Abstract
The alcohol research field has amassed an impressive number of gene expression datasets spanning key brain areas for addiction, species (humans as well as multiple animal models), and stages in the addiction cycle (binge/intoxication, withdrawal/negative affect, and preoccupation/anticipation). These data have improved our understanding of the molecular adaptations that eventually lead to dysregulation of brain function and the chronic, relapsing disorder of addiction. Identification of new medications to treat alcohol use disorder (AUD) will likely benefit from the integration of genetic, genomic, and behavioral information included in these important datasets. Systems pharmacology considers drug effects as the outcome of the complex network of interactions a drug has rather than a single drug-molecule interaction. Computational strategies based on this principle that integrate gene expression signatures of pharmaceuticals and disease states have shown promise for identifying treatments that ameliorate disease symptoms (called in silico gene mapping or connectivity mapping). In this Review, we suggest that gene expression profiling for in silico mapping is critical to improve drug repurposing and discovery for AUD and other psychiatric illnesses. We highlight studies that successfully apply gene mapping computational approaches to identify or repurpose pharmaceutical treatments for psychiatric illnesses. Furthermore, we address important challenges that must be overcome to maximize the potential of these strategies to translate to the clinic and improve healthcare outcomes.
Keywords: Systems pharmacology, network medicine, in silico gene mapping, connectivity map, L1000, LINCS, alcoholism, alcohol dependence, gene expression, transcriptome
Introduction
Developing more effective pharmacotherapies to treat disease is an important goal in public health. This is especially true for complex psychiatric diseases like Alcohol Use Disorder (AUD), where there are limited pharmaceutical treatment options. We use AUD throughout this Review for consistency as this is the terminology used in the current edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM5), but this does not exclude previous DSM version diagnostic criteria or preclinical/clinical trials based on those. AUD is a chronic, relapsing disease that devastates individuals, families, and society, and is a major public health problem. Though recovery is possible regardless of disease severity, there are few pharmaceutical treatments available to aid in the recovery process. There are several points of intervention along the time course of AUD where pharmacotherapies might be effective, including AUD initiation (initial alcohol use), development (sporadic intermittent alcohol use; the binge-intoxication phase), progression (regular use), early abstinence (the withdrawal - negative affect stage) or protracted abstinence (the preoccupation - anticipation (craving) stage) (Koob et al. 2009; Kreek et al. 2002). Sleep disturbances are a key contributor to relapse in abstinence and therefore offer another target for treatment (Brower 2015; Miller et al. 2017). Therapeutic interventions at any point along this continuum could improve the health of the individual.
Pharmaceutical treatments can either be developed de novo for a specific drug target, repurposed, or rescued. While the usage and definition of the terminology “drug repurposing” and “drug rescue” can be complex (Langedijk et al. 2015), here we define drug repurposing as finding a novel clinical use for an approved drug and drug rescue as finding a clinical use for a stalled drug (whether the drug is in development but not yet approved or failed for one indication but could be useful for another disease or patient subgroup; Phase 2 or beyond). Drug repurposing (also referred to as drug repositioning) is appealing since it reduces the overall costs of drug development and expedites the availability of treatments to those who need them (Nosengo 2016). Drug repurposing has largely centered around side effect data, and, while this approach has been somewhat successful for brain diseases, there is a great need for improved strategies for drug selection. De novo drug development has traditionally relied on target identification through basic research. Over four decades of alcohol research has identified key neurotransmitter systems and brain regions that contribute to the various stages of AUD pathology and represent potential targets for pharmaceutical development. Despite these advancements, there has been sparse translational success clinically. There are only three FDA approved drugs for AUD: naltrexone (oral: ReVia®, injectable: Vivitrol®), acamprosate (Campral®) and disulfiram (Antabuse®), the most recent of which, acamprosate, was approved in 2004. This gap between advances in basic research (conducted primarily at academic institutions) and pharmaceutical development (primarily undertaken by industry, e.g., pharmaceutical companies) has been dubbed the “valley of death” (Butler 2008).
The explosion of both the quantity and availability of various types of molecular datasets (e.g., gene sequence/genotype, gene expression, epigenetic marks, metabolic measures) and computational strategies to exploit them, offers new solutions to this problem and is moving disease diagnosis and treatment into the molecular realm. Many computational (or in silico) strategies exist, and all are concerned with finding the “similarity” between diseases and drugs. The computational strategies highlighted in this Review involve integrating molecular profiles of a disease state with those of pharmaceuticals to predict effective treatments. Molecular profiles can be derived from multiple molecular phenotypes, including gene expression, protein targets (see the issue in this article for proteome targets in the accumbens by Clyde Hodge and colleagues), genetic variants (single nucleotide polymorphisms (SNPs)), and others, though the focus of this Review will be on gene expression. Another approach to computational repurposing uses crystal structures of receptors to conduct structure-based ligand discovery (Heusser et al. 2013). In this Review, we focus on the aforementioned computational approaches and will not discuss structure-based ligand discovery in detail, but interested readers are referred to a review (Howard et al. 2014).
Traditional Approach
Drug Development
Disease-related drug development begins with mechanistic studies of target identification followed by validation (see the Review in this issue by Ciccocioppo and colleagues for an indepth discussion of target validation), preclinical and clinical trials, and FDA review. Typically, a single cellular or molecular target is sourced from the results of much neurobiological research (Fig 1). Despite clear scientific evidence for its involvement in disease pathology at multiple levels of analysis (e.g., molecular, neuropharmacological, neurocircuitry, behavior), the single target approach has largely been a failure for brain diseases (Hutson et al. 2017). A striking example of this is for Huntington’s disease, where the single causative gene (HTT) has been known since 1993 (MacDonald et al. 1993). Despite this single, well-validated target, no drug nor therapeutic options have been developed as treatments. One example of this for AUD is the corticotrophin releasing factor (CRF) system, which has tremendous research support for its involvement in AUD pathology, yet CRF inhibitors have produced disappointing results in double-blind, placebo controlled trials (Kwako et al. 2015; Pomrenze et al. 2017; Schwandt et al. 2016).
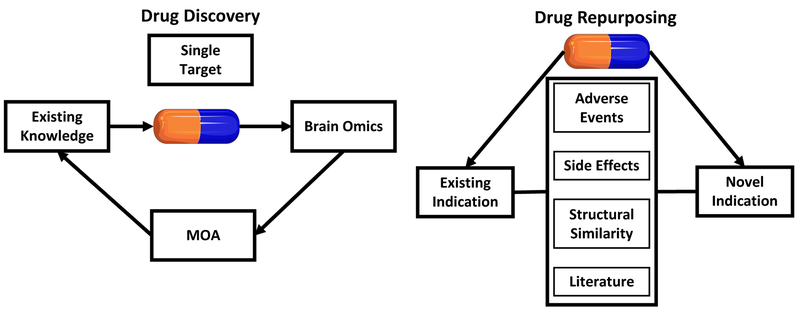
Fig1.
Traditional approach to drug discovery and drug repurposing: Existing knowledge of a disease state (built upon basic science) is applied to select a compound designed for a single target (chosen for its involvement in a disease process) and these are tested in vitro and/or in vivo. Brain gene expression levels (Brain Omics) are measured for drugs that ameliorate disease phenotype which helps further elucidate the mechanisms of action (MOAs) of drugs and suggests other molecules that can be targeted by candidate drugs. Traditionally, drug repurposing (finding new indications for existing compounds) has been largely based on side effect data, adverse events, existing literature or structural similarity between compounds used to treat different diseases (the idea being that the compound of one disease might be able to treat another since it shares structural similarity with compounds used to treat that disease). Drug repurposing efforts would benefit greatly if there was a system established to report positive side effects as is the case for “adverse events”. Capsule images from http://smart.servier.com/category/general- items/drugs-and-treatments/. Servier Medical Art by Servier is licensed under CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/)
Despite the vital insights gained from neurobiological research (both in humans and animal models), these findings have not translated into therapeutic success. There are a number of possible reasons for this, including genetically heterogeneous human populations and the complexities of alcohol’s many targets (Most et al. 2014; Pomrenze et al. 2017). The brain is highly complex, and psychiatric diseases are characterized by numerous symptoms. Reducing this complexity to a single target is appealing for its simplicity but perhaps misguided, and expecting modulation of a single gene or molecule to ameliorate all symptoms of complex diseases is likely to produce disappointing results.
Targets (molecules) do not work in isolation, but function as part of a system (or network) to accomplish biological functions. The hypothesis that a disease state represents a shift from normal physiological homeostasis and can be thought of as a network perturbation has been proposed and described in detail, and is attractive for several reasons (Barabasi et al. 2011; Chen and Butte 2013; Jacunski and Tatonetti 2013; Kolodkin et al. 2012; Silbersweig and Loscalzo 2017; Silverman and Loscalzo 2013). First, there could be many network perturbations that lead to the same disease classification, which fits with the heterogeneous patient populations we observe in AUD. Secondly, the other side of this argument is that if a disease represents a perturbed state of a biological network, there could be multiple pharmacological intervention points to reverse those perturbations and return the system to homeostasis. Targeting the network at several points might be more efficient (or even necessary) to shift the system back to normal homeostasis. This also provides a basis for polypharmacology (the use of drug combinations to treat a disease) and could guide the selection of drug combinations, which will not be discussed in depth in this Review, but interested readers are referred to (Ryall and Tan 2015) for more information. For these reasons, we and others propose that to maximize the likelihood of successful treatment for complex disorders, it is imperative to “drug the network” rather than focus solely on single targets (see Computational Approaches section below).
Drug Repurposing
Traditionally, getting a drug to market takes 13–15 years and costs 2–3 billion dollars on average (Nosengo 2016). Many drugs that are currently FDA approved could be beneficial for diseases other their original indication. Additionally, pharmaceutical companies have invested considerable resources into developing drugs that passed initial safety trials but failed in efficacy trials (sometimes referred to as shelved compounds) that are waiting for a suitable indication (Nosengo 2016). Often, successful drug repurposing has been serendipitous (Fig 1). There are many examples spanning a variety of conditions, from the classic example of sildenafil (Viagra®), a PDE5 inhibitor being developed for hypertension, that was repurposed for erectile dysfunction (Goldstein et al. 1998), to bimatoprost (Lumigan® / Latisse®), a prostaglandin analog, that was repurposed for a cosmetic application as it was noticed to lengthen and darken the eyelashes as a side effect of those using it to treat glaucoma (Tosti et al. 2004).
Drug repurposing has also been successful for brain diseases. For example, buprenorphine, a mixed partial agonist opioid receptor modulator, was originally used for pain relief and was repurposed to treat opiate dependence (Jasinski et al. 1978). Ropinirole (Requip), a dopamine agonist used an anti-Parkinson’s agent, was repurposed for treatment of both Restless Legs Syndrome and SSRI-induced sexual dysfunction (Cheer et al. 2004; Worthington et al. 2002). Additional examples include bupropion (depression to smoking cessation) (Lief 1996), dimethyl fumarate (psoriasis to multiple sclerosis) (Bomprezzi 2015), and guanfacine (hypertension to ADHD) (Strange 2008).
Several FDA approved or shelved compounds have shown promise in treating AUD and many are currently undergoing human lab testing or are in clinical trials (ClinicalTrials.gov_AUD), including gabapentin, topiramate, varenicline, ABT-436, mifepristone (RU-486), citicoline, baclofen, nalmefene, and others (Litten et al. 2016; Lyon 2017) (Table 1). Gabapentin (Neurontin) was initially used as an anti-epileptic, then later approved for neuropathic pain and amyotrophic lateral sclerosis. Baclofen (Liorsel) is a GABAB receptor agonist, was originally made as an anti-epileptic with disappointing results, but showed remarkable effectiveness for treating spasticity in many conditions, especially for spinal cord injury, cerebral palsy, and multiple sclerosis. As mentioned, it is being considered for treatment of AUD (with mixed findings) (Farokhnia et al. 2017).
Table 1:
Drugs with Potential to be Repurposed for AUD
| Drug Name | Target | Original Indication |
Novel Indication |
Clinical Trial | Refs |
|---|---|---|---|---|---|
| Quetiapine (Seroquel) | Atypical Antipsychotic Antagonist: D1 and D2 dopamine receptors, alpha 1 and alpha 2 adrenoreceptors, and 5-HT1A and 5-HT2 serotonin receptors, histamine H1 receptor (and others) |
Schizophrenia (Scz) Bipolar disorder (BD) Major depressive disorder (MDD) (along with an SSRI) | AUD and Bipolar; Sleep disturbances in abstinence; Schizophrenia and SUD; AUD and anxiet |
NCT00457197 NCT00114686 NCT00223249 NCT00550394 NCT00434876 NCT00156715 NCT00352469 |
(Jensen et al. 2008; Schotte et al. 1996) (Litten et al. 2012) |
| Aripiprazole (Abilify) | Atypical antipsychotic Antagonist: D1 and D2 dopamine receptors, alpha 1 and alpha 2 adrenoreceptors, and 5-HT1A and 5-HT2 serotonin receptors, histamine H1 receptor (and others) |
Scz BD MDD (along with an SSRI) | AUD and Bipolar | NCT02918370 | (Anton et al. 2008; Kenna et al. 2009; Martinotti et al. 2009; Martinotti et al. 2007; Shapiro et al. 2003; Voronin et al. 2008) |
| Duloxetine (Cymbalta) | Serotonin–norepinephrine reuptake inhibitor (SNRI) | MDD GAD Muscle pain Peripheral neuropathy | AUD | NCT00929344 | (Bymaster et al. 2001) |
| Venlafaxine (Effexor) | SNRI | MDD GAD Panic disorder Social anxiety disorder | AUD and anxiety | NCT00248612 | (Bymaster et al. 2001; Ciraulo et al. 2013; Upadhyaya et al. 2001) |
| Rolipram | Phosphodiesteras e-4 inhibitor | Shelved compound; Phase 3 for MDD (Fleischhacker et al, 1992) | AUD | (Bell et al. 2017; Dominguez et al. 2016; Gong et al. 2017; Hu et al. 2011; Liu et al. 2017; Ray et al. 2014; Wen et al. 2012) | |
| Ibudilast | Phosphodiesterase inhibitor | Asthma (Japan) | AUD | NCT02025998 | (Bell et al. 2015; Crews et al. 2017; Ray et al. 2017; Ray et al. 2014) |
| Fenofibrate (Tricor) | Fibrate PPARα agonist | Hypercholesterolemia Hypertriglyceridemia | AUD | NCT02158273 | (Blednov et al. 2015; Blednov et al. 2016a; b; Ferguson et al. 2014; Haile and Kosten 2017; Karahanian et al. 2014; Rivera-Meza et al. 2017) |
| Gabapentin (Neurontin) | Anticonvulsant binds to the α2δ subunit of the voltagedependent calcium channels (Pregabalin is structurally related to gabapentin) | Seizures Restless leg syndrome Postherpetic neuralgia Shingles | AUD; Abstinence initiation in AUD; AUD (in combination with naltrexone); Sleep disturbances in AUD; AUD (in combination with flumazenil for withdrawal and relapse prevention); AUD (in combination with lorazepam for withdrawal); Comorbid alcohol and opioid abuse |
NCT02771925 NCT01141049 NCT00391716 NCT00183196 NCT01014533 NCT00262639 NCT03274167 NCT00011297 NCT03205423 NCT02252536 |
(Geisler and Ghosh 2014; Guglielmo et al. 2012; Litten et al. 2016; Mason et al. 2014a; Mason et al. 2014b; Nunes 2014) |
| Pregabalin (Lyrica) | Anticonvulsant binds to the α2δ subunit of the voltagedependent calcium channels | Epilepsy Neuropathic pain Fibromyalgia Generalized anxiety disorder (GAD) | AUD; AUD and PTSD |
NCT03256253 NCT02884908 NCT00929344 |
(Guglielmo et al. 2012; Li et al. 2011) |
| Topiramate (Topamax) | Anticonvulsant Blocks voltagegated Na+ Channels, PAMs of subunits of the GABAA Receptor Modulates AMPA/kainite glutamate receptors. Blocks carbonic anhydrase (CA) CA I1 and CA IV |
Epilepsy Migraines | AUD; AUD and PTSD; AUD and Borderline Personality Disorder; AUD and BD; AUD and cocaine dependence; AUD and nicotine dependence |
NCT01135602 NCT01145677 NCT01749215 NCT00769158 NCT00463775 NCT00210925 NCT00572117 NCT00223639 NCT00884884 NCT00006205 NCT00571246 NCT03120468 NCT00802412 NCT00448825 NCT00329407 NCT00862563 NCT01182766 NCT00300742 NCT00167245 NCT02371889 NCT01764685 NCT01087736 NCT01408641 NCT00550394 NCT03018704 |
(Guglielmo et al. 2015; Ray and Bujarski 2016; Shank et al. 2000) |
| Varenicline (Chantix and Champix) | α7 nicotinic acetylcholine receptor agonist α4β2, α3β4, and α6β2 subtype s partial agonist weak agonist on the α3β2 containing receptors | Smoking cessation | AUD; AUD and tobacco dependence; AUD and cocaine dependence; AUD, SCZ and nicotine dependence |
NCT01071187 NCT01146613 NCT00705523 NCT00846859 NCT00873535 NCT01553136 NCT01347112 NCT01151813 NCT01169610 NCT00727103 NCT01011907 NCT01092702 NCT01286584 NCT01592695 NCT02698215 |
(Falk et al. 2015; Litten et al. 2013) |
| ABT-436 | highly selective vasopressin V1B receptor antagonist | Shelved compound; Phase 2 for MDD (NCT01741142) | AUD | NCT01613014 | (Ryan et al. 2017) |
| Mifepristone (RU-486) (Mifeprex) | glucocorticoid and progesterone receptor antagonist | Abortifacient Hyperglycemia Diabetes mellitus | AUD |
NCT02243709 NCT02179749 NCT02989662 NCT01548417 |
(Donoghue et al. 2016; Howland 2013; Lyon 2017; Vendruscolo et al. 2012; Vendruscolo et al. 2015) |
| Citicoline (Cebroton, Ceraxon, Cidilin, Citifar, Cognizin, more) | membrane permeability enhancer glutathione transferase stimulant (overthe-counter nutritional supplement) | Stroke Alzheimer’s disease Senile dementia Parkinson’s disease Attentiondeficit/hyperactiv ity disorder (ADHD) Glaucoma | AUD; AUD and BD |
NCT02074735 NCT02582905 |
(Secades and Lorenzo 2006; Wignall and Brown 2014) |
| Baclofen (Lioresal) | Central nervous system depressant; skeletal muscle relaxant GABAB receptor agonist |
Spastic movement disorders (commonly for spinal cord injury, cerebral palsy, and multiple sclerosis) | AUD; Alcohol withdrawal; AUD and Hepatitis C; AUD with liver disease; AUD and anxiety disorders |
NCT02596763 NCT03034408 NCT03293017 NCT01008280 NCT02511886 NCT01711125 NCT01751386 NCT00877734 NCT00614328 NCT01266655 NCT01980706 NCT01738282 NCT01002105 NCT00802035 NCT02835365 NCT01604330 NCT02723383 NCT01076283 NCT01937364 NCT00525252 NCT02107352 NCT02771925 |
(Bell et al. 2017; Borro et al. 2016; Colombo et al. 2004; Farokhnia et al. 2017; Geisel et al. 2016; Imbert et al. 2015; Litten et al. 2016; Liu and Wang 2017; Lyon 2017; Mirijello et al. 2015; Morley et al. 2014; Muller et al.2015; Ponizovsky et al. 2015; Rigal et al. 2015; Rolland et al. 2015a; Rolland et al. 2015b; Weibel et al. 2015) |
| Nalmefene (Selincro) | Antagonist of the μ-opioid receptor weak partial agonist of the κ-opioid receptor | Antidote for opioid overdose Approved in Europe for AUD | AUD; AUD with cirrhosis; AUD and tobacco dependence; AUD and Borderline Personality Disorder; AUD and opioid use disorder |
NCT00811720 NCT01969617 NCT00812461 NCT00811941 NCT02824354 NCT02382276 NCT02364947 NCT02197598 NCT02679469 NCT02372318 NCT02195817 NCT02492581 NCT00000450 NCT00000437 NCT02752503 NCT03034408 NCT03279562 |
(Litten et al. 2016; Naudet 2016; Naudet et al. 2016; Soyka 2016; Soyka et al. 2016) |
Computational Approaches
The generation and accumulation of publicly accessible, high-throughput genomic datasets make it possible to integrate large-scale drug and disease signatures at the molecular level to predict compounds with the potential to treat a disease based on multiple targets (e.g. gene networks). These data-rich resources include public repositories (primary archives), integrative databases, and value-added databases (these tools are designed to process, analyze and annotate complex information from primary data sources to lower the computational barriers to access primary data). A selection of these resources is summarized in Table 2.
Table 2:
Data Resources
| Primary Repositories | |||
|---|---|---|---|
| Name | Description | URL | Ref |
| Gene Expression Omnibus | Public functional genomics data repository for array- and sequencebased data. | ncbi.nlm.nih.gov/geo/ | (Barrett et al. 2013) |
| ArrayExpress | Public functional genomics data repository for array- and sequencebased data. | ebi.ac.uk/arrayexpress/ | (Kolesnikov et al. 2015) |
| ParkDB | Repository for gene expression datasets related to Parkinson’s disease (PD) | www2.cancer.ucl.ac.uk/Parkinson_Db2/ | (Taccioli et al. 2011) |
| Integrative Databases | |||
| HUGO Gene Nomenclature Committee (HGNC) database | Repository of HGNC-approved gene nomenclature, gene families and associated resources including links to genomic, proteomic and phenotypic information. | genenames.org/ | (Gray et al. 2015) |
| Online Mendelian Inheritance in Man (OMIM) | Publicly available dataset of human genes and genetic disorders and traits, with particular focus on the molecular relationship between genetic variation and phenotypic expression. | omim.org/ | (Amberger and Hamosh 2017) |
| UK Brain Expression Consortium | Publicly available dataset of geneotyping and gene expression data from 134 brains from individuals free of neurodegenerative disorders (up to twelve brain regions). | ukbec.wordpress.com/braineac.org/ | |
| ENCODE (Encyclopedia of DNA Elements) | Integrates multiple technologies and approaches in a collective effort to discover and define the functional elements encoded in the human genome, including genes, transcripts, and transcriptional regulatory regions, together with their attendant chromatin states and DNA methylation patterns. | encodeproject.org/ | (Consortium 2011) |
| Genotype–Tissue Expression (GTEx) project | Collection and analysis of multiple human tissues from donors who are also densely genotyped, to assess genetic variation within their genomes. By analyzing global RNA expression within individual tissues and treating the expression levels of genes as quantitative traits, variations in gene expression that are highly correlated with genetic variation can be identified as expression quantitative trait loci, or eQTLs. | gtexportal.org/home/ | (Consortium 2013) |
| Depression Genes and Networks (DGN) cohort | RNA sequencing data and analyses from 922 genotyped individuals, providing information regarding the regulatory consequences of genetic variation | dags.stanford.edu/dgn/ | (Battle et al. 2014) |
| Psychiatric Genomics Consortium (PGC) | Psychiatric Genomics Consortium (PGC) unites investigators around the world to conduct meta- and megaanalyses of genome-wide genomic data for psychiatric disorders. There are samples from more than 900,000 individuals (and growing) collected by over 800 investigators from 38 countries. | med.unc.edu/pgc | (O’Donovan 2015) |
| Library of Integrated Network-Based Cellular Signatures LINCS-L1000 | Publicly available dataset from the Broad Institute. The 1.3M L1000 cellular signatures catalog transcriptional responses of human cells to chemical and genetic perturbation. A total of 27,927 perturbagens have been profiled to produce 476,251 expression signatures. About half of those signatures make up the Touchstone (reference) dataset generated from testing well-annotated genetic and small-molecular perturbagens in a core panel of cell lines. | clue.io/ | (Subramanian 2017) |
| Connectivity Map (CMap) | Publicly available dataset from the Broad Institute. Connectivity Map Build 02 includes data from 7,056 Affymetrix microarrays, for 1,309 small-molecule compounds, and 6,100 treatment instances in 5 human cell lines. | broadinstitute.org/cmap/ | (Lamb et al. 2006) |
| Added-Value Databases and Tools | |||
| Genes | |||
| Enrichr | Web tool for gene set enrichment analysis | amp.pharm.mssm.edu/Enrichr/ | (Kuleshov et al. 2016) |
| NetworkAnalyst | Web tool for performing various common and complex meta-analyses of gene expression data | networkanalyst.ca/ | (Xia et al. 2015) |
| Database for Annotation, Visualization and Integrated Discovery (DAVID) | Web tool for gene set enrichment analysis | david.ncifcrf.gov/ | (Huang da et al. 2009) |
| GeneMANIA | Web tool for generating hypotheses about gene function, analyzing gene lists and prioritizing genes for functional assays. There is also a GeneMANIA Cytoscape plugin. | genemania.org/ | (Montojo et al. 2014) |
| WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) | Web tool for gene set enrichment analysis. | webgestalt.org | (Wang et al. 2013) |
| PubMatrix | Web tool that allows simple text based mining of the NCBI literature search service PubMed using any two lists of keywords terms, resulting in a frequency matrix of term cooccurrence. | pubmatrix.irp.nia.nih.gov/ | (Becker et al. 2003) |
| Ingenuity Pathway Analysis (IPA®) | Tool for analyzing and visualizing data from ‘omics experiments | qiagenbioinformatics.com/products/ingenuity-pathwayanalysis/ | (Kramer et al. 2014) |
| Gene-Set Enrichment Analysis (GSEA) | Web tool for determining whether an apriori defined set of genes shows statistically significant, concordant differences between two biological states (phenotypes). | broadinstitute.org/gsea/ | (Subramanian et al. 2005) |
| MetaXcan | Algorithm that allows imputation of gene expression z-scores based on GWAS summary statistics. | github.com/hakyimlab/MetaXcan | (Barbeira et al. 2016) |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | Database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies. (So et al. 2017) used KEGG to download the Anatomical Therapeutic Classification (ATC) for drugs | genome.jp/kegg/ | (Ogata et al. 1999) |
| GeneGO’s Metacore | Integrated software suite for functional analysis of experimental data based on a curated database of human proteinprotein, protein-DNA interactions, transcription factors, signaling and metabolic pathways, disease and toxicity, and the effects of bioactive molecules. Suite contains tools for data visualization, mapping and exchange, multiple networking algorithms and filters. | portal.genego.com/ | (Ekins et al. 2006) |
| GeneWeaver | Curated repository of genomic experimental results from published genome-wide association studies, quantitative trait locus, microarray, RNA-sequencing and mutant phenotyping studies with an accompanying tool set for dynamic integration of these data sets, enabling users to identify genefunction associations across diverse experiments, species, conditions, behaviors or biological processes. | geneweaver.org/ | (Baker et al. 2012) |
| GeneCards | Database of human genes that provides genomic, proteomic, transcriptomic, genetic and functional information on all known and predicted human genes. Developed and maintained by the Crown Human Genome Center at the Weizmann Institute of Science. | genecards.org | (Stelzer et al. 2016) |
| TransFind | Web tool for predicting transcriptional regulators for gene sets | transfind.sys-bio.net/ | (Kielbasa et al. 2010) |
| JASPAR | Web tool for predicting transcriptional regulators for gene sets | jaspar.genereg.net/ | (Mathelier et al. 2016) |
| TRANSFAC (TRANScription FACtor database) | Web tool for predicting transcriptional regulators for gene sets | generegulation.com/pub/databases.html | (Matys et al. 2006) |
| Proteins | |||
| STRING | Web tool / database that provides a critical assessment and integration of protein-protein interactions, including direct (physical) as well as indirect (functional) associations. | string-db.org | (Szklarczyk et al. 2015) |
| iRefWeb | Web tool / database that integrates data on protein-protein interactions (PPI) consolidated from major public databases. | wodaklab.org/iRefWeb/ | (Turinsky et al. 2014) |
| Hippie | Web tool to generate reliable and meaningful human protein-protein interaction networks. | cbdm-01.zdv.unimainz.de/~mschaefer/hippie/ | (Alanis-Lobato et al. 2017) |
| Drugs | |||
| sscMap | Java application that performs connectivity mapping tasks using the CMap build 02 data. Users can add custom collections of reference profiles. | purl.oclc.org/NET/sscMap | (Zhang and Gant 2009) |
| Searchable Platform Independent Expression Database Webtool (SPIEDw) | Web tool used for querying publically available gene expression data (including the CMap build 02 drug data). | spied.org.uk/cgibin/HGNC-SPIED3.1.cgi | (Williams 2012) |
| Drug-Set Enrichment Analysis (DSEA) | Web tool for identifying shared pathways whose genes are upregulated (or downregulated) by the drugs in the set. | dsea.tigem.it/ | (Napolitano et al. 2016) |
| ChemBioServer | Web tool for mining and filtering chemical compounds used in drug discovery | bioserver-3.bioacademy.gr/Bioserver/ChemBioServer/ | (Athanasiadis et al. 2012) |
| Mode of Action by NeTwoRk Analysis (Mantra 2.0) | Web tool for the analysis of the Mode of Action (MoA) of novel drugs and the identification of known and approved candidates for “drug repositioning” using CMap drug data. | mantra.tigem.it/ | (Carrella et al. 2014) |
| Comparative Toxicogenomics Database (CTD) | Publicly available dataset describing relationships between chemicals, genes, and human diseases | ctdbase.org/ | (Davis et al. 2017) |
| MEDication Indication resource (MEDI) | Compiled from four public medication resources, including RxNorm, Side Effect Resource 2 (SIDER2), Wikipedia and MedlinePlus. A random subset of the extracted indications was also reviewed by physicians. The MEDI high-precision subset (MEDIHPS), only includes drug indications found in RxNorm or in at least two of the other three sources, with an | vumc.org/cpm/centerprecision-medicineblog/medi-ensemblemedication-indicationresource | (Wei et al. 2013) |
| ClinicalTrials.gov | Contains information about clinical trials. | ClinicalTrials.gov | |
| National Institute for Occupational Safety and Health List of Antineoplastic and Other Hazardous Drugs | Contains drugs known to be toxic, according to published literature | cdc.gov/niosh/topics/hazdrug/default.html | (Traynor 2014) |
| DrugBank | Bioinformatics and cheminformatics resource that combines detailed drug data with comprehensive drug target information. | DrugBank.ca/ | (Wishart et al. 2006) |
| STITCH | Database that includes information on chemical-protein interactions. The interactions include direct (physical) and indirect (functional) associations; they stem from computational prediction, from knowledge transfer between organisms, and from interactions aggregated from other (primary) databases. Currently it has 9,643,763 proteins from 2,031 organisms. | stitch.embl.de/ | (Szklarczyk et al. 2016) |
| PharmGKB | Repository for pharmacogenetic and pharmacogenomic data, and curators provide integrated knowledge in terms of gene summaries, pathways, and annotated literature. | pharmgkb.org | (Owen et al. 2007) |
| SuperTarget | Added-value database that integrates information about drugs, proteins and side effects from other databases to form drug-protein, protein-protein and drug-side-effect relationships and includes annotation about the source, ID’s, physical properties, references and much more. | insilico.charite.de/supertarget/ | (Hecker et al. 2012) |
| KEGG Drug | Comprehensive drug information resource for approved drugs in Japan, USA, and Europe unified based on the chemical structures and/or the chemical components, and associated with target, metabolizing enzyme, and other molecular interaction network information. | genome.jp/kegg/drug/ | (Ogata et al. 1999) |
| AUD-Specific | |||
| INIA Texas Gene Expression Database (ITGED) | Contains the top statistical results from genomic studies focusing on models of excessive alcohol consumption. | inia.icmb.utexas.edu/ | |
| Ethanol-Related Gene Resource (ERGR) | Contains more than 30 large datasets from literature and 21 mouse QTLs from public database (see data summary). These data are from 5 organisms (human, mouse, rat, fly and worm) and produced by multiple approaches (expression, association, linkage, QTL, literature search etc) | bioinfo.uth.edu/ERGR/ | (Guo et al. 2009) |
| Gene Network | Contains large collections of genotypes (e.g., SNPs) and phenotypes that are obtained from groups of related individuals, including human families, experimental crosses of strains of mice and rats, and organisms as diverse as Drosophila melanogaster, Arabidopsis thaliana, and barley. | genenetwork.org/webqtl/main.py | (Mulligan et al. 2017) |
There are two essential datasets from these resources that are required to match disease and drug: (1) measurements of a molecular phenotype induced by a disease state and (2) measurements of the same molecular phenotype induced by drugs. Obtaining this type of reliable drug library is not trivial. Surprisingly, attaining a list of approved drugs and their indications is not a straightforward task. These difficulties are the result of poor data storage and electronic retrieval mechanisms, complex and rapidly changing nomenclature (drugs can be called by their common name, chemical name, simplified molecular-input line-entry system (SMILES), International Chemical Identifier (InChI)), and legal issues surrounding off-label advertisement of pharmaceuticals. Fortunately, many of these challenges have been overcome largely by the pioneering work by a collaborative effort of the Broad Institute and funding from a National Institutes of Health Common Fund (https://commonfund.nih.gov/lincs). They have compiled the Library of Integrated Network-based Cellular Signatures (LINCS-L1000) database which contains gene expression responses to genetic and pharmacologic manipulation across a diverse set of human cell lines (Subramanian et al. 2017). They also maintain a repurposing hub that contains over 5,000 manually-curated drugs that are either FDA approved or in clinical trials (Corsello et al. 2017). The availability of these tremendous resources is a primary reason we focus on gene expression as the molecular phenotype, as other molecular responses to drugs are not as well characterized in such a systematic manner or as accessible for analysis. With the two required datasets described above, there are three main steps to proceed from gene networks to candidate compounds (see Fig 2), which then can be tested in a preclinical animal model or human laboratory study:
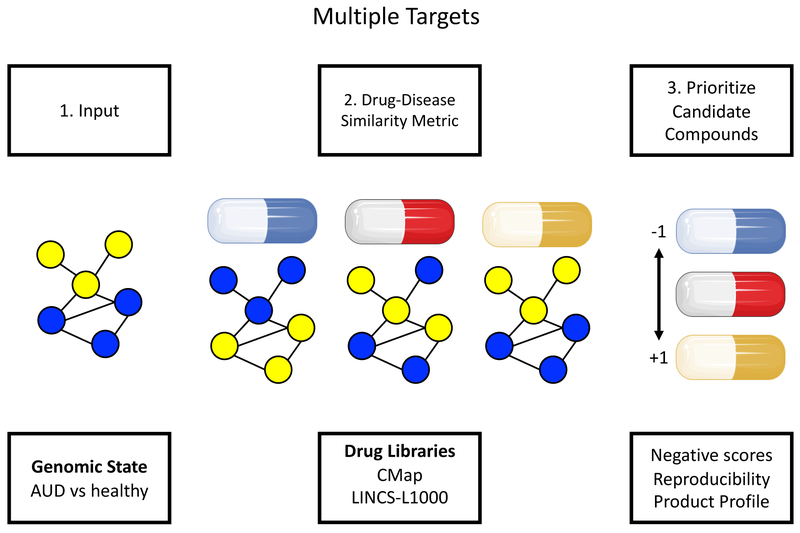
Fig2.
Computational approach to drug discovery and drug repurposing: Disease state can be either acquired (disease or substance of abuse changes gene networks and these changes drive disease) or predisposed (genetic variants cause disruptions in gene networks). The goal of in silico gene mapping is to integrate the targets (gene networks) of disease and drugs to find a drug (or combination of drugs) that affect similar targets as the disease. Drugs that oppose the disease-state’s molecular disruption (many targets) are chosen as candidate compounds to ameliorate disease phenotype. There are 3 steps to go from gene expression datasets to candidate compounds: (1) generate an input genomic signature or network. Shown is a gene-gene coexpression network of genes related to a disease state: nodes = genes, edges = gene-gene expression correlation, yellow = up-regulated genes, blue = down-regulated genes), (2) compare the disease signature to those induced by drugs to identify drugs that would reverse the disease signature. Shown are the effects of 3 different drugs in the reference database (e.g., LINCS-L1000) on the disease-related genes that served as the input, (3) prioritize candidate compounds for in vivo testing. The blue drug that received a perfect negative score would be prioritized since it down-regulated the genes that were up-regulated in the disease state and up-regulated the genes that were down-regulated in the disease state. The yellow drug would be predicted to mimic or worsen the disease state. Had the input been a desirable biological state (e.g., the gene expression profile of patients with AUD who had prolonged recovery vs those who relapsed quickly after ceasing alcohol consumption), then the yellow drug would be prioritized since it is predicted to mimic the beneficial biological state. Capsule images from http://smart.servier.com/category/general-items/drugs-and-treatments/. Servier Medical Art by Servier is licensed under CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/). AUD = alcohol use disorder.
Generate an input signature that captures the genomic state of interest (gene expression differences between disease and healthy state, for example).
High-throughput identification of compounds using an in silico screen (similarity metric).
Prioritize candidate compounds.
The details of each step are described below.
Generate an input signature that captures the genomic state of interest.
The purpose of the signature is to capture the molecular changes that are the most relevant to the biological state of interest at a given point in time. There are many different options for constructing an input signature. Applying such approaches to brain diseases is still in its infancy and understanding the optimal input parameters is a major challenge (see the Challenges section below). Genetic variation (genotyping or exome / whole genome sequencing data) has been the primary approach used for genomic medicine / precision medicine for cancer (Letai 2017). A functional genomic measure, such as gene expression can also be used. This is referred to as in silico gene mapping, gene mapping, or connectivity mapping, the latter named after one of the first characterizations of the method using the Broad Institute’s database called the Connectivity Map (CMap) (Lamb et al. 2006). Importantly, the AUD research field has generated an incredible library of gene expression data that spans multiple species (human, mouse, monkey, rat), conditions/treatments (genetic predisposition, various acute or chronic ethanol exposures or paradigms), various brain regions, and isolated cell types (including microglia and astrocytes) (Table 2).
High-throughput identification of compounds using an in silico screen (similarity metric).
At their core, the various approaches used for in silico gene mapping aim to compare drug and disease signatures. If an effect size measure (such as fold change) is available, a correlation coefficient could be calculated, to reflect the correlation between gene expression changes between drug and vehicle and those between disease and normal. Positive correlations would indicate that the drug mimics the disease’s effects on transcription levels, while negative correlations would indicate that the drug reverses it. An alternative approach is to use an enrichment score to assess the overlap between two lists of differentially expressed genes, such as the hypergeometric statistic or the rank-based gene set enrichment analysis (GSEA; corresponds to a weighted Kolmogorov-Smirnov) (Subramanian et al. 2005). For example, list A contains the top differentially expressed genes between disease and healthy samples, and list B contains the top differentially expressed genes between drug and vehicle samples. The hypergeometric statistic would give the probability of the overlap between list A and list B (the genes changed by both drug and disease). GSEA, the approach implemented by the Connectivity Map (CMap) and LINCS – L1000, avoids using arbitrary cut-offs (the p – value which designates differential expression between two conditions or treatments) by considering all of the genes in an experiment. Ranked methods for the hypergeometric test have also been described and offer the same benefits as GSEA (Plaisier et al. 2010).
Prioritize candidate compounds.
Regardless of which statistical test is chosen, the output of the previous step will include a list of predicted compounds with a corresponding similarity score (also called a connectivity score). Because only a handful of drugs can be tested in vivo, this list must be filtered to select the most promising candidate compounds. The working hypothesis is that negative scores would predict reversal of gene expression from disease back to normal state. However, this hypothesis is rarely tested directly (see Challenges), though it does have some support (Chen et al. 2017; Delahaye-Duriez et al. 2016; Wagner et al. 2015). Regardless, drugs with either the highest absolute value or the most negative similarity scores should be prioritized, as these reflect the drugs that affect the most disease-related genes.
Beyond the sign (+/−) and magnitude of the connectivity score, there are additional practical considerations for prioritizing candidate compounds (Oprea and Overington 2015). For example, any identified high-priority candidate drugs for AUD treatment would also benefit from having 1) known oral dosing data available, 2) have little or no safety warnings (especially regarding liver toxicity), 3) have low abuse liability, 4) low drug-drug interaction potential, 5) negligible cytotoxic actions 6) high brain penetrability, among others. These considerations alone will assist in narrowing the pool of potentially “testable” compounds considerably, if the information is available (which is frequently not the case). Upon first glance, it might seem that the challenge is selecting only a few compounds from hundreds of candidates generated by in silico screens to test clinically or pre-clinically. However, this is not the case. Meeting the ideal practical considerations outlined above could eliminate virtually all candidate compounds (Oprea and Overington 2015). In that case, medicinal chemistry approaches could be used to modify the chemical structure to suit the desired product profile.
Drug prioritization should also depend on the reliability of the analytical results. That is, the connectivity score should be reproducible for strong candidates. This is especially important to consider since small changes in the genes that makeup the input signature can result in the identification of different candidate compounds. The above mentioned CMap database utilizes a statistical measure of reliability (permutation test) to achieve this goal. Stricter statistical measures have also been developed for CMap. For example, the statistically significant connectivity map (ssCMap) was developed (Zhang and Gant 2009), which includes a measure of stability by removing single genes from the input in a systematic manner and assessing reproducibility (McArt and Zhang 2011). However, for larger datasets, such as LINCS-L1000, implementation of permutations tests becomes computationally expensive and less straightforward. Currently, the web app for querying the LINCS-L1000 data (https://clue.io/l1000-query) uses the “sig_gutc” tool (Subramanian et al. 2017) to summarize the connectivity scores and provide a measure of reliability. Each compound has been profiled under multiple experimental conditions (different cell lines, drug doses, and exposure time points). To attain a compound-level analysis, sig_gutc reports a summary score of the distribution of scores for a compound across all experiments. The tool then ranks the connectivity score between the query signature and the compound signature, based upon the compound’s pre-computed distribution of connectivity scores with the other hundreds of thousands of signatures in the LINCS-L1000 database. This provides a measure of the likelihood of a connectivity score for a drug given that drug’s connectivity with the database as a whole, thus mitigating false positives from drugs with widespread effects on transcription. However, an appropriate statistical framework with which to interpret LINCS-L1000 results needs to be developed.
Application to Brain Diseases
While initially used in cancer research (for review see (Chen and Butte 2016)), these computational repurposing strategies have also been applied to brain diseases, albeit in a more limited capacity. However, it should be noted that although not used as widely, the studies using these computational approaches for drug discovery for brain diseases have provided promising leads for variety of disease states. Because there are few applications so far for psychiatric disorders, this Review includes the use of in silico gene mapping strategies for any disease in which brain is the primary affected organ, for which there have been 19 studies so far to the best of our knowledge (Table 3).
Table 3:
Studies Applying Systems Pharmacology Approaches to Brain Diseases
| Disease / Condition |
Organism/ Model |
Tissue | Input | Similarity Metric |
Key Findings | Validation | Measure of Reliability / Other Prioritization Methods |
Public resources used |
Ref |
|---|---|---|---|---|---|---|---|---|---|
| Alzheimer’s disease (AD) |
Human | Hippocampus | DEGs- Top 500 up-regulated Top 500 down-regulated genes based on fold change (unadjusted P-value ≤ 0.05) Calculated 3 different ways (Limma , Limma -ChDir, and mAP-KL) for 5 independent studies (all hippocampus) |
KS-like statistic Correlation coefficient | Proposed 27 candidate drugs. Highlighted potential roles of PKC, HDAC, ARG and GSK3 in mechanism of AD. | None | Negative scores CMap (permuted results P-value < 0.05) SPIEDw (results with significance value > 2) sscMap treatment set score normalized to unity with a tolerance of one false connection among all possible drugs (P < 1/1309, strictest parameterization) LINCS-L1000 (mean connectivity score across the four cell lines in which the perturbagen connected most strongly to the query (best score 4) < −0.9)) Systematically varied calculation of DEGs for input (x5 hippocampal datasets) and algorithms. Combined score across all. |
GEO CMap (build 2) sscMap SPIEDw LINCS-L1000 Enrichr ChemBioServer NetworkAnalyst Mode of Action by NeTwoRk Analysis (Mantra 2.0) | (Siavelis et al. 2016) |
| AD | Human | Entorhinal cortex Hippocampus Middle temporal gyrus Posterior cingulate cortex Superior frontal gyrus Visual cortex | DEGs - Top 400 up-regulated Top 400 down-regulated genes based on fold change (FDR<1%) |
KS-like statistic | Gene signature induced by thioridazine was similar with AD gene signatures in EC, HIP, MTG, and SFG. On the contrary, gene signatures induced by 2 histone deacetylase (HDAC) inhibitors vorinostat and trichostatin A were anticorrelated with AD gene signature in HIP and PC, which indicated that these drugs could possibly reverse the AD gene signature. | None | Positive scores (mechanistic insight) Negative scores (candidate pharmacotherapeutics) | GEO DAVID TransFind CMap KEGG | (Chen et al. 2013) |
| AD | Human | Hippocampus Cerebral cortex | DEGs - Hippocampus: 40 DEGs (40 genes reported by Hata, R. et al 2001; top 20 up and top 20 down) Cerebral cortex: 25 genes with FC > 5 reported by Ricciarelli, R. et al 2004; 11 up and 14 down) |
KS-like statistic | No genes in common between these two query signatures, but both yielded negative connectivity scores with the two independent instances of 4,5-dianilinophthalimide (DAPH). This strengthened the candidacy of DAPH as a potential AD therapeutic. |
in silico (Literature: DAPH was found to reverse the formation of fibrils (a variety of DAPH analogs have been synthesized as potential treatments for AD) |
Negative scores Permutation p-value |
CMap | (Lamb et al. 2006) |
| AD / Cognition enhancers | Human | Hippocampus Cerebral cortex | DEGs - Hippocampus: 40 DEGs (40 genes reported by Hata, R. et al 2001; top 20 up and top 20 down) Cerebral cortex: 25 genes with FC > 5 reported by Ricciarelli, R. et al 2004; 11 up and 14 down) |
KS-like statistic | No genes in common between these two query signatures, but both queries resulted in a common list of negative connections that were given higher confidence. They used an integrative chemoinformatics approach combining CMap with QSAR/VS hits to emphasize connections from the CMap that one would not choose otherwise. |
in vitro (validated binding to 5HT6R, also that raloxifene binds to 5HT6R) in silico (Literature: raloxifene given at a dose of 120 mg/day, but not 60 mg/day, led to reduced risk of cognitive impairment in postmenopausal women) |
Negative scores (They mention statistically significant negative drugs but do not provide the threshold used.) Note: No summarization across multiple experiments for same compound (doses, timepoints, cell lines) |
CMap | (Hajjo et al. 2012) |
| Parkinson’s disease (PD) | Human | Substantia nigra | DEGs- Top 20, 50, 100, 200 or all (535) DE genes from integrated approach. 2 GEO datasets: FC > 1.5 FDR<10% ParkDB (human): Consistent up- or down-regulation across different experiments (P ≤ 0.01) |
KS-like statistic | Top 20 genes from their integrated approach for prioritizing genes outperformed the top 100, 200, 500, and all (536) DEGs for the input signature. Performance was measured by how many (out of the top 50) molecules returned by CMap from their approach were enriched with therapeutic molecules for PD. One candidate, alvespimycin (17-DMAG), was found to be neuroprotective in an in vitro rotenone model of PD. |
in vitro (neuroblastoma cell line) in silico (Enrichment score) |
Negative scores Integrated approach (NOTE: simply using the DEGs from the GEO datasets performed as well as or better than their integrated approach for all but the top 20 genes from their integrated approach). |
CMap GEO ParkDB OMIM CTD | (Gao et al. 2014) |
| Huntington’s disease (HD) | Human | Caudate nucleus | DEGs - Top 100 (absolute FC) 8 up-regulated, 92 downregulated (P < 0.05) |
KS-like statistic | Using a gene signature for HD, CMap identified potential therapeutic agents with multiple modes of action and validated 2 (deferoxamine and chlorzoxazone) in vivo (ameliorated neurodegeneration in flies expressing a mutant HTT fragment). |
in vitro luminescent caspase-activation assay of HTT-induced apoptosis in a PC12 cell line 7 / 12 drugs with negative connectivity scores reduced HTT-induced apoptosis. Chlorzoxazone, copper sulphate, deferoxamine, felbinac, oligomycin and primidone did so in a dosedependent manner. Chemicals with positive connectivity scores had little effect on caspase activation. in vitro High-throughput recording of HTT103Q aggregation in PC12 cells using Cellomics imaging technology. Of the 7, deferoxamine and oligomycin, significantly altered the formation of HTT103Qcontaining inclusion bodies. in vivo Deferoxamine and chlorzoxazone ameliorated neurodegeneration in flies expressing a mutant HTT fragment (a widely studied fruit fly model of mutant HTT toxicity) (Steffan et al. 2001) |
Negative scores | CMap ArrayExpress | (Smalley et al. 2016) |
| Memorymodulating compounds | Human | Saliva | Intragenic SNPs associated with aversive memory recall | Not specified | They used genomic information related to aversive memory—a trait central to posttraumatic stress disorder—to identify several potential drug targets and compounds. In a subsequent pharmacological study with one of the identified compounds, diphenhydramine, they found a druginduced reduction of aversive memory. These findings indicate that genomic information can be used as a starting point for the identification of memory-modulating compounds |
in vivo A single administration of diphenhydramine (50 mg) compared with placebo significantly reduced delayed recall of aversive, but not of positive or neutral, pictures in a double-blind, placebocontrolled study in healthy volunteers |
Not specified | Ingenuity Pathway Analysis (IPA) (note: not public) | (Papassoti ropoulos et al. 2013) |
| Motivation to exercise | Mouse (4 models of motivation to exercise and controls) | Striatum | DEGs - Top 287 genes up-regulated Top 235 genes downregulated in selected lines vs controls (FDR<5%) |
KS-like statistic | Mice from 4 lines selected for wheel running (and 4 non-selected lines) were allowed full access to a running wheel for 6 days. On day 7, half of the high runners and half of the low runners were blocked from wheel access and striatum taken at the time of maximum wheel running and submitted for RNA sequencing. LINCS identified the protein kinase C δ inhibitor, rottlerin, the tyrosine kinase inhibitor, Linifanib and the delta-opioid receptor antagonist 7-benzylidenenaltrexone as potential compounds that mimic the transcriptional signature of the increased motivation to run. | None | Positive scores (to mimic the high motivational state for exercise) Considered both compounds tested across all cell lines and those only tested in neuronal cell lines No statistical p-value mentioned |
DAVID LINCS-L1000 | (Saul et al. 2017) |
| Schizophrenia Major depressive disorder Bipolar disorder Alzheimer’s disease Anxiety disorders Autistic spectrum disorders Attention deficit hyperactivity disorder | Human | 10 brain areas available in GTEx: Anterior cingulate cortex (BA24) Caudate (basal ganglia) Cerebellar hemisphere Cerebellum Cortex Frontal cortex (BA9) Hippocampus Hypothalamus Nucleus accumbens (basal ganglia) Putamen (basal ganglia) | GWAS summary statistics converted to transcriptomic profiles Top K DEGs where K was varied for 50, 100, 250, 500. Results were averaged across each K. |
5 methods: KS-like statistic Spearman correlation with all or with K differentially expressed genes Pearson correlation with all or with K differentially expressed genes |
They imputed transcriptome profiles for 7 psychiatric conditions based on GWAS summary statistics and compared these to druginduced changes in gene expression (CMap) to find possible treatments and found that the top 15 predicted compounds were enriched with known drug-indication pairs. | in silico | Negative Scores Permutation test (shuffled the diseaseexpression z-scores and compared them to drug transcriptomic profiles. Performed 100 permutations for each drug–disease pair and combined the distribution of ranks under the null across all drug–disease pairs, such that the null distribution was derived from 347,800 ranks under H0). |
CMap GTEx MetaXcan KEGG ClinicalTrials.gov MEDication Indication resource (MEDI) Psychiatric Genomics Consortium(PGC) | (So et al. 2017) |
| Morphine Tolerance | Rat | Whole brain | DEGs - Morphine-tolerant + saline versus morphine-tolerant + LPS Placebo-control + saline versus placebo-control + LPS Placebo-control + saline versus morphine-tolerant + saline rats (No p-value threshold reported) |
KS-like statistic | Here, LINCS-L1000 gene knockdown and overexpression experiments were used to inform mechanism of action. Response to LPS was altered during morphine tolerance and indicated that VPS28 may be one of the genes responsible for the alterations associated with morphine tolerance. | None | Genetic perturbation experiments only Positive scores Negative scores |
LINCS-L1000 Query App (apps.lincscloud. org/query) (now deprecated. Instead use clue.io/l1000-query) |
(Chang et al. 2017) |
| Binge-like drinking (important risk factor for AUD) | Mouse (HDID-1 and HS/Npt controls) | Prefrontal cortex Nucleus accumbens core Nucleus accumbens shell Bed nucleus of the stria terminalis Basolateral amygdala Central nucleus of the amygdala Ventral tegmental area Ventral striatum | DEGs - Top 100 up-regulated Top 100 down-regulated genes from each brain area (based on fold change, unadjusted p < 0.05) Differentially expressed landmark genes from each brain area (unadjusted p < 0.05) (landmark genes are those whose expression is directly measured in the L1000 assay) |
KS-like statistic | Many anti-inflammatory compounds had highly negative connectivity scores across brain areas, providing additional evidence for a neuroimmune component in regulating ethanol intake. The top 2 candidates, terreic acid and pergolide, were behaviorally validated to decrease ethanol intake and blood alcohol levels. |
in vivo The top 2 candidates, terreic acid and pergolide, were behaviorally validated to decrease ethanol intake and blood alcohol levels. |
Negative scores Integrated approach Sig_gutc tool (see text) |
LINCS-L1000 GEO | Ferguson et al, 2017 |
| Traumatic Brain Injury (TBI) | Rat | Perilesional cortex Thalamus | DEGs - Top 4964 in perilesional cortex Top 1966 in thalamus (FDR<5%) |
Not reported | The study highlighted tubulins, Nfe2l2, Nfkb2, and S100a4 as target genes modulated by compounds with a high LINCS connectivity score relative to the TBI-sig. Their data suggested that desmethylclomipramine, an active metabolite of the antidepressant clomipramine, is a promising TBI treatment candidate. |
in silico 2/11 top compounds had previously been investigated in epileptogenesis models in vivo. |
Negative scores | LINCS-L1000 GEO IPA GSEA MsigDB | (Lipponen et al. 2016) |
| Ischemic stroke | Rat | Brain (whole hemisphere) | DEGs - Genes with FC1 > 1.5, FC2 > 1.2 and RR > 0 FC1: fold change between middle cerebral artery occlusion (MCAO) and Sham (FC1) FC2: fold change between MCAO and XST where Ci, Mi, and Xi are the average expressions of gene i in control group, MCAO group, and XST treatment group, respectively. |
KS-like statistic | This study sought to investigate the mechanism of action of a Chinese medicine called Xuesaitong injection (XST), a prescription drug made of Panax notoginseng) that is used for treating stroke in China. They looked at positive scores. Inhibition of inflammatory response and coagulation were identified as the major mechanisms involved in the protective effects of XST. | None | Positive scores Permutation p-values < 0.05 |
CMap | (Wang et al. 2015) |
| Epilepsy | Human | Cerebellar cortex Temporal cortex Frontal cortex Occipital cortex Hippocampus Thalamus White matter Medulla Putamen | Epilepsy-associated coexpression module (M30) | Fischer’s Exact Test | They used post-mortem human brain samples from healthy individuals from the UK Brain Expression Consortium (UKBEC) dataset to build gene co-expression networks (modules) and integrated modules with whole-exome-sequencing (WES) studies data of rare de novo mutations in those with epileptic encephalopathy (EE). A single module was selected: M30. The M30 genes’ functional expression for 3 epilepsies suggested downregulation of the network as a common mechanism. They used CMap to identify drugs that could up-regulate the M30 genes. Valproic acid (VPA), a widely used antiepileptic drug, was the drug most significantly predicted to up-regulate the genes in M30 (toward health). | None However, confirmed in vitro that VPA upregulates 51% of the M30 genes in neurons (FDR < 10%), replicating and strengthening the VPAsignature in the cancer cell lines from CMap |
Tested the overlap of M30 genes with the list of genes upregulated by a drug using one-tail
Fischer’s Exact Test (FET) (in order to prioritize drugs predicted to reverse the downregulation of M30 genes
observed in epileptic hippocampi). BH corrected P values for multiple hypotheses testing Included only 152 drugs with ≥ 10 DEGs (FDR < 10%) No summarization across cell lines, doses, timepoints for each drug |
UK Brain Expression Consortium Genotype-Tissue Expression (GTEx) project STRING WebGestalt GeneMANIA Hippie iRefWeb HUGO Gene Nomenclature Committee database CMap | (Delahaye-Duriez et al. 2016) |
| Epilepsy | Mouse (pilocarpine-induced chronic epilepsy) | Hippocampus | DEGs - FDR <0.05 & fold change ≥2 (929 up, 1,164 down) |
KS-like Statistic (used query app on lincscloud.org) | The authors queried the LINCSL1000 database with an epilepsy signature consisting of the top DEGs between the hippocampus of a model of epileptic mice and control mice. They identified 123 compounds with negative scores beyond a chosen threshold (mean of best 4 ≤ −85). These 123 compounds were enriched with compounds known to have antiepileptic effects. Despite a diverse set of mechanisms of action, these compounds targeted similar biological pathways and were better at reversing pathways affected by epilepsy than common antiepileptic compounds. After filtering for practical exclusion criteria, 36 compounds remained. Of these, sitagliptin was confirmed to have antiepileptic activity in vivo. |
in silico (the 123 compounds with LINCS mean connectivity score threshold of −85 or less were 6-fold more enriched with antiepileptic drugs (7/123) than the drugs in the LINCS database as a whole (203/19,767)) in vivo (Sitagliptin produced a dosedependent reduction in seizure scores) in the 6 Hz psychomotor seizure mouse model of pharmacoresistant epilepsy) |
Negative scores Inclusion criterion: LINCS mean connectivity score threshold of −85 (123 compounds) Exclusion criteria: toxicity, parenteral route of administration, lack of animal or human dosage data, or BBB-impermeability (36 compounds remained). This was the only study on the table that used any practical considerations to filter compounds. No published evidence of BBBimpermeability for any of the drugs |
LINCS-L1000 Drug-Set Enrichment Analysis (DSEA) Gene-Set Enrichment Analysis (GSEA) | (Mirza et al. 2017) |
| Nerve regeneration | Rat | Dorsal root ganglion neurons 13 separate studies: a total of 382 gene expression datasets (microarray) related to nerve injury | (1) PPI network consisting of 280 genes (2) regeneration-associated co-expression modules |
KS-like statistic | The authors identified a transcriptional program observed after peripheral, but not central, nerve injury. They used the regenerationassociated modules to query CMap (2 different inputs) and tested the top 3 candidates that emerged from the intersection of the 2 queries (ambroxol, lasalocid, and disulfiram). Only ambroxol enhanced axonal outgrowth of DRG neurons in vitro and it also increased optic nerve (ON) regeneration in mice after a crush injury, albeit a modest improvement. |
in vitro ambroxol, lasalocid, and disulfiram were tested, but only ambroxol enhanced axonal outgrowth of DRG neurons in vivo ambroxol increased optic nerve (ON) regeneration in mice after a crush injury, albeit a modest improvement. |
Positive scores Permutation p-value |
CMap GEO PubMatrix DAVID JASPAR TRANSFAC STRING ENCODE | (Chandran et al. 2016) |
| CNS injury | Human MCF7 breast adenocarcinoma cells | NA | DEGs - absolute fold change ≥1.5 p <0.05 (10 up and 12 down) 6 hr treatment of F05 (5 μM; a compound the group previously identified to promote neurite growth in vitro and in vivo) vs vehicle (DMSO, 0.05%) |
KS-like statistic | The authors used the transcriptional signature of a compound known to induce neurite growth (F05) as a “seed” to identify other compounds with this same property. Remarkably, despite no chemical similarity to F05, a group of piperazine phenothiazine antipsychotics had similar effects on gene expression and were found to promote neurite growth in vitro at least partially through antagonism of calmodulin signaling, independent of dopamine receptor antagonism. |
in vitro ¾ piperazine phenothiazine antipsychotics (but none of the other classes of antipsychotics) significantly enhanced neurite growth of dissociated hippocampal neurons and rat retinal ganglion cells on a substrate of a mixture of chondroitin sulfate proteoglycans (CSPGs) (glial scar proteins) |
Positive scores | CMap | (Johnstone et al. 2012) |
| CNS injury / neurodegenerat ion | Mouse (Ascl1-EGFPBac transge nic reporter mouse line) | Subventricular zone (SVZ) of the dentate gyrus microdomains Regionspecific neural stem cells (NSCs) and their immediate progeny Transient amplifying cells (TAPs) | DEG’s - 1.8-fold change FDR < 5% |
Pearson correlation (via SPIED webbased tool) | This study aimed to identify compounds that could direct germinal activity in the subventricular zone (SVZ), which would have therapeutic potential in nerve injury / neurodegenerative or demyelination diseases. They examined NSC lineages in the SVZ microdomains (dorsal versus ventral/lateral) and identified small molecules that direct the fate of these cells toward neurogenic as opposed to oligodendrogenic lineages. LY-294002, an inhibitor of PI3K/Akt, induces transcriptional changes that promote oligodendrogenesis. AR-A014418, an inhibitor of GSK3β induces transcriptional changes that promote rejuvenation of the adult SVZ. |
in vivo Infusion of AR-A014418 or CHIR99021 in the adult (P90) dSVZ lateral ventricle dramatically stimulated the germinal activity of the adult dSVZ CHIR99021, a GSK3β inhibitor, showed regenerative potential in a neuropathological context in a model of premature injury that leads to diffuse oligodendroglial and neuronal loss throughout the cortex |
Positive scores | CMap GeneGO Metacore for Process Networks SPIEDw | (Azim et al. 2017) |
| Down Syndrome (DS) |
Human Mouse (3 models of DS: Dp16, Ts65Dn and Ts1Cje and controls) |
Human: Second trimester amniocytes Induced pluripotent stem cells (iPSCs) Neurons derived from iPSCs Post-mortem human fetal cerebellum and cerebrum Mouse: Developing forebrain (E15.5) |
DEGs - BH-FDR < 5% and 20% Because of the limited number of differentially regulated genes at FDR 20%, they used the top 1% up- and down-regulated genes |
KS-like statistic | The authors used a human/mouse integrated approach to identify 17 high priority molecules predicted to reverse pathway changes in both human cells and mouse models. They tested the effects of apigenin, one of the molecules predicted by the CMap to treat dysregulated pathways in DS, on human amniocytes derived from fetuses with DS and on the Ts1Cje mouse model of DS. Apigenin treatment reduced oxidative stress in DS amniocytes and improved some aspects of brain morphogenesis, gene expression and postnatal behavior in the Ts1Cje mouse model (manuscript in preparation). | None – currently undergoing in vitro and in vivo testing (not yet published) | Negative scores Threshold: −0.7 or less Integrated approach (multiple inputs) |
GSEA DAVID IPA GEO CMap | (Guedj et al. 2016) |
| Down Syndrome | Human | Second trimester amniotic fluid | DEGs - FDR<5% (414 probes individually differentially expressed between trisomy 21 and controls |
KS-like statistic | This is one of the first studies to demonstrate that transcriptional profiling of RNA in uncultured amniotic fluid provides molecular insights into developmental disorders in the living human fetus. They found 4 compounds with average connectivity scores >0.7 (indicating a high correlation with the DS molecular signature), and 9 compounds with average connectivity scores less than −0.7 (indicating a high negative correlation) (NSC-5255229, celastrol, calmidazolium, NSC-5109870, dimethyloxalylglycine, NSC-5213008, verapamil, HC toxin, and felodipine). The 4 compounds that most mimic the DS phenotype were related to potassium and calcium signaling or oxidation which further supports the importance of oxidative stress and ion transport as functional classes involved in DS. | None | Positive scores (mechanistic insight) Negative scores (candidate pharmacotherapeutics) |
GSEA DAVID CMap (build 1.0) | (Slonim et al. 2009) |
Regarding the construction of the input genomic signature, the studies fall into two main categories: those that use genotype data (i.e., SNPs related to a disease phenotype discovered from genome-wide association studies (GWAS)) and those that use gene expression data. Gene expression measurement technology (RNA sequencing or microarray) provides the expression levels of all genes in the genome simultaneously, supplying a functional genomic readout of the effects of the combination of the genetic variants that could be contributing to disease. Gene expression is by far the primary input used by the studies in Table 3, the idea being to compare the gene expression levels between disease and healthy tissue and to use the top differentially expressed genes as the input signature, as this is thought to best capture the molecular differences driving disease phenotypes. However, there is no consensus on the optimal threshold or number of differentially expressed genes to use. Differentially expressed genes can be subdivided into groups of genes with highly correlated expression levels. Indeed, several studies incorporate gene co-expression networks or protein-protein interaction networks to refine the genomic input signature (Chandran et al. 2016; Delahaye-Duriez et al. 2016; Gao et al. 2014). One study compared the performance of using only differentially expressed genes between Parkinson’s and normal brain to query CMap versus a combined approach that used both differential expression and gene coexpression network information (Gao et al. 2014). They calculated the number of known Parkinson’s therapeutics in the top 50 ranked compounds from each approach. Using the top 20 genes from the combined method outperformed using the top 20 genes from differential expression alone. They were not able to assess the performance of using only the gene coexpression network as a query for lack of up-regulated genes in the coexpression module. Interestingly, using more than the top 20 genes from the combined approach led to a decrease in performance. Since gene coexpression network modules are driven by variability in the data, and cell type is a major contributor to gene expression variability, it is possible that network based approaches could be more useful for diseases that primarily affect a specific cell type (like is the case for Parkinson’s disease). One downside of using gene expression data is that human brain tissue can only be obtained postmortem and the transcriptional signature can be confounded by a lifetime with the disease or pharmaceutical management of the disease (see Challenges and Future Directions).
Genotype/gene sequence data, on the other hand, is readily available, easy to attain, and is relatively static throughout the patient’s lifetime, but it is not without its drawbacks. Many genes contribute to the genetic risk of most complex psychiatric disorders, each contributing a small effect. A minority of disease-associated SNPs are mapped to protein-coding regions of the genome, and there are few drugs that specifically target particular gene products. Despite these challenges, Papassotiropoulos et al, (2013) used intragenic SNPs related to aversive memory performance to select the antihistamine, diphenhydramine, as a potential drug that would reduce aversive memory recall (Papassotiropoulos et al. 2013). This was verified in a double-blind, placebo-controlled, cross-over study in which a single administration of diphenhydramine (50 mg) compared with placebo significantly reduced delayed recall of aversive, but not of positive or neutral, pictures.
Most disease-associated SNPs, however, occur in non-coding regions and their impact on disease outcome is difficult to causally link to gene expression, although this is an active area of research and databases exist trying to relate SNPs with gene expression in a variety of tissues including brain (e.g., Genotype-Tissue Expression Project; https://www.gtexportal.org/home/). One study approached this problem in an innovative way for psychiatric illnesses and could be considered a hybrid approach since the authors inferred gene expression data from genotype data (So et al. 2017). The approach relied on an algorithm called MetaXcan (Barbeira et al. 2016), that incorporated GTEx data to build statistical models for predicting expression levels from SNPs in a reference transcriptome data set, and these prediction models were used to impute the expression z-scores (i.e., z-statistics derived from association tests of expression changes with disease status) based on GWAS summary statistics. Transcriptome profiles were imputed for 7 psychiatric conditions based on GWAS summary statistics and compared to drug-induced changes in gene expression using CMap to identify potential treatment candidates. Novel compounds were not tested; however, it was promising that the top 15 predicted compounds for some of the psychiatric disorders were enriched with known and predicted psychiatric medications according to several drug-disease indication measures (Anatomical Therapeutic Chemical (ATC) codes, ClinicalTrials.gov, MEDication Indication (MEDI) resource).
Once the input genomic signature is defined, it can be compared to a database of drug signatures. Most of the studies in Table 3 (13/19) use the original CMap database. The benefit of CMap is that it is smaller and simpler to perform statistics to assess a connectivity score’s reliability. However, the trade-off is fewer drugs and cell lines, the latter of which is especially important for brain diseases since CMap contains no brain cell lines, whereas LINCS-L1000 contains 2 brain cell lines with considerable data, NEU and NPC. Unfortunately, at this time, these cell lines are not included in the implementation on their query app at clue.io, however, the LINCS-L1000 datasets can also be downloaded from Gene Expression Omnibus (GEO) (accession numbers GSE70138 and GSE92742). Efforts are underway to include more brain cell lines in the LINCS-L1000 database to facilitate its relevance to brain diseases (RDM, personal communication).
To compare the disease and drug signatures the KS-like statistic (as described by (Lamb et al. 2006; Subramanian et al. 2017) is the most frequently used similarity metric, although several studies also use Spearman or Pearson correlation coefficients (Azim et al. 2017; Siavelis et al. 2016; So et al. 2017) or Fischer’s Exact Test (Delahaye-Duriez et al. 2016). Most studies operate under the transcriptional “reversal hypothesis”, which assumes that drugs with negative connectivity scores (i.e., with gene expression signatures that revert the disease’s effects on gene expression to the control state) would ameliorate disease phenotype. Five of the 19 studies outlined in Table 3 have functionally validated this hypothesis, in that the candidate compounds ameliorated some disease phenotype when tested behaviorally (though none have confirmed that the beneficial effects of the compound were due to the restoration of gene expression to the “normal” state) (Chandran et al. 2016; Ferguson et al. 2017; Mirza et al. 2017; Papassotiropoulos et al. 2013; Smalley et al. 2016).
These studies provide a functional rationale for prioritizing negatively-scoring compounds, i.e., those that have opposing effects on gene expression associated with the disease state. However, in addition to reflecting gene expression changes that drive the disease or represent deleterious aspects of a disease state, the differentially expressed genes between disease and healthy samples could also reflect protective homeostatic compensations within the system. Because some of the differentially expressed genes might be beneficial, it is reasonable to also consider drugs with high positive connectivity scores.
The rationale for the reversal hypothesis was tested directly utilizing a gene expression signature comprised of the top 100 differentially expressed genes identified in Huntington’s disease (HD). Data were obtained from the caudate nucleus from disease vs sex-and age-matched human controls followed by CMap query (Smalley et al. 2016). The top 12 positive and negative scoring compounds were tested in in vitro caspase-activation assays to assess the degree to which they modulated mutant huntingtin (HTT)-induced apoptosis in a PC12 cells. None of the positive scoring compounds affected caspase activity, while 7/12 negative scoring compounds decreased caspase activity, two of which had neuroprotective effects in vivo in a drosophila model of HD. This outcome supports the “reversal hypothesis”. However, because the caspase activity was approaching 100% (i.e. a ceiling effect), the ability to observe increased caspase activity precluded experimental outcomes predicted from positive scores that might mimic/worsen disease phenotypes (the converse of the reversal hypothesis).
One study does provide in vivo validation of the converse of the reversal hypothesis: that drugs with positive scores (i.e., with gene expression signatures that are similar the disease’s effects on gene expression) would mimic the state of interest. Azim and colleagues (2017) sought to identify small molecules to mobilize endogenous stem cells and direct their fate as a therapy for neurodegenerative and demyelinating disorders (Azim et al. 2017). These studies used the transcriptional signatures of neural stem cells (NSCs) in the ventral/lateral subventricular zone (SVZ) of the dentate gyrus which give rise to interneurons of the olfactory bulb and cortical areas, and of NSCs in the dorsal SVZ which give rise to glutamatergic neurons and oligodendrocytes. The authors prioritized positively-scoring compounds with the hopes that that would reproduce the lineage-specific transcriptional signatures. Indeed, the most promising candidates, LY-294002, an inhibitor of PI3K/Akt, promoted development of oligodendrocytes, and AR-A014418, an inhibitor of GSK3β, rejuvenated the NSC lineage. Furthermore, another GSK3β inhibitor promoted regeneration in a mouse model of hypoxic brain injury, by recruiting new oligodendrocytes and glutamatergic neurons into the cortex.
In addition to gene expression signatures of drug perturbation, the LINCS-L1000 database also catalogs gene expression response to genetic perturbation. One study utilized this resource and compared the input genomic signature to those of gene knockdown or overexpression in LINCS-L1000 to gain mechanistic insight into how morphine tolerance alters response to lipopolysaccharide (LPS), and found that VPS28 may be one of the genes responsible for the alterations associated with morphine tolerance (Chang et al. 2017). In addition to looking at the negatively-correlated drugs for treatment candidates, several studies also analyzed the positively-correlated drugs for mechanistic insight into the disease, as these would be predicted to produce similar effects on gene expression and mimic or worsen disease phenotype (Chen et al. 2013; Slonim et al. 2009).
Challenges and Future Directions
The CMap and LINCS-L1000 databases contain multiple experiments for the same compound. It is clear that the compound’s effects on gene expression are greatly affected by variables such as cell line, dose, and time point at which gene expression is assayed (Chen et al. 2017). Some researchers make no attempt to summarize across cell lines, doses, and time points to attain a composite compound-level view, which could lead to spurious results, particularly if the cell line is vastly different from the cellular makeup of the tissue used to generate the genomic signature used as the input query. For these reasons, we propose that using multiple expression datasets, algorithm parameter settings, and methods for prioritizing compounds (as taken by (Ferguson et al. 2017; Gao et al. 2014; Guedj et al. 2016; Siavelis et al. 2016)) are critical to identify an effective drug candidate, at least until proper gold standard datasets exist with which to benchmark the optimal settings (see below).
An important limitation of in silico gene mapping approaches is that they rely on comparisons of brain gene expression data to gene expression data from cell culture, and are therefore constrained by the same limitations of any in vitro system. Brain gene expression is a complex combination of direct and indirect expression changes occurring in multiple cell types and brain regions. Even if brain-relevant cell lines were included in the expression profiles of LINCS-L1000 or other databases of drug-related transcriptomes, it is unclear how relevant in vitro results are to the biology of an intact organism, which is why in vivo experimental validation is critical. Only 6 of the 19 studies for brain disease listed in Table 3 performed in vivo validation of the proposed pharmaceutical candidates (Azim et al. 2017; Chandran et al. 2016; Ferguson et al. 2017; Mirza et al. 2017; Papassotiropoulos et al. 2013; Smalley et al. 2016), and none directly tested the underlying assumption of in silico connectivity mapping. Specifically, it is important to address the following question: if a candidate compound is effective in treating a given disease phenotype, was it the result of a reversal in expression of disease-related genes by the compound? This is difficult to assess given the complexity of the regulation of gene expression. Parameters such as drug dose and treatment times are critical for determining meaningful gene expression changes. Therefore, a range of doses and time points would need to be measured, and although the cost of whole genome sequencing is decreasing, to do this with the required samples sizes would be cost prohibitive. In addition to L1000 technology, the development of less expensive sequencing techniques, such TagSeq, improve feasibility to test this hypothesis which will provide important mechanistic insight into in silico gene mapping approaches (Lohman et al. 2016; Meyer et al. 2011).
Each of the 6 studies discussed in the previous paragraph used a different approach to identify a candidate compound that ameliorated disease phenotype when tested (Azim et al. 2017; Chandran et al. 2016; Ferguson et al. 2017; Mirza et al. 2017; Papassotiropoulos et al. 2013; Smalley et al. 2016), and it is critical to identify the approach(s) with the greatest predictive accuracy. In other words, which choices at each of the main steps outlined above are the most likely to identify compounds that actually ameliorate the disease state? It is currently difficult to address this question because of the low throughput nature of behavioral testing and the non-existence of gold standard data with which to benchmark various approaches.
A benchmark approach requires a gold standard dataset comprised of two components from the same population: (1) gene expression and/or genotyping data. Ideally, gene expression data would be obtained from multiple brain areas, cell types (single cell or cell-type transcriptomes) and tissue types (peripheral blood mononuclear cells (PBMCs), liver, gut microbiome, etcetera), and (2) drug response. This should be the same measurement for each drug and there would ideally be a large range of drug effects. This continuous variable would lend itself to correlation analysis (rather than a binary measure of 0: drug was ineffective and 1: drug was effective). Drugs that are known to affect the phenotype (true positives) and drugs that are known to not effect phenotype (true negatives) should be present to assess how well the approach can discriminate (assign high scores to the true positives and poor scores to the true negatives). A benchmarking test-case scenario using this ideal gold standard dataset would systematically vary the input, algorithm, and prioritization scoring choice and assess the outputs for their predictability (Table 4).
Table 4:
Benchmarking Test Case
| 1. Input | 2. Similarity Metric | 3. Prioritization |
|---|---|---|
|
Tissue: Whole brain Brain regions Cell type-specific transcriptome Single cell transcriptome Genes: SNPs Differentially-expressed (top 50, 100 →500) Coexpression modules |
Correlation Enrichment (hypergeometric, Fischer’s, modified KS statistic) Pattern matching | Statistical methods Negative score vs absolute value of score Threshold (for example, −90 cut off) Median Combination of above |
One caveat to this benchmarking strategy outlined here, is that it is unreasonable to assume that all compounds with therapeutic potential would be identified by in silico gene mapping. The best way to evaluate these approaches would be to take a heuristic testing strategy and select a few compounds nominated from various combinations at each of the 3 steps to test behaviorally, but as mentioned previously, behavioral testing is low throughput and this would be resource-intensive.
As discussed before, the affected tissue (brain) is not available for testing until postmortem, which certainly poses a problem if computational approaches that rely on gene expression measures are to be incorporated into drug repurposing / personalized medicine endeavors for brain diseases. Moreover, analysis of postmortem brain expression is plagued with the “chicken and the egg” conundrum. Meaning that it is impossible to know if the observed gene expression changes are the cause or the effect (due to years of alcohol use, for example) of the disease. This is one reason why animal models are key in studying brain diseases, and using animal models with high predictive validity for selecting therapeutic compounds is one way around this problem. Another option would be to identify a surrogate for brain gene expression, and this is an active area of research. Great hope has arrived with the discovery of inducible pluripotent stem cells (iPSC’s) that can be differentiated into various neuronal types (Takahashi and Yamanaka 2006). There are also methods that skip the induced pluripotent steps allowing direct conversion into functional neurons, called induced neurons, or iN’s (for review see (Drouin-Ouellet et al. 2017)). Although these cell models hold promise for improving treatment of psychiatric diseases (Oni et al. 2016; Stern et al. 2017), the protocols are long and tedious to produce adult-like neurons and the relevance to in silico gene mapping remains unexplored. Another surrogate for brain gene expression could be in peripherally accessible cell types, like PBMCs, especially considering the impressive evidence for immune involvement in AUD (for reviews see (Crews et al. 2017; de Timary et al. 2017; Mayfield and Harris 2017)). Another option that does not require access to brain tissue is imputing gene expression from GWAS summary data as discussed above (So et al. 2017). However, the latter approach has yet to be validated in vivo and will likely improve as the databases used to make the imputations improve, for example, by increasing samples in GTEx to better detect eQTLs. The GTEx Consortium plans to include up to 1,000 donors in the final data release and collect complementary molecular data on subsets of samples, including epigenetic and protein data (Consortium et al. 2017). Using GTEx data to impute transcriptomes for diverse groups of people should be approached with caution, as the donors are currently 83.7% European American and 15.1% African American (with the v7 Release) (Consortium et al. 2017).
Conclusion
The benefits of using computational strategies to transition to a more molecularly-informed healthcare system are numerous. For example, diagnoses could be more precise and treatments more successful. Patients diagnosed with the same disease often represent a heterogeneous mixture of different underlying disorders, since there are numerous molecular disruptions that could lead to similar clinical presentations. This is especially true for AUD and other brain diseases, where a molecular readout of the affected organ is limited. It is no surprise, then, that the standard treatments fail for many because of incomplete knowledge regarding the underlying cause of a patient’s disruptive symptoms. As we become more advanced in our ability to construct and interpret a molecular signature underlying disease symptoms, healthcare will advance towards personalized medicine, where each patient is treated to his or her individual profile.
The systems pharmacology approaches discussed in this Review have two main beneficial outcomes that should be considered independently. The first is an effective treatment and the other is mechanistic insight. It might be that the effectiveness of a compound is understood before its mechanism of action. However, progressing promising pharmaceutical treatment should not wait for the full understanding of the mechanism, as the mechanism underlying some of the most long-standing and successful treatments in medicine are still poorly understood (Letai 2017). In fact, as suggested by (Hajjo et al. 2012), one of the main benefits of this approach is to identify potentially therapeutic compounds without necessarily understanding the underlying target-specific mechanism.
Much hope has been placed on information contained within large genomic datasets and network approaches to drive clinical treatment towards personalized medicine and revolutionize healthcare. And, indeed, bioinformatics approaches have shown some success for identifying novel treatments for brain diseases. However, this research is still in its infancy, and many questions remain to be answered if these high expectations are to be met. Here, we have proposed the steps required for in silico gene mapping for the purpose of drug discovery and repurposing, reviewed state-of-the-art applications of these approaches to brain diseases, and highlighted some of the critical challenges facing the field. Success relies on the integration of enormous amounts of sequence and phenotype data from public and private sector sources. Ultimately, it will take a collaborative effort from academia, industry and government to advance drug development and repurposing for AUD (Litten et al. 2014).
Acknowledgements
Supported by funding from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) of the National Institutes of Health (NIH): AA024332 to LF and AA020926 to RDM, and to the Integrative Neuroscience Initiative on Alcoholism (INIA)-Neuroimmue consortium AA012404 to RAH.
References
- Alanis-Lobato G, Andrade-Navarro MA, Schaefer MH (2017) HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res 45: D408–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Hamosh A (2017) Searching Online Mendelian Inheritance in Man (OMIM): A Knowledgebase of Human Genes and Genetic Phenotypes. Curr Protoc Bioinformatics 58: 1 2 1–1 2 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton RF, Kranzler H, Breder C, Marcus RN, Carson WH, Han J (2008) A randomized, multicenter, double-blind, placebo-controlled study of the efficacy and safety of aripiprazole for the treatment of alcohol dependence. J Clin Psychopharmacol 28: 5–12. [DOI] [PubMed] [Google Scholar]
- Athanasiadis E, Cournia Z, Spyrou G (2012) ChemBioServer: a web-based pipeline for filtering, clustering and visualization of chemical compounds used in drug discovery. Bioinformatics 28: 3002–3. [DOI] [PubMed] [Google Scholar]
- Azim K, Angonin D, Marcy G, Pieropan F, Rivera A, Donega V, Cantu C, Williams G, Berninger B, Butt AM, Raineteau O (2017) Pharmacogenomic identification of small molecules for lineage specific manipulation of subventricular zone germinal activity. PLoS Biol 15: e2000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ (2012) GeneWeaver: a web-based system for integrative functional genomics. Nucleic Acids Res 40: D1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Gulbahce N, Loscalzo J (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira A, Shah KP, Torres JM, Wheeler HE, Torstenson ES, Edwards T, Garcia T, Bell GI, Nicolae D, Cox NJ, Im HK (2016) MetaXcan: Summary Statistics Based Gene-Level Association Method Infers Accurate PrediXcan Results. bioRxiv. [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A (2013) NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41: D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C, Haudenschild CD, Beckman KB, Shi J, Mei R, Urban AE, Montgomery SB, Levinson DF, Koller D (2014) Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res 24: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Hosack DA, Dennis G Jr., Lempicki RA, Bright TJ, Cheadle C, Engel J (2003) PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics 4: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Hauser SR, Liang T, Sari Y, Maldonado-Devincci A, Rodd ZA (2017) Rat animal models for screening medications to treat alcohol use disorders. Neuropharmacology 122: 201–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Lopez MF, Cui C, Egli M, Johnson KW, Franklin KM, Becker HC (2015) Ibudilast reduces alcohol drinking in multiple animal models of alcohol dependence. Addict Biol 20: 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Benavidez JM, Black M, Ferguson LB, Schoenhard GL, Goate AM, Edenberg HJ, Wetherill L, Hesselbrock V, Foroud T, Harris RA (2015) Peroxisome proliferator-activated receptors alpha and gamma are linked with alcohol consumption in mice and withdrawal and dependence in humans. Alcohol Clin Exp Res 39: 136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Black M, Benavidez JM, Stamatakis EE, Harris RA (2016a) PPAR Agonists: I. Role of Receptor Subunits in Alcohol Consumption in Male and Female Mice. Alcohol Clin Exp Res 40: 553–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Black M, Benavidez JM, Stamatakis EE, Harris RA (2016b) PPAR Agonists: II. Fenofibrate and Tesaglitazar Alter Behaviors Related to Voluntary Alcohol Consumption. Alcohol Clin Exp Res 40: 563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomprezzi R (2015) Dimethyl fumarate in the treatment of relapsing-remitting multiple sclerosis: an overview. Ther Adv Neurol Disord 8: 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borro P, Leone S, Testino G (2016) Liver Disease and Hepatocellular Carcinoma in Alcoholics: The Role of Anticraving Therapy. Curr Drug Targets 17: 239–51. [DOI] [PubMed] [Google Scholar]
- Brower KJ (2015) Assessment and treatment of insomnia in adult patients with alcohol use disorders. Alcohol 49: 417–27. [DOI] [PubMed] [Google Scholar]
- Butler D (2008) Translational research: crossing the valley of death. Nature 453: 840–2. [DOI] [PubMed] [Google Scholar]
- Bymaster FP, Dreshfield-Ahmad LJ, Threlkeld PG, Shaw JL, Thompson L, Nelson DL, Hemrick-Luecke SK, Wong DT (2001) Comparative affinity of duloxetine and venlafaxine for serotonin and norepinephrine transporters in vitro and in vivo, human serotonin receptor subtypes, and other neuronal receptors. Neuropsychopharmacology 25: 871–80. [DOI] [PubMed] [Google Scholar]
- Carrella D, Napolitano F, Rispoli R, Miglietta M, Carissimo A, Cutillo L, Sirci F, Gregoretti F, Di Bernardo D (2014) Mantra 2.0: an online collaborative resource for drug mode of action and repurposing by network analysis. Bioinformatics 30: 1787–8. [DOI] [PubMed] [Google Scholar]
- Chandran V, Coppola G, Nawabi H, Omura T, Versano R, Huebner EA, Zhang A, Costigan M, Yekkirala A, Barrett L, Blesch A, Michaelevski I, Davis-Turak J, Gao F, Langfelder P, Horvath S, He Z, Benowitz L, Fainzilber M, Tuszynski M, Woolf CJ, Geschwind DH (2016) A Systems-Level Analysis of the Peripheral Nerve Intrinsic Axonal Growth Program. Neuron 89: 956–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SL, Huang W, Mao X, Sarkar S (2017) NLRP12 Inflammasome Expression in the Rat Brain in Response to LPS during Morphine Tolerance. Brain Sci 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheer SM, Bang LM, Keating GM (2004) Ropinirole: for the treatment of restless legs syndrome. CNS Drugs 18: 747–54; discussion 755–6. [DOI] [PubMed] [Google Scholar]
- Chen B, Butte AJ (2013) Network medicine in disease analysis and therapeutics. Clin Pharmacol Ther 94: 627–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Butte AJ (2016) Leveraging big data to transform target selection and drug discovery. Clin Pharmacol Ther 99: 285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Ma L, Paik H, Sirota M, Wei W, Chua MS, So S, Butte AJ (2017) Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets. Nat Commun 8: 16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Guan Q, Nie ZY, Jin LJ (2013) Gene expression profile and functional analysis of Alzheimer’s disease. Am J Alzheimers Dis Other Demen 28: 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciraulo DA, Barlow DH, Gulliver SB, Farchione T, Morissette SB, Kamholz BW, Eisenmenger K, Brown B, Devine E, Brown TA, Knapp CM (2013) The effects of venlafaxine and cognitive behavioral therapy alone and combined in the treatment of co-morbid alcohol use-anxiety disorders. Behav Res Ther 51: 729–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo G, Addolorato G, Agabio R, Carai MA, Pibiri F, Serra S, Vacca G, Gessa GL (2004) Role of GABA(B) receptor in alcohol dependence: reducing effect of baclofen on alcohol intake and alcohol motivational properties in rats and amelioration of alcohol withdrawal syndrome and alcohol craving in human alcoholics. Neurotox Res 6: 403–14. [DOI] [PubMed] [Google Scholar]
- Consortium EP (2011) A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT (2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, Nih/Nci, Nih/Nhgri, Nih/Nimh, Nih/Nida, Biospecimen Collection Source Site N, Biospecimen Collection Source Site R, Biospecimen Core Resource V, Brain Bank Repository-University of Miami Brain Endowment B, Leidos Biomedical-Project M, Study E, Genome Browser Data I, Visualization EBI, Genome Browser Data I, Visualization-Ucsc Genomics Institute UoCSC, Lead a, Laboratory DA, Coordinating C, management NIHp, Biospecimen c, Pathology, e QTLmwg, Battle A, Brown CD, Engelhardt BE, Montgomery SB (2017) Genetic effects on gene expression across human tissues. Nature 550: 204–213.29022597 [Google Scholar]
- Corsello SM, Bittker JA, Liu Z, Gould J, McCarren P, Hirschman JE, Johnston SE, Vrcic A, Wong B, Khan M, Asiedu J, Narayan R, Mader CC, Subramanian A, Golub TR (2017) The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med 23: 405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Lawrimore CJ, Walter TJ, Coleman LG Jr. (2017) The role of neuroimmune signaling in alcoholism. Neuropharmacology 122: 56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ (2017) The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res 45: D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Timary P, Starkel P, Delzenne NM, Leclercq S (2017) A role for the peripheral immune system in the development of alcohol use disorders? Neuropharmacology 122: 148–160. [DOI] [PubMed] [Google Scholar]
- Delahaye-Duriez A, Srivastava P, Shkura K, Langley SR, Laaniste L, Moreno-Moral A, Danis B, Mazzuferi M, Foerch P, Gazina EV, Richards K, Petrou S, Kaminski RM, Petretto E, Johnson MR (2016) Rare and common epilepsies converge on a shared gene regulatory network providing opportunities for novel antiepileptic drug discovery. Genome Biol 17: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez G, Dagnas M, Decorte L, Vandesquille M, Belzung C, Beracochea D, Mons N (2016) Rescuing prefrontal cAMP-CREB pathway reverses working memory deficits during withdrawal from prolonged alcohol exposure. Brain Struct Funct 221: 865–77. [DOI] [PubMed] [Google Scholar]
- Donoghue K, Rose A, Coulton S, Milward J, Reed K, Drummond C, Little H (2016) Doubleblind, 12 month follow-up, placebo-controlled trial of mifepristone on cognition in alcoholics: the MIFCOG trial protocol. BMC Psychiatry 16: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin-Ouellet J, Pircs K, Barker RA, Jakobsson J, Parmar M (2017) Direct Neuronal Reprogramming for Disease Modeling Studies Using Patient-Derived Neurons: What Have We Learned? Front Neurosci 11: 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Bugrim A, Brovold L, Kirillov E, Nikolsky Y, Rakhmatulin E, Sorokina S, Ryabov A, Serebryiskaya T, Melnikov A, Metz J, Nikolskaya T (2006) Algorithms for network analysis in systems-ADME/Tox using the MetaCore and MetaDrug platforms. Xenobiotica 36: 877–901. [DOI] [PubMed] [Google Scholar]
- Falk DE, Castle IJ, Ryan M, Fertig J, Litten RZ (2015) Moderators of Varenicline Treatment Effects in a Double-Blind, Placebo-Controlled Trial for Alcohol Dependence: An Exploratory Analysis. J Addict Med 9: 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farokhnia M, Schwandt ML, Lee MR, Bollinger JW, Farinelli LA, Amodio JP, Sewell L, Lionetti TA, Spero DE, Leggio L (2017) Biobehavioral effects of baclofen in anxious alcohol-dependent individuals: a randomized, double-blind, placebo-controlled, laboratory study. Transl Psychiatry 7: e1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson LB, Most D, Blednov YA, Harris RA (2014) PPAR agonists regulate brain gene expression: relationship to their effects on ethanol consumption. Neuropharmacology 86: 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson LB, Ozburn AR, Ponomarev I, Metten P, Reilly M, Crabbe JC, Harris RA, Mayfield RD (2017) Genome-Wide Expression Profiles Drive Discovery of Novel Compounds that Reduce Binge Drinking in Mice. Neuropsychopharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Zhao G, Fang JS, Yuan TY, Liu AL, Du GH (2014) Discovery of the neuroprotective effects of alvespimycin by computational prioritization of potential anti-Parkinson agents. FEBS J 281: 1110–22. [DOI] [PubMed] [Google Scholar]
- Geisel O, Hellweg R, Muller CA (2016) Serum levels of brain-derived neurotrophic factor in alcohol-dependent patients receiving high-dose baclofen. Psychiatry Res 240: 177–180. [DOI] [PubMed] [Google Scholar]
- Geisler BP, Ghosh A (2014) Gabapentin treatment for alcohol dependence. JAMA Intern Med 174: 1201. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Lue TF, Padma-Nathan H, Rosen RC, Steers WD, Wicker PA (1998) Oral sildenafil in the treatment of erectile dysfunction. Sildenafil Study Group. N Engl J Med 338: 1397404. [DOI] [PubMed] [Google Scholar]
- Gong MF, Wen RT, Xu Y, Pan JC, Fei N, Zhou YM, Xu JP, Liang JH, Zhang HT (2017) Attenuation of ethanol abstinence-induced anxiety- and depressive-like behavior by the phosphodiesterase-4 inhibitor rolipram in rodents. Psychopharmacology (Berl). [DOI] [PubMed] [Google Scholar]
- Gray KA, Yates B, Seal RL, Wright MW, Bruford EA (2015) Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 43: D1079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedj F, Pennings JL, Massingham LJ, Wick HC, Siegel AE, Tantravahi U, Bianchi DW (2016) An Integrated Human/Murine Transcriptome and Pathway Approach To Identify Prenatal Treatments For Down Syndrome. Sci Rep 6: 32353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmo R, Martinotti G, Clerici M, Janiri L (2012) Pregabalin for alcohol dependence: a critical review of the literature. Adv Ther 29: 947–57. [DOI] [PubMed] [Google Scholar]
- Guglielmo R, Martinotti G, Quatrale M, Ioime L, Kadilli I, Di Nicola M, Janiri L (2015) Topiramate in Alcohol Use Disorders: Review and Update. CNS Drugs 29: 383–95. [DOI] [PubMed] [Google Scholar]
- Guo AY, Webb BT, Miles MF, Zimmerman MP, Kendler KS, Zhao Z (2009) ERGR: An ethanol-related gene resource. Nucleic Acids Res 37: D840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile CN, Kosten TA (2017) The peroxisome proliferator-activated receptor alpha agonist fenofibrate attenuates alcohol self-administration in rats. Neuropharmacology 116: 364370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjo R, Setola V, Roth BL, Tropsha A (2012) Chemocentric informatics approach to drug discovery: identification and experimental validation of selective estrogen receptor modulators as ligands of 5-hydroxytryptamine-6 receptors and as potential cognition enhancers. J Med Chem 55: 5704–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker N, Ahmed J, von Eichborn J, Dunkel M, Macha K, Eckert A, Gilson MK, Bourne PE, Preissner R (2012) SuperTarget goes quantitative: update on drug-target interactions. Nucleic Acids Res 40: D1113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusser SA, Howard RJ, Borghese CM, Cullins MA, Broemstrup T, Lee US, Lindahl E, Carlsson J, Harris RA (2013) Functional validation of virtual screening for novel agents with general anesthetic action at ligand-gated ion channels. Mol Pharmacol 84: 670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard RJ, Trudell JR, Harris RA (2014) Seeking structural specificity: direct modulation of pentameric ligand-gated ion channels by alcohols and general anesthetics. Pharmacol Rev 66: 396–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howland RH (2013) Mifepristone as a therapeutic agent in psychiatry. J Psychosoc Nurs Ment Health Serv 51: 11–4. [DOI] [PubMed] [Google Scholar]
- Hu W, Lu T, Chen A, Huang Y, Hansen R, Chandler LJ, Zhang HT (2011) Inhibition of phosphodiesterase-4 decreases ethanol intake in mice. Psychopharmacology (Berl) 218: 331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Hutson PH, Clark JA, Cross AJ (2017) CNS Target Identification and Validation: Avoiding the Valley of Death or Naive Optimism? Annu Rev Pharmacol Toxicol 57: 171–187. [DOI] [PubMed] [Google Scholar]
- Imbert B, Alvarez JC, Simon N (2015) Anticraving Effect of Baclofen in Alcohol-Dependent Patients. Alcohol Clin Exp Res 39: 1602–8. [DOI] [PubMed] [Google Scholar]
- Jacunski A, Tatonetti NP (2013) Connecting the dots: applications of network medicine in pharmacology and disease. Clin Pharmacol Ther 94: 659–69. [DOI] [PubMed] [Google Scholar]
- Jasinski DR, Pevnick JS, Griffith JD (1978) Human pharmacology and abuse potential of the analgesic buprenorphine: a potential agent for treating narcotic addiction. Arch Gen Psychiatry 35: 501–16. [DOI] [PubMed] [Google Scholar]
- Jensen NH, Rodriguiz RM, Caron MG, Wetsel WC, Rothman RB, Roth BL (2008) N-desalkylquetiapine, a potent norepinephrine reuptake inhibitor and partial 5-HT1A agonist, as a putative mediator of quetiapine’s antidepressant activity. Neuropsychopharmacology 33: 2303–12. [DOI] [PubMed] [Google Scholar]
- Johnstone AL, Reierson GW, Smith RP, Goldberg JL, Lemmon VP, Bixby JL (2012) A chemical genetic approach identifies piperazine antipsychotics as promoters of CNS neurite growth on inhibitory substrates. Mol Cell Neurosci 50: 125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahanian E, Quintanilla ME, Fernandez K, Israel Y (2014) Fenofibrate--a lipid-lowering drug--reduces voluntary alcohol drinking in rats. Alcohol 48: 665–70. [DOI] [PubMed] [Google Scholar]
- Kenna GA, Leggio L, Swift RM (2009) A safety and tolerability laboratory study of the combination of aripiprazole and topiramate in volunteers who drink alcohol. Hum Psychopharmacol 24: 465–72. [DOI] [PubMed] [Google Scholar]
- Kielbasa SM, Klein H, Roider HG, Vingron M, Bluthgen N (2010) TransFind--predicting transcriptional regulators for gene sets. Nucleic Acids Res 38: W275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, Megy K, Pilicheva E, Rustici G, Tikhonov A, Parkinson H, Petryszak R, Sarkans U, Brazma A (2015) ArrayExpress update--simplifying data submissions. Nucleic Acids Res 43: D1113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin A, Boogerd FC, Plant N, Bruggeman FJ, Goncharuk V, Lunshof J, Moreno-Sanchez R, Yilmaz N, Bakker BM, Snoep JL, Balling R, Westerhoff HV (2012) Emergence of the silicon human and network targeting drugs. Eur J Pharm Sci 46: 190–7. [DOI] [PubMed] [Google Scholar]
- Koob GF, Kenneth Lloyd G, Mason BJ (2009) Development of pharmacotherapies for drug addiction: a Rosetta stone approach. Nat Rev Drug Discov 8: 500–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J Jr., Tugendreich S (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ, LaForge KS, Butelman E (2002) Pharmacotherapy of addictions. Nat Rev Drug Discov 1: 710–26. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwako LE, Spagnolo PA, Schwandt ML, Thorsell A, George DT, Momenan R, Rio DE, Huestis M, Anizan S, Concheiro M, Sinha R, Heilig M (2015) The corticotropin releasing hormone-1 (CRH1) receptor antagonist pexacerfont in alcohol dependence: a randomized controlled experimental medicine study. Neuropsychopharmacology 40: 1053–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR (2006) The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313: 1929–35. [DOI] [PubMed] [Google Scholar]
- Langedijk J, Mantel-Teeuwisse AK, Slijkerman DS, Schutjens MH (2015) Drug repositioning and repurposing: terminology and definitions in literature. Drug Discov Today 20: 1027–34. [DOI] [PubMed] [Google Scholar]
- Letai A (2017) Functional precision cancer medicine-moving beyond pure genomics. Nat Med 23: 1028–1035. [DOI] [PubMed] [Google Scholar]
- Li Z, Taylor CP, Weber M, Piechan J, Prior F, Bian F, Cui M, Hoffman D, Donevan S (2011) Pregabalin is a potent and selective ligand for alpha(2)delta-1 and alpha(2)delta-2 calcium channel subunits. Eur J Pharmacol 667: 80–90. [DOI] [PubMed] [Google Scholar]
- Lief HI (1996) Bupropion treatment of depression to assist smoking cessation. Am J Psychiatry 153: 442. [DOI] [PubMed] [Google Scholar]
- Lipponen A, Paananen J, Puhakka N, Pitkanen A (2016) Analysis of Post-Traumatic Brain Injury Gene Expression Signature Reveals Tubulins, Nfe2l2, Nfkb, Cd44, and S100a4 as Treatment Targets. Sci Rep 6: 31570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litten RZ, Fertig JB, Falk DE, Ryan ML, Mattson ME, Collins JF, Murtaugh C, Ciraulo D, Green AI, Johnson B, Pettinati H, Swift R, Afshar M, Brunette MF, Tiouririne NA, Kampman K, Stout R, Group NS (2012) A double-blind, placebo-controlled trial to assess the efficacy of quetiapine fumarate XR in very heavy-drinking alcohol-dependent patients. Alcohol Clin Exp Res 36: 406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litten RZ, Ryan M, Falk D, Fertig J (2014) Alcohol medications development: advantages and caveats of government/academia collaborating with the pharmaceutical industry. Alcohol Clin Exp Res 38: 1196–9. [DOI] [PubMed] [Google Scholar]
- Litten RZ, Ryan ML, Fertig JB, Falk DE, Johnson B, Dunn KE, Green AI, Pettinati HM, Ciraulo DA, Sarid-Segal O, Kampman K, Brunette MF, Strain EC, Tiouririne NA, Ransom J, Scott C, Stout R, Group NS (2013) A double-blind, placebo-controlled trial assessing the efficacy of varenicline tartrate for alcohol dependence. J Addict Med 7: 277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litten RZ, Wilford BB, Falk DE, Ryan ML, Fertig JB (2016) Potential medications for the treatment of alcohol use disorder: An evaluation of clinical efficacy and safety. Subst Abus 37: 286–98. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang LN (2017) Baclofen for alcohol withdrawal. Cochrane Database Syst Rev 8: CD008502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hao PD, Yang MF, Sun JY, Mao LL, Fan CD, Zhang ZY, Li DW, Yang XY, Sun BL, Zhang HT (2017) The phosphodiesterase-4 inhibitor roflumilast decreases ethanol consumption in C57BL/6J mice. Psychopharmacology (Berl) 234: 2409–2419. [DOI] [PubMed] [Google Scholar]
- Lohman BK, Weber JN, Bolnick DI (2016) Evaluation of TagSeq, a reliable low-cost alternative for RNAseq. Mol Ecol Resour 16: 1315–1321. [DOI] [PubMed] [Google Scholar]
- Lyon J (2017) More Treatments on Deck for Alcohol Use Disorder. JAMA 317: 2267–2269. [DOI] [PubMed] [Google Scholar]
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Srinidhi CLL, Barnes G, Taylor SA, James M, Groot N, MacFarlane H, Jenkins B, Anderson MA, Wexler NS, Gusella JF (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell 72: 971–83. [DOI] [PubMed] [Google Scholar]
- Martinotti G, Di Nicola M, Di Giannantonio M, Janiri L (2009) Aripiprazole in the treatment of patients with alcohol dependence: a double-blind, comparison trial vs. naltrexone. J Psychopharmacol 23: 123–9. [DOI] [PubMed] [Google Scholar]
- Martinotti G, Di Nicola M, Janiri L (2007) Efficacy and safety of aripiprazole in alcohol dependence. Am J Drug Alcohol Abuse 33: 393–401. [DOI] [PubMed] [Google Scholar]
- Mason BJ, Goodell V, Shadan F (2014a) Gabapentin treatment for alcohol dependence--reply. JAMA Intern Med 174: 1201–2. [DOI] [PubMed] [Google Scholar]
- Mason BJ, Quello S, Goodell V, Shadan F, Kyle M, Begovic A (2014b) Gabapentin treatment for alcohol dependence: a randomized clinical trial. JAMA Intern Med 174: 70–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, Lenhard B, Sandelin A, Wasserman WW (2016) JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44: D110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J, Harris RA (2017) The Neuroimmune Basis of Excessive Alcohol Consumption. Neuropsychopharmacology 42: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArt DG, Zhang SD (2011) Identification of candidate small-molecule therapeutics to cancer by gene-signature perturbation in connectivity mapping. PLoS One 6: e16382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV (2011) Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol Ecol 20: 3599–616. [DOI] [PubMed] [Google Scholar]
- Miller MB, DiBello AM, Carey KB, Borsari B, Pedersen ER (2017) Insomnia severity as a mediator of the association between mental health symptoms and alcohol use in young adult veterans. Drug Alcohol Depend 177: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirijello A, D’Angelo C, Ferrulli A, Vassallo G, Antonelli M, Caputo F, Leggio L, Gasbarrini A, Addolorato G (2015) Identification and management of alcohol withdrawal syndrome. Drugs 75: 353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza N, Sills GJ, Pirmohamed M, Marson AG (2017) Identifying new antiepileptic drugs through genomics-based drug repurposing. Hum Mol Genet 26: 527–537. [DOI] [PubMed] [Google Scholar]
- Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q (2014) GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res 3: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley KC, Baillie A, Leung S, Addolorato G, Leggio L, Haber PS (2014) Baclofen for the Treatment of Alcohol Dependence and Possible Role of Comorbid Anxiety. Alcohol Alcohol 49: 654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Most D, Ferguson LB, Harris RA (2014) Molecular basis of alcoholism. Handb Clin Neurol 125: 89–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CA, Geisel O, Pelz P, Higl V, Kruger J, Stickel A, Beck A, Wernecke KD, Hellweg R, Heinz A (2015) High-dose baclofen for the treatment of alcohol dependence (BACLAD study): a randomized, placebo-controlled trial. Eur Neuropsychopharmacol 25: 1167–77. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Mozhui K, Prins P, Williams RW (2017) GeneNetwork: A Toolbox for Systems Genetics. Methods Mol Biol 1488: 75–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano F, Sirci F, Carrella D, di Bernardo D (2016) Drug-set enrichment analysis: a novel tool to investigate drug mode of action. Bioinformatics 32: 235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naudet F (2016) Comparing Nalmefene and Naltrexone in Alcohol Dependence: Is there a Spin? Pharmacopsychiatry 49: 260–261. [DOI] [PubMed] [Google Scholar]
- Naudet F, Palpacuer C, Boussageon R, Laviolle B (2016) Evaluation in alcohol use disorders - insights from the nalmefene experience. BMC Med 14: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosengo N (2016) Can you teach old drugs new tricks? Nature 534: 314–6. [DOI] [PubMed] [Google Scholar]
- Nunes EV (2014) Gabapentin: a new addition to the armamentarium for alcohol dependence? JAMA Intern Med 174: 78–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan MC (2015) What have we learned from the Psychiatric Genomics Consortium. World Psychiatry 14: 291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oni EN, Halikere A, Li G, Toro-Ramos AJ, Swerdel MR, Verpeut JL, Moore JC, Bello NT, Bierut LJ, Goate A, Tischfield JA, Pang ZP, Hart RP (2016) Increased nicotine response in iPSC-derived human neurons carrying the CHRNA5 N398 allele. Sci Rep 6: 34341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea TI, Overington JP (2015) Computational and Practical Aspects of Drug Repositioning. Assay Drug Dev Technol 13: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RP, Klein TE, Altman RB (2007) The education potential of the pharmacogenetics and pharmacogenomics knowledge base (PharmGKB). Clin Pharmacol Ther 82: 472–5. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Gerhards C, Heck A, Ackermann S, Aerni A, Schicktanz N, Auschra B, Demougin P, Mumme E, Elbert T, Ertl V, Gschwind L, Hanser E, Huynh KD, Jessen F, Kolassa IT, Milnik A, Paganetti P, Spalek K, Vogler C, Muhs A, Pfeifer A, de Quervain DJ (2013) Human genome-guided identification of memory-modulating drugs. Proc Natl Acad Sci U S A 110: E4369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisier SB, Taschereau R, Wong JA, Graeber TG (2010) Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res 38: e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomrenze MB, Fetterly TL, Winder DG, Messing RO (2017) The Corticotropin Releasing Factor Receptor 1 in Alcohol Use Disorder: Still a Valid Drug Target? Alcohol Clin Exp Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponizovsky AM, Rosca P, Aronovich E, Weizman A, Grinshpoon A (2015) Baclofen as add-on to standard psychosocial treatment for alcohol dependence: a randomized, double-blind, placebo-controlled trial with 1 year follow-up. J Subst Abuse Treat 52: 24–30. [DOI] [PubMed] [Google Scholar]
- Ray LA, Bujarski S (2016) Mechanisms of topiramate effects: refining medications development for alcoholism. Addict Biol 21: 183–4. [DOI] [PubMed] [Google Scholar]
- Ray LA, Bujarski S, Shoptaw S, Roche DJ, Heinzerling K, Miotto K (2017) Development of the Neuroimmune Modulator Ibudilast for the Treatment of Alcoholism: A Randomized, Placebo-Controlled, Human Laboratory Trial. Neuropsychopharmacology 42: 1776–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Roche DJ, Heinzerling K, Shoptaw S (2014) Opportunities for the development of neuroimmune therapies in addiction. Int Rev Neurobiol 118: 381–401. [DOI] [PubMed] [Google Scholar]
- Rigal L, Legay Hoang L, Alexandre-Dubroeucq C, Pinot J, Le Jeunne C, Jaury P (2015) Tolerability of High-dose Baclofen in the Treatment of Patients with Alcohol Disorders: A Retrospective Study. Alcohol Alcohol 50: 551–7. [DOI] [PubMed] [Google Scholar]
- Rivera-Meza M, Munoz D, Jerez E, Quintanilla ME, Salinas-Luypaert C, Fernandez K, Karahanian E (2017) Fenofibrate Administration Reduces Alcohol and Saccharin Intake in Rats: Possible Effects at Peripheral and Central Levels. Front Behav Neurosci 11: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland B, Labreuche J, Duhamel A, Deheul S, Gautier S, Auffret M, Pignon B, Valin T, Bordet R, Cottencin O (2015a) Baclofen for alcohol dependence: Relationships between baclofen and alcohol dosing and the occurrence of major sedation. Eur Neuropsychopharmacol 25: 1631–6. [DOI] [PubMed] [Google Scholar]
- Rolland B, Valin T, Langlois C, Auffret M, Gautier S, Deheul S, Danel T, Bordet R, Cottencin O (2015b) Safety and drinking outcomes among patients with comorbid alcohol dependence and borderline personality disorder treated with high-dose baclofen: a comparative cohort study. Int Clin Psychopharmacol 30: 49–53. [DOI] [PubMed] [Google Scholar]
- Ryall KA, Tan AC (2015) Systems biology approaches for advancing the discovery of effective drug combinations. J Cheminform 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan ML, Falk DE, Fertig JB, Rendenbach-Mueller B, Katz DA, Tracy KA, Strain EC, Dunn KE, Kampman K, Mahoney E, Ciraulo DA, Sickles-Colaneri L, Ait-Daoud N, Johnson BA, Ransom J, Scott C, Koob GF, Litten RZ (2017) A Phase 2, Double-Blind, Placebo-Controlled Randomized Trial Assessing the Efficacy of ABT-436, a Novel V1b Receptor Antagonist, for Alcohol Dependence. Neuropsychopharmacology 42: 1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul MC, Majdak P, Perez S, Reilly M, Garland T, Jr., Rhodes JS (2017) High motivation for exercise is associated with altered chromatin regulators of monoamine receptor gene expression in the striatum of selectively bred mice. Genes Brain Behav 16: 328–341. [DOI] [PubMed] [Google Scholar]
- Schotte A, Janssen PF, Gommeren W, Luyten WH, Van Gompel P, Lesage AS, De Loore K, Leysen JE (1996) Risperidone compared with new and reference antipsychotic drugs: in vitro and in vivo receptor binding. Psychopharmacology (Berl) 124: 57–73. [DOI] [PubMed] [Google Scholar]
- Schwandt ML, Cortes CR, Kwako LE, George DT, Momenan R, Sinha R, Grigoriadis DE, Pich EM, Leggio L, Heilig M (2016) The CRF1 Antagonist Verucerfont in Anxious Alcohol-Dependent Women: Translation of Neuroendocrine, But not of Anti-Craving Effects. Neuropsychopharmacology 41: 2818–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secades JJ, Lorenzo JL (2006) Citicoline: pharmacological and clinical review, 2006 update. Methods Find Exp Clin Pharmacol 28 Suppl B: 1–56. [PubMed] [Google Scholar]
- Shank RP, Gardocki JF, Streeter AJ, Maryanoff BE (2000) An overview of the preclinical aspects of topiramate: pharmacology, pharmacokinetics, and mechanism of action. Epilepsia 41 Suppl 1: S3–9. [PubMed] [Google Scholar]
- Shapiro DA, Renock S, Arrington E, Chiodo LA, Liu LX, Sibley DR, Roth BL, Mailman R (2003) Aripiprazole, a novel atypical antipsychotic drug with a unique and robust pharmacology. Neuropsychopharmacology 28: 1400–11. [DOI] [PubMed] [Google Scholar]
- Siavelis JC, Bourdakou MM, Athanasiadis EI, Spyrou GM, Nikita KS (2016) Bioinformatics methods in drug repurposing for Alzheimer’s disease. Brief Bioinform 17: 322–35. [DOI] [PubMed] [Google Scholar]
- Silbersweig D, Loscalzo J (2017) Precision Psychiatry Meets Network Medicine: Network Psychiatry. JAMA Psychiatry 74: 665–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EK, Loscalzo J (2013) Developing new drug treatments in the era of network medicine. Clin Pharmacol Ther 93: 26–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonim DK, Koide K, Johnson KL, Tantravahi U, Cowan JM, Jarrah Z, Bianchi DW (2009) Functional genomic analysis of amniotic fluid cell-free mRNA suggests that oxidative stress is significant in Down syndrome fetuses. Proc Natl Acad Sci U S A 106: 9425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley JL, Breda C, Mason RP, Kooner G, Luthi-Carter R, Gant TW, Giorgini F (2016) Connectivity mapping uncovers small molecules that modulate neurodegeneration in Huntington’s disease models. J Mol Med (Berl) 94: 235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Chau CK, Chiu WT, Ho KS, Lo CP, Yim SH, Sham PC (2017) Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat Neurosci 20: 1342–1349. [DOI] [PubMed] [Google Scholar]
- Soyka M (2016) Nalmefene for the treatment of alcohol use disorders: recent data and clinical potential. Expert Opin Pharmacother 17: 619–26. [DOI] [PubMed] [Google Scholar]
- Soyka M, Friede M, Schnitker J (2016) Comparing Nalmefene and Naltrexone in Alcohol Dependence: Are there any Differences? Results from an Indirect Meta-analysis - Comment to Naudet. Pharmacopsychiatry 49: 261–262. [DOI] [PubMed] [Google Scholar]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D (2016) The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics 54: 1 30 1–1 30 33. [DOI] [PubMed] [Google Scholar]
- Stern S, Santos R, Marchetto MC, Mendes AP, Rouleau GA, Biesmans S, Wang QW, Yao J, Charnay P, Bang AG, Alda M, Gage FH (2017) Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BC (2008) Once-daily treatment of ADHD with guanfacine: patient implications. Neuropsychiatr Dis Treat 4: 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, Lahr DL, Hirschman JE, Liu Z, Donahue M, Julian B, Khan M, Wadden D, Smith IC, Lam D, Liberzon A, Toder C, Bagul M, Orzechowski M, Enache OM, Piccioni F, Johnson SA, Lyons NJ, Berger AH, Shamji AF, Brooks AN, Vrcic A, Flynn C, Rosains J, Takeda DY, Hu R, Davison D, Lamb J, Ardlie K, Hogstrom L, Greenside P, Gray NS, Clemons PA, Silver S, Wu X, Zhao WN, Read-Button W, Wu X, Haggarty SJ, Ronco LV, Boehm JS, Schreiber SL, Doench JG, Bittker JA, Root DE, Wong B, Golub TR (2017) A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171: 1437–1452 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M (2016) STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 44: D380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli C, Tegner J, Maselli V, Gomez-Cabrero D, Altobelli G, Emmett W, Lescai F, Gustincich S, Stupka E (2011) ParkDB: a Parkinson’s disease gene expression database. Database (Oxford) 2011: bar007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–76. [DOI] [PubMed] [Google Scholar]
- Tosti A, Pazzaglia M, Voudouris S, Tosti G (2004) Hypertrichosis of the eyelashes caused by bimatoprost. J Am Acad Dermatol 51: S149–50. [DOI] [PubMed] [Google Scholar]
- Traynor K (2014) NIOSH revamps hazardous drugs update. Am J Health Syst Pharm 71: 2099100. [DOI] [PubMed] [Google Scholar]
- Turinsky AL, Razick S, Turner B, Donaldson IM, Wodak SJ (2014) Navigating the global protein-protein interaction landscape using iRefWeb. Methods Mol Biol 1091: 315–31. [DOI] [PubMed] [Google Scholar]
- Upadhyaya HP, Brady KT, Sethuraman G, Sonne SC, Malcolm R (2001) Venlafaxine treatment of patients with comorbid alcohol/cocaine abuse and attention-deficit/hyperactivity disorder: a pilot study. J Clin Psychopharmacol 21: 116–8. [DOI] [PubMed] [Google Scholar]
- Vendruscolo LF, Barbier E, Schlosburg JE, Misra KK, Whitfield TW, Logrip ML, Rivier C, Repunte-Canonigo V, Zorrilla EP, Sanna PP, Heilig M, Koob GF (2012) Corticosteroid-dependent plasticity mediates compulsive alcohol drinking in rats. J Neurosci 32: 7563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo LF, Estey D, Goodell V, Macshane LG, Logrip ML, Schlosburg JE, McGinn MA, Zamora-Martinez ER, Belanoff JK, Hunt HJ, Sanna PP, George O, Koob GF, Edwards S, Mason BJ (2015) Glucocorticoid receptor antagonism decreases alcohol seeking in alcohol-dependent individuals. J Clin Invest 125: 3193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronin K, Randall P, Myrick H, Anton R (2008) Aripiprazole effects on alcohol consumption and subjective reports in a clinical laboratory paradigm--possible influence of self-control. Alcohol Clin Exp Res 32: 1954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Cohen N, Kelder T, Amit U, Liebman E, Steinberg DM, Radonjic M, Ruppin E (2015) Drugs that reverse disease transcriptomic signatures are more effective in a mouse model of dyslipidemia. Mol Syst Biol 11: 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B (2013) WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 41: W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yu Y, Yang J, Zhao X, Li Z (2015) Dissecting Xuesaitong’s mechanisms on preventing stroke based on the microarray and connectivity map. Mol Biosyst 11: 3033–9. [DOI] [PubMed] [Google Scholar]
- Wei WQ, Cronin RM, Xu H, Lasko TA, Bastarache L, Denny JC (2013) Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc 20: 954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel S, Lalanne L, Riegert M, Bertschy G (2015) Efficacy of High-Dose Baclofen for Alcohol Use Disorder and Comorbid Bulimia: A Case Report. J Dual Diagn 11: 203–4. [DOI] [PubMed] [Google Scholar]
- Wen RT, Zhang M, Qin WJ, Liu Q, Wang WP, Lawrence AJ, Zhang HT, Liang JH (2012) The phosphodiesterase-4 (PDE4) inhibitor rolipram decreases ethanol seeking and consumption in alcohol-preferring Fawn-Hooded rats. Alcohol Clin Exp Res 36: 2157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall ND, Brown ES (2014) Citicoline in addictive disorders: a review of the literature. Am J Drug Alcohol Abuse 40: 262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G (2012) A searchable cross-platform gene expression database reveals connections between drug treatments and disease. BMC Genomics 13: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34: D668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington JJ 3rd, Simon NM, Korbly NB, Perlis RH, Pollack MH, Anxiety Disorders Research P (2002) Ropinirole for antidepressant-induced sexual dysfunction. Int Clin Psychopharmacol 17: 307–10. [DOI] [PubMed] [Google Scholar]
- Xia J, Gill EE, Hancock RE (2015) NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 10: 823–44. [DOI] [PubMed] [Google Scholar]
- Zhang SD, Gant TW (2009) sscMap: an extensible Java application for connecting small-molecule drugs using gene-expression signatures. BMC Bioinformatics 10: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]