Abstract
The extent to which cells in normal tissues accumulate mutations throughout life is poorly understood. Some mutant cells expand into clones that can be detected by genome sequencing. We mapped mutant clones in normal esophageal epithelium from nine donors (age range 20 to 75 years). Somatic mutations accumulated with age and were mainly caused by intrinsic mutational processes. We found strong positive selection of clones carrying mutations in 14 cancer genes, with tens to hundreds of clones per square centimeter. In middle-aged and elderly donors, clones with cancer-associated mutations covered much of the epithelium, with NOTCH1 and TP53 mutations affecting 12 to 80% and 2 to 37% of cells, respectively. Unexpectedly, the prevalence of NOTCH1 mutations in normal esophagus was several times higher than in esophageal cancers. These findings have implications for our understanding of cancer and ageing.
Somatic mutations occur in healthy cells throughout life (1–3). Most of these mutations do not alter cell behaviour and accumulate passively (4). Occasionally, however, a key gene is altered in a way that provides mutant cells with a competitive advantage, leading to the formation of persistent mutant clones. Such clones are thought to be the origin of cancer and have also been linked to other diseases (5, 6). Despite its importance, understanding the extent of somatic mutation in normal tissues has been challenging due to the difficulties of identifying mutations present in small numbers of cells.
The most highly mutated normal tissue reported to date is sun-exposed human skin. Deep targeted sequencing of sun-exposed skin from four middle-aged individuals revealed large numbers of mutant clones under positive selection, with around a quarter of skin cells carrying cancer-driving mutations (7). Since most mutations were caused by ultraviolet light, it is unclear whether aged sun-exposed skin represents a special case due to a lifetime exposure to a powerful mutagen. This question motivated us to investigate the mutational landscape of esophageal epithelium, a tissue with a similar structure but a very different exposure to mutagens. Like the skin, esophageal epithelium consists of layers of keratinocytes. Cells are shed from the surface throughout life and are replaced by proliferation. In addition, both the skin and the upper- and mid-esophagus develop squamous cell cancers.
We performed ultra-deep targeted sequencing of 844 small samples of normal esophageal epithelium from nine deceased organ transplant donors, ranging from 20 to 75 years of age (Table S1). None of the donors had a known history of esophageal or other chronic disease and none was taking prescription medication for gastroesophageal reflux. Four of the nine donors had a history of cigarette smoking. Upper- and mid-esophageal epithelium was separated from the underlying stroma and cut into a contiguous grid of 2mm2 samples, allowing us to map clones that spanned multiple samples (Methods S1). Each sample was examined under a dissecting microscope and no lesions were seen. Histology and wholemount confocal imaging of adjacent tissue were also normal (Fig. S1, S2). Deep targeted sequencing of 74 cancer genes was performed on each sample to a median on-target coverage after duplicate removal of 870x (Methods S2). 21 samples that were found to be dominated by large clones from the targeted sequencing data were also whole-genome sequenced to a median coverage of 37x. This captures the state of the genome of the cell whose progeny subsequently colonized the sample.
Detection of mutations in normal esophagus
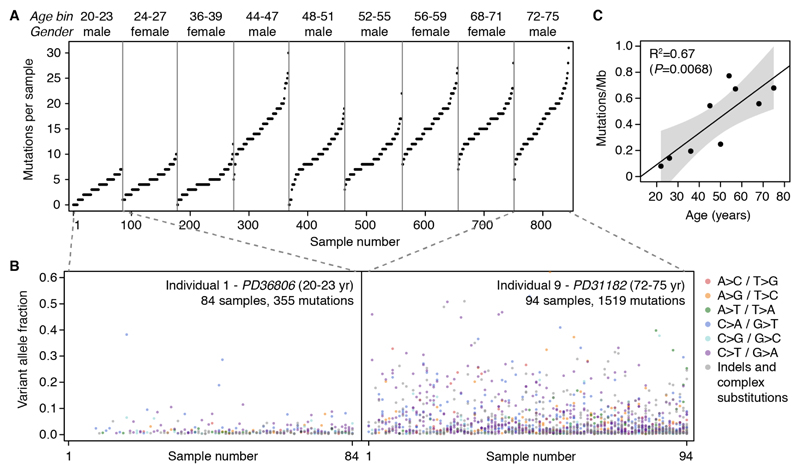
To detect mutations present in only a small fraction of each sample from deep-targeted sequencing data we used the ShearwaterML algorithm (7, 8). This algorithm uses the observed error rates per site from a large collection of normal samples to build a site-specific error model for every type of change in every targeted site (Methods S3, Fig. S3). In this dataset, we identified 8,919 somatic coding mutations across 844 samples from all donors (total area ~17cm2). Of these, 6,935 were considered independent events after merging mutations shared by nearby samples into single clones (Methods S3.3) (Table S2). Most sites in the genome display error rates below 1e-4 errors per base, which enables accurate identification of mutations at low allele frequencies (Methods S3.2, Fig. S3-S4). The median allele frequency of the mutations detected by ShearwaterML was 1.6%, with a third of all mutations below 1% (Fig. S3). Since the fraction of sequencing reads that carry a mutation is a function of the fraction of mutant cells within a sample and of the local copy number, we can integrate allele frequencies and sample areas to obtain approximate estimates of the sizes of detectable mutant clones in normal esophagus, which ranged from 0.01 to over 8 mm2 (Methods S5).
The number of mutations identified per sample and their allele frequencies varied markedly across individuals, with both the number of detectable mutations and the sizes of mutant clones roughly increasing with donor age (Fig. 1A-B). To better understand the passive rate of accumulation of mutations in healthy esophagus, we can estimate the mean number of mutations per cell in each individual by integrating allele frequencies (Methods S5) (7). These are conservative lower-bound estimates as they are limited to mutations present in detectable clones. On average, healthy cells in the esophageal epithelium carry at least several hundred mutations per cell in people in their twenties, rising to over 2,000 mutations per cell late in life (Fig. 1C). Similar estimates were obtained from the whole-genome sequencing data (Methods S5.1). These estimates of the mutation rate in normal esophagus are broadly comparable to the mutation rates reported in human stem cells of the colon, small intestine and liver based on sequencing of clonal organoids (9).
Fig. 1. Detection of somatic mutations in normal esophagus.
(A) Number of mutations detected per sample across the 844 samples from the 9 transplant donors (sorted by age). Donor age is shown as 4-year bins to increase sample anonymity. (B) Variant allele fraction (VAF) of the mutations detected in the youngest and oldest donor, colored by mutation type. The VAF is the fraction of sequencing reads reporting a mutation within a sample. (C) Scatter plot of donor age and the estimated mean mutation burden per cell for each donor. The fitted line, R-square value and P-value were obtained by linear regression.
Widespread positive selection driving clonal growth
In middle-aged individuals, the number of mutations per cell is approximately ten times lower in normal esophagus than in sun-exposed skin (7), a difference partially due to the high degree of UV-damage sustained by the skin. Given this, we anticipated that the frequency of cancer-driver mutations in esophagus would be much lower than that in skin. Unexpectedly, however, analysis of the frequency and size of mutant clones revealed a higher density of cancer-associated mutations in normal esophagus than in sun-exposed skin, suggesting that there is stronger positive selection of clones with mutations in cancer-associated genes.
To formally quantify the extent of selection driving clonal expansions in normal esophagus, we estimated the ratio of non-synonymous (dN) to synonymous (dS) mutation rates (dN/dS) across genes, which is a widely-used measure of selection. We used dNdScv, an implementation of dN/dS for somatic data that controls for trinucleotide mutational signatures, sequence composition and variable mutation rates across genes (4) (Methods S6). This method has been shown to reliably identify genes under positive selection in cancer and normal tissues (4, 7). In the context of this experiment, dN/dS ratios reveal how much more (or less) likely it is for a non-synonymous mutation to reach a detectable clone size compared to a synonymous mutation (Methods S6.2).
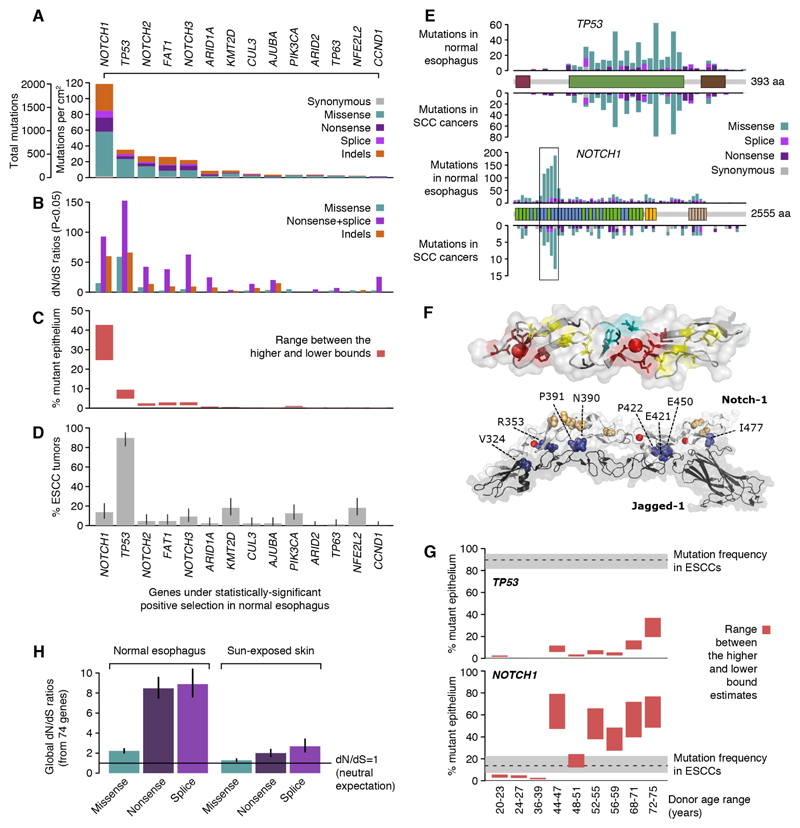
This analysis revealed strong evidence of selection driving clonal expansions in normal esophagus. At the gene-level, we detected significant positive selection in 14 of the genes that we sequenced (Fig. 2A-D) (Table S3). This means that mutation of these genes confers a competitive advantage on mutant cells relative to neighboring cells. Sorted by mutation frequency, the list comprises NOTCH1, TP53, NOTCH2, FAT1, NOTCH3, ARID1A, KMT2D, CUL3, AJUBA, PIK3CA, ARID2, TP63, NFE2L2 and CCND1. Interestingly, the five most frequently mutated genes in normal esophagus also dominated the mutational landscape in sun-exposed skin. Many of the positively selected genes play a role in keratinocyte differentiation through NOTCH signaling e.g. NOTCH1, NOTCH3, and TP53 (10–13); through redox cellular stress e.g. NFE2L2, TP63 and CUL3 (14–17) or through epigenetic regulation e.g. KMT2D (18). Tilting cell fate balance away from differentiation toward proliferation may confer a competitive advantage to mutant cells in normal esophageal epithelium (19).
Fig. 2. Widespread positive selection of cancer-associated mutations in normal esophagus.
(A) Number of mutations detected in each of the 14 genes found under positive selection. (B) Observed-to-expected ratios for missense substitutions, truncating (nonsense and essential splice site) substitutions and indels. Observed-to-expected ratios for substitutions are dN/dS ratios. Only ratios with P<0.05 are shown. (C) Estimated percentage of cells carrying a mutation in each gene (Methods S5.3). (D) Percentage of ESCCs with a non-synonymous substitution or an indel in each gene. Error bars depict 95% Poisson confidence intervals. (E) Distribution of mutations within TP53 and NOTCH1 in normal esophagus (above the gene domains diagram) and in SCC cancers from TCGA (below). EGF8-12 region is boxed. (F) Consequences of NOTCH1 missense mutations. (Top panel) Most affect structural residues in EGF domains (shown in stick form, PDB 2VJ3): calcium-binding consensus residues (red), hydrophobic interdomain packing residues (teal), cysteine residues which form disulphide bonds (yellow), conserved glycines (black). Calcium ions shown as red spheres. (Bottom panel) Other residues affected by missense mutations (≥4 per residue) in the EGF8-12 region are shown in space filling representation. Many are predicted to disrupt the Notch receptor/ligand binding interface (shown in deep blue and labelled with residue number), while others are distal (colored wheat) (PDB 5UK5). (G) Estimated percentage of mutant epithelium per donor compared to ESCC mutation frequency. (H) dN/dS values estimated from all 74 target genes together in normal esophagus and sun-exposed skin (7).
At least 11 of the 14 genes found under positive selection in normal esophagus are canonical drivers of esophageal squamous cell carcinomas (ESCC) (Methods S6.5) (20–22). Their presence in normal epithelium suggests that they might act as early ESCC drivers, leading to the expansion of persisting clones that could undergo further mutation and malignant transformation. The landscape of selection in normal squamous epithelium of the esophagus more closely resembles that of ESCCs than esophageal adenocarcinomas (EACs) (21), consistent with the fact that ESCCs typically develop from the squamous epithelium of upper- and mid-esophagus, while EACs evolve from epithelium close to the stomach junction and are associated with Barrett’s metaplasia.
Colonization of the epithelium by NOTCH1-mutant clones
One unexpected observation was the very high prevalence of NOTCH1 mutations in normal esophagus (Fig. 2A-C). Across the nine donors, we detected 2,055 coding mutations in NOTCH1, of which over 98% were non-synonymous, with an average of ~120 different NOTCH1 mutations per cm2 of normal esophagus (Fig. 2A). NOTCH1 acts as an oncogene in different leukemias but has a mutation pattern consistent with a tumor suppressor gene in squamous carcinomas (SCCs) of the skin, head and neck, esophagus and lung (23). As in SCCs, mutations in NOTCH1 in normal esophagus were enriched for truncating mutations (dN/dS>50), including stop-gains, essential splice site mutations and indels (Fig. 2B). Missense mutations were also frequent in NOTCH1, and they were concentrated in five of the 36 extracellular epidermal growth factor (EGF) repeat domains, EGF8-12 (Fig. 2E). These EGF repeats contain the binding domains for the Notch1 ligands Jagged and Delta. The most recurrent codon alterations occurred at sites predicted to affect structural residues (calcium-binding motifs, cysteine residues, interdomain packing) or the contact surface with Notch1 ligands (Fig. 2F) (Supplementary Text) (23, 24). The large number of positively-selected NOTCH1 mutations provides structural and functional insights into this key regulatory protein.
Integrating the allele fractions of the mutations and allowing for the possibility that mutations may affect one or two alleles per cell, we can estimate the fraction of mutant cells in a tissue for any given gene (Methods S5.3). On average across the nine donors, 25 to 42% of the cells in normal esophagus harbored NOTCH1 mutations (Fig. 2C). There was a large increase in the frequency of NOTCH1 mutant clones with age. Approximately 30 to 80% of normal esophagus was NOTCH1-mutant in five of the six middle-aged or elderly individuals compared to 1 to 6% in the three individuals under 40 years of age (Fig. 2G). This observation is consistent with data from experimental mouse models showing that transgenic inhibition of Notch signaling in a small fraction of cells confers clonal advantage and enables these clones to colonize the normal esophageal epithelium (19, 25).
This observation has potentially important implications. The NOTCH1 gene has been widely assumed to be a driver in ESCCs because it is mutated in ~10% of tumors (21, 26) (Fig. 2D,G). The observation that, in middle-aged individuals, NOTCH1 is typically mutated in 30 to 80% of the normal esophageal epithelium suggests that NOTCH1 mutations may be less frequent in cancers than in the background of normal tissue from which the cancers develop. This raises questions about the role of NOTCH1 in the development of ESCCs.
The case of NOTCH1 contrasts with that of TP53 (Fig. 2G), which is mutated in over 90% of ESCCs but in a minority of cells in the normal esophageal epithelium. TP53 is the second most frequently mutated gene in normal esophagus, with ~35 mutations per square centimeter and strong positive selection for both truncating and missense mutations (dN/dS ratios ~150 and ~50, respectively; Fig. 2A,B). As in cancer genomes, the missense mutations mostly affect the central DNA-binding domain (Fig. 2E). Across the nine donors, 5 to 10% of the epithelium carried a TP53 mutation, a fraction that appeared to increase with age, with the oldest donor having TP53 mutations in 20 to 35% of cells (Fig. 2G).
In summary, we found an unexpectedly high density of driver mutations in normal esophagus and positive selection acting on most of the main drivers of ESCC. Combining the 74 genes studied, global dN/dS ratios for missense and protein-truncating (nonsense and essential splice site) mutations were around 2.2 and 8.6, respectively, with the enrichment of non-synonymous mutations increasing rapidly with clone size (Fig. 2H, Fig. S5B). This suggests that approximately 55% of all missense mutations and 88% of all truncating mutations identified in this dataset were actively driven to detectable clone sizes by positive clonal selection. Overall, using dN/dS ratios, and considering substitutions and indels in the 14 genes under significant selection, we estimate that there are 3,915 (CI95%: 3,829-3,988) positively-selected driver mutations in the ~17 cm2 of normal esophageal epithelium sequenced in this study, of which 52% are in genes other than NOTCH1 (Methods S6.3). This number is comparable to the yield of driver mutations obtained from sequencing over 1,000 cancer genomes (4).
Variation of the mutational and selective landscape across donors
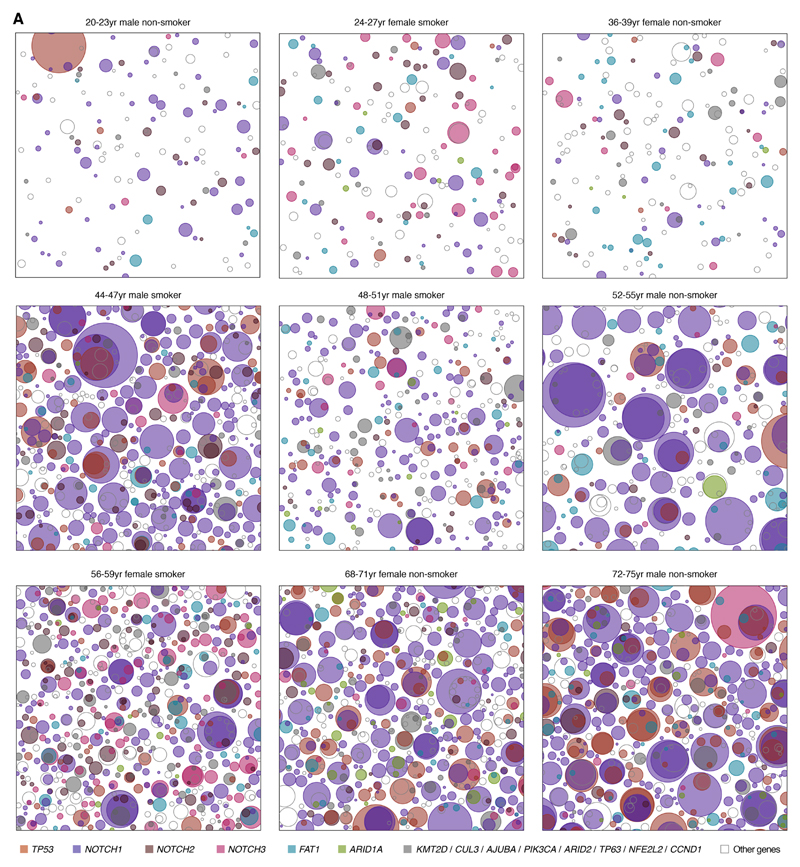
The patterns of somatic evolution varied greatly across the nine individuals in this study showed large differences in mutation density, clone sizes and overall driver frequency (Fig. 3). Age is by far the strongest risk factor in ESCC, with cancer incidence rising near-geometrically with age (27, 28). We used mixed effect regression models to evaluate the association between the mutation landscape and age, while controlling for other risk factors such as gender and smoking status (Methods S7). Despite the modest cohort size, this analysis revealed a significant increase in the number of mutations per sample (P-value=0.009) and clone sizes (P-value=0.027) with age. This is consistent with the significant increase in mutation burden with age described in Fig. 1C by standard linear regression (P-value=0.0068, R2=0.67). We also noted that the two heavy smokers in the cohort appear to have a higher number of mutations than expected for their age (Fig. 1A, Fig. 3, Methods S7). However, larger cohorts will be needed to reliably study the impact of behavioral risk factors on the mutational landscape in the esophagus.
Fig. 3. Variation of the mutational landscape across the 9 donors.
Representative patchwork plots from each donor. Each panel is a schematic representation of the mutant clones in an average 1 cm2 of normal esophageal epithelium from each donor. To generate each figure, a number of samples from the donor are randomly selected to amount to 1 cm2 of tissue and all clones detected are represented as circles randomly distributed in space. The density and size of the clones are inferred from the sequencing data and the nesting of clones and subclones is inferred from the data when possible and randomly allocated otherwise (Methods S5.4).
Despite the dominant effect of age, there are unexplained differences across individuals, including differences in the strength of selection on different genes across individuals, as suggested by the different colored clones in Fig. 3. To formally quantify differences in selection pressure per gene across donors, while removing the effect of variable mutation rates and signatures across individuals, we used an extension of dNdScv that compares two dN/dS ratios (Methods S6.4). This confirmed that there are significant differences in the driver landscape across donors (Fig. S5C-E). For example, across individuals, NOTCH1 is mutated 5 times more frequently than NOTCH3. Yet, in one donor, we detected nearly the same number of mutations in NOTCH1 and NOTCH3 (Fig. S5D, q-value=5e-13, Likelihood-Ratio Test). Similarly, the oldest donor showed a 2-fold relative enrichment in TP53 mutations compared to other individuals (q-value<1e-15, Likelihood-Ratio Test; Fig. S5E), consistent with the observation that 20 to 37% of normal esophageal epithelium was TP53 mutant in this donor (Fig. 2G). It is unclear whether the variation in the driver landscape across donors reflects differences in exposure to environmental factors, the genetic background of each individual or both. Nevertheless, differences in mutation rates, clone sizes and driver preferences may have implications for understanding inter-individual variation in cancer risk.
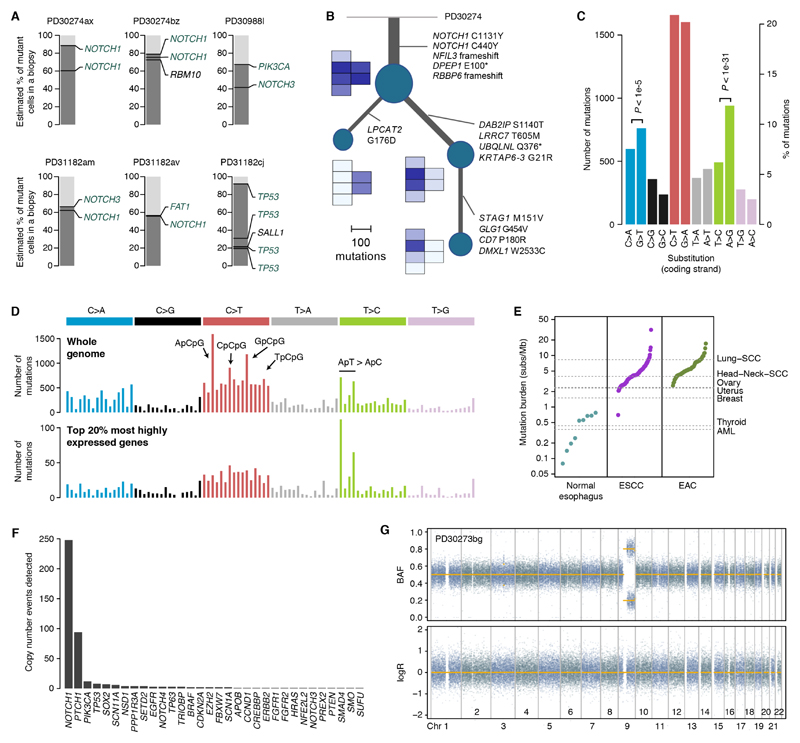
Given the large increase in driver mutant clones with age, many clones are expected to acquire more than one driver mutation over the course of a lifetime. Although the small clone sizes limit our ability to determine which mutations within a sample occur in the same cells, 25 samples had sufficiently large clones for us to confidently group mutations (Methods S5.5) (Fig. 4A and Fig. S6). Most cases (14/25) were examples of NOTCH1 bi-allelic inactivation by two mutations. We also observed examples of clones carrying mutations in NOTCH1 and FAT1, NOTCH1 and NOTCH3, and PIK3CA and NOTCH3. In the oldest donor (aged 72-75 years), whose samples showed an enrichment of TP53 mutations, we found a large clone, measuring over 4 mm2, with a founder heterozygous TP53 mutation and three separate subclones each carrying a second TP53 mutation (Fig. 4A). In a large clone extending over six samples and measuring over 8.5 mm2, we were able to integrate whole-genome data and spatial information to reconstruct its phylogenetic history (Methods S5.6). The tree shows that the ancestor cell underwent a large clonal expansion after losing both copies of NOTCH1, followed by branching evolution with two subclones dominating spatially distinct areas (Fig. 4B).
Fig. 4. Phylogenetic and mutational patterns in normal esophagus.
(A) Representation of mutations co-occurring in the same clones using the pigeonhole principle (see Supplementary material 5.5). (B) Phylogenetic reconstruction of the evolution of a large clone overlapping six samples using whole-genome sequencing data and spatial information. A small heatmap of the six affected samples is shown next to each node in the tree, depicting the mean VAF of the mutations in each node. (C) Number of substitutions per mutation type as mapped to the coding (untranscribed) strand from all donors. P-values reflect transcription strand asymmetry (exact Poisson test). (D) 96-mutation-class barplot depicting the number of mutations in each of the possible 96 trinucleotides (strand independent). The top panel shows the whole-genome plot aggregating all 21 whole-genomes. The bottom panel shows the spectrum for mutations occurring in the transcribed region of the top 20% most highly expressed genes. (E) Mutation burden in normal esophagus and in ESCC and EAC tumors (every point corresponds to a donor, sorted by mutation burden). (F) Number of copy number events detected in each gene across the 844 samples using the targeted data. (G) Representative LogR and B-allele frequency (BAF) scatter plots for heterozygous SNPs from whole genome data showing a copy-neutral LOH event affecting NOTCH1 (sample PD30273bg shown).
The whole-genome mutational landscape in normal esophagus
To better understand the contribution of different mutational processes and the extent of structural variation in normal esophagus, we performed whole-genome sequencing of 21 samples dominated by a major clone. Across all donors, C>T/G>A mutations dominate the spectra with a clear excess of mutations at CpG dinucleotides (Fig. 4C,D; Fig. S7). These changes result from the deamination of 5-methylcytosine into thymine, and are believed to occur spontaneously throughout life (29, 30). Signature analysis revealed that the pattern of mutations largely resembles a combination of COSMIC mutational signatures 1 and 5 (30) (Methods S4, Fig. S7,8). Both signatures have been shown to dominate the accumulation of mutations in normal tissues such as colon, small intestine and liver during life (9).
In addition, we observed two other mutational processes. There was a considerable rate of C>A/G>T changes with a modest but significant transcription-strand bias. Multiple mechanisms can lead to these types of changes, including smoking. Although four of the nine individuals were smokers, we did not observe a clear signature of tobacco-induced mutations (COSMIC signature 4) (Methods S4). We also observed considerable variation in T>C changes across the 21 whole-genomes, with a strong transcription strand asymmetry (Fig. 4D, Fig. S7,8). Stratification of the mutation spectra by gene expression level revealed that highly transcribed genes are targets of a process of transcription-coupled mutagenesis that induces T>C changes preferentially at ApT sites in the transcribed strand, a phenomenon previously described in liver cancers (31) (related to COSMIC signature 16) (Fig. 4D, Fig. S8).
Overall most mutations seem to be generated by intrinsic mutational processes associated with age or transcription (30, 31), without clear evidence of external mutagenic processes. Interestingly, we found no evidence of COSMIC signatures 2 and 13 in the targeted or the whole-genome data. These signatures are believed to be caused by APOBEC cytidine deaminases and contribute large numbers of mutations in esophageal cancers (20, 21, 32). This partially explains the observation that the mutation burden in normal esophagus is approximately an order of magnitude lower than the median mutation burden of ESCC and EAC cancers (Fig. 4E). The rarity of APOBEC mutagenesis in normal esophagus may suggest that this is acquired later in the evolution of ESCC or that ESCCs are more likely to evolve from rare clones displaying APOBEC mutagenesis.
Esophageal cancers are characterized by large numbers of copy number changes and structural rearrangements (21, 33). To explore the extent of copy number changes in normal esophagus, we first analyzed the deep targeted sequencing data. We used a copy number detection algorithm designed to identify low-frequency subclonal loss of heterozygosity (LOH) on targeted data, exploiting the statistical phasing of heterozygous SNPs to detect small allelic imbalances (7) (Methods S3.4). NOTCH1 loss was the most frequent copy number change identified, although caution must be exercised because statistical power varies across genes and donors. NOTCH1 LOH was detected in nearly 30% of all samples (Fig. 4F) and in virtually all of the samples with a single high-frequency NOTCH1 mutation, confirming that loss of NOTCH1 is typically bi-allelic. PTCH1 sits on the same arm of chromosome 9 as NOTCH1 and is often lost together with NOTCH1. We also detected less frequent but recurrent whole-chromosome 3 gains, which lead to the duplication of PIK3CA/SOX2/TP63, an event observed in approximately half of ESCCs (21) (Fig. 4F). Several instances of TP53 LOH were also detected, largely concentrated in the oldest donor.
Copy number analysis of the 21 whole-genomes confirmed that segmental loss of NOTCH1 is typically mediated by copy-neutral LOH without detectable rearrangements (Fig. 4G, Fig. S9, Methods S3.5.2, Table S4). Such events may be generated by mitotic homologous recombination. Events varied in size, from whole-arm losses to focal events (Fig. 4G, Fig. S9,10). With the exception of copy-neutral LOH events in NOTCH1 and an instance of chromosome 3 gain, the 21 genomes appeared largely diploid, without evidence of other copy number changes that may be expected to accumulate by chance over time (Fig. 4G, Fig. S9, Methods S3.5.2, Table S4). The rarity of copy number changes in large clones, none of which had TP53 mutations, suggests that the background rate of copy number changes is low in normal cells of the esophagus or that such changes are negatively selected. Either way, this represents a major difference between normal esophageal cells and ESCCs, suggesting that structural changes may occur late in the evolution of esophageal cancers (33).
Discussion
These data have unveiled a hidden world of somatic mutation and clonal competition in normal esophagus. We have detected thousands of mutations per cell, hundreds of positively selected clones per square centimeter, and clones with cancer-associated mutations colonizing most of the esophageal epithelium with age, all without grossly detectable changes in histology.
The higher frequency of cancer-associated mutations in normal esophagus than in sun-exposed skin is unexpected, particularly given the lower mutation rate in the esophagus. Although we found most of the common drivers of ESCC already under selection in normal esophageal epithelium, key differences remain between the genomes of cells in mutant clones in aging normal epithelium and cancer cells. These include a mutation burden approximately ten times lower than that in many ESCCs, no evidence of APOBEC mutagenesis and an apparent lack of chromosomal instability. Further, although clones carrying cancer-driver mutations are widespread, the average number of driver mutations per cell in normal esophagus is much lower than that in cancer cells (Fig. 2C), a result consistent with the multi-stage theory of carcinogenesis (27, 28, 34). Larger scale genomic studies of normal tissues in healthy individuals and of premalignant lesions of different grades will help refine our understanding of the transition from normal to cancer (3, 28, 33, 35).
An unexpected observation is the high frequency of NOTCH1 mutation in aged normal esophagus compared to ESCCs. This suggests that ESCCs are more likely to evolve from cells in the epithelium without NOTCH1 mutations. In contrast, TP53 mutations, which are several-fold less frequent than NOTCH1 mutations, are almost ubiquitous in ESCCs, suggesting that cancers arise from the small fraction of TP53 mutant cells. Cancer risk may therefore vary across the aging epithelium depending on the colonizing mutations present. Interventions that decrease the proportion of mutations at a higher risk of transformation in normal epithelium may thus be beneficial.
We note that, even if they do not contribute to carcinogenesis, drivers of benign clonal expansions could still appear as recurrently mutated genes in cancer genomes owing to their high mutation frequency in the normal cells from which tumors evolve. Better understanding of the mutational landscape in normal tissues may thus help refine current catalogs of cancer-driver genes, with important implications for early diagnosis and targeted therapy.
Positive selection of mutant clones has now been observed during normal aging in blood, sun-exposed skin and esophageal epithelium (7, 36). This opens the theoretical possibility of clonal selection across tissues as a contributing factor to tissue and organismal aging (4, 37, 38). Somatic mutation has long been recognized as a possible factor contributing to aging, with mutations and other forms of damage deleterious to the carrying cells passively accumulating during life and progressively reducing cellular fitness (39). Widespread positive selection of mutant clones may be an additional contributory factor in ageing as it can greatly accelerate the accumulation of functional mutations and altered phenotypes. Throughout life, somatic mutations increasing cellular fitness can spread and even dominate tissues, independently of their cost to the organism. If the selected mutations negatively impact tissue function, the physiological integrity of the organism will decline, a hallmark of the aging process.
This study emphasizes how little we know about somatic evolution within normal tissues, a fundamental process that is likely to take place to varying degrees in every tissue of every species. Better understanding of the extent of somatic mutation and selection across tissues, in health and disease promises to provide insights into the origins of cancer and aging.
Supplementary Material
Acknowledgments
We are very grateful to the families of deceased donors for their consent and to the Cambridge Biorepository for Translational Medicine for access to human tissue. Funding: I.M. is funded by Cancer Research UK (C57387/A21777). P.J.C. is a Wellcome Trust Senior Clinical Fellow. This work was funded by a Cancer Research UK programme grant to P.H.J., (C609/A17257), an MRC Centenary Grant, Welcome Trust core funding to the Wellcome Sanger Institute and an MRC grant-in-aid to the MRC Cancer Unit.
Footnotes
Author contributions: PHJ initiated the project. PHJ and JCF designed the experiments. IM led data analysis with help from ARJL, FA, AC and MH and advice from MRS and PJC. PAH analyzed the structural implications of NOTCH1 mutations. KS-P and KM collected the samples. JCF, AW and KM performed experiments. RCF contributed to a pilot study. IM, JCF and PHJ wrote the paper.
Competing interests: M.R.S. is on the Scientific Advisory Board of GRAIL. The other authors declare no competing interests.
Data and materials availability: Sequencing data are deposited in EGA (EGAD00001004158, EGAD00001004159).
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at https://urldefense.proofpoint.com/v2/url?u=http-3A__www.sciencemag.org_&d=DwICaQ&c=D7ByGjS34AllFgecYw0iC6Zq7qlm8uclZFI0SqQnqBo&r=DRIw5xG6YQMmEU1qw8ZX5Q&m=F45emOAiNpHTsMEILWi8FEpCHGrYme24WPJIW86saMg&s=6RdLKh1tqpEY7uDsIPdGLbXc2uB03Y3e2T7FV9pRzDM&e=.
The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B, et al. Cancer genome landscapes. Science (New York, N.Y.) 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nature reviews. Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 4.Martincorena I, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041.e1021. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anglesio MS, et al. Cancer-Associated Mutations in Endometriosis without Cancer. The New England journal of medicine. 2017;376:1835–1848. doi: 10.1056/NEJMoa1614814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaiswal S, et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. The New England journal of medicine. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science (New York, N.Y.) 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstung M, Papaemmanuil E, Campbell PJ. Subclonal variant calling with multiple samples and prior knowledge. Bioinformatics (Oxford, England) 2014;30:1198–1204. doi: 10.1093/bioinformatics/btt750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohashi S, et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology. 2010;139:2113–2123. doi: 10.1053/j.gastro.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto K, et al. Reduction of NOTCH1 expression pertains to maturation abnormalities of keratinocytes in squamous neoplasms. Laboratory investigation; a journal of technical methods and pathology. 2012;92:688–702. doi: 10.1038/labinvest.2012.9. [DOI] [PubMed] [Google Scholar]
- 12.Lefort K, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes & development. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yugawa T, et al. Regulation of Notch1 gene expression by p53 in epithelial cells. Molecular and cellular biology. 2007;27:3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhaduri A, et al. Network Analysis Identifies Mitochondrial Regulation of Epidermal Differentiation by MPZL3 and FDXR. Developmental cell. 2015;35:444–457. doi: 10.1016/j.devcel.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamanaka RB, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Science signaling. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nature genetics. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Molecular and cellular biology. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopkin AS, et al. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS genetics. 2012;8:e1002829. doi: 10.1371/journal.pgen.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcolea MP, et al. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nature cell biology. 2014;16:615–622. doi: 10.1038/ncb2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. American journal of human genetics. 2015;96:597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawada G, et al. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology. 2016;150:1171–1182. doi: 10.1053/j.gastro.2016.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Nowell CS, Radtke F. Notch as a tumour suppressor. Nature reviews. Cancer. 2017;17:145–159. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 24.Luca VC, et al. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science (New York, N.Y.) 2017;355:1320–1324. doi: 10.1126/science.aaf9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alcolea MP, Jones PH. Cell competition: winning out by losing notch. Cell cycle (Georgetown, Tex.) 2015;14:9–17. doi: 10.4161/15384101.2014.988027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 27.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. British journal of cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science (New York, N.Y.) 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 29.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nature genetics. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haradhvala NJ, et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell. 2016;164:538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang J, et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nature communications. 2017;8 doi: 10.1038/ncomms15290. 15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross-Innes CS, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nature genetics. 2015;47:1038–1046. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brash DE. Cancer. Preprocancer. Science (New York, N.Y.) 2015;348:867–868. doi: 10.1126/science.aac4435. [DOI] [PubMed] [Google Scholar]
- 35.Martinez P, et al. Evolution of Barrett's esophagus through space and time at single-crypt and whole-biopsy levels. Nature communications. 2018;9 doi: 10.1038/s41467-017-02621-x. 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. The New England journal of medicine. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith JM. Review Lectures on Senescence. I. The Causes of Ageing. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1962;157:115–127. doi: 10.1098/rspb.1962.0066. [DOI] [PubMed] [Google Scholar]
- 38.Risques RA, Kennedy SR. Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS genetics. 2018;14:e1007108. doi: 10.1371/journal.pgen.1007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.