monte Carlo sampling of the uncertainty bounds based on the peak-to-peak noise within each spectrum.
Cell volume determination by NMR spectroscopy
Cell volume was determined by measurement of the partitioning of trifluoroacetate (TFA) across the plasma membrane as previously described58. Approximately 75 million cells were trypsinized, counted, pelleted and then resuspended in 300 µl of RPMI medium containing 2 mM sodium trifluoroacetate. A known volume of the cell suspension (and therefore known cell number) was then transferred to a 5 mm NMR tube. One-dimensional 19F NMR spectra were obtained on a Avance IIIHD spectrometer operating at 400 MHz (25 °C) equipped with a SmartProbe using a 30-degree excitation pulse of 6 μs, relaxation delay of 3 s, sweepwidth 302 ppm, and acquisition time of 1.15 s (262144 complex points). After four dummy scans, 256 transients were obtained over 18 mins. Spectra were recorded until the resonance intensities corresponding to intracellular and extracellular TFA stabilized (~40 mins). During this period, visible sedimentation of cells in the NMR tube was negligible. 2 Hz line broadening and zerofilling to 128K complex points was applied prior to Fourier transformation. Deconvolution of the 19F resonances corresponding to intra- and extracellular TFA was performed using the dcon facility in TopSpin. Relative volumes were then used to calculate cell volume.
Supplementary Material
Acknowledgments
We thank all members of the Anastasiou lab for valuable discussions and input throughout this work, and particularly J. Macpherson and N. Bevan for technical help. We are grateful to L. Cantley for advice during early stages of this work. We acknowledge S. O'Callaghan for the algorithm to correct for natural isotope abundance in metabolomics data. We thank J. Kleinjung for advice with statistical methods, M. Howell for advice and help with cell proliferation and viability measurements, J. Cerveira for help with flow cytometry measurements, and A. Gould for comments on the manuscript. We are grateful to the staff at the Medical Research Council National Biomedical NMR Centre at the Francis Crick Institute where NMR data were obtained. We apologise to the numerous authors whose primary work we couldn’t cite due to space constrains. This work was funded by the MRC (MC_UP_1202/1) and by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001033), the UK Medical Research Council (FC001033) and the Wellcome Trust (FC001033) to DA.
Footnotes
Code Availability
The computer code used in this study is available from the corresponding author upon reasonable request.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Accession codes and relevant web links can be found in the respective legends and methods sections.
Author Contributions
PCD and TJR performed NMR experiments; FG generated HIF1α mutant cell lines and wrote scripts for metabolomics data analysis and visualisation; AJ and PN performed respiration experiments; MG assisted with the development of LC-MS analytical methods; GD generated and characterised cell lines; MSdS and JIM assisted with and advised on metabolomics experiments; AJW did flux modelling; GP and NO’R synthesised DM-[13C5]-αKG; SC and DH synthesised MOG and advised on chemistry; CHB contributed to the large-scale DMOG sensitivity screen; KC performed experiments and analysed data; LF and DA designed and performed experiments, analysed data and wrote the manuscript. All authors reviewed and commented on the manuscript.
Competing Financial Interests Statement
The authors declare no competing interests.
References
- 1.Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36:1302–1315. doi: 10.15252/embj.201696151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Still ER, Yuneva MO. Hopefully devoted to Q: targeting glutamine addiction in cancer. Br J Cancer. 2017;116:1375–1381. doi: 10.1038/bjc.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zdzisinska B, Zurek A, Kandefer-Szerszen M. Alpha-Ketoglutarate as a Molecule with Pleiotropic Activity: Well-Known and Novel Possibilities of Therapeutic Use. Arch Immunol Ther Exp (Warsz) 2017;65:21–36. doi: 10.1007/s00005-016-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anastasiou D, Cantley LC. Breathless cancer cells get fat on glutamine. Cell Res. 2012;22:443–446. doi: 10.1038/cr.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang L, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Ivan M, Kaelin WG., Jr The EGLN-HIF O2-Sensing System: Multiple Inputs and Feedbacks. Mol Cell. 2017;66:772–779. doi: 10.1016/j.molcel.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan MC, Holt-Martyn JP, Schofield CJ, Ratcliffe PJ. Pharmacological targeting of the HIF hydroxylases--A new field in medicine development. Mol Aspects Med. 2016;47–48:54–75. doi: 10.1016/j.mam.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin L, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27:257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marino G, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–725. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Taniguchi CM, et al. Cross-talk between hypoxia and insulin signaling through Phd3 regulates hepatic glucose and lipid metabolism and ameliorates diabetes. Nat Med. 2013;19:1325–1330. doi: 10.1038/nm.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aragones J, et al. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat Genet. 2008;40:170–180. doi: 10.1038/ng.2007.62. [DOI] [PubMed] [Google Scholar]
- 15.Olenchock BA, et al. EGLN1 Inhibition and Rerouting of alpha-Ketoglutarate Suffice for Remote Ischemic Protection. Cell. 2016;164:884–895. doi: 10.1016/j.cell.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eltzschig HK, Bratton DL, Colgan SP. Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat Rev Drug Discov. 2014;13:852–869. doi: 10.1038/nrd4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011;40:4364–4397. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- 18.Cunliffe CJ, Franklin TJ, Hales NJ, Hill GB. Novel inhibitors of prolyl 4-hydroxylase. 3. Inhibition by the substrate analogue N-oxaloglycine and its derivatives. J Med Chem. 1992;35:2652–2658. doi: 10.1021/jm00092a016. [DOI] [PubMed] [Google Scholar]
- 19.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 20.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 21.Hamada S, et al. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors. Bioorg Med Chem Lett. 2009;19:2852–2855. doi: 10.1016/j.bmcl.2009.03.098. [DOI] [PubMed] [Google Scholar]
- 22.Baader E, Tschank G, Baringhaus KH, Burghard H, Gunzler V. Inhibition of prolyl 4-hydroxylase by oxalyl amino acid derivatives in vitro, in isolated microsomes and in embryonic chicken tissues. Biochem J. 1994;300(Pt 2):525–530. doi: 10.1042/bj3000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraisl P, Aragones J, Carmeliet P. Inhibition of oxygen sensors as a therapeutic strategy for ischaemic and inflammatory disease. Nat Rev Drug Discov. 2009;8:139–152. doi: 10.1038/nrd2761. [DOI] [PubMed] [Google Scholar]
- 24.Leite de Oliveira R, et al. Gene-targeting of Phd2 improves tumor response to chemotherapy and prevents side-toxicity. Cancer Cell. 2012;22:263–277. doi: 10.1016/j.ccr.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 26.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Sun RC, Denko NC. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 2014;19:285–292. doi: 10.1016/j.cmet.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halestrap AP. The SLC16 gene family - structure, role and regulation in health and disease. Mol Aspects Med. 2013;34:337–349. doi: 10.1016/j.mam.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Escuredo J, et al. Monocarboxylate transporters in the brain and in cancer. Biochim Biophys Acta. 2016;1863:2481–2497. doi: 10.1016/j.bbamcr.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson RF. The perturbation treatment of chemical reactivity. Angew Chem Int Ed. 1973;12:36–56. [Google Scholar]
- 32.Sekine N, et al. Low lactate dehydrogenase and high mitochondrial glycerol phosphate dehydrogenase in pancreatic beta-cells. Potential role in nutrient sensing. J Biol Chem. 1994;269:4895–4902. [PubMed] [Google Scholar]
- 33.Broer S, et al. Characterization of the high-affinity monocarboxylate transporter MCT2 in Xenopus laevis oocytes. Biochem J. 1999;341(Pt 3):529–535. doi: 10.1042/0264-6021:3410529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan J, et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhdanov AV, Okkelman IA, Collins FW, Melgar S, Papkovsky DB. A novel effect of DMOG on cell metabolism: direct inhibition of mitochondrial function precedes HIF target gene expression. Biochim Biophys Acta. 2015;1847:1254–1266. doi: 10.1016/j.bbabio.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Rendina AR, et al. Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism. Biochemistry. 2013;52:4563–4577. doi: 10.1021/bi400514k. [DOI] [PubMed] [Google Scholar]
- 37.Kell DB. Finding novel pharmaceuticals in the systems biology era using multiple effective drug targets, phenotypic screening and knowledge of transporters: where drug discovery went wrong and how to fix it. FEBS J. 2013;280:5957–5980. doi: 10.1111/febs.12268. [DOI] [PubMed] [Google Scholar]
- 38.Gong L, Goswami S, Giacomini KM, Altman RB, Klein TE. Metformin pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet Genomics. 2012;22:820–827. doi: 10.1097/FPC.0b013e3283559b22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gan L, et al. Metabolic targeting of oncogene MYC by selective activation of the proton-coupled monocarboxylate family of transporters. Oncogene. 2016;35:3037–3048. doi: 10.1038/onc.2015.360. [DOI] [PubMed] [Google Scholar]
- 40.Pertega-Gomes N, et al. Monocarboxylate transporter 2 (MCT2) as putative biomarker in prostate cancer. Prostate. 2013;73:763–769. doi: 10.1002/pros.22620. [DOI] [PubMed] [Google Scholar]
- 41.Christen S, et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell Rep. 2016;17:837–848. doi: 10.1016/j.celrep.2016.09.042. [DOI] [PubMed] [Google Scholar]
- 42.Avril N. GLUT1 expression in tissue and (18)F-FDG uptake. J Nucl Med. 2004;45:930–932. [PubMed] [Google Scholar]
- 43.Kulkarni A, et al. Glucose Metabolism and Oxygen Availability Govern Reactivation of the Latent Human Retrovirus HTLV-1. Cell Chem Biol. 2017;24:1377–1387 e1373. doi: 10.1016/j.chembiol.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daberkow RL, White BR, Cederberg RA, Griffin JB, Zempleni J. Monocarboxylate transporter 1 mediates biotin uptake in human peripheral blood mononuclear cells. J Nutr. 2003;133:2703–2706. doi: 10.1093/jn/133.9.2703. [DOI] [PubMed] [Google Scholar]
- 45.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 46.Hopkins AL, Mason JS, Overington JP. Can we rationally design promiscuous drugs? Curr Opin Struct Biol. 2006;16:127–136. doi: 10.1016/j.sbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, et al. EglN2 associates with the NRF1-PGC1alpha complex and controls mitochondrial function in breast cancer. EMBO J. 2015;34:2953–2970. doi: 10.15252/embj.201591437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Ran FA, et al. Nature Protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allen A, Kwagh J, Fang J, Stanley CA, Smith TJ. Biochemistry. 2004;43:14431–14443. doi: 10.1021/bi048817i. [DOI] [PubMed] [Google Scholar]
- 52.Frezza C, Cipolat S, Scorrano L. Nature Protocols. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 53.Zhang T, Creek DJ, Barrett MP, Blackburn G, Watson DG. Evaluation of Coupling Reversed Phase, Aqueous Normal Phase, and Hydrophilic Interaction Liquid Chromatography with Orbitrap Mass Spectrometry for Metabolomic Studies of Human Urine. Anal Chem. 2012;84:1994–2001. doi: 10.1021/ac2030738. [DOI] [PubMed] [Google Scholar]
- 54.Schleucher J, et al. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J Biomol NMR. 1994;4:301–306. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- 55.Bax A, Summers MF. H-1 and C-13 Assignments from Sensitivity-Enhanced Detection of Heteronuclear Multiple-Bond Connectivity by 2d Multiple Quantum Nmr. Journal of the American Chemical Society. 1986;108:2093–2094. doi: 10.1021/ja00268a061. [DOI] [Google Scholar]
- 56.Hwang TL, Shaka AJ. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson. 1995;112:275–279. doi: 10.1006/jmra.1995.1047. [DOI] [Google Scholar]
- 57.Dalvit C, Fogliatto G, Stewart A, Veronesi M, Stockman B. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J Biomol NMR. 2001;21:349–359. doi: 10.1023/a:1013302231549. [DOI] [PubMed] [Google Scholar]
- 58.London RE, Gabel SA. Biochemistry. 1989;28:2378–2382. doi: 10.1021/bi00432a006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.