Summary
CRISPR-Cas systems provide prokaryotes with sequence-specific immunity against viruses and plasmids, based on DNA acquired from these invaders, known as spacers. Surprisingly, many archaea possess spacers that match chromosomal genes of related species, including those encoding core housekeeping genes. By sequencing genomes of environmental archaea isolated from a single site, we demonstrate that inter-species spacers are common. We show experimentally by mating Haloferax volcanii and Haloferax mediterranei, that spacers are indeed acquired chromosome-wide, although a preference for integrated mobile elements and nearby regions of the chromosome exists. Inter-species mating induces increased spacer acquisition and may result in interactions between the acquisition machinery of the two species. Surprisingly, many of the spacers acquired following inter-species mating target self-replicons along with those originating from the mating partner, indicating that the acquisition machinery cannot distinguish self from non-self under these conditions. Engineering the chromosome of one species to be targeted by the other's CRISPR-Cas reduces gene exchange between them substantially. Thus, spacers acquired during inter-species mating could limit future gene transfer, resulting in a role for CRISPR-Cas systems in microbial speciation.
Keywords: CRISPR, archaea, horizontal gene transfer, lateral gene transfer, adaptation, acquisition, speciation, mobile genetic elements
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas (CRISPR-associated proteins) systems provide acquired heritable immunity to bacteria and archaea against invasion by selfish DNA elements. CRISPR loci are composed of partially palindromic repeats interspersed by short unique DNA spacers and multiple cas genes that encode proteins involved in the immune response. These systems can acquire DNA fragments from foreign selfish elements and integrate them as spacers into the CRISPR arrays 1,2. Subsequently, One or several spacer arrays in a prokaryotic cell can be transcribed and processed into small CRISPR RNA (crRNA) molecules, which then together with the Cascade (CRISPR-associated complex for antiviral defense) complex hybridize with nucleotide sequences in the invader and direct its degradation by Cas endonucleases 1,3.
CRISPR-Cas loci are considered to be primarily anti-viral defense systems, and in some lineages whose viruses have been studied, such as the Sulfolobales, that is reflected by the vast majority of spacers matching viral sequence 4,5. Strikingly, however, the vast majority of CRISPR spacers in bacteria and archaea that have database hits match integrated proviruses rather than lytic bacteriophages or archaeal viruses 6. This can be attributed to the under-sampling of virus sequence space, but may also indicate that excision from host chromosomes represents a preferred opportunity for spacer incorporation.
We previously observed that multiple archaea belonging to diverse clades have CRISPR spacers that match chromosomal housekeeping genes of related species, rather than selfish elements 7. This raises the question of how such spacers were acquired and whether they can affect gene exchange dynamics between species, when present. Notably, halophilic archaea can undergo a mating by cell fusion process involving cytoplasmic bridges 8, which can efficiently occur between cells from different species 9. Similar cytoplasmic bridges between cells have also been observed in multiple other archaeal lineages, such as members of Sulfolobales 10, Thermococcales 11, nanoarchaea and Thermoplasmatales 12. Here we test the hypothesis that spacers can be acquired naturally during inter-species mating from partner chromosomes. We use genome sequences of 15 haloarchaea isolated from the same coastal site to show that inter-species spacer acquisition is common within a natural ecosystem, and that the spacers can inform us of the environmental network of gene exchange. We then demonstrate directly that haloarchaea acquire spacers from the mating partner chromosomes, but also from self-replicons during mating. Finally, we examine the consequences of such spacer acquisition events and show that CRISPR-Cas targeting reduces the frequency of gene exchange via fusion across species, thus restricting horizontal gene transfer across species.
Results
Inter-species targeting is pervasive in halophilic archaea
Previous surveys of CRISPR spacers in bacteria and archaea indicated a dominance for spacers that match viruses known to infect the CRISPR-Cas containing organism or related species 7,13–16. When we compared the spacers from all halophilic archaea (class Halobacteria) in the CRISPRdb database, to the NCBI database using a sequence similarity search 5.3 % had significant database matches. Surprisingly, most haloarchaeal CRISPR arrays had spacers that matched genes in other haloarchaeal species (Supplementary Table 1). Such cross-targeting spacers were nearly as abundant as spacers matching viral sequences (Supplementary Tables 2). Most of the spacers with non-viral hits matched genes found on the main chromosomes, while relatively few matched plasmid-encoded genes or transposable elements, and included known housekeeping functions (Supplementary Fig. 1 and Supplementary Table 2). In conclusion, spacers that match chromosomal loci in other species are fairly abundant in haloarchaea.
To explore whether this unusual pattern of inter-species targeting is also common in nature, we obtained draft genomes of 15 different strains, belonging to four different genera, isolated from the same small sampling site (see Methods). Out of these 15 genomes, 11 had both CRISPR arrays and cas genes of type I-B CRISPR systems. Of the 1104 spacers in these arrays, only 35 had significant BLASTN matches (3.3%), as is generally the case in both archaea and bacteria 6. Notably, five of these isolates, belonging to the genera Haloferax and Haloarcula, had spacers that targeted other strains from the same site, with a total of 16 inter-species spacers, 13 of which were perfect matches along the entire spacer length (Table 1, Supplementary Table 3). Most of the spacers matched genes on contigs inferred to be parts of the main chromosomes (see Methods), rather than plasmids, and some had sequence identity to known house-keeping (Supplementary Tables 4 and 5). Nevertheless, some of these chromosomal targets (4/13) were less than 20Kb away from recombinase genes, indicating that they either target an island or provirus or a chromosomal locus just flanking it (Supplementary Table 6). These results are in agreement with a large survey of spacers in bacteria and archaea, showing that integrated mobile elements represent the most common CRISPR targets 6.
Table 1. CRISPR spacers of haloarchaeal strains isolated in the summers of 2012/2014 from Atlit.
Genus assignment was based on the 16S rRNA and the polB I gene sequences.
| Strain ID | Year Isolated | Genus of strain | Spacers that match haloarchaea1 | Spacers that match viruses | Total spacers | cas genes presence |
|---|---|---|---|---|---|---|
| 19N | 2012 | Haloferax | 1 | 0 | 154 | + |
| 48N | 2012 | Haloferax | 1* | 2 | 92 | + |
| 47N | 2012 | Haloferax | 3 (1) | 1 | 70 | + |
| 24N | 2012 | Haloferax | 4 (1) | 0 | 109 | + |
| 105R | 2014 | Haloferax | 4 (1) | 0 | 109 | + |
| 109R | 2014 | Haloferax | 4 (1) | 0 | 109 | + |
| 7R | 2014 | Haloarcula | 7 (1)** | 0 | 111 | + |
| 47R | 2014 | Haloarcula | 0 | 3 | 109 | + |
| 120R | 2014 | Haloarcula | 0 | 2 | 30 | + |
| 31R | 2014 | Halobellus | 0 | 0 | 43 | + |
| 38R | 2014 | Halobellus | 0 | 0 | 0 | - |
| 26R | 2014 | Halorubrum | 0 | 3 | 168 | + |
| 8R | 2014 | Halorubrum | 0 | 0 | 0 | - |
| 9R | 2014 | Halorubrum | 0 | 0 | 0 | - |
| 28R | 2014 | Halorubrum | 0 | 0 | 0 | - |
In parentheses number of spacers also matching self.
*denotes a spacer with a single mismatch to the protospacer (length 37-40bp), ** indicates two such spacers in the respective genome.
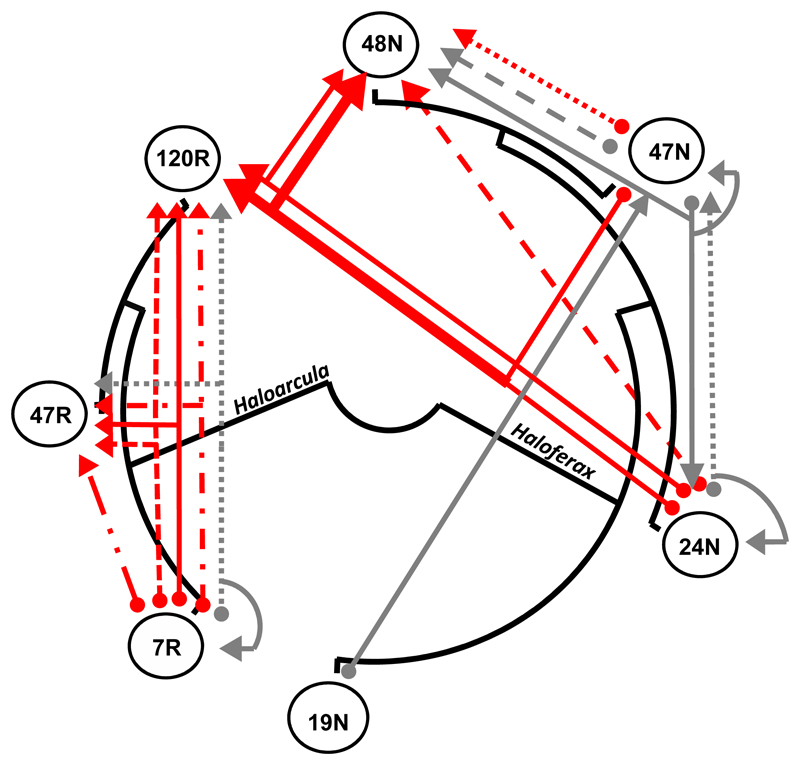
Of the 13 cases of perfect spacer-protospacer (target) identity, nearly all were within-genus matches (Fig. 1), while one Haloferax spacer matched both a sequence in another Haloferax strain and a Haloarcula strain. Additionally, a single spacer in Haloferax strain 24N only matched a gene in Haloracula strain 120R. About a third of the spacers (4/13) that matched chromosomal contigs were in the 3 first (leader-proximal) positions in their respective arrays, and therefore likely to be fairly recent acquisitions, and not necessarily selected for retention 7. Chromosome-matching spacers were enriched over 3 fold in these positions compared to their general occurrence in these arrays (hypergeometric p = 0.03), indicating that although they are a small subset of the total spacers in these haloarchaea, they nevertheless constitute a substantial fraction of recent acquisition events.
Fig. 1. A Simplified representation of perfect inter-species matches between CRISPR spacers and DNA sequences in the genomes of environmental Atlit isolates.
The phylogenetic relationships between isolates, that either target other strains or are targeted by them, are marked in black. Each arrow represents a spacer pointing to the isolate/s it matches. Branching arrows indicate that a spacer has more than one target, including the possibility of self-targeting (circular reference). Gray colored arrows have non-active corresponding interference PAM sequences while red arrows represent spacers with PAMs previously shown to be active in Haloferax or Haloarcula. Thicker arrows lines indicate a spacer common to both 24N and 47N that targets two genomes. Multiple independent spacers for a given genomic target in the same strain are marked with a different line pattern.
Three of the 13 perfect match spacers were also self-targeting spacers (Fig.1; Supplementary Table 4), i.e. matching a sequence within the same genome that is outside the CRISPR array. Since having such “auto-immune” spacers is generally considered to be highly deleterious to the organism 17–22, it was surprising to observe them in recently isolated strains that are presumably fit. However, target DNA degradation (known as “interference”) by type I CRISPR-Cas systems also requires an appropriate protospacer adjacent motif (PAM) sequence. When we inferred the PAM sequences associated with these self-targeting spacers, we observed that all three spacers had PAMs that were previously shown in the same genera as unable to confer efficient interference: in Haloferax volcanii 22 the GGC PAM observed in 24N, and the GAT observed in 47N are considered to be inefficient in conferring interference, as was the CCG PAM observed in 7R when previously tested in Haloarcula hispanica 23. In contrast, the most abundant PAM sequence for all the other cross-targeting spacers was TTC, previously shown to be interference-proficient in both Haloferax and Haloarcula (Supplementary Table 4, 22,23). Thus, we conclude that these self-targeting spacers are tolerated in the isolates because they are inactive due to the incompatibility of their PAM with the interference complexes of these type I CRISPR-Cas systems. Alignment of the protospacers from isolates and related genomes from the NCBI database showed that these interference-inactive PAMs in those cases are also conserved in genomes that have no such CRISPR self-targeting (Supplementary Fig. 2). Thus, in all likelihood, these inactive PAMs are the product of spacer integration with non-canonical PAMs (see below) rather than subsequent PAM mutations that evade auto-immunity 17.
Acquisition of new inter-species spacers during haloarchaeal mating
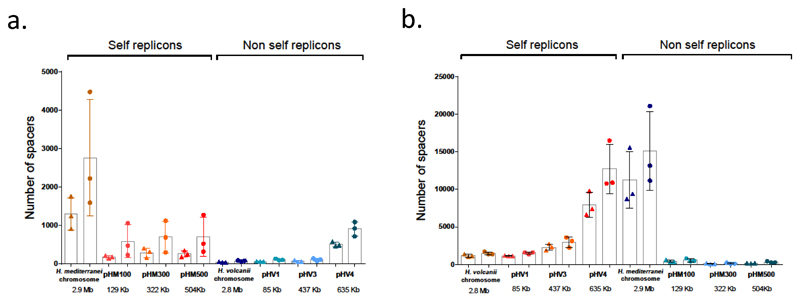
The multiple occurrences of inter-species spacers in haloarchaeal genomes raised the question of how such spacers were acquired. One obvious possibility is mating between species by cell fusion 9, during which the entire gene content (both plasmids and chromosomes) of each mating partner is exposed to the other 8, thereby providing an opportunity for acquisition of such cross-species anti-chromosomal spacers. We tested the mating hypothesis experimentally using the two model haloarchaea H. volcanii and Haloferax mediterranei, which can mate fairly efficiently (only 3.5 times lower than within species mating) 9 despite being quite distant genetically. Both of these species possess active CRISPR-Cas systems of subtype I-B, encoded on the large plasmids pHV4 and pHM500 (for H. volcanii and H. mediterranei respectively) 24, 25–27. Importantly, spacer acquisition has not been shown for either species, and under normal growth conditions, mRNA levels of cas1 and cas2, the key genes in spacer acquisition are extremely low in both species (13.9 RPKM in H. volcanii and 30.7 RPKM in H. mediterranei) 28. After mating H. volcanii and H. mediterranei and selecting for mated cells we obtained about 200 colonies from which total DNA was extracted. These colonies are not clonal, since they begin from mating products that are heterozygous cells, containing both parental genotypes (chromosomes and plasmids), and later these cells give rise to different recombinant cells that contain chimeric genomes with loci spanning the selectable markers 9. We then performed PCR amplification on the leader ends of each CRISPR array in the two species (six arrays in H. mediterranei and three in H. volcanii) to determine which spacers were acquired (see Methods). We also separately performed shot-gun community sequencing of the same DNA (Supplementary Fig. 3) to gain an estimate of how well were individual CRISPR arrays represented in the mating products. Sequence data analysis revealed substantial acquisition in all arrays in the mating products, with the exception of array E in H. volcanii (Supplementary Fig. 4; Supplementary Table 7). Curiously, the vast majority of H. mediterranei spacers were derived from its own replicons. Nevertheless, some H. mediterranei spacers matched the H. volcanii replicons, primarily targeting H. volcanii's plasmid pHV4 (Fig. 2). H. volcanii cells acquired more spacers from the H. mediterranei chromosome than from all three H. mediterranei plasmids combined (Fig. 2B), in agreement with findings from the environmental genomes (see above), and unlike previous observations in bacteria, where acquisition was strongly biased toward plasmid DNA 29.
Fig. 2. Number of spacers acquired during mating between H. volcanii and H. mediterranei.
Three independent biological replicates were performed. For each replicon, the mean of unique spacers (no pattern) or total spacers (dotted pattern) is shown. Error bars represent standard error of the mean. A. H. mediterranei spacer acquisitions. B. H. volcanii spacer acquisitions.
Spacers that target the chromosome are acquired genome-wide
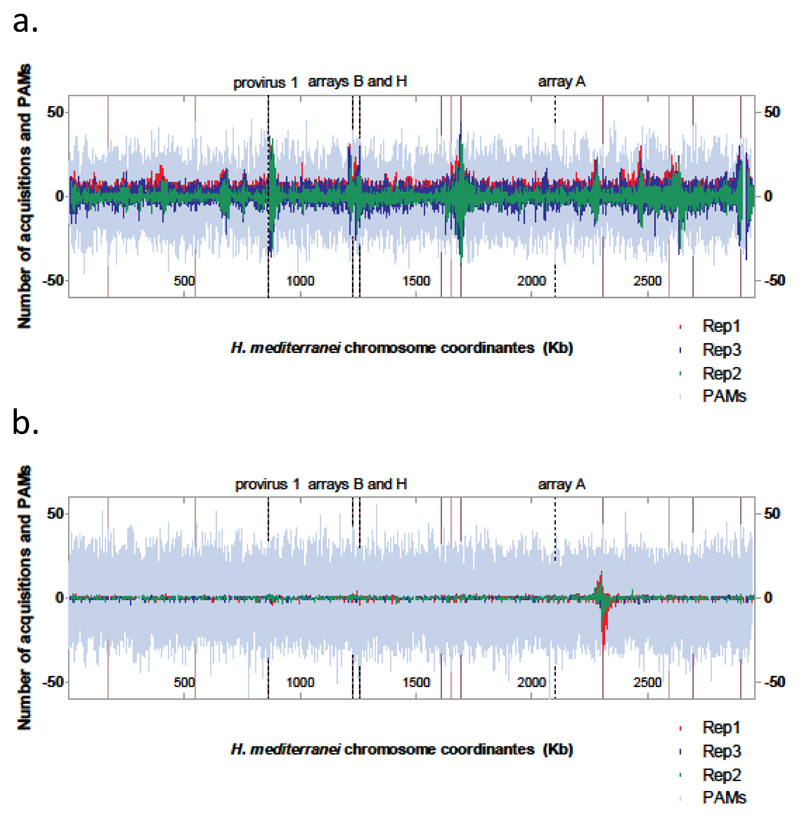
Spacers were acquired by H. volcanii from the entire H. mediterranei chromosome, as seen in the environmental genomes (Fig. 3A; Supplementary Fig. 5A). However, we did observe regions with a higher density of matching spacers (regions that were more than three times higher than neighboring bins) next to putative mobile genetic elements (MGEs, Fig. 3A), which we inferred based on the presence of genes encoding integrases, site-specific recombinases and transposases. One such element, previously referred to as provirus 1, but lacking detectable capsid genes (see Methods), has been previously shown as capable of excising from the genome 27,30. To test whether this island may excise during our inter-species mating experiments we used inverse PCR to detect its circular (excised) form in the DNA extracted from the same samples that were processed for identification of spacer acquisition. Indeed, we could clearly observe the circular form of that element, which rarely exits the genome of H. mediterranei under normal growth conditions 27, but was dominant in the between-species mating experiments (Supplementary Fig. 6). Thus, MGEs that excise from the chromosome may be preferred substrates for spacer acquisition in Haloferax. Additional loci of increased acquisition were found close to active CRISPR arrays such as array B and H on the H. mediterranei chromosome, and C and D on pHV4 (Fig. 3A; Supplementary Fig. 5), in agreement with previous studies in bacteria 31,32 and archaea 33. We observed an apparent “no-acquisition zone” in the region between 2899120-2922023 in the H. mediterranei genome that hinted that this region has been deleted from the genomes. PCR analysis confirmed the suspicion that this locus had already been deleted in the parental strain WR646 prior to the mating experiments.
Fig. 3. Spacers acquired from the H. mediterranei chromosome by either H. volcanii or H. mediterranei CRISPR-Cas.
The H. mediterranei genome was divided into equally sized bins, and the number of unique spacers per bin is represented on the Y axis and colored by biological replicate. PAMs per bin are marked in grey (TAC for H. volcanii acquisitions, TTC for H. mediterranei's). Dotted lines mark CRISPR arrays and provirus 1. Purple lines mark locations of genes encoding integrases and recombinases and those of genes encoding transposases are denoted by pink lines. A. H. volcanii PAM signature and spacer acquisition (2458 bp bins). B. H. mediterranei PAM signature and spacer acquisition (1160 bp bins). Different sized bins were chosen to reflect the fact that the TTC PAM is more than twice as abundant as the PAM in the H. mediterranei chromosome.
In both species, which have roughly similar genome sizes, even when accounting for natural plasmids, many spacers were acquired against self replicons (chromosomes and plasmids). In H. mediterranei spacers against self replicons outnumbered those derived from the mating partner about three-fold (Fig. 2), while In H. volcanii spacers against self replicons, primarily from pHV4, were approximately as abundant as those obtained from the H. mediterranei replicons, primarily from the major chromosome of the latter species. In terms of acquisition from self replicons both species acquired more spacers from their respective plasmids than from their chromosomes, when normalizing for replicon length (Supplementary Fig. 7).
Interestingly, while H. volcanii acquired many spacers from the putative MGEs of H. mediterranei, the latter archaeon only showed a hot-spot of acquisition against its own chromosome close to a putative MGE (Fig. 3B; Supplementary Fig. 5B). This implies that the two different CRISPR-Cas systems have different acquisition preferences, even when acquiring from the same replicon.
Within-species mating results in lower levels of spacer acquisition
Given that CRISPR spacer acquisition was induced by inter-species mating, we examined whether mating would also induce acquisition when cells belong to the same species. We therefore performed within-species mating experiments in H. volcanii. Experiments comparing between-species (excluding mediterranei-derived spacers from the calculation, Supplementary Table 8) to within-species mating, revealed much reduced spacer acquisition in the volcanii-volcanii mating compared to volcanii-mediterranei mating. Thus, mating between species leads to subsequent auto-immunity against self-replicons that would not otherwise emerge.
We also tested the effect of “nutritional competence” 34, the ability of H. volcanii to take up foreign DNA on spacer acquisition. When H. volcanii cells were incubated with high molecular weight H. mediterranei DNA, we observed no acquisition of spacers derived from H. mediterranei, and a low level of spacers against self-replicons, comparable to that observed in within-species mating (Supplementary Table 8). We thus conclude that nutritional competence is unlikely to be a major source of spacer acquisition in Haloferax.
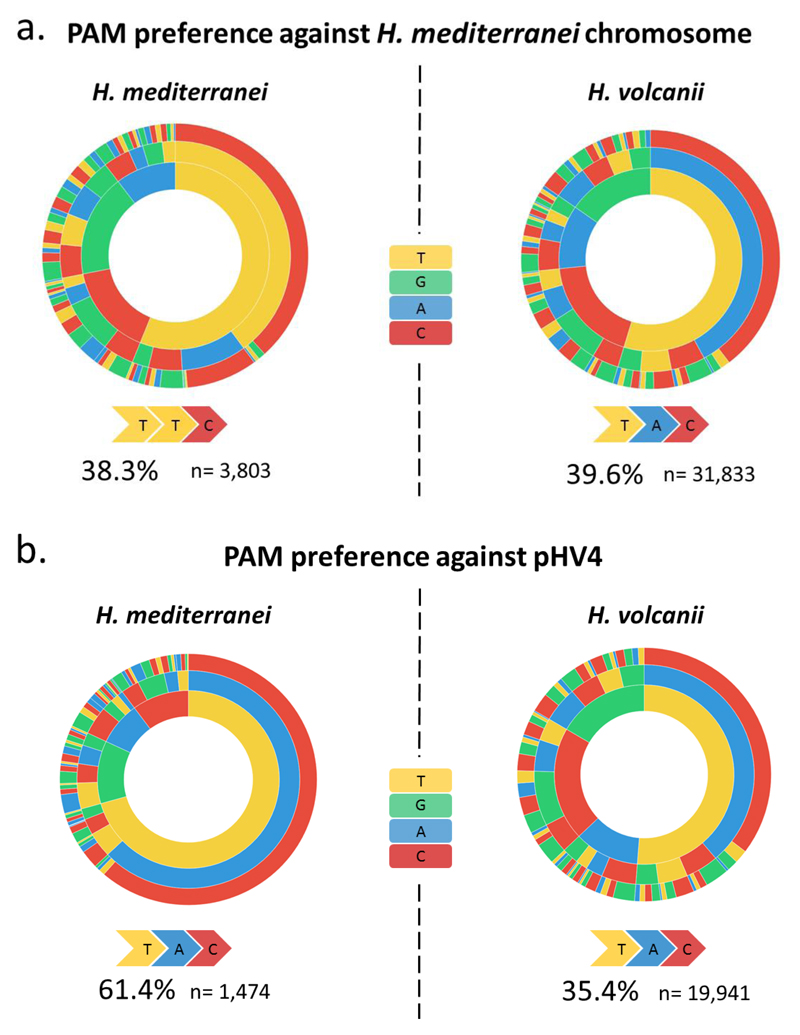
Protospacer adjacent motifs suggest an interaction between acquisition machineries of the two CRISPR-Cas systems during inter-species mating
The fact that two active CRISPR-Cas systems come into contact during inter-species mating creates an opportunity for them to interact functionally. However, these I-B systems are highly divergent with only 68% and 37% identity between their Cas1 and Cas2 proteins respectively. It is therefore not surprising that their respective leader sequences that are critical for spacer integration also differ (Supplementary Fig. 8). To investigate whether one system could have incorporated spacers produced by the biochemical machinery of the other, we first identified the PAM sequences of each CRISPR-Cas system based on the newly acquired spacer data (Supplementary Fig. 9). The two systems had different PAM signatures: for H. mediterranei the preferred PAM sequence was TTC while for H. volcanii it was TAC. While the TAC PAM was observed for H. volcanii acquisitions from all replicons, the H. mediterranei TTC PAM was only observed for spacers acquired from its own replicons (Fig 4; Supplementary Table 9). In contrast, the spacers acquired by H. mediterranei from H. volcanii replicons instead showed the TAC signature, indicating that they were most likely incorporated into H. mediterranei arrays by the H. volcanii acquisition machinery. In agreement with this conclusion, the pattern of acquisition against pHV4 was also similar between species (Supplementary Fig. 5C and D).
Fig. 4. Protospacer Adjacent Motif (PAM) preference for each of the species against different replicons.
Sequences of the three bases upstream for each of the individual unique protospacer were extracted from the acquisition data. The relative abundances of these three-base PAMs were calculated, and are represented in a PAM wheel for each of the species by the spacer target. The favorable PAM (the one with the highest frequency) is marked for each PAM wheel. “n” represents the total number of spacers accounted for in the chart. A. PAM wheels for each species based on spacers acquired from H. mediterranei chromosome. B. PAM wheels for each species based on spacers acquired from H. volcanii natural plasmid pHV4.
Another interesting feature of these PAMs in both Haloferax species was a large fraction of acquired spacers that had non-preferred PAM sequences (Fig 4.), although this fraction was lower when examining only spacers that were observed more than once (non-singletons, Supplementary Fig. 9B). This pattern of acquisition is more noisy than other archaeal CRISPR-Cas systems (33, 35), but similar results were obtained in E. coli (29), and could explain some of the self-targeting spacers with interference-inactive PAMs that we observed in the environmental Haloferax isolates (see above).
Acquisition PAMs must be able to also mediate effective interference so that CRISPR-Cas can function as an adaptive immune system. It has been experimentally shown in H. volcanii that the CRISPR-Cas system is able to lead to CRISPR-mediated degradation of artificially transformed plasmids 26. However, in a screen for nucleotide motifs that could serve as efficient PAMs for DNA degradation in H. volcanii, TAC was not identified as an active PAM 26. Since a specific spacer-PAM combination can sometimes be inactive even when either component is individually active 36, we tested the TAC PAM in H. volcanii in an inhibition of transformation assay (Supplementary Table 10). Indeed, a spacer targeting a sequence with the TAC PAM yielded over 100-fold inhibition of plasmid transformation, confirming that this PAM is efficient in mediating degradation of invading DNA by the CRISPR-Cas system of H. volcanii.
Inter-species CRISPR targeting has a negative effect on mating success
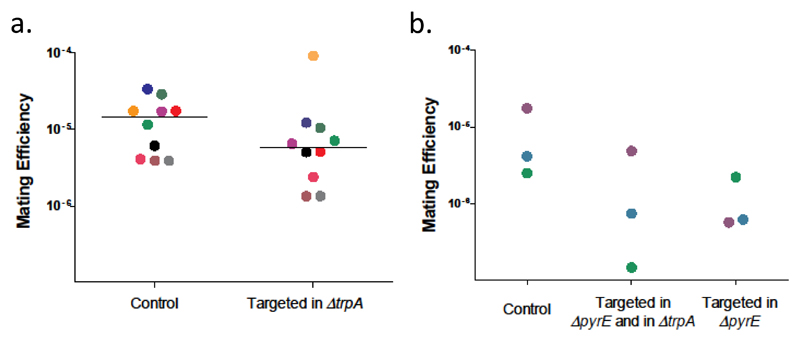
Our previous experiments clearly showed that spacers can be acquired from another chromosome during inter-species mating by fusion, a mechanism of horizontal gene transfer (HGT) that facilitates the transfer of large plasmids, the emergence of heterozygous fused cells and inter-species recombinant hybrids 9. However, such spacer acquisition from the other species' genome can reduce the success of future inter-species mating: if a CRISPR-Cas system starts degrading the other genome, this could potentially cause cells to sense DNA damage and separate prematurely, reducing the chances of plasmid exchange and/or recombination between chromosomal loci. To test this hypothesis, we planted a 40 bp sequence that is efficiently targeted by H. volcanii CRISPR and mediates interference 22 into the H. mediterranei genome generating a targeted H. mediterranei strain (Supplementary Fig. 10). We then performed mating assays crossing H. volcanii with the targeted H. mediterranei strain and as a control we did a parallel experiment crossing H. volcanii with an isogenic non-targeted H. mediterranei strain. In 9 out of 10 biological replicates we observed a substantial decrease in mating efficiency in comparison to the non-targeted control [nearly 2.5 fold median reduction, P < 0.002, Wilcoxon signed paired samples rank test (Fig. 5A)].
Fig. 5. Inter-species mating is reduced by CRISPR-Cas targeting.
H. volcanii was mated with either H. mediterranei strain WR646 ("Control") or with an isogenic strain engineered to contain a validated H. volcanii CRISPR spacer+PAM sequence ("Targeted"), thereby allowing H. volcanii to target it during mating (Supplementary Fig. 10). Mating efficiency was calculated as the number of CFUs on the mating plates (mating products) divided by the average number of CFU for each parental strain and are shown for the targeted H. mediterranei strain and the targeting-free control. Each independent biological replicate is marked by a different color. A. Mating when the target is in the ΔtrpA region; the median for each group shown as a line. 9 out of the 10 repeats showed a significant decrease in mating efficiency in comparison to the control. P < 0.002, Wilcoxon signed-rank test comparison of paired samples. B. Mating with H. mediterranei targeted in both ΔpyrE2 and ΔtrpA regions and only in ΔpyrE2.
To rule out the possibility that mating efficiency is affected by the location of the spacer in the H. mediterranei genome, which was in the selectable marker region [ΔtrpA(704395)], we created two more targeted strains: one in which the spacer+PAM sequence was inserted into the ΔpyrE2(299911) region, and another that had two specific insertions of that 40 bp sequence, in both ΔpyrE2 and ΔtrpA regions. We observed the same trend of reduction in mating products using both strains in comparison to the non-targeted control (see Fig. 5B). These results clearly indicate that targeting of partner chromosomes by the CRISPR-Cas machinery can reduce HGT by mating across species, even when the selection marker itself is far from the targeted locus.
Discussion
Here we show spacer acquisition from another species' chromosome following inter-species mating in archaea. These experiments can explain, at least in part, the observation of anti-chromosomal spacers in archaeal genomes. The phenomenon of spacers that mostly match chromosomal loci has also been observed in bacteria, in the genomes of pathogenic Neisseria species 43. Notably, while some of those Neisseria spacers matched genomic islands, others were identical to core housekeeping genes, similar to our observations in haloarchaea. Neisseria are naturally competent and experience frequent HGT and recombination, in resemblance to Haloferax 44, which could provide the opportunity for spacer acquisition from islands as well as other genomic loci. Indeed although spacers in our experiments were acquired from the entire main chromosome, there were obvious hotspots in islands, as observed in Neisseria, and more recently in Pectobacterium atrosepticum 36. Our results provide direct evidence that spacers are frequently derived from selfish elements that are incapable of a lytic lifecycle. They also represent strong experimental support for the view that CRISPR-Cas systems play an important role in controlling integrative selfish elements 7, and thereby modulate genome content. This role is complementary to, and may sometimes exceed the importance of, anti-viral defense, especially in the many prokaryotic lineages that have relatively low exposure to viruses but experience frequent DNA exchange.
Unlike many bacterial CRISPR-Cas systems, such as I-F systems36,45, where the key nuclease-helicase cas3 is fused to the acquisition gene cas2 and the single polypeptide produced is involved in both functions ,46, in type I-B systems this is not the case. Consequently there can be robust constitutive interference while the acquisition machinery remains tightly repressed, and such is the case in Haloferax. Indeed based on our findings, such regulation may actually be required, due to a lack of a preference for non-self replicons, or for plasmids compared to the main chromosome. H. volcanii like the vast majority of archaea lacks RecBCD, which is responsible for the bias that greatly reduces the acquisition of chromosome-derived spacers in Bacteria 35. The biases that we did observe seemed to favor acquisition from integrated selfish mobile elements, similar to work in Pyrococcus furiosus showing acquisition that was biased in favor of rolling circle replication plasmids37. In both cases spacer acquisition spanned the entire element, without a particular sharp increase at an exposed end, as was reported for viral injection 8.
Another key observation is hat self-targeting spacers are acquired naturally in Haloferax, as a byproduct of inter-species mating. While this represents collateral damage incurred due to CRISPR-Cas activity, such accidents may nevertheless have profound effects on genome dynamics. Acquisition of a spacer targeting an endogenous plasmid gene can lead to DNA degradation and result in either loss of the targeting activity by mutational events, or in the deletion of the target region 19,30. Which of these scenarios dominates will probably be determined by the cost or benefit that this plasmid provides in a given environment. Such semi-random deletion processes will yield a population of cells that carry plasmids differing in their gene content, thereby increasing the genotypic variation within the meta-population37. Furthermore, since the costs and benefits of plasmid-encoded genes will vary with environmental fluctuations, this process will increase the chances that a transiently fit genotype will emerge, and thus will benefit the overall population fitness.
Acquisition from the mating partner's chromosome, could affect horizontal gene transfer (HGT) between species. We show that once a species' CRISPR-Cas system effectively targets another's genome, the frequency of productive mating events between them drops substantially. While the 2.5 fold reduction we observed may seem small, the decrease in mating that we observed is similar to the reduction in mating efficiency noted when cross-species mating (i.e. mating between H. volcanii with H. mediterranei wild type cells) is compared to a situation when the mating partners are both H. volcanii (3.5 fold median reduction)11. Importantly, the effect of CRISPR-Cas that we observed reduced gene exchange globally and not locally at the CRISPR-targeted site: both our selectable marker loci were not close to the targeting site.
In summary, spacer acquisition from chromosomes, whose targets may occasionally be genuine housekeeping genes, can be a common side effect of CRISPR-Cas activity that is primarily directed against selfish elements, especially islands. Such accidental acquisition however, could then have global impacts on gene exchange, and increase genetic separation between lineages, contributing to speciation.
Methods
Identification of haloarchaeal CRISPR spacers that match sequences in sequence databases
All 1161 high confidence (confirmed) spacers from pre-existing haloarchaeal genomes were downloaded from the CRISPRdb website (http://crispr.u-psud.fr/crispr/database, see 1 and compared using NCBI BLASTN (last GenBank version-18/6/16) to Halobacteria (taxid:183963) and viruses (taxid:10239) in the NCBI database. Self-hits (hits of spacers against themselves within the CRISPR array context) were filtered out by removing all 100% identity hits where the organism as well as the locus of the spacer and its matching sequence were the same. We examined only hits that met all the following criteria: Coverage ≥ 0.5, Score ≥40 bits and E-value ≤ 0.001.
Analysis of spacers in environmental isolates from the Atlit seashore
Isolates were collected from evaporation puddles, less than 100 square meters in area, on the coast of Atlit, Israel, in the summers of 2012 and 2014. This rocky shore has much evaporation in summer, due to direct sunlight, resulting in small tidal evaporation pools that are hypersaline, and often exhibit a visible salt crust. Since this site is so small and experiences westerly winds daily, cells from one pool can come into contact with those from other pools and potentially even mate by cell fusion 2,3. Isolates were sequenced by Illumina 2 × 250 base paired end whole genome sequencing. Genus identity was determined according to 16S rRNA and the polB genes sequences. Raw reads were first trimmed with Cutadapt v1.9.14 to remove adaptor sequences and bases with a Phred score lower than 20. Genomes were assembled using SPAdes v3.7.0 5 with kmer sizes 21,33,55,77,99, and 127. Assembly quality was checked using Quast v2.36, coding sequences and annotations were predicted using Prokka v1.11 7 ignoring contigs that were shorter than 200 base pairs. CRISPR arrays were identified using the CRISPR Recognition Tool (CRT) v1.1. 8 and CRISPR-finder from the CRISPRdb website 1. We extracted all confirmed array locations and spacers for each isolate. Cross-targeting spacers were identified using a BLASTN search against the isolates combined genome files, followed by manual elimination in cases of shared spacers or CRISPR arrays using the CRISPR array coordinates in the targeted isolate. We also searched traces of degenerate repeat sequences (up to 10 mismatches) in proximity to the protospacer in order to eliminate cases of “false hits”. We designated “self” targeting spacers, those spacers from confirmed arrays that had match to their own genome in locations that did not map to other confirmed/hypothetical CRISPR arrays. Protospacer location was estimated as chromosomal in cases where conserved housekeeping genes were present on the same contig (DNA and RNA polymerase subunits and genes encoding rRNA and core ribosomal proteins), otherwise the location was assumed to be plasmid. Isolates' confirmed spacers were also compared using NCBI BLASTN to all viruses in the NCBI database with the criteria: Coverage ≥ 0.5, Bit Score ≥40 and E-value ≤ 0.001. All “self”-targeted isolates genomes had fully intact open reading frames of all cas genes known to be involved in degrading targeted DNA (“interference“) (i.e. cas3, 5, 6, 7 and 8). Three Haloferax isolates (105R, 109R, 24N), had identical CRISPR arrays despite variable genomic content, and thus were treated as a single CRISPR genotype in subsequent analysis.
Culture Conditions
H. volcanii and H. mediterranei cells were routinely grown as described in 9.
Haloferax strain construction
Strain construction were performed according to the protocol described in 10,11.
Mating experiments
The mating experiments were conducted using an H. volcanii strain lacking the ability to synthesize thymidine H729 (∆hdrB [2754021]) and an H. mediterranei strain that is unable to synthesize tryptophan and uracil WR646 (∆trpA [704395], ∆pyrE2 [299911], Supplementary Table S11). Liquid cultures of both parental strains were grown to an O.D600 of ~1.8. The parental strains were then mixed in 1:1 ratio and applied to a nitrocellulose 0.45µm filters using a Swinnex 25mm filter holder. The filter with the mating products was transferred to a rich medium plate (Hv-YPC with thymidine) for 24 hours for phenotypic expression. The cells were then re-suspended and washed in Hv-Ca (Haloferax volcanii casamino acids) broth before plated on Hv-Ca media containing tryptophan (H. volcanii and H. mediterranei mating products were selected using the pyrE2 and hdrB chromosomal markers).The mating “within species” in H. volcanii was performed using H729 (∆hdrB [2754021]) and WR536(∆trpA [302281], ∆pyrE2 [301751]) selecting on the pyrE2 and hdrB markers.
Nutritional competence experiments
H. volcanii strain H729 cells were grown for a week in YPC-HV broth containing extracted DNA from H. mediterranei strain WR646 (50ng/µl). DNA from that culture was then extracted and used for detection of new spacer's in arrays C and D of H. volcanii as described below.
Community DNA sequencing of mating products
About 200 colonies of mating products (derived from H. volcanii strain H729 and H. mediterranei strain WR646) from each of three independent mating experiments were suspended in Hv-Ca broth media prior to DNA extraction using the DNA spooling protocol described in 10. Purified DNA was sent for metagenomic sequencing using the Nextera XT protocol and the Illumina NextSeq500 sequencing platform at the Center for Genomic Research, University of Illinois, Chicago, USA. After trimming of adaptors and low quality bases by Trimomatic 12, between 1.8 to 2.1 million high-quality sequence reads were obtained for each biological replicate and matched, using blastn, to reference sequences of H. volcanii and H. mediterranei, including all their natural plasmids; to improve accuracy, the Blast e-value threshold used for mapping reads was set to 1e-60. Reads mapping to more than one locus were ignored. Reads counts were normalized per kb of sequence.
Detection of acquisition of new spacers
About 200 colonies of mating products (using H. volcanii strain H729 and H. mediterranei strain WR646 as the parental strains for H. volcanii-H. mediterranei mating, WR510 and UG453 for H. mediterranei within-species mating, and H729 and WR536 for H. volcanii within-species mating) were suspended together in Hv-Ca broth media before extracting their DNA using the DNA spooling protocol as described previously 10. The extracted DNA was used as template for PCR for both species CRISPR arrays using specific primers amplifying the region between the leader and the third spacer in the array for arrays A-H, and the region between leader and the end of the first spacer in array I (see primers list Supplementary Table 12). When analyzed by agarose gel electrophoresis, longer PCR products indicate new spacer-repeat acquisitions. To obtain visible acquisition bands in the agarose gel, we extracted the approximated elongated length region from the gel, isolated DNA and amplified the fragment through another cycle of PCR. New acquisition events could then be detected via the presence of a higher band. PCR products were then sent for processing and Illumina amplicon sequencing (240,000-290,000 reads per array per biological repeat) at the Center for Genomic Research, University of Illinois, USA. Briefly, the elongated PCR product was enriched using Ampure beads size-selection; sample specific barcodes and Illumina adaptors were added by PCR; and the resulting products were purified, pooled, and paired-ends sequenced on a MiSeq Illumina platform. Notably, even after these consecutive steps of size selection many reads still represented amplicons derived from no acquisition amplifications.
Initial data processing
Paired-end raw Illumina reads were quality-filtered (Q>20) and merged using PEAR (paired-end read merger, [9]), yielding, for most samples, 240000-290000 high-quality sequences; samples which yielded over 300000 seqs per sample were subsampled randomly, using VSEARCH, to 280000 seqs/sample. Biological replicates within each array were then converted to a single fasta file using QIIME's multiple_split_libraries_fastq.py script 13, followed by de-replication, abundance sorting and clustering (99% threshold) using VSEARCH 14. Pairwise identity at the clustering step was defined as [matching columns]/[alignment length] (set in VSERACH as –iddef 1), which ensures terminal gaps are not ignored and sequences of different length do not cluster together. The length distribution of the clustered sequences is similar to that of the raw sequences; arrays A-H show a peak at 250 bp (size of the original fragment, containing 3 repeats), with progressively smaller peaks at 320 bp (corresponding to one new acquisition) and 390 bp (2 new acquistions). For array I, the original fragment size was 320 bp (since the forward primer, for technical reasons, was located at the beginning of the leader sequence), with corresponding peaks at 390 and 460 bp. The raw reads were then mapped back to the clusters, again with a 99% identity threshold, to create a table presenting the abundance of each cluster in each biological repeat.
Spacer extraction
The centroid sequence of each cluster was used for extraction of acquired new spacer sequences. To identify true acquisition events while excluding DNA rearrangement events (which may also result in an extended PCR product), a custom made R script based on the 'Biostrings' and 'tools' R packages was used to count the number of repeats in each centroid sequence, allowing up to 2 mismatches per repeat. Fragments containing more than 3 repeats (for arrays A-H) or more than 1 repeat (array I) were tagged as putative acquisitions, and the sequence between the 2 repeats closest to the leader end was extracted. “False positives”, which are the result of rearrangement of spacers within or between arrays rather than canonical acquisition, were eliminated by screening the extracted putative spacers against all original CRISPR arrays from both species, allowing up to 5 mismatches per spacer; spacers with matches in existing CRISPR arrays were excluded from further analysis.
To facilitate comparison of CRISPR activity between arrays, we also counted the number of sequences containing the original number of repeats (3 for arrays A-H; 1 for array I). All spacer sequences, positions and PAM's are provided (Supplementary Tables 13 and 14).
Mapping new spacers to genomic location
In order to establish the protospacer location for each new acquisition, we used the blastn-short program 15 at an E-value of .0001 against a file containing genome sequences of both H. mediterranei and H. volcanii, including their natural plasmids. Blast results were refined using Custom made R scripts based on the 'stringr' package. In brief, no more than 3 mismatches between the spacer and the protospacer were allowed; in cases of multiple matches, the location with the highest score was preserved (while conserving abundance information); and spacers with equally high scores across multiple locations were excluded. Furthermore, spacers that were aligned only partially were removed when the alignment length was shorter than the total spacer length by five or more bases.
Quantification of spacer acquisition events
Using the mapping approach described above we quantified total acquisition events for each spacer and the mean number of events across biological replicates for each spacer was calculated. 4 spacers that showed a highly aberrant pattern when comparing the 3 biological replicates (abundance in one sample more than 1000-fold higher than in the other 2 samples) were corrected for by replacing the aberrantly high value with the average counts of that spacer in the other 2 biological replicates.
PAM determination
The ten upstream bases from each unique protospacer were extracted, while adjusting for the rare cases where there was a misalignment within the first five base pairs of the spacer. We observed that only the final three bases contained a non-random PAM sequence. The relative abundance of these three-base PAMs was calculated, and represented in a PAM-wheel 16 using SunburstR, Yaml, and Rcpp R libraries. In cases where a single unique spacer was aligned in multiple locations, (usually corresponding to transposable element repeat sequences that appear multiple times throughout the genome), those matches were removed from the quantitative analysis, since in those cases the protospacer they were acquired from cannot be unambiguously determined.
Annotation of newly acquired spacer targets
The location of protospacers was annotated for each of the acquired spacers using the publically available H. volcanii 17 and H. mediterranei 18 genomes [NCBI GenBank accession files 19] CP001868-1871 and CP001953-1957.
Bioinformatic analysis of provirus 1
Proteins sequences from the predicted island regions (HFX_0898-0929) were submitted to the HHpred server for remote homology detection and structure prediction 20 to rule out proteins with significant similarity to known capsid genes. Such approaches have been shown to be sensitive in detecting novel viral capsid proteins in archaeal genomes 21.
Plasmid invader tests
The H. volcanii strain H119 was transformed with the invader plasmid pTA409-PAM28-P1.1, which carries the PAM TAC upstream of spacer 1 of CRISPR locus P1 22. As a control H. volcanii cells were transformed with the vector pTA409. Plasmids were passaged through E. coli GM121 cells to avoid methylation and subsequently introduced into H. volcanii using the PEG method. The transformations were repeated four times and plated on Hv-Ca plates without uracil to ensure selection for the plasmids. Transformations with at least a 100-fold reduction in transformation rate are defined as successful interference reactions 22.
Supplementary Material
Acknowledgments
The authors wish to thank Rotem Sorek and Anat Herskovits for their helpful comments and insights, and Hua Xiang for providing sequence data and provirus annotations. The authors thank Stefan Green of the University of Illinois at Chicago, for his continued expert help in challenging sequencing projects and Eugene Koonin (NIH) for helpful discussions. Funding: Deutsche Forschungsgemeinschaft [MA1538/16-2]; Israel Science Foundation [535/15]; Binational science Foundation [2013061] and partial support by the Constantiner Institute; European Research Council (grant ERC-AdG 787514).
Footnotes
Data availability
Haloarchaeal environmental isolates genomes are available at GenBank cccessions [PSYS00000000, PSYT00000000, PSYU00000000, PSYV00000000, PSYW00000000, PSYX00000000, PSYY00000000, QEQI00000000, QEQJ00000000, QPLN00000000, QPLO00000000, QPLP00000000, QPLQ00000000, QPLR00000000, QPLS00000000, QPLT00000000, QPLU00000000, QXIJ00000000, QXIK00000000, and QPLV00000000].
Code availability
All scripts relating to new spacer acquisition and analysis had been deposited in https://github.com/leahfa/CrispR-analysis.
Author Contributions
U.G. and I.T.-G conceived the study; U.G. I.T-G, and A.M. designed experiments; S.M.S. assembled and annotated genome sequences; I.T.-G, A.N., N.A-P designed and constructed strains; I.T.-G, S.Y., K.E., Y.S., A-E.S., and M.Z. performed experiments; L.R, S.M, I.T.G, and U.G. analyzed data; U.G. and I.T.-G. wrote the manuscript, L.R., S.M.S. and A.M. commented and made critical revisions to the manuscript.
Competing interests
The authors declare no competing interests.
ORCID Uri Gophna: https://orcid.org/0000-0001-5129-5652
References
- 1.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009;37:23–8. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]
- 5.Held NL, Herrera A, Quiroz HC, Whitaker RJ. CRISPR associated diversity within a population of Sulfolobus islandicus. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shmakov SA, et al. The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes. MBio. 2017;8:e01397–17. doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodt A, Lurie-Weinberger MN, Gophna U. CRISPR loci reveal networks of gene exchange in archaea. Biol Direct. 2011;6:65. doi: 10.1186/1745-6150-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenshine I, Tchelet R, Mevarech M. The mechanism of DNA transfer in the mating system of an archaebacterium. Science. 1989;245:1387–1389. doi: 10.1126/science.2818746. [DOI] [PubMed] [Google Scholar]
- 9.Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 2012;22:1444–1448. doi: 10.1016/j.cub.2012.05.056. [DOI] [PubMed] [Google Scholar]
- 10.Schleper C, Holz I, Janekovic D, Murphy J, Zillig W. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J Bacteriol. 1995;177:4417–26. doi: 10.1128/jb.177.15.4417-4426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuwabara T, et al. Thermococcus coalescens sp. nov., a cell-fusing hyperthermophilic archaeon from Suiyo Seamount. Int J Syst Evol Microbiol. 2005;55:2507–2514. doi: 10.1099/ijs.0.63432-0. [DOI] [PubMed] [Google Scholar]
- 12.Burstein D, et al. New CRISPR–Cas systems from uncultivated microbes. Nature. 2016;542:237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haft DH, Selengut J, Mongodin EF, Nelson KE, White O. A Guild of 45 CRISPR-Associated (Cas) Protein Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 15.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 16.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 17.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc Natl Acad Sci U S A. 2011;108:20136–41. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vercoe RB, et al. Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selle K, Klaenhammer TR, Barrangou R. CRISPR-based screening of genomic island excision events in bacteria. Proc Natl Acad Sci U S A. 2015;112:8076–81. doi: 10.1073/pnas.1508525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, et al. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44:e34–e34. doi: 10.1093/nar/gkv1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer S, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–33365. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Wang R, Xiang H. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 2014;42:7226–35. doi: 10.1093/nar/gku389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mojica FJM, Juez G, Rodriguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993;9:613–621. doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- 25.Brendel J, et al. A complex of cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (CRISPR)-derived RNAs (crRNAs) in haloferax volcanii. J Biol Chem. 2014;289:7164–7177. doi: 10.1074/jbc.M113.508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer S, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–63. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, et al. Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J Bacteriol. 2013;195:867–875. doi: 10.1128/JB.01688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Artieri CG, et al. Cis-regulatory evolution in prokaryotes revealed by interspecific archaeal hybrids. Sci Rep. 2017;7:3986. doi: 10.1038/s41598-017-04278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, et al. Activation of a dormant replication origin is essential for Haloferax mediterranei lacking the primary origins. Nat Commun. 2015;6:8321. doi: 10.1038/ncomms9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy A, et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staals RHJ, et al. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR–Cas system. Nat Commun. 2016;7:12853. doi: 10.1038/ncomms12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiimori M, et al. Role of free DNA ends and protospacer adjacent motifs for CRISPR DNA uptake in Pyrococcus furiosus. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chimileski S, Dolas K, Naor A, Gophna U, Papke RT. Extracellular DNA metabolism in Haloferax volcanii. Front Microbiol. 2014;5:57. doi: 10.3389/fmicb.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erdmann S, Le Moine Bauer S, Garrett RA. Inter-viral conflicts that exploit host CRISPR immune systems of S ulfolobus. Mol Microbiol. 2014;91:900–917. doi: 10.1111/mmi.12503. [DOI] [PubMed] [Google Scholar]
- 36.Maier L-K, et al. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biol. 2013;10:865–74. doi: 10.4161/rna.24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Sanchez M-J, et al. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol. 2012;85:1057–1071. doi: 10.1111/j.1365-2958.2012.08172.x. [DOI] [PubMed] [Google Scholar]
- 1.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 2012;22:1444–1448. doi: 10.1016/j.cub.2012.05.056. [DOI] [PubMed] [Google Scholar]
- 3.Chimileski S, Franklin MJ, Papke R. Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer. BMC Biol. 2014;12:65. doi: 10.1186/s12915-014-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10. [Google Scholar]
- 5.Bankevich A, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 8.Bland C, et al. CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allers T, Barak S, Liddell S, Wardell K, Mevarech M. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl Environ Microbiol. 2010;76:1759–1769. doi: 10.1128/AEM.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of Additional Selectable Markers for the Halophilic Archaeon Haloferax volcanii Based on the leuB and trpA Genes. Appl Environ Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bitan-Banin G, Ortenberg R, Mevarech M. Development of a Gene Knockout System for the Halophilic Archaeon Haloferax volcanii by Use of the pyrE Gene. J Bacteriol. 2003;185:772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tom Hall. BioEdit Sequence Alignment Editor for Windows 95/98/NT/XP/Vista/7. [Accessed: 9th February 2017];2013 Available at: http://www.mbio.ncsu.edu/BioEdit/bioedit.html.
- 16.Leenay RT, et al. Identifying and Visualizing Functional PAM Diversity across CRISPR-Cas Systems. Mol Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman AL, et al. The Complete Genome Sequence of Haloferax volcanii DS2, a Model Archaeon. PLoS One. 2010;5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han J, et al. Complete genome sequence of the metabolically versatile halophilic archaeon Haloferax mediterranei, a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) producer. J Bacteriol. 2012;194:4463–4. doi: 10.1128/JB.00880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmermann L, et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2017 doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Makarova KS, et al. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles. 2014;18:877–93. doi: 10.1007/s00792-014-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer S, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–63. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.