Biomass constitutes an appealing alternative to fossil resources for the production of materials and energy. The abundance and attractiveness of vegetal biomass come along with challenges pertaining to the intricacy of its structure, evolved during billions of years to face and resist abiotic and biotic attacks.
KEYWORDS: fungi, LPMO, peroxidase, catalase, redox enzymes, hydrogen peroxide, Fenton reaction, lignocellulose
SUMMARY
Biomass constitutes an appealing alternative to fossil resources for the production of materials and energy. The abundance and attractiveness of vegetal biomass come along with challenges pertaining to the intricacy of its structure, evolved during billions of years to face and resist abiotic and biotic attacks. To achieve the daunting goal of plant cell wall decomposition, microorganisms have developed many (enzymatic) strategies, from which we seek inspiration to develop biotechnological processes. A major breakthrough in the field has been the discovery of enzymes today known as lytic polysaccharide monooxygenases (LPMOs), which, by catalyzing the oxidative cleavage of recalcitrant polysaccharides, allow canonical hydrolytic enzymes to depolymerize the biomass more efficiently. Very recently, it has been shown that LPMOs are not classical monooxygenases in that they can also use hydrogen peroxide (H2O2) as an oxidant. This discovery calls for a revision of our understanding of how lignocellulolytic enzymes are connected since H2O2 is produced and used by several of them. The first part of this review is dedicated to the LPMO paradigm, describing knowns, unknowns, and uncertainties. We then present different lignocellulolytic redox systems, enzymatic or not, that depend on fluxes of reactive oxygen species (ROS). Based on an assessment of these putatively interconnected systems, we suggest that fine-tuning of H2O2 levels and proximity between sites of H2O2 production and consumption are important for fungal biomass conversion. In the last part of this review, we discuss how our evolving understanding of redox processes involved in biomass depolymerization may translate into industrial applications.
INTRODUCTION
One of the main pillars of the Earth's carbon cycle is the depolymerization of complex plant biomass (1), and a complete understanding of this process is of utmost interest for fundamental biology and crucial for the emerging bioeconomy (2, 3). The structural intricacy of this raw material, primarily composed of cellulose, various hemicelluloses, and lignin, is mirrored by the complexity of the network of enzymatic and chemical reactions developed by microorganisms to decompose it. This network is far from fully understood. Until recently, degradation of the recalcitrant polysaccharides in plant biomass was thought to be mainly achieved by an arsenal of hydrolytic enzymes called glycoside hydrolases (GHs) (4). In some ecosystems, the enzymatic decomposition process is thought to be supported by Fenton chemistry, i.e., transition metal-driven in situ generation of H2O2-derived hydroxyl radicals, which are among the most powerful oxidizing species found on Earth (5) and are able to unspecifically oxidize both polysaccharides and lignin in plant biomass (6).
Several decades ago, Elwyn Reese and colleagues proposed that nonhydrolytic proteins were involved in cellulose decomposition. This proposal is known as the C1-Cx postulate in which the C1 factor (nonhydrolytic proteins) acts as an enhancing protein for the Cx factor (hydrolytic enzymes) (7). In 1974, Eriksson and colleagues noticed that cellulose degradation by culture filtrates of white-rot fungi was more efficient in the presence of O2, leading to the hypothesis that oxidative processes contribute to cellulose conversion (8). In 2005, studying enzymatic chitin degradation, Vaaje-Kolstad et al. showed that a 21-kDa protein named chitin-binding protein, or CBP21, by Suzuki et al. (9), drastically enhanced the efficiency of classical (hydrolytic) chitinases (10). Five years later, in 2010, i.e., 60 years after Reese's postulate, it was shown that CBP21 represents a new class of enzymes that carry out oxidative cleavage of polysaccharides (11). These enzymes, today known as lytic polysaccharide monooxygenases (LPMOs) (12), are mono-copper redox enzymes (13, 14) that hydroxylate the C-1 or C-4 carbons of scissile glycosidic bonds (11, 15–18) in an O2- and reductant-dependent manner (11).
As illustrated by the many different names they have been given over the years, LPMOs still retain many secrets. After having been considered sluggish fungal glycoside hydrolases belonging to family GH61 (19, 20) or noncatalytic bacterial carbohydrate binding modules (CBMs) belonging to family CBM33, the 2010 discovery led to gathering these proteins of diverse origins under the common acronym LPMO (12) or, alternatively, polysaccharide monooxygenase (PMO) (14). Today, LPMOs are classified as auxiliary activities (AA) (21) in the database of carbohydrate-active enzymes (CAZy) (22, 23), where they form families AA9, -10, -11, -13, -14, and -15. LPMOs are today seen as key frontline weapons in the warfare between attackers (e.g., fungi and bacteria) and defenders (e.g., plants) (24).
LPMOs are unique in the sense that they are able to attack polysaccharides that are organized in recalcitrant structures (e.g., crystalline cellulose or chitin; hemicellulose-cellulose complexes) (25). While canonical glycoside hydrolases (GHs) interact with single polysaccharide chains, meaning that a decrystallization penalty needs to be paid (26), LPMOs act on surfaces; that is, they cleave a polysaccharide chain while this chain is in a crystalline context (11). By doing so, LPMOs render a relatively inaccessible substrate tractable to further depolymerization by GHs (27–30). In this connection, real-time atomic force microscopy studies have shown that LPMOs are relatively immobile on the cellulose surface and that cellulase-catalyzed substrate turnover is higher after LPMO treatment (31). In a very interesting study using in situ imaging, it has also been shown that the progression of hydrolases and the boosting effect of LPMOs are dependent on the type of plant tissue (30).
A recent milestone in the field concerns the discovery that H2O2 can drive LPMO reactions in the absence of O2 (32, 33). In fact, it has been claimed that H2O2 is the preferred, and perhaps even the only, cosubstrate of LPMOs (32, 34), which contrasts with established paradigms and raises questions as to whether LPMOs should be classified as monooxygenases. Importantly, H2O2 is also a reaction product or substrate in several other enzyme-catalyzed lignocellulolytic reactions, in particular, in lignin conversion (35). Thus, H2O2 may play a central role in the reaction networks of biomass conversion. Interestingly, H2O2 and reactive oxygen species (ROS) in general are increasingly considered metabolites with a variety of possible (regulatory) functions beyond simply being oxidants (36, 37). It is thus worth considering whether H2O2 could be a central regulatory metabolite in biomass conversion, the levels of which are temporally and spatially regulated by the actions of substrate-specialized (and thus localized) H2O2-producing and -consuming enzymes.
In this context, we wish to first introduce the concept of LPMO catalysis by presenting the monooxygenase (MO; O2-based) and peroxygenase (PO; H2O2-based) reaction paradigms and discuss the knowns and unknowns. Then, a critical retrospect on LPMO literature is carried out with the aim of shedding new light on previously reported results. The second part of this review describes other H2O2-producing or -consuming systems encountered during lignocellulose conversion and discusses potential and proven interconnections in the light of available biochemical and multi-omics data. The last part of this review focuses on how our improving understanding of natural biomass conversion translates into the design of better industrial biorefining processes, today and tomorrow.
RETROSPECT ON LPMO RESEARCH
Introduction to LPMO Catalysis: Knowns and Unknowns
The present review does not aim to summarize all aspects of LPMOs, such as structural aspects and putative mechanistic routes, since these have been comprehensively covered in other recent reviews (12, 17, 18, 24, 38–42). Nevertheless, the recent discovery of the peroxygenase activity of LPMOs (32, 34, 43, 44) shatters a widely accepted paradigm for LPMO catalysis that laid the foundations for previous discussions and analyses. Therefore, a clarification of what has been explicitly proven and what remains hypothetical is required.
A conserved catalytic core embedded in an evolutionarily divergent binding surface.
The uniqueness of LPMOs comes in part from the fact that they can carry out the oxidative cleavage of a glycosidic chain embedded in a crystalline lattice, an unprecedented ability in the world of CAZymes. To do so, LPMOs have to bring their active site close to the crystalline surface at the right location (45). Importantly, there is growing evidence that LPMOs also act on noncrystalline substrates (46–48) or copolymeric structures (25, 49–51), indicating that substrate crystallinity is not a sine qua non condition for LPMO catalysis to occur. Numerous crystallographic structures (39, 52) show that LPMOs, irrespective of their phylogenetic origins and substrate specificities, display rather flat, solvent-exposed substrate-binding surfaces that include two conserved histidines coordinating a single copper atom, also known as a histidine brace (13). The histidine brace is the only totally conserved feature across the LPMO superfamily (Fig. 1). Other surface-exposed residues involved in substrate binding (45, 52–58) and/or the second coordination shell of the active-site copper are often relatively conserved within phenotypic subgroups and therefore probably dictate complementarity with the target substrate (39). Despite a few studies aimed at unraveling the determinants of oxidative regioselectivity (56, 59, 60) and substrate specificity (61), these determinants remain largely unknown.
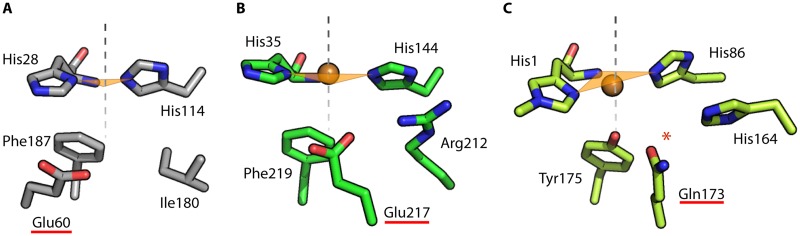
FIG 1.
LPMO active sites. The figure shows a close-up view of the catalytic center of SmAA10A (also known as CBP21; PDB accession number 2BEM) (A), ScAA10C (also known as CelS2; PDB accession number 4OY7) (B), and TaAA9A (also known as TaGH61A; PDB accession number 2YET) (C), which are representatives of bacterial chitin-active, bacterial cellulose-active, and fungal cellulose-active LPMOs, respectively. The gray dotted line shows the axis defined as axial, and the orange triangle represents the equatorial plane defined by the three copper-coordinating nitrogens in the histidine brace (best visible in panel C). The red star indicates the location of an oxygen species observed in the neutron structural studies by O'Dell et al. (69), and the Glu/Gln potentially interacting with this oxygen species is underlined. Note that Phe187, Phe219, and Tyr175 are in equivalent positions in their respective proteins, namely, the proximal axial coordination position. The distal axial position is solvent exposed and will be occupied by substrate upon binding (52).
Some cellulose-active LPMOs are regiospecific (i.e., exclusively oxidizing the C-1 or C-4 carbon), while others display a lack of specificity, oxidizing both C-1 and C-4 carbons. Notably, data on enzyme-substrate complexes (45, 52) show that the C-1 and C-4 carbons are both close to the copper site, meaning that minor variation in substrate positioning could lead to a change in oxidative regioselectivity, a notion that is supported by various studies (56, 60). It is intriguing that some LPMOs seemingly are somewhat ambiguous when it comes to substrate binding and positioning, while accurate assembly of the catalytic complex is crucial to control the very powerful redox chemistry and to prevent off-pathway reactions, as explained in “Oxidative self-inactivation of LPMOs,” below.
The O2 reaction mechanism(s): oxidase and monooxygenase activities.
The seminal study by Vaaje-Kolstad et al. unraveled the oxidative activity of the chitin-active CBP21, AA10A, from the bacterium Serratia marcescens (SmAA10A) by showing that aldonic acids were released from chitin under aerobic conditions and in the presence of reductant. Using mass spectrometry and labeled oxygen (18O2), it was shown that the introduced oxygen atom was derived from O2 (11), and this was later also shown for a cellulose-active fungal LPMO, AA9E, from Neurospora crassa (NcAA9E-CBM1, also known as NcPMO-08760) (16). The combined use of mass spectrometry and labeled water (H218O) showed that the detected aldonic acids result from spontaneous hydrolysis of a lactone form (11). The lactone form has been proposed to arise from a spontaneous elimination reaction that happens upon hydroxylation of the C-1 carbon (16) (Fig. 2, right side). A similar mechanism is envisaged for C-4-oxidizing LPMOs, but in this case the product is a ketoaldose, which is hydrated to a gem-diol (14, 62).
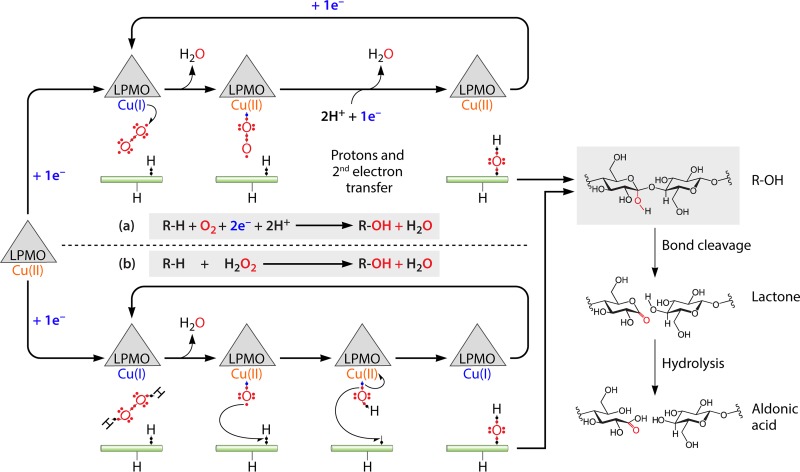
FIG 2.
Comparison of O2-based (a) and H2O2-based (b) reaction pathways. To enter both pathways, a reduction is necessary to reduce Cu(II) to Cu(I). In pathway a, transfer of a second electron and of 2 protons is necessary to complete the catalytic cycle. In pathway b, H2O2 alone is sufficient to complete a reaction cycle. Detailed potential catalytic pathways involving O2 (17, 18) or H2O2 (32) as a cosubstrate have been described elsewhere. Both pathways are thought to generate a hydroxylated end product (either at C-1 or C-4). The right-hand side of the figure shows a C-1-hydroxylated product undergoing a spontaneous elimination reaction that leads to cleavage of the glycosidic bond and formation of a lactone, which is hydrolyzed to become an aldonic acid.
LPMOs were assigned as monooxygenases based on (i) the monooxygenated nature of the reaction product, (ii) the apparent dependency of the reaction on O2 and reducing equivalents, and (iii) similarities with other copper-dependent enzymes known as monooxygenases, in particular, methane monooxygenase (13, 14). This logical reasoning has been widely accepted by the scientific community. However, it has recently been pointed out that while the apparent O2 dependency shows that O2 can be the source of the incorporated oxygen, this dependency is not a proof stricto sensu that O2 is the cosubstrate of LPMOs since, in a reducing environment, O2-derived intermediates such as superoxide or H2O2 will be generated (32).
Among the knowns, it has unambiguously been established that LPMOs are mono-copper enzymes (13, 14) which can catalyze the monooxygenation of several carbohydrate substrates (11, 13, 14, 25, 46, 47, 62–65). It is also well known that the LPMO-Cu(II) state is the resting inactive state and that reduction of the copper precedes catalytic action. It is also established that in the absence of substrate and presence of reductant, LPMOs can act as oxidases, i.e., that they are able to carry out the reduction of O2 (15), leading to H2O2 formation (62, 66). Importantly, this implies that even in reactions with substrate, non-substrate-bound LPMOs that become reduced may generate H2O2, which could fuel H2O2-driven reactions catalyzed by substrate-bound LPMOs (see below for more details).
While the ability of LPMOs to produce H2O2 is well established, it is not clearly established whether the superoxide species resulting from O2 single-electron reduction [by LPMO-Cu(I)] is released in the reaction mixture before undergoing spontaneous disproportionation, as suggested by some density functional theory (DFT) calculations (15), or if H2O2 is produced in the LPMO active site. The latter scenario was recently suggested by Span et al., who noted that addition of superoxide dismutase (SOD) to an AA9 LPMO reaction mixture had no effect on H2O2 production (67). This would suggest that a second electron and two protons have to be delivered to the LPMO active site to complete the two-electron reduction of O2 to H2O2. Span et al. further showed that a second-shell glutamine (Fig. 1) likely contributes to keeping the superoxide bound to the active site. In support of the latter observations, analysis of LPMO structural diversity shows that a conserved second-shell glutamate or glutamine is pointing toward the active site in all LPMOs, suggesting a key role in LPMO catalysis (Fig. 1) (39). Such a role of this glutamine residue is also supported by recent quantum mechanics/molecular mechanics (QM/MM) studies (Fig. 1C) (68). Accordingly, recent neutron structures of NcAA9D from Neurospora crassa (also known as NcPMO-01050) indicate that His157 and Gln166 (equivalent to His164 and Gln173 shown in Fig. 1C) interact with an equatorially bound oxygen species (69). Similar equatorial binding of an oxygen species has also been observed in a neutron structure of a bacterial AA10 LPMO (70). Mutational studies have shown that Glu60 in SmAA10A (54) (Fig. 1A) and Gln151 in Thielavia terrestris AA9E (TtAA9E) (19) (equivalent to Gln173 in Thermoascus aurantiacus AA9A [TaAA9A]) (Fig. 1C) are important for activity on chitin and cellulose degradation, respectively. While this indicates that the studies on oxygen activation discussed above are relevant for understanding LPMO catalysis, generally, great caution is needed when observations made under substrate-free conditions are transposed to productive conditions (i.e., in the presence of substrate; see below). Of note, the acid/base chemistry that may be exerted by second-sphere amino acid residues such as Glu60 in SmAA10A or His164 in TaAA9A (Fig. 1A) during O2 reduction and/or catalysis is not understood, and no pKa values for these residues have been reported.
One of the major challenges in understanding LPMO catalysis pertains to the insoluble nature of natural substrates that constitutes a hurdle for the use of most standard experimental approaches. In this respect, a landmark study has reported the crystallographic resolution of the first AA9 LPMO-oligosaccharide complex revealing interactions between the enzyme and a soluble substrate (52). This study showed that substrate binding, expectedly, shields the copper ion from the solvent and that the catalytic oxygen species must bind in the equatorial position. The latter was supported by the presence of a chloride ion in the equatorial copper coordination position in the enzyme-substrate complex, which could mimic a reactive oxygen species. Frandsen et al. also confirmed previous observations by Borisova et al. (71) that substrate binding leads to changes in copper coordination, as was also reported, more recently, for the chitin-active SmAA10A (45), leading to the suggestion that substrate binding increases the catalytic competence of the enzyme. This would make sense since it would help the enzyme to unleash its oxidative power only if substrate is present. Despite claims indicating the opposite, still very little is known about polysaccharide cleavage by LPMOs. A key point to be taken from the work by Frandsen et al. and others is that the active-site environment of an LPMO in solution is different from that of an LPMO bound to a crystalline polysaccharide surface. Notably, confinement of the active site upon crystalline polysaccharide binding seems to restrict access to reactants, as indicated by simulations suggesting formation of a gated tunnel connecting the active-site cavity to the bulk solvent (45).
There is clear and growing evidence (crystallographic, biochemical, and computational) for activation of O2 in the absence of substrate, but there is no strong experimental proof for such activation in the presence of a bound substrate (see reference 44 for a very recent discussion). One particularly intriguing issue concerns the fact that the monooxygenase reaction paradigm (R—H + O2 + 2e− + 2H+ → R—OH + H2O) requires that two electrons and two protons are recruited during catalysis. In the case of an LPMO, the first electron can be stored in the form of Cu(I), but the second electron either has to be stored by the enzyme or timely supplied when required. In the mono-copper and otherwise cofactor-free LPMO, a second electron could be derived from transient residue-derived radicals, such as radicals observed for the modified tyrosine in galactose oxidase (72, 73). A tyrosine, located in the axial direction (Fig. 1C), is conserved across fungal LPMOs and may play such a role, whereas a phenylalanine is usually found at the equivalent position in bacterial LPMOs. Alternatively, some researchers have proposed an electron transport chain or channel that would allow delivery of an electron to the substrate-bound LPMO (17, 18) (Fig. 2, option a). The existence of such an electron channel has so far not been established, and there are no conserved sequence features among LPMOs that could support the existence of such a channel.
It is worth noting that the monooxygenase reaction also requires the proper supply of two protons and that nothing is known about how this could happen. Knowledge about second-sphere residues' acid/base chemistry and proton networks (67) will be useful to elucidate this.
The H2O2 reaction mechanism(s): peroxygenase and peroxidase activities.
Recently, it has been proposed that the monooxygenase paradigm applied to LPMOs may need to be revised based on experiments showing that H2O2 can efficiently drive LPMO reactions. Importantly, inhibition of LPMO catalysis by a H2O2-scavenging enzyme (horseradish peroxidase [HRP]) under standard reaction conditions (i.e., excess of O2 and reductant) suggested that O2 is a precursor molecule for the true cosubstrate, H2O2 (32, 33). Accordingly, it was also shown that H2O2-driven reactions are much faster than O2-driven reactions and that the enzymes prefer H2O2 over O2 in competition experiments. The ability of H2O2 to promote fast AA9 LPMO catalysis has recently been confirmed in independent experiments by Hangasky et al. (44). Bissaro et al. (32, 33) concluded that H2O2 is not just an alternative to O2 but that it is the catalytically relevant cosubstrate of the polysaccharide oxidation by different LPMOs, which would thus act as peroxygenases (R—H + H2O2 → R—OH + H2O). This conclusion has been debated in the field (44, 74) and still needs more proof, but the ability of H2O2 to drive fast AA9 and AA10 LPMO reactions is now well proven.
The proposed H2O2-based mechanism entails an initial reduction from LPMO-Cu(II) to Cu(I), termed the priming reduction. The reduced enzyme reacts in a controlled and substrate-associated manner with H2O2 to unleash the intrinsic oxidative power of the latter. This leads to hydroxylation of the substrate, concomitant release of a water molecule, and regeneration of the LPMO-Cu(I) state, which can enter a new catalytic cycle (Fig. 2, option b). It is important to note that the redox state of the LPMO has not been experimentally monitored along the reaction. The concept of the priming reduction has mainly been deduced from the observation that suprastoichiometric amounts of oxidized products (relative to the reductant) were generated when the AA10 LPMO was supplied with H2O2, indicating that a reduced LPMO catalyzes multiple turnovers (32). Of note, such substoichiometric consumption of reductant is not compatible with the originally proposed O2-dependent reaction mechanism (Fig. 2).
The nature of the most likely reactive oxygen species emerging during catalysis remains a matter of discussion for both the O2 and the H2O2 mechanisms (32, 42, 44, 74). One route proposed for the H2O2 reaction mechanism (32) involves a [CuO+] core intermediate as the species catalyzing the hydrogen atom abstraction from the glycosyl unit (Fig. 2). Earlier QM/MM calculations, in the working frame of an O2 reaction mechanism, also suggested that the [CuO+] intermediate would be the relevant catalytic species (68, 75, 76). Still, other mechanisms, such as a mechanism involving the formation of a H2O2-derived hydroxyl radical as oxidant, cannot be excluded (32, 44). While mechanistic details remain to be elucidated, it is worth noting that the H2O2 mechanism solves the conundrum of second-electron delivery discussed above. H2O2 carries both the protons and reducing equivalents necessary for LPMO catalysis to occur.
Prompted by the discovery of H2O2-driven LPMO catalysis in 2016 (32, 33), several follow-up studies have recently appeared. A detailed kinetic study of the peroxygenation of chitin by SmAA10A yielded a catalytic constant of 6.7 s−1 and a Km of 2.8 μM, suggesting high affinity for H2O2 (34). The resulting catalytic efficiency, in the range of 106 M−1 · s−1, is similar to catalytic efficiencies typically reported for peroxygenases (77). In another study, Breslmayr et al. used 2,6-dimethoxyphenol as chromogenic substrate and H2O2 as a cosubstrate to assess the peroxidase activity of several AA9s, yielding catalytic rates varying from 0.9 to 18.6 s−1 (43). Two different QM/MM studies have shown that the peroxygenase reaction is plausible from a theoretical point of view, with low overall energy barriers and involving a [CuO+] intermediate as oxidant (74, 78).
Using a fungal AA9 active on cellohexaose, Hangasky et al. found much higher reactivity with H2O2 as a cosubstrate (4.75 to 15.25 s−1) than with O2 (0.28 s−1) (44), thus confirming the findings by Bissaro et al. Interestingly, despite a difference of four orders of magnitude in the catalytic efficiencies observed with H2O2 (106 M−1 · s−1) (34) and O2 (102 M−1 · s−1) (44), based on additional experiments, these authors concluded that O2 may still be the most relevant and natural cosubstrate.
Regarding the stoichiometry of the reaction, data obtained for H2O2-driven LPMO reactions support a 1:1 molar ratio between consumed H2O2 and produced oxidized sugars for both AA9 and AA10 cellulose-active (32, 44) and AA10 chitin-active (34) LPMOs. No equivalent data are available for O2-driven reactions although one may expect a similar 1:1 stoichiometry in a system where nonproductive events are minimized (e.g., reduction of O2 to water). All in all, accumulating experimental evidence suggests that, in the presence of substrate, AA9 and AA10 LPMOs react much more efficiently with H2O2 than with O2. The question of whether LPMOs at all use O2 directly, that is, without prior reduction to H2O2, remains to be settled. Importantly, as described in the following sections, there are plenty of sources of H2O2 in lignocellulolytic environments.
The carbohydrate intermediate species.
Little is known about the carbohydrate intermediates potentially occurring along the reaction pathway since the substrate radical resulting from hydrogen abstraction and the subsequent hydroxylated product have never been experimentally observed. It is not known either whether the molecular rearrangement induced by the destabilizing hydroxylation, which leads to glycosidic bond cleavage and lactone formation, occurs spontaneously in the reaction mixture or in an enzyme-assisted manner. Recent calculations, however, suggest that hydroxylation-induced glycosidic bond cleavage can occur in the absence of enzyme (74). This mechanistic ambiguity is at the origin of the debate on the relevance of the term “lytic” in the LPMO acronym and explains why some choose to call these enzymes PMOs (18). The term lytic is meant to indicate the fact that bond cleavage occurs, which contrasts with most other monooxygenases, which tend to catalyze oxy-functionalization but not cleavage of their substrates.
Oxidative self-inactivation of LPMOs.
A key aspect of LPMO catalysis pertains to operational stability, a parameter that is of high importance in industrial, biological, and chemical contexts. When analyzing the literature, one can observe that a correlation can be established between poor substrate binding and rapid enzyme inactivation (56, 79–81). We know today that this enzyme inactivation is due to oxidative self-inactivation of the LPMO and that oxidative damage of the enzyme is confined to the active site, notably the copper-coordinating histidines (32). These suicide reactions can be prevented by productive substrate binding (32, 34, 44). In studies with H2O2 as a cosubstrate, Kuusk et al. showed that the rate of inactivation of SmAA10A in the absence of substrate was about 1,000 times lower than the rate of substrate cleavage in reactions with substrate (34).
In general, it is thus extremely important to have full control of the reduction state of the LPMO during its handling (i.e., protein extraction, purification, storage, and reaction setup) since accidental reduction of the copper center in the absence of the proper substrate and in the presence of O2 or H2O2 will lead to enzyme oxidative self-inactivation and thus to nonfunctional protein. It must be noted that an oxidatively damaged LPMO will still look normal on an SDS-PAGE gel and that the damage thus may remain undetected and lead to false conclusions as to the activity of the enzyme in question. To cope with such issues, several precautions could be envisaged, such as using of metal chelators (e.g., EDTA) to remove the copper ion while the enzyme is not used, working under anaerobic conditions, or avoiding reducing conditions. Research pertaining to the control of these inactivation processes will likely be a topic of investigation in the near future, notably by seeking inspiration from the fields of peroxidases and peroxygenases that face similar problems (82).
What is in the name?
As mentioned in the introduction, over time, LPMOs have had several names. Today, the term LPMO is widely accepted, and most researchers name their LPMOs as XxLPMOnX or XxAAnX, where Xx indicates the source microbe (e.g., Nc for Neurospora crassa), n indicates the LPMO family according to CAZy (currently 9, 10, 11, 13, 14, or 15), and X is a capital letter that is assigned to multiple LPMOs from a certain organism, often by the order of their functional characterization or the order of their gene numbers in the genome. Considering the recent discoveries on the role of H2O2, the term LPMO may need revision. While including peroxygenase (PO) in the name (e.g., LPPO) may appear premature, the existing term monooxygenase (MO), which by definition implies O2 as a cosubstrate, seems incorrect. Informal discussions among scholars have led to the suggestion to use the more general term oxidase to refer to LPMOs. The term LPO, for lytic polysaccharide oxidase, entails a simple change relative to LPMO, does not assume the nature of the cosubstrate, and includes the indisputable ability of LPMOs to generate H2O2 (which is an oxidase reaction). On the other hand, the term oxidase does not reflect the fact that an oxygen atom is incorporated into the final product, and in that sense “oxygenase” appears more appropriate. Discussions, debates, and scientific progress should allow sorting out this issue in the near future.
In the following sections, we revisit previously published studies on LP(M)Os to pinpoint overlooked incoherencies or unexplained phenomena that find sense in the frame of the peroxygenase paradigm for LPMO action.
A Wide Diversity of Reductants
LPMO reductants and associated catalytic rates.
It has been demonstrated that LPMOs can be activated by a wide diversity of reductants (Table 1). These include organic compounds such as ascorbic acid (AscA) (11), cysteine (48, 65), reduced glutathione (11), and a wide range of plant- and fungus-derived phenols (65, 83), as well as lignin and fragments thereof (84–86). Functional reductants also include enzymatic systems such as cellobiose dehydrogenase (CDH) (see subsection “The CDH Case: a Multifunctional Redox Partner,” below) or photocatalytic systems (87, 88) (see subsection “Stimulation of LPMO Activity by Photocatalytic Systems,” below). The ability of CDH to drive fungal AA9 LPMO reactions was detected early in the development of the field (14, 64). It should be noted that an equivalent natural enzymatic redox partner has not yet been found in bacteria.
TABLE 1.
Diversity of reductants promoting LPMO activity and associated apparent catalytic ratesa
| Enzyme (concn)b | Electron-supplying system (concn)f | Reaction conditions |
Quantification method (fraction)j | Observed oxidative rate (min−1)l | Reference | |||
|---|---|---|---|---|---|---|---|---|
| Substrate(s) (concn)g | Bufferh | T (°C) | Mixing (rpm)i | |||||
| SmAA10A (1 μM)c | Reduced glutathione (1 mM) | β-Chitin (0.45 g/liter) | Tris-HCl (20 mM, pH 8.0) | 37 | Thermomixer (1,000) | UHPLC-UV (total) | 1.28 | 11 |
| ScAA10C-CBM2 (1 μM)d | AscA (2 mM) | PASC (2 g/liter) | Am-Ac (20 mM, pH 6.0) | 50 | Thermomixer (900) | HPAEC-PAD (soluble) | 0.22* | 351 |
| LsAA9A (1 μM) | AscA (5 mM) | FRET substrate (10–100 μM)d | BT-HCl (20 mM, pH 7.0) | 37 | 96-Well MP | FRET (total) | 6.6 | 52 |
| NcAA9C-CBM1 (1.47 μM) | AscA (1 mM) | XG14 (0.2 mM) | Am-Ac (25 mM, pH 8.0) | 40 | Thermomixer (600) | HPAEC-PAD (total) | 3.6 | 46 |
| AscA (1 mM) | Cellopentaose (0.2 mM) | Am-Ac (25 mM, pH 8.0) | 40 | Thermomixer (600) | HPAEC-PAD (total) | 1.8 | 46 | |
| AscA (1 mM) | Tamarind XG (5 g/liter) | Na-P (5 mM, pH 8.0) | 50 | Thermomixer (1,000) | DNS | 6.6 | 46 | |
| AscA (1 mM) | PASC (5 g/liter) | Na-P (5 mM, pH 8.0) | 50 | Thermomixer (1,000) | DNS | 6.6 | 46 | |
| NcAA9C-CBM1 (4 μM) | AscA (2 mM) | Tamarind XG (5 g/liter) | Na-P (40 mM, pH 6.5) | 50 | Thermomixer (1,000) | DNS | 6.0 | 71 |
| AscA (2 mM) | PASC (5 g/liter) | Na-P (40 mM, pH 6.5) | 50 | Thermomixer (1,000) | DNS | 2.4 | 71 | |
| VcAA10B-X-Y-CBM73 (2 μM)e | AscA (1 mM) | β-Chitin nanofibers (5 g/liter) | BTm-HCl (50 mM, pH 6.8) | 37 | Thermomixer (800) | UHPLC-UV (soluble) | 2.7 | 429 |
| SmAA10A (1 μM) | AscA (0.5 mM) | β-Chitin (10 g/liter) | BTm-HCl (50 mM, pH 6.0) | 40 | Thermomixer (1,000) | UHPLC-UV (soluble) | 4.17* | 102 |
| AscA (1 mM) | β-Chitin (10 g/liter) | BTm-HCl (50 mM, pH 6.0) | 40 | Thermomixer (1,000) | UHPLC-UV (soluble) | 6.6* | 102 | |
| AscA (2 mM) | β-Chitin (10 g/liter) | BTm-HCl (50 mM, pH 6.0) | 40 | Thermomixer (1,000) | UHPLC-UV (soluble) | 9.72* | 102 | |
| AscA (5 mM) | β-Chitin (10 g/liter) | BTm-HCl (50 mM, pH 6.0) | 40 | Thermomixer (1,000) | UHPLC-UV (soluble) | 13.2* | 102 | |
| CfAA10-CBM2 (1 μM) | AscA (1 mM) | PASC (0.3 g/liter) | Na-P (50 mM, pH 6.0) | 37 | Thermomixer (150) | HPAEC-PAD (soluble) | 0.49 | 80 |
| AscA (1 mM) | Avicel (0.3 g/liter) | Na-P (50 mM, pH 6.0) | 37 | Thermomixer (150) | HPAEC-PAD (soluble) | 0.35 | 80 | |
| AscA (1 mM) | BMCC (0.3 g/liter) | Na-P (50 mM, pH 6.0) | 37 | Thermomixer (150) | HPAEC-PAD (soluble) | 0.78 | 80 | |
| TbAA10-CBM2 (1 μM) | AscA (1 mM) | PASC (0.3 g/liter) | Na-P (50 mM, pH 6.0) | 37 | Thermomixer (150) | HPAEC-PAD (soluble) | 0.23 | 80 |
| AscA (1 mM) | Avicel (0.3 g/liter) | Na-P (50 mM, pH 6.0) | 37 | Thermomixer (150) | HPAEC-PAD (soluble) | 0.10 | 80 | |
| AscA (1 mM) | BMCC (0.3 g/liter) | Na-P (50 mM, pH 6.0) | 37 | Thermomixer (150) | HPAEC-PAD (soluble) | 0.32 | 80 | |
| CjAA10A-CBM5-CBM73 (0.5 μM) | AscA (1 mM) | α-Chitin (10 g/liter) | BTp-HCl (20 mM, pH 7.2) | 37 | Thermomixer (1,000) | UHPLC (soluble) | 1.43* | 79 |
| TrAA9A-CBM1 (4 μM) | AscA (0.4 mM) | PASC (4 g/liter) | Na-Ac (10 mM, pH 5.0) | 37 | Mixing in miniplate well | Oxygen consumptionk | 1.26 | 104 |
| TtAA9E (4 μM) | AscA (0.4 mM) | PASC (4 g/liter) | Na-Ac (10 mM, pH 5.0) | 37 | Mixing in miniplate well | Oxygen consumption | 0.88 | 104 |
| ThtAA9A (4 μM) | AscA (0.4 mM) | PASC (4 g/liter) | Na-Ac (10 mM, pH 5.0) | 37 | Mixing in miniplate well | Oxygen consumption | 0.93 | 104 |
| ThtAA9B-CBM1 (0.27 μM) | AscA (1 mM) | RAC (2.8 g/liter) | Am-Ac (50 mM, pH 5.0) | 50 | HOTSR (20) | HPAEC-PAD (total) | 0.28*m | 91 |
| ScAA10C-CBM2 (0.5 μM) | AscA (1 mM) | Avicel (10 g/liter), H2O2 (200 μM) | Na-P (50 mM, pH 7.0) | 40 | Magnetic stirring | HPAEC-PAD (soluble) | 82.4*n | 32 |
| AscA (1 mM) | Avicel (10 g/liter) | Na-P (50 mM, pH 7.0) | 40 | Magnetic stirring | HPAEC-PAD (soluble) | 3.2*n | 32 | |
| PcAA9D (0.5 μM) | AscA (1 mM) | Avicel (10 g/liter), H2O2 (100 μM) | Na-P (50 mM, pH 7.0) | 40 | Magnetic stirring | HPAEC-PAD (soluble) | 15.6*o | 32 |
| AscA (1 mM) | Avicel (10 g/liter) | Na-P (50 mM, pH 7.0) | 40 | Magnetic stirring | HPAEC-PAD (soluble) | 2.1*o | 32 | |
| SmAA10A (50 nM) | AscA (100 μM) | CNW (sat), H2O2 | Na-Ac (50 mM, pH 6.1) | 25 | Static | 14C radioactivity | 402 | 34 |
| ThtAA9E (50 nM) | AscA (2 mM) | Cellohexaose (1 mM), H2O2 (100 μM) | MES/MOPS (100 mM, pH 6.5) | 40 | NR | HPAEC-PAD (total) | 916p | 44 |
| ThtAA9E (1 μM) | AscA (2 mM) | Cellohexaose (sat) | MES/MOPS (100 mM, pH 6.5) | 40 | NR | HPAEC-PAD (total) | 10q | 373 |
| SmAA10A (1 μM) | Lactose (3 mM)/MtCDH (1.5 μM) | β-Chitin (10 g/liter) | BTm-HCl (25 mM, pH 6.0) | 40 | Thermomixer (1,000) | UHPLC-UV (total) | 3.3 | 102 |
| TtAA9E (2.22 μM) | Chl (1.6 mM)/visible light + AscA (2 mM) | PASC (7.5 g/liter) | Cit-P (100 mM, pH 6.3) | 50 | Thermomixer (1,000) | HPAEC-PAD (total) | 33 | 87 |
| ScAA10C-CBM2 (0.5 μM) | Chl (0.5 mM)/visible light + AscA (1 mM) | Avicel (10 g/liter) | Na-P (50 mM, pH 7.0) | 40 | Magnetic stirring | HPAEC-PAD (soluble) | 96*n | 33 |
| ScAA10C-CBM2 (1 μM) | H2O/V-TiO2 (5 g/liter)/visible light | Avicel (10 g/liter) | Na-P (50 mM, pH 6.0) | 40 | Magnetic stirring | HPAEC-PAD (soluble) | 0.28* | 88 |
| H2O + MeOH/V-TiO2 (5 g/liter)/visible light | Avicel (10 g/liter) | Na-P (50 mM, pH 6.0) | 40 | Magnetic stirring | HPAEC-PAD (soluble) | 0.78* | 88 | |
| NcAA9C-CBM1 (1.25 μM) | Fungal- and plant-derived phenols (1 mM) | MCC (25 g/liter) | K-P (50 mM, pH 6.0) | 30 | Thermomixer (800) | HPLC-ED40 (soluble) | NRr | 83 |
| ThtAA9A, ThtAA9B-CBM1, and ThtAA9C (2.5, 5, and 2.5 mg/g substrate) | Plant-derived phenols (1 mM) | RAC (1.5 g/liter) | Am-Ac (50 mM, pH 5.0) | 50 | HOTSR (20) | HPAEC-PAD (soluble) | NRr | 65 |
| TtAA9E (1 μM) | Lignin (1 mg of HMW + 2 mM LMW) | PASC (7.5 g/liter) | Cit-P (20 mM, pH 5.9) | 50 | Thermomixer (1,000) | HPAEC (soluble) | NRs | 86 |
The table contains apparent rates for only full-length enzymes. Note that this table should not be used to draw conclusions concerning the substrate specificities of LPMOs since assays with varying substrates were carried out under highly varying conditions and since the listed rates are apparent and not true kinetic parameters. If one assumes that the H2O2-mechanism is valid, production of H2O2 was likely the rate-limiting step in many of the reported experiments with solid substrates, but not, for example, in the experiments with cellohexaose carried out by Hangasky et al. (44, 373). See the text for details.
Cf, Cellulomonas fimi; Cj, Cellvibrio japonicus; Ls, Lentinus similis; Tht, Thermothelomyces thermophila (previously Myceliophthora thermophila); Nc, Neurospora crassa; Pc, Phanerochaete chrysosporium; Sc, Streptomyces coelicolor; Sm, Serratia marcescens; Ta, Thermoascus aurantiacus; Tb, Thermobispora bispora; Tr, Trichoderma reesei; Tt, Thielavia terrestris.
Also known as CBP21.
Also known as CelS2.
Also known as Vibrio cholerae colonization factor, GbpA (GlcNAc binding protein A). GbpA is a four-domain protein where X and Y denote unknown domains related to the flagellin protein p5 and pilus-binding chaperone FimC, respectively (430).
Complex electron supplying systems include light-driven oxidation of water, catalyzed by vanadium-doped titanium dioxide (V-TiO2), light-excited chlorophyllin (Chl) in the presence of ascorbic acid (AscA), lactose oxidation catalyzed by cellobiose dehydrogenase from Myriococcum thermophilum (MtCDH), and mixture of high-molecular-weight (HMW) and low-molecular-weight (LMW) lignins.
CNW, chitin nanowhiskers; FRET, Förster resonance energy transfer; (B)MCC, (bacterial) microcrystalline cellulose; PASC, phosphoric acid-swollen cellulose; RAC, regenerated amorphous cellulose; sat, saturating concentration; XG, xyloglucan; XG14, a 14-mer xyloglucan.
Am-Ac, ammonium acetate buffer; BT, Bis-Tris; BTm, Bis-Tris-methane; BTp, Bis-Tris-propane; Cit-P, citrate phosphate buffer; K-P, potassium phosphate buffer; MES, morpholineethanesulfonic acid; MOPS, morpholinepropanesulfonic acid; Na-P, sodium phosphate buffer.
HOTSR, head-over-tail Stuart rotator; MP, microplate; NR, not reported.
DNS, dinitrosalicylic acid assay for concentration of reducing ends; HPAEC-PAD, high-performance anion-exchange chromatography with pulsed amperometric detection; HPLC-ED40, high-performance liquid chromatograph equipped with a Dionex ED40 electrochemical detector; UHPLC, ultra-high-performance liquid chromatography.
Value independent of the extent of solubilization of oxidized products and reflects thus total LPMO activity.
Values marked with an asterisk were calculated by us on the basis of progress curves reported in the original article.
An approximate molecular weight of 31 kDa, which does not account for glycosylations, was considered to convert the rate from micromolars per minute to per minute.
Estimated based on product quantity released after a 2-min reaction.
Estimated based on product quantity released after a 3-min reaction.
Apparent rate values 285, 444, and 647 min−1 were also determined as with [H2O2] = 12.5, 25, and 50 μM, respectively (44).
Determined at ambient O2. Note that Hangasky et al. have determined a full set of kinetic parameters for ThtAA9E at different O2 and cellohexaose concentrations (373).
In a 24-h reaction.
In a 12-h reaction.
The highly surface-exposed active sites of LPMOs are unusual and may explain the apparent absence of reductant specificity. Nevertheless, taking into account the fact that the reductants listed above display very different sizes and topologies, as well as electrostatic or hydrophobicity properties, it appears intriguing that they can all directly reduce the LPMO copper center. Of note, redox partner diversity has also been observed for cytochrome P450 monooxygenases (89). Both for P450 monooxygenases (90) and LPMOs (83) a correlation between the reduction potential of the redox partner and the reduction rate of the enzyme has been established. Also, it has been reported that an increase in pH led to a decrease in reduction potential of the reductant and to an increase in AA9 LPMO initial rates (91).
Importantly, no correlation has ever been established between the rate of reduction of LPMOs (per millisecond range) and the apparent catalytic rate (per minute range), which is orders of magnitude lower. Assuming the O2-based mechanism, this remarkable discrepancy in rates indicates that transfer of the second electron, the rate of which cannot be measured directly, is rate limiting and affected by the reductant type. Alternatively, in the H2O2-based mechanism, priming reduction of the copper may not be rate limiting, and the dependency of the LPMO catalytic rate on the reductant may reflect different potentials of each reducing system to generate and/or accumulate H2O2. Table 1 shows an overview of available kinetic data for a large diversity of LPMO substrates and reductants. Importantly, with a few exceptions, the apparent enzyme rates are low and fall in a relatively narrow window roughly between 1 and 10 min−1. Much higher LPMO rates (in the range of 1 to 10 per second) have been obtained in two settings: (i) when a photocatalytic system is used (87) (see also subsection “Stimulation of LPMO Activity by Photocatalytic Systems,” below) and (ii) when H2O2 is used to drive the reaction (32, 34, 44). Determination of LPMO rates is generally complicated because of the inactivation processes discussed above. In reactions with added H2O2, the LPMO catalytic rate depends on the H2O2 supply rate, but saturation kinetics may not be reached before inactivation phenomena occur, which is a common problem encountered in the field of H2O2-using enzymes (82). Of note, when available kinetic data for LPMOs are evaluated (Table 1), it is important to consider that in most studies only the carbohydrate substrate concentration was controlled, whereas neither the identity nor the quantity of the oxygen-containing cosubstrate was known or controlled.
Almost all rates listed in Table 1 are apparent rates and not true kinetic parameters. As a consequence, the listed rates for various substrates cannot be used to draw any conclusions as to the substrate specificity of LPMOs. In fact, whereas LPMOs are thought to have evolved primarily to attack crystalline substrates (11, 29), the true substrate preferences of LPMOs, e.g., in terms of kcat/Km values, remain unknown.
ROS as reductants.
It has been shown that superoxide (O2˙−), the product of O2 single-electron reduction (Fig. 3), can reduce and thus activate AA10 LPMOs, while H2O2 cannot (32). Therefore, superoxide constitutes a possible electron shuttle between a reductant and the LPMO active site, as has been shown for myeloperoxidases (92). Many photosystems display the appropriate reduction potential to catalyze production of superoxide (93). Also, O2˙− can be produced by a wide range of oxidases (94) and can emerge in reactions involving semiquinones (95), which are intermediates between hydroquinones and quinones, all abundant in biomass-degrading ecosystems (96). Notably, O2˙− will spontaneously disproportionate to H2O2 in protic solvents such as H2O, decreasing its lifetime as an LPMO reductant but generating an LPMO cosubstrate. This disproportionation process can be accelerated by ascorbic acid (97, 98) or by phenolics present in biomass (see subsection “Nonenzymatic Production and Use of H2O2,” below). Notably, in the H2O2-dependent mechanism, only a priming reduction is needed, meaning that small amounts of O2˙− could be sufficient to activate LPMOs.
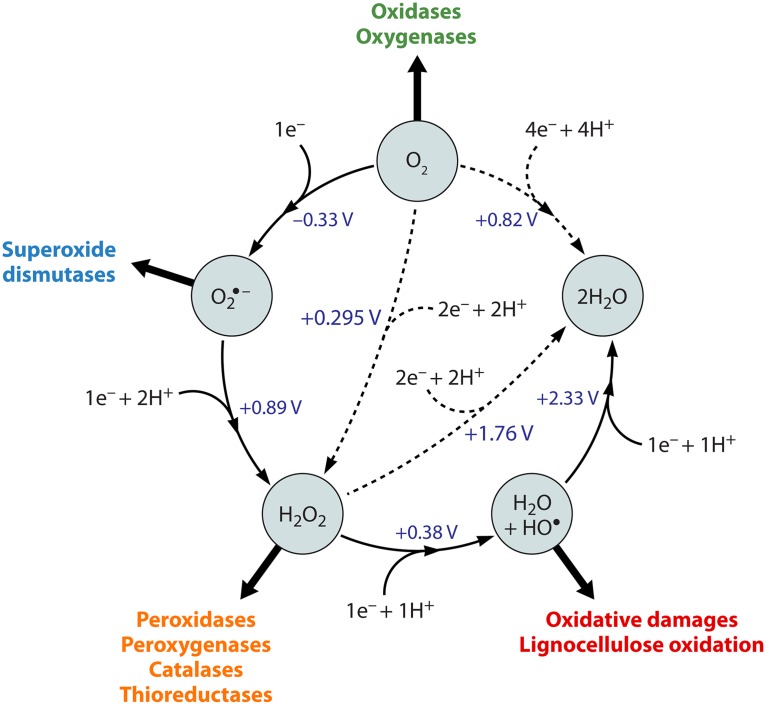
FIG 3.
The reduction cycle of reactive oxygen species (ROS) from molecular oxygen (O2) to water. O2 can undergo a single-electron reduction leading to the formation of superoxide (O2˙−), which can be further reduced to H2O2, either spontaneously, enzymatically, or by small organic reductants. H2O2 can also be generated via a two-electron reduction of O2. H2O2 can enter pathways leading either to the formation of a hydroxyl radical after a single-electron reduction [e.g., by Fe(II) or by Cu(I), i.e., Fenton reactions] or to the production of two H2O molecules via a two-electron reduction. H2O molecules can also be obtained by a direct four-electron reduction of O2. Reduction potentials are indicated in the figure (at pH 7 versus SHE) (99, 100). Each ROS can also be the substrate of other chemical or enzymatic reactions, as indicated by the large filled arrows.
To complicate things, it is known that reduced LPMOs can catalyze the single-electron reduction of O2 into O2˙− (15), which eventually results in H2O2. In light of this, it is worth noting the single electron reduction potentials of H2O2 (E0 = +0.38 V) and O2 (−0.33 V) (Fig. 3) (99, 100), which suggests that single-electron transfer from LPMO-Cu(I) would be more thermodynamically favorable for H2O2 than for O2. Indeed, the 18O competition experiments alluded to above (32) clearly showed that AA10 LPMOs prefer to react with H2O2 rather than with O2 when presented with both. Having this in mind, in a biological context, prereduction of O2 to H2O2 via an energetically easier two-electron reduction process (+0.295 V) catalyzed by enzymes evolved in nature for this purpose (e.g., flavin adenine dinucleotide [FAD]-dependent oxidases) (99, 101) (Fig. 4) represents an appealing and efficient strategy to provide H2O2 and fuel LPMO reactions. Indeed, such enzymes are found together with LPMOs in biomass-degrading ecosystems (see Insights into the Network of Lignocellulolytic Redox Reactions).
FIG 4.
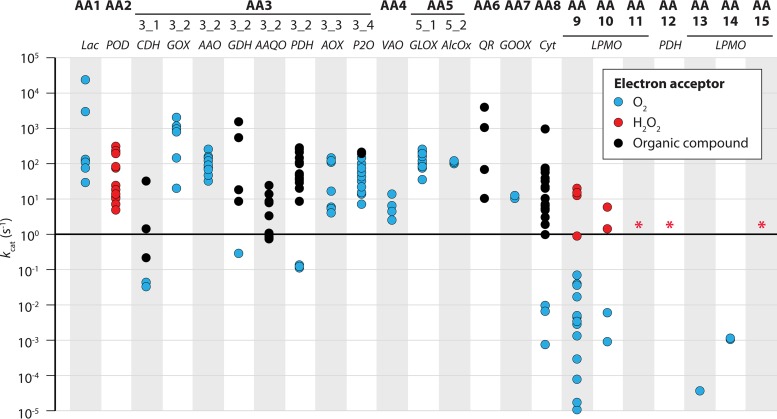
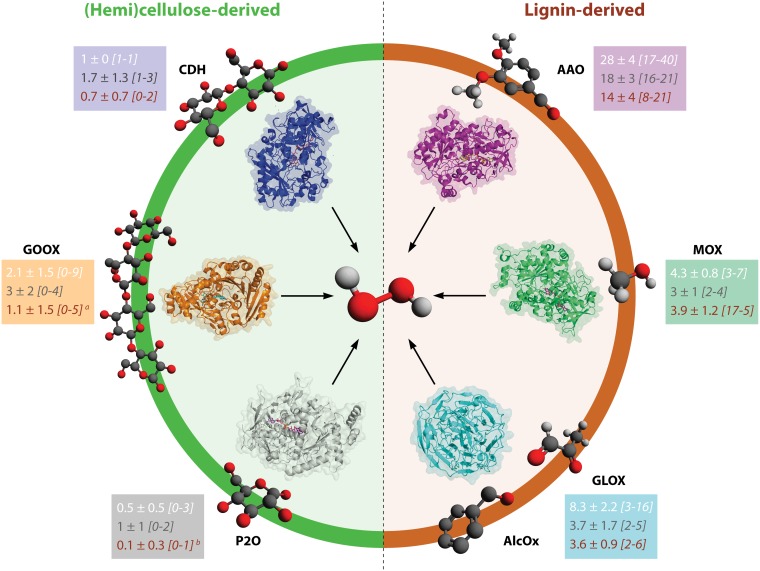
Catalytic constants of auxiliary activities. Auxiliary activities comprise redox enzymes involved in biomass conversion and include the LPMOs (21). For each AA family, apparent catalytic constants collected from the literature are indicated by dots that are colored depending on whether the final electron acceptor is O2, H2O2, or an organic compound/protein, as indicated. Details on the sources of the displayed data, including a reference to the correct publication for each data point, are provided in Table S1 in the supplemental material. Red stars indicate the absence of kinetic data. Abbreviations (and associated references for corresponding data) are as follows: Lac, laccase (380–383); POD, peroxidase (380, 384–388); CDH, cellobiose dehydrogenase (102, 128–130, 389–392); GOX, glucose oxidase (167, 393–398); AAO, aryl alcohol oxidase (166, 399, 400); GDH, glucose dehydrogenase (401, 402); AAQO, aryl alcohol quinone oxidoreductase (403); PDH, pyranose dehydrogenase (404–407); AOX, alcohol oxidase (177, 408–410); P2O, pyranose 2-oxidase (181, 411–413); VAO, vanillyl alcohol oxidase (186, 187); GLOX, glyoxal oxidase (170, 414); AlcOx, alcohol oxidase (172); QR, quinone reductase (415–418); GOOX, gluco-oligosaccharide oxidase (173, 174, 419, 420); Cyt, cytochrome b (83, 102, 124, 130, 353, 389, 391, 421); LPMO, lytic polysaccharide monooxygenase (AA9, (43, 44, 66, 67, 103, 422, 423); AA10, (32, 34, 56); AA13, (424); AA14 (25); PDH, pyranose dehydrogenase.
How much reducing power is needed to fuel LPMOs?
An intriguing aspect of LPMO biochemistry pertains to observed dose-response relationships for the reductant (102). In most experiments published so far under standard conditions (aerobic, reductant, and no added H2O2) molar enzyme/reductant ratios were in the 1:10,000 range (Table 1). One may wonder why variation in such a large excess of reductant influences the catalytic rate of the LPMO, knowing that LPMO reduction is a fast process (83). One possible answer is that the reductant, in addition to reducing the LPMO, is involved in the generation of the H2O2 whose availability is rate limiting for the reaction. Alternatively, in an O2-based mechanism, the reductant concentration could affect the delivery rate for the second electron (see above).
H2O2 may be generated from O2 by reduced LPMOs in solution (62, 66, 103) or by reactions involving O2 and reductant. As to the latter option, Gusakov et al. have shown that O2 consumption in the absence of LPMO increased when the concentration of AscA increased (104). It is important to note that generated H2O2 may engage in all kinds of redox reactions, for example, those involving free metals (105) or phenolic compounds present in the lignocellulosic substrate. In this respect, LPMO experimentalists have certainly noticed variability and reproducibility issues when they compared different batches of substrate or used different batches of reductant.
If one wishes to control LPMO reactions, it is of utmost importance to control the levels of reductant and oxygen species, which, considering all the possible reactions, is a major challenge. One way of reaching such control is to run reactions anaerobically, with small amounts of reductant and a steady, slow supply of H2O2 (32). When reactions are run with O2, current data indicate that, instead of using small-molecule reductants, it is better to use a source of reducing equivalents that is less prone to uncontrollable auto-oxidation, such as a dehydrogenase and its substrate (102) or photocatalytic systems (33, 87, 88). Such systems are discussed below.
Why LPMOs Do Not Seem To Produce H2O2 in the Presence of Substrate
The binding of an LPMO to its target substrate is thought to be controlled by structural properties of the enzyme surface (45, 52) and, in the case of multimodular LPMOs, by appended CBMs (12, 39). Substrate binding has also been suggested to be influenced by other players of the LPMO reaction since cyanide and chloride, both mimics of superoxide, increase binding (52, 55). Current data suggest that binding is strengthened by formation of a ternary complex with substrate and an oxygen species.
Available data show that the extent of LPMO binding varies a lot. For AA10 chitin-active LPMOs, the bound fraction has been reported to lie between 80% or more (54, 81) and down to 40% (106) or 19% (107). Regarding cellulose-active LPMOs, one can find qualitative estimations for bound fractions spanning from ca. 100% bound (for a full-length enzyme with CBM) (80) to 40% (108). Thus, in most cases, a significant fraction of the LPMO is not bound to the substrate and is free in solution.
As noted above, H2O2 accumulates in LPMO reactions that lack substrate (62, 66, 103). This is usually considered a futile reaction, also known as an uncoupling reaction. In the presence of the appropriate substrate, such LPMO-mediated H2O2 production is not observed, and this is usually attributed to the fact that the catalytically competent LPMO acts on the substrate rather than being engaged into the uncoupling reaction. However, one may wonder why the unbound fractions of LPMOs apparently do not produce H2O2. For instance, in the case of the fungal NcAA9C-CBM1, no H2O2 was detected in reaction mixtures containing 5 mM Glc6 (62) although 14% of the enzymes were probably free in solution, given a Kd (dissociation constant) value for this substrate of 0.81 mM (71). One possible explanation is that reduced LPMOs are never free in solution because they bind much more strongly to the substrate than suggested by the Kd value, which was determined in the absence of reductant. This explanation is supported by a recent report showing that LPMO-Cu(I) binds cellulose more strongly than LPMO-Cu(II) (109), but also in this case, binding did not seem complete. In any case, strong and even 100% binding of the reduced LPMO cannot account for the complete absence of H2O2 since, under the conditions used, H2O2 will also be produced by non-LPMO-catalyzed reactions involving the reductant, O2, and transition metals in solution.
An obvious alternative explanation for these observations follows from the H2O2 mechanism, which dictates that H2O2 produced by non-LPMO-catalyzed reactions or by unbound LPMOs is readily consumed by bound LPMOs carrying out H2O2-driven catalysis on the substrate. In this scenario, the very low H2O2 concentrations observed in LPMO reaction mixtures containing substrate are hiding production and consumption fluxes (Fig. 5).
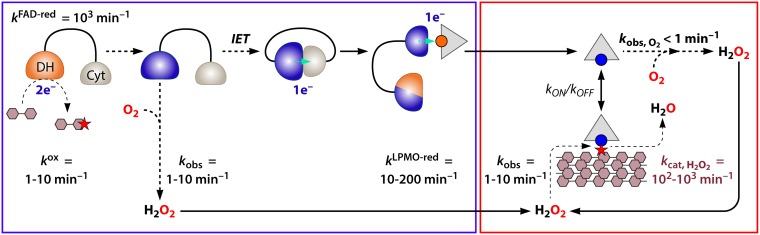
FIG 5.
On the disappearance of H2O2 during LPMO catalysis, illustrated using CDH as an LPMO-independent H2O2-generating system. Dotted arrows indicate chemical reactions, whereas solid arrows indicate diffusion. IET stands for internal electron transfer; blue and orange indicate reduced and oxidized states, respectively. The blue frame shows a system generating reducing equivalents that both produce H2O2 and serve to reduce the LPMO. The reduced LPMO (red frame) is in equilibrium between the bound and unbound forms and will either generate H2O2 (uncoupling reaction, kobs, O2) or oxidize a polysaccharide. Here, the blue frame depicts a CDH system (see subsection “The CDH Case: a Multifunctional Redox Partner” for a detailed explanation), but this system could be replaced by several alternative reducing systems. In the figure, from left to right, rates are shown as follows: for cellobiose oxidation with O2 being the electron acceptor (102); for the reduction (kFAD-red) of the flavin (FAD)-containing DH domain (83, 124, 391); for LPMO reduction (83, 102, 124); for H2O2 generation by CDH (102, 117, 119, 129–131). The apparent rates of LPMO-catalyzed reactions are given in Tables 1 and S1. Note that LPMO reduction (kLPMO-red) by CDH-Cyt obeys pseudo-first-order kinetics and depends on the LPMO concentration (102). Thus, for comparative purposes, the kLPMO-red range is given for 1 μM LPMO (a common concentration in most published assays). Note that the catalytic constant for H2O2-driven polysaccharide oxidation by LPMOs (kcat, H2O2) is much higher than the apparent rate constant, derived from standard reactions without added H2O2 (kobs). In the latter reactions H2O2 generation is rate limiting for LPMO action.
The CDH Case: a Multifunctional Redox Partner?
It is now well established that fungal CDHs, which are more common in white-rot fungi than brown-rot fungi (110), can promote AA9 LPMO activity (14, 64). Genomic cooccurrence and coexpression of CDH along with AA9 LPMOs is often observed (18, 83). Knocking out the cdh gene leads to lower efficiency of the cellulolytic secretome (14) and has also been shown to promote putatively compensatory mechanisms by the fungus, such as the secretion of additional β-glucosidases as well as AA3_2 flavo-oxidases (111). A plethora of roles have been proposed for CDHs throughout the last decades, one of them being reduction of transition metals [e.g., Fe(III)] and generation of H2O2 to drive hydroxyl radical-generating Fenton reactions (112–116) (see subsection “Nonenzymatic Production and Use of H2O2,” below). Of note, the H2O2 production ability varies between CDHs and is pH and substrate dependent (117, 118) but is relatively low (119–122) compared to that of classical oxidases (Fig. 4; see also Table S1 in the supplemental material). This low rate led scholars to adopt the name cellobiose dehydrogenase (CDH) instead of the initial cellobiose oxidase (CBO) (123).
Today, there is strong evidence that CDH constitutes a natural redox partner for AA9 LPMOs (55, 124, 125) although the exact mode of interplay still needs to be fully elucidated, including the second-electron conundrum in the O2-based LPMO mechanism. Considering the recent doubts concerning this mechanism, it is worthwhile to revisit some of the available kinetic data for CDHs and their interplay with LPMOs.
CDHs are bi-modular redox enzymes belonging to the superfamily of glucose-methanol-choline (GMC) oxidoreductase (EC 1.1.99.18) containing a flavin adenine dinucleotide (FAD)-dependent dehydrogenase domain (DH; AA3_1 subfamily) and a cytochrome domain (Cyt; family AA8) connected by a flexible linker allowing mobility between the two domains. The DH domain constitutes the catalytic part of the enzyme where a two-electron oxidation of the substrate (cellobiose and several other oligosaccharides) reduces the flavin cofactor (FAD + 2e− + 2H+ → FADH2). Reoxidation of the flavin may happen by reduction of a two-electron acceptor (e.g., dichlorophenolindophenol [DCPIP], benzoquinone, or O2) or by sequential single-electron transfer to the Cyt domain (126). It is known that the reduced Cyt domain can transfer electrons to AA9 and AA10 LPMOs (102, 124). The existence of a CDH binding site on a fungal AA9 LPMO has been suggested (127), but theoretical considerations (42) and lack of sequence conservation in the proposed docking site (125), as well as studies of interactions between CDH and an AA9 LPMO by computational modeling (124) or nuclear magnetic resonance (NMR) (55), rather support direct electron transfer at the copper site.
The reoxidation of the reduced DH by direct (from the FAD) (Fig. 5) and indirect (via Cyt) (not shown in Fig. 5) reduction of O2 is slow, with observed rates typically being in the order of 10−1 to 10−2 and 10−2 to 10−3 s−1, respectively (128–130) (Fig. 4). It has been shown that the reduced Cyt domain reacts much faster with the LPMO (AA9) than with atmospheric O2 (47, 83, 124). This is logical when the reduction potential of Cyt (E0 = +93 to 163 mV versus standard hydrogen electrode [SHE]) (131) is compared with the potential for the thermodynamically challenging single-electron reduction of O2 (E0 ≈ −330 mV versus SHE) and the reduction potential of LPMO-Cu(II), which is much more prone to reduction (E0 ≈ +250 mV versus SHE). Note that the total spin is conserved for the reduced cytochrome reaction with both LPMO-Cu(II) and O2. Under standard conditions (i.e., 1 μM LPMO) the LPMO reduction rate by reduced Cyt lies in the range of 10 to 200 min−1 (83, 102, 124), which is one to two orders of magnitude faster than reported LPMO catalytic rates (Fig. 5 and Table 1). Therefore, the initial reduction step probably does not constitute a rate-limiting step in CDH-driven LPMO catalysis. However, CDH does play a rate-limiting role under certain conditions, as it has been shown that increasing the amount of CDH activity in a reaction increases AA10 LPMO activity (102).
Taken together, the existing data on the interactions between CDH and AA9 and AA10 LPMOs may seem to suggest that delivery of the second electron is rate limiting. However, this reasoning requires reconsideration in light of a possible role of H2O2 in LPMO catalysis. Incubation of CDH with a substrate (e.g., lactose) in the absence of an electron acceptor will lead to the production of H2O2, whereas such production seemingly does not happen when the reaction mixture also contains an LPMO and its substrate. The common explanation for the disappearance of H2O2 when the LPMO is present requires that the reducing equivalents acquired by CDH upon lactose oxidation are preferentially transferred to the LPMO rather than to O2. Such a conclusion, however, is questionable given the abundance of O2 (ca. 250 μM) versus that of LPMO (1 μM), suggesting that the balance between both processes may be less tilted in favor of LPMO reduction than usually thought. Moreover, it has been shown that the rate of H2O2 production by the lactose/CDH system in the absence of LPMO plus substrate (1 to 10 min−1) (see Table S1 in the supplemental material) is remarkably similar to the LPMO (AA10) oxidative rate when the latter is fueled by the lactose/CDH system (Table 1) (102, 128). This observation may, of course, be due to coincidence but does suggest that the rate-limiting step in CDH-driven LPMO catalysis is the formation of H2O2, which does not accumulate because it is consumed by the LPMO.
Comparing the high reduction rate of the FAD domain (by electrons derived from oligosaccharide oxidation) with all other rates of subsequent reactions in the CDH system (Fig. 5) shows that the rate of reoxidation of CDH is determining the overall turnover rate and that this rate is driven by the nature of the electron acceptor and its ability to accept electrons from CDH (Table S1) (125). In the CDH-LPMO systems, there is a mismatch between the (relatively high) electron-donating capacity of CDH and the (relatively low) electron consumption by the LPMO. Thus, electrons will inevitably be routed from the reduced dehydrogenase domain toward O2, leading to slow H2O2 generation. The similar rates of H2O2 generation by the CDH and of CDH-driven LPMO catalysis strongly suggest that generation of H2O2 is the rate-limiting step in CDH-LPMO systems. The observation that CDH-fueled LPMO systems are inhibited by a peroxidase competing for H2O2 fully supports this view (32, 44). With hindsight, it is interesting that the poor oxidase activity (i.e., slow H2O2 generation) and the more efficient dehydrogenase activity (i.e., reducing equivalent generation) of CDHs may both be biologically relevant.
Stimulation of LPMO Activity by Photocatalytic Systems
In 2016, two photocatalytic systems were reported to promote LPMO activity. First, Cannella et al. showed that AA9 LPMOs can be fueled by the combined use of a pigment (e.g., chlorophyllin) and a reductant (e.g., AscA) when exposed to (low-intensity) visible light (87). This approach resulted in an impressive boost in LPMO activity, reaching rates that were 10- to 100-fold faster than those of reference experiments under standard conditions. The authors of this study proposed that high redox potential electrons, generated by the photoexcited pigment, would be at the origin of LPMO activation and the rate enhancement. In this system, the reductant would merely serve to regenerate the electron pool of the pigment.
It has been proposed (33) that the generation of ROS could be the underlying reason for the activity boost observed by Cannella et al. (87). Several photosystems encountered in nature perform the single-electron reduction of molecular oxygen to superoxide, which requires a low reduction potential, i.e., a high reducing strength (O2 + 1e− | O2˙−; E0 = −0.33 V) (Fig. 3). Bissaro et al. showed that superoxide is formed when chlorophyllin is exposed to (high-intensity) light (in the absence of reductant) and that superoxide can activate an AA10 LPMO (33). When a reductant (AscA) is added to the system, a dramatic boost in LPMO activity was observed (33), thus confirming the original work by Cannella et al. (87). However, based on several experiments, Bissaro et al. claimed that this boost was correlated to the fact that AscA accelerated the conversion of superoxide into H2O2 (33), which will speed up LPMO catalysis (32, 34, 44). In a follow-up study, the idea of ROS being involved in the photocatalytically promoted LPMO activity was dismissed, mainly on the basis of the absence of effects of the addition of catalase on AA9 LPMO activity (132). It could be argued that the lack of a catalase effect could be due to the micromolar affinity for H2O2 of LPMOs (32, 34), which likely enables these enzymes to compete with catalases that have apparent Km values in the millimolar range (see “The housekeeping role of catalases,” below). The issue remains controversial, and a direct comparison of the two studies is complicated, primarily due to the use of different light intensities (33, 87). More work is required to decipher the underlying mechanism of light/pigment-driven LPMO catalysis.
In the same year, 2016, it was also shown that light-driven oxidation of water, catalyzed by vanadium-doped titanium dioxide (V-TiO2), can provide the reducing equivalents that LPMOs need to oxidize polysaccharides, thus alleviating the need for externally added electron donors (88). This proof of concept yielded much lower LPMO rates than the chlorophyllin system described above (tested for both AA9 and AA10) (Table 1). With hindsight, it is likely that the light-driven LPMO activity observed in this study reflected light-driven production of H2O2, which is a known ability of photoexcited TiO2-based photocatalysts (133–136). Indeed, the ability of TiO2 to catalyze light-driven peroxygenase reactions has recently been demonstrated using the unspecific peroxygenase from Agrocybe aegerita as a model enzyme (137).
INSIGHTS INTO THE NETWORK OF LIGNOCELLULOLYTIC REDOX REACTIONS
Our understanding of biomass conversion in natural environments, notably by fungi (4), is constantly challenged and improved, as illustrated by the relatively recent discovery of LPMOs (10, 11). As reviewed by Berrin et al. (138) and others (139), several studies conducted during the past few years have reported biomass-dependent upregulation of LPMO expression or secretion by many fungi: Hypocrea jecorina (140), Myceliophthora thermophila (141), Schizophyllum commune, Phanerochaete chrysosporium, Gloeophyllum trabeum (142), Aspergillus nidulans (143), Phanerochaete carnosa (144, 145), Postia placenta (146), Ceriporiopsis subvermispora (147), Pycnoporus coccineus (148), Phlebia radiata (149), Podospora anserina (111), and Neurospora crassa (150). Beyond an array of well-known hydrolases, fungi tend to coexpress/cosecrete a plethora of other oxidoreductases along with LPMOs, and many of these generate or consume H2O2. The ability of H2O2 to efficiently drive LPMO catalysis sheds new light on the potential interplay between the different enzymatic and nonenzymatic elements of lignocellulolytic enzyme systems. In the next paragraphs, we describe different redox enzyme systems thought to be involved in depolymerization of lignocellulose, followed by a discussion of their spatial and temporal interconnections (Fig. 6).
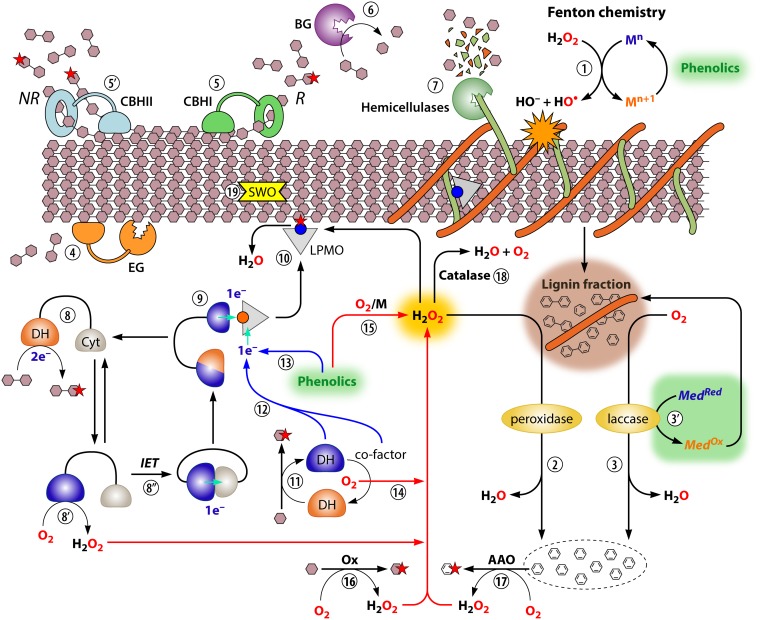
FIG 6.
An integrative view on reactions happening during lignocellulolysis. The enzymes presented here do not always all cooccur in genomes or secretomes and may act sequentially, depending on the microorganism or the nature of the biomass. The main nonenzymatic weapon is constituted by Fenton reaction-derived hydroxyl radicals (step 1). The lignin fraction, which constitutes a physical barrier, can be modified and to some extent depolymerized via the action of enzymes such as peroxidases (step 2) or laccases (step 3). Note that the peroxidases and laccases are also involved in repolymerization of lignin (not shown). These enzymes can also catalyze the oxidation of mediators that may be involved in lignin oxidation (step 3′; shown only for laccases). The main cellulases are endoglucanases (EGs) (e.g., Cel7B) acting internally (step 4), cellobiohydrolases CBHI (e.g., Cel7A) and CBHII (e.g., Cel6A) acting, respectively, from reducing (R) and nonreducing (NR) chain ends and primarily releasing cellobiose (steps 5 and 5′), which is further hydrolyzed to glucose by β-glucosidases (BG) (step 6). The various cellulases will also release minor amounts of products carrying an oxidation at C-1 or C-4 that was introduced by an LPMO (the presence of such oxidations is indicated by a red star). A wide diversity of hemicellulases (261, 425) and possibly pectinases (426, 427) acts on the hemicellulosic and pectin fractions, respectively (step 7). CDH oxidizes cello-oligosaccharides (step 8), and acquired reducing equivalents can be used to generate H2O2 (step 8′) or be transferred to the cytochrome (Cyt) domain (step 8″), which then reduces LPMOs (step 9). Once reduced, LPMOs can oxidize the cellulose (step 10), provided that the cosubstrate H2O2 (or O2) is present. (As noted elsewhere in this review, the question of whether O2 can act as a cosubstrate without prior reduction to H2O2 is still under debate [44].) LPMOs can also be activated by single-domain dehydrogenases and/or noncovalently bound reduced cofactor (step 11 to 12) or by reduced phenolics (step 13). Single-domain dehydrogenases (step 14) and reduced phenolics (especially in the presence of transition metals [M]) (step 15) can also lead to the production of H2O2 under aerobic conditions. Several oxidases (Ox) such as methanol oxidase, glyoxal oxidase, copper radical oxidase, or diverse oligosaccharide oxidases can generate H2O2 (step 16) to fuel the different H2O2-consuming systems (here, a secreted pyranose 2-oxidase is shown) (see subsection “The Function of Lignocellulolytic Oxidoreductases”). Aryl-alcohol oxidases (AAO) oxidize lignin-derived compounds to generate H2O2 (step 17). Catalase acts as a safety belt by converting excess H2O2 into H2O and O2 (step 18). Expansins/swollenins (SWO) may contribute to lignocellulolysis by loosening the plant cell wall structure, also called amorphogenesis (step 19) (282) although their mode of action remains unknown. Note that for the sake of simplicity the stoichiometry of reactions is not taken into account. See Table S1 in the supplemental material and Fig. 7 for an overview of known lignocellulolytic redox enzyme activities.
The Function of Lignocellulolytic Oxidoreductases
From an evolutionary perspective.
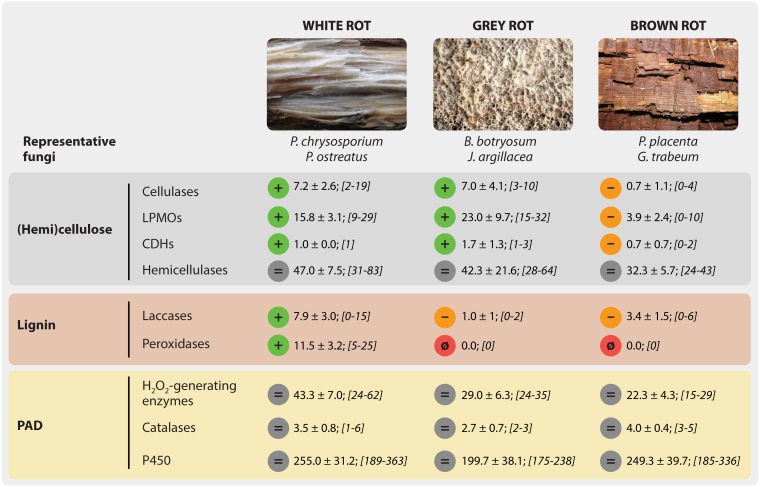
Our current understanding of the role of different oxidoreductases involved in lignocellulose conversion is essentially derived from studies on two kinds of living laboratories, namely, wood-decaying basidiomycetes commonly classified as white-rot or brown-rot fungi. The difference in appearance of the rotted wood is a direct consequence of the depolymerization strategy adopted by either fungus, which reflects the enzymatic arsenal that the fungi deploy (Fig. 7). Notably, fungi with hybrid phenotypes, sometimes called gray rot, have been described previously (151). With the exception of hemicellulases, which are equally abundant in the genomes of both types of fungi, the two fungal types have quite different enzyme arsenals. In particular, white-rot fungi have more laccases, cellulases, and lignin-active peroxidases than brown-rot fungi. It has been proposed that the first wood-rotting fungi appeared through acquisition of ligninolytic peroxidases (110) by ancestral basidiomycetes, whereas the later transition from white-rot to brown-rot decay among fungi involved the loss of lignin peroxidases but maintenance of oxidases (152–154).
FIG 7.
Enzymatic features of wood-decaying basidiomycetes. The phylum of Basidiomycota contains 32% of all described fungi and contains mostly saprotrophic fungi, including most wood-decaying fungi. Brown rots represent approximately only 6% of the latter but dominate in boreal forests, where they are associated with conifer wood. Here, we report average numbers of genes for different classes of enzymes in white-rot, brown-rot, and gray-rot fungi. For each group of fungi, the plus and minus symbols indicate whether the number of genes is higher or lower than that of at least one of the other two groups. An equal sign indicates that the numbers are similar in all three groups, whereas the null symbol (ø) indicates the absence of any gene encoding a given type of (known) oxidoreductase. For each species, the number of genes encoding the major cellulases (GH6 and GH7), hemicellulases (GH10, -11, -16, -51, -62, and -74), CDH (AA3_1), laccases (AA1_1), peroxidases (AA2), and main secreted H2O2-generating CAZymes (AA3_2, AA3_3 and AA3_4, AA5_1, and AA7) were retrieved from Riley et al. (151). The list of individual fungi is provided in the legend of Fig. 8. The numbers of putative catalases and P450s (secreted and cytosolic) were obtained from the MycoCosm online database (258). For each species, the total number of genes corresponding to each enzyme category was calculated, and then an average value ±95% confidence interval was calculated; the interval of minimum to maximum values is provided in brackets for each phenotypic subgroup. PAD, prooxidant, antioxidant, and detoxifying enzymes (which notably include GMC oxidoreductases). (The rotting-wood pictures were obtained from Wikimedia Commons. The white-rot photo, by Jerzy Opiola, is licensed under the Creative Commons Attribution-Share Alike 4.0 International license [https://creativecommons.org/licenses/by-sa/4.0/legalcode]; the gray-rot photo, by James K. Lindsey, is licensed under the Creative Commons Attribution-Share Alike 2.5 Generic license [https://creativecommons.org/licenses/by-sa/2.5/deed.en]; and the brown-rot photo is licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license [https://creativecommons.org/licenses/by-sa/3.0/deed.en].)
The oxidases that are maintained in all wood-decaying fungi are well-known H2O2 producers and belong to the superfamilies of glucose-methanol-choline (GMC) oxidoreductases (AA3) or copper radical oxidases (CRO; AA5). Among AA3s, aryl-alcohol oxidases (AAO), glucose oxidases (GOX), and alcohol oxidases (AOX) are phylogenetically the most related enzymes, followed by pyranose 2-oxidase (P2O) and CDH, which share the oldest ancestor with other GMC oxidoreductases (155). In white-rot fungi, an obvious role for these oxidases is to fuel lignin peroxidases with H2O2. In brown-rot fungi, which lack lignin peroxidases, H2O2 may be used to drive the Fenton systems that are unique for these fungi. Importantly, Fenton systems may not be the only H2O2 sink since there is a strong cooccurrence of AA9 LPMOs and AA3-encoding genes in brown-rot fungi (83), and we know now that LPMOs efficiently use H2O2 to catalyze the oxidative cleavage of polysaccharides. Interestingly, cooccurrence of lpmo and cdh genes is more scarce in brown-rot fungi than the strong correlation found in white-rot fungi (83), suggesting that alternative LPMO activation systems exist in brown-rot fungi, as detailed below.
Laccases.
Along with peroxidases (discussed below), laccases are major contributors of ligninolysis in white-rot fungi (156, 157). Also known as benzenediol:O2 oxidoreductases (EC 1.10.3.2; AA1 CAZy family) belonging to the multicopper oxidase superfamily (73), they use O2 as a cosubstrate and can directly oxidize a wide range of phenolic substrates (156). The resulting reduced form of the laccase catalyzes the reduction of O2 to H2O (158) while the oxidized (poly)phenols get involved in depolymerization, cross-linking, or internal reactions such as ring cleavage or quinone formation. The reduction potential of laccases, ca. +0.8 V (compared to >+1 V for lignin-active peroxidases), does not allow activity on nonphenolic moieties (E0 > +1.3 V). This limitation is overcome by the so-called laccase-mediator systems (159, 160). In short, the mediator (e.g., a phytophenolic) is oxidized by the laccase and acts as an electron shuttle by diffusing out of the laccase active site to further oxidize substrates, such as nonphenolic lignin subunits, otherwise not directly tractable by the laccase itself (161). The interplay between laccases and LPMOs is not clear yet. It is clear that soluble lignin fragments emerging from laccase action, which still contain phenolic groups and reducing power, can activate AA9 LPMOs (86, 162). Under certain conditions, the competition of laccases for oxygen may be relevant.
Lignin-active peroxidases.
Peroxidases are so far the only known type of ligninolytic enzymes relying on the use of H2O2. Peroxidases belong to four independently evolved superfamilies (163), spanning all kingdoms of life. The largest family, the peroxidase-catalase superfamily, contains three families (called families I to III) (163) (PeroxiBase database [peroxibase.toulouse.inra.fr]). Three types of lignin-modifying peroxidases, all belonging to family II (164), have been identified thus far, namely, the lignin (LiPs), manganese (MnPs), and versatile (VPs) peroxidases. All of these enzymes belong to family AA2 in CAZy.
LiPs, MnPs, and VPs all use H2O2 as an oxidant but employ very different strategies to act on lignin (165). While LiP, with a high redox potential (E0′ ≈ +1.2 V versus SHE at pH 3) can directly oxidize nonphenolic aromatic compounds, MnP uses an indirect pathway relying on the oxidation of Mn(II) to Mn(III), which is released from the enzyme. Mn(III) is then chelated by organic compounds (e.g., oxalate or malate) and can act as a diffusible oxidizing agent on phenolic (but not on nonphenolic) substrates. VP, with a very high redox potential (E0′ > 1.4 V versus SHE), shows features that are common to LiP and MnP and can oxidize both nonphenolic and phenolic compounds. It is worth noting that despite, or because of, its indirect mode of action, MnP is often the most abundant lignin-active peroxidase found in white-rot fungal secretomes.
Peroxidases compete with LPMOs for H2O2 and may thus inhibit LPMO activity under certain conditions, as has been shown by several authors (32, 44).
A diversity of enzymatic H2O2 producers.
In lignocellulolytic systems, H2O2 can be produced by several extracellular enzymes, most of which belong to the GMC oxidoreductase superfamily: cellobiose dehydrogenases (AA3_1; see above); aryl alcohol oxidase (AAO) (AA3_2; EC 1.1.3.7) (166), glucose oxidase (GOX) (AA3_2; EC 1.1.3.4) (167), methanol oxidase (MOX) (AA3_3; EC 1.1.3.13) (168), and pyranose 2-oxidase (P2O) (AA3_4; EC 1.1.3.10) (169). H2O2 may also be generated by extracellular copper radical oxidases (CRO), which include glyoxal oxidase (GLOX) (AA5_1; EC 1.2.3.15) (170, 171) and alcohol oxidase (AlcOx) (AA5_2; EC 1.1.3.13) (172), and by gluco-oligosaccharide oxidases (GOOX) (AA7, EC 1.1.3.−) (173, 174) (Fig. 8). H2O2-generating enzymes are widely distributed in both white-rot and brown-rot fungi (Fig. 7) (155, 175), indicating that H2O2 will be generated during biomass conversion, regardless of the decomposition strategy.
FIG 8.
Extracellular H2O2 producers encountered during fungal wood decay. The figure shows enzyme structures and associated representative substrates, derived from (hemi)cellulose or lignin fractions, for the CDH (AA3_1) from Phanerochaete chrysosporium (PDB accession number 1KDG; dark blue), an AAO (AA3_2) from Pleurotus eryngii (PDB accession number 5OC1; magenta), an MOX (AA3_3) from Pichia pastoris (PDB accession number 5HSA; green), a P2O (AA3_4) from Phanerochaete chrysosporium (PDB accession number 4MIF; gray), an AlcOx (AA5_2) from Colletotrichum graminicola (PDB accession number 5C86; light blue), and a GOOX (AA7) from Sarocladium strictum (PDB accession number 1ZR6; orange). Note that there is no structure available for GLOX (AA5_1). The average numbers of genes (±95% confidence interval) found in the genomes of white-, gray-, and brown-rot fungi are indicated in the colored boxes (from top to bottom, respectively) and were retrieved from Riley et al. (151). The interval of minimum to maximum values of actual (varying) gene numbers per genome is indicated in brackets. The white-rot species include the following: Auricularia subglabra, Ceriporiopsis subvermispora, Dichomitus squalens, Fomitiporia mediterranea, Galerina marginata, Heterobasidion annosum, Phanerochaete carnosa, Phanerochaete chrysosporium, Pleurotus ostreatus, Punctularia strigosozonata, Stereum hirsutum, and Trametes versicolor. The gray-rot species are Botryobasidium botryosum, Jaapia argillacea, and Schizophyllum commune; the brown-rot species are Coniophora puteana, Dacryopinax sp., Fomitopsis pinicola, Gloeophyllum trabeum, Postia placenta, Serpula lacrymans, and Wolfiporia cocos. aP. placenta and Dacryopinax sp. are the only brown-rot fungi containing GOOX-encoding genes (3 and 5 genes, respectively). bG. trabeum is the only brown-rot fungus containing a P2O gene (1 gene). It should be noted that the dichotomy between lignin and (hemi)cellulose-derived substrates is not always clear-cut since substrates such as veratryl alcohol can be synthesized as a secondary metabolite de novo from glucose (428).
These H2O2-producing enzymes act on many different compounds derived from lignocellulose (see Table S1 in the supplemental material). MOX, which seems equally abundant in white- and brown-rot fungi (155), oxidizes methanol, which is a product of the demethoxylation of lignin-derived phenolics (176) and has been suggested to be the main H2O2 supplier of Fenton chemistry during brown-rot decay (177). AAO, one of the most frequent GMC oxidases in white-rot fungi (155) (Fig. 8), is an extracellular enzyme acting preferentially on fungal metabolites such as 4-methoxylated benzoyl alcohols (166). These aromatic alcohols are the products of lignin-derived aldehydes or acids that have been reduced by aryl-alcohol dehydrogenases (EC 1.1.1.90) and aryl aldehyde dehydrogenases (EC 1.2.1.30) (178). GOXs, which are structurally and sequentially closely related to AAO and found both intra- and extracellularly, are not widespread in wood decayers, and their role in lignocellulolysis is not clear. P2O is found at the hyphal periplasmic space or in the secretome (putatively upon cell lysis) and has a broader substrate specificity than GOX as it can catalyze the oxidation of several aldo-pyranoses at the C-2 position (179, 180). Although efficient at reducing O2, P2O has also been shown to reduce quinones and complexed metals (Fig. 4 and Table S1) (181, 182). The involvement of P2O in lignocellulose conversion is supported by immunocytochemical and substrate-dependent gene regulation studies (169, 183–185).
Vanillyl-alcohol oxidases (AA4) are intracellular FAD-dependent enzymes that act on activated aromatic alcohols such as 4-hydroxybenzyl alcohols, leading to the concomitant production of H2O2 (186, 187). As intracellular enzymes, AA4s are not thought to be directly involved in lignocellulolysis but in the metabolism of lignin-derived compounds (188, 189).
GLOX (AA5_1) is widely distributed among wood decayers. The enzyme acts mainly on aldehydes that are released during lignin and carbohydrate processing (190) and is considered to be physiologically coupled to lignin-active peroxidases (149, 170, 175, 190, 191). The substrate specificity of the AA5 family has recently been expanded by the characterization of two alcohol oxidases (constituting a subclade of AA5_2) active on aliphatic primary alcohols (e.g., butan-1-ol, benzyl, and cinnamyl alcohol) (172). The authors of the study speculated that these AA5_2 enzymes may have a role in plant cell wall depolymerization even though the identity of the natural substrate has not yet been determined.
Gluco-oligosaccharide oxidase (GOOX) activity was reported more than 25 years ago (192). GOOX enzymes, classified as AA7s, have hitherto received less attention than other H2O2 suppliers in fungal secretomes. GOOXs are secreted enzymes, sharing some substrate specificity with CDH, P2O, and GOX (173). They are primarily active on gluco-oligosaccharides, but activity on xylo-oligosaccharides has also been described (174). Very little is known about their biological role. Some researchers have proposed that GOOX may fuel lignin-active peroxidases, which is the usual proposal when it comes to identifying a H2O2 sink in lignocellulolytic enzyme systems. Today, LPMOs provide an alternative H2O2 sink. Interestingly, considering the substrate specificities of GOOX and LPMOs, the two enzymes could be acting in close proximity, which could lead to better reaction control. AA7s have been detected together with AA9s in the secretome of S. commune, a gray-rot fungus, when they colonize artichoke stalk (142).
Several members of the GMC oxidoreductase superfamily have now been shown to activate LPMOs (83, 193). As described above (see subsection “The CDH Case: a Multifunctional Redox Partner”), CDH is a known redox partner that, by means of its two-domain structure, plays the dual role of reducing the LPMO and supplying H2O2. Interestingly, the Cyt domain is not absolutely required for GMC oxidoreductases to drive LPMO reactions as single-domain DHs, such as the glucose dehydrogenase (GDH) or aryl-alcohol quinone oxidoreductases (AAQO) (subfamily AA3_2), can also drive AA9 LPMO activity (193). On the other hand, single-domain strict oxidases such as the GOX or AAO are not able to reduce or activate the LPMO (32, 193). The way in which GDH or AAQO reduces the LPMO remains unclear and may involve mediators, such as superoxide or cofactors. Interestingly, some fungal secretomes contain many H2O2-generating oxidases as well as LPMOs while lacking both CDH and any usual H2O2 consumer (e.g., peroxidase) (83, 143). In light of the recent insights into LPMO functionality, these oxidases can be viewed as H2O2-generating partners of LPMOs although they may be fulfilling other roles that remain to be discovered. In this respect, it is worth noting that knocking out cdh genes in Podospora anserina did not alter its growth on lignocellulose but led to increased production of (H2O2-generating) flavo-oxidases (AA3_2) and CRO (AA5_1) along with β-glucosidases (111). It is possible that LPMOs are reduced by other factors, such as redox mediators, and that the oxidases then fuel the reaction by delivering H2O2. Indeed, it has been shown that glucose oxidases, which alone cannot drive LPMO reaction, do so very well upon reduction of the LPMO by a reductant (32). Notably, the natural environment of wood-decaying fungi is rich in phenolic compounds, which can reduce LPMOs (83).
As shown in Fig. 4, H2O2-producing oxidases (AA3_2, _3, _4, AA4, AA5, and AA7) display, in general, catalytic constants spanning two orders of magnitude, from 10 to 300 s−1, approximately (see Table S1 in the supplemental material for details). On the other hand, the H2O2-producing ability of CDH is much lower and lies in the range of 10−2 to 10−1 s−1. LPMOs present the lowest capacity to reduce O2 into H2O2, with reported catalytic rates ranging between 10−4 and 10−2 s−1.
Whether the relatively low H2O2 production rate displayed by CDH would be a way to control LPMO action during white-rot decay is an interesting possibility. The necessity of a match between H2O2 production and H2O2 consumption in lignocellulolytic enzyme systems has been argued in the past by Philip Kersten, who noticed that the H2O2 generator GLOX drives the lignin peroxidase action and that the much higher catalytic efficiency of GLOX (than that of the peroxidase) may explain why it is yet a minor component of the lignocellulolytic broth (170). It is also worth noting that H2O2 has been proposed as a limiting factor in lignocellulolysis by P. chrysosporium (194, 195) or limiting the action of MnP during compost lignin conversion by Agaricus bisporus (196). Overall, it is tempting to think that, in order to match and control H2O2 fluxes between emitters and receptors, nature has coevolved catalytic efficiencies and enzyme secretion levels. However, such correlations remain to be clearly demonstrated.
The housekeeping role of catalases.
As highlighted above, H2O2 appears to be a central molecule produced and used by several enzymatic systems involved in lignocellulolysis. H2O2 is per se a rather stable molecule (197) but can be destabilized. Indeed, in a complex environment, such as biomass-decomposing litter, free reduced metals are likely to be found, and these may react with H2O2, leading to the generation of hydroxyl radicals by Fenton chemistry. Beyond metal-catalyzed reduction of H2O2, direct reduction by reductants is also a possibility (Fig. 3) although it will occur at much lower rates. For instance, the second-order rate constant for the reaction of H2O2 with the strong reductant ascorbate is only 1.03 M−1 · s−1 at pH 7.0 (198).
The generation of hydroxyl radicals and other ROS exposes the system's components to irreversible and destructive oxidative damage. Housekeeping enzymatic activities referred to as catalase activities have evolved to avoid such damage (199–201). Catalase activity, i.e., the enzymatic disproportionation of H2O2 into O2 and H2O (2H2O2 → 2H2O + O2), is found in all kingdoms of life and can be carried out by three groups of proteins, namely, by typical (also known as monofunctional) catalases (EC 1.11.1.6), by some members of the peroxidase-catalase family (EC 1.11.1.6/7), and by nonheme manganese catalases (a minor group) (EC 1.11.1.6). Typical catalases, which constitute the predominant group (199), are heme iron-dependent enzymes with high kcat (104 to 105 s−1) but rather poor apparent Km values for H2O2 (3 to 103 mM range) (200, 201). Similar values apply to the bifunctional peroxidase-catalases (103 to 104 s−1 and 3.7 to 8 mM for the catalase activity) (202). These values differ strongly from values for peroxidases, i.e., enzymes that use H2O2 as oxidant, which show lower catalytic rates (10−1 to 103 s−1) but also much lower Km values (101 to 103 μM) (203–206). Typical catalases display a deeply buried active site, to which H2O2 molecules gain access via a long channel, the properties of which (shape, size, and composition) are optimized to increase the ratio of H2O2 to H2O and, thus, reactivity (207, 208). Variations in channel features, impacting inlet and outlet fluxes, may explain the wide range of observed catalytic efficiencies (199, 202).
The kinetic properties of catalases, with their high, noncompetitive Km values, seem adapted to keeping the level of H2O2 low enough to avoid oxidative damage without inhibiting H2O2-dependent enzymes with their much more competitive Km values. High catalytic efficiencies displayed by catalases may, on the other hand, be related to the necessity of a fast response in the case of H2O2 accumulation. This scenario would obviously benefit lignocellulosic redox enzyme systems. It has been shown that catalases are secreted simultaneously with H2O2-consuming enzymes, such as peroxidases, during the biodegradation of spruce wood by the white-rot fungus Phanerochaete chrysosporium (209). The brown-rot fungus Postia placenta secretes catalases along with GHs in a later phase of wood degradation, possibly as a means to protect the GHs from residual Fenton reagents used in the earlier phases of the brown-rot process (146) (see subsection “Enzyme Production during Lignocellulose Depolymerization,” below). The role of catalases in H2O2 regulation may translate into antagonistic effects, as observed with knockout of catalase genes in Podospora anserina (210, 211). Bourdais et al. (210) showed that the five P. anserina catalases play different roles and that there may be a trade-off between the need for protection and the need for H2O2 to increase the efficiency of lignocellulose depolymerization efficiency. The multiplicity of catalase genes in microbial genomes suggests a need for flexibility and underpins a need for further research on the roles of these enzymes during biomass conversion.
The dynamic aspect of H2O2 fluxes renders the determination of real H2O2 levels encountered in biological systems rather challenging. Impressively, values reported in the literature span approximately seven orders of magnitude. Intracellular levels are thought to be extremely low to protect from lethal ROS-induced damage, i.e., in the low-nanomolar range (212, 213). Conversely, extracellular levels are higher and may be bacteriostatic (micromolar) or even bactericidal (millimolar) (214). Some pathogenic microorganisms, such as some Streptococcus species (215) or the rice blast fungus Magnaporthe oryzae (216), can resist millimolar concentrations of H2O2. In the 1 to 100 μM range, effects with different amplitudes can be observed on fungal growth depending on the region considered, with the hyphae being for instance more resistant than the conidia (217–219). Plants can tolerate up to several millimolars of H2O2 (100 μM to 200 mM) (220, 221). Importantly, H2O2 produced intracellularly is likely to be used intracellularly while H2O2 produced extracellularly is consumed extracellularly, suggesting that both systems are partly disjoint. Indeed, it has been shown that H2O2 transmembrane transport in Saccharomyces cerevisiae and bacteria is very limited (222), and this is consistent with the (potentially) enormous difference in the concentrations encountered on both sides of the cellular membrane.
It is known that plants trigger a defense mechanism called oxidative burst when they are under threat (223), which implies production of ROS and, notably, H2O2 (224, 225). Interestingly, for hitherto unexplained reasons, necrotrophic fungal pathogens such as Botrytis cinerea or Colletotrichum graminicola take advantage of this response to proliferate (226, 227). These microorganisms can express multiple LPMOs and peroxidases (24, 228), and one may wonder if the pathogens harness plant-generated H2O2 to fuel their degradative redox enzyme machinery. The ambivalent role of H2O2 is also highlighted by the fact that some plants use catalases as defense against pathogenic fungi such as Aspergillus flavus (229). Another indication for a role of H2O2 and H2O2-producing enzymes in pathogenicity is provided by two independent studies showing that Botrytis cinerea became less virulent upon deletion of genes encoding either superoxide dismutase (SOD) (230) or GLOX (231). Membrane-bound GLOX has also been implicated in pathogenesis for crop pest Fusarium species (232).
Nonenzymatic Production and Use of H2O2
In ecological niches where biomass decomposition occurs, several nonenzymatic reactions can generate or consume H2O2. This additional layer of complexity needs to be taken into account in order to understand lignocellulose conversion.
Nonenzymatic sources of H2O2.
The reduction of O2 into H2O2, possibly via superoxide, O2˙−, as an intermediate (Fig. 3) requires compounds with low enough reduction potentials. Many plant-derived phenols, also called phytophenolics, have this capacity and are abundant in decomposing matter (233). Phenolics are also known metabolites of brown-rot fungi (234), and it has been shown that the activity of some microbial extracellular enzymes is modulated by these compounds (235). Like many reducing agents with antioxidant properties, phenolics can also act as prooxidants, e.g., by reducing transition metals that subsequently engage in damaging oxidative processes such as Fenton chemistry (236). What happens in a biological system depends on the concentration of the chemicals in question, the presence of transition metals (Fe and Cu) and of O2, and the reduction potentials of the derived radical(s). Phytophenolics have different structures, which affect their anti- and prooxidant properties (237). Importantly, the H2O2 production potential of phenolics is also pH dependent (238). As outlined herein, several phytophenolics promote LPMO activity with different efficiencies (65, 83), and one may question if their role is only to reduce the copper site or also to provide H2O2. The role of phytophenolics during lignocellulolysis remains rather unclear, but their potential importance is well documented.
Fenton-type chemistry: a sink for H2O2.
Regarding nonenzymatic H2O2 sinks, the chelator-mediated Fenton (CMF) reaction (6, 239), notably used by brown-rot and certain LiP-deficient white-rot fungi, is considered central. Fenton reactions, i.e., the reaction of H2O2 with a reduced transition metal, generate hydroxyl radicals, which are thought to be involved in an initial, nonenzymatic pretreatment of lignocellulose. The CMF system finds biological relevance when one considers that enzymes are usually too big (20 to 100 Å in diameter assuming a globular shape [240]) to penetrate wood pores (10 to 40 Å in diameter; up to 100 Å in some cases [239, 241, 242]). On the other hand, such a chemical strategy is rather dangerous for the microorganism, and, therefore, the hydroxyl radicals have to be generated at a safe distance. Due to the diffusion-limited and nonspecific character of their reactivity, with a half-life of nanoseconds (239), hydroxyl radicals have to be generated in situ, i.e., in wood pores, to be both efficient and not harmful for the fungus. To perform such spatial control, it has been proposed that the pH dependency of metal reduction and concentration gradients of metal chelators such as oxalic acid or 3,4-dihydroxybenzoic acid (DHBA) play a major role (243), as has been reviewed elsewhere (234).
In the gradient scenario, low pH (≈2) and a high concentration of oxalate (a fungal metabolite) at close proximity of the fungal hyphae keep Fe(III) in a stable chelated form that can diffuse into wood pores. Once within the wood, the combined effect of higher pH (≈5.5 to 6) and lower oxalate concentration leads to release of Fe(III), which can then be reduced to Fe(II) by wood-penetrating redox compounds such as catechol or hydroxamic acids (244), oxalate (245), or, potentially, by low-molecular-weight peptides (234, 246). Redox cycling involving low-molecular-weight metabolites such as hydroquinones (247) or involutin (248) has also been proposed to be involved in iron reduction. The reduced transition metal can then react with H2O2 (249, 250) via the well-known Fenton reaction reported in 1894 (251), which produces a hydroxyl radical and a hydroxide ion (Fig. 6). CDH has often been linked to Fenton chemistry, both as a producer of H2O2 and because of its ability to reduce iron (112–116); such a role, however, seems questionable because CDH likely cannot penetrate wood pores. Of note, superoxide can also play the role of reductant via the Haber-Weiss reaction (252).
The control of H2O2 levels, which directly impact the efficiency of the CMF reaction, is a key parameter to consider. In brown-rot fungi the CMF system is thought to be fueled with H2O2 by the action of GMC oxidoreductases and/or copper radical oxidases (155) (see subsection “The Function of Lignocellulolytic Oxidoreductases,” above). Little is known about the actual H2O2 concentrations needed. In vitro studies of Fenton reactions often use high (millimolar) H2O2 concentrations and time scales that do not seem compatible with in vivo conditions.
While Fenton chemistry, with its seemingly clear biological relevance, is almost by default mentioned as an H2O2 sink in discussions of fungal biomass-related enzymology, it is important to note that the CMF system is just one way of exploiting the oxidative power of combining H2O2 with a reductant. Reactions between the reductant and H2O2 may take place in many locations, in a controlled or noncontrolled manner, and be beneficial or detrimental. For example, it has been suggested that such chemistry occurs in the termite gut, which seemingly lacks LPMOs and peroxidases but contains many H2O2-generating and iron-reducing enzymes (253). Similarly, Fenton-type reactions were shown to occur in the midgut of a leaf-feeding caterpillar (254). It is also worth noting that the use of a Fenton-like system during biomass decomposition has recently been described for a bacterium (255).
Enzyme Production during Lignocellulose Depolymerization
Connections between two enzymes are likely biologically relevant if (i) the product (a compound or reducing equivalents) of one enzyme is used by another and (ii) the enzymes are cooccurring in a common environment. The availability of next-generation sequencing methods and developments in adjacent omics technologies now offer possibilities to get insight into enzyme systems involved in lignocellulose degradation. The first white-rot fungus genome (from P. chrysosporium) was published in 2004 (256). Since the takeoff in fungal genomics in 2009 (257), more than 1,000 fungal genomes have been hitherto sequenced, mostly under the flag of the 1,000 Fungal Genomes Project (258) led by the U.S. Department of Energy's Joint Genome Institute. Lignocellulose conversion involves many players (Fig. 6 and Table 2), and genomes alone are not sufficient to elucidate how the orchestra of lignocellulose-active enzymes is regulated in time and space. With the emergence of multi-omics data, the reconstruction of biomolecular reaction networks involved in plant biomass decomposition becomes a possibility (259, 260). Based on accumulating omics data, we briefly discuss below (i) the orchestration of enzymatic machineries in simultaneous or sequential depolymerization of lignin and (hemi)cellulose and (ii) the interplay between Fenton chemistry and enzymes in brown-rot decay. The regulation and properties of cellulases and hemicellulases have been extensively covered by other investigators in comprehensive reviews (261–264).
TABLE 2.
Current overview of plant cell wall-degrading enzymesa
| Substrate and enzyme activity(ies)b | Common abbreviation(s)c | CAZy family(ies) |
|---|---|---|
| Lignin | ||
| Laccases | LAC or Lcc | AA1 |
| Versatile, lignin, and manganese peroxidases | VP, LiP, and MnP | AA2 |
| Aryl-alcohol oxidases | AAO | AA3_2 |
| Methanol oxidase | MOX | AA3_3 |
| Glyoxal oxidase | GLOX | AA5_1 |
| 1,4-Benzoquinone reductase | QR | AA6 |
| Hemicellulose | ||
| β-Mannosidase | MANB | GH2 |
| β-1,4-Endo-mannanases | MAN | GH5_7, GH5_26 |
| β-1,4-Galactosidase | AGA, AGL | GH27, GH36 |
| β-1,4-Galactosidase | LAC, BGA, GAL | GH2, GH35 |
| Galactomannan acetyl esterase | GMAE | |
| β-Xylosidases | XYL | GH3, GH39, GH43, GH52 |
| β-1,4-Endo-xylanases | XYN, XLN | GH10, GH11 |
| Arabinoxylan arabinofuranohydrolase | AXH | GH62 |
| α-l-Arabinofuranosidases | ABF | GH43, GH51, GH54, GH62 |
| α-Glucuronidases | AGU | GH67, GH115 |
| Feruloyl esterases | FAE | CE1 |
| Acetyl esterases | AE | CE1–CE7, CE12, CE16 |
| 4-O-Methyl glucuronoyl methylesterases | GE/GCE | CE15 |
| Xyloglucan transferase/hydrolases | XTH | GH12, GH16, GH74 |
| α-Xylosidases | AXL/XYL | GH31 |
| Galactose 6-oxidase | GalOx | AA5_2 |
| Lytic xylan oxidase (LPMO) | LPMO14 | AA14 |
| Cellulose | ||
| β-Glucosidases | β-Glu, BG, BGL | GH1, 3 |
| Endoglucanases | EG | GH5_5, GH5_7, GH5_9, GH5_12, GH5_45, GH5_48 GH5_74, GH5_131, GH5_148 |
| Cellobiohydrolases | Cel, CBH | GH6, GH7 |
| Cellobiose dehydrogenase | CDH | AA8-AA3_1-(CBM1) |
| Glucose 1-oxidase | GOx | AA3_2 |
| Pyranose 2-oxidase | P2O | AA3_4 |
| Gluco-oligosaccharide oxidases | GOOX | AA7 |
| Fungal LPMOs | LPMO9 | AA9 |
| Bacterial LPMOs | LPMO10 | AA10 |
| PQQ-dependent pyranose dehydrogenased | PDH | AA8-AA12-CBM1 |
The table summarizes the main enzymatic activities that are thought to be involved in lignocellulose conversion. They have been ordered according to the class of substrate they act on, namely, lignin, hemicelluloses, pectin or cellulose. For each type of enzyme we provide the most common abbreviation(s) and the CAZy (sub)family (21). Catalases, cytochrome P450s and other non-CAZymes are not included.
See Rytioja et al. (263) for a detailed list of pectin-active CAZymes (GH2, -3, -28, -35, -43, -51, -53, -54, -62, -78, -88, -93 and -105; PL1, -3, -4, -9, and -11; and CE1, -8, and -12), which should be updated with several recently discovered pectin-active CAZyme families (GH137 to GH143 [427], GH145 [431], and GH146 and GH147 [432]).
For historical reasons (evolving terminology and enzyme reclassification), several abbreviations may be found for the same activity. Early names are often kept by habit but also for optimal tracking of the literature. A more standardized approach consists in using the CAZy (sub)family preceded by initials, in italics, of the microorganism of origin. For example, SmAA10A would be the first characterized AA10 from Serratia marcescens (historically known as CBP21). Alternatively, the two-letter code indicating the CAZy class maybe replaced by a three- or four-letter abbreviation that indicates what the enzyme does. For example, the GH5 family contains both cellulases and mannanases; hence, one may find TrCel5A and TrMan5A, which indicate a GH5 endo-cellulase and a GH5 endo-mannanase from Trichoderma reesei, respectively.
PQQ, pyrroloquinoline quinone.
Insights from recent multi-omics studies.
It is known that some microorganisms have evolved enzymatic arsenals that are specifically adapted to a given habitat (265–267) while others have kept a broad array of activities (110, 263, 268, 269), ensuring adapted responses to diverse environments (270). In 2017, a large-scale study analyzed 22 transcriptomics data sets from 10 species of plant-decaying basidiomycetes displaying a white-, gray-, or brown-rot phenotype (271). Among 328 consistently differentially expressed genes, the authors could define a core set of 50 CAZymes involved in the degradation of plant polysaccharides. Among these, five enzyme types were found upregulated in at least 18 of the data sets: β-glucosidase, β-xylosidase, endo-xylanase, endo-mannanase, and α-glucuronidase. This result highlights the point that enzymatic depolymerization of hemicellulose is common among all fungal types. According to this same meta-analysis, the most prevalent additional common enzyme types were cellulases, hemicellulases, and pectinases (271). Oxidoreductases were not present in these sets of commonly employed enzymes, and this is logical since different wood-decaying fungi employ different oxidative strategies (Fig. 7).
With this global, but static, enzymatic picture being drawn, we need to consider the temporal fate of the lignin, hemicellulose, and cellulose fractions in order to understand which enzymes may or may not be physiologically linked. Owing to the high recalcitrance of lignin, it has been thought for decades to be the last biopolymer to be degraded, eventually leading to the production of humic substances. However, we know today that several scenarios can take place, depending on both the fungus and the type of substrate. In contrast to most white-rot fungi, which generally (but not always) carry out simultaneous degradation of lignin and cellulose (110, 272–274), fungi such as Phanerochaete carnosa (144, 145) and Ceriporiopsis subvermispora first attack lignin and then cellulose (275). For instance, when grown on aspen wood, C. subvermispora employs a strategy whereby oxidative lignin-active enzymes are first produced (AAO and MnP) and (hemi)cellulolytic enzymes are secreted later (GH5_5, GH12, and GH45 endoglucanases [EGs], GH6 cellobiohydrolase II [CBHII], GH7 CBHI, EG, GH10 xylanase, CE1 acetyl/feruloyl esterase and CE15 glucuronoyl esterases, LPMOs, and CDH) (147). During the ligninolytic phase, genes involved in lipid metabolism (e.g., encoding desaturases) are upregulated along with genes directly involved in lignin modification (MnP) (276), in agreement with the hypothesis that a connection exists between lipid peroxidation products and oxidation of nonphenolic structures present in lignin (277–279).
When the white-rot P. chrysosporium is grown on spruce wood, expression of genes encoding ligninolytic enzymes (MOX and GLOX) is upregulated, whereas cellulase genes are expressed in a more constitutive manner (209). Earlier reports have shown that CDH expression is also induced early, particularly by products of cellulase action (280). In a recent comprehensive study, Miyauchi et al. used an integrative omics approach to show that the saprophytic white-rot fungus Pycnoporus coccineus simultaneously secretes GHs, esterases, and oxidoreductases when grown on various lignocellulosic substrates (148). In this study, peroxidases (AA2), CDH (AA3_1-AA8), AAO (AA3_2), GLOX (AA5_1), and LPMOs (AA9) were highly upregulated, highlighting that H2O2-dependent peroxidases, H2O2-producing enzymes, and LPMOs cooccur during lignocellulose breakdown.
Studies of the transcriptomes of Aspergillus niger and Trichoderma reesei, which are used in the industry for the production of enzymatic cocktails, during growth on steam-exploded sugarcane bagasse revealed large temporal differences between the two fungi and showed the importance of redox enzymes in both (281). A. niger showed fast upregulation of CAZyme genes and a relatively early decay in expression levels, whereas the situation was the opposite for T. reesei. A total of 53% and 39%, respectively, of all AA-encoding genes were induced by the biomass for A. niger and T. reesei (out of a total of 68 and 33, respectively). Several extracellular H2O2-generating enzymes (AA3_2 and AA3_3) were highly upregulated in both fungi, as well as several AA9s, highlighting the importance of these enzymes. Interestingly, next to catalases and glutathione S-transferases, with putative housekeeping functions, and AA9 LPMOs, only a single known H2O2-consuming enzyme, a putative peroxidase, was transcribed. Of note, the T. reesei genome does not contain any cdh gene whereas the A. niger genome contains only one that was not induced (281).
The results of comparative studies of the secretome of the white-rot fungus Dichomitus squalens grown on woody (aspen and spruce) and nonwoody (wheat bran and cotton seed hulls) biomasses further support the notion that one main role of H2O2-generating enzymes could be to fuel other enzymes (LPMOs) than peroxidases. In this study, H2O2-consuming peroxidases were expressed at much lower levels on nonwoody (lignin-poor) substrates whereas the secretome pattern of H2O2-supplying enzymes (AA3_3 and AA5_1) was not affected by the nature of the substrate (270). AA9 LPMOs were secreted on all substrates, and their expression patterns were found to cluster with different sets of CAZymes, suggesting that LPMOs target different structures.
Several other enzymes or proteins with putative roles in biomass conversion appear in studied secretomes and transcriptomes. One example is the family of expansins, which, by changing the cell wall structure through a hitherto unknown mechanism, may affect the wall's susceptibility to enzymatic degradation (282, 283). Another class of enzymes worth mentioning are the P450 monooxygenases, which are abundant and largely nonsecreted but have been detected in the secretomes of white-rot fungi (110, 152, 175, 276, 284) and in the hyphal front of brown-rot fungi (146). P450s are ubiquitous heme enzymes, catalyzing a wide range of redox reactions, and their potential role in biomass conversion remains obscure. Most interestingly, in 2018, Reisky et al. described P450s catalyzing the oxidative demethylation of carbohydrates present in algal polysaccharides, likely decreasing the recalcitrance of this marine biomass (285). A role of P450s in lignocellulose conversion is plausible since they are more abundant in wood-decaying basidiomycetes than in non-wood-decaying relatives (286).
How do brown-rot fungi deal with ROS?
The simultaneous use of potentially harmful ROS (generated in the Fenton reaction) and enzymes by brown-rot fungi is intriguing. As discussed herein, the feasibility of this system is usually ascribed to a spatial partition between the generation of nonselective ROS and GHs (see subsection “Nonenzymatic Production and Use of H2O2,” above). Recent studies suggest that the brown-rot fungi Postia placenta, Serpula lacrymans, and Gloeophyllum trabeum (also) use separation in time (146, 287). Expression data indicate that these fungi use a two-step mechanism where oxidative and hydrolytic actions are temporally separated by differential gene expression (146). Zhang et al. showed that oxidative processes (i.e., H2O2 and Fe2+-generating enzymatic systems) involved in Fenton reactions are first triggered at the hyphal front, constituting thus a sort of pretreatment, which is followed by expression of (hemi)cellulases at a later stage. It is worth noting that expansins and pectinases (GH28) were also expressed during this early phase, in agreement with their potential action in initial loosening of the plant cell wall. Importantly, AAO and superoxide dismutase, which are H2O2 suppliers, were also expressed during the later stage of the process along with hydrolytic enzymes. The authors of this study noted that H2O2 produced by AAO must be directed toward a pathway that remains to be identified (the usual recipients, lignin peroxidases, are absent in brown rots). Interestingly, a single AA9 LPMO was also expressed during the later stage of biodegradation, and this enzyme may use the AAO-generated H2O2.
In general, the simultaneous secretion of enzymes with complementary (H2O2 producers versus H2O2 consumers) or competitive (different H2O2 consumers) impacts on H2O2 fluxes highlights that fungi possess regulatory systems and/or have evolved well-balanced enzymatic arsenals that allow the maximizing of enzyme efficiency while minimizing inhibition or oxidative damage. One particularly intriguing question is whether fungi have systems for generating proximity between redox partners. For example, proximity between the sites of production and consumption of H2O2 could be one way to minimize damage caused by side reactions involving free H2O2. This is discussed further below.
On abundance and importance.
When omics data are assessed, an important point of discussion concerns the implications of the numbers. Why are there so many LPMO- and GMC oxidoreductase-encoding genes? Why are there so few CDH genes? Why are there so few catalase genes? These questions underpin the complexity of analyzing genomic, transcriptomics, or secretomics data since the number of genes and proteins does not necessarily reflect their importance. While abundance is likely a sign of importance, importance is not necessarily reflected in abundance. This is why quantities (of genes, of transcripts, and of secreted proteins) may need to be weighted by the catalytic efficiency of a given biocatalyst, something that is rarely done.
The abundance of LPMO genes may reflect the need to face several different recalcitrant structures on copolymeric plant cell walls (288, 289), as beautifully illustrated by the recent discovery of an LPMO that specifically acts on copolymeric cellulose-xylan structures (25). On the other hand, white-rot fungi harbor only a single, but conserved, CDH gene. While it could be envisioned that a single CDH enzyme could serve as a common reductant and H2O2 generator for any kind of LPMOs, the question of proximity between these two enzymes with potentially different targets is more cumbersome. The diversity of H2O2-generating enzymes, beyond CDH, may reflect a need to produce this central ROS at different specific locations during plant material decomposition.
IMPLEMENTATION OF REDOX STRATEGIES INTO BIOREFINING PROCESSES
The first two sections of this review highlighted the observation that oxidative processes form an essential part of biomass degradation in nature. In this section, we will illustrate how biorefining processes can leverage our current knowledge on these oxidative processes. Microorganisms do achieve complete biomass conversion, but these biological processes occur on relatively long timescales (weeks and months) due to physiological and environmental constraints. From a biotechnological standpoint, one of the main challenges consists of not only mimicking but also speeding up the microbial strategies at reasonable cost (290). Here, we provide a nonexhaustive overview of the (possible) impact of the most recent advances in our understanding of the role of LPMOs and other oxidoreductases on industrial biomass conversion. We discuss oxidative pretreatments of biomass, consider the composition of lignocellulolytic enzyme cocktails and the contribution of oxidoreductases, and end by addressing the implementation in bioprocess design.
Oxidative Pretreatments
In the lignocellulose-based biorefinery concept, the biomass is first rendered more accessible to enzymes by pretreatment methods (291, 292) and then enzymatically depolymerized to platform molecules (e.g., mono- and oligosaccharides) (293), which can, in turn, be converted into value-added products by microorganisms. As several comprehensive reviews on pretreatment technologies are available, here we address only pretreatment methods utilizing oxidative processes. Pretreatment technologies using oxidative processes can be classified into biological (294), biochemical, and (physico)chemical processes: biological pretreatment methods use a biomass-degrading microorganism (295), biochemical pretreatment methods use oxidative enzymes, and (physico)chemical pretreatment methods utilize oxidizing agents for the generation of reactive oxygen species (ROS).
Biological pretreatment.
The most common biological pretreatments include treating the biomass with lignin-degrading bacteria, white-rot fungi, or brown-rot fungi (294). Such treatments primarily target lignin, which is generally considered the most important hindrance for efficient depolymerization of the polysaccharides. One promising strategy is the use of microbes that can depolymerize lignin and take up the resulting products, thus acting as microbial sinks. Uptake of lignin-derived compounds prevents repolymerization reactions and thereby improves overall biomass delignification (296, 297). Some white-rot fungal species preferentially target lignin during the onset of biomass conversion, which renders them a suitable choice for selective biological delignification. For instance, pretreatment (during 18 days) of several biomass feedstocks by Ceriporiopsis subvermispora resulted in a 2- to 3-fold increase in saccharification efficiency compared to the level with untreated raw material (298). Similarly, pretreatment of bamboo culms with the white-rot basidiomycete Punctularia sp. improved the subsequent enzymatic release of total sugars by 60% (299). A very recent study showed impressive improvements in cellulose hydrolysis yields from corn stover when it was pretreated (for 28 days) with the white-rot fungus Physisporinus vitreus: saccharification of the pretreated material with a commercial cellulase cocktail from Trichorderma longibrachiatum gave an overall hydrolysis yield of cellulose of 92%, in contrast to 26% without pretreatment. The extent of the beneficial effect was proportional to the duration of the pretreatment, and the obtained maximum yields were similar to what could be obtained after thermochemical pretreatment (300). In order to reduce long biological pretreatment times, often associated with sugar loss, Brethauer et al. proposed an alternative approach that consists of simultaneous saccharification and fermentation combined with in situ pretreatment by a white-rot fungus (Irpex lacteus) (301).
Biochemical pretreatment.
While biological pretreatment has produced some success, it usually requires long incubation times (ca. 20 to 90 days) and, depending on the species used, reduces overall yields through sugar consumption by the microorganism (294). Sugar yield loss may be circumvented by using biochemical approaches inspired by the oxidative strategies employed by these organisms. Lignin-active enzymes, such as peroxidases and oxidases, from white-rot fungi may be used to generate free radicals that nonspecifically attack lignin. For instance, Asgher et al. have shown that pretreatment of sugarcane bagasse with a secretome from Pleurotus ostreatus led to saccharification yields (ca. 70% glucose yields) similar to those of chemical pretreatment with 4% NaOH (302). In principle, enzymatic delignification is advantageous because one uses mild conditions that require low energy input and may give high yields. However, so far, this approach still suffers from long reaction times (several weeks) compared to those with classical physico-chemical treatments (303). The cost of the enzymes and their cofactors is also a limiting factor.
Mimicking brown-rot fungi by using Fenton (304) and chelator-mediated Fenton (CMF) (243) treatments to generate lignin-destroying hydroxyl radicals from H2O2 close to the biomass is an attractive scenario (305). Indeed, Fenton-type reactions have been successfully used to pretreat cotton fibers (306), garden biomass (307), rice straw (308), steam-exploded poplar (309), Miscanthus (310), switchgrass (310), corn stover (310), or wheat straw (310), allowing up to 5-fold improvements in saccharification yields (310). However, these Fenton reaction-based treatments require large amounts of chemicals (typically in the 0.1 to 10 M range for H2O2) and may lead to considerable mass loss, while special care must be taken to ensure innocuousness of the reaction mixture prior to proceeding with downstream steps.
(Physico)chemical oxidative pretreatment.
In addition to biological and biochemical pretreatment strategies that are inspired by natural processes, some known (physico)chemical pretreatment methods employ reactive oxygen species. The most common of these methods are ozonolysis (using ozone at ambient temperature and pressure for lignin removal) and wet oxidation (using air/oxygen with water or H2O2 at elevated temperatures for lignin and hemicellulose removal) (291). Although these methods are time-efficient and give good subsequent saccharification yields, they require high energy input (e.g., high pressure and temperature) and/or harsh conditions (e.g., high concentrations of chemicals such as hydrogen peroxide and acid) and usually lead to the generation of inhibitory compounds.
One may wonder if a future, deeper understanding of microbial strategies will allow development of natural-process-inspired biochemical/biological pretreatments that are cost- and time-efficient as well as environmentally friendly.
Design of Enzymatic Cocktails: the Effect of Oxidoreductases
Although the costs of enzymes used for lignocellulose saccharification have decreased nearly 20-fold during the past two decades (41), enzymes are still a major cost factor in biorefining (311–313), especially for the more recalcitrant of substrates. Today, the most common commercial products for saccharification are enzyme cocktails produced by heavily modified versions of the filamentous ascomycetes Trichoderma reesei (also known as Hypocrea jecorina) (314), Aspergillus niger, and Myceliophthora thermophila. In recent years, it has become more apparent that enzyme mixtures need to be customized for each target substrate (depending on the biomass type and the pretreatment method it has been subjected to), just as in nature (see Insights into the Network of Lignocellulolytic Redox Reactions, above).
Enzymes may be produced on site (312) or purchased from commercial enzyme producers (315, 316). In this respect, one has to keep in mind that the composition of hitherto available commercial enzyme blends has been optimized under conditions which may be suboptimal for the given application. As an example, Novozymes launched a hemicellulase preparation (Cellic HTec series) to complement their commercial cellulase preparation (Cellic CTec series) to be used on substrates rich in xylan.
In the past years, the rise in awareness of the key role played by oxidative enzymes in biomass conversion has prompted industrialists and scholars to investigate their synergy with canonical (hemi)cellulases. In the following paragraphs, we review the reported effects of LPMOs, lignin-active oxidoreductases, detoxifying enzymes, and CDHs on the conversion of industrially relevant biomasses.
The effect of LPMOs.
The performance of cellulolytic enzyme cocktails produced by Novozymes experienced a 10-fold improvement between 2000 and 2012, which in part is due to the inclusion of LPMOs (41, 317). The effect of LPMOs on the efficiency of cellulolytic cocktails has been assessed in several studies. In an early study, before the oxidative nature of LPMO activity was described, Harris et al. had demonstrated that the secretome of an engineered T. reesei strain expressing the GH61A from Thermoascus aurantiacus (today known as TaAA9A) reduced the enzyme loading needed to reach 90% conversion of pretreated corn stover 2-fold compared to the level with a natural T. reesei secretome (19). Also, a relative increase of 30% in hydrolysis yield (from 69% to 89%) was measured. Interestingly, this positive effect was not observed on pure cellulose (19), which, with hindsight, can be explained by the lack of reducing power to drive the LPMO reaction (in pretreated corn stover lignin fulfills this role). The results obtained by Harris et al. were similar to the results obtained with CBP21 in 2005, when LPMO activity was first observed in studies of a chitinolytic enzyme system (10, 318).
Subsequent to the discovery of the LPMO activity, including the need for reducing power and oxygen (11), several studies assessed the effects of supplementing enzyme cocktails with LPMOs. In an early study, Cannella et al. noted a 25% difference in the saccharification efficiency of pretreated wheat straw in a comparison of LPMO-deficient Celluclast and LPMO-containing Cellic CTec2 (84). In 2014, Hu et al. showed that spiking of Celluclast with TaAA9A increased hydrolysis yields for corn stover, poplar, or lodgepole pine by up to 25%, depending on the pretreatment method employed (319). In 2015, Müller et al. showed that supplementation of a mixture of Celluclast and Novozym 188 (a β-glucosidase) with TaAA9A improved glucose yields by up to 32% (i.e., from 64% to 85% of maximum theoretical yield) for steam-exploded birchwood (320), whereas a similar study later showed a 22% relative increase (from 63% to 77%) for sulfite-pulped Norway spruce (321). Importantly, these beneficial effects were not observed under anaerobic conditions or in the absence of sufficient reducing power (320, 321). In 2016, Scott et al. also showed the beneficial effect of TaAA9A addition to a mixture of Celluclast and β-glucosidase on the conversion of steam-exploded wheat straw, leading to a 43% relative increase in saccharification yield (from 39% to 56%) (322). Another study has shown that addition of an AA9 from Penicillium oxalicum to a commercial cellulase cocktail from the same fungus increased cellulose conversion by 33% (from 67% to 89% yield) and 31% (from to 63% to 82%) for alkali-pretreated wheat straw and corn stover, respectively (323). The effect of the LPMO was less when washed substrates were used, which is likely due to a lack of reducing power (323).
LPMOs are estimated to make up ca. 15% (wt/wt) of the composition of Cellic CTec2 from Novozymes (320). Interestingly, Hu et al. have shown that the amount of LPMO (AA9) required to reach optimal hydrolysis depends on the biomass concentration, with high-solid loading requiring less LPMO (324). This important finding likely relates to the most recent discoveries on LPMOs that, among other things, show that activated LPMOs must bind to substrate in order to avoid self-destructive off-pathway processes (32, 34, 44). From our own unpublished work, we have strong indications that, due to unfavorable process conditions, only a fraction of the LPMOs in commercial cellulase cocktails is actually used, whereas a large fraction is subject to oxidative self-inactivation (see below).
Although the beneficial effect of hemicellulases on biomass conversion has been studied for decades, the potential synergy between hemicellulases and oxidative enzymes such as LPMOs is, for obvious reasons, a fairly recent matter of investigation. As alluded to above, LPMO activity on various hemicelluloses (xyloglucan, glucomannan, and xylan) has been detected. The impact of these hemicellulolytic activities on biomass processing and the extent of direct synergies between hemicellulases and hemicellulose-active LPMOs in hemicellulose conversion remain largely unknown. These effects are difficult to study because of multiple enzyme activities and synergistic effects that are hard to deconvolute unless one uses an extensive analytical toolbox (for example, simple measuring of reducing sugars does not show which reactions are actually happening). Another complication lies in the fact that complex formation between cellulose and hemicelluloses may affect the activity of LPMOs on each of the components. For example, the first xylan-active LPMO ever described showed this activity only for xylan that was grafted onto cellulose (49). Studies with mixtures of phosphoric acid-swollen cellulose (PASC) and hemicelluloses have shown that hemicelluloses restrict enzyme access to the cellulose and that LPMO activity on these hemicelluloses increases cellulose conversion (50, 51).
It has been shown that coincubation of an LPMO, AA9, from Chaetomium globosum (CgAA9) with a xylanase led to a 30% increase in reducing sugar yield from an insoluble xylan preparation compared to level with the LPMO-free reference reaction (325). The same authors also showed that addition of CgAA9 to Celluclast led to a 10% to 20% increase in solubilized reducing sugar obtained from pretreated rice straw. Another recent study showed that a mixture of a xylanase (GH10A) and an LPMO (AA9A) from the fungus Gloeophyllum trabeum (GtGH10A and GtAA9A, respectively) improves the performance of Celluclast (final ratio of Celluclast/GtGH10A/GtAA9A of 60:20:20, wt/wt/wt) in the saccharification of wheat straw (326). Interestingly, the presence of the GH10 was necessary to unlock the full potential of the AA9. Notably, these interesting studies remain somewhat inconclusive since LPMO products were not analyzed, and it is thus not clear whether the observed effects are due, at least in part, to oxidative cleavage of xylan.
A potentially major breakthrough came in 2018 when Couturier et al. described the first member of a new LPMO family, AA14 (25). This AA14 acts specifically on copolymeric assemblies of cellulose and xylan, cleaving xylan chains that glue cellulose fibrils together and likely promoting the dissociation of hemicellulose from cellulose. While the studied enzyme was only a minor component of the fungal secretome when the fungus was grown on pine and poplar, it played a significant role in the overall biomass conversion efficiency. It will be exciting to see if additional LPMOs specifically acting on certain copolymeric structures in plant cell walls will be discovered and to what extent these enzymes can improve the efficiency of cellulase cocktails.
The effect of lignin-active oxidoreductases.
Lignin constitutes a hurdle for efficient biomass conversion into fuels and chemicals, and, in spite of recent progress (327), biorefineries have thus far overcome this issue mainly by using harsh thermochemical pretreatments. As discussed above, redox enzymes such as lignin peroxidases (AA2) or laccases (AA1) could be part of oxidative pretreatment strategies, and such lignin-active oxidoreductases may be beneficial when added to commercial enzymatic cocktails (328). To the best of our knowledge, the potential of such redox enzymes has not yet been harnessed in industrial settings, which is likely due in part to the inherent complexity of redox enzyme systems, which require cofactors and are subject to damaging off-pathway redox processes.
A few recent studies have addressed the possible contribution of such redox enzymes at lab scale. Although laccases have been studied for decades (mainly on model substrates), their effect on real biomass decomposition is still poorly understood. Laccases have been used as a delignifying pretreatment tool (329) and/or for decreasing the toxicity of lignin-derived compounds, which could hamper enzymatic saccharification and/or subsequent fermentation steps (330–332). Regarding the direct addition of laccases into cellulolytic cocktails, Rytioja et al. found a 1.5-fold increase in reducing sugar yield (from 5.3% to 8%) when a fungal laccase (from M. thermophila) was added to a mix of cellulases acting on sugar beet pulp while no effect was observed on lignin-rich wheat bran (333) (Note the low yields, which are due to the absence of pretreatment.) Very recently, Singh et al. studied the effect of a bacterial laccase (from Amycolatopsis sp. strain 75iv3) on saccharification of steam-exploded poplar by Celluclast and found an 8% increase in cellulose hydrolysis yield (from 61% to 66%), which they ascribed to the lignin depolymerization action of the laccase and/or to its detoxification effect (334). On the other hand, Brenelli et al. found that addition of laccases from M. thermophila (MtL) or Trametes villosa (TvL) had a negative effect on the efficiency of the LPMO-containing Cellic CTec2 cocktail during conversion of various biomasses, with a 49% decrease in hydrolysis yield on steam-exploded sugarcane bagasse (162). Interestingly, the addition of laccases led to considerable consumption of O2, which may inhibit LPMO action and may explain the lower glucose yields. Laccases are indeed relatively efficient O2 consumers (Fig. 3; see also Table S1 in the supplemental material). It would be interesting to supply laccase-containing reaction mixtures with H2O2, which is a cosubstrate for LPMOs but not for laccases.
All in all, while effects of adding laccases to cellulolytic enzyme cocktails have been observed, the nature of these effects remains largely unknown. The effects may vary from detoxification effects to a direct effect on lignin structure.
Lignin peroxidases have mainly been explored as oxidative pretreatment tool (see subsection “Oxidative Pretreatments,” above). Studies assessing the potential synergy between peroxidases and cellulolytic cocktails are scarce. A patent from Novozymes (335) describes the beneficial effects of peroxidases that are ascribed to the removal of excess H2O2 that may induce oxidative damage. In light of current knowledge, this concept likely needs some revision since peroxidases will compete with LPMOs for H2O2 (32, 44). In a rare public report, Salvachúa et al. showed that the addition of a fungal peroxidase (DyP from Irpex lacteus) to a cellulase cocktail (Celluclast complemented with commercial β-glucosidase, β-xylosidase, and xylanases) improved saccharification of wheat straw to an extent (between 16% and 41% relative increase) that was dependent on whether the biomass had undergone biological pretreatment and/or alkali washing (336). Another study showed that combining a commercial cellulase cocktail from T. longibrachiatum with the versatile peroxidase from P. vitreus, supplemented with a glucose oxidase as H2O2 generator, led to a 14% increase in the yield of released glucose from raw corn stover (from 85% to 97%) (300). In light of recent findings on LPMOs, this effect could also be due to promotion of LPMO activity by the generated H2O2.
The effect of prooxidant, antioxidant, and detoxifying enzymes.
Prooxidant, antioxidant, and detoxifying (PAD) enzymes include SOD, catalase, and aldo-keto reductase (AKR). These enzymes are thought to be involved in securing the stability of redox systems and, in some cases, in converting potential lignin-derived inhibitory compounds. The (potential) roles of these enzymes in biomass processing are intriguing and complex and have not yet been investigated in sufficient depth.
A study published in 2017 has addressed the role of an AKR from the termite Coptotermes gestroi (CgAKR-1) on the conversion efficiency of sugarcane bagasse (337). Beyond a role in the detoxification of yeast fermentation inhibitors (e.g., furfural and 5-hydroxymethylfurfural [HMF]), it was shown that supplementation of Celluclast with CgAKR-1 led to a 33% increase (after 24 h) in reducing sugar yields during saccharification of phosphoric acid-pretreated sugarcane bagasse. Interestingly, CgAKR-1 catalyzed the NADPH-dependent reduction of O2 to H2O2, and the improvement in saccharification was correlated with H2O2 generation. The authors of this study proposed that H2O2 was involved in oxidation of the lignin fraction, which could improve substrate access for cellulolytic enzymes (337). An effect on LPMO activity could also be envisaged although LPMO concentrations in Celluclast are low (320).
Scott et al. have demonstrated that the addition of a catalase to the Cellic CTec3 cocktail decreases the rate of enzyme inactivation during conversion of pretreated wheat straw with, however, only modest effects (ca. 10% relative increase) on final glucose yields (322). They found a larger catalase effect when working with a cocktail of Celluclast (80%), TaAA9A (10%), and β-glucosidase (10%); in this case, catalase addition increased the final glucose yield from ca. 56% to 68% of the theoretical maximum (i.e., a 20% relative increase) (322). Based on these and additional observations for reaction mixtures with added H2O2, the authors proposed that the catalase protects the cellulolytic enzymes from getting inactivated by H2O2-derived species (e.g., hydroxyl radicals). H2O2 was proposed to originate from biotic and abiotic reactions, and its production/accumulation was proposed to depend on parameters such as O2 availability, temperature, pH, dry matter, and metal content (for further discussion, see below and “The housekeeping role of catalases,” above). Of note, such a beneficial effect of catalase addition had been observed as early as 1992 in a study of the synergism between P. chrysosporium CDH (PcCDH) and a cellulase mixture (338). In this study, the inhibitory effects of high concentrations of CDH, attributed to high H2O2 production rates leading to oxidative inactivation of cellulases, were alleviated by the addition of a catalase, which led to a 25% increase in the hydrolysis yield of microcrystalline cellulose.
Superoxide dismutases (SODs) have received massive attention in the past decades since they convert superoxide, which is toxic for cells, to H2O2 that is further reduced by H2O2-scavenging enzymes. SODs have been implemented in many health- and cosmetics-related applications (339) but have not made a breakthrough in the field of biomass conversion. However, a few patents (340, 341) mention the use of SOD in enzymatic mixtures for biomass conversion. Although SODs are usually intracellular enzymes, Rashid et al. have recently reported the existence of two extracellular SODs from the bacterium Sphingobacterium sp. strain T2 that displays ligninolytic activity (342). The mode of action of these SODs is not established, but the authors proposed that the enzymes may be involved in the generation of hydroxyl radicals acting as lignin oxidants. The role of SOD is complex because the enzyme removes one ROS species, superoxide, while generating another, hydrogen peroxide. As alluded to above, H2O2 can engage in a wide variety of reactions, varying from beneficial (improved LPMO activity and well-controlled Fenton chemistry) to highly detrimental. Interestingly, it has been shown that SOD can be beneficial for AA10 LPMO activity under conditions that generate a constant flux of superoxide, an effect that was ascribed to the beneficial effect of continuous in situ generation of H2O2 (33).
The effect of CDHs.
In pre-LPMO times, it was shown that addition of PcCDH to a mixture of cellulases from Trichoderma viride promoted saccharification of microcrystalline cellulose (338). Nevertheless, the use of CDH to improve industrial lignocellulose conversion has so far been little discussed, and the patent literature shows seemingly conflicting results (343, 344). While an early patent, from 2010, by Sweeney et al. describes a negative effect of CDH (343), in 2013, Sigoillot et al. claimed that spiking an enzyme cocktail from T. reesei with a CDH from the fungus Pycnoporus cinnabarinus would lead to an increase in total sugar release (344). In a related publication, Bey et al. showed that the addition of this CDH to the T. reesei GC220 enzyme cocktail (Genecor-Danisco) supplemented with a β-glucosidase (Novozym 188) doubled sugar release from wheat straw (345). The authors suggested that differences in reported CDH effects could be due to differences between the enzymes, such as the absence or presence of a CBM (345). With hindsight, variation in the H2O2 production ability and the interplay with H2O2-dependent enzymes present in the enzyme mixture could be seen as another explanation for the observations since H2O2 can represent both a beneficial and a harmful factor. A recent study showed that addition of a CBM-free CDH from Volvariella volvacea to a secretome from T. reesei D-86271 leads to a CDH dose-dependent increase in overall hydrolysis yield for both filter paper and delignified wheat straw (346).
The effect of CBMs.
The efficiency of carbohydrate-active enzymes is affected by the presence of carbohydrate binding modules (CBMs) (347, 348). The substrate affinity provided by CBMs brings the catalytic domain into close proximity of the substrate, a property that may be particularly beneficial in the case of hardly accessible and nondiffusible substrates such as the copolymeric plant cell wall. Of note, it has been shown that the strong substrate binding enabled by CBMs becomes less beneficial, and even negative, at higher substrate concentrations, which lead to an increased frequency of enzyme-polysaccharide association events (349, 350). These observations are particularly relevant in industrial settings, where high-solid loadings are desirable.
As of August 2018, 83 families of CBMs have been included in the CAZy database, and LPMOs are appended to several of these (12). While there are many natural LPMOs without a CBM, deletion of the CBM from LPMOs with a CBM is deleterious for enzyme efficiency (56, 79, 80, 351, 352), and this deleterious effect may be substrate dependent (71, 80). Today, there are clear indications that, in the absence of substrate, LPMOs provided with electrons and their oxygen-containing cosubstrate suffer from autoxidation of key amino acids in their catalytic center (32). Substrate affinity provided by attached CBMs may postpone or prevent such oxidative self-inactivation.
Interestingly, a recent study of activation of an AA9 LPMO by a pyranose dehydrogenase (PDH) belonging to the AA12 family (353) showed that deletion of the CBM from this three-domain CDH analogue (AA8-AA12-CBM1) reduced the overall efficiency of the PDH-LPMO system (354). This relatively preliminary result is potentially of great importance because it could reflect a proximity effect. The presence of the cellulose-binding CBM could ensure that reduction of the LPMO and/or production of its cosubstrate H2O2, both catalyzed by the PDH, happen close to the LPMO substrate, thus preventing off-pathway reactions in the LPMO.
The effect of GHs.
Since LPMO stability is affected by substrate availability, the interplay between GHs and LPMOs needs attention in the designing of enzyme cocktails. GHs could play an indirect role in stabilizing LPMOs by removing obstacles from the polysaccharide surfaces, including chains already oxidized by LPMOs, and thus increasing the available (crystalline) surface area for LPMO binding. Details of the interplay between GHs and LPMOs remain remarkably unexplored. We predict that increasing insights into LPMO functionality, including LPMO stability, will lead to adjustments in the composition of GHs in enzyme cocktails for biomass processing.
The Impact of Oxygen Dependency on Bioprocessing Strategies
As discussed above, LPMOs are major players in the saccharification of several biomasses of industrial relevance. Until recently, LPMO reactions were thought to be driven by O2 (only), and this O2 dependency poses several challenges to industrial application. Aeration, i.e., dissolution and homogeneous dispersion of oxygen, at an industrial scale is expensive. Capital and operational costs for aeration are considered major hurdles for the development of large-scale (e.g., 100 to 1,000 m3) aerobic fermentation processes (355, 356). As recently underlined by Humbird et al., the aeration-related costs are even more critical for low-margin and high-volume production commodities such as biofuels (357). Furthermore, at an industrial scale, high substrate loadings (>10%, wt/wt) are necessary to lower water consumption and maximize the sugar concentration after saccharification, leading to viscous and heterogeneous slurries and increasing the technological challenges and costs associated with mixing and aeration (358–360). In the case of lignin-poor biomasses, LPMO action requires not only O2 but also addition of reducing agents (320, 321), which adds costs. In oxygen-driven processes, reductants need to be supplied in equimolar amounts (two electrons per LPMO reaction [11]). Importantly, the use of O2 can lead to the waste of valuable reducing equivalents via unproductive, and sometimes harmful, oxidative reactions.
The peroxygenase nature of LPMOs may offer a more cost-efficient and process-friendly alternative for LPMO activation at large scale since, in this case, the cosubstrate is a pumpable liquid, and stoichiometric amounts of reductant are not needed. Indeed, efficient, H2O2-fueled saccharification with the LPMO-containing cocktail Cellic CTec2 has been demonstrated for Avicel, sulfite-pulped Norway spruce, and steam-exploded birch wood (361). Importantly, and well known from work with peroxygenases (362), tight regulation of H2O2 supply is required to avoid side reactions that deplete reducing power or inactivate the enzymes (361). In other words, conditions should be such that the concentration of H2O2 is kept at a minimum (likely in the low-micromolar range) (32, 34). Indeed, Scott et al. have shown that the batch-wise addition of H2O2 to a saccharification reactor, leading to temporarily high H2O2 concentrations, has negative effects on overall process efficiency (322).
As an alternative to supplementing external H2O2, H2O2 may be generated in situ, for example, using glucose oxidase (32), a widely used enzyme in industry (363), or via electrochemical or photocatalytic reduction of molecular oxygen (88, 364). These last approaches, however, would not solve the problem of aeration although the demands for aeration could differ due to different kinetics of the various alternatives.
An important aspect of biomass conversion concerns the main overall strategy adopted to optimize potential synergies and minimize the effects of inhibiting compounds generated during the various processing steps. Two main approaches are usually considered which differ in that the processes of biomass saccharification and microbial conversion of sugars into added-value products are run sequentially (separate hydrolysis and fermentation, or SHF) or simultaneously (simultaneous saccharification and fermentation, or SSF). So far, SSF approaches have been considered incompatible with harnessing the enzymatic power of LPMOs since the fermenting microorganisms either compete for the supplied O2 or require anaerobic conditions (365, 366). Activation of LPMOs with H2O2, at low, nonharmful concentrations, under anaerobic conditions opens up new possibilities for SSF.
While the H2O2-driven saccharification of a relatively clean substrate such as Avicel works well, with seemingly stoichiometric incorporation of the cosubstrate into oxidized sugars (32, 44), the situation becomes more complicated when complex biomass is used. Lignin-containing biomass contains lots of redox-active components that may engage in reactions with H2O2, as clearly shown in a recent study by Müller et al. (361).
Finally, one issue for future studies concerns the temporal orchestration of enzyme additions. Inspired by extensive knowledge of glycoside hydrolases (367) and LPMOs and by increased knowledge of natural strategies (see Insights into the Network of Lignocellulolytic Redox Reactions, above), the impact of simultaneous or sequential and timely addition of various enzyme types at different stages of biomass conversion may be worth (re)investigating. Orchestrating the concert of oxidative and hydrolytic activities will certainly be one of the future challenges on the road toward more efficient and sustainable processes of biomass conversion.
CONCLUDING REMARKS
The controlled decomposition of biomass in general and of lignocellulose in particular involves a wide diversity of enzymatic activities and chemical reactions, which are probably not all identified yet and whose interconnections are far from clear. Here, we have reviewed enzymes, processes, and possible interconnections while focusing on LPMOs and the potentially central role of hydrogen peroxide. Based on the discovery of the peroxygenase activity of LPMOs (32, 33) and subsequent supporting studies (34, 43, 44, 74), we have revisited established views on the LPMO paradigm and provided alternative H2O2-based interpretations of literature data.
The question as to what extent LPMO activity directly driven by O2 is relevant remains open. It is worth noting the history of the discovery of GH mechanisms. Initially, GHs were thought to employ one of two main mechanisms (368, 369), but as research progressed, a variety of family-dependent mechanistic features was discovered (370–372). There may be mechanistic variations and not yet discovered alternative mechanisms within the LPMO superfamily. In the case of GHs, variations in mechanism are often related to the nature of the target substrate. The same could very well apply to LPMOs, especially since current (limited) data indicate that binding of the two substrates (i.e., the polysaccharide and the oxygen-delivering cosubstrate) is interdependent (34, 373). Considering the importance of ternary complex formation and the effect of substrate binding on the confinement of the otherwise exposed catalytic center (45), it is possible that substrate-dependent variations in the catalytic mechanism occur, as also recently suggested by Simmons et al. (57).
Regardless of the true mechanism(s) of LPMOs, the efficient use of H2O2 by fungal LPMOs (AA9) is compatible with their ecological context. Above, we have stressed the Janus-type role played by H2O2 in lignocellulose conversion, being a product of oxidases and of the oxidation of organic compounds, being a substrate of peroxidases, peroxygenases, and catalases or in Fenton-type reactions, but also being a potentially damaging entity, primarily through its reaction with transition metals. Several authors have pointed out the intriguing genomic cooccurrence and coexpression of H2O2-generating enzymes (e.g., strict oxidases) along with GHs and LPMOs in the absence of classical, known H2O2 sinks. Considering H2O2-driven catalysis by LPMOs, which are abundantly encoded in genomes and abundantly expressed during growth on biomass, the cooccurrence of these enzymes now has a plausible explanation. It appears reasonable to speculate that, in order to tame ROS, dedicated enzymatic tools have evolved to deal efficiently with the respective chemistries of their reduction, resulting in optimized cooperation between enzymes. While oxidases, often FAD-dependent enzymes, appear equipped to deal with the reduction of O2, to provide in fine H2O2, LPMOs are clearly much more efficient in reducing H2O2 than in reducing O2 directly.
The role of nonenzymatic entities such as phytophenolics in the redox processes occurring during biomass conversion is undeniable and highly relevant, both biologically and industrially (19, 83, 86, 374, 375). For example, it is clear that lignin and fragments thereof affect LPMO catalysis. Furthermore, it is obvious that ROS such as H2O2 will react with some of these nonenzymatic redox entities. Finally, the role of seemingly uncontrollable Fenton chemistry remains intriguing. Integrating these chemical aspects, which do not appear from omics studies, into our understanding of lignocellulolysis constitutes a considerable future challenge.
Another future challenge concerns the need for more insight into spatial and temporal aspects. The impacts of simultaneous versus sequential decomposition and the temporal and spatial regulation of enzyme expression still need more attention (261). Another remaining issue concerns the possible existence of backup mechanisms, as illustrated by the study of a Δcdh strain of Podospora anserina, whose growth was not affected due to the activation of alternative strategies (111) (see above for details).
The remarkable diversity of H2O2-generating enzymes encountered during lignocellulose conversion raises interesting questions. As pointed out above, the widely accepted idea that these enzymes are general partners of peroxidases may need revision. Notably, if general H2O2 production, rather than substrate oxidation, is the main purpose of these enzymes, why then are there so many different enzymes employed to generate an identical product? Noting the wide variation in substrate specificity among these enzymes, which act on a variety of (hemi)cellulose- or lignin-derived compounds, one may wonder if this wide spectrum of H2O2-producing enzymes has evolved to ensure that H2O2 is produced only at specific locations, i.e., locations where the oxidase substrate is present and an enzyme such as an LPMO or a peroxidase is ready to use the produced H2O2. Such a strategy would reduce hazardous bulk production of H2O2 and satisfy the need for proximity between redox partners, minimizing undesired competition between enzymes and off-pathway reactions. Such proximity effects may be crucial for optimizing enzyme system efficiency and may also be achieved by appending CBMs to interacting redox partners, as alluded to above (354).
H2O2 is a ubiquitous molecule. It is a relatively stable carrier of reducing equivalents and of oxygen atoms and is sometimes referred to as a master hormone, notably in plant metabolism (220). Compared to atmospheric O2, which constitutes a hardly controllable and infinite input in air-exposed environments, H2O2 is a liquid compound offering a much wider concentration range that nature can tightly regulate. This is illustrated by the wide diversity of H2O2-using enzymes, with catalytic efficiencies and substrate affinities spanning several orders of magnitude. Interestingly, within the context of biomass conversion, Robert Blanchette pointed out in 1991 that oxygen concentrations are extremely low within decaying tree trunks, leading to the suggestion that “some mechanism must be operative that delivers necessary oxygen for oxidative reactions involved in lignin degradation” (272). In ancient times, before the Great Oxidation Event (GOE; controversially dated around 2,450 and 800 million years ago [376]), H2O2 was highly abundant on Earth while O2 was not (377). This has led to the suggestion that P450 cytochromes were initially peroxygenases and that their monooxygenase activity evolved later (378). LPMOs are ancient enzymes, putatively already in use more than 400 million years ago (153, 379). Given their ability to harness H2O2, one may wonder if LPMOs were already present in pre-GOE times, and, if so, on which substrate they were acting. Cytochrome P450s use H2O2 only at high (millimolar) concentrations and with low total turnover numbers, in what is referred to as a shunt reaction that is a putative remainder of ancestral function. In contrast, LPMOs operate in an efficient and stable manner when supplied with low concentrations of H2O2.
The regulation of the network of lignocellulolytic reactions, involving hydrolases, oxidoreductases, other enzymes, and nonenzymatic entities, is essential for the stability and efficiency of microbial systems and, thus, for the global carbon cycle. Next to the discovery and understanding of individual players, there is a clear need to better understand their interconnection in a biological situation, which eventually may inspire the design of improved, substrate-adapted industrial processes. We hope that the present overview of known and potential players in lignocellulolysis will help in targeting future research in this field.
ACKNOWLEDGMENTS
We thank current and past members of our groups for their contributions to our research on LPMOs and Roland Ludwig and coworkers at BOKU, University of Natural Resources and Life Sciences (Vienna, Austria), for inspiring collaborations on redox enzymes.
The production of this review was supported by the Research Council of Norway through grants 243663, 240967, and 262853.
We are not aware of any affiliations, memberships, funding, or financial interests that might be perceived as affecting the objectivity of this review.
Biographies

Bastien Bissaro was born in 1988 in the south of France (Toulouse area). He holds an M.Sc. degree in biochemical engineering (2011) and a Ph.D. in enzyme engineering (2014) from the National Institute of Applied Sciences (INSA) of Toulouse. In 2014, he obtained an AgreenSkills fellowship (Marie Curie action) and funding from the French National Institute for Agricultural Research (INRA) to study LPMOs in the group of Professor V. Eijsink at the Norwegian University of Life Sciences (NMBU). Dr. Bissaro is currently working at NMBU as a postdoctoral fellow with Dr. Å. K. Røhr aiming at deciphering the LPMO H2O2 reaction mechanism discovered in 2016. Since 2011, his main scientific interests have focused on the mechanism, engineering, and evolution of CAZymes related to biomass conversion and valorization.

Anikó Várnai holds an M.Sc. degree in bioengineering from the Budapest University of Technology and Economics in Hungary (2007) and obtained a Ph.D. degree in biotechnology at the University of Helsinki in collaboration with the VTT Technical Research Centre of Finland. After completing her Ph.D. in 2012, she moved to Professor Vincent Eijsink's group at the Norwegian University of Life Sciences. Her research interest is biomass valorization, with particular focus on understanding the role various enzyme components play, both individually and in combination with other enzyme components, in nature and in industrial biomass conversion processes and applying that knowledge for process optimization to maximize conversion yields. She has 10+ years of experience in enzymatic biomass conversion with cellulose- and hemicellulose-active hydrolases, esterases, and redox enzymes.

Åsmund K. Røhr holds an M.Sc. degree in physical biochemistry from the University of Oslo (UiO) in Norway (2001). In 2010, he obtained a Ph.D. at the same university, working with in situ spectroscopy, protein crystallography/radiation damage, and computational chemistry. This work was continued during a 2012-2014 postdoc period at the UiO and the European Synchrotron Radiation Facility. Between 2008 and 2012, Dr. Røhr also worked as Research Manager for the company Keep-it Technologies AS located in Oslo, Norway. In 2015, Dr. Røhr established a team at the Norwegian University of Life Sciences (NMBU) in Ås, Norway, where the main focus is directed towards understanding the function and mechanisms of oxidative enzymes in biomass decomposition.

Vincent G. H. Eijsink holds an M.Sc. degree from Wageningen University in the Netherlands (1986). After obtaining a Ph.D. degree at the University of Groningen, The Netherlands (1991), and a short postdoc at the same university working on enzyme stability engineering, he moved to the Norwegian University of Life Sciences (NMBU) in Ås, Norway, where he became professor in 1997 and established an independent research program. The Eijsink group has a primary focus on fundamental and applied enzymology related to biomass conversion. Early work mainly concerned the conversion of chitin, whereas today lignocellulosic biomass is central. The group is best known for being the first to demonstrate the synergistic effects between chitinases and proteins today known as LPMOs, published in 2005, and for the discovery of LPMO activity, published in 2010.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MMBR.00029-18.
REFERENCES
- 1.Glass NL, Schmoll M, Cate JHD, Coradetti S. 2013. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol 67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 2.Chundawat SPS, Beckham GT, Himmel ME, Dale BE. 2011. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng 2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- 3.Scarlat N, Dallemand J-F, Monforti-Ferrario F, Nita V. 2015. The role of biomass and bioenergy in a future bioeconomy: policies and facts. Environ Dev 15:3–34. doi: 10.1016/j.envdev.2015.03.006. [DOI] [Google Scholar]
- 4.Cragg SM, Beckham GT, Bruce NC, Bugg TDH, Distel DL, Dupree P, Etxabe AG, Goodell BS, Jellison J, McGeehan JE, McQueen-Mason SJ, Schnorr K, Walton PH, Watts JEM, Zimmer M. 2015. Lignocellulose degradation mechanisms across the tree of life. Curr Opin Chem Biol 29:108–119. doi: 10.1016/j.cbpa.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gligorovski S, Strekowski R, Barbati S, Vione D. 2015. Environmental implications of hydroxyl radicals (˙OH). Chem Rev 115:13051–13092. doi: 10.1021/cr500310b. [DOI] [PubMed] [Google Scholar]
- 6.Arantes V, Goodell B. 2014. Current understanding of brown-rot fungal biodegradation mechanisms: a review. ACS Symp Series 1158:1–21. [Google Scholar]
- 7.Reese ET, Siu RG, Levinson HS. 1950. The biological degradation of soluble cellulose derivatives and its relationship to the mechanism of cellulose hydrolysis. J Bacteriol 59:485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriksson K-E, Pettersson B, Westermark U. 1974. Oxidation: An important enzyme reaction in fungal degradation of cellulose. FEBS Lett 49:282–285. doi: 10.1016/0014-5793(74)80531-4. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki K, Suzuki M, Taiyoji M, Nikaidou N, Watanabe T. 1998. Chitin binding protein (CBP21) in the culture supernatant of Serratia marcescens 2170. Biosci Biotechnol Biochem 65:128–135. doi: 10.1271/bbb.62.128. [DOI] [PubMed] [Google Scholar]
- 10.Vaaje-Kolstad G, Horn SJ, van Aalten DMF, Synstad B, Eijsink VGH. 2005. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J Biol Chem 280:28492–28497. doi: 10.1074/jbc.M504468200. [DOI] [PubMed] [Google Scholar]
- 11.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, Eijsink VGH. 2010. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 12.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. 2012. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5:45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen J-CN, Johansen KS, Krogh KBRM, Jørgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A 108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips CM, Beeson WT, Cate JH, Marletta MA. 2011. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol 6:1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 15.Kjaergaard CH, Qayyum MF, Wong SD, Xu F, Hemsworth GR, Walton DJ, Young NA, Davies GJ, Walton PH, Johansen KS, Hodgson KO, Hedman B, Solomon EI. 2014. Spectroscopic and computational insight into the activation of O2 by the mononuclear Cu center in polysaccharide monooxygenases. Proc Natl Acad Sci U S A 111:8797–8802. doi: 10.1073/pnas.1408115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beeson WT, Phillips CM, Cate JHD, Marletta MA. 2012. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc 134:890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- 17.Walton PH, Davies GJ. 2016. On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr Opin Chem Biol 31:195–207. doi: 10.1016/j.cbpa.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Beeson WT, Vu VV, Span EA, Phillips CM, Marletta MA. 2015. Cellulose degradation by polysaccharide monooxygenases. Annu Rev Biochem 84:923–946. doi: 10.1146/annurev-biochem-060614-034439. [DOI] [PubMed] [Google Scholar]
- 19.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, Xu F, Cherry J, Larsen S, Lo Leggio L. 2010. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry 49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson J, Saloheimo M, Siika-aho M, Tenkanen M, Penttilä M, Tjerneld F. 2001. Homologous expression and characterization of Cel61A (EG IV) of Trichoderma reesei. Eur J Biochem 268:6498–6507. doi: 10.1046/j.0014-2956.2001.02605.x. [DOI] [PubMed] [Google Scholar]
- 21.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrissat B. 1991. A classification of glycosyl hydrolases based sequence similarities amino acid. Biochem J 280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen KS. 2016. Lytic polysaccharide monooxygenases: the microbial power tool for lignocellulose degradation. Trends Plant Sci 21:926–936. doi: 10.1016/j.tplants.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Couturier M, Ladevèze S, Sulzenbacher G, Ciano L, Fanuel M, Moreau C, Villares A, Cathala B, Chaspoul F, Frandsen KE, Labourel A, Herpoël-Gimbert I, Grisel S, Haon M, Lenfant N, Rogniaux H, Ropartz D, Davies GJ, Rosso MN, Walton PH, Henrissat B, Berrin JG. 2018. Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat Chem Biol 14:306–310. doi: 10.1038/nchembio.2558. [DOI] [PubMed] [Google Scholar]
- 26.Beckham GT, Matthews JF, Peters B, Bomble YJ, Himmel ME, Crowley MF. 2011. Molecular-level origins of biomass recalcitrance: decrystallization free energies for four common cellulose polymorphs. J Phys Chem B 115:4118–4127. doi: 10.1021/jp1106394. [DOI] [PubMed] [Google Scholar]
- 27.Villares A, Moreau C, Bennati-Granier C, Garajova S, Foucat L, Falourd X, Saake B, Berrin J-G, Cathala B. 2017. Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci Rep 7:40262. doi: 10.1038/srep40262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vermaas JV, Crowley MF, Beckham GT, Payne CM. 2015. Effects of lytic polysaccharide monooxygenase oxidation on cellulose structure and binding of oxidized cellulose oligomers to cellulases. J Phys Chem B 119:6129–6143. doi: 10.1021/acs.jpcb.5b00778. [DOI] [PubMed] [Google Scholar]
- 29.Eibinger M, Ganner T, Bubner P, Rosker S, Kracher D, Haltrich D, Ludwig R, Plank H, Nidetzky B. 2014. Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J Biol Chem 289:35929–35938. doi: 10.1074/jbc.M114.602227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chabbert B, Habrant A, Herbaut M, Foulon L, Aguié-Béghin V, Garajova S, Grisel S, Bennati-Granier C, Gimbert-Herpoël I, Jamme F, Réfrégiers M, Sandt C, Berrin JG, Paës G. 2017. Action of lytic polysaccharide monooxygenase on plant tissue is governed by cellular type. Sci Rep 7:17792. doi: 10.1038/s41598-017-17938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eibinger M, Sattelkow J, Ganner T, Plank H, Nidetzky B. 2017. Single-molecule study of oxidative enzymatic deconstruction of cellulose. Nat Commun 8:894. doi: 10.1038/s41467-017-01028-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bissaro B, Røhr ÅK, Müller G, Chylenski P, Skaugen M, Forsberg Z, Horn SJ, Vaaje-Kolstad G, Eijsink VGH. 2017. Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat Chem Biol 13:1123–1128. doi: 10.1038/nchembio.2470. [DOI] [PubMed] [Google Scholar]
- 33.Bissaro B, Rohr AK, Skaugen M, Forsberg Z, Horn SJ, Vaaje-Kolstad G, Eijsink V. 2016. Fenton-type chemistry by a copper enzyme: molecular mechanism of polysaccharide oxidative cleavage. bioRxiv doi: 10.1101/097022. [DOI]
- 34.Kuusk S, Bissaro B, Kuusk P, Forsberg Z, Eijsink VGH, Sørlie M, Väljamäe P. 2018. Kinetics of H2O2-driven degradation of chitin by a bacterial lytic polysaccharide monooxygenase. J Biol Chem 293:523–531. doi: 10.1074/jbc.M117.817593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forney LJ, Reddy CA, Tien M, Aust SD. 1982. The involvement of hydroxyl radical derived from hydrogen peroxide in lignin degradation by the white rot fungus Phanerochaete chrysosporium. J Biol Chem 257:11455–11462. [PubMed] [Google Scholar]
- 36.Murphy MP, Holmgren A, Larsson N-G, Halliwell B, Chang CJ, Kalyanaraman B, Rhee SG, Thornalley PJ, Partridge L, Gems D, Nyström T, Belousov V, Schumacker PT, Winterbourn CC. 2011. Unraveling the biological roles of reactive oxygen species. Cell Metab 13:361–366. doi: 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickinson BC, Chang CJ. 2011. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol 7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frandsen KEH, Lo Leggio L. 2016. Lytic polysaccharide monooxygenases: a crystallographer's view on a new class of biomass-degrading enzymes. IUCrJ 3:448–467. doi: 10.1107/S2052252516014147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaaje-Kolstad G, Forsberg Z, Loose JS, Bissaro B, Eijsink VG. 2017. Structural diversity of lytic polysaccharide monooxygenases. Curr Opin Struct Biol 44:67–76. doi: 10.1016/j.sbi.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Span EA, Marletta MA. 2015. The framework of polysaccharide monooxygenase structure and chemistry. Curr Opin Struct Biol 35:93–99. doi: 10.1016/j.sbi.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Johansen KS. 2016. Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem Soc Trans 44:143–149. doi: 10.1042/BST20150204. [DOI] [PubMed] [Google Scholar]
- 42.Meier KK, Jones SM, Kaper T, Hansson H, Koetsier MJ, Karkehabadi S, Solomon EI, Sandgren M, Kelemen B. 2018. Oxygen activation by Cu LPMOs in recalcitrant carbohydrate polysaccharide conversion to monomer sugars. Chem Rev 118:2593–2635. doi: 10.1021/acs.chemrev.7b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breslmayr E, Hanžek M, Hanrahan A, Leitner C, Kittl R, Šantek B, Oostenbrink C, Ludwig R. 2018. A fast and sensitive activity assay for lytic polysaccharide monooxygenase. Biotechnol Biofuels 11:79. doi: 10.1186/s13068-018-1063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hangasky JA, Iavarone AT, Marletta MA. 2018. Reactivity of O2 versus H2O2 with polysaccharide monooxygenases. Proc Natl Acad Sci U S A 115:4915–4920. doi: 10.1073/pnas.1801153115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bissaro B, Isaksen I, Vaaje-Kolstad G, Eijsink VGH, Røhr ÅK. 2018. How a lytic polysaccharide monooxygenase binds crystalline chitin. Biochemistry 57:1893–1906. doi: 10.1021/acs.biochem.8b00138. [DOI] [PubMed] [Google Scholar]
- 46.Agger JW, Isaksen T, Várnai A, Vidal-Melgosa S, Willats WGT, Ludwig R, Horn SJ, Eijsink VGH, Westereng B. 2014. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci U S A 111:6287–6292. doi: 10.1073/pnas.1323629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vu VV, Beeson WT, Span EA, Farquhar ER, Marletta MA. 2014. A family of starch-active polysaccharide monooxygenases. Proc Natl Acad Sci U S A 111:13822–13827. doi: 10.1073/pnas.1408090111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo Leggio L, Simmons TJ, Poulsen JCN, Frandsen KEH, Hemsworth GR, Stringer MA, Von Freiesleben P, Tovborg M, Johansen KS, De Maria L, Harris PV, Soong CL, Dupree P, Tryfona T, Lenfant N, Henrissat B, Davies GJ, Walton PH. 2015. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun 6:5961. doi: 10.1038/ncomms6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frommhagen M, Sforza S, Westphal AH, Visser J, Hinz SWA, Koetsier MJ, Van Berkel WJH, Gruppen H, Kabel MA. 2015. Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase. Biotechnol Biofuels 8:101. doi: 10.1186/s13068-015-0284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nekiunaite L, Petrović DM, Westereng B, Vaaje-Kolstad G, Hachem MA, Várnai A, Eijsink VGH. 2016. FgLPMO9A from Fusarium graminearum cleaves xyloglucan independently of the backbone substitution pattern. FEBS Lett 590:3346–3356. doi: 10.1002/1873-3468.12385. [DOI] [PubMed] [Google Scholar]
- 51.Kojima Y, Várnai A, Ishida T, Sunagawa N, Petrovic DM, Igarashi K, Jellison J, Goodell B, Alfredsen G, Westereng B, Eijsink VGH, Yoshida M. 2016. A lytic polysaccharide monooxygenase with broad xyloglucan specificity from the brown-rot fungus Gloeophyllum trabeum and its action on cellulose-xyloglucan complexes. Appl Environ Microbiol 82:6557–6572. doi: 10.1128/AEM.01768-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frandsen KEH, Simmons TJ, Dupree P, Poulsen J-CN, Hemsworth GR, Ciano L, Johnston EM, Tovborg M, Johansen KS, von Freiesleben P, Marmuse L, Fort S, Cottaz S, Driguez H, Henrissat B, Lenfant N, Tuna F, Baldansuren A, Davies GJ, Lo Leggio L, Walton PH. 2016. The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases. Nat Chem Biol 12:298–303. doi: 10.1038/nchembio.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aachmann FL, Sørlie M, Skjåk-Bræk G, Eijsink VGH, Vaaje-Kolstad G. 2012. NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc Natl Acad Sci U S A 109:18779–18784. doi: 10.1073/pnas.1208822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaaje-Kolstad G, Houston DR, Riemen AHK, Eijsink VGH, van Aalten DMF. 2005. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J Biol Chem 280:11313–11319. doi: 10.1074/jbc.M407175200. [DOI] [PubMed] [Google Scholar]
- 55.Courtade G, Wimmer R, Røhr ÅK, Preims M, Felice AKG, Dimarogona M, Vaaje-Kolstad G, Sørlie M, Sandgren M, Ludwig R, Eijsink VGH, Aachmann FL. 2016. Interactions of a fungal lytic polysaccharide monooxygenase with β-glucan substrates and cellobiose dehydrogenase. Proc Natl Acad Sci U S A 113:5922–5927. doi: 10.1073/pnas.1602566113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forsberg Z, Bissaro B, Gullesen J, Dalhus B, Vaaje-Kolstad G, Eijsink VGH. 2018. Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J Biol Chem 293:1397–1412. doi: 10.1074/jbc.M117.817130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmons TJ, Frandsen KEH, Ciano L, Tryfona T, Lenfant N, Poulsen JC, Wilson LFL, Tandrup T, Tovborg M, Schnorr K, Johansen KS, Henrissat B, Walton PH, Lo Leggio L, Dupree P. 2017. Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates. Nat Commun 8:1064. doi: 10.1038/s41467-017-01247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kruer-Zerhusen N, Alahuhta M, Lunin VV, Himmel ME, Bomble YJ, Wilson DB. 2017. Structure of a Thermobifida fusca lytic polysaccharide monooxygenase and mutagenesis of key residues. Biotechnol Biofuels 10:243. doi: 10.1186/s13068-017-0925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vu Van V, Beeson WT, Phillips CM, Cate JHD, Marletta MA. 2014. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc 136:562–565. doi: 10.1021/ja409384b. [DOI] [PubMed] [Google Scholar]
- 60.Danneels B, Tanghe M, Joosten HJ, Gundinger T, Spadiut O, Stals I, Desmet T. 2017. A quantitative indicator diagram for lytic polysaccharide monooxygenases reveals the role of aromatic surface residues in HjLPMO9A regioselectivity. PLoS One 12:e0178446. doi: 10.1371/journal.pone.0178446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Book AJ, Yennamalli RM, Takasuka TE, Currie CR, Phillips GN Jr, Fox BG. 2014. Evolution of substrate specificity in bacterial AA10 lytic polysaccharide monooxygenases. Biotechnol Biofuels 7:109. doi: 10.1186/1754-6834-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isaksen T, Westereng B, Aachmann FL, Agger JW, Kracher D, Kittl R, Ludwig R, Haltrich D, Eijsink VGH, Horn SJ. 2014. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem 289:2632–2642. doi: 10.1074/jbc.M113.530196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forsberg Z, Vaaje-Kolstad G, Westereng B, Bunæs AC, Stenstrøm Y, MacKenzie A, Sørlie M, Horn SJ, Eijsink VG. 2011. Cleavage of cellulose by a CBM33 protein. Protein Sci 20:1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langston JA, Shaghasi T, Abbate E, Xu F, Vlasenko E, Sweeney MD. 2011. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol 77:7007–7015. doi: 10.1128/AEM.05815-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frommhagen M, Koetsier MJ, Westphal AH, Visser J, Hinz SWA, Vincken J-P, van Berkel WJH, Kabel MA, Gruppen H. 2016. Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity. Biotechnol Biofuels 9:186. doi: 10.1186/s13068-016-0594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kittl R, Kracher D, Burgstaller D, Haltrich D, Ludwig R. 2012. Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol Biofuels 5:79. doi: 10.1186/1754-6834-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Span EA, Suess DLM, Deller MC, Britt RD, Marletta MA. 2017. The role of the secondary coordination sphere in a fungal polysaccharide monooxygenase. ACS Chem Biol 12:1095–1103. doi: 10.1021/acschembio.7b00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hedegård ED, Ryde U. 2017. Multiscale modelling of lytic polysaccharide monooxygenases. ACS Omega 2:536–545. doi: 10.1021/acsomega.6b00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O'Dell WB, Agarwal PK, Meilleur F. 2017. Oxygen activation at the active site of a fungal lytic polysaccharide monooxygenase. Angew Chemie Int Ed Engl 129:785–788. doi: 10.1002/ange.201610502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bacik JP, Mekasha S, Forsberg Z, Kovalevsky AY, Vaaje-Kolstad G, Eijsink VGH, Nix JC, Coates L, Cuneo MJ, Unkefer CJ, Chen JC-H. 2017. Neutron and atomic resolution X-ray structures of a lytic polysaccharide monooxygenase reveal copper-mediated dioxygen binding and evidence for N-terminal deprotonation. Biochemistry 56:2529–2532. doi: 10.1021/acs.biochem.7b00019. [DOI] [PubMed] [Google Scholar]
- 71.Borisova AS, Isaksen T, Dimarogona M, Kognole AA, Mathiesen G, Várnai A, Røhr ÅK, Payne CM, Sørlie M, Sandgren M, Eijsink VGH. 2015. Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. J Biol Chem 290:22955–22969. doi: 10.1074/jbc.M115.660183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whittaker JW. 2005. The radical chemistry of galactose oxidase. Arch Biochem Biophys 433:227–239. doi: 10.1016/j.abb.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 73.Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber-Emmons MT, Kjaergaard CH, Hadt RG, Tian L. 2014. Copper active sites in biology. Chem Rev 114:3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang B, Johnston EM, Li P, Shaik S, Davies GJ, Walton PH, Rovira C. 2018. QM/MM studies into the H2O2-dependent activity of lytic polysaccharide monooxygenases: evidence for the formation of a caged hydroxyl radical intermediate. ACS Catal 8:1346–1351. doi: 10.1021/acscatal.7b03888. [DOI] [Google Scholar]
- 75.Kim S, Ståhlberg J, Sandgren M, Paton RS, Beckham GT. 2014. Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proc Natl Acad Sci U S A 111:149–154. doi: 10.1073/pnas.1316609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bertini L, Breglia R, Lambrughi M, Fantucci P, De Gioia L, Borsari M, Sola M, Bortolotti CA, Bruschi M. 2018. Catalytic mechanism of fungal lytic polysaccharide monooxygenases investigated by first-principles calculations. Inorg Chem 57:86–97. doi: 10.1021/acs.inorgchem.7b02005. [DOI] [PubMed] [Google Scholar]
- 77.Hofrichter M, Ullrich R. 2014. Oxidations catalyzed by fungal peroxygenases. Curr Opin Chem Biol 19:116–125. doi: 10.1016/j.cbpa.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 78.Hedegård ED, Ryde U. 2018. Molecular mechanism of lytic polysaccharide monooxygenases. Chem Sci 9:3866–3880. doi: 10.1039/C8SC00426A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Forsberg Z, Nelson CE, Dalhus B, Mekasha S, Loose JSM, Crouch LI, Røhr ÅK, Gardner JG, Eijsink VGH, Vaaje-Kolstad G. 2016. Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus. J Biol Chem 291:7300–7312. doi: 10.1074/jbc.M115.700161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crouch LI, Labourel A, Walton PH, Davies GJ, Gilbert HJ. 2016. The contribution of non-catalytic carbohydrate binding modules to the activity of lytic polysaccharide monooxygenases. J Biol Chem 291:7439–7449. doi: 10.1074/jbc.M115.702365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loose JSM, Arntzen MØ, Bissaro B, Ludwig R, Eijsink VGH, Vaaje-Kolstad G. 2018. Multi-point precision binding of substrate protects LPMOs from self-destructive off-pathway processes. Biochemistry 57:4114–4124. doi: 10.1021/acs.biochem.8b00484. [DOI] [PubMed] [Google Scholar]
- 82.Valderrama B, Ayala M, Vazquez-Duhalt R. 2002. Suicide inactivation of peroxidases and the challenge of engineering more robust enzymes. Chem Biol 9:555–565. doi: 10.1016/S1074-5521(02)00149-7. [DOI] [PubMed] [Google Scholar]
- 83.Kracher D, Scheiblbrandner S, Felice AKG, Breslmayr E, Preims M, Haltrich D, Eijsink VGH, Ludwig R. 2016. Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 352:1098–1101. doi: 10.1126/science.aaf3165. [DOI] [PubMed] [Google Scholar]
- 84.Cannella D, Hsieh CWC, Felby C, Jørgensen H. 2012. Production and effect of aldonic acids during enzymatic hydrolysis of lignocellulose at high dry matter content. Biotechnol Biofuels 5:26. doi: 10.1186/1754-6834-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dimarogona M, Topakas E, Olsson L, Christakopoulos P. 2012. Lignin boosts the cellulase performance of a GH-61 enzyme from Sporotrichum thermophile. Bioresour Technol 110:480–487. doi: 10.1016/j.biortech.2012.01.116. [DOI] [PubMed] [Google Scholar]
- 86.Westereng B, Cannella D, Wittrup Agger J, Jørgensen H, Larsen Andersen M, Eijsink VGH, Felby C. 2015. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep 5:18561. doi: 10.1038/srep18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cannella D, Möllers KB, Frigaard N-U, Jensen PE, Bjerrum MJ, Johansen KS, Felby C. 2016. Light-driven oxidation of polysaccharides by photosynthetic pigments and a metalloenzyme. Nat Commun 7:11134 doi: 10.1038/ncomms11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bissaro B, Forsberg Z, Ni Y, Hollmann F, Vaaje-Kolstad G, Eijsink VGH. 2016. Fueling biomass-degrading oxidative enzymes by light-driven water oxidation. Green Chem 18:5357–5366. doi: 10.1039/C6GC01666A. [DOI] [Google Scholar]
- 89.Lewis DFV, Hlavica P. 2000. Interactions between redox partners in various cytochrome P450 systems: functional and structural aspects. Biochim Biophys Acta 1460:353–374. doi: 10.1016/S0005-2728(00)00202-4. [DOI] [PubMed] [Google Scholar]
- 90.Eyer C, Backes W. 1992. Relationship between the rate of reductase-cytochrome P450 complex formation and the rate of first electron transfer. Arch Biochem Biophys 293:231–240. doi: 10.1016/0003-9861(92)90390-I. [DOI] [PubMed] [Google Scholar]
- 91.Frommhagen M, Westphal AH, Hilgers R, Koetsier MJ, Hinz SWA, Visser J, Gruppen H, van Berkel WJH, Kabel MA. 2018. Quantification of the catalytic performance of C1-cellulose-specific lytic polysaccharide monooxygenases. Appl Microbiol Biotechnol 102:1281–1295. doi: 10.1007/s00253-017-8541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kettle AJ, Anderson RF, Hampton MB, Winterbourn CC. 2007. Reactions of superoxide with myeloperoxidase. Biochemistry 46:4888–4897. doi: 10.1021/bi602587k. [DOI] [PubMed] [Google Scholar]
- 93.Hayyan M, Hashim MA, Alnashef IM. 2016. Superoxide ion: generation and chemical implications. Chem Rev 116:3029–3085. doi: 10.1021/acs.chemrev.5b00407. [DOI] [PubMed] [Google Scholar]
- 94.Chaiyen P, Fraaije MW, Mattevi A. 2012. The enigmatic reaction of flavins with oxygen. Trends Biochem Sci 37:373–380. doi: 10.1016/j.tibs.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 95.Anusevicius Z, Ramanavicius A, Sarlauskas J. 1998. Some aspects of electron-transfer reaction of ascorbate with quinones. Chem Pap 52:643–649. [Google Scholar]
- 96.Song Y, Buettner GR. 2010. Thermodynamic and kinetic considerations for the reaction of semiquinone radicals to form superoxide and hydrogen peroxide. Free Radic Biol Med 49:919–962. doi: 10.1016/j.freeradbiomed.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Scarpa M, Stevanatos R, Vigiinos P, Rig A. 1983. Superoxide ion as active intermediate in the autoxidation of ascorbate by molecular oxygen. J Biol Chem 258:6695–6697. [PubMed] [Google Scholar]
- 98.Nishikimi M. 1975. Oxidation of ascorbic acid with superoxide anion generated by the xanthine-xanthine oxidase system. Biochem Biophys Res Commun 63:463–468. doi: 10.1016/0006-291X(75)90710-X. [DOI] [PubMed] [Google Scholar]
- 99.Klinman JP. 2007. How do enzymes activate oxygen without inactivating themselves? Acc Chem Res 40:325–333. doi: 10.1021/ar6000507. [DOI] [PubMed] [Google Scholar]
- 100.Wood PM. 1988. The potential diagram for oxygen at pH 7. Biochem J 253:287–289. doi: 10.1042/bj2530287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mattevi A. 2006. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem Sci 31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 102.Loose JSM, Forsberg Z, Kracher D, Scheiblbrandner S, Ludwig R, Eijsink VGH, Vaaje-Kolstad G. 2016. Activation of bacterial lytic polysaccharide monooxygenases with cellobiose dehydrogenase. Protein Sci 25:2175–2186. doi: 10.1002/pro.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bennati-Granier C, Garajova S, Champion C, Grisel S, Haon M, Zhou S, Fanuel M, Ropartz D, Rogniaux H, Gimbert I, Record E, Berrin J-G. 2015. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol Biofuels 8:90. doi: 10.1186/s13068-015-0274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gusakov AV, Bulakhov AG, Demin IN, Sinitsyn AP. 2017. Monitoring of reactions catalyzed by lytic polysaccharide monooxygenases using highly-sensitive fluorimetric assay of the oxygen consumption rate. Carbohydr Res 452:156–161. doi: 10.1016/j.carres.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 105.Boatright WL. 2016. Oxygen dependency of one-electron reactions generating ascorbate radicals and hydrogen peroxide from ascorbic acid. Food Chem 196:1361–1367. doi: 10.1016/j.foodchem.2015.07.141. [DOI] [PubMed] [Google Scholar]
- 106.Vaaje-Kolstad G, Bunæs AC, Mathiesen G, Eijsink VGH. 2009. The chitinolytic system of Lactococcus lactis ssp. lactis comprises a nonprocessive chitinase and a chitin-binding protein that promotes the degradation of α- and β-chitin. FEBS J 276:2402–2415. doi: 10.1111/j.1742-4658.2009.06972.x. [DOI] [PubMed] [Google Scholar]
- 107.Nakagawa YS, Kudo M, Loose JSM, Ishikawa T, Totani K, Eijsink VGH, Vaaje-Kolstad G. 2015. A small lytic polysaccharide monooxygenase from Streptomyces griseus targeting α- and β-chitin. FEBS J 282:1065–1079. doi: 10.1111/febs.13203. [DOI] [PubMed] [Google Scholar]
- 108.Forsberg Z, Røhr AK, Mekasha S, Andersson KK, Eijsink VGH, Vaaje-Kolstad G, Sørlie M. 2014. Comparative study of two chitin-active and two cellulose-active AA10-type lytic polysaccharide monooxygenases. Biochemistry 53:1647–1656. doi: 10.1021/bi5000433. [DOI] [PubMed] [Google Scholar]
- 109.Kracher D, Andlar M, Furtmüller PG, Ludwig R. 2018. Active-site copper reduction promotes substrate binding of fungal lytic polysaccharide monooxygenase and reduces stability. J Biol Chem 293:1676–1687. doi: 10.1074/jbc.RA117.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Dueñas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, St John F, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 111.Tangthirasunun N, Navarro D, Garajova S, Chevret D, Tong LCH, Gautier V, Hyde KD, Silar P, Berrin J-G. 2017. Inactivation of cellobiose dehydrogenases modifies the cellulose degradation mechanism of Podospora anserina. Appl Environ Microbiol 83:e02716-16. doi: 10.1128/AEM.02716-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mason MG, Nicholls P, Wilson MT. 2003. Rotting by radicals–the role of cellobiose oxidoreductase? Biochem Soc Trans 31:1335–1336. doi: 10.1042/bst0311335. [DOI] [PubMed] [Google Scholar]
- 113.Hyde SM, Wood PM. 1997. A mechanism for production of hydroxyl radicals by the brown-rot fungus Coniophora puteana: Fe(III) reduction by cellobiose dehydrogenase and Fe(II) oxidation at a distance from the hyphae. Microbiology 143:259–266. doi: 10.1099/00221287-143-1-259. [DOI] [PubMed] [Google Scholar]
- 114.Ludwig R, Harreither W, Tasca F, Gorton L. 2010. Cellobiose dehydrogenase: A versatile catalyst for electrochemical applications. ChemPhysChem 11:2674–2697. doi: 10.1002/cphc.201000216. [DOI] [PubMed] [Google Scholar]
- 115.Henriksson G, Johansson G, Pettersson G. 2000. A critical review of cellobiose dehydrogenases. J Biotechnol 78:93–113. doi: 10.1016/S0168-1656(00)00206-6. [DOI] [PubMed] [Google Scholar]
- 116.Henriksson G, Zhang L, Li J, Ljungquist P, Reitberger T, Pettersson G, Johansson G. 2000. Is cellobiose dehydrogenase from Phanerochaete chrysosporium a lignin degrading enzyme? Biochim Biophys Acta 1480:83–91. doi: 10.1016/S0167-4838(00)00096-0. [DOI] [PubMed] [Google Scholar]
- 117.Pricelius S, Ludwig R, Lant N, Haltrich D, Guebitz GM. 2009. Substrate specificity of Myriococcum thermophilum cellobiose dehydrogenase on mono-, oligo-, and polysaccharides related to in situ production of H2O2. Appl Microbiol Biotechnol 85:75–83. doi: 10.1007/s00253-009-2062-0. [DOI] [PubMed] [Google Scholar]
- 118.Ludwig R, Ortiz R, Schulz C, Harreither W, Sygmund C, Gorton L. 2013. Cellobiose dehydrogenase modified electrodes: advances by materials science and biochemical engineering. Anal Bioanal Chem 405:3637–3658. doi: 10.1007/s00216-012-6627-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nutt A, Salumets A, Henriksson G, Sild V, Johansson G. 1997. Conversion of O species by cellobiose dehydrogenase (cellobiose oxidase) and glucose oxidase—a comparison. Biotechnol Lett 19:379–384. doi: 10.1023/A:1018315320696. [DOI] [Google Scholar]
- 120.Morpeth FF, Jones GD. 1986. Resolution, purification and some properties of the multiple forms of cellobiose quinone dehydrogenase from the white-rot fungus Sporotrichum pulverulentum. Biochem J 236:221–226. doi: 10.1042/bj2360221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mason MG, Wilson MT, Ball A, Nicholls P. 2002. Oxygen reduction by cellobiose oxidoreductase: the role of the haem group. FEBS Lett 518:29–32. doi: 10.1016/S0014-5793(02)02633-9. [DOI] [PubMed] [Google Scholar]
- 122.Mason MG, Nicholls P, Divne C, Hallberg BM, Henriksson G, Wilson MT. 2003. The heme domain of cellobiose oxidoreductase: a one-electron reducing system. Biochim Biophys Acta 1604:47–54. doi: 10.1016/S0005-2728(03)00023-9. [DOI] [PubMed] [Google Scholar]
- 123.Wilson MT, Hogg N, Jones GD. 1990. Reactions of reduced cellobiose oxidase with oxygen. Is cellobiose oxidase primarily an oxidase? Biochem J 270:265–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tan T-C, Kracher D, Gandini R, Sygmund C, Kittl R, Haltrich D, Hällberg BM, Ludwig R, Divne C. 2015. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat Commun 6:7542. doi: 10.1038/ncomms8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kracher D, Ludwig R. 2016. Cellobiose dehydrogenase: An essential enzyme for lignocellulose degradation in nature—a review. Die Bodenkultur 67:145–163. doi: 10.1515/boku-2016-0013. [DOI] [Google Scholar]
- 126.Igarashi K, Momohara I, Nishino T, Samejima M. 2002. Kinetics of inter-domain electron transfer in flavocytochrome cellobiose dehydrogenase from the white-rot fungus Phanerochaete chrysosporium. Biochem J 365:521–526. doi: 10.1042/bj20011809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li X, Beeson WT, Phillips CM, Marletta MA, Cate JHD. 2012. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 20:1051–1061. doi: 10.1016/j.str.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sygmund C, Santner P, Krondorfer I, Peterbauer CK, Alcalde M, Nyanhongo GS, Guebitz GM, Ludwig R. 2013. Semi-rational engineering of cellobiose dehydrogenase for improved hydrogen peroxide production. Microb Cell Fact 12:38. doi: 10.1186/1475-2859-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hyde SM, Wood PM. 1996. Kinetic and antigenic similarities for cellobiose dehydrogenase from the brown rot fungus Coniophora puteana and the white rot fungus Phanerochaete chrysosporium. FEMS Microbiol Lett 145:439–444. doi: 10.1111/j.1574-6968.1996.tb08613.x. [DOI] [Google Scholar]
- 130.Kremer SM, Wood PM. 1992. Production of Fenton's reagent by cellobiose oxidase from cellulolytic cultures of Phanerochaete chrysosporium. Eur J Biochem 208:807–814. doi: 10.1111/j.1432-1033.1992.tb17251.x. [DOI] [PubMed] [Google Scholar]
- 131.Sygmund C, Kracher D, Scheiblbrandner S, Zahma K, Felice AKG, Harreither W, Kittl R, Ludwig R. 2012. Characterization of the two Neurospora crassa cellobiose dehydrogenases and their connection to oxidative cellulose degradation. Appl Environ Microbiol 78:6161–6171. doi: 10.1128/AEM.01503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Möllers KB, Mikkelsen H, Simonsen TI, Cannella D, Johansen KS, Bjerrum MJ, Felby C. 2017. On the formation and role of reactive oxygen species in light-driven LPMO oxidation of phosphoric acid swollen cellulose. Carbohydr Res 448:182–186. doi: 10.1016/j.carres.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 133.Hashimoto K, Irie H, Fujishima A. 2005. TiO2 photocatalysis: a historical overview and future prospects. Jpn J Appl Phys 44:8269–8285. doi: 10.1143/JJAP.44.8269. [DOI] [Google Scholar]
- 134.Maciá-Agulló JA, Corma A, Garcia H. 2015. Photobiocatalysis: the power of combining photocatalysis and enzymes. Chemistry 21:10940–10959. doi: 10.1002/chem.201406437. [DOI] [PubMed] [Google Scholar]
- 135.Sheng H, Ji H, Ma W, Chen C, Zhao J. 2013. Direct four-electron reduction of O2 to H2O on TiO2 surfaces by pendant proton relay. Angew Chemie Int Ed Engl 52:9686–9690. doi: 10.1002/anie.201304481. [DOI] [PubMed] [Google Scholar]
- 136.Ran J, Zhang J, Yu J, Jaroniec M, Qiao SZ. 2014. Earth-abundant cocatalysts for semiconductor-based photocatalytic water splitting. Chem Soc Rev 43:7787–7812. doi: 10.1039/C3CS60425J. [DOI] [PubMed] [Google Scholar]
- 137.Zhang W, Fernández-Fueyo E, Ni Y, van Schie M, Gacs J, Renirie R, Wever R, Mutti FG, Rother D, Alcalde M, Hollmann F. 2018. Selective aerobic oxidation reactions using a combination of photocatalytic water oxidation and enzymatic oxyfunctionalizations. Nat Catal 1:55–62. doi: 10.1038/s41929-017-0001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Berrin J, Rosso M, Abou Hachem M. 2017. Fungal secretomics to probe the biological functions of lytic polysaccharide monooxygenases. Carbohydr Res 448:155–160. doi: 10.1016/j.carres.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 139.Monclaro AV, Filho EXF. 2017. Fungal lytic polysaccharide monooxygenases from family AA9: Recent developments and application in lignocelullose breakdown. Int J Biol Macromol 102:771–778. doi: 10.1016/j.ijbiomac.2017.04.077. [DOI] [PubMed] [Google Scholar]
- 140.Bengtsson O, Arntzen MØ, Mathiesen G, Skaugen M, Eijsink VGH. 2016. A novel proteomics sample preparation method for secretome analysis of Hypocrea jecorina growing on insoluble substrates. J Proteomics 131:104–112. doi: 10.1016/j.jprot.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 141.Dos Santos HB, Bezerra TMS, Pradella JGC, Delabona P, Lima D, Gomes E, Hartson SD, Rogers J, Couger B, Prade R. 2016. Myceliophthora thermophila M77 utilizes hydrolytic and oxidative mechanisms to deconstruct biomass. AMB Express 6:103. doi: 10.1186/s13568-016-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhu N, Liu J, Yang J, Lin Y, Yang Y, Ji L, Li M, Yuan H. 2016. Comparative analysis of the secretomes of Schizophyllum commune and other wood-decay basidiomycetes during solid-state fermentation reveals its unique lignocellulose-degrading enzyme system. Biotechnol Biofuels 9:42. doi: 10.1186/s13068-016-0461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nekiunaite L, Arntzen MØ, Svensson B, Vaaje-Kolstad G, Abou Hachem M. 2016. Lytic polysaccharide monooxygenases and other oxidative enzymes are abundantly secreted by Aspergillus nidulans grown on different starches. Biotechnol Biofuels 9:187. doi: 10.1186/s13068-016-0604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.MacDonald J, Doering M, Canam T, Gong Y, Guttman DS, Campbell MM, Master ER. 2011. Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl Environ Microbiol 77:3211–3218. doi: 10.1128/AEM.02490-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Macdonald J, Master ER. 2012. Time-dependent profiles of transcripts encoding lignocellulose-modifying enzymes of the white rot fungus Phanerochaete carnosa grown on multiple wood substrates. Appl Environ Microbiol 78:1596–1600. doi: 10.1128/AEM.06511-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang J, Presley GN, Hammel KE, Ryu J-S, Menke JR, Figueroa M, Hu D, Orr G, Schilling JS. 2016. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc Natl Acad Sci U S A 113:10968–10973. doi: 10.1073/pnas.1608454113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hori C, Gaskell J, Igarashi K, Kersten P, Mozuch M, Samejima M, Cullen D. 2014. Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism's strategy for degrading lignocellulose. Appl Environ Microbiol 80:2062–2070. doi: 10.1128/AEM.03652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Miyauchi S, Navarro D, Grisel S, Chevret D, Berrin JG, Rosso MN. 2017. The integrative omics of white-rot fungus Pycnoporus coccineus reveals co-regulated CAZymes for orchestrated lignocellulose breakdown. PLoS One 12:e0175528. doi: 10.1371/journal.pone.0175528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kuuskeri J, Häkkinen M, Laine P, Smolander OP, Tamene F, Miettinen S, Nousiainen P, Kemell M, Auvinen P, Lundell T. 2016. Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: Growth on spruce wood and decay effect on lignocellulose. Biotechnol Biofuels 9:192. doi: 10.1186/s13068-016-0608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Benz JP, Chau BH, Zheng D, Bauer S, Glass NL, Somerville CR. 2014. A comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations. Mol Microbiol 91:275–299. doi: 10.1111/mmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee Y-H, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie X, Kües U, Hibbett DS, Hoffmeister D, Högberg N, Martin F, Grigoriev IV, Watkinson SC. 2011. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 153.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Choi C, Cichocki N, Clum A, Colpaert J, Copeland A, Costa MD, Doré J, Floudas D, Gay G, Girlanda M, Henrissat B, Herrmann S, Hess J, Högberg N, Johansson T, Khouja H-R, LaButti K, Lahrmann U, Levasseur A, Lindquist EA, Lipzen A, Marmeisse R, Martino E, Murat C, Ngan CY, Nehls U, Plett JM, Pringle A, Ohm RA, Perotto S, Peter M, Riley R, Rineau F, Ruytinx J, Salamov A, Shah F, Sun H, Tarkka M, Tritt A, Veneault-Fourrey C, Zuccaro A, Tunlid A, Grigoriev IV, Hibbett DS, Martin F, Martin F. 2015. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 47:410–415. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- 154.Ruiz-Dueñas FJ, Lundell T, Floudas D, Nagy LG, Barrasa JM, Hibbett DS, Martínez AT. 2013. Lignin-degrading peroxidases in Polyporales: an evolutionary survey based on 10 sequenced genomes. Mycologia 105:1428–1444. doi: 10.3852/13-059. [DOI] [PubMed] [Google Scholar]
- 155.Ferreira P, Carro J, Serrano A, Martinez AT. 2015. A survey of genes encoding H2O2-producing GMC oxidoreductases in 10 Polyporales genomes. Mycologia 107:1105–1119. doi: 10.3852/15-027. [DOI] [PubMed] [Google Scholar]
- 156.Baldrian P. 2006. Fungal laccases—occurrence and properties. FEMS Microbiol Rev 30:215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- 157.Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G. 2010. Laccases: a never-ending story. Cell Mol Life Sci 67:369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bento I, Silva CS, Chen Z, Martins LO, Lindley PF, Soares CM. 2010. Mechanisms underlying dioxygen reduction in laccases. Structural and modelling studies focusing on proton transfer. BMC Struct Biol 10:28. doi: 10.1186/1472-6807-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Barreca AM, Fabbrini M, Galli C, Gentili P, Ljunggren S. 2003. Laccase/mediated oxidation of a lignin model for improved delignification procedures. J Mol Catal B Enzym 26:105–110. doi: 10.1016/j.molcatb.2003.08.001. [DOI] [Google Scholar]
- 160.Crestini C, Jurasek L, Argyropoulos DS. 2003. On the mechanism of the laccase-mediator system in the oxidation of lignin. Chemistry 9:5371–5378. doi: 10.1002/chem.200304818. [DOI] [PubMed] [Google Scholar]
- 161.Eggert C, Temp U, Eriksson KE. 1996. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl Environ Microbiol 62:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brenelli L, Squina FM, Felby C, Cannella D. 2018. Laccase-derived lignin compounds boost cellulose oxidative enzymes AA9. Biotechnol Biofuels 11:10. doi: 10.1186/s13068-017-0985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zámocký M, Hofbauer S, Schaffner I, Gasselhuber B, Nicolussi A, Soudi M, Pirker KF, Furtmüller PG, Obinger C. 2015. Independent evolution of four heme peroxidase superfamilies. Arch Biochem Biophys 574:108–119. doi: 10.1016/j.abb.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zámocký M, Janeèek Š, Obinger C. 2017. Fungal Hybrid B heme peroxidases—unique fusions of a heme peroxidase domain with a carbohydrate-binding domain. Sci Rep 7:9393. doi: 10.1038/s41598-017-09581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pollegioni L, Tonin F, Rosini E. 2015. Lignin-degrading enzymes. FEBS J 282:1190–1213. doi: 10.1111/febs.13224. [DOI] [PubMed] [Google Scholar]
- 166.Guillén F, Martínez AT, Martínez MJ. 1992. Substrate specificity and properties of the aryl-alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem 209:603–611. doi: 10.1111/j.1432-1033.1992.tb17326.x. [DOI] [PubMed] [Google Scholar]
- 167.Kelley RL, Reddy CA. 1986. Purification and characterization of glucose oxidase from ligninolytic cultures of Phanerochaete chrysosporium. J Bacteriol 166:269–274. doi: 10.1128/jb.166.1.269-274.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nishida A, Eriksson KE. 1987. Formation, purification, and partial characterization of methanol oxidase, a H2O2-producing enzyme in Phanerochaete chrysosporium. Biotechnol Appl Biochem 9:325–338. [Google Scholar]
- 169.Daniel G, Volc J, Kubatova E. 1994. Pyranose oxidase, a major source of H2O2 during wood degradation by Phanerochaete chrysosporium, Trametes versicolor, and Oudemansiella mucida. Appl Environ Microbiol 60:2524–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kersten PJ. 1990. Glyoxal oxidase of Phanerochaete chrysosporium: Its characterization and activation by lignin peroxidase. Proc Natl Acad Sci U S A 87:2936–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kersten PJ, Kirk TK. 1987. Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J Bacteriol 169:2195–2201. doi: 10.1128/jb.169.5.2195-2201.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yin D, Urresti S, Lafond M, Johnston EM, Derikvand F, Ciano L, Berrin J-G, Henrissat B, Walton PH, Davies GJ, Brumer H. 2015. Structure–function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family. Nat Commun 6:10197. doi: 10.1038/ncomms10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lee MH, Lai WL, Lin SF, Hsu CS, Liaw SH, Tsai YC. 2005. Structural characterization of glucooligosaccharide oxidase from Acremonium strictum. Appl Environ Microbiol 71:8881–8887. doi: 10.1128/AEM.71.12.8881-8887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vuong TV, Vesterinen A-H, Foumani M, Juvonen M, Seppälä J, Tenkanen M, Master ER. 2013. Xylo- and cello-oligosaccharide oxidation by gluco-oligosaccharide oxidase from Sarocladium strictum and variants with reduced substrate inhibition. Biotechnol Biofuels 6:148. doi: 10.1186/1754-6834-6-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, Hammel KE, Vanden Wymelenberg A, Gaskell J, Lindquist E, Sabat G, Splinter BonDurant S, Larrondo LF, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro AG, Lavín JL, Oguiza JA, Master E, Henrissat B, Coutinho PM, Harris P, Magnuson JK, Baker SE, Bruno K, Kenealy W, Hoegger PJ, Kües U, Ramaiya P, Lucas S, Salamov A, Shapiro H, Tu H, Chee CL, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev IV, Brettin T, Rokhsar D, Berka R, Cullen D. 2009. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci U S A 106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ander P, Eriksson MER, Eriksson K. 1985. Methanol production from lignin-related substances by Phanerochaete chrysosporium. Physiol Plant 65:317–321. doi: 10.1111/j.1399-3054.1985.tb02402.x. [DOI] [Google Scholar]
- 177.Daniel G, Volc J, Filonova L, Plíhal O, Kubátová E, Halada P. 2007. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl Environ Microbiol 73:6241–6253. doi: 10.1128/AEM.00977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hernández-Ortega A, Ferreira P, Martínez AT. 2012. Fungal aryl-alcohol oxidase: a peroxide-producing flavoenzyme involved in lignin degradation. Appl Microbiol Biotechnol 93:1395–1410. doi: 10.1007/s00253-011-3836-8. [DOI] [PubMed] [Google Scholar]
- 179.Giffhorn F. 2000. Fungal pyranose oxidases: Occurrence, properties and biotechnical applications in carbohydrate chemistry. Appl Microbiol Biotechnol 54:727–740. doi: 10.1007/s002530000446. [DOI] [PubMed] [Google Scholar]
- 180.Janssen FW, Ruelius HW. 1968. Carbohydrate oxidase, a novel enzyme from Polyporus obtusus. II. Specificity and characterization of reaction products. Biochim Biophys Acta 167:501–510. [DOI] [PubMed] [Google Scholar]
- 181.Leitner C, Volc J, Haltrich D. 2001. Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor. Appl Environ Microbiol 67:3636–3644. doi: 10.1128/AEM.67.8.3636-3644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pisanelli I, Kujawa M, Spadiut O, Kittl R, Halada P, Volc J, Mozuch MD, Kersten P, Haltrich D, Peterbauer C. 2009. Pyranose 2-oxidase from Phanerochaete chrysosporium-Expression in E. coli and biochemical characterization. J Biotechnol 142:97–106. doi: 10.1016/j.jbiotec.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 183.Volc J, Kubátová E, Daniel G, Prikrylová V. 1996. Only C-2 specific glucose oxidase activity is expressed in ligninolytic cultures of the white rot fungus Phanerochaete chrysosporium. Arch Microbiol 165:421–424. doi: 10.1007/s002030050348. [DOI] [PubMed] [Google Scholar]
- 184.Nyanhongo GS, Gübitz G, Sukyai P, Leitner C, Haltrich D, Ludwig R. 2007. Oxidoreductases from Trametes spp. in biotechnology: a wealth of catalytic activity. Food Technol Biotechnol 45:250–268. [Google Scholar]
- 185.de Koker TH, Mozuch MD, Cullen D, Gaskell J, Kersten PJ. 2004. Isolation and purification of pyranose 2-oxidase from Phanerochaete chrysosporium and characterization of gene structure and regulation. Appl Environ Microbiol 70:5794–5800. doi: 10.1128/AEM.70.10.5794-5800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fraaije MW, Veeger C, van Berkel WJ. 1995. Substrate specificity of flavin-dependent vanillyl-alcohol oxidase from Penicillium simplicissimum. Evidence for the production of 4-hydroxycinnamyl alcohols from 4-allylphenols. Eur J Biochem 234:271–277. [DOI] [PubMed] [Google Scholar]
- 187.Fraaije MW, Van Den Heuvel RHH, Roelofs JCAA, Van Berkel WJH. 1998. Kinetic mechanism of vanillyl-alcohol oxidase with short-chain 4-alkylphenols. Eur J Biochem 253:712–719. doi: 10.1046/j.1432-1327.1998.2530712.x. [DOI] [PubMed] [Google Scholar]
- 188.Ewing TA, Fraaije MW, Mattevi A, Van Berkel WJH. 2017. The VAO/PCMH flavoprotein family. Arch Biochem Biophys 632:104–117. doi: 10.1016/j.abb.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 189.Fraaije MW, Pikkemaat M, Van Berkel WJH. 1997. Enigmatic gratuitous induction of the covalent flavoprotein vanillyl-alcohol oxidase in Penicillium simplicissimum. Appl Environ Microbiol 63:435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Daou M, Faulds CB. 2017. Glyoxal oxidases: their nature and properties. World J Microbiol Biotechnol 33:87. doi: 10.1007/s11274-017-2254-1. [DOI] [PubMed] [Google Scholar]
- 191.Whittaker MM, Kersten PJ, Nakamura N, Sanders-Loehr J, Schweizer ES, Whittaker JW. 1996. Glyoxal oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J Biol Chem 271:681–687. doi: 10.1074/jbc.271.2.681. [DOI] [PubMed] [Google Scholar]
- 192.Lin SF, Yang TY, Inukai T, Yamasaki M, Tsai YC. 1991. Purification and characterization of a novel glucooligosaccharide oxidase from Acremonium strictum T1. Biochim Biophys Acta 1118:41–47. doi: 10.1016/0167-4838(91)90439-7. [DOI] [PubMed] [Google Scholar]
- 193.Garajova S, Mathieu Y, Beccia MR, Bennati-Granier C, Biaso F, Fanuel M, Ropartz D, Guigliarelli B, Record E, Rogniaux H, Henrissat B, Berrin J-G. 2016. Single-domain flavoenzymes trigger lytic polysaccharide monooxygenases for oxidative degradation of cellulose. Sci Rep 6:28276. doi: 10.1038/srep28276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kirk TK, Tien M, Johnsrud SC, Eriksson KE. 1986. Lignin degrading activity of Phanerochaete chrysosporium Burds.: comparison of cellulase-negative and other strains. Enzyme Microb Technol 8:75–80. doi: 10.1016/0141-0229(86)90074-8. [DOI] [Google Scholar]
- 195.Buswell JA, Mollet B, Odier E. 1984. Ligninolytic enzyme production by Phanerochaete chrysosporium under conditions of nitrogen sufficiency. FEMS Microbiol Lett 25:295–299. doi: 10.1111/j.1574-6968.1984.tb01475.x. [DOI] [Google Scholar]
- 196.Vos AM, Jurak E, Pelkmans JF, Herman K, Pels G, Baars JJ, Hendrix E, Kabel MA, Lugones LG, Wösten HAB. 2017. H2O2 as a candidate bottleneck for MnP activity during cultivation of Agaricus bisporus in compost. AMB Express 7:124. doi: 10.1186/s13568-017-0424-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Roth EM, Shanley ES. 1953. Stability of pure hydrogen peroxide. Ind Eng Chem 45:2343–2349. doi: 10.1021/ie50526a053. [DOI] [Google Scholar]
- 198.Rodriguez-Lopez JN, Gilabert MA, Tudela J, Thorneley RNF, Garcia-Canovas F. 2000. Reactivity of horseradish peroxidase compound II toward substrates: Kinetic evidence for a two-step mechanism. Biochemistry 39:13201–13209. doi: 10.1021/bi001150p. [DOI] [PubMed] [Google Scholar]
- 199.Zamocky M, Furtmüller PG, Obinger C. 2008. Evolution of catalases from bacteria to humans. Antioxid Redox Signal 10:1527–1548. doi: 10.1089/ars.2008.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Switala J, Loewen PC. 2002. Diversity of properties among catalases. Arch Biochem Biophys 401:145–154. doi: 10.1016/S0003-9861(02)00049-8. [DOI] [PubMed] [Google Scholar]
- 201.Díaz A, Valdés V-J, Rudiño-Piñera E, Horjales E, Hansberg W. 2009. Structure-function relationships in fungal large-subunit catalases. J Mol Biol 386:218–232. doi: 10.1016/j.jmb.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 202.Smulevich G, Jakopitsch C, Droghetti E, Obinger C. 2006. Probing the structure and bifunctionality of catalase-peroxidase (KatG). J Inorg Biochem 100:568–585. doi: 10.1016/j.jinorgbio.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 203.Mishra S, Imlay J. 2012. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys 525:145–160. doi: 10.1016/j.abb.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hrycay EG, Bandiera SM. 2015. Monooxygenase, peroxidase and peroxygenase properties and reaction mechanisms of cytochrome P450 enzymes. Adv Exp Med Biol 851:1–61. doi: 10.1007/978-3-319-16009-2_1. [DOI] [PubMed] [Google Scholar]
- 205.Singh R, Wiseman B, Deemagarn T, Jha V, Switala J, Loewen PC. 2008. Comparative study of catalase-peroxidases (KatGs). Arch Biochem Biophys 471:207–214. doi: 10.1016/j.abb.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 206.Busse N, Wagner D, Kraume M, Czermak P. 2013. Reaction kinetics of versatile peroxidase for the degradation of lignin compounds. Am J Biochem Biotechnol 9:365–394. doi: 10.3844/ajbbsp.2013.365.394. [DOI] [Google Scholar]
- 207.Domínguez L, Sosa-Peinado A, Hansberg W. 2010. Catalase evolved to concentrate H2O2 at its active site. Arch Biochem Biophys 500:82–91. doi: 10.1016/j.abb.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 208.Hansberg W, Salas-Lizana R, Domínguez L. 2012. Fungal catalases: Function, phylogenetic origin and structure. Arch Biochem Biophys 525:170–180. doi: 10.1016/j.abb.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 209.Korripally P, Hunt CG, Houtman CJ, Jones DC, Kitin PJ, Cullen D, Hammel KE. 2015. Regulation of gene expression during the onset of ligninolytic oxidation by Phanerochaete chrysosporium on spruce wood. Appl Environ Microbiol 81:7802–7812. doi: 10.1128/AEM.02064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bourdais A, Bidard F, Zickler D, Berteaux-Lecellier V, Silar P, Espagne E. 2012. Wood utilization is dependent on catalase activities in the filamentous fungus Podospora anserina. PLoS One 7:e29820. doi: 10.1371/journal.pone.0029820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zintel S, Bernhardt D, Rogowska-Wrzesinska A, Osiewacz HD. 2011. PaCATB, a secreted catalase protecting Podospora anserina against exogenous oxidative stress. Aging (Albany NY) 3:768–781. doi: 10.18632/aging.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. 2011. ROS signaling: the new wave? Trends Plant Sci 16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 213.Hancock J, Desikan R, Harrison J, Bright J, Hooley R, Neill S. 2006. Doing the unexpected: proteins involved in hydrogen peroxide perception. J Exp Bot 57:1711–1718. doi: 10.1093/jxb/erj180. [DOI] [PubMed] [Google Scholar]
- 214.D'Autréaux B, Toledano MB. 2007. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol 8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 215.Xu Y, Itzek A, Kreth J. 2014. Comparison of genes required for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis. Microbiology 160:2627–2638. doi: 10.1099/mic.0.082156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Samalova M, Meyer AJ, Gurr SJ, Fricker MD. 2014. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol 201:556–573. doi: 10.1111/nph.12530. [DOI] [PubMed] [Google Scholar]
- 217.Ivanova AE, Aslanidi KB, Karpenko YV, Belozerskaya TA. 2005. The effect of hydrogen peroxide on the growth of microscopic mycelial fungi isolated from habitats with different levels of radioactive contamination. Microbiology 74:655–663. doi: 10.1007/s11021-005-0120-x. [DOI] [PubMed] [Google Scholar]
- 218.Gessler NN, Aver'yanov AA, Belozerskaya TA. 2007. Reactive oxygen species in regulation of fungal development. Biochemistry (Mosc) 72:1091–1109. doi: 10.1134/S0006297907100070. [DOI] [PubMed] [Google Scholar]
- 219.Videira A, Kasuga T, Tian C, Lemos C, Castro A, Glass NL. 2009. Transcriptional analysis of the response of Neurospora crassa to phytosphingosine reveals links to mitochondrial function. Microbiology 155:3134–3141. doi: 10.1099/mic.0.029710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Slesak I, Libik M, Karpinska B, Karpinski S, Miszalski Z. 2007. The role of hydrogen peroxide in regulation of plant metabolism and cellular signalling in response to environmental stresses. Acta Biochim Pol 54:39–50. [PubMed] [Google Scholar]
- 221.Neill S, Desikan R, Hancock J. 2002. Hydrogen peroxide signalling. Curr Opin Plant Biol 5:388–395. doi: 10.1016/S1369-5266(02)00282-0. [DOI] [PubMed] [Google Scholar]
- 222.Bienert GP, Schjoerring JK, Jahn TP. 2006. Membrane transport of hydrogen peroxide. Biochim Biophys Acta 1758:994–1003. doi: 10.1016/j.bbamem.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 223.Heller J, Tudzynski P. 2011. Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu Rev Phytopathol 49:369–390. doi: 10.1146/annurev-phyto-072910-095355. [DOI] [PubMed] [Google Scholar]
- 224.Torres MA, Jones JDG, Dangl JL. 2006. Reactive oxygen species signaling in response to pathogens. Plant Physiol 141:373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Torres MA. 2010. ROS in biotic interactions. Physiol Plant 138:414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 226.Govrin EM, Levine A. 2000. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10:751–757. doi: 10.1016/S0960-9822(00)00560-1. [DOI] [PubMed] [Google Scholar]
- 227.Jwa N-S, Hwang BK. 2017. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front Plant Sci 8:1687. doi: 10.3389/fpls.2017.01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.O'Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, Altmüller J, Alvarado-Balderrama L, Bauser CA, Becker C, Birren BW, Chen Z, Choi J, Crouch JA, Duvick JP, Farman MA, Gan P, Heiman D, Henrissat B, Howard RJ, Kabbage M, Koch C, Kracher B, Kubo Y, Law AD, Lebrun MH, Lee YH, Miyara I, Moore N, Neumann U, Nordström K, Panaccione DG, Panstruga R, Place M, Proctor RH, Prusky D, Rech G, Reinhardt R, Rollins JA, Rounsley S, Schardl CL, Schwartz DC, Shenoy N, Shirasu K, Sikhakolli UR, Stüber K, Sukno SA, Sweigard JA, Takano Y, Takahara H, Trail F, Van Der Does HC, Voll LM, Will I, Young S, Zeng Q, Zhang J, Zhou S, Dickman MB, Schulze-Lefert P, Ver Loren Van Themaat E, Ma LJ, Vaillancourt LJ. 2012. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Magbanua ZV, De Moraes CM, Brooks TD, Williams WP, Luthe DS. 2007. Is catalase activity one of the factors associated with maize resistance to Aspergillus flavus? Mol Plant Microbe Interact 20:697–706. doi: 10.1094/MPMI-20-6-0697. [DOI] [PubMed] [Google Scholar]
- 230.Rolke Y, Liu S, Quidde T, Williamson B, Schouten A, Weltring K-M, Siewers V, Tenberge KB, Tudzynski B, Tudzynski P. 2004. Functional analysis of H2O2-generating systems in Botrytis cinerea: the major Cu-Zn-superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol Plant Pathol 5:17–27. doi: 10.1111/j.1364-3703.2004.00201.x. [DOI] [PubMed] [Google Scholar]
- 231.Aichinger C, Schreier P, Leuthner B, Adamczewski M, Hillebrand S, Kuck K, Van Kan J, Visser J, Stefanato F, Kahmann R. July 2003. Fungal glyoxal oxidases. US patent 20,030,140,370.
- 232.Song XS, Xing S, Li HP, Zhang JB, Qu B, Jiang JH, Fan C, Yang P, Liu JL, Hu ZQ, Xue S, Liao YC. 2016. An antibody that confers plant disease resistance targets a membrane-bound glyoxal oxidase in Fusarium. New Phytol 210:997–1010. doi: 10.1111/nph.13806. [DOI] [PubMed] [Google Scholar]
- 233.Min K, Freeman C, Kang H, Choi S-U, Min K, Freeman C, Kang H, Choi S-U. 2015. The regulation by phenolic compounds of soil organic matter dynamics under a changing environment. Biomed Res Int 2015:825098. doi: 10.1155/2015/825098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Arantes V, Milagres AMF, Filley TR, Goodell B. 2011. Lignocellulosic polysaccharides and lignin degradation by wood decay fungi: the relevance of nonenzymatic Fenton-based reactions. J Ind Microbiol Biotechnol 38:541–555. doi: 10.1007/s10295-010-0798-2. [DOI] [PubMed] [Google Scholar]
- 235.Hoostal MJ, Bouzat JL. 2008. The modulating role of dissolved organic matter on spatial patterns of microbial metabolism in Lake Erie sediments. Microb Ecol 55:358–368. doi: 10.1007/s00248-007-9281-7. [DOI] [PubMed] [Google Scholar]
- 236.Sakihama Y, Cohen MF, Grace SC, Yamasaki H. 2002. Plant phenolic antioxidant and prooxidant activities: phenolics-induced oxidative damage mediated by metals in plants. Toxicology 177:67–80. doi: 10.1016/S0300-483X(02)00196-8. [DOI] [PubMed] [Google Scholar]
- 237.de Graft-Johnson J, Nowak D. 2016. Effect of selected plant phenolics on Fe2+-EDTA-H2O2 system mediated deoxyribose oxidation: molecular structure-derived relationships of anti- and pro-oxidant actions. Molecules 22:59. doi: 10.3390/molecules22010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Akagawa M, Shigemitsu T, Suyama K. 2003. Production of hydrogen peroxide by polyphenols and polyphenol-rich beverages under quasi-physiological conditions. Biosci Biotechnol Biochem 67:2632–2640. doi: 10.1271/bbb.67.2632. [DOI] [PubMed] [Google Scholar]
- 239.Goodell B, Jellison J, Liu J, Daniel G, Paszczynski A, Fekete F, Krishnamurthy S, Jun L, Xu G. 1997. Low molecular weight chelators and phenolic compounds isolated from wood decay fungi and their role in the fungal biodegradation of wood. J Biotechnol 53:133–162. doi: 10.1016/S0168-1656(97)01681-7. [DOI] [Google Scholar]
- 240.Erickson HP. 2009. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online 11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Flournoy DS, Kirk TK, Highley TL. 1991. Changes in pore structure and cell wall pore volume in wood decayed by brown-rot and white-rot fungi. Holzforschung 45:457–463. doi: 10.1515/hfsg.1991.45.5.383. [DOI] [Google Scholar]
- 242.Arantes V, Saddler JN. 2010. Access to cellulose limits the efficiency of enzymatic hydrolysis: the role of amorphogenesis. Biotechnol Biofuels 3:4. doi: 10.1186/1754-6834-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Xu G, Goodell B. 2001. Mechanisms of wood degradation by brown-rot fungi: chelator-mediated cellulose degradation and binding of iron by cellulose. J Biotechnol 87:43–57. doi: 10.1016/S0168-1656(00)00430-2. [DOI] [PubMed] [Google Scholar]
- 244.Arantes V, Milagres AMF. 2006. Degradation of cellulosic and hemicellulosic substrates using a chelator-mediated Fenton reaction. J Chem Technol Biotechnol 81:413–419. doi: 10.1002/jctb.1417. [DOI] [Google Scholar]
- 245.Schmidt CJ, Whitten BK, Nicholas DD. 1981. A proposed role for oxalic acid in non-enzymatic wood decay by brown-rot fungi. Proc Am Wood Preserv Assoc 77:157–163. [Google Scholar]
- 246.Wang W, Gao PJ. 2003. Function and mechanism of a low-molecular-weight peptide produced by Gloeophyllum trabeum in biodegradation of cellulose. J Biotechnol 101:119–130. doi: 10.1016/S0168-1656(02)00321-8. [DOI] [PubMed] [Google Scholar]
- 247.Suzuki MR, Hunt CG, Houtman CJ, Dalebroux ZD, Hammel KE. 2006. Fungal hydroquinones contribute to brown rot of wood. Environ Microbiol 8:2214–2223. doi: 10.1111/j.1462-2920.2006.01160.x. [DOI] [PubMed] [Google Scholar]
- 248.Shah F, Schwenk D, Nicolás C, Persson P, Hoffmeister D, Tunlid A. 2015. Involutin is an Fe3+ reductant secreted by the ectomycorrhizal fungus Paxillus involutus during Fenton-based decomposition of organic matter. Appl Environ Microbiol 81:8427–8433. doi: 10.1128/AEM.02312-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Halliwell B, Gutteridge JM. 1992. Biologically relevant metal ion-dependent hydroxyl radical generation. An update. FEBS Lett 307:108–112. doi: 10.1016/0014-5793(92)80911-Y. [DOI] [PubMed] [Google Scholar]
- 250.Henry WP. 2003. Non-enzymatic iron, manganese, and copper chemistry of potential importance in wood decay. Wood Deterior Preserv 845:175–195. doi: 10.1021/bk-2003-0845.ch010. [DOI] [Google Scholar]
- 251.Fenton HJH. 1894. LXXIII. Oxidation of tartaric acid in presence of iron. J Chem Soc Trans 65:899–910. [Google Scholar]
- 252.Halliwell B. 1978. Superoxide-dependent formation of hydroxyl radicals in the presence of iron chelates: is it a mechanism for hydroxyl radical production in biochemical systems? FEBS Lett 92:321–326. doi: 10.1016/0014-5793(78)80779-0. [DOI] [PubMed] [Google Scholar]
- 253.Franco Cairo JPL, Carazzolle MF, Leonardo FC, Mofatto LS, Brenelli LB, Gonçalves TA, Uchima CA, Domingues RR, Alvarez TM, Tramontina R, Vidal RO, Costa FF, Costa-Leonardo AM, Paes Leme AF, Pereira GAG, Squina FM. 2016. Expanding the knowledge on lignocellulolytic and redox enzymes of worker and soldier castes from the lower termite Coptotermes gestroi. Front Microbiol 7:1518. doi: 10.3389/fmicb.2016.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Barbehenn R, Dodick T, Poopat U, Spencer B. 2005. Fenton-type reactions and iron concentrations in the midgut fluids of tree-feeding caterpillars. Arch Insect Biochem Physiol 60:32–43. doi: 10.1002/arch.20079. [DOI] [PubMed] [Google Scholar]
- 255.Ma J, Zhang K, Huang M, Hector SB, Liu B, Tong C, Liu Q, Zeng J, Gao Y, Xu T, Liu Y, Liu X, Zhu Y. 2016. Involvement of Fenton chemistry in rice straw degradation by the lignocellulolytic bacterium Pantoea ananatis Sd-1. Biotechnol Biofuels 9:211. doi: 10.1186/s13068-016-0623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Martinez D, Larrondo LF, Putnam N, Sollewijn Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 257.Ohm RA, Riley R, Salamov A, Min B, Choi IG, Grigoriev IV. 2014. Genomics of wood-degrading fungi. Fungal Genet Biol 72:82–90. doi: 10.1016/j.fgb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 258.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Samal A, Craig JP, Coradetti ST, Benz JP, Eddy JA, Price ND, Glass NL. 2017. Network reconstruction and systems analysis of plant cell wall deconstruction by Neurospora crassa. Biotechnol Biofuels 10:225. doi: 10.1186/s13068-017-0901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Horta MAC, Filho JAF, Murad NF, De Oliveira Santos E, Dos Santos CA, Mendes JS, Brandão MM, Azzoni SF, De Souza AP. 2018. Network of proteins, enzymes and genes linked to biomass degradation shared by Trichoderma species. Sci Rep 8:1341. doi: 10.1038/s41598-018-19671-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Amore A, Giacobbe S, Faraco V. 2013. Regulation of cellulase and hemicellulase gene expression in fungi. Curr Genomics 14:230–249. doi: 10.2174/1389202911314040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Ståhlberg J, Beckham GT. 2015. Fungal Cellulases. Chem Rev 115:1308–1448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- 263.Rytioja J, Hildén K, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR. 2014. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol Mol Biol Rev 78:614–649. doi: 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kubicek CP, Starr TL, Glass NL. 2014. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol 52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 265.Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, Porcel BM, Couloux A, Aury J-M, Ségurens B, Poulain J, Anthouard V, Grossetete S, Khalili H, Coppin E, Déquard-Chablat M, Picard M, Contamine V, Arnaise S, Bourdais A, Berteaux-Lecellier V, Gautheret D, de Vries RP, Battaglia E, Coutinho PM, Danchin EG, Henrissat B, Khoury REL, Sainsard-Chanet A, Boivin A, Pinan-Lucarré B, Sellem CH, Debuchy R, Wincker P, Weissenbach J, Silar P. 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol 9:R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Battaglia E, Benoit I, van den Brink J, Wiebenga A, Coutinho PM, Henrissat B, de Vries RP. 2011. Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: a highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genomics 12:38. doi: 10.1186/1471-2164-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier JM, Quévillon E, Sharon A, Simon A, Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun MH, Dickman M. 2011. Genomic analysis of the necrotrophic fungal pathogens sclerotinia sclerotiorum and botrytis cinerea. PLoS Genet 7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Pel HJ, De Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, De Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, Van Den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, Van Dijck PWM, Van Dijk A, Dijkhuizen L, Driessen AJM, D'Enfert C, Geysens S, Goosen C, Groot GSP, De Groot PWJ, Guillemette T, Henrissat B, Herweijer M, Van Den Hombergh JPTW, Van Den Hondel CAMJJ, Van Der Heijden RTJM, Van Der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, Van Kuyk PA, Lauber J, Lu X, Van Der Maarel MJEC, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, Van Peij NNME, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, Van De Vondervoort PJJ, Wedler H, Wösten HAB, Zeng AP, Van Ooyen AJJ, Visser J, Stam H. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 269.Benoit I, Culleton H, Zhou M, DiFalco M, Aguilar-Osorio G, Battaglia E, Bouzid O, Brouwer CPJM, El-Bushari HBO, Coutinho PM, Gruben BS, Hildén KS, Houbraken J, Barboza LAJ, Levasseur A, Majoor E, Mäkelä MR, Narang HM, Trejo-Aguilar B, Van Den Brink J, VanKuyk PA, Wiebenga A, McKie V, McCleary B, Tsang A, Henrissat B, De Vries RP. 2015. Closely related fungi employ diverse enzymatic strategies to degrade plant biomass. Biotechnol Biofuels 8:107. doi: 10.1186/s13068-015-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Rytioja J, Hildén K, Di Falco M, Zhou M, Aguilar-Pontes MV, Sietiö OM, Tsang A, de Vries RP, Mäkelä MR. 2017. The molecular response of the white-rot fungus Dichomitus squalens to wood and non-woody biomass as examined by transcriptome and exoproteome analyses. Environ Microbiol 19:1237–1250. doi: 10.1111/1462-2920.13652. [DOI] [PubMed] [Google Scholar]
- 271.Peng M, Aguilar-Pontes MV, Hainaut M, Henrissat B, Hildén K, Mäkelä MR, de Vries RP. 2018. Comparative analysis of basidiomycete transcriptomes reveals a core set of expressed genes encoding plant biomass degrading enzymes. Fungal Genet Biol 112:40–46. doi: 10.1016/j.fgb.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 272.Blanchette RA. 1991. Delignification by wood-decay fungi. Annu Rev Phytopathol 29:381–403. doi: 10.1146/annurev.py.29.090191.002121. [DOI] [Google Scholar]
- 273.Vanden Wymelenberg A, Gaskell J, Mozuch M, Kersten P, Sabat G, Martinez D, Cullen D. 2009. Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl Environ Microbiol 75:4058–4068. doi: 10.1128/AEM.00314-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Vanden Wymelenberg A, Gaskell J, Mozuch M, Sabat G, Ralph J, Skyba O, Mansfield SD, Blanchette RA, Martinez D, Grigoriev I, Kersten PJ, Cullen D. 2010. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl Environ Microbiol 76:3599–3610. doi: 10.1128/AEM.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Otjen L, Blanchette R, Effland M, Leatham G. 1987. Assessment of 30 white rot basidiomycetes for selective lignin degradation. Holzforschung 41:343–349. doi: 10.1515/hfsg.1987.41.6.343. [DOI] [Google Scholar]
- 276.Fernandez-Fueyo E, Ruiz-Duenas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, Polanco R, Tello M, Honda Y, Watanabe T, Watanabe T, Ryu JS, Kubicek CP, Schmoll M, Gaskell J, Hammel KE, St John FJ, Vanden Wymelenberg A, Sabat G, Splinter BonDurant S, Syed K, Yadav JS, Doddapaneni H, Subramanian V, Lavin JL, Oguiza JA, Perez G, Pisabarro AG, Ramirez L, Santoyo F, Master E, Coutinho PM, Henrissat B, Lombard V, Magnuson JK, Kues U, Hori C, Igarashi K, Samejima M, Held BW, Barry KW, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Riley R, Salamov AA, Hoffmeister D, Schwenk D, Hadar Y, Yarden O, de Vries RP, Wiebenga A, Stenlid J, Eastwood D, Grigoriev IV, Berka RM, Blanchette RA, Kersten P, Martinez AT, Vicuna R, Cullen D. 2012. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc Natl Acad Sci U S A 109:5458–5463. doi: 10.1073/pnas.1119912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hammel KE, Cullen D. 2008. Role of fungal peroxidases in biological ligninolysis. Curr Opin Plant Biol 11:349–355. doi: 10.1016/j.pbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 278.Paszczynski A, Huynh V-B, Crawford R. 1985. Enzymatic activities of an extracellular, manganese-dependent peroxidase from Phanerochaete chrysosporium. FEMS Microbiol Lett 29:37–41. doi: 10.1111/j.1574-6968.1985.tb00831.x. [DOI] [Google Scholar]
- 279.Kapich AN, Jensen KA, Hammel KE. 1999. Peroxyl radicals are potential agents of lignin biodegradation. FEBS Lett 461:115–119. doi: 10.1016/S0014-5793(99)01432-5. [DOI] [PubMed] [Google Scholar]
- 280.Hori C, Suzuki H, Igarashi K, Samejima M. 2012. Transcriptional response of the cellobiose dehydrogenase gene to cello- and xylooligosaccharides in the basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol 78:3770–3773. doi: 10.1128/AEM.00150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Borin GP, Sanchez CC, de Santana ES, Zanini GK, dos Santos RAC, de Oliveira Pontes A, de Souza AT, Menegaldo Tavares Soares Dal'Mas RM, Riaño-Pachón DM, Goldman GH, Oliveira JVC. 2017. Comparative transcriptome analysis reveals different strategies for degradation of steam-exploded sugarcane bagasse by Aspergillus niger and Trichoderma reesei. BMC Genomics 18:501. doi: 10.1186/s12864-017-3857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Cosgrove DJ. 2015. Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25:162–172. doi: 10.1016/j.pbi.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Nikolaidis N, Doran N, Cosgrove DJ. 2014. Plant expansins in bacteria and fungi: Evolution by horizontal gene transfer and independent domain fusion. Mol Biol Evol 31:376–386. doi: 10.1093/molbev/mst206. [DOI] [PubMed] [Google Scholar]
- 284.Couturier M, Navarro D, Chevret D, Henrissat B, Piumi F, Ruiz-Dueñas FJ, Martinez AT, Grigoriev IV, Riley R, Lipzen A, Berrin JG, Master ER, Rosso MN. 2015. Enhanced degradation of softwood versus hardwood by the white-rot fungus Pycnoporus coccineus. Biotechnol Biofuels 8:216. doi: 10.1186/s13068-015-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Reisky L, Büchsenschütz HC, Engel J, Schweder T, Hehemann J-H, Bornscheuer UT. 2018. P450 monooxygenases catalyze the oxidative demethylation of algal carbohydrates. Nat Chem Biol 14:342–344. doi: 10.1038/s41589-018-0005-8. [DOI] [PubMed] [Google Scholar]
- 286.Syed K, Nelson DR, Riley R, Yadav JS. 2013. Genomewide annotation and comparative genomics of cytochrome P450 monooxygenases (P450s) in the polypore species Bjerkandera adusta, Ganoderma sp. and Phlebia brevispora. Mycologia 105:1445–1455. doi: 10.3852/13-002. [DOI] [PubMed] [Google Scholar]
- 287.Presley GN, Schilling JS. 2017. Distinct growth and secretome strategies for two taxonomically divergent brown rot fungi. Appl Environ Microbiol 83:e02987-16. doi: 10.1128/AEM.02987-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Grantham NJ, Wurman-Rodrich J, Terrett OM, Lyczakowski JJ, Stott K, Iuga D, Simmons TJ, Durand-Tardif M, Brown SP, Dupree R, Busse-Wicher M, Dupree P. 2017. An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat Plants 3:859–865. doi: 10.1038/s41477-017-0030-8. [DOI] [PubMed] [Google Scholar]
- 289.Simmons TJ, Mortimer JC, Bernardinelli OD, Pöppler A-C, Brown SP, deAzevedo ER, Dupree R, Dupree P. 2016. Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid-state NMR. Nat Commun 7:13902. doi: 10.1038/ncomms13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Martínez ÁT, Ruiz-Dueñas FJ, Martínez MJ, del Río JC, Gutiérrez A. 2009. Enzymatic delignification of plant cell wall: from nature to mill. Curr Opin Biotechnol 20:348–357. doi: 10.1016/j.copbio.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 291.Kumar AK, Sharma S. 2017. Recent updates on different methods of pretreatment of lignocellulosic feedstocks: a review. Bioresour Bioprocess 4:7. doi: 10.1186/s40643-017-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Alvira P, Tomás-Pejó E, Ballesteros M, Negro MJ. 2010. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: a review. Bioresour Technol 101:4851–4861. doi: 10.1016/j.biortech.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 293.Gupta VK, Kubicek CP, Berrin JG, Wilson DW, Couturier M, Berlin A, Filho EXF, Ezeji T. 2016. Fungal enzymes for bio-products from sustainable and waste biomass. Trends Biochem Sci 41:633–645. doi: 10.1016/j.tibs.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 294.Sindhu R, Binod P, Pandey A. 2016. Biological pretreatment of lignocellulosic biomass—an overview. Bioresour Technol 199:76–82. doi: 10.1016/j.biortech.2015.08.030. [DOI] [PubMed] [Google Scholar]
- 295.Vasco-Correa J, Ge X, Li Y. 2016. Biological pretreatment of lignocellulosic biomass, p 561–585. In Mussatto SI. (ed), Biomass fractionation technologies for a lignocellulosic feedstock based biorefinery. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 296.Salvachúa D, Karp EM, Nimlos CT, Vardon DR, Beckham GT. 2015. Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem 17:4951–4967. doi: 10.1039/C5GC01165E. [DOI] [Google Scholar]
- 297.Salvachúa D, Katahira R, Cleveland NS, Khanna P, Resch MG, Black BA, Purvine SO, Zink EM, Prieto A, Martínez MJ, Martínez AT, Simmons BA, Gladden JM, Beckham GT. 2016. Lignin depolymerization by fungal secretomes and a microbial sink. Green Chem 18:6046–6062. doi: 10.1039/C6GC01531J. [DOI] [Google Scholar]
- 298.Wan C, Li Y. 2011. Effectiveness of microbial pretreatment by Ceriporiopsis subvermispora on different biomass feedstocks. Bioresour Technol 102:7507–7512. doi: 10.1016/j.biortech.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 299.Suhara H, Kodama S, Kamei I, Maekawa N, Meguro S. 2012. Screening of selective lignin-degrading basidiomycetes and biological pretreatment for enzymatic hydrolysis of bamboo culms. Int Biodeterior Biodegrad 75:176–180. doi: 10.1016/j.ibiod.2012.05.042. [DOI] [Google Scholar]
- 300.Kong W, Fu X, Wang L, Alhujaily A, Zhang J, Ma F, Zhang X, Yu H. 2017. A novel and efficient fungal delignification strategy based on versatile peroxidase for lignocellulose bioconversion. Biotechnol Biofuels 10:218. doi: 10.1186/s13068-017-0906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Brethauer S, Robert Lawrence S, Michael Hans-Peter S. 2017. Enhanced simultaneous saccharification and fermentation of pretreated beech wood by in situ treatment with the white rot fungus Irpex lacteus in a membrane aerated biofilm reactor. Bioresour Technol 237:135–138. doi: 10.1016/j.biortech.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 302.Asgher M, Ahmad Z, Iqbal HMN. 2013. Alkali and enzymatic delignification of sugarcane bagasse to expose cellulose polymers for saccharification and bio-ethanol production. Ind Crops Prod 44:488–495. doi: 10.1016/j.indcrop.2012.10.005. [DOI] [Google Scholar]
- 303.Sanchez O, Sierra R, Carlos J. 2011. Delignification process of agro-industrial wastes an alternative to obtain fermentable carbohydrates for producing fuel, 111–154. In Manzanera M. (ed), Alternative fuel. InTech Europe, Rijeka, Croatia. [Google Scholar]
- 304.Arantes V, Jellison J, Goodell B. 2012. Peculiarities of brown-rot fungi and biochemical Fenton reaction with regard to their potential as a model for bioprocessing biomass. Appl Microbiol Biotechnol 94:323–338. doi: 10.1007/s00253-012-3954-y. [DOI] [PubMed] [Google Scholar]
- 305.Enoki A, Itakura S, Tanaka H. 1997. The involvement of extracelluar substances for reducing molecular oxygen to hydroxyl radical and ferric iron to ferrous iron in wood degradation by wood decay fungi. J Biotechnol 53:265–272. doi: 10.1016/S0168-1656(97)01682-9. [DOI] [Google Scholar]
- 306.Jain P, Vigneshwaran N. 2012. Effect of Fenton's pretreatment on cotton cellulosic substrates to enhance its enzymatic hydrolysis response. Bioresour Technol 103:219–226. doi: 10.1016/j.biortech.2011.09.110. [DOI] [PubMed] [Google Scholar]
- 307.Bhange VP, William SP, Sharma A, Gabhane J, Vaidya AN, Wate SR. 2015. Pretreatment of garden biomass using Fenton's reagent: influence of Fe2+ and H2O2 concentrations on lignocellulose degradation. J Environ Heal Sci Eng 13:12. doi: 10.1186/s40201-015-0167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Jung YH, Kim HK, Park HM, Park YC, Park K, Seo JH, Kim KH. 2015. Mimicking the Fenton reaction-induced wood decay by fungi for pretreatment of lignocellulose. Bioresour Technol 179:467–472. doi: 10.1016/j.biortech.2014.12.069. [DOI] [PubMed] [Google Scholar]
- 309.Oliva JM, Manzanares P, Ballesteros I, Negro MJ, González A, Ballesteros M. 2005. Application of Fenton's reaction to steam explosion prehydrolysates from poplar biomass. Appl Biochem Biotechnol 121–124:887–899. [DOI] [PubMed] [Google Scholar]
- 310.Kato DM, Elía N, Flythe M, Lynn BC. 2014. Pretreatment of lignocellulosic biomass using Fenton chemistry. Bioresour Technol 162:273–278. doi: 10.1016/j.biortech.2014.03.151. [DOI] [PubMed] [Google Scholar]
- 311.Aden A, Foust T. 2009. Technoeconomic analysis of the dilute sulfuric acid and enzymatic hydrolysis process for the conversion of corn stover to ethanol. Cellulose 16:535–545. doi: 10.1007/s10570-009-9327-8. [DOI] [Google Scholar]
- 312.Humbird D, Davis R, Tao L, Kinchin C, Hsu D, Aden A, Schoen P, Lukas J, Olthof B, Worley M, Sexton D, Dudgeon D. 2011. Process design and economics for biochemical conversion of lignocellulosic biomass to ethanol: dilute-acid pretreatment and enzymatic hydrolysis of corn stover. Technical report. National Renewable Energy Laboratory, Golden, CO: https://www.nrel.gov/docs/fy11osti/47764.pdf. [Google Scholar]
- 313.Joelsson E, Erdei B, Galbe M, Wallberg O. 2016. Techno-economic evaluation of integrated first- and second-generation ethanol production from grain and straw. Biotechnol Biofuels 9:1. doi: 10.1186/s13068-015-0423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Bischof RH, Ramoni J, Seiboth B. 2016. Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb Cell Fact 15:106. doi: 10.1186/s12934-016-0507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Verardi A, Bari De I, Ricca E, Calabrò V. 2012. Hydrolysis of lignocellulosic biomass: current status of processes and technologies and future perspectives, p 95–122. In Lima MAP. (ed), Bioethanol. InTech Europe, Rijeka, Croatia. [Google Scholar]
- 316.Li S, Yang X, Yang S, Zhu M, Wang X. 2012. Technology prospecting on enzymes: application, marketing and engineering. Comput Struct Biotechnol J 2:e201209017. doi: 10.5936/csbj.201209017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Harris PV, Xu F, Kreel NE, Kang C, Fukuyama S. 2014. New enzyme insights drive advances in commercial ethanol production. Curr Opin Chem Biol 19:162–170. doi: 10.1016/j.cbpa.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 318.Eijsink VGH, Vaaje-Kolstad G, Vårum KM, Horn SJ. 2008. Towards new enzymes for biofuels: lessons from chitinase research. Trends Biotechnol 26:228–235. doi: 10.1016/j.tibtech.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 319.Hu J, Arantes V, Pribowo A, Gourlay K, Saddler JN. 2014. Substrate factors that influence the synergistic interaction of AA9 and cellulases during the enzymatic hydrolysis of biomass. Energy Environ Sci 7:2308. doi: 10.1039/C4EE00891J. [DOI] [Google Scholar]
- 320.Müller G, Várnai A, Johansen KS, Eijsink VGH, Horn SJ. 2015. Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnol Biofuels 8:187. doi: 10.1186/s13068-015-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Chylenski P, Petrović DM, Müller G, Dahlström M, Bengtsson O, Lersch M, Siika-aho M, Horn SJ, Eijsink VGH. 2017. Enzymatic degradation of sulfite-pulped softwoods and the role of LPMOs. Biotechnol Biofuels 10:177. doi: 10.1186/s13068-017-0862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Scott BR, Huang HZ, Frickman J, Halvorsen R, Johansen KS. 2016. Catalase improves saccharification of lignocellulose by reducing lytic polysaccharide monooxygenase-associated enzyme inactivation. Biotechnol Lett 38:425–434. doi: 10.1007/s10529-015-1989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Du J, Song W, Zhang X, Zhao J, Liu G, Qu Y. 2018. Differential reinforcement of enzymatic hydrolysis by adding chemicals and accessory proteins to high solid loading substrates with different pretreatments. Bioprocess Biosyst Eng 41:1153–1163. doi: 10.1007/s00449-018-1944-x. [DOI] [PubMed] [Google Scholar]
- 324.Hu J, Chandra R, Arantes V, Gourlay K, Susan van Dyk J, Saddler JN. 2015. The addition of accessory enzymes enhances the hydrolytic performance of cellulase enzymes at high solid loadings. Bioresour Technol 186:149–153. doi: 10.1016/j.biortech.2015.03.055. [DOI] [PubMed] [Google Scholar]
- 325.Kim IJ, Youn HJ, Kim KH. 2016. Synergism of an auxiliary activity 9 (AA9) from Chaetomium globosum with xylanase on the hydrolysis of xylan and lignocellulose. Process Biochem 51:1445–1451. doi: 10.1016/j.procbio.2016.06.017. [DOI] [Google Scholar]
- 326.Sanhueza C, Carvajal G, Soto-Aguilar J, Lienqueo ME, Salazar O. 2018. The effect of a lytic polysaccharide monooxygenase and a xylanase from Gloeophyllum trabeum on the enzymatic hydrolysis of lignocellulosic residues using a commercial cellulase. Enzyme Microb Technol 113:75–82. doi: 10.1016/j.enzmictec.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 327.Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE. 2014. Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- 328.Falade AO, Nwodo UU, Iweriebor BC, Green E, Mabinya LV, Okoh AI. 2017. Lignin peroxidase functionalities and prospective applications. Microbiologyopen 6:e00394. doi: 10.1002/mbo3.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kudanga T, Le Roes-Hill M. 2014. Laccase applications in biofuels production: Current status and future prospects. Appl Microbiol Biotechnol 98:6525–6542. doi: 10.1007/s00253-014-5810-8. [DOI] [PubMed] [Google Scholar]
- 330.Jönsson LJ, Palmqvist E, Nilvebrant NO, Hahn-Hägerdal B. 1998. Detoxification of wood hydrolysates with laccase and peroxidase from the white-rot fungus Trametes versicolor. Appl Microbiol Biotechnol 49:691–697. doi: 10.1007/s002530051233. [DOI] [Google Scholar]
- 331.Jurado M, Prieto A, Martínez-Alcalá Á, Martínez ÁT, Martínez MJ. 2009. Laccase detoxification of steam-exploded wheat straw for second generation bioethanol. Bioresour Technol 100:6378–6384. doi: 10.1016/j.biortech.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 332.Fillat Ú, Ibarra D, Eugenio M, Moreno A, Tomás-Pejó E, Martín-Sampedro R. 2017. Laccases as a potential tool for the efficient conversion of lignocellulosic biomass: a review. Fermentation 3:17. doi: 10.3390/fermentation3020017. [DOI] [Google Scholar]
- 333.Rytioja J, Hildén K, Mäkinen S, Vehmaanperä J, Hatakka A, Mäkelä MR. 2015. Saccharification of lignocelluloses by carbohydrate active enzymes of the white rot fungus Dichomitus squalens. PLoS One 10:e0145166. doi: 10.1371/journal.pone.0145166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Singh R, Hu J, Regner MR, Round JW, Ralph J, Saddler JN, Eltis LD. 2017. Enhanced delignification of steam-pretreated poplar by a bacterial laccase. Sci Rep 7:42121. doi: 10.1038/srep42121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Xu F, Quinlan J. December 2009. Methods for increasing enzymatic hydrolysis of cellulosic material. US patent 20,100,159,509 A1.
- 336.Salvachúa D, Prieto A, Martínez ÁT, Martínez MJ. 2013. Characterization of a novel dye-decolorizing peroxidase (DyP)-type enzyme from Irpex lacteus and its application in enzymatic hydrolysis of wheat straw. Appl Environ Microbiol 79:4316–4324. doi: 10.1128/AEM.00699-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Tramontina R, Franco Cairo JPL, Liberato MV, Mandelli F, Sousa A, Santos S, Rabelo SC, Campos B, Ienczak J, Ruller R, Damásio ARL, Squina FM. 2017. The Coptotermes gestroi aldo-keto reductase: a multipurpose enzyme for biorefinery applications. Biotechnol Biofuels 10:4. doi: 10.1186/s13068-016-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Bao W, Renganathan V. 1992. Cellobiose oxidase of Phanerochaete chrysosporium enhances crystalline cellulose degradation by cellulases. FEBS Lett 302:77–80. doi: 10.1016/0014-5793(92)80289-S. [DOI] [PubMed] [Google Scholar]
- 339.Bafana A, Dutt S, Kumar S, Ahuja PS. 2011. Superoxide dismutase: an industrial perspective. Crit Rev Biotechnol 31:65–76. doi: 10.3109/07388551.2010.490937. [DOI] [PubMed] [Google Scholar]
- 340.Scharf ME, Sethi A. May 2015. Termite superoxide dismutases and glutathione peroxidases for biomass conversion. WIPO patent WO2015069308A1.
- 341.Pereira GAG, Cairo JPL, Squina FM, Carazzole MF, Mandelli F, Pradella JGC, Alvares TM, Leonardo FC, Costa FF, Leonardo AMC, Tramontina R, Brenelli De Paiva LB, Gonçalves TA, Robl D. January 2017. Composition of lignocellulolytic enzymes, enzymatic conversion method and superoxide dismutase expression vector. WIPO patent WO2017011885A1 https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2017011885.
- 342.Rashid GMM, Taylor CR, Liu Y, Zhang X, Rea D, Fülöp V, Bugg TDH. 2015. Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation. ACS Chem Biol 10:2286–2294. doi: 10.1021/acschembio.5b00298. [DOI] [PubMed] [Google Scholar]
- 343.Sweeney M, Vlasenko E, Abbate E. July 2010. Methods for increasing hydrolysis of cellulosic material in the presence of cellobiose dehydrogenase. WIPO patent WO2010080532A1 https://worldwide.espacenet.com/publicationDetails/biblio?CC=WO&NR=2010080532A1&KC=A1&FT=D#.
- 344.Sigoillot J-C, Berrin JG, Bey M. April 2013. Compositions comprising cellobiose dehydrogenase from Pycnoporus cinnabarinus and their use for the degradation of lignocellulosic biomass. WIPO patent WO2013004377A2.
- 345.Bey M, Berrin JG, Poidevin L, Sigoillot JC. 2011. Heterologous expression of Pycnoporus cinnabarinus cellobiose dehydrogenase in Pichia pastoris and involvement in saccharification processes. Microb Cell Fact 10:113. doi: 10.1186/1475-2859-10-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Chen K, Liu X, Long L, Ding S. 2017. Cellobiose dehydrogenase from Volvariella volvacea and its effect on the saccharification of cellulose. Process Biochem 60:52–58. doi: 10.1016/j.procbio.2017.05.023. [DOI] [Google Scholar]
- 347.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Guillén D, Sánchez S, Rodríguez-Sanoja R. 2010. Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol 85:1241–1249. doi: 10.1007/s00253-009-2331-y. [DOI] [PubMed] [Google Scholar]
- 349.Várnai A, Siika-Aho M, Viikari L. 2013. Carbohydrate-binding modules (CBMs) revisited: Reduced amount of water counterbalances the need for CBMs. Biotechnol Biofuels 6:30. doi: 10.1186/1754-6834-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Jensen MS, Fredriksen L, MacKenzie AK, Pope PB, Leiros I, Chylenski P, Williamson AK, Christopeit T, Østby H, Vaaje-Kolstad G, Eijsink VGH. 2018. Discovery and characterization of a thermostable two-domain GH6 endoglucanase from a compost metagenome. PLoS One 13:e0197862. doi: 10.1371/journal.pone.0197862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Forsberg Z, Mackenzie AK, Sørlie M, Røhr ÅK, Helland R, Arvai AS, Vaaje-Kolstad G, Eijsink VGH. 2014. Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc Natl Acad Sci U S A 111:8446–8451. doi: 10.1073/pnas.1402771111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Arfi Y, Shamshoum M, Rogachev I, Peleg Y, Bayer EA. 2014. Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc Natl Acad Sci U S A 111:9109–9114. doi: 10.1073/pnas.1404148111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Takeda K, Matsumura H, Ishida T, Samejima M, Ohno H, Yoshida M, Igarashi K, Nakamura N. 2015. Characterization of a novel PQQ-dependent quinohemoprotein pyranose dehydrogenase from Coprinopsis cinerea classified into auxiliary activities family 12 in carbohydrate-active enzymes. PLoS One 10:e0115722. doi: 10.1371/journal.pone.0115722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Várnai A, Umezawa K, Yoshida M, Eijsink VGH. 2018. The pyrroloquinoline-quinone dependent pyranose dehydrogenase from Coprinopsis cinerea (CcPDH) drives lytic polysaccharide monooxygenase (LPMO) action. Appl Environ Microbiol 84:e00156-18. doi: 10.1128/AEM.00156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Crater J, Galleher C, Lievense J. 2017. Consultancy on large-scale submerged aerobic cultivation process design—final technical report. National Renewable Energy Laboratory, Golden, CO: https://www.nrel.gov/docs/fy17osti/67963.pdf. [Google Scholar]
- 356.Hannon JR, Bakker A, Lynd LR, Wyman CE. 2007. Comparing the scale-up of aerobic and anaerobic biological processes, abstr 649a. Proc Annu Meet Am Inst Chem Eng, Salt Lake City, UT, 4 to 9 November 2007. [Google Scholar]
- 357.Humbird D, Davis R, McMillan JD. 2017. Aeration costs in stirred-tank and bubble column bioreactors. Biochem Eng J 127:161–166. doi: 10.1016/j.bej.2017.08.006. [DOI] [Google Scholar]
- 358.Viikari L, Vehmaanperä J, Koivula A. 2012. Lignocellulosic ethanol: From science to industry. Biomass Bioenergy 46:13–24. doi: 10.1016/j.biombioe.2012.05.008. [DOI] [Google Scholar]
- 359.Jørgensen H, Vibe-Pedersen J, Larsen J, Felby C. 2007. Liquefaction of lignocellulose at high-solids concentrations. Biotechnol Bioeng 96:862–870. doi: 10.1002/bit.21115. [DOI] [PubMed] [Google Scholar]
- 360.Koppram R, Tomás-Pejó E, Xiros C, Olsson L. 2014. Lignocellulosic ethanol production at high-gravity: challenges and perspectives. Trends Biotechnol 32:46–53. doi: 10.1016/j.tibtech.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 361.Müller G, Chylenski P, Bissaro B, Eijsink VGH, Horn SJ. 2018. The impact of hydrogen peroxide supply on LPMO activity and overall saccharification efficiency of a commercial cellulase cocktail. Biotechnol Biofuels 11:209. doi: 10.1186/s13068-018-1199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Hernandez K, Berenguer-Murcia A, Rodrigues CR, Fernandez-Lafuente R. 2012. Hydrogen peroxide in biocatalysis. A dangerous liaison. Curr Org Chem 16:2652–2672. [Google Scholar]
- 363.Bankar SB, Bule MV, Singhal RS, Ananthanarayan L. 2009. Glucose oxidase—an overview. Biotechnol Adv 27:489–501. doi: 10.1016/j.biotechadv.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 364.Bormann S, Gomez Baraibar A, Ni Y, Holtmann D, Hollmann F. 2015. Specific oxyfunctionalisations catalysed by peroxygenases: opportunities, challenges and solutions. Catal Sci Technol 5:2038–2052. doi: 10.1039/C4CY01477D. [DOI] [Google Scholar]
- 365.Cannella D, Jørgensen H. 2014. Do new cellulolytic enzyme preparations affect the industrial strategies for high solids lignocellulosic ethanol production? Biotechnol Bioeng 111:59–68. doi: 10.1002/bit.25098. [DOI] [PubMed] [Google Scholar]
- 366.Müller G, Kalyani DC, Horn SJ. 2017. LPMOs in cellulase mixtures affect fermentation strategies for lactic acid production from lignocellulosic biomass. Biotechnol Bioeng 114:552–559. doi: 10.1002/bit.26091. [DOI] [PubMed] [Google Scholar]
- 367.Van Dyk JS, Pletschke BI. 2012. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes-Factors affecting enzymes, conversion and synergy. Biotechnol Adv 30:1458–1480. doi: 10.1016/j.biotechadv.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 368.Koshland DE., Jr 1953. Stereochemistry and the mechanism of enzymatic reactions. Biol Rev Camb Philos Soc 28:416–436. doi: 10.1111/j.1469-185X.1953.tb01386.x. [DOI] [Google Scholar]
- 369.Sinnott ML. 1990. Catalytic mechanism of enzymic glycosyl transfer. Chem Rev 90:1171–1202. doi: 10.1021/cr00105a006. [DOI] [Google Scholar]
- 370.Vuong TV, Wilson DB. 2010. Glycoside hydrolases: catalytic base/nucleophile diversity. Biotechnol Bioeng 107:195–205. doi: 10.1002/bit.22838. [DOI] [PubMed] [Google Scholar]
- 371.Rye CS, Withers SG. 2000. Glycosidase mechanisms. Curr Opin Chem Biol 4:573–580. doi: 10.1016/S1367-5931(00)00135-6. [DOI] [PubMed] [Google Scholar]
- 372.Bissaro B, Monsan P, Fauré R, O'Donohue M. 2015. Glycosynthesis in a waterworld: New insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem J 467:17–35. doi: 10.1042/BJ20141412. [DOI] [PubMed] [Google Scholar]
- 373.Hangasky JA, Marletta MA. 2018. A random-sequential kinetic mechanism for polysaccharide monooxygenases. Biochemistry 57:3191–3199. doi: 10.1021/acs.biochem.8b00129. [DOI] [PubMed] [Google Scholar]
- 374.Jönsson LJ, Martín C. 2016. Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol 199:103–112. doi: 10.1016/j.biortech.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 375.Zhai R, Hu J, Saddler JN. 2018. Extent of enzyme inhibition by phenolics derived from pretreated biomass Is significantly influenced by the size and carbonyl group content of the phenolics. ACS Sustain Chem Eng 6:3823–3829. doi: 10.1021/acssuschemeng.7b04178. [DOI] [Google Scholar]
- 376.Kump LR. 2008. The rise of atmospheric oxygen. Nature 451:277–278. doi: 10.1038/nature06587. [DOI] [PubMed] [Google Scholar]
- 377.McKay CP, Hartman H. 1991. Hydrogen peroxide and the evolution of oxygenic photosynthesis. Orig Life Evol Biosph 21:157–163. doi: 10.1007/BF01809444. [DOI] [PubMed] [Google Scholar]
- 378.Joo H, Lin Z, Arnold FH. 1999. Laboratory evolution of peroxide-mediated cytochrome P450 hydroxylation. Nature 399:670–673. doi: 10.1038/21395. [DOI] [PubMed] [Google Scholar]
- 379.Sabbadin F, Hemsworth GR, Ciano L, Henrissat B, Dupree P, Tryfona T, Marques RDS, Sweeney ST, Besser K, Elias L, Pesante G, Li Y, Dowle AA, Bates R, Gomez LD, Simister R, Davies GJ, Walton PH, Bruce NC, McQueen-Mason SJ. 2018. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat Commun 9:756. doi: 10.1038/s41467-018-03142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Kersten PJ, Kalyanaraman B, Hammel KE, Reinhammar B, Kirk TK. 1990. Comparison of lignin peroxidase, horseradish peroxidase and laccase in the oxidation of methoxybenzenes. Biochem J 268:475–480. doi: 10.1042/bj2680475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Saparrat MCN, Guillén F, Arambarri AM, Martínez AT, Martínez MJ. 2002. Induction, isolation, and characterization of two laccases from the white rot basidiomycete Coriolopsis rigida. Appl Environ Microbiol 68:1534–1540. doi: 10.1128/AEM.68.4.1534-1540.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Palmieri G, Cennamo G, Faraco V, Amoresano A, Sannia G, Giardina P. 2003. Atypical laccase isoenzymes from copper supplemented Pleurotus ostreatus cultures. Enzyme Microb Technol 33:220–230. doi: 10.1016/S0141-0229(03)00117-0. [DOI] [Google Scholar]
- 383.Fukushima Y, Kirk TK. 1995. Laccase component of the Ceriporiopsis subvermispora lignin-degrading system. Appl Environ Microbiol 61:872–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Tuisel H, Sinclair R, Bumpus JA, Ashbaugh W, Brock BJ, Aust SD. 1990. Lignin peroxidase H2 from Phanerochaete chrysosporium: Purification, characterization and stability to temperature and pH. Arch Biochem Biophys 279:158–166. doi: 10.1016/0003-9861(90)90476-F. [DOI] [PubMed] [Google Scholar]
- 385.Lundell T, Wever R, Floris R, Harvey P, Hatakka A, Brunow G, Schoemaker H. 1993. Lignin peroxidase L3 from Phlebia radiata. Pre-steady-state and steady-state studies with veratryl alcohol and a non-phenolic lignin model compound 1-(3,4-dimethoxyphenyl)-2-(2-methoxyphenoxy)propane-1,3-diol. Eur J Biochem 211:391–402. [DOI] [PubMed] [Google Scholar]
- 386.Miki Y, Calviño FR, Pogni R, Giansanti S, Ruiz-Dueñas FJ, Martínez MJ, Basosi R, Romero A, Martínez AT. 2011. Crystallographic, kinetic, and spectroscopic study of the first ligninolytic peroxidase presenting a catalytic tyrosine. J Biol Chem 286:15525–15534. doi: 10.1074/jbc.M111.220996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Kuan I-C, Johnson KA, Tiens M. 1993. Kinetic analysis of manganese peroxidase. The reaction with manganese complexes. J Biol Chem 268:20064–20070. [PubMed] [Google Scholar]
- 388.Fernández-Fueyo E, Ruiz-Dueñas FJ, Martínez AT. 2014. Engineering a fungal peroxidase that degrades lignin at very acidic pH. Biotechnol Biofuels 7:114. doi: 10.1186/1754-6834-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Rogers MS, Jones GD, Antonini G, Wilson MT, Brunori M. 1994. Electron transfer from Phanerochaete chrysosporium cellobiose oxidase to equine cytochrome c and Pseudomonas aeruginosa cytochrome c-551. Biochem J 298:329–334. doi: 10.1042/bj2980329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Jones GD, Wilson MT. 1988. Rapid kinetic studies of the reduction of cellobiose oxidase from the white-rot fungus Sporotrichum pulverulentum by cellobiose. Biochem J 256:713–718. doi: 10.1042/bj2560713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Igarashi K, Yoshida M, Matsumura H, Nakamura N, Ohno H, Samejima M, Nishino T. 2005. Electron transfer chain reaction of the extracellular flavocytochrome cellobiose dehydrogenase from the basidiomycete Phanerochaete chrysosporium. FEBS J 272:2869–2877. doi: 10.1111/j.1742-4658.2005.04707.x. [DOI] [PubMed] [Google Scholar]
- 392.Kracher D, Zahma K, Schulz C, Sygmund C, Gorton L, Ludwig R. 2015. Inter-domain electron transfer in cellobiose dehydrogenase: modulation by pH and divalent cations. FEBS J 282:3136–3148. doi: 10.1111/febs.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Gibson QH, Swoboda BEP, Massey V. 1964. Kinetics and mechanism of action of glucose oxidase. J Biol Chem 239:3927–3934. [PubMed] [Google Scholar]
- 394.Kalisz HM, Hecht HJ, Schomburg D, Schmid RD. 1991. Effects of carbohydrate depletion on the structure, stability and activity of glucose oxidase from Aspergillus niger. Biochim Biophys Acta 1080:138–142. doi: 10.1016/0167-4838(91)90140-U. [DOI] [PubMed] [Google Scholar]
- 395.van Stroe-Biezen SAM, Janssen APM, Janssen LJJ. 1994. A kinetic study of soluble glucose oxidase using a rotating-disc electrode. Bioelectrochem Bioenerg 33:55–60. doi: 10.1016/0302-4598(94)87032-2. [DOI] [Google Scholar]
- 396.Rando D, Kohring G-W, Giffhorn F. 1997. Production, purification and characterization of glucose oxidase from a newly isolated strain of Penicillium pinophilum. Appl Microbiol Biotechnol 48:34–40. doi: 10.1007/s002530051011. [DOI] [Google Scholar]
- 397.Simpson C, Jordaan J, Gardiner NS, Whiteley C. 2007. Isolation, purification and characterization of a novel glucose oxidase from Penicillium sp. CBS 120262 optimally active at neutral pH. Protein Expr Purif 51:260–266. doi: 10.1016/j.pep.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 398.Witt S, Singh M, Kalisz HM. 1998. Structural and kinetic properties of nonglycosylated recombinant Penicillium amagasakiense glucose oxidase expressed in Escherichia coli. Appl Environ Microbiol 64:1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Ferreira P, Medina M, Guillen F, Martinez MJ, van Berkel WJH, Martinez AT. 2005. Spectral and catalytic properties of aryl-alcohol oxidase, a fungal flavoenzyme acting on polyunsaturated alcohols. Biochem J 389:731–738. doi: 10.1042/BJ20041903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Couturier M, Mathieu Y, Li A, Navarro D, Drula E, Haon M, Grisel S, Ludwig R, Berrin JG. 2016. Characterization of a new aryl-alcohol oxidase secreted by the phytopathogenic fungus Ustilago maydis. Appl Microbiol Biotechnol 100:697–706. doi: 10.1007/s00253-015-7021-3. [DOI] [PubMed] [Google Scholar]
- 401.Piumi F, Levasseur A, Navarro D, Zhou S, Mathieu Y, Ropartz D, Ludwig R, Faulds CB, Record E. 2014. A novel glucose dehydrogenase from the white-rot fungus Pycnoporus cinnabarinus: production in Aspergillus niger and physicochemical characterization of the recombinant enzyme. Appl Microbiol Biotechnol 98:10105–10118. doi: 10.1007/s00253-014-5891-4. [DOI] [PubMed] [Google Scholar]
- 402.Sygmund C, Klausberger M, Felice AK, Ludwig R. 2011. Reduction of quinones and phenoxy radicals by extracellular glucose dehydrogenase from Glomerella cingulata suggests a role in plant pathogenicity. Microbiology 157:3203–3212. doi: 10.1099/mic.0.051904-0. [DOI] [PubMed] [Google Scholar]
- 403.Mathieu Y, Piumi F, Valli R, Aramburu JC, Ferreira P, Faulds CB, Record E. 2016. Activities of secreted aryl alcohol quinone oxidoreductases from Pycnoporus cinnabarinus provide insights into fungal degradation of plant biomass. Appl Environ Microbiol 82:2411–2423. doi: 10.1128/AEM.03761-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Sygmund C, Kittl R, Volc J, Halada P, Kubátová E, Haltrich D, Peterbauer CK. 2008. Characterization of pyranose dehydrogenase from Agaricus meleagris and its application in the C-2 specific conversion of d-galactose. J Biotechnol 133:334–342. doi: 10.1016/j.jbiotec.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 405.Krondorfer I, Lipp K, Brugger D, Staudigl P, Sygmund C, Haltrich D, Peterbauer CK. 2014. Engineering of pyranose dehydrogenase for increased oxygen reactivity. PLoS One 9:e91145. doi: 10.1371/journal.pone.0091145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Graf MMH, Weber S, Kracher D, Kittl R, Sygmund C, Ludwig R, Peterbauer C, Haltrich D. 2017. Characterization of three pyranose dehydrogenase isoforms from the litter-decomposing basidiomycete Leucoagaricus meleagris (syn. Agaricus meleagris). Appl Microbiol Biotechnol 101:2879–2891. doi: 10.1007/s00253-016-8051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Kujawa M, Volc J, Halada P, Sedmera P, Divne C, Sygmund C, Leitner C, Peterbauer C, Haltrich D. 2007. Properties of pyranose dehydrogenase purified from the litter-degrading fungus Agaricus xanthoderma. FEBS J 274:879–894. doi: 10.1111/j.1742-4658.2007.05634.x. [DOI] [PubMed] [Google Scholar]
- 408.de Oliveira BV, Teixeira GS, Reis O, Barau JG, Teixeira PJPL, do Rio MCS, Domingues RR, Meinhardt LW, Paes Leme AF, Rincones J, Pereira GAG. 2012. A potential role for an extracellular methanol oxidase secreted by Moniliophthora perniciosa in Witches' broom disease in cacao. Fungal Genet Biol 49:922–932. doi: 10.1016/j.fgb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 409.Koch C, Neumann P, Valerius O, Feussner I, Ficner R. 2016. Crystal structure of alcohol oxidase from Pichia pastoris. PLoS One 11:e0149846. doi: 10.1371/journal.pone.0149846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Chakraborty M, Goel M, Chinnadayyala SR, Dahiya UR, Ghosh SS, Goswami P. 2014. Molecular characterization and expression of a novel alcohol oxidase from Aspergillus terreus MTCC6324. PLoS One 9:e95368. doi: 10.1371/journal.pone.0095368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Artolozaga MJ, Kubátová E, Volc J, Kalisz HM. 1997. Pyranose 2-oxidase from Phanerochaete chrysosporium—further biochemical characterisation. Appl Microbiol Biotechnol 47:508–514. doi: 10.1007/s002530050964. [DOI] [PubMed] [Google Scholar]
- 412.Danneel HJ, Rössner E, Zeeck A, Giffhorn F. 1993. Purification and characterization of a pyranose oxidase from the basidiomycete Peniophora gigantea and chemical analyses of its reaction products. Eur J Biochem 214:795–802. doi: 10.1111/j.1432-1033.1993.tb17982.x. [DOI] [PubMed] [Google Scholar]
- 413.Maresová H, Veèerek B, Hradská M, Libessart N, Beèka S, Saniez M-H, Kyslík P. 2005. Expression of the pyranose 2-oxidase from Trametes pubescens in Escherichia coli and characterization of the recombinant enzyme. J Biotechnol 120:387–395. doi: 10.1016/j.jbiotec.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 414.Daou M, Piumi F, Cullen D, Record E, Faulds CB. 2016. Heterologous production and characterization of two glyoxal oxidases from Pycnoporus cinnabarinus. Appl Environ Microbiol 82:4867–4875. doi: 10.1128/AEM.00304-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Jensen KA Jr, Ryan ZC, Wymelenberg Vanden A, Cullen D, Hammel KE. 2002. An NADH: Quinone oxidoreductase active during biodegradation by the brown-rot basidiomycete Gloeophyllum trabeum. Appl Environ Microbiol 68:2699–2703. doi: 10.1128/AEM.68.6.2699-2703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Qi W, Jellison J. 2004. Induction and catalytic properties of an intracellular NADH-dependent 1,4-benzoquinone reductase from the brown-rot basidiomycete Gloeophyllum trabeum. Int Biodeterior Biodegrad 54:53–60. doi: 10.1016/j.ibiod.2004.02.001. [DOI] [Google Scholar]
- 417.Lee S-S, Moon D-S, Choi HT, Song H-G. 2007. Purification and characterization of an intracellular NADH: quinone reductase from Trametes versicolor. J Microbiol 45:333–338. [PubMed] [Google Scholar]
- 418.Brock BJ, Gold MH. 1996. 1,4-Benzoquinone reductase from the basidiomycete Phanerochaete chrysosporium: spectral and kinetic analysis. Arch Biochem Biophys 331:31–40. doi: 10.1006/abbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 419.Kulys J, Tetianec L, Schneider P. 2001. Specificity and kinetic parameters of recombinant Microdochium nivale carbohydrate oxidase. J Mol Catal B Enzym 13:95–101. doi: 10.1016/S1381-1177(00)00233-2. [DOI] [PubMed] [Google Scholar]
- 420.Xu F, Golightly EJ, Fuglsang CC, Schneider P, Duke KR, Lam L, Christensen S, Brown KM, Jørgensen CT, Brown SH. 2001. A novel carbohydrate:acceptor oxidoreductase from Microdochium nivale. Eur J Biochem 268:1136–1142. doi: 10.1046/j.1432-1327.2001.01982.x. [DOI] [PubMed] [Google Scholar]
- 421.Samejima M, Eriksson K-EL. 1992. A comparison of the catalytic properties of cellobiose: quinone oxidoreductase and cellobiose oxidase from Phanerochaete chrysosporium. Eur J Biochem 207:103–107. doi: 10.1111/j.1432-1033.1992.tb17026.x. [DOI] [PubMed] [Google Scholar]
- 422.Patel I, Kracher D, Ma S, Garajova S, Haon M, Faulds CB, Berrin JG, Ludwig R, Record E. 2016. Salt-responsive lytic polysaccharide monooxygenases from the mangrove fungus Pestalotiopsis sp. NCi6. Biotechnol Biofuels 9:108. doi: 10.1186/s13068-016-0520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Petrović DM, Bissaro B, Chylenski P, Skaugen M, Sørlie M, Jensen MS, Aachmann FL, Courtade G, Várnai A, Eijsink VGH. 4 July 2018. Methylation of the N-terminal histidine protects a lytic polysaccharide monooxygenase from auto-oxidative inactivation. Protein Sci doi: 10.1002/pro.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Nekiunaite L, Isaksen T, Vaaje-Kolstad G, Abou Hachem M. 2016. Fungal lytic polysaccharide monooxygenases bind starch and β-cyclodextrin similarly to amylolytic hydrolases. FEBS Lett 590:2737–2747. doi: 10.1002/1873-3468.12293. [DOI] [PubMed] [Google Scholar]
- 425.Shallom D, Shoham Y. 2003. Microbial hemicellulases. Curr Opin Microbiol 6:219–228. doi: 10.1016/S1369-5274(03)00056-0. [DOI] [PubMed] [Google Scholar]
- 426.Willats WGT, Mccartney L, Mackie W, Knox JP. 2001. Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47:9–27. doi: 10.1023/A:1010662911148. [DOI] [PubMed] [Google Scholar]
- 427.Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, Terrapon N, Buffetto F, Nepogodiev S, Xiao Y, Field RA, Zhu Y, O'Neill MA, Urbanowicz BR, York WS, Davies GJ, Abbott DW, Ralet MC, Martens EC, Henrissat B, Gilbert HJ. 2017. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544:65–70. doi: 10.1038/nature21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Lundquist K, Kirk TK. 1978. De novo synthesis and decomposition of veratryl alcohol by a lignin-degrading basidiomycete. Phytochemistry 17:1676. doi: 10.1016/S0031-9422(00)94674-0. [DOI] [Google Scholar]
- 429.Loose JSM, Forsberg Z, Fraaije MW, Eijsink VGH, Vaaje-Kolstad G. 2014. A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett 588:3435–3440. doi: 10.1016/j.febslet.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 430.Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AFM, Svergun DI, Eijsink VGH, Chatterjee NS, van Aalten DMF. 2012. The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog 8:e1002373. doi: 10.1371/journal.ppat.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Munoz-Munoz J, Cartmell A, Terrapon N, Henrissat B, Gilbert HJ. 2017. Unusual active site location and catalytic apparatus in a glycoside hydrolase family. Proc Natl Acad Sci U S A 114:4936–4941. doi: 10.1073/pnas.1701130114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet MC, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. 2018. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]