The clinical and epidemiological threat of the growing antimicrobial resistance in Gram-negative pathogens, particularly for β-lactams, the most frequently used and relevant antibiotics, urges research to find new therapeutic weapons to combat the infections caused by these microorganisms. An essential previous step in the development of these therapeutic solutions is to identify their potential targets in the biology of the pathogen.
KEYWORDS: peptidoglycan, murein, recycling, pathogenesis, β-lactamase regulation, lysozyme, inflammation, NOD receptors, bacterial secretion system, flagellum, penicillin binding proteins, peptidoglycan recognition proteins
SUMMARY
The clinical and epidemiological threat of the growing antimicrobial resistance in Gram-negative pathogens, particularly for β-lactams, the most frequently used and relevant antibiotics, urges research to find new therapeutic weapons to combat the infections caused by these microorganisms. An essential previous step in the development of these therapeutic solutions is to identify their potential targets in the biology of the pathogen. This is precisely what we sought to do in this review specifically regarding the barely exploited field analyzing the interplay among the biology of the peptidoglycan and related processes, such as β-lactamase regulation and virulence. Hence, here we gather, analyze, and integrate the knowledge derived from published works that provide information on the topic, starting with those dealing with the historically neglected essential role of the Gram-negative peptidoglycan in virulence, including structural, biogenesis, remodeling, and recycling aspects, in addition to proinflammatory and other interactions with the host. We also review the complex link between intrinsic β-lactamase production and peptidoglycan metabolism, as well as the biological costs potentially associated with the expression of horizontally acquired β-lactamases. Finally, we analyze the existing evidence from multiple perspectives to provide useful clues for identifying targets enabling the future development of therapeutic options attacking the peptidoglycan-virulence interconnection as a key weak point of the Gram-negative pathogens to be used, if not to kill the bacteria, to mitigate their capacity to produce severe infections.
INTRODUCTION
During the last few decades, the problem of antimicrobial resistance in nosocomial pathogens has continued to grow due to several factors, such as the development of new medical treatments usually entailing immunosuppression and/or invasive procedures, all of which lead to increased life expectancies but also to longer periods of hospital admission and an obvious increase in the probability of acquiring opportunistic infections (1, 2). Moreover, the historically erroneous politics of antibiotic use in several geographical areas, as well as the progressive appearance and horizontal dissemination of resistance determinants on plasmids and/or transposons, have decisively contributed to worsening this scenario. The worldwide emergence and spread of the so-called successful high-risk clones, mainly in species such as Pseudomonas aeruginosa, Acinetobacter baumannii, or certain Enterobacteriaceae, which are highlighted thanks to their multiresistance patterns and excellent capacity for inter- and intrahospital dissemination, add even more severity to the situation (3, 4).
Nevertheless, the issue of antibiotic resistance is not negligible in community-acquired or common infectious diseases, given that the strains as well as the horizontally transmitted determinants are often selected and maintained thanks to the presence of antibiotics in the food chain, besides the above-mentioned wrong antibiotic prescription, misuse, and self-medication practices (5–9). Obviously, all the cited circumstances are even more serious in the case of the β-lactams, since they are the most used antibiotics in clinical practice (10–12), and in the case of the species of interest for this review, Gram-negative pathogens. This is due to the high degree of intrinsic β-lactam resistance and the outstanding capacity for additional resistance development in several classic representatives of nosocomial species, such as the above-mentioned P. aeruginosa, A. baumannii, and some Enterobacteriaceae, but also in emerging ones, such as the members of the Burkholderia cepacia complex (BCC), Stenotrophomonas maltophilia, or Achromobacter xylosoxidans, among others (13).
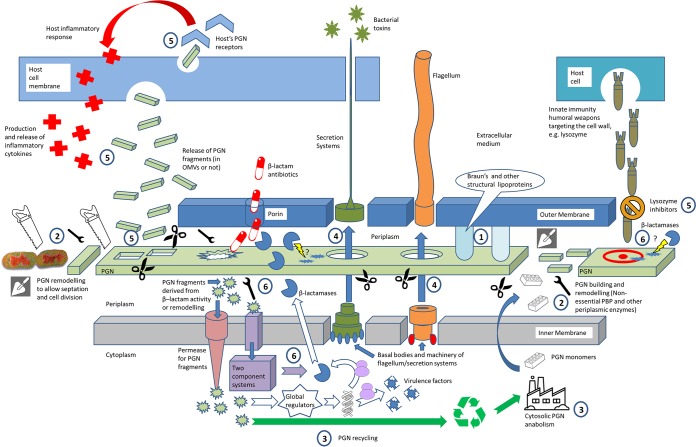
Therefore, in this worrying conjuncture, new therapeutic solutions are urgently needed if not to kill the microorganism at least to make the infections less harmful for the patient. This would entirely agree with the concept of antivirulence therapies, which are envisaged as excellent and still barely exploited remaining weapons in the described panorama of antibiotic arsenal reduction (14) to be used along with the novel approaches oriented toward the blockade and/or reversion of the pathways leading to β-lactam resistance (15, 16). Nevertheless, the essential previous step for the development of new therapies is the finding of targets, ergo, weak points to be attacked in the biology of the pathogen. In this sense, the general connection between resistance and virulence is a classically interesting interplay to be approached in order to find therapeutic targets, since it is generally accepted that antibiotic resistance often entails a biologic cost that usually dampens virulence, a circumstance which we could take advantage of (17). In this work, we only collaterally address this already reviewed general interplay but add another specific ingredient, introducing as the starring actor the biology of the peptidoglycan (PGN) of Gram-negative bacteria, which also has β-lactam resistance implications, since the regulation of β-lactamases is often intimately linked to PGN recycling (18, 19). Traditionally, and in contrast to the PGN of Gram-positive bacteria, the PGN of Gram-negative bacteria has usually been neglected as a key element in pathogenesis. Obviously, this circumstance is explained by the greater width of the PGN of Gram-negative bacteria and the lack of an outer membrane in Gram-negative bacteria, with studies classically highlighting the Gram-positive bacterial cell wall in terms of its importance for antibiotic resistance, survival against immune system weapons, and virulence (20–22). Nevertheless, there is a notable amount of published evidence (much of it very recent) that identifies several targets in the Gram-negative PGN biology (related or not to the regulation or effects of β-lactamase production) and that provides clues enabling the future development of therapeutic options attacking the above-mentioned PGN-virulence/fitness interplay. As can be observed in Fig. 1, we have organized these targets into different sections to ease the reader's task: (i) purely structural-architectural targets, (ii) periplasmic enzymes involved in PGN building and remodeling/recycling (nonessential penicillin binding proteins [PBPs] and others), (iii) cytoplasmic actors involved in the biosynthesis and recycling of PGN fragments, (iv) targets related to the local modification of PGN to allow for the correct assembly of flagella and secretion systems, (v) elements involved in the interaction of PGN with the host and the elicited response (both of which consider the mechanisms of PGN fragment release but also the host receptors and immune system elements), and (vi) the interplay among PGN metabolism, β-lactamase production regulation (including intrinsic and horizontally acquired enzymes), and fitness/virulence. Thus, as can be deduced from these lines, the PGN is an element that shows a very wide array of implications to finally influence fitness and virulence beyond its obvious architectural role, a circumstance that we wanted to reflect in the title with the concept “peptidoglycan biology.” However, and additionally, it has to be noted that several of the reviewed targets could probably fall into more than one of the cited categories and could fall into even more if we think of the pleiotropic nature of some of them, and hence, these categories should be taken only as a part of a sensu lato organization with nonexclusive but just schematizing purposes.
FIG 1.
Overview of the peptidoglycan (PGN)-related components and processes from Gram-negative pathogens proved to influence bacterial fitness/virulence and pathogenesis in the host. The numbers indicate the different contexts in which these targets can be found, and they are directly related to the sections of this review: 1, structural targets; 2, building and remodeling of PGN in the periplasm, affecting morphogenesis, cell division, and turnover of PGN fragments; 3, biogenesis and recycling of PGN (cytosol); 4, opening of gaps in the PGN layer to allow the insertion of secretion systems and/or flagella; 5, release of PGN fragments, interaction with host receptors, and the elicited response; 6, interplay among β-lactamase regulation, PGN metabolism, and fitness/virulence.
DEALING WITH CELL WALL STRUCTURE: PEPTIDOGLYCAN-ASSOCIATED LIPOPROTEINS AS MAJOR VIRULENCE DETERMINANTS
The purpose of this review is obviously not to delve into the structural aspects of Gram-negative PGN, and therefore, this section only superficially assesses the architectural issue, for which several excellent reviews, including more exhaustive descriptions and analysis, may be consulted (23, 24). Briefly, the Gram-negative PGN (also called murein or sacculus) is a polymer composed of glycan chains (N-acetylglucosamine [NAcGlc]–N-acetylmuramic acid [NAcMur] disaccharides) and cross-linked peptides (initially, pentapeptides bound to the NAcMur units) that forms a mesh-like layer outside the cytoplasmic membrane. It constitutes the solid basis on which the outer membrane relies and gets punctually linked through Braun's lipoprotein (which has the only covalent bond to PGN) and other murein lipoproteins (such as the PGN-associated lipoprotein [Pal]), all of which provide shape, structure, and the capacity to counteract the osmotic pressure, as well as a certain degree of resistance to the diffusion of molecules (25–27). Therefore, the proper synthesis and maintenance of the PGN, but also a correct architecture, are essential not only for bacterial virulence but also for viability, since, for instance, defects in the structural lipoproteins (or their anchoring to PGN) usually drive the outer membrane to a loss of integrity, leading to periplasmic protein leakage, vesicle formation, blebbing, etc (25, 28). As mentioned in the introduction, the PGN of Gram-positive bacterial species has classically been bestowed with great importance in terms of resistance against the action of immune weapons or certain antibiotics, but also as a key virulence factor often related to the inflammatory response elicited in the host (20–22, 29–32). However, given the protection exerted by the outer membrane and the anchored lipopolysaccharide (LPS) and its thinner width, the PGN of Gram-negative species has not been given such a protagonistic role, or at least a role not as important as that of the Gram-positive PGN. Nevertheless, many works have studied the Gram-negative PGN from the point of view of its structure and organization, and different contradictory models regarding the architecture and orientation of glycan chains, as well as the nature of a mono- or multilayer, have been proposed (24, 33). In Escherichia coli, for instance, a planar layer that could occasionally include sizable regions of multilayered PGN is mostly accepted. However, some data regarding the width of PGN layers from different Gram-negative bacteria have been published and have indicated, for instance, that the PGN of P. aeruginosa shows less than one-half of the thickness of that of E. coli (23, 24, 33, 34). In any case, in this section, only the purely structural PGN targets affecting virulence are reviewed, and then all the elements related to the biosynthesis, maintenance, remodeling, and recycling of PGN are analyzed in the following sections. Therefore, some examples of the strictly architecture-related role of PGN (i.e., elements that physically build the static PGN structure) can be found, and they are displayed in Table 1. For instance, the disruption of the PGN-associated lipoprotein (Pal) of Burkholderia cenocepacia (belonging to the BCC) has recently been linked to a major impairment in the virulence of this species in the Galleria mellonella larva infection model, together with a dampened capacity for host cell attachment and a weaker elicitation of inflammatory cytokine secretion (35). In a similar context, some papers have related the defects in yersinia Braun's lipoprotein to an important virulence impairment in murine models of infection, which suggested that the studied knockout (KO) mutants are potential vaccine candidates for bubonic and pneumonic plague (36–38). Similarly, in Salmonella enterica serovar Typhimurium, the inactivation of two murein lipoproteins caused a very significant impairment of virulence, in terms of a drastic reduction of invasion/cytotoxicity in cell culture and of mortality in murine models (39) (Table 1). And again, similarly to yersiniae, S. enterica serovar Typhimurium mutants with mutations in some PGN structural elements, such as Braun's lipoprotein, have recently been tested as vaccine candidates, with promising results (40). Interestingly, and still related to Braun's lipoprotein, Hernández and colleagues have shown in Salmonella that the effects of bile could induce the selection of some modifications leading to an increased resistance to this natural detergent (41). These changes in the PGN structure, detected during exposure to sublethal concentrations of sodium deoxycholate, consisted of a lower degree of Braun's lipoprotein binding to PGN and reduced levels of muropeptide cross-linking by l-(meso)-diaminopimelyl-d-(meso)-diaminopimelic acid peptide bridges. These structural changes to the PGN were shown to be similar to those observed in a triple l,d-transpeptidase mutant (YbiS, ErfK, YcfS), which also led to enhanced resistance to bile (41) (Table 1).
TABLE 1.
Targets related to the static PGN structure/architecture
| Target | Role(s) | Species | Effect(s) of target disruption | Reference(s) |
|---|---|---|---|---|
| Pal | A peptidoglycan-associated lipoprotein anchoring the outer membrane to PGN | B. cenocepacia | Reduction of virulence in a Galleria mellonella model of circa 90-fold; impaired host cell attachment and reduced stimulation of proinflammatory cytokine secretion | 35 |
| Lpp1, Lpp2 | Structural murein lipoproteins | S. enterica serovar Typhimurium | Defective in invasion of macrophages and cytotoxicity for macrophages | 39 |
| Lpp | Braun's lipoprotein | Yersiniae | Decreased proinflammatory effects and mortality rates in murine/rat models of infection, decreased survival in murine macrophages, impaired tissue dissemination capacity, and elicitation of an inflammatory response | 36–38, 42 |
| YbiS, ErfK, YcfS | l,d-Transpeptidases participating in the anchoring of Braun's lipoproteins to PGN | S. enterica | Changes in PGN structure leading to higher resistance to bile salts | 41 |
As can be concluded from the previous lines, given that PGN is essentially composed of sugar chains and stem peptides, it is not easy to find structural building elements, other than the above-mentioned lipoproteins, that may be considered static targets. As shown above, modification of these accessory structural proteins entails diverse consequences beyond the sensu stricto PGN probably related to the breakdown of outer membrane integrity, causing permeability barrier loss and an impaired performance of the anchored LPS (42). The direct consequences of these circumstances would be a decrease in the capacity to resist certain immune weapons of the host but also a decrease in adherence and even motility, although some contradictory findings do exist, like the above-mentioned increase in the resistance against bile achieved thanks to a weaker degree of linkage with Braun's lipoprotein (39, 41, 43). Still, regarding these lipoproteins, besides their structural roles, they have also been proposed to be elements contributing to pathogenesis during sepsis, since, once they are released (probably within outer membrane vesicles [OMVs]), they seem to elicit potent inflammatory responses in the host (44, 45). Therefore, they could also be considered relevant pathogen-associated molecular patterns (PAMPs), but as explained above, since they are only accessory elements to the bona fide PGN basic structure, we have not included them in the section dedicated to the interaction of PGN with host receptors (see below). In fact, the pattern recognition receptors (PRRs) for the PGN-associated lipoproteins are the Toll-like receptors (TLRs), mainly TLR2 (39, 46, 47), and not the receptors typical for the PGN fragments (namely, NOD receptors; see below).
Finally, the proteins participating in the anabolic pathways of the sugars and peptides and those responsible for their assembly and incorporation into the sacculus also influence the final architecture of the PGN, but since they participate in different dynamic reactions and processes, they are dissected in the next sections.
THE BUILDING, REMODELING, AND RECYCLING OF THE PEPTIDOGLYCAN: INDISPENSABLE TASKS FOR BACTERIAL VIRULENCE
Besides the structural point of view displayed in the section above, in which the PGN is shown to be a kind of skeletal support for the cell, we have to take into account the fact that this bacterial structure is far away from being a static element and therefore continuously needs the participation of many enzymes for its building, maintenance, remodeling, and recycling, including such an elemental process as cell division. An excellent review of these issues and their link to β-lactamase regulation has recently been published by Dik and colleagues (19). But in our work, for the sake of clarity, given that we relate the cited topics to virulence in a review for the first time, the participating actors will be separated into three subsections: nonessential PBPs, other periplasmic enzymes, and finally, cytoplasmic enzymes participating in the above-mentioned processes.
Penicillin Binding Proteins: When Not Indispensable for Viability, Essential for Full Virulence
Following the structure of the PGN described above, the enzymes that catalyze the reactions indispensable for its building are the largely studied penicillin binding proteins (PBPs). There are mainly two reactions: the polymerization of the glycan strand (N-acetylmuramic acid [NAcMur]–N-acetylglucosamine [NAcGlc]), namely, transglycosylation, and the cross-linking between the glycan chains through the stem peptides bound to NAcMur units (transpeptidation) (48). The latter reaction involves a necessary step of terminal d-Ala scission from the preexisting pentapeptide performed by the PBP before the final cross-linkage is allowed.
A large amount of information has been published on the topic of PBPs (48–51), and the reason that some PBPs are considered essential is due to the reactions that they perform, as their knockout mutants are inviable because the polymer cannot be properly synthesized, leading to autolysis. In a few words, the PBPs have been classified as high-molecular-mass PBPs (HMM-PBPs) and low-molecular-mass PBPs (LMM-PBPs). The C-terminal domain of HMM-PBPs typically shows transpeptidase activity, whereas the N-terminal domain can perform glycosyltransferase activity (hence, allowing the elongation of the glycan chains; these PBPs are classified as class A PBPs) or other different functions usually related to cell morphogenesis (in this case, the proteins are classified as class B PBPs). The HMM-PBPs were classically considered essential, but this is changing, as will be shown below for E. coli or P. aeruginosa, for instance (48). Meanwhile, the LMM-PBPs (also called class C PBPs) have been considered nonessential, as they perform more specific activities and potentially secondary activities with regard to the described main function of PGN building. Some examples could be assistance with septation (i.e., septum formation, which is the concerted invagination of the cytoplasmic membrane, PGN, and outer membrane prior to cell separation), the turnover of certain fragments to initiate recycling, the scission of the terminal d-Ala from pentapeptides to block the possibility of cross-linking, and the cleavage of cross-links between stem peptides (endopeptidase activity) for remodeling (48, 50, 52). As mentioned before, the deletion of essential PBPs is not compatible with cell viability, and they have obviously been studied and are considered very good candidates for therapeutic targets. Nevertheless, given the large amount of published information on these facts, which is not specifically linked to virulence, we will not explore the topic further (50). Thus, we review the influence of the disruption of certain nonessential PBPs on fitness and virulence, and in this context, the identified targets are summarized in Table 2. The complexity of the topic must be emphasized, since most Gram-negative species differ in the number and denomination of PBPs. In E. coli, for instance, 12 PBPs have been described: 3 class A PBPs (PBP1a, PBP1b, and PBP1c), 2 class B PBPs (PBP2 and PBP3), and 7 LMM-PBPs. Meanwhile, in P. aeruginosa we find only 8 PBPs (HMM-PBPs PBP1a, -1b, -2, -3, and -3a/3x and LMM-PBPs PBP4, -5/6, and -7) and in Neisseria meningitidis we find 4 PBPs (2 HMM-PBPs and 2 LMM-PBPs) (48, 50), to cite some examples. Among this wide range of possibilities, it has to be highlighted that most of the information connecting the nonessential PBPs to fitness/virulence implications has been published in E. coli-based studies. In this regard, almost 20 years ago, a very interesting work assessing the effects of different single or combined inactivations of the PBP genes was published (53); surprisingly, mutants with almost all the PBP combinations, including mutants with up to eight deleted PBP genes, grew almost as well as the wild-type strain, with the only exception being the mutant with the combination PBP1a-PBP1b deletion, which was not viable. Other effects were also reported, such as the essential role of PBP3 for septation and cell division, with the inactivation of this enzyme causing the filamentous growth of the mutant in long chains of unseparated cells (Table 2) (25, 53). More recently, some papers have further investigated the topic of cell division in this species, unraveling the participation of some additional actors, such as PBP4 and PBP7 (with a secondary role), besides the major periplasmic amidases AmiA, AmiB, and AmiC, which will be analyzed below (see Table 3) (54). Interestingly, PBP4 and PBP7 were also suggested to contribute to the morphogenesis and production of biofilms (54). Nevertheless, none of these works has addressed whether the alterations in morphology/septation mentioned here could have direct effects on virulence. Of course, what could be expected is an impaired motility (with obvious negative consequences for dissemination and, hence, for virulence), since flagellar movement would not be able to propel the long filaments of unseparated cells as efficiently as it does individual bacteria. Some other plausible negative effects for pathogenesis could be deduced from the aberrant morphologies (in terms of altered contours, cell branching, or increased diameters, for instance) potentially interfering with the host cell's adhesion obtained, for instance, by Nelson and Young (55) in PBP5 knockout mutants (Table 2). However, apart from these assumptions, no additional effects on virulence have been experimentally proved, not even in more recent work (56), where the LMM-PBPs of E. coli PBP6 and PBP6b have been related to the correct location of the septal protein FtsZ to allow proper cell division and avoid the insertion of inert PGN at unusual positions, leading to branching (56). Finally, what has recently been proposed for E. coli is that the redundancy in PBPs with carboxypeptidase activity (PBP4, PBP4b, PBP5, PBP6, PBP7, and AmpH) allows for the correct cell shape under a wide array of different environmental conditions; therefore, for instance, PBP6b (dacD) has been shown to be the most important PBP for morphogenesis under acidic growth conditions (Table 2) (57).
TABLE 2.
Penicillin binding proteins described to have implications on fitness/virulence
| Target | Role(s) | Species | Effect(s) of target disruption | Reference |
|---|---|---|---|---|
| PBP3 (ftsL) | A transpeptidase and major protein of the divisome, the cellular division complex | E. coli | Growth in elongated chains of unseparated cells; probable impairment of motility | 53 |
| PBP4 (dacB) and PBP7 (pbpG) | Endopeptidases collaborating with the periplasmic N-acetylmuramyl-l-alanine amidases (AmiA, AmiB, and AmiC) for cell septation; unspecified auxiliary roles in maintaining regular cellular morphology and enhancing the formation of biofilms; participation during daughter cell separation after cell division, when the amidases or lytic transglycosylases are absent | E. coli | Aberrant morphologies with growth in elongated chains of unseparated cells; impaired formation of biofilms; probable impairment of motility | 54 |
| PBP5 (dscA) | A major carboxypeptidase cleaving the terminal d-Ala from pentapeptides, making them unavailable for transpeptidation, therefore participating in the fine-tuning of cross-linking in PGN, which affects morphology | E. coli | Increased cell diameter, aberrant contour and morphology (branching), as well as alterations in surface uniformity and overall topology of the peptidoglycan saccules; probable impairment of host cell attachment | 55 |
| PBP6 (dacC) and PBP6b (dacD) | Carboxypeptidases collaborating with the septal protein FtsZ to allow correct septation, cell division, and overall cell shape | E. coli | Mislocalization of cytosolic FtsZ, leading to abnormal septation events and derived cell branching | 56 |
| PBP6b (dacD) | A major carboxypeptidase contributing to cell morphogenesis under acidic conditions | E. coli | Aberrant morphologies and increased length of cells at pH 5.0 | 57 |
| AmpH | A bifunctional d,d-endopeptidase and d,d-carboxypeptidase showing weak β-lactamase activity and an unknown exact role in the cell wall | E. coli | Uneven contours and aberrant and asymmetric constrictions between dividing cells in AmpH-AmpC double mutants | 255 |
| PBP1a (ponA) and PBP2 (pbpA) | A major transpeptidase-transglycosylase (PBP1a) and a transpeptidase involved in cell elongation (PBP2) | P. aeruginosa | Impairment of swarming motility in a PBP1a single mutant; impairment in in vitro competition experiments and biofilm formation in double mutants | 52 |
| PBP1b (mrcB) | A major transpeptidase-transglycosylase | P. aeruginosa | A decrease in biofilm formation capacity when deleted together with PBP2 | 52 |
| PBP7/8 (pbpG) | An endopeptidase cleaving cross-bridges between stem peptides | A. baumannii | A decrease in survival in a rat soft tissue infection model and a rat pneumonia model; a significant increase in bacterial killing in 90% human serum in vitro; a greater prevalence of coccobacillary forms than wild-type forms | 62 |
| PBP1a (mrcA) | A transpeptidase-transglycosylase mainly involved in cell wall synthesis during elongation that works together with LpoA, a lipoprotein activator located in the outer membrane indispensable for PBP1a performance | V. cholerae | Growth deficiencies in minimal medium and increased susceptibility to deoxycholate and bile in a PBP1a knockout mutant (but also an LpoA knockout mutant), which also showed a decreased capacity in competition assays with the wild type both in vitro and in an infant mouse small intestine model, which were found only if the bacteria were used during stationary phase | 63 |
TABLE 3.
Periplasmic elements other than penicillin binding proteins related to PGN biology
| Target | Role(s) | Species | Effect(s) of target disruption | Reference(s) |
|---|---|---|---|---|
| Targets related to cell septation/division | ||||
| AmiA, AmiB, AmiC | Periplasmic N-acetylmuramyl-l-alanine amidases that cleave stem peptides from glycan chains on PGN | E. coli | Formation of abnormal septa, causing growth in long chains of unseparated cells | 64, 65 |
| YebA, EnvC, and NlpD | LytM factors participating through their peptidase activity in cell elongation and division | H. influenzae | Alterations in division with irregular cell architecture and massive membrane blebbing for YebA and NlpD knockout mutants; a reduction in the amount of periplasmic proteins and an impaired capacity to adhere to epithelial cells, to form biofilms, and to resist the bactericidal power of serum with EnvC deletion | 76 |
| AmiC | A periplasmic N-acetyl-muramyl-l-alanine amidase that cleaves stem peptides from glycan chains on PGN | Burkholderia spp. | In daughter cells, an inability to separate, growing in filaments and losing motility, all of which leads to an inability to survive in the rat model and colonize the insect gut | 68, 69 |
| AmiC | A periplasmic N-acetylmuramyl-l-alanine amidase that cleaves the peptide side chains linked to the glycan strands on PGN and whose activity is potentiated by the presence of the regulator NlpD | X. campestris | Impaired daughter cell separation, aberrant cell and colony morphology, and impaired T3SS performance | 70 |
| AmiA | A periplasmic N-acetylmuramoyl-l-alanyl amidase essential for daughter cell separation | H. pylori | Appearance of long chains of unseparated cells and impaired motility; a reduced capacity for colonization of the stomach in a mouse model | 66 |
| AmiB | A periplasmic N-acetylmuramyl–l-alanine amidase involved in stem peptide cleavage from PGN chains | P. aeruginosa | Filamentous growth with a marked deficiency in the invagination of the inner membrane and increased permeability of the outer membrane | 67 |
| PBP3SAL | A specialized PGN synthase enabling formation of the division septum and promoting cell division in the acidic intraphagosomal environment | S. enterica serovar Typhimurium | Induction of PBP3SAL during infection favoring higher bacterial loads in murine models of infection when a wild type was compared with a knockout mutant | 71 |
| AmiA, AmiC, and SufI and the pathways for their correct expression (Cpx system) and export to the periplasm (Tat) | Periplasmic N-acetylmuramoyl-l-alanyl amidases that cleave stem peptides from glycan chains on PGN (AmiA and AmiC) and a divisomal transpeptidase (SufI) | S. Typhimurium (also E. coli) | For AmiA and AmiC, impairment in septation and separation of daughter cells (filamentation) and virulence attenuation in a BALB/c mouse infection model; for SufI, increased sensitivity to detergents and cationic antimicrobial peptides and impaired motility | 72–75 |
| Modification of PGN related to morphogenesis | ||||
| PgdA | An N-deacetylase for PGN modification | H. pylori | Decreased resistance of PGN to lytic activity of lysozyme; double mutant (PgdA-PatA) impaired for mouse colonization | 81 |
| PatA | A putative O-acetyltransferase for PGN modification | H. pylori | Decreased resistance of the PGN to lytic activity of lysozyme; double mutant (PgdA-PatA) impaired for mouse colonization | 81 |
| Csd1 to Csd3 and CcmA | Periplasmic endopeptidase homologues essential to allow a decrease in PGN cross-linking levels | H. pylori | In single mutants, a curved instead of a helical shape, which was related to a decrease in colonization capacity in mouse stomach model competitions | 80 |
| Ape1 | An O-acetylpeptidoglycan esterase responsible for de-O-acetylation of PGN | C. jejuni | Changes in PGN biochemistry; defects in virulence-associated features, including motility, biofilm formation, sodium deoxycholate resistance, adhesion, invasion, intracellular survival, induction of IL-8 release, and impairment of chick colonization | 79 |
| Pgp1 | A d,l-carboxypeptidase involved in maintenance of cell shape, cleaving monomeric tripeptides to dipeptides | C. jejuni | Loss of helical shape; increase in the level of stimulation of NOD-1 receptors and derived induction of IL-8 release; decreased motility and biofilm formation; deficient for chick colonization | 77 |
| Pgp2 | An l,d-carboxypeptidase involved in maintenance of cell shape, converting PGN tetrapeptides into tripeptides, which in turn are substrates for Pgp1 | C. jejuni | Loss of helical shape; defective in motility on semisolid agar and biofilm formation; reduced fitness in chick colonization model | 78 |
| LtgA and LtgD | Nonessential, nonredundant lytic transglycosylases | N. gonorrhoeae | Decreased envelope integrity, leading to increased susceptibility to lysozyme and neutrophil killing | 82 |
| PGN-degrading enzymes related to sacculus remodeling to allow elongation and recycling | ||||
| MltB | A membrane-bound lytic murein transglycosylase participating in PGN degradation | N. meningitidis | Inability to cause systemic infection in an infant rat model | 84 |
| MtgA | A biosynthetic transglycosylase | Brucella spp. | Upregulation during infection; in a knockout mutant, lower virulence in a mouse infection model | 85, 86 |
| MltE | A membrane-bound lytic transglycosylase | Erwinia amylovora | Upregulation during infection; in a knockout mutant, reduced virulence and growth in a pear model | 85, 87 |
| 90_A18ORF1 | A soluble lytic murein transglycosylase | Haemophilus influenzae | Upregulation of the 90_A18ORF1 gene during infection (a KO mutant is not available) | 85, 88 |
| Ipx10.11 | A lytic murein transglycosylase | Pseudomonas syringae pv. tomato | Upregulation during infection; in a knockout mutant, impaired virulence in an Arabidopsis thaliana infection model | 85, 89 |
| Ddc | A d-alanine–d-alanine carboxypeptidase | A. baumannii | Hypersusceptibility to serum and polymyxin B; defective in intramacrophage survival; a drastically reduced capacity for survival in a mouse bloodstream infection model | 83 |
| ShyA, ShyB, and ShyC | Periplasmic proteins containing M23 family peptidase domains; for ShyA, a d,d-endopeptidase preferentially cleaving cross-links between tetrapeptides and located in the lateral cell wall; for ShyC, preferential location in the septum | V. cholerae | In a ShyA and ShyB double mutant, a significant growth deficiency; in a shyC mutant with depletion of ShyA, dramatic impairment of cell elongation rates and a significant increase in cell width, which would presumably affect attachment to host tissue | 90 |
Moreover, a very recent work by Chen and colleagues (52) revealed that the unique PBP essential for the growth of P. aeruginosa is PBP3 (ftsI) and characterized the effects on fitness/virulence that the deletion of the rest of the PBPs could have on this species. Thus, what was observed is that P. aeruginosa tolerates the single deletion of the rest of the nonessential PBPs, with these single deletions not having a significant impact on fitness. Conversely, the simultaneous deletion of PBP1a (ponA) and PBP2 (pbpA) caused a significant reduction of in vitro competitiveness (Table 2) (52). The same deletion combination or that of PBP2 plus PBP1b (mrcB) also drove a significant reduction of biofilm formation, suggesting that PBP2 disruption could directly or indirectly affect the virulence phenotype mainly if it was combined with the loss of PBP1a or PBP1b (Table 2). This was supported by the attention-calling increase of circa 30% in pyocyanin secretion when PBP2 was disrupted, potentially caused by the stress derived from the above-mentioned gene deletion. Interestingly, the inactivation of PBP1a alone severely impaired the swarming motility of the mutant, although the basis for this circumstance is not yet clear. A direct involvement of the protein in this kind of movement or, alternatively, a collateral effect derived from the presumptive stress caused by the PGN alteration that the deletion of PBP1a would entail is a plausible explanation (58). In this regard, although in the past motility has been seen to be a very important feature of P. aeruginosa virulence (59), paradoxically, these effects were not translated into significant differences when testing the single PBP knockout mutants in a Caenorhabditis elegans model (52) (Table 2). In this context, regarding PBPs, it should be taken into account that the deletion of a target could potentially entail not only consequences for the derived virulence phenotype but also some polymorphisms, an issue that has barely been studied to date. In this sense, and still in P. aeruginosa, it has been described that some amino acid changes in PBP3 (R504H, A539T, V465A, or F533L, all of which are in the specific binding site for meropenem) lead to resistance, but their potential consequences on fitness/virulence have never been assessed (60). Therefore, analysis of these kinds of polymorphisms usually leading to increased resistance (another example could be changes in AmpC amino acids [61]) for their potential negative consequences on fitness/virulence is a new horizon that is worth further exploration for future therapy development.
Meanwhile, in A. baumannii it has been shown that PBP7/8 decisively contributes to resistance to complement-mediated bactericidal activity, although the basis for this circumstance is still unknown. Therefore, the deletion of this protein was translated into dramatic reductions in virulence in soft tissue and rat lung infection models and in survival after incubation with human serum and, finally, was related to an increased prevalence of coccoidal forms, suggesting an effect of PBP7/8 on morphogenesis (62) (Table 2). Besides, in Vibrio cholerae, PBP1a has been shown to be very important for pathogenesis, since its deletion led to a reduced competition capacity both in vitro and in mouse models, as well as increased susceptibility to detergents, but only when using bacteria in stationary phase. Hence, this effect of PBP1a deletion, which apparently affects virulence, can be bypassed if cells are in exponential growth. Interestingly, given that this protein works together with lipoprotein LpoA, located in the outer membrane, the deletion of the latter caused the same phenotypes (Table 2) (63).
Thus, to conclude this section, although few virulence-specific conclusions regarding the nonessential PBPs have been published, many inferences can be drawn, if not because of a direct dampening of pathogenesis factors at least because of abnormalities in cell shape or motility, which are obviously also important for full virulence performance.
Periplasmic Actors Other than Penicillin Binding Proteins
Besides the above-mentioned role of the nonessential PBPs in the process of sacculus building, other enzymatic actors that have different functions and that are located in the periplasm have been shown to have fitness/virulence implications when disrupted. In this regard, the types of activities that these actors perform can be subdivided, as displayed in Table 3: cell septation and division-related events, modifications of mature PGN to allow cell shaping, and finally, the PGN-degrading enzymes related to remodeling to allow an increase in cell size or the release of fragments to be internalized into the cytosol for recycling.
Periplasmic elements involved in septation and cell division.
With regard to PGN enzymes related to cell septation, the role of the periplasmic N-acetylmuramyl-l-alanine amidases (cleaving the bond between the NAcMur units and the stem peptides and, hence, disconnecting the links among glycan chains) in the process, mainly in E. coli, is well-known (64, 65). In this regard, the AmiA, AmiB, and AmiC enzymes have been proposed to be essential for correct continued PGN synthesis during cell division, assisting with the formation of a proper septum and, hence, avoiding filamentation, which has also been found in Salmonella (64, 65). With regard to other species, it has been shown that AmiA of Helicobacter pylori is essential for the separation of daughter cells. Therefore, its inactivation leads to the formation of long chains of unseparated cells and a consequent obvious impairment of motility and colonization capacity in the mouse stomach model (66) (Table 3). AmiB of P. aeruginosa is also involved in cell separation and has been proven to be essential for bacterial viability, since its deletion entailed negative consequences for outer membrane integrity and impermeability levels (67). Meanwhile, in the genus Burkholderia, it has been shown that the inactivation of the periplasmic AmiC amidase causes the loss of daughter cell separation capacity, obviously entailing a severe impairment for motility, which was translated into the impossibility of causing infection in the rat model or colonizing the insect gut (68, 69). Similar results for AmiC of the phytopathogen Xanthomonas campestris have been reported, together with negative effects on type III secretion system (T3SS) performance, obviously entailing an impairment of virulence (70). Nevertheless, at present it remains to be seen whether this impairment of the T3SS function could be related to defects in the insertion of its machinery through the PGN layer (caused by the AmiC absence), as will be displayed later for other proteins.
Besides the cited amidases, very recently the implication that a specialized enzyme takes part in the intracellular division of S. enterica serovar Typhimurium and therefore is probably not negligible for pathogenesis in the host has been proposed (71). More specifically, this enzyme (namely, PBP3SAL) is 63% identical to PBP3 and, in contrast to the latter, is not essential (a lack of PBP3 renders cells unable to grow at neutral pH) but exerts a positive effect on septation and cell replication in acidic environments, such as those inside phagosomes. In fact, PBP3SAL expression has been shown to be induced during infection in the murine model, which has been related to the higher bacterial loads of the wild-type strain than of PBP3SAL knockout mutants (71). Still in Salmonella (but also in E. coli), the coordinate roles of AmiA, AmiC, and SufI (also named FtsP) have been shown to be very important for full virulence. These three proteins have been demonstrated to be essential for septation, with AmiC and SufI being located in the divisome, the complex of at least 15 proteins that allows the formation of the septum. Interestingly, the defects not only in these proteins but also in the pathways that they use to be sufficiently expressed (through the envelope stress-sensing two-component system CpxA-CpxR) and/or exported to the periplasm (through the twin arginine translocation [Tat] system) have been shown to be essential for full virulence, entailing the multiple effects from the deletion of genes encoding these proteins, including defects in motility and filamentation, an increase in susceptibility to detergents, etc. (Table 3) (72–75). Nevertheless, the implication of these pathways in a wide variety of processes, probably not directly related to septation or divisome formation, also must be taken into account as the cause of the cited virulence impairments.
Finally, in Haemophilus influenzae, three lysostaphin-like metalloproteases, also called LytM factors, YebA, EnvC, and NlpD, have been studied in depth (76). These factors, widely described in Gram-negative bacteria, participate in cell division by modulating the cleavage and remodeling of PGN, and in the specific case of H. influenzae, they have been reported to affect the outer membrane composition when disrupted, with negative consequences, such as altered cell structure, a loss of periplasmic proteins, and a reduced capacity for cell adhesion, survival against serum, and biofilm formation (Table 3) (76).
Periplasmic elements involved in morphogenesis.
With regard to periplasmic enzymes related to cell morphogenesis, there are some recent works about two enzymes (namely, the carboxypeptidases Pgp1 and Pgp2) in Campylobacter jejuni shown to be essential for correct cell shaping. In fact, the loss of the typical helical shape of this species has been linked to the loss of virulence in a chick infection model, besides additional features linked to pathogenesis reduction (Table 3) (77, 78). In the same species, it has been shown that an increase in susceptibility to bile, together with a decrease in many other parameters leading to a less virulent outcome in the chick infection model, arises as a result of abnormally high levels of PGN O-acetylation (caused by inactivation of the Ape1 esterase) (see Table 3 for further details) (79). Similarly, with regard to morphogenesis, it has also been shown that a helical shape is essential for full virulence in another digestive tract pathogen, Helicobacter pylori, since the disruption of different endopeptidases (Csd1, Csd2, Csd3, and CcmA; Table 3), which cause relaxation of the PGN cross-linking level, leads to a curved instead of a helical cell morphology, finally causing a decreased capacity for mouse stomach colonization (although no great effects on motility or resistance to stomach environment stresses were observed) (80). Still, in H. pylori and also with regard to resistance to external aggressions, the deletion of two PGN-modifying enzymes (the N-deacetylase PgdA and the O-acetyltransferase PatA) has been shown to increase the susceptibility to lysozyme, probably because the loss of these enzymes entails the inability to modify the PGN in order to avoid enzymatic recognition by lysozyme (81). More specifically, their combined inactivation led to a PGN circa 5 times more susceptible to lysozyme-mediated in vitro degradation than the wild-type purified PGN. The modification of PGN to avoid the action of certain immune proteins has been largely described in Gram-positive bacteria (mainly thanks to changes in the chemical properties/structure of the PGN molecules theoretically able to bind the active site of the protein), but through this work it was shown to be also used as a very notable strategy by certain Gram-negative species (81). In fact, the loss of the cited enzymes rendered strains with a very significant reduction in the capacity for stomach colonization in a murine model (81) (Table 3).
The decrease in resistance to lysozyme as a consequence of alterations in enzymes involved in PGN biology has also been observed in other species, such as Neisseria gonorrhoeae (Table 3). More specifically, the lytic transglycosylases LtgA and LtgD from the gonococcus were demonstrated to contribute to the maintenance of envelope integrity, limiting exposure to the lysozyme and other antimicrobial proteins from neutrophil granules (82). Besides these and the previously mentioned data regarding PGN modifications affecting the activity of some host elements targeting the cell wall (lysozyme, bile, etc.), the large topic of the Gram-negative PGN interaction with the host (including receptors, inflammatory implications, and the specific mechanisms that some bacteria display to inhibit the activity of lysozyme) will be more widely approached below.
Periplasmic peptidoglycan-degrading enzymes involved in remodeling and recycling.
Finally, dealing with enzymes related to PGN degradation for remodeling (mainly to allow elongation) and/or recycling purposes, some examples in different species can be cited. Although in some cases the exact role of the involved elements has not yet been elucidated, the consequences for fitness/virulence have been described (Table 3). In this context, in A. baumannii, for instance, the d-Ala–d-Ala-carboxypeptidase-encoding gene ddc has been identified by Subashchandrabose and colleagues (83) to be one of those essential for bloodstream infection development. More specifically, the disruption of this target has been shown to cause hypersensitivity to complement and antimicrobial peptides as well as impaired survival within murine macrophages (83). Moreover, in N. meningitidis the periplasmic lytic transglycosylase MltB, allegedly contributing to PGN turnover, was shown to be indispensable for the development of systemic infection in an infant rat model, presumably because of certain alterations to the cell wall composition or even because of the elicited metabolic disorders caused by its inactivation (84). Still in this genus, but specifically in the gonococcus, the nonessential lytic transglycosylases LtgA and LtgD, besides having a crucial role in proinflammatory PGN fragment release to the extracellular medium (the issue is reviewed below), have been shown to be very important for envelope integrity, since their disruption leads to increased susceptibility to lysozyme and neutrophil killing (82). Other lytic transglycosylases with implications for fitness/virulence are the MltE (Erwinia spp.), 90_A18ORF1 (H. influenzae), and Ipx10.11 (Pseudomonas syringae) elements, although very little information regarding the molecular basis has been published (Table 3) (85–89).
Finally, in V. cholerae, some interesting targets presumably related to cell wall remodeling to allow growth have been identified, namely, the periplasmic proteins ShyA, ShyB, and ShyC, all of which show M23 family peptidase domains (90). In their work, Dörr and colleagues reported several findings, such as the nature of ShyA as a d,d-endopeptidase with preferred activity cleaving cross-links between tetrapeptides in a key step to allow cell elongation (90). Moreover, although none of these three proteins was shown to be essential per se, the double deletion of ShyA and ShyB caused severe growth deficiencies, and the depletion of ShyA in a ShyC mutant rendered cells with increased widths and significant defects in elongation rates (the double ShyA and ShyC KO mutant was not viable). Finally, the locations of ShyA and ShyC were shown to be different: the first is in the lateral cell wall, whereas the latter is preferentially in the septum, although its specific role during septation has never been studied (Table 3) (90).
Cytosolic Enzymes Working on Peptidoglycan Biosynthesis and Recycling
This section reviews the role of cytoplasmic enzymes involved in the pathways of biogenesis of the PGN monomers (NAcGlc-NAcMur-pentapeptides) that have been shown to also influence fitness/virulence. These are displayed in Table 4. Obviously, this is not a review of the anabolic pathways leading to PGN unit synthesis/recycling, but to learn more about the issue, there are interesting reviews (19, 91–94). In a few words, the cytosolic biogenesis pathways of the elements constituting the PGN could be separated into reactions allowing the synthesis of the sugars (NAcMur and NAcGlc); the synthesis of the amino acids and the stem peptide itself (initially a pentapeptide), thanks to the sequential incorporation of l-alanine (l-Ala), d-glutamic acid (d-Glu), meso-diaminopimelic acid (m-DAP), and the d-alanine–d-alanine (d-Ala–d-Ala) dimer; and of course, the reactions of the linkages among these components. However, we should also consider the events that allow the transit of the PGN monomers (disaccharide-pentapeptides) across the inner membrane, specifically, through the lipid I and II cycles and flippase action, which release the PGN monomers into the periplasm to be incorporated into the nascent PGN. Moreover, the elements that participate in cytosolic recycling processes and that allow for the reuse of some of the cited compounds (disaccharide peptides, peptides, and monosaccharides), once they are released from the PGN, instead of their de novo synthesis, are also assessed here (19, 93).
TABLE 4.
Cytosolic targets involved in PGN biosynthesis and recycling
| Target | Role(s) | Species | Effect(s) of target disruption | Reference(s) |
|---|---|---|---|---|
| LdcA | An l,d-carboxypeptidase cleaving the terminal d-alanine from tetrapeptides proceeding from cell wall turnover, releasing tripeptides which enter the recycling pathway to finally provide UDP-NAcMur-pentapeptides | E. coli | A dramatic decrease in the degree of stem peptide cross-linking, leading to weakening of the cell wall and to bacteriolysis in the stationary phase of growth in liquid culture | 96 |
| UppP | An undecaprenyl pyrophosphate phosphatase essential for synthesis of undecaprenyl phosphate | Burkholderia spp. | Increased susceptibility to lysozyme, hypotonic and hypertonic shock, and centrifugal force; decreased capacity to colonize the stinkbug gut | 95 |
| Mpl-1 | A UDP-N-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-diaminopimelate ligase involved in PGN recycling | N. meningitidis | An inability to cause systemic infection in an infant rat model | 84 |
| AmpD | An N-acetylmuramyl-l-alanine amidase involved in PGN recycling | N. meningitidis | An inability to cause systemic infection in an infant rat model; hyperexpression during infection in wild-type strains | 84 |
| MurA | An enolpyruvyl transferase acting in the cytosolic transformation of UDP-NAcGlc to UDP-NAcMur | P. aeruginosa | Impaired growth in minimal medium, changes in colony morphology, dampened capacity for lung colonization in a murine model of intratracheal infection, and decreased survival against macrophage-mediated killing | 101 |
| MurD | A UDP-N-acetylmuramyl l-alanyl-d-glutamate-diaminopimelate ligase allowing the cytosolic building of the stem peptide in the PGN precursor UDP-NAcMur pentapeptide | P. aeruginosa | Impaired growth in minimal medium, changes in colony morphology, dampened capacity for lung colonization in a murine model of intratracheal infection, and decreased survival against macrophage-mediated killing; decreased pathogenesis in a plant infection model | 101 |
| MurF | A UDP-N-acetylmuramyl l-alanyl-d-glutamyl-diaminopimelate-d-alanyl-d-alanine ligase allowing the cytosolic building of the stem peptide in the PGN precursor UDP-NAcMur pentapeptide | P. aeruginosa | Impaired growth in minimal medium, changes in colony morphology, abnormally elongated cells, and decreased survival against macrophage-mediated killing; decreased pathogenesis in a plant infection model | 101 |
| DapA | A dihydrodipicolinate synthase responsible for the synthesis of m-DAP and lysine | Serratia marcescens | Upregulation during infection; in KO mutants, greater sensitivity to hypotonic shock, an aberrant irregular elliptical shape, and disruption of the cell wall structure, as well as increased cytotoxicity and hemolysin production and a reduced capacity for attachment to surfaces | 85, 100 |
To start with, although UppP is not a strictly cytosolic protein but an inner membrane phosphatase that allows the synthesis of undecaprenyl phosphate (the basis for the lipid I and II structures), which, in turn, allows the export of the PGN monomers to the periplasm, UppP could be included in this section since it allows the final steps of anabolism before the new monomers are added to the PGN. It has been reported that defects in this phosphatase in the genus Burkholderia are responsible for increased susceptibilities to lysozyme and other PGN stresses, such as hyper- and hypo-osmotic shocks and centrifugal force, rendering strains with impaired virulence and an impaired capacity to infect the stinkbug gut (Table 4) (95). In a step before the above-mentioned lipid I and II formation, in E. coli the LdcA protein was first identified to be essential for the transformation of stem tetrapeptides into tripeptides, once they have entered the cytosol (Table 4) (96). The tripeptides can then be used for recycling, allowing the final addition of the terminal d-Ala–d-Ala (thanks to MurF) to finally constitute the UDP-NAcMur-pentapeptides to be incorporated into the PGN. Nevertheless, when LdcA is deleted, the precursors formed are the UDP-NAcMur tetrapeptides, which are incorporated into PGN but which do not allow transpeptidation, since the scission of the terminal d-Ala of the pentapeptides is required for this process (13). Therefore, LdcA deletion results in a low degree of cross-linked stem peptides, which finally leads to the weakening of the PGN structure and to bacteriolysis in the stationary growth phase in liquid culture (96, 97).
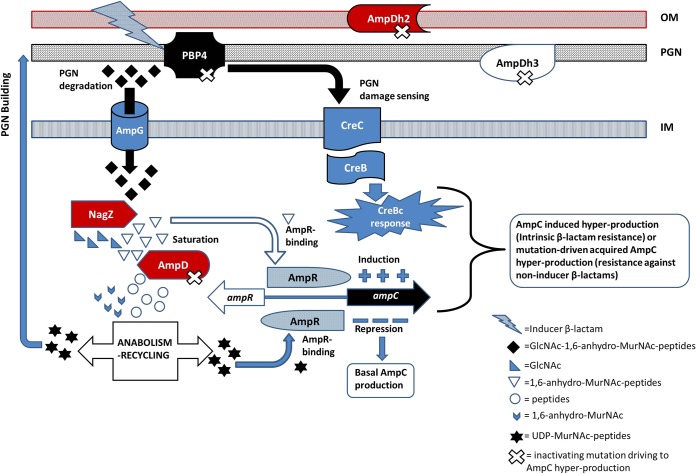
In N. meningitidis, in its turn, Mpl-1 and AmpD, two cytoplasmic enzymes involved in PGN recycling, were shown to be indispensable for the development of systemic infection in an infant rat model, although the exact molecular basis for the virulence attenuation linked to the inactivation of Mpl-1 or AmpD was not ascertained (Table 4) (84). More specifically, Mpl-1 allows the ligation of the above-mentioned recycled tripeptides (l-Ala–d-Glu–m-DAP) to UDP-NAcMur, whereas AmpD cleaves the bond between anhydro-NAcMur and stem peptides, once these fragments are internalized into the cytosol after release from the PGN (93). Since the role of the AmpD amidases (in this review, those from P. aeruginosa, Neisseria spp., Ralstonia solanacearum, or S. enterica serovar Typhimurium, for instance, are cited [84, 85, 98, 99]), apart from recycling, is intimately linked to β-lactamase regulation, its influence on fitness/virulence will be analyzed later in the section approaching the topic of the intrinsic β-lactamases (see Table 7).
TABLE 7.
Elements simultaneously involved in regulation of expression of intrinsic β-lactamases and PGN metabolism
| Target | Role(s) | Species | Effect(s) of target disruption | Reference(s) |
|---|---|---|---|---|
| AmpR | A LysR-type global transcriptional regulator controlling the expression of ampC and other genes related to virulence, quorum sensing, biofilm formation, etc. | P. aeruginosa | A significant reduction of the ability to kill Caenorhabditis elegans with AmpR inactivation; with the G154R AmpR mutation, prevalent in the ST175 high-risk clone, reduced virulence in the same model of infection | 267 |
| CreBC/CreD | For CreBC, a two-component system and global metabolic regulator involved in the response against stress or biofilm growth, among others; for CreD an inner membrane effector protein of the CreBC system | P. aeruginosa | A decrease in fitness in in vitro competition experiments compared with that of the wild type, mainly in the presence of β-lactams; a decrease in the formation of biofilms, mainly in the presence of β-lactams; decrease of exoS expression, especially in the presence of ceftazidime | 276 |
| AmpD-AmpDh2-AmpDh3 | N-Acetyl-anhydromuramyl-l-alanine amidases that are involved in PGN turnover and recycling as well as AmpC repression and that are cytoplasmic (AmpD) or periplasmic and involved in stem peptide cleavage (AmpDh2 and AmpDh3) | P. aeruginosa | Severely compromised growth rates, motility, and cytotoxicity in the triple mutant; repression in key virulence factors, such as protease LasA, phospholipase C, or T3SS components; decreased virulence of circa 100-fold in Galleria mellonella model | 223 |
| NagZ, AmpG, and AmpD-AmpDh2-AmpDh3 | Key elements allowing PGN recycling (see the text for further details) | P. aeruginosa | Increased susceptibility to lysozyme and human PGRPs; if PGN blockade is combined with AmpC hyperproduction (for the AmpD-AmpDh2-AmpDh3 triple mutant), an even higher increase in susceptibility to lysozyme and PGRP2 specifically | 203 |
| CreBC | A two-component system and global metabolic regulator involved in the response against stress | S. maltophilia | A decrease in secreted protease activity | 277 |
| AmpD | A cytosolic 1,6-anhydro-N-acetylmuramyl– l-alanine amidase cleaving the stem peptide from 1,6-anhydro-N-acetylmuramic acid in a key step for PGN recycling | Salmonella enterica serovar Typhimurium | Upregulation during infection; in a knockout mutant, a decreased capacity to invade macrophages and more potent induction of the nitric oxide response of bacterial killing; decreased competitiveness in a BALB/c mouse model of infection | 85, 99 |
| AmpD | A cytosolic 1,6-anhydro-N-acetylmuramyl–l-alanine amidase involved in PGN recycling | N. meningitidis | An inability to cause systemic infection in an infant rat model; in wild-type strains, hyperexpression of AmpD during infection | 84 |
| AmpD | A cytosolic 1,6-anhydro-N-acetylmuramyl–l-alanine amidase cleaving the stem peptide from 1,6-anhydro-N-acetylmuramic acid in a key step for PGN recycling | Ralstonia solanacearum | Upregulation during infection; in a knockout mutant, reduced virulence in eggplant and tomato models | 85, 98 |
| NagZxc | A β-N-acetylglucosaminidase that cleaves the bond between 1,6-anhydro-N-acetylmuramic acid and N-acetylglucosamine | X. campestris | Reduced average lesion areas in cabbage infection model | 232 |
| AmpGxc | A specific permease for PGN fragments containing the disaccharide N-acetylmuramic acid–N-acetylglucosamine | X. campestris | Slightly increased average lesion areas in cabbage infection model | 232 |
| AmpC | A class C β-lactamase (noninducible due to the lack of the AmpR regulator in E. coli) | E. coli | In AmpH-AmpC double mutants, uneven contours and aberrant and asymmetric constrictions between dividing cells | 255 |
Some other examples of cytosolic targets, such as DapA from Serratia marcescens, involved in the synthesis of m-DAP, were previously gathered by Cloud-Hansen et al. (85). DapA was reported to intervene in swarming motility and envelope architecture, and its inactivation led to an altered cell wall structure and increased susceptibility to osmotic shock but, paradoxically, also to increased hemolysin production and cytotoxicity (85, 100), although the basis for these observations remains elusive.
Some other enzymes, such as those encoded by P. aeruginosa genes murA, murD, and murF, act during the last cytosolic steps of synthesis of the PGN precursor UDP-NAcMur-pentapeptide (101) (Table 4). More specifically, MurA intervenes in UDP-NAcGlc transformation into UDP-NAcMur, whereas MurD and MurF intervene in the ligation of d-Glu to UDP-NAcMur-l-Ala and the final d-Ala–d-Ala, respectively, to constitute the UDP-NAcMur-pentapeptide (102). They have very recently been described to dampen bacterial fitness and virulence when disrupted, probably because their inactivation entails disorders in PGN metabolism and a derived stress, although the exact effects have not been studied. In any case, what has been proved is that mutants with a single knockout of the cited genes are significantly impaired in their growth in minimal medium and display changes in colony morphology, and in the specific case of the murF mutant, abnormally elongated cells are observed by microscopy. In addition, all the cited mutants are more susceptible to macrophage-mediated killing and dampened in terms of growth in the lungs of intratracheally infected mice, intriguingly, with the exception of the murF mutant, which shows wild-type behavior. Also paradoxically, the murA mutant was the only one not impaired in terms of the pathogenesis elicited in a plant model (101) (Table 4).
THE PEPTIDOGLYCAN, A BARRIER TO BE LOCALLY MODIFIED IN ORDER TO ALLOW THE FULL PERFORMANCE OF ESSENTIAL VIRULENCE FACTORS
Again, from a partially structural point of view, the PGN has to be considered a barrier to be opened in order to allow the correct assembly, anchoring, and performance of structures that are considered very important for virulence, such as the flagella and secretion systems (which are indispensable for motility and the delivery of toxins, respectively) (59, 103). Therefore, the enzymes responsible for opening local gaps in the PGN linked to the insertion of these elements have been described in different Gram-negative species as targets to dampen motility and toxicity, also highlighting the strict control under which they usually work; since these enzymes have autolytic activities, they could lead to cell lysis if they work in a dysregulated manner (103–105). Table 5 displays different examples of PGN enzymes essential for the correct assembly of secretion systems and flagella, together with the species and the data regarding the specific role and the effects of inactivation of each target.
TABLE 5.
Targets related to the proper assembly and performance of essential virulence factors across the peptidoglycan
| Target | Role(s) | Species | Effect(s) of target disruption | Reference(s) |
|---|---|---|---|---|
| Secretion systems | ||||
| ExeAB | PGN binding and remodeling to allow secretin ExeD insertion and T2SS multimerization, assembly, and performance | A. hydrophila | Loss of aerolysin secretion and of lipase activity in culture supernatants | 107–109 |
| HpaH | A lytic transglycosylase required for T3SS assembly | X. campestris | Attenuated for pathogenicity in a plant model | 116 |
| HrpH, HopP1, and HopAJ1 | Lytic transglycosylases coregulated by the T3SS to allow the translocation of effector proteins into plant cells | P. syringae | Attenuated for pathogenicity in a tobacco plant model | 115 |
| L0045 | A putative lytic transglycosylase critical for T3SS | Enterohemorrhagic E. coli | T3SS impairment | 114 |
| EtgA | A lytic transglycosylase required for T3SS assembly | Enterohemorrhagic E. coli | Attenuated for T3SS activity and red blood cell lysis | 112, 113 |
| MltE | A lytic transglycosylase required for T6SS assembly | Enteroaggregative E. coli | Loss of T6SS function and, consequently, cytotoxicity impairment | 119 |
| VirB operon | A transglycosylase with activity that allows the formation of the T pilus, a subassembly of the vir T4SS | A. tumefaciens | Loss of tumorigenesis in a Kalanchoe daigremontiana plant model | 117 |
| AtlA | A transglycosylase with activity that allows the assembly of T4SS | N. gonorrhoeae | Loss of allolysis in E. coli | 118 |
| TagX | A l,d-endopeptidase required for T6SS assembly | Acinetobacter spp. | Loss of T6SS function and, therefore, cytotoxicity impairment; a decrease in bacterial killing capacity in E. coli coculture assays | 120 |
| PtlE | A peptidoglycanase responsible for local PGN degradation during Ptl secretion complex assembly | B. pertussis | Decreased release of pertussis toxin, leading to reduced effects (such as focal adhesion loss) on CHO cells | 121 |
| Flagellum | ||||
| FlgJ | A β-N-acetylglucosaminidase required for degradation of PGN to allow assembly of flagella | S. enterica and other beta- and gammaproteobacteria | Nonmotile phenotype | 122 |
| Sltf | A lytic transglycosylase interacting with FlgJ to allow the penetration of the nascent flagellar structure across the PGN | Rhodobacter sphaeroides | Nonmotile phenotype | 123, 124 |
| MltD | A lytic transglycosylase contributing to maturation of PGN for the proper anchoring and functionality of the flagellar motor | H. pylori | Nonmotile phenotype | 125 |
Secretion Systems
To cite some examples regarding the secretion systems, it must be stated that most information is related to the insertion and assembly of the systems that directly inject the effector proteins into the cytoplasm of the host cells: the type III, type IV, and type VI secretion systems (T3SS, T4SS, and T6SS, respectively). However, for the release of toxins to the extracellular medium without injection, the information on enzymes responsible for opening windows in the PGN to insert the type II secretion system (T2SS) machinery is much scarcer (and nonexistent for the type I and V secretion systems, to our knowledge) and is linked to only a few species, such as Aeromonas hydrophila. In this species, it has been shown that the assembly of the secretion channel ExeD (which is also called secretin and which is essential to enable the rest of the T2SS apparatus [106]) in the outer membrane is dependent on the inner membrane ExeAB complex (107–109) (Table 5). More specifically, it has been proposed that ExeA shows a PGN-binding region that could open pores in the PGN to allow the transport and assembly of ExeD (107–109). But interestingly and very recently, it has been shown that alterations in the degree of PGN cross-linking, achieved through the hyperexpression of the d,d-carboxypeptidase PBP5, allow the assembly of ExeD and, therefore, of T2SS in an ExeAB-independent manner (110).
In the case of T3SS, the linked PGN-degrading enzymes have been studied in-depth in Escherichia coli (mainly in enterohemorrhagic strains, with the EtgA enzyme being identified as a key actor in the process) and in Pseudomonas and some closely related genera, such as Xanthomonas (111–116) (Table 5).
In the case of T4SS and T6SS, which are involved not only in eukaryotic cell killing but also in the injection of toxins to eliminate bacterial competence, the species with which the most studies have been performed are quite diverse and heterogeneous: H. pylori, Agrobacterium tumefaciens, Neisseria gonorrhoeae, Acinetobacter spp., enteroaggregative E. coli, and even Bordetella pertussis, in whose pertussis toxin secretion apparatus (which belongs to T4SS and which comprises the products of the nine ptl [pertussis toxin liberation] genes) a peptidoglycanase (PGNase) activity linked to PtlE has been described (117–121).
Either way, what seems to be a general trend among the different space-making autolysins, which is the name given to these enzymes in a very interesting review on the topic (103), is their obvious PGN-degrading activity: they usually display the same enzymatic profile, breaking the bonds between the NAcMur and the NAcGlc disaccharide units of the glycan chains, in which case they are considered lytic muramidases or transglycosylases, although some exceptions do exist, such as the l,d-endopeptidase activity attributed to TagX of Acinetobacter spp. (120). Another general trend is the impairment of the toxicity and virulence of the different knockout mutants studied in these works, as is obvious for a dampened system of injection of effector proteins in the host cells (Table 5).
Flagella
In the case of enzymes that are indispensable for allowing the correct assembly of the flagellar structure through the PGN and, hence, that are essential for correct motility, given that the bodies of the described secretion systems and flagellar apparatus are similar, it could be expected that the profiles of the PGN-degrading enzymes involved would also be alike and would thus be considered typical lytic transglycosylases (122–124). Nevertheless, it has been shown that FlgJ, widely found in beta- and gammaproteobacteria, shows β-N-acetylglucosaminidase activity instead (122), although the final role may be the same, similar to what was previously found for the glucosaminidase Auto of the Gram-positive bacterium Listeria monocytogenes, shown to be indispensable for virulence and motility (29, 125). In the case of MltD from H. pylori, it has been shown that its activity is indispensable not only for correct assembly but also for the appropriate localization of the flagellar motor protein MotB to the bacterial pole to allow motility (Table 5) (125).
The motility itself has an obvious utility for bacteria, as it allows the cells to move in order to expand infection, and hence, a flagellum that cannot be correctly inserted into the PGN would dampen motility and, consequently, virulence (59). Nevertheless, it is important to state that flagella are more than simple motility motors, as they have been described to contribute to adherence to surfaces, to help with differentiation into biofilms, and to assist with the secretion of certain effector molecules, but also to allow further penetration through tissue structures or to activate phagocytosis to gain entry into eukaryotic cells (59, 126). Moreover, and in an opposite sense, it has been described that the loss of flagellar motility is usually selected in P. aeruginosa during chronic infection in cystic fibrosis patients, since it seems to entail an impairment of phagocytosis, an issue which adds complexity to the role of flagella during pathogenesis (127). Therefore, proper flagellar anchoring and performance can have wider implications for virulence than solely motility, which makes this topic a very promising antivirulence target worthy of study from a multidisciplinary point of view.
INTERACTION OF PEPTIDOGLYCAN WITH THE HOST
Release of Peptidoglycan Fragments as an Inflammation-Mediated Pathogenesis Mechanism
As opposed to what happens with Gram-positive bacteria, given the presence of the outer membrane and the lipopolysaccharide wrapping the Gram-negative PGN, it was classically thought to have a low degree of interaction with the host receptors (which will be reviewed below), at least when the bacteria are intact. Nevertheless, in a few Gram-negative bacteria, some fragments of their PGN have been typically described to be somehow actively delivered to the extracellular medium to exert diverse biological functions, generally entailing a strong inflammatory response. The targets related to this phenomenon are displayed in Table 6. Indeed, the role of B. pertussis tracheal cytotoxin (TCT) as a virulence factor heavily relies on its proinflammatory power: TCT is a muramyl peptide (specifically, the 921-Da fragment N-acetylglucosaminyl-1,6-anhydro-N-acetylmuramyl-l-alanyl-d-glutamyl-meso-diaminopimelyl–d-alanine) derived from PGN, thanks to the activity of still uncharacterized periplasmic lytic enzymes, which causes damage to the respiratory epithelia and extrusion of ciliated cells through the generation of increased levels of nitric oxide and the induction of a massive inflammatory response (128–130). Similar features have been described for PGN-derived fragments of N. gonorrhoeae and N. meningitidis (85, 131, 132). More specifically, these PGN fragments are referred to as PGN-derived cytotoxin (PGCT), which is a monomer identical to the TCT cited above. In N. gonorrhoeae, the main enzymes responsible for the generation of the PGCT were initially identified to be the lytic transglycosylases LtgA and LtgD (133, 134). Interestingly, what has also been shown for N. gonorrhoeae is that, in comparison with other nonpathogenic species of the genus (such as Neisseria sicca and Neisseria mucosa), the gonococcus shows a poorly efficient PGN recycling machinery (probably because of a less functional AmpG permease, the door to the cytosol for the PGN fragments to be recycled). This circumstance has been associated with the higher degree of PGN fragment release (and, hence, of PGCT) in N. gonorrhoeae than in other species from the genus. In fact, the blockade of cytosolic doors for PGN fragments through the inactivation of AmpG (but also of the Opp-MppA system, specific for oligopeptides) has been shown to increase the release of PGN fragments to the extracellular medium in E. coli or Vibrio fischeri, for instance (132, 135–137). Besides, the periplasmic l,d-carboxypeptidase LdcA, also of N. gonorrhoeae, has recently been shown to play a key role in the generation of fragments detectable by human NOD1 and NOD2 receptors (our intracellular specific PGN receptors, which will be reviewed below) and the derived responses (138) (Table 6).
TABLE 6.
Targets related to host detection and response against PGN
| Target | Role(s) | Species | Effect(s) of target disruption | Reference(s) |
|---|---|---|---|---|
| Targets involved in PGN fragment release | ||||
| AtlA and LtgA | Lytic transglycosylases allowing the release of the peptidoglycan-derived cytotoxin | N. gonorrhoeae | Reduction in monomeric released PGN | 133 |
| LtgD | A lytic transglycosylase contributing, together with AtlA and LtgA, to the release of the peptidoglycan-derived cytotoxin | N. gonorrhoeae | Absence of PGN monomer release and, instead, active release of large soluble fragments in an LtgA-LtgD double mutant | 134 |
| LdcA | A periplasmic serine protease l,d-carboxypeptidase cleaving the tetrapeptide that therefore provides tripeptide stems and that is also capable of breaking specific peptide cross-bridges (endopeptidase activity) | N. gonorrhoeae | Elimination of NOD1 and NOD2 activation by soluble PGN products from N. gonorrhoeae when LdcA is disrupted, presumably leading to a significant decrease in the inflammatory response | 138 |
| Slt | A periplasmic lytic transglycosylase that is necessary for the release of PGN fragments and whose expression is induced in vivo during infection | H. pylori | Reduction of the PGN fragment release and, consequently, of IL-8 delivery by the host | 142, 143, 145 |
| SltY | A periplasmic lytic transglycosylase homologue of H. pylori Slt | S. flexneri | Effects presumably similar to those derived from H. pylori Slt inactivation | 146 |
| MppA | A periplasmic binding protein allowing the entrance of oligopeptides into the cytosol | S. flexneri | Strong attenuation of a knockout mutant in nasal and intravenous infections in mice; increased PGN fragment release to the extracellular medium; increased NOD-1-mediated activation of the NF-κB route | 148 |
| AmpG | A permease specific for PGN fragments containing the disaccharide N-acetylmuramic acid–N-acetylglucosamine | S. flexneri | Increased PGN fragment release to the extracellular medium; increased NOD-1-mediated activation of the NF-κB route | 148 |
| AmpG | A permease specific for PGN fragments containing the disaccharide N-acetylmuramic acid–N-acetylglucosamine | V. fischeri | A circa 100-fold increase in net PGN monomer release | 137 |
| LtgA, LtgD, and LtgY | Lytic transglycosylases | V. fischeri | Very poor accumulation of PGN monomers in culture supernatants; increased susceptibility of the symbiont host squid Euprymna scolopes to superinfection | 137 |
| Host PGN receptors | ||||
| NOD1 | Detection of PGN fragments with a terminal m-DAP, promoting the secretion of inflammatory cytokines in response | Homo sapiens | Contradictory results, depending on the work; in some studies, a key role of the receptor in the innate immune defense against Gram-negative bacteria and the contrary finding in others | 174–176, 179, 180–183 |
| NOD2 | Detection of MDP, promoting the secretion of inflammatory cytokines in response | Homo sapiens | Contradictory results depending on the work; in some studies, a key role of the receptor in the innate immune defense against Gram-negative bacteria and the contrary finding in others | 173–175, 180–184 |
| NLRP1 | Detection of MDP and activation of the inflammasome in response | Homo sapiens | Pyroptosis, acute lung injury, and morbidity in a mouse model with activation of NLRP | 185 |
| Hexokinase-NLRP3 | Detection of N-acetylglucosamine and activation of an inflammasome in response | Homo sapiens | Expression of IL-1β and IL-18, generation of reactive oxygen species, pyroptosis, and acute lung injury with activation of NLRP3 in a mouse model | 189–191, 195, 196 |
| PGRP2 | A peptidoglycan recognition protein with amidase activity; controversial reports regarding its alleged bactericidal capacity and inflammation regulatory capacity after PGN detection | Homo sapiens | In some studies with PGRP2 deletion in mice, a better outcome for the host after infection (linked to a decrease in the inflammatory response); protection against infection in other studies | 170, 245–248 |
| Bacterial lysozyme inhibitors | ||||
| Ivy | An inhibitor of vertebrate lysozyme (c-type lysozyme inhibitor) | E. coli | Increased in vitro susceptibility to lysozyme plus lactoferrin and saliva | 207, 208 |
| MliC | A membrane-bound inhibitor of c-type lysozyme | E. coli | Increased susceptibility to lysozyme and serum and decreased virulence in chicken infection model | 205, 209 |
| Ivy | An inhibitor of vertebrate lysozyme (c-type lysozyme inhibitor) | Edwardsiella tarda | Impaired tissue dissemination, resistance to serum, replication within macrophages, and overall virulence in a turbot (Scophthalmus maximus) model | 211 |
| MliC | A membrane-bound inhibitor of c-type lysozyme | Edwardsiella tarda | Impaired tissue dissemination, resistance to serum, replication within macrophages, and overall virulence in a turbot (Scophthalmus maximus) model | 212 |
| Ivy | An inhibitor of vertebrate lysozyme (c-type lysozyme inhibitor) | Yersinia pestis | Hypersusceptibility to lysozyme and polymorphonuclear phagocytes | 210 |
| MliC | A membrane-bound inhibitor of c-type lysozyme | S. enterica serovar Typhi | A reduction in intramacrophage survival | 204 |
| PliC | A periplasmic inhibitor of c-type lysozyme | S. enterica serovar Enteritidis | Hypersusceptibility to lysozyme plus lactoferrin | 204 |
| PliG | A periplasmic lysozyme inhibitor of g-type lysozyme | E. coli | Hypersusceptibility to g-type lysozyme | 219, 220 |
| PliI | A periplasmic inhibitor of i-type lysozyme | Aeromonas hydrophila | Decreased tolerance against Tapes japonica shell lysozyme | 218 |
| LipA and LipB | Potential c-type lysozyme inhibitors with structural similarities to MliC/PliC proteins | Moraxella catarrhalis | Hypersusceptibility to human lysozyme plus apolactoferrin | 222 |
Moreover, the range of Gram-negative species in which the release of PGN fragments is considered an active mechanism to damage the host and, hence, in which the cited PGN fragments could also be considered bona fide toxins is getting amplified and includes, for instance, Haemophilus influenzae, in which some PGN fragments and specific modifications to them have been shown to induce different degrees of inflammation and brain edema in animal models of meningitis (139–141). Still in this context, in H. pylori the delivery of PGN fragments into host cells via OMVs and via the cag type IV secretion system has been proven, leading to host tissue self-injury mediated by an excessive inflammatory response (142, 143). In fact, it has been proposed that H. pylori does not display a significantly productive PGN recycling pathway (which resembles that in the already mentioned case of gonococcus), which supports the presence of major PGN degradation fragments readily available in the periplasm and, consequently, in the extracellular medium, being detected by our specialized receptors to elicit the release of inflammatory cytokines, such as interleukin-8 (IL-8). Linked to this, it has been demonstrated that the lytic transglycosylase Slt is necessary for IL-8 secretion in this context and, moreover, that the production of Slt is enhanced in vivo in human gastric biopsy specimens, suggesting that the release of PGN fragments occurs actively and that the triggered inflammation is a desired effect for the pathogen (142, 144, 145). Finally, Shigella flexneri shares some of the last cited features: (i) its SltY lytic transglycosylase (a homologue of H. pylori Slt) is upregulated during infection (144, 146), and (ii) when PGN recycling is blocked (by inactivating either ampG or mppA), the release of PGN fragments increases, in turn leading to a higher NOD1-mediated NF-κB activation of inflammatory cytokine genes in comparison with that in the wild type. Nevertheless and intriguingly, only the mutant with a mutation in MppA was shown to be impaired for virulence in murine models (147, 148), once more showing the high level of complexity of the interplay that we review here.
The biologically active PGN fragments reviewed in this section, derived from active degradation during bacterial growth, are qualitatively and quantitatively different from those released from lysed bacteria, which, however, can also stimulate the innate immune system through our PGN-specific receptors (85). Therefore, this classic concept of the Gram-negative PGN as a structural element with no additional biological functions is probably becoming obsolete (144). For this reason, maybe not only the cited species (B. pertussis, N. meningitidis, N. gonorrhoeae, H. pylori, or S. flexneri) but also others should be considered active PGN fragment deliverers; these other PGN fragment deliverers may also release PGN fragments by means of OMVs, for instance, during regular growth and not only after cell lysis (149–151). This concept could be supported by the fact that some Gram-negative species have been shown to upregulate certain genes related to PGN metabolism during infection, such as the mentioned MtgA from Brucella spp. or MltE of Erwinia amylovora (85) (Tables 3, 4, and 6).
In this sense, it has to be noted that some works have recently shown in E. coli but also in other more niche-specific species, such as some periodontal pathogens (Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia), the association between their released OMVs and the activation of NOD1 and/or NOD2 in the host, which strongly suggests that the presence of PGN fragments within the cited vesicles potentially contributes to the inflammatory response in the infected tissues (152–154). In fact, in recent years OMVs have been recognized to be effective virulence factor delivery systems, and hence, the presence of certain PGN fragments within OMVs could contribute to this role (155–157). Moreover, to conclude and as additional evidence that Gram-negative PGN biology interacts with pathogenesis (in this case, with a close relation to OMVs), some studies have directly related PGN dynamics to the modulation of OMV production levels, which would have obvious consequences for virulence (158, 159). In short, it has been proposed that modulation of the PGN-outer membrane cross-linking level through the localized fine-tuning of PGN degradation and synthesis (therefore involving different actors, such as endopeptidases; nonessential PBPs; and the Lpp lipoprotein, which cross-links the PGN with the outer membrane) is a very tightly regulated mechanism that Gram-negative bacteria use to modulate vesiculation and contributes to a better adaptation of the bacterium to stress. For instance, the authors suggested that an inverse relationship does exist between OMV production and the PGN-Lpp cross-linking level (158, 159). Nevertheless, quantification of the potential effects derived from this PGN biology-driven OMV level modulation on virulence has not been performed yet.
Host Receptors Involved in the Detection of Gram-Negative Peptidoglycan: Potential Targets To Reduce the Self-Injury-Mediated Pathogenesis of Infections
After mentioning some relevant examples of biologically active Gram-negative PGN fragments and the enzymes involved (an excellent overview of their principal effects on the host was presented by Boneca in his work of 2005 [144]), a brief overview of their human-specific receptors seems suitable (Table 6). A very recent review on the topic, which focused only on P. aeruginosa, was published in 2018 and could probably be seen as a nice reference for the rest of the Gram-negative bacteria (160). In this sense, the main response after the detection of PGN by host cells is the induction of an inflammatory response. This is mainly mediated by the cytoplasmic receptors NOD1 and NOD2, although other actors may take part in Gram-negative PGN sensing, as will be mentioned below. NOD1 recognizes PGN fragments with a terminal meso-diaminopimelic acid (m-DAP), such as NAcGlc–NAcMur–l-Ala–d-isoGlu–m-DAP (GM-triDAP) or NAcMur–l-Ala–d-isoGlu–m-DAP (M-triDAP), specific for Gram-negative bacteria. NOD2 is a more general sensor, as it can recognize muramyl dipeptide (MDP), which is common to Gram-positive bacteria. NOD2 was classically thought not to recognize muropeptides containing m-DAP, although it has recently been shown to be able to bind these fragments even better if m-DAP is not at a terminal position (161–163). In any case, the recognition of their PGN ligands by these professional receptors triggers the activation of the innate immune response, mainly through the NF-κB pathway, but also that of the AP-1 transcriptional regulator, enhancing the expression of inflammatory cytokines, such as IL-1β, IL-6, IL-8, and tumor necrosis factor alpha (161, 164, 165). Interestingly, in this context, it has been described that modifications in the PGN composition trigger different levels of activation of NOD receptors, eliciting differential levels of inflammation, and that this strategy is used by P. aeruginosa during chronic infection in cystic fibrosis patients to lower immune detection and, consequently, ameliorate chronic persistence (166). Related to this, in the in vitro context it has recently been described that the amidation of m-DAP from the stem peptides in the Gram-negative PGN structure would lead to the evasion of detection through NOD1 (167). Nevertheless, to our knowledge, no studies of the potential existence of this mechanism in natural strains exist, and it could be worth performing them in order to gain knowledge of unknown strategies that bacteria can use to avoid immunity mainly in the chronic infection context.
Still related to these topics, it is generally accepted that an excessive level of triggered inflammation leads to self-injury in the host tissues, constituting the pathogenic basis linked to PGN detection classically attributed to Gram-positive bacteria but increasingly to certain Gram-negative bacteria. In fact, different models of PGN-induced inflammatory pathologies (arthritis, uveitis, etc.) have been developed (168–170), and NOD1 and/or NOD2 alterations have been linked to chronic inflammatory diseases, such as asthma, Crohn's disease, or Blau syndrome, among others (161, 165, 168, 171).
Regarding the real impact of these receptors on the innate defense against Gram-negative bacteria, some contradictory results have been published. For instance, NOD2 has been shown to be dispensable for the innate macrophage response against Yersinia enterocolitica or for the control of Coxiella burnetii infection (172, 173), as have both NOD1 and NOD2 for the proper resolution of Brucella abortus infection in a mouse model (174). In the same sense, the inactivation of NOD1 or NOD2 did not increase the susceptibility of mice to polymicrobial sepsis (175).
On the contrary, in several other models of infection, these receptors have been shown to take part in the innate response against various Gram-negative species: for instance, NOD1 against P. aeruginosa (176), H. pylori (177), Shigella flexneri (147), Campylobacter jejuni (178), or Legionella pneumophila (179). In a model of infection with Legionella pneumophila, NOD1 knockout mice were especially impaired in bacterial clearance, neutrophil recruitment, or inflammatory cytokine production, for instance (179). Moreover, NOD1 and NOD2 together have repeatedly been shown to play an important role in the defense against other Gram-negative pathogens, such as N. gonorrhoeae (180), S. enterica serovar Typhimurium (181, 182), and Chlamydophila pneumoniae (183), and in the general neutrophil response (184).
Besides NOD1 and NOD2, some additional cytosolic receptors have been suggested to take part in Gram-negative PGN sensing, although the amount of information on the topic and evidence of real biological significance are still scarce. For instance, NLRP1 is allegedly capable of sensing certain PGN fragments, such as the MDP (common to both Gram-positive and Gram-negative species) and of promoting inflammatory cytokine secretion (mainly IL-1β and IL-18) in response (through inflammasome activation). It has been connected with the pathology of acute lung infections, always related to overinflammatory self-injuries (185–189). Moreover, in a recent work, Wolf and colleagues (189) have described a very novel mechanism of PGN fragment sensing: the inhibition of our enzyme hexokinase by the NAcGlc proceeding from bacterial PGN causes its dissociation from mitochondrial outer membranes, which in turn activates the NLRP3 inflammasome, all of which lead to the expression of IL-1β and IL-18 (189, 190). In addition, the activation of NLRP1 or NLRP3 inflammasomes has been linked not only to inflammatory cytokine secretion but also to pyroptosis (a programmed cell death mode intimately linked to inflammatory processes) of host cells, therefore contributing to pathogenesis during infection (160, 185, 190, 191–193). In fact, in some other works, as happens with NOD receptors, the involvement of NLRPs and inflammasomes has been linked to certain chronic inflammatory diseases (194). However, as stated before, the real impact of Gram-negative PGN detection through NLRP1 and/or NLRP3 on the pathology of infection has yet not been truly quantified. However, what has recently been shown in several works is that NLRP1 and/or NLRP3 activation (mediated or not by PGN sensing, a circumstance not analyzed enough for the moment) decisively contributes to inflammation and pyroptosis-mediated acute tissue injury in different models (190, 191, 195, 196) (Table 6). Therefore, these works support the idea of self-injury caused by an overinflammatory response to PGN detection to be one of the bases for the pathogenesis of infection, proposing the tuning of the inflammatory response as a strategy for future treatments (197). In fact, in an organism not classically considered to be an active releaser of PGN fragments, such as P. aeruginosa, the detection of these delivered signals has been linked to the virulence of the infection (involving NOD and probably NLRP receptors), once more supporting the idea of the Gram-negative PGN having a considerable and probably neglected importance during infectious disease pathogenesis, with, in this case, importance specifically being related to the detection of and response by the host (176, 186).
Lysozyme Inhibitors: Targets To Increase the Activity of the Innate Immune System
Besides the information regarding the detection of PGN by the host and the elicited response that has been provided, another kind of interaction with the host must be approached: that of the bacteria with the innate immune proteins targeting the PGN and, more specifically here, with the lysozyme. In this regard, it must be stated that several bacterial species seem to have some active mechanisms to avoid the alleged bactericidal power of this humoral weapon that enzymatically cleaves the β-1,4 linkages between NAcMur and NAcGlc in the murein sacculus, finally leading to osmotic cell lysis (198). These bacterial mechanisms are generically called lysozyme inhibitors. We will try to highlight the most important lysozyme inhibitor-related findings for Gram-negative pathogens in the next lines and in Table 6, but for an exhaustive review on the topic, including findings for nonhuman pathogens and inhibitors not only of lysozyme but also of other muramidases that bacteria inject into competitors, an excellent review by Callewaert and colleagues (199) can be consulted.
First, it is worth mentioning that some works have suggested that the muramidase activity of lysozyme is not necessary for its bactericidal power, since it displays membrane permeabilization action, therefore actually working as a cationic peptide (198, 200–202). Additionally, it has to be stated that many works have proposed that lysozyme generally has low activity per se against Gram-negative bacteria, unless the outer membrane is permeabilized by some other innate immune compound (198, 203). However, different bacterial inhibitors of the three types of lysozyme that exist in the animal kingdom (chicken or conventional [c-type], goose [g-type], and invertebrate [i-type] lysozymes) have been described (204). The Gram-negative species in which these inhibitors have been studied more often is probably E. coli, in which Ivy and MliC (each one of which belongs to a different protein family) were first described to be inhibitors of c-type lysozyme (205, 206). An Ivy KO mutant showed hypersusceptibility to in vitro treatment with lysozyme and permeabilizers (such as lactoferrin) and also to saliva (207, 208), and similar features were found for an E. coli MliC mutant (205). Nonetheless, the importance of these two E. coli inhibitors has been assessed in additional assays (a serum resistance assay and a subcutaneous chicken infection model) with controversial results: disruption of mliC clearly decreased the in vivo virulence, whereas the absence of Ivy produced no effect. Interestingly, the double KO mutant showed behavior similar to that of the wild type, suggesting the existence of additional inhibitors and a regulatory interaction in the expression of these different elements (209). Meanwhile, the disruption of mliC was shown to reduce the intramacrophage survival of S. enterica serovar Typhi, similar to what happened with S. enterica serovar Enteritidis homologue (PliC) inactivation, which caused increased lysozyme susceptibility in the presence of lactoferrin (205). Therefore, the importance of these inhibitors is not always the same and heavily depends on the species that we are considering. In P. aeruginosa, for instance, the deletion of Ivy or MliC homologues has been shown to produce no effect with regard to lysozyme susceptibility, as happened for MliC of Yersinia pestis, which was not required for lysozyme resistance and the development of plague (203, 205, 210). On the contrary, the inactivation of Ivy in Y. pestis caused hypersusceptibility to lysozyme, neutrophil-mediated killing, and virulence attenuation in a rat infection model (210). However, a closely related species, such as Yersinia pseudotuberculosis, did not require Ivy either to counteract the lysozyme activity or for virulence (210). On the contrary, in Edwardsiella tarda, an unusual human pathogen, both Ivy and MliC have been shown to be necessary for efficient tissue dissemination, resistance to serum, replication within macrophages, and overall virulence (211, 212). The presence of homologues of MliC/PliC in other Gram-negative pathogens has been reported, although to date, their impact on virulence has not been studied: Aeromonas hydrophila (213), Salmonella enterica serovar Typhimurium (214), Brucella abortus (215), etc. (199).
Finally, it has to be mentioned that some works suggest that the true physiological function of the Ivy proteins is to control the autolytic activity of lytic transglycosylases within the periplasm of Gram-negative bacteria and that the inhibition of exogenous lysozyme by Ivy is a fortuitous coincidence, which could add more controversy to the real role of lysozyme inhibitors in Gram-negative bacteria (216). In any case, in general terms, the above-mentioned inhibitors are quite specific and do not show significant cross inhibition against other lysozyme types, with the exception of Ivy, which weakly inhibits the g-type lysozyme (217, 218). Moreover, much scarcer information regarding the inhibitors of the other two types of lysozyme (i-type or g-type lysozymes) has been published. In the case of the invertebrate type, the PliI protein of Aeromonas hydrophila has been shown to contribute to resistance in the presence of an outer membrane permeabilizer (218). In the same work, the existence of similar i-type inhibitors in Serratia marcescens and Proteus mirabilis was proposed (218). Besides, PliG has been proposed to be a g-type inhibitor in E. coli and shows no activity against the other lysozyme types (219). Deletion of pliG increased the in vitro susceptibility to the g-type lysozyme (220) but showed no effect on serum resistance or virulence in a chick infection model (Table 6) (209).
Therefore, to conclude, as can be seen in these last few paragraphs, the impact and importance of the lysozyme inhibitors are complex issues, since they are quite variable, depending on the species and conditions. Nevertheless, what does seem clear is that they have at least some degree of participation in virulence. This would be also supported by the fact that the levels of expression of some inhibitors seem to be tightly regulated and under the control of two-component systems, presumably responding to the stress on PGN that the presence of lysozyme entails. This has been proposed for E. coli (the Rcs two-component system regulon includes the Ivy- and MliC-encoding genes [221]) and Moraxella catarrhalis. In this last species, the two-component system MesRS has been shown to finely tune several genes necessary for infection development; among them are the genes encoding the proteins LipA and LipB, which show structural homology to bacterial lysozyme inhibitors. These two proteins were shown to inhibit human lysozyme activity in vitro and in saliva, and a double KO mutant was impaired for resistance to lysozyme in the presence of permeabilizers (Table 6) (222).
THE REGULATION OF INTRINSIC β-LACTAMASES AND PEPTIDOGLYCAN METABOLISM: INTIMATELY LINKED PROCESSES INFLUENCING VIRULENCE
Overview of Interaction between Intrinsic β-Lactamase Regulation and Peptidoglycan Metabolism
As shown in the previous sections, the amount of information on structure, metabolism, host detection, and the inflammation implications for the PGN-virulence interplay is notable. However, many fewer works have linked PGN biology to virulence together with the production of β-lactamase regulation, mainly if we consider only the intrinsic β-lactamases. Indeed, it is tempting to speculate that the production of an intrinsic β-lactamase, at least at regular levels, should not entail any biological cost, but what about when the enzyme is overproduced? Very few works have tried to answer this question (203, 223). Either way, the targets that we have identified in the contexts mentioned above are displayed in Table 7.
Although the array of mechanisms of intrinsic β-lactamase regulation in Gram-negative bacteria is very wide (13), one of the most extended models and, consequently, the model for which a higher number of virulence-related publications exists is for AmpC from P. aeruginosa or certain Enterobacteriaceae, among other species. Several elements take part in this mechanism (which is intimately linked to PGN recycling), with the most important probably being AmpG, NagZ, AmpD, and AmpR, although additional actors have been described (13, 19, 224). For the sake of clarity, Fig. 2 shows a scheme of this regulatory pathway with P. aeruginosa as a model, which is taken from the work of Juan et al. (13), and is a useful reference on this specific topic of intrinsic β-lactamase regulation. Briefly, the products of PGN remodeling released during growth or degradation, thanks to the effects of β-lactams, reach the cytosol through the permease AmpG (specific for NAcGlc-1,6-anhydromuropeptides). Once there, these PGN fragments are processed by the β-N-acetylglucosaminidase NagZ to generate 1,6-anhydromuropeptides, which under regular conditions are metabolized by the N-acetylanhydromuramyl-l-alanine amidase AmpD to release the stem peptides, which enter the recycling circuit to finally generate UDP-NAcMur-pentapeptides (therefore, AmpD is a very important enzyme for PGN recycling). A certain proportion of these last elements, besides being exported to the nascent PGN, bind the LysR-type transcriptional regulator AmpR, and this complex is a repressor of ampC expression (allowing only basal levels of AmpC production) (225). On the contrary, when the amount of PGN fragments is higher because of the action of an inducer β-lactam, such as cefoxitin or imipenem, AmpD may get saturated; the 1,6-anhydromuropeptides, not metabolized by AmpD, could displace the UDP-NAcMur-pentapeptides from AmpR; and the new complex is an inducer of ampC expression (18, 226). Similarly, the mutational inactivation of AmpD (usually selected during β-lactam treatments) would presumably lead to the accumulation of 1,6-anhydromuropeptides and lead to ampC stable hyperexpression, resulting in clinically relevant levels of β-lactam resistance (13, 227, 228). Besides these phenomena, the nonessential penicillin binding protein 4 (PBP4) has been shown to act as a kind of sentinel for PGN damage against inducer β-lactams. Under these conditions or when its encoding gene (dacB) gets disrupted by mutation (which is very usually selected during treatment; in fact, it was the first cause of β-lactam resistance among P. aeruginosa clinical strains [229]), the situation is sensed by CreC (which belongs to the two-component system CreBC [BlrAB]), which in turn activates a complex and not fully understood parallel response that finally leads to enhancement of the effectiveness of AmpC production and the derived resistance level (Fig. 2) (230). Interestingly, whatever the route of AmpC hyperproduction, it has been shown that it is always AmpR dependent, since this transcriptional regulator finally controls the levels of ampC expression (13). Among the actors cited to be involved in the process, AmpG and NagZ have been shown to be indispensable for AmpC hyperproduction, since they allow, respectively, the physical entrance into the cytosol and an essential biochemical transformation (scission of NAcGlc) to obtain future ampC-inducing signals (via AmpR binding) (229, 231). Interestingly in this regard, in a closely related species such as Xanthomonas campestris, NagZ has been shown to be indispensable not only to allow a significant expression of its intrinsic class A AmpR-regulated β-lactamase but also for its full pathogenicity (Table 7) (232). This work revealed a novel connection between β-lactamase regulation, PGN biology, and virulence never found in any other species. Intriguingly, in the same work, an AmpG mutant was found to be more virulent than the wild type. Whether this last fact is related to an increased release of PGN fragments into the extracellular medium, as described before for N. gonorrhoeae, remains to be elucidated (136). It is also unknown why the cleavage of NAcGlc could be essential for virulence in X. campestris (232), but it opens up interesting horizons in the interplay with β-lactamase virulence that may be exportable to other species of clinical interest.
FIG 2.
Overview of intrinsic β-lactamase induction and overexpression mechanisms (under the control of the LysR-type regulator and CreBC system) and their relatedness to PGN metabolism, taking P. aeruginosa AmpC as a model.
Besides the effects of these two elements (AmpG and NagZ), the roles of AmpD (and its homologues) (233, 234) in β-lactamase production but also in virulence have been investigated in some species (Table 7). To start with, in S. enterica serovar Typhimurium, although it has lost its ampC and ampR genes during evolution, the inactivation of AmpD has been shown to cause a dramatic decrease in virulence (99). More specifically, the reduced capacity for macrophage invasion and for intracellular growth is likely linked to the more potent activation of the bactericidal inducible nitric oxide synthase in the macrophage that was measured. An AmpD knockout mutant also showed impaired competitiveness in a BALB/c mouse model of infection. Interestingly, in the same work, the authors demonstrated that an AmpG mutant behaved like a wild-type strain in the cited parameters, and then they concluded that, more than the impairment of PGN recycling (obviously achieved through AmpG disruption but also through the disruption of AmpD), the basis for virulence attenuation was the cytoplasmic accumulation of PGN fragments derived from the absence of an active AmpD. In this regard, the authors suggested that, as well as a signaling role for β-lactamase induction through AmpR binding, some PGN fragments could also bind to other regulators, such as SinR or SpvR, finally modulating the expression of genes related to virulence (99). A similar hypothesis (although opposite) could be formulated for the above-cited case of X. campestris, in which NagZ would be essential to allow a reaction indispensable for the generation of PGN-derived inducing signals that would work as activators of virulence genes after binding to unknown regulators (232).
Interestingly, and as an almost unique feature in the microbial world (a few exceptions do exist, such as Yersinia enterocolitica [235]), P. aeruginosa shows two additional homologues of the cytosolic AmpD amidase, namely, AmpDh2 and AmpDh3 (19, 233, 234, 236, 237). Other species, such as members of the family Enterobacteriaceae, show only one additional homologue (AmiD). AmpDh2 has been shown to be anchored to the outer membrane with the active site oriented toward the periplasm and to be the orthologue of the above-mentioned AmiD, whereas AmpDh3 is a periplasmic amidase (233, 234, 236, 237). Therefore, in contrast to AmpD, AmpDh2 and AmpDh3 have been proposed to start the reactions of PGN turnover already in the periplasm (233, 234). Moreover, the presence of two additional homologues of AmpD has been interpreted to be an advantage for P. aeruginosa, since this feature would allow the selection of AmpD-inactivated mutants showing increased levels of constitutive AmpC production and β-lactam resistance but still retaining full fitness and virulence levels, since the mutants would still maintain two active homologues (237). Moreover, it was shown that through the consecutive inactivation of the AmpD homologues, growing levels of AmpC production were achieved, reaching constitutive derepression of the chromosomal cephalosporinase in the triple AmpD-AmpDh2-AmpDh3 knockout mutant (236, 237). The high biological cost recently demonstrated for the triple AmpD mutant supports the idea that this mutant is unable to compete in the natural environment, although the existence of natural double mutants (AmpD-AmpDh2 or AmpD-AmpDh3, both with high levels of AmpC production, in contrast to AmpDh2-AmpDh3, which showed wild-type levels) needs to be further investigated (223). In this regard, it was shown that the inactivation of the three amidases, besides AmpC derepression and PGN recycling impairment, caused a dramatic effect on fitness and pathogenicity, severely compromising growth rates, motility, and cytotoxicity, with the effect on cytotoxicity likely being achieved thanks to the repression of key genes, such as those for protease LasA, phospholipase C, or T3SS components (223) (Table 7). Further, the two circumstances that were shown to be indispensable for obtaining this virulence-attenuated phenotype were AmpC derepression in a background of PGN recycling impairment, and hence, other pathways triggering similar levels of AmpC production and resistance, such as those involving dacB disruption, showed no biological cost, even under conditions of CreBC system deletion (223). Although the energetic burden of AmpC derepression or of the PGN recycling blockade or the total loss of amidase activity per se was ruled out as the molecular basis for this attenuated phenotype, the molecular basis for this attenuated phenotype has not yet been ascertained. Therefore, the hypothesis of recycling impairment plus derepressed AmpC-derived accumulation of PGN fragments potentially acting as virulence inhibitory signals (allegedly through binding to transcriptional regulators), similar to that proposed by Folkesson and colleagues (99), still needs to be studied.
Finally, an additional gene in P. aeruginosa with potential virulence implications that has recently been shown to take part in PGN recycling and whose disruption causes increased levels of ampC expression (circa 20-fold compared with that of the wild type) and derived increases in β-lactam MICs (1.5-fold for ceftazidime, for instance) is mpl, which encodes a UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase and which allows the cytosolic ligation of the new UDP-NAcMur units with recycled tripeptides (238, 239). Although its inactivation has been reported in strains from patients with chronic infections (cystic fibrosis) but also acute infections (ventilator-associated pneumonia), with strains with inactivated mpl showing a progressive loss of virulence during the development of disease and with mpl inactivation clearly being linked to selection during β-lactam treatment, the direct cause-effect relationship in terms of mpl disruption influencing virulence has not yet been established (240, 241). Thus, although the attenuation of certain virulence features during cystic fibrosis and other chronic infections is a classically accepted trait for many species of opportunistic Gram-negative bacteria (242) and a general inverse correlation between the levels of antibiotic resistance and virulence is often established (also in acute infections) (17), the identified targets simultaneously related to intrinsic β-lactamase production, PGN metabolism, and virulence are still scarce.
Interplay among β-Lactamase Production, Peptidoglycan Recycling, and Susceptibility to Innate Immune Proteins Targeting the Cell Wall
Further, the analysis of a set of P. aeruginosa mutants very similar to those used in the above-mentioned work by Pérez-Gallego and colleagues (223) has recently revealed that, besides fitness and virulence attenuation, PGN recycling impairment (together or not with AmpC derepression) determines a great increase in bacterial susceptibility to innate immune weapons targeting the PGN, such as lysozyme and the four human PGN recognition proteins (PGRPs) (203). Lysozyme (also known as muramidase or N-acetylmuramide glycanhydrolase) is especially abundant in secretions and in neutrophil granules and catalyzes the hydrolysis of 1,4-beta linkages between NAcMur and NAcGlc. Therefore, as stated above, its bactericidal activity was initially linked to the PGN-degrading capacity, although some evidence of its nonenzymatic action has also been reported, with a detergent-like activity of lysozyme being suggested (200, 201). Besides, the PGRPs were initially described to be PGN binding, but in the case of PGRP1, -3, and -4, they do not show any PGNase activity. However, they have bactericidal power through their induction of a complex response mainly based on overactivation of the two-component system CpxA-CpxR (entailing oxidative, thiol, and metal stresses), finally leading to bacterial suicide (243, 244). Conversely, PGRP2 shows amidase activity (and, hence, a certain PGNase capacity), but its alleged bactericidal power or its implication in the regulation of inflammation in response to PGN detection is still controversial (170, 245–247). In fact, in some work with knockout mice, PGRP2 seems to be protective against the pathogen (S. Typhimurium infection [246]), whereas in other work, it looks like a proinflammatory actor, finally leading to a worse outcome for the host (P. aeruginosa keratitis, as described by Gowda et al. [245]), which indicates the complexity of these innate immune compound roles (170, 248) (Table 6). Besides, PGRP2 has also been linked to certain chronic pathologies allegedly unrelated to infections, hence adding even more interest to this protein's capacities and potential interplays (249, 250).
Either way, the work of Torrens and colleagues (203) showed that, once the permeability barrier is overcome, the activity of lysozyme and the four human PGRPs is dramatically enhanced when inhibiting key peptidoglycan recycling components (such as the 3 above-mentioned AmpD homologues, AmpG, or NagZ), indicating a decisive protective role for cell wall recycling against the innate immune aggressions cited earlier (Table 7). Even though the exact way in which PGN recycling could exert this protective role remains elusive, some hypotheses can be formulated: it has been described that PGRP1, -3, and -4, besides overactivating CpxA-CpxR, exert their activity by affecting the biology of PGN by dampening the initial steps of its anabolic pathways, and thus, in this situation, PGN recycling would become essential to avoid perturbation of the metabolism and of the cell wall itself that the activity of the above-cited innate proteins could entail as bactericidal mediators. Obviously, under these circumstances, PGN recycling would become a key protection (203). Furthermore, it was specifically shown for lysozyme and PGRP2 that an even higher bactericidal activity level was achieved by blocking PGN recycling and simultaneously overexpressing AmpC (which happens in the AmpD-AmpDh2-AmpDh3 triple knockout mutant). This phenotype was found to be associated with a circa 30% decrease in the amount of PGN per cell (203). The reasons why the PGN blockade together with AmpC hyperproduction negatively influences the resistance of P. aeruginosa to lysozyme and PGRP2 have not been determined to date, although they are presumably linked to the reduction in the PGN amount per cell mentioned above. It has been accepted that P. aeruginosa has a PGN relatively thinner (approximately 30%) than that of other Gram-negative bacteria (33, 34), and this could be the basis for the increase in susceptibility to lytic aggressions, such as those exerted by lysozyme and PGRP2, especially if it affects certain regions likely to be essential for counteracting osmotic pressure (33, 34, 203). The explanations for these results could involve the energetic burden of producing very high levels of AmpC, added to a direct effect of the β-lactamase on PGN physiology, likely due to residual peptidase activity reminiscent of its potential PBP ascendance, which has been widely reported in the past (Table 7) (48, 251–254). In fact, at least in E. coli, AmpC has been shown to somehow contribute to its regular morphology, since its inactivation, if it occurs together with that of LMM-PBP AmpH, rendered aberrant morphologies, which could somehow support the idea of a β-lactamase directly acting on the PGN structure and/or composition (255, 256). It still needs to be elucidated whether these effects are exclusive to the AmpC-type β-lactamases (and, consequently, linked to a potential exclusive remaining activity) or could be achieved by any kind of β-lactamases.
The Effect of Global Regulators: Controlling Not Only Intrinsic β-Lactamase Production but Also Virulence
The previously cited LysR-type regulators (denominated AmpR, in most cases), whose activity allegedly occurs through the binding of different PGN fragments, have been shown to be indispensable for allowing the expression of significant levels of intrinsic β-lactamases, such as AmpC from P. aeruginosa and some Enterobacteriaceae, PenA and AmpC from BCC, and L1 and L2 from S. maltophilia (13, 257, 258). These circumstances have been known for decades for some species (259, 260), for which even a detailed characterization of the structure of the regulator but also the PGN fragments capable of binding to it have recently been published. In this sense, AmpR from P. aeruginosa and its PGN-derived activating ligands during induction have been studied in depth (18, 19, 225). Nevertheless, the knowledge regarding the capacity of LysR-type regulators to regulate the expression of wider features of the bacteria, in addition to β-lactamase production, is more recent. In P. aeruginosa, for instance, AmpR has been shown to regulate not only the expression of ampC but also the expression of a repertoire of more than 500 genes; it is a positive promoter of virulence factors relevant in acute infections, while it is a repressor of biofilm formation, a key driver of chronic infections. The above-mentioned array also includes genes involved in iron acquisition, heat shock, and oxidative stress responses, among others (261–264). Moreover, Sánchez-Diener and colleagues have recently reported that the inactivation of AmpR but also some specific polymorphisms strongly linked to certain high-risk clones of P. aeruginosa (clones with the Gly154Arg substitution, which is very prevalent in sequence type 175 [ST175] isolates, and isolates that constitutively promote ampC expression [265, 266]) entail significant impairments of virulence in a Caenorhabditis elegans model (Table 7) (267). Even though a very high number of studies regarding the control of virulence gene expression under the control of LysR family regulators in a wide range of species were published decades ago (268–272), it is noteworthy that, except for the works mentioned in this section, no link to the regulation of intrinsic β-lactamases or PGN metabolism has yet been found. Therefore, this interplay among PGN metabolism, β-lactamase production, and virulence, all of which are under the control of a common LysR-type regulator, is still barely studied and is worthy of more investigation in order to find future therapeutic targets.
With regard to global transcriptional regulators other than those of the LysR type, two-component systems could be good candidates to coordinately control the expression of genes related to virulence, β-lactamase production, and/or PGN metabolism. These systems consist of a first element (sensor), anchored to the cytosolic membrane, which is usually a histidine kinase that autophosphorylates when activated by an appropriate stimulus. The second element (cytosolic) is activated through phosphorylation, performed by the sensor, and acts as a transcription factor over distinct sets of genes to respond to certain environmental conditions and/or punctual stresses (273). Examples of these systems are BlrAB (where Blr stands for “β-lactam resistance”), which has been described to be the regulator of the multiple β-lactamases in Aeromonas spp. (274), or CreBC (where Cre stands for “carbon source responsive”) of E. coli, which has been proven to be not only a β-lactamase regulator but also a global one involved in metabolic control (275). Similarly, the previously mentioned CreBC system from P. aeruginosa (276) has been shown to be an essential actor in the response against β-lactams, entailing a complex metabolic response based on hyperproduction of its effector inner membrane protein, CreD, and on a better output for a given AmpC production level, improving its effectiveness and derived resistance. For these reasons, it has been considered a global regulator especially important during stresses (such as exposure to β-lactams): disruption of the system significantly impaired the fitness of the mutants in competition experiments and biofilm formation. Interestingly, among the genes with decreased expression (circa 2-fold) after CreBC inactivation, mainly during exposure to ceftazidime, there was exoS, encoding an exotoxin injected through the T3SS in the host cells. Nevertheless, the real impact of this hypoexpression has not yet been determined (276); in any case, the impact seems minor, since inactivated mutants with mutations in CreBC have recently been shown to display wild-type levels of virulence in the G. mellonella model (223). Similarly, the study of the same system in S. maltophilia has very recently provided data also linked to virulence, since a significant decrease in secreted protease activity was observed in CreBC double KO mutants but also in a CreC single mutant. Besides, an increase in swimming motility was also documented and shown to be CreD upregulation dependent (Table 7) (277). As happened with LysR-type regulators, many works relate the function of two-component systems to the control of pathogenesis in bacteria (278), but apart from the works cited in this section, no additional relatedness to PGN metabolism and/or β-lactamase regulation has been reported. This is the contrary to what happens with Gram-positive bacteria, where the volume of information on the topic is very important (279).
To conclude, some other different kinds of regulators have been shown to control intrinsic β-lactamase production in certain Gram-negative bacteria, presumably within complex regulatory networks also affecting features of the bacteria that could potentially include virulence (13). In this sense, it has been published that Elizabethkingia meningoseptica harbors an intrinsic class B β-lactamase (BlaB) which is not controlled by any known regulator but which is capable of increasing its production in response to stress conditions likely resembling those that bacteria find when causing infections, which suggests a coordinate regulation of β-lactamase production and virulence (280). Similarly, in Ralstonia pickettii, its intrinsic β-lactamases (OXA-22 and OXA-60) have been shown to be under the control of the barely studied ORF-RP3 element, to which global regulatory capabilities have been attributed, since it affects the lag phase of bacterial growth as well as the survival capacity against some stresses, such as pH changes, heat exposure, and osmolarity (281). Finally, in Caulobacter crescentus, the peculiar environment of the CAU-1 β-lactamase gene suggests that its regulation is under the control of a gene linked to the ArsR family of transcriptional regulators, which have been shown to be responsive against the stressing concentrations caused by heavy metals (282). Additionally, and more interestingly, these ArsR regulators have also been reported to control the expression of virulence genes in species such as Vibrio cholerae and Mycobacterium spp. (283, 284). Nevertheless, it still needs to be studied whether these clues suppose that there are more virulence genes within the cited regulatory networks and whether these models could be extended to more species for the future discovery of antivirulence targets.
EFFECTS OF HORIZONTALLY ACQUIRED β-LACTAMASES ON FITNESS AND VIRULENCE
The issue of the potential fitness and virulence costs derived from the expression of horizontally acquired β-lactamases has been widely studied mainly in Enterobacteriaceae but also in other Gram-negative bacteria, but with the information available, we are not able to draw any general conclusions. Moreover, the potential influence of these acquired enzymes (the genes for which are usually carried in integrons located on plasmids) on virulence has to be analyzed from different points of view: besides the alleged energetic burden that the massive production of the enzyme (or even the replication of the large natural plasmids carrying the genes) could entail or even the potential effects of the β-lactamase on the PGN, other factors have to be taken into account, such as the potential codification of virulence genes in the mentioned plasmids (which could, consequently, even increase the virulence of the strain) or the influence of some genes carried by the plasmid on the expression of chromosomal ones (48, 251–253, 285, 286). In this sense, it has been reported that ampR (as mentioned above, a transcriptional regulator of intrinsic β-lactamases), which is usually cotransferred on the same plasmid with some AmpC-type β-lactamases, can also regulate some virulence-related features in the host. More specifically, ampR from a plasmid containing the DHA-1 enzyme in a clinical strain of K. pneumoniae was shown to be involved in capsule synthesis and the derived serum resistance, biofilm formation, type 3 fimbrial gene expression, adhesion to cells in culture, as well as colonization of the gastrointestinal tract in mice (287). These results add strength to the previously mentioned idea on the wide pleiotropic power of ampR (and other LysR regulators) to regulate not only intrinsic or horizontally acquired β-lactamases expression but also many other bacterial processes, including virulence. Some other works proposed a correlation between the expression of acquired β-lactamases and increased levels of virulence. In this regard, Sahly and coworkers studied more than 200 clinical isolates of K. pneumoniae (producers of different extended-spectrum β-lactamases [ESBL] versus nonproducers) and found a positive association between increased adhesion, invasion capacities, and ESBL production. The molecular basis for this observation was that the acquisition of some ESBL-carrying plasmids appeared to upregulate the expression of some chromosomal genes (those encoding fimbrial adhesins, for instance), finally leading to increased invasiveness (285). In the same sense, Schaufler and colleagues studied seven clinical isolates from pandemic E. coli lineages (such as B2-ST131) carrying plasmids harboring CTX-M-type ESBLs and demonstrated (using a C. elegans infection model) that the expression of these plasmids did not dampen fitness but increased virulence (286). The presence of different virulence-related genes in the cited plasmids (finO or traT, for instance) but also the presumptive influence of an ESBL-positive plasmid on chromosomally encoded virulence-associated features would support the conclusions of this work (286). In this sense, the work of Ramirez and colleagues also collected information supporting the idea of a clear cotransfer of resistance determinants and virulence genes on the same plasmids specifically in K. pneumoniae (288). Another recent work with K. pneumoniae allows us to draw similar conclusions: a significant association between ESBL carriage and increased biofilm production and serum resistance was documented (289).
In an important number of publications, conversely, no significant increase/decrease in fitness or virulence was observed for different species carrying horizontally acquired β-lactamases. For instance, the reference strain of P. aeruginosa PAO1 was transformed with a vector containing a cloned blaIMP metallo-β-lactamase gene, and its virulence in terms of invasion of MDCK cells in culture and in a mouse bacteremia model did not significantly change (290). Similarly, in E. coli, the production of the β-lactamase CTX-M-1 did not seem to affect virulence: Dubois et al. (291) reported the isolation from a patient with neonatal meningitis of an E. coli strain harboring three different plasmids, one of which produced the aforementioned β-lactamase. The curation of these plasmids did not alter the incidence of meningitis in a neonatal mouse model, hence suggesting that the production of the cited β-lactamase does not influence the virulence of E. coli. Furthermore, even though the presence of virulence-related genes (iss, aer, and iroN) in the cited plasmids was documented, their curation did not impair the pathogenesis capacity (291).
Furthermore, given the widespread presence of horizontal β-lactamases in successful or epidemic clones from different Gram-negative bacteria (mainly Enterobacteriaceae), it is tempting to speculate that their carriage does not represent an important handicap for fitness and virulence or, at least, that under antibiotic pressure their carriage is positively selected as it adds more advantages than drawbacks for the bacteria. Some examples of this would be an ST131 strain (of the O25:H4 serotype) of E. coli carrying the ESBL CTX-M-15, which has emerged internationally as a successful multidrug-resistant strain (292). The latter β-lactamase and other CTX-M-type variants are also very prevalent in other virulent clones of E. coli, such as Shiga toxin-producing serotypes O104:H4 and O26:H11, and even in strains from farm animals (293, 294). Some other works have tried to understand the virulence capabilities of successful clones, such as the above-mentioned ST131 strain carrying the CTX-M or NDM β-lactamase (295), revealing a very complex interplay and balance between the expression of β-lactamases and derived resistance and levels of virulence. Therefore, what seems plausible is that certain β-lactamases and virulence genes can successfully coexist, mainly in specific clones of internationally spread strains and possibly as a result of gradual coevolution processes intimately linked to antibiotic pressure.
Finally, several works have demonstrated that the expression of acquired β-lactamases could entail a handicap for the bacterium, specifically, for fitness and/or virulence, as published by Lavigne and coworkers (296). In this paper, however, the authors reported that not all the plasmids harboring the acquired β-lactamases and not all the strains responded equally to these alleged biological costs. In this regard, assessing the virulence of the clinical strains and their plasmid-cured derivatives in a C. elegans model, it was shown that epidemic strains, such as the ST258 strain, harbor KPC-carrying plasmids associated with a biological cost lower than that for plasmids from nonepidemic strains (296). Another example is the recent work of Göttig and colleagues (297), in which the expression of the carbapenemase NDM-1 in E. coli or K. pneumoniae did not, apparently, significantly change the cytotoxicity in cell culture or virulence in the G. mellonella larva model. Nevertheless, and intriguingly, a significant fitness cost was observed in pairwise competition assays, where the wild-type strains of the cited species significantly outcompeted their respective transformants (obtained through transformation of the natural plasmid or a construct with the NDM-1 gene cloned on a vector), with the conclusion of this work being that efficient horizontal gene transfer probably has a higher impact on the dissemination of NDM-1 than potential fitness or virulence changes (297). Similarly, Marciano et al. reported a significant fitness cost associated with cloned SME-1 β-lactamase expression in E. coli, according to the results of growth and competition assays (298). Some more evidences in this paper were a decrease in culture viability (for the strain harboring a vector with the cloned SME-1 versus that for the wild type) and an increase in the rate of plasmid loss compared to the rate of loss of isogenic plasmids carrying a different β-lactamase gene (blaTEM-1). Moreover, the exchange of the SME-1 signal sequence for the TEM-1 one alleviated the fitness cost in the strain transformed with this new construct, and therefore, the authors suggested that the SME-1 signal sequence was the most responsible for the fitness cost, potentially due to the accumulation of the SME-1 signal sequences (when SME-1 is exported to the periplasm) resulting in inner membrane pore formation that dampened viability. The authors concluded that the fitness costs associated with some β-lactamases (whatever is the basis of this cost) may be the origin of their limited dissemination in comparison with the level of dissemination of others (298). In this sense, additional explanations for the virulence and fitness costs associated with certain β-lactamases were formulated some years ago: Fernández et al., using E. coli both for in vitro assays and in a murine model of systemic infection, linked the reported costs to quantitative changes in the composition of the PGN (a decrease in the level of cross-linking of the muropeptides and an increase in the average length of the glycan chains), presumably due to the residual dd-endopeptidase activity of the acquired enzymes (OXA-10, OXA-24, and SFO-1) (299). In fact, the potential residual activity of certain β-lactamases, due to or not due to their presumed PBP ascendance, has been proposed in the past and in this review (48, 251–255) and could still have some barely studied effects on the PGN (presumably, a weakening of its structure) that could entail a handicap for the bacterium under certain conditions (203, 223). These findings support the importance of the interaction between β-lactamases and PGN metabolism and the potentially associated biological costs and, as mentioned before, may even provide information on the epidemiology of β-lactamases and help explain the low incidence of some of them in specific pathogens, such as Salmonella spp. In this regard, several studies have highlighted the effect of β-lactamases on the fitness and virulence of S. enterica serovar Typhimurium. For instance, Cordeiro et al. observed that the synthesis of VIM-2 entailed a very important reduction in the growth rate, motility, and invasiveness in cell culture in this species, together with alterations in the micro- and macroscopic morphology of the cells, which the authors related to the extremely low number of reports of Salmonella isolates carrying acquired metallo-β-lactamases (300). Meanwhile, Morosini and coworkers associated the unregulated (in the absence of AmpR) expression of ampC in Salmonella with changes in colony morphology and a high reduction in the growth rate and invasiveness in cell culture, concluding that the expression of the AmpC in this species probably represented an almost unbearable cost (253). In fact, very few reports of Salmonella strains harboring AmpC-type β-lactamases have been published (301, 302). Nevertheless, some studies proposed that to make the AmpC-type enzyme production in Salmonella bearable, more than a low expression level of the enzyme (kept under the control of an active AmpR regulator) is needed, and the essential point could be the presence of additional encoded functions in the plasmid carrying the enzyme. This would compensate for the biological cost of AmpC-type enzyme overexpression (specifically, the CMY-7 β-lactamase in the previous work [303]).
In conclusion, and as reviewed in the work of Beceiro et al. (17), the issue of the antibiotic resistance/virulence interplay is still a controversial and very complex topic that is still a long way from being completely understood. With this review and specifically this section, we provide more data not only to support this idea of complexity but also to make it clear that the interplay between PGN–β-lactamases and virulence does exist and, thus, is a potential target to be exploited in the future.
CONCLUDING REMARKS: DELINEATING NEW THERAPEUTIC STRATEGIES BASED ON THE INTERPLAY BETWEEN PEPTIDOGLYCAN BIOLOGY, β-LACTAMASES, AND VIRULENCE
As stated throughout this review, much evidence of PGN being indispensable for virulence, in connection or not with the regulation of β-lactamases, has been gathered. Therefore, it is hoped that the existence of different bacterial targets leading to an attenuation of fitness/virulence could be deduced in this context, and a hope for alternatives to the current scenario of an almost useless antibiotic arsenal may exist. In fact, some current projects try to take advantage of targets in the β-lactamase–PGN biology interplay intended to block or revert the bacterial mechanisms leading to resistance to β-lactams but also to exert some bactericidal power. In this sense, several inhibitors of elements in PGN metabolism are currently being studied. These inhibitors affect, for instance, NagZ, AmpG, the septal protein FtsZ, PBP1a-PBP1b, PBP2, and even the membrane-bound lytic transglycosylases, with the inhibitors showing diverse levels of success as treatments or adjuvants (16, 229, 231, 304–311). Nevertheless, with the connection to virulence that we reviewed here, a new, additional horizon could be opened to, if not kill the bacteria or block/revert the β-lactam resistance, at least attenuate its pathogenesis in the host to ameliorate the clinical outcome (hence, the field of antivirulence therapies) (14). Nevertheless, in spite of all the evidence in this review, we are still a long way from using the identified weak points as antivirulence targets. The answers to many questions are still unknown and need to be resolved prior to finding therapeutic solutions; for instance, we need to know whether some PGN-derived fragments can act as signals to attenuate virulence (through binding to a transcriptional regulator, similar to what happens with AmpR-AmpC or two-component system models or not) or even whether the same β-lactamases can have a residual activity that could negatively affect PGN under certain conditions. Interestingly, the future development of these therapies could be approached from different perspectives, which improves the possibilities of success: the development of therapies based on the reduction of inflammation in response to PGN detection by our immune system (197), drugs intended to weaken the cell wall in order to make our innate immune weapons more effective in attacking the PGN (such as lysozyme and PGRPs), or drugs that block the motility/cytotoxicity of the pathogen to avoid the dissemination of the infection are only some of the possibilities that the issues that we have reviewed here could open. Hence, although the information dissected here clearly identifies PGN to be a structure essential for the virulence of Gram-negative bacteria, we have to take into account the fact that not all the actors playing a role on its biosynthetic, degradation, or recycling pathways have the same importance and the same chance to be used as therapeutic targets, and this is even more marked if we compare different species. According to this and since we have reached the so-called era of antibiotic resistance, perhaps we should change our conception of antimicrobial treatments and adapt them more specifically to the features of specific pathogens and their virulence factors (312). Thus, to conclude, although much work is still needed, the clues shown here clearly display weak points in the biology of Gram-negative bacteria (the PGN–β-lactamase–virulence interplay) to be exploited as targets for the design of novel therapeutic options in the future.
ACKNOWLEDGMENTS
This work was supported by the Ministerio de Economía y Competitividad of Spain and the Instituto de Salud Carlos III, cofinanced by the European Regional Development Fund (ERDF; A way to achieve Europe) through the Spanish Network for the Research in Infectious Diseases (RD12/0015 and RD16/0016), and grants CP12/03324, PI15/00088, and PI15/02212. I.M.B. is funded by the project SOIB Jove: Qualificats Sector Públic (JQ-SP 18/17), cofinanced by SOIB, Garantía Juvenil, and the European Social Fund.
We declare no competing financial interests.
C.J. and A.O. designed the content and structure of the review. C.J., G.T., and I.M.B. performed the literature review and wrote the initial draft. A.O. critically revised the manuscript, providing additional contributions. All authors reviewed and approved the final version.
Biographies

Carlos Juan completed his Ph.D. (2007) on the molecular mechanisms of Pseudomonas aeruginosa β-lactam resistance at the Hospital Son Dureta (Palma, Spain). Afterwards, he obtained two competitive Spanish grants to work in the field of infection and immunity at the University of the Balearic Islands (UIB) and at the Hospital Universitari Son Espases-IdISBa (Palma). He also worked as an invited researcher at the Karolinska Institute (Stockholm, Sweden) in the group of Dr. Staffan Normark, expanding his fields of expertise to bacterial pathogenesis-related techniques. He has also been Associate Professor of Immunology at UIB since 2011 in the Biochemistry Degree and Master in Advanced Microbiology programs. Therefore, his research moved from the basic molecular mechanisms of antibiotic resistance in nosocomial Gram-negative pathogens (a field in which he still collaborates) to projects investigating the links between resistance-virulence, pathogenesis mechanisms, and host-pathogen interactions, in which the study of the peptidoglycan always played a starring role, altogether with the final goal of finding new therapeutic targets and strategies against Gram-negative bacteria.

Gabriel Torrens (Palma, Spain, 1987) obtained his bachelor of biological sciences and master's degree in advanced microbiology at the University of the Balearic Islands, Palma. After finishing his studies, he started working in the field of antibiotic resistance and the pathogenicity of bacterial infections at the University Hospital Son Espases, where he is now a Ph.D. candidate working on cell wall recycling in Pseudomonas aeruginosa and its link with β-lactamase induction and virulence. He is skilled in the use of liquid chromatography techniques learned in the Severo Ochoa Molecular Biology Center in Madrid, Spain, as well as in the Umeå Centre for Microbial Research at the University of Umeå, Umeå, Sweden. His interests lie in bacterial cell wall and infection biology, transcriptomics, development of novel antimicrobial therapies, and liquid chromatography-mass spectrometry approaches to understanding antimicrobial resistance.

Isabel Maria Barceló (Palma, Spain, 1991) received her bachelor of biochemistry degree and master in advanced microbiology at the University of the Balearic Islands (UIB), Palma (2009 to 2015). Her initial research projects, performed at the Hospital Son Espases (Palma) and Health Research Institute of the Balearic Islands (IdISBa), were focused on the study of the interactions of new β-lactam combinations with penicillin binding proteins (PBPs) in relevant nosocomial Gram-negative pathogens, such as Pseudomonas aeruginosa and Acinetobacter baumannii. She currently holds a JoTReS grant from the Government of the Balearic Islands to work on her Ph.D. at the Hospital Son Espases/IdISBa in this same research line, expanded to other combinations and pathogens. Within this project she is also collaborating with the group of Dr. Bartolomé Moyá from the Center for Pharmacometrics & Systems Pharmacology, University of Florida.

Antonio Oliver (Madrid, Spain, 1973) has a doctorate in pharmacy (Complutense University of Madrid, 2002) and a specialist degree in clinical microbiology (Hospital Ramón y Cajal, Madrid, 2001). Since 2002 he has worked as a clinical microbiologist in the Department of Microbiology of the Hospital Son Espases (formerly the Hospital Son Dureta) in Palma de Mallorca. Since 2003 Dr. Oliver has been an Associate Professor of Microbiology in the University of the Balearic Islands and is the Principal Investigator of the Antimicrobial Resistance and Pathogenicity of Bacterial Infections Group at the Health Research Institute of the Balearic Islands (IdISBa). Dr. Oliver is a member of the Scientific Committee of the Spanish Network for Research in Infectious Diseases (REIPI) and an ESCMID fellow. His main research interest includes the investigation of the mechanisms of adaptation and antimicrobial resistance in bacterial pathogens, with a particular focus on Pseudomonas aeruginosa infections and the development of novel strategies to combat them. Within this research line, Dr. Oliver has published over 200 manuscripts in peer-reviewed journals.
REFERENCES
- 1.Adler A, Friedman ND, Marchaim D. 2016. Multidrug-resistant Gram-negative bacilli: infection control implications. Infect Dis Clin North Am 30:967–997. doi: 10.1016/j.idc.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 2.George MP, Masur H, Norris KA, Palmer SM, Clancy CJ, McDyer JF. 2014. Infections in the immunosuppressed host. Ann Am Thorac Soc 11:S211–S220. doi: 10.1513/AnnalsATS.201401-038PL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 4.Livermore DM. 2012. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med 27:128–142. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goossens H, Sprenger MJW. 1998. Community acquired infections and bacterial resistance. BMJ 317:654–657. doi: 10.1136/bmj.317.7159.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta KC, Dargad RR, Borade DM, Swami OC. 2014. Burden of antibiotic resistance in common infectious diseases: role of antibiotic combination therapy. J Clin Diagn Res 8:ME05–ME08. doi: 10.7860/JCDR/2014/8778.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kariuki S, Gordon MA, Feasey N, Parry CM. 2015. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 33:C21–C29. doi: 10.1016/j.vaccine.2015.03.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Duin D, Paterson D. 2016. Multidrug resistant bacteria in the community: trends and lessons learned. Infect Dis Clin North Am 30:377–390. doi: 10.1016/j.idc.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, Zhu Y, Lu NH. 2017. Novel and effective therapeutic regimens for Helicobacter pylori in an era of increasing antibiotic resistance. Front Cell Infect Microbiol 7:168. doi: 10.3389/fcimb.2017.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong KF, Schneper L, Mathee K. 2010. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. Acta Pathol Microbiol Immunol Scand 118:1–36. doi: 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacVane SH. 2017. Antimicrobial resistance in the intensive care unit: a focus on Gram-negative bacterial infections. J Intensive Care Med 32:25–37. doi: 10.1177/0885066615619895. [DOI] [PubMed] [Google Scholar]
- 12.Baur D, Gladstone BP, Burkert F, Carrara E, Foschi F, Döbele S, Tacconelli E. 2017. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis 17:990–1001. doi: 10.1016/S1473-3099(17)30325-0. [DOI] [PubMed] [Google Scholar]
- 13.Juan C, Torrens G, González-Nicolau M, Oliver A. 2017. Diversity and regulation of intrinsic β-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol Rev 41:781–815. doi: 10.1093/femsre/fux043. [DOI] [PubMed] [Google Scholar]
- 14.Dickey SW, Cheung GYC, Otto M. 2017. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PubMed] [Google Scholar]
- 15.Mark BL, Vocadlo DJ, Oliver A. 2011. Providing β-lactams a helping hand: targeting the AmpC β-lactamase induction pathway. Future Microbiol 6:1415–1427. doi: 10.2217/fmb.11.128. [DOI] [PubMed] [Google Scholar]
- 16.Lamers RP, Burrows LL. 2016. Pseudomonas aeruginosa: targeting cell-wall metabolism for new antibacterial discovery and development. Future Med Chem 8:975–992. doi: 10.4155/fmc-2016-0017. [DOI] [PubMed] [Google Scholar]
- 17.Beceiro A, Tomás M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev 26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M, Dhar S, De Benedetti S, Hesek D, Boggess B, Blázquez B, Mathee K, Mobashery S. 2016. Muropeptides in Pseudomonas aeruginosa and their role as elicitors of β-lactam-antibiotic resistance. Angew Chem Int Ed Engl 55:6882–6886. doi: 10.1002/anie.201601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dik DA, Fisher JF, Mobashery S. 2018. Cell-wall recycling of the Gram-negative bacteria and the nexus to antibiotic resistance. Chem Rev 118:5952–5984. doi: 10.1021/acs.chemrev.8b00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreillon P, Majcherczyk PA. 2003. Proinflammatory activity of cell-wall constituents from gram-positive bacteria. Scand J Infect Dis 35:632–641. doi: 10.1080/00365540310016259. [DOI] [PubMed] [Google Scholar]
- 21.Sriskandan S, Cohen J. 1999. Gram-positive sepsis. Mechanisms and differences from gram-negative sepsis. Infect Dis Clin North Am 13:397–412. doi: 10.1016/S0891-5520(05)70082-9. [DOI] [PubMed] [Google Scholar]
- 22.Fedtke I, Götz F, Peschel A. 2004. Bacterial evasion of innate host defenses—the Staphylococcus aureus lesson. Int J Med Microbiol 294:189–194. doi: 10.1016/j.ijmm.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Quintela JC, Caparrós M, de Pedro MA. 1995. Variability of peptidoglycan structural parameters in gram-negative bacteria. FEMS Microbiol Lett 125:95–100. doi: 10.1111/j.1574-6968.1995.tb07341.x. [DOI] [PubMed] [Google Scholar]
- 24.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 25.den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev 32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 26.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harbor Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Pedro MA, Cava F. 2015. Structural constraints and dynamics of bacterial cell wall architecture. Front Microbiol 6:449. doi: 10.3389/fmicb.2015.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol 184:754–759. doi: 10.1128/JB.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabanes D, Dussurget O, Dehoux P, Cossart P. 2004. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol Microbiol 51:1601–1614. doi: 10.1111/j.1365-2958.2003.03945.x. [DOI] [PubMed] [Google Scholar]
- 30.Flores-Kim J, Darwin AJ. 2014. Regulation of bacterial virulence gene expression by cell envelope stress responses. Virulence 5:835–851. doi: 10.4161/21505594.2014.965580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown S, Santa Maria JP, Walker S. 2013. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. 2008. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol Rev 32:386–408. doi: 10.1111/j.1574-6976.2007.00097.x. [DOI] [PubMed] [Google Scholar]
- 33.Turner R, Vollmer W, Foster S. 2014. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol 91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matias VR, Al-Amoudi A, Dubochet J, Beveridge TJ. 2003. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol 185:6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennehy R, Romano M, Ruggiero A, Mohamed YF, Dignam SL, Mujica-Troncoso C, Callaghan M, Valvano MA, Berisio R, McClean S. 2017. The Burkholderia cenocepacia peptidoglycan-associated lipoprotein is involved in epithelial cell attachment and elicitation of inflammation. Cell Microbiol 19:e12691. doi: 10.1111/cmi.12691. [DOI] [PubMed] [Google Scholar]
- 36.van Lier CJ, Sha J, Kirtley ML, Cao A, Tiner BL, Erova TE, Cong Y, Kozlova EV, Popov VL, Baze WB, Chopra AK. 2014. Deletion of Braun lipoprotein and plasminogen-activating protease-encoding genes attenuates Yersinia pestis in mouse models of bubonic and pneumonic plague. Infect Immun 82:2485–2503. doi: 10.1128/IAI.01595-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sha J, Agar SL, Baze WB, Olano JP, Fadl AA, Erova TE, Wang S, Foltz SM, Suarez G, Motin VL, Chauhan S, Klimpel GR, Peterson JW, Chopra AK. 2008. Braun lipoprotein (Lpp) contributes to virulence of yersiniae: potential role of Lpp in inducing bubonic and pneumonic plague. Infect Immun 76:1390–1409. doi: 10.1128/IAI.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agar SL, Sha J, Baze WB, Erova TE, Foltz SM, Suarez G, Wang S, Chopra AK. 2009. Deletion of Braun lipoprotein gene (lpp) and curing of plasmid pPCP1 dramatically alter the virulence of Yersinia pestis CO92 in a mouse model of pneumonic plague. Microbiology 155:3247–3259. doi: 10.1099/mic.0.029124-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sha J, Fadl AA, Klimpel GR, Niesel DW, Popov VL, Chopra AK. 2004. The two murein lipoproteins of Salmonella enterica serovar Typhimurium contribute to the virulence of the organism. Infect Immun 72:3987–4003. doi: 10.1128/IAI.72.7.3987-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erova TE, Kirtley ML, Fitts EC, Ponnusamy D, Baze WB, Andersson JA, Cong Y, Tiner BL, Sha J, Chopra AK. 2016. Protective immunity elicited by oral immunization of mice with Salmonella enterica serovar Typhimurium Braun lipoprotein (Lpp) and acetyltransferase (MsbB) mutants. Front Cell Infect Microbiol 6:148. doi: 10.3389/fcimb.2016.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernández SB, Cava F, Pucciarelli MG, García-Del Portillo F, de Pedro MA, Casadesús J. 2015. Bile-induced peptidoglycan remodelling in Salmonella enterica. Environ Microbiol 17:1081–1089. doi: 10.1111/1462-2920.12491. [DOI] [PubMed] [Google Scholar]
- 42.Sha J, Kirtley ML, van Lier CJ, Wang S, Erova TE, Kozlova EV, Cao A, Cong Y, Fitts EC, Rosenzweig JA, Chopra AK. 2013. Deletion of the Braun lipoprotein-encoding gene and altering the function of lipopolysaccharide attenuate the plague bacterium. Infect Immun 81:815–828. doi: 10.1128/IAI.01067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uhlich GA, Gunther NW IV, Bayles DO, Mosier DA. 2009. The CsgA and Lpp proteins of an Escherichia coli O157:H7 strain affect HEp-2 cell invasion, motility, and biofilm formation. Infect Immun 77:1543–1552. doi: 10.1128/IAI.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellman J, Roberts JD Jr, Tehan MM, Allaire JE, Warren HS. 2002. Bacterial peptidoglycan-associated lipoprotein is released into the bloodstream in gram-negative sepsis and causes inflammation and death in mice. J Biol Chem 277:14274–14280. doi: 10.1074/jbc.M109696200. [DOI] [PubMed] [Google Scholar]
- 45.Fadl AA, Sha J, Klimpel GR, Olano JP, Niesel DW, Chopra AK. 2005. Murein lipoprotein is a critical outer membrane component involved in Salmonella enterica serovar Typhimurium systemic infection. Infect Immun 73:1081–1096. doi: 10.1128/IAI.73.2.1081-1096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilhelmsen K, Mesa KR, Prakash A, Xu F, Hellman J. 2012. Activation of endothelial TLR2 by bacterial lipoprotein upregulates proteins specific for the neutrophil response. Innate Immun 18:602–616. doi: 10.1177/1753425911429336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ihalin R, Eneslätt K, Asikainen S. 2018. Peptidoglycan-associated lipoprotein of Aggregatibacter actinomycetemcomitans induces apoptosis and production of proinflammatory cytokines via TLR2 in murine macrophages RAW 264.7 in vitro. J Oral Microbiol 10:1442079. doi: 10.1080/20002297.2018.1442079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–358. doi: 10.1111/j.1574-6976.2008.00105.x (Erratum, 32:556, doi:.) [DOI] [PubMed] [Google Scholar]
- 49.Mohamed YF, Valvano MA. 2014. A Burkholderia cenocepacia MurJ (MviN) homolog is essential for cell wall peptidoglycan synthesis and bacterial viability. Glycobiology 24:564–576. doi: 10.1093/glycob/cwu025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egan AJF, Biboy J, van't Veer I, Breukink E, Vollmer W. 2015. Activities and regulation of peptidoglycan synthases. Philos Trans R Soc Lond B Biol Sci 370:20150031. doi: 10.1098/rstb.2015.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao H, Patel V, Helmann JD, Dörr T. 2017. Don't let sleeping dogmas lie: new views of peptidoglycan synthesis and its regulation. Mol Microbiol 106:847–860. doi: 10.1111/mmi.13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W, Zhang YM, Davies C. 2017. Penicillin-binding protein 3 is essential for growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e01651-16. doi: 10.1128/AAC.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181:3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Priyadarshini R, Popham DL, Young KD. 2006. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J Bacteriol 188:5345–5355. doi: 10.1128/JB.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson DE, Young KD. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol 182:1714–1721. doi: 10.1128/JB.182.6.1714-1721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potluri LP, de Pedro MA, Young KD. 2012. Escherichia coli low molecular weight penicillin binding proteins help orient septal FtsZ, and their absence leads to asymmetric cell division and branching. Mol Microbiol 84:203–224. doi: 10.1111/j.1365-2958.2012.08023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peters K, Kannan S, Rao VA, Biboy J, Vollmer D, Erickson SW, Lewis RJ, Young KD, Vollmer W. 2016. The redundancy of peptidoglycan carboxypeptidases ensures robust cell shape maintenance in Escherichia coli. mBio 7:e00819-16. doi: 10.1128/mBio.00819-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun 66:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cabot G, Zamorano L, Moyà B, Juan C, Navas A, Blázquez J, Oliver A. 2016. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob Agents Chemother 60:1767–1778. doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berrazeg M, Jeannot K, Ntsogo Enguéné VY, Broutin I, Loeffert S, Fournier D, Plésiat P. 2015. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255. doi: 10.1128/AAC.00825-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russo TA, MacDonald U, Beanan JM, Olson R, MacDonald IJ, Sauberan SL, Luke NR, Schultz LW, Umland TC. 2009. Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J Infect Dis 199:513–521. doi: 10.1086/596317. [DOI] [PubMed] [Google Scholar]
- 63.Dörr T, Möll A, Chao MC, Cava F, Lam H, Davis BM, Waldor MK. 2014. Differential requirement for PBP1a and PBP1b in in vivo and in vitro fitness of Vibrio cholerae. Infect Immun 82:2115–2124. doi: 10.1128/IAI.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Priyadarshini R, de Pedro MA, Young KD. 2007. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol 189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Höltje JV. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol 41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- 66.Chaput C, Ecobichon C, Pouradier N, Rousselle JC, Namane A, Boneca IG. 2016. Role of the N-acetylmuramoyl-l-alanyl amidase, AmiA, of Helicobacter pylori in peptidoglycan metabolism, daughter cell separation, and virulence. Microb Drug Resist 22:477–486. doi: 10.1089/mdr.2016.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yakhnina AA, McManus HR, Bernhardt TG. 2015. The cell wall amidase AmiB is essential for Pseudomonas aeruginosa cell division, drug resistance, and viability. Mol Microbiol 97:957–973. doi: 10.1111/mmi.13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hunt TA, Kooi C, Sokol PA, Valvano MA. 2004. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect Immun 72:4010–4022. doi: 10.1128/IAI.72.7.4010-4022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee JB, Byeon JH, Jang HA, Kim JK, Yoo JW, Kikuchi Y, Lee BL. 2015. Bacterial cell motility of Burkholderia gut symbiont is required to colonize the insect gut. FEBS Lett 589:2784–2790. doi: 10.1016/j.febslet.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 70.Yang LC, Gan YL, Yang LY, Jiang BL, Tang JL. 2018. Peptidoglycan hydrolysis mediated by the amidase AmiC and its LytM activator NlpD is critical for cell separation and virulence in the phytopathogen Xanthomonas campestris. Mol Plant Pathol 19:1705–1718. doi: 10.1111/mpp.12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castanheira S, Cestero JJ, Rico-Pérez G, García P, Cava F, Ayala JA, Pucciarelli MG, García-Del Portillo F. 2017. A specialized peptidoglycan synthase promotes Salmonella cell division inside host cells. mBio 8:e01685-17. doi: 10.1128/mBio.01685-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samaluru H, SaiSree L, Reddy M. 2007. Role of SufI (FtsP) in cell division of Escherichia coli: evidence for its involvement in stabilizing the assembly of the divisome. J Bacteriol 189:8044–8052. doi: 10.1128/JB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mickael CS, Lam PKS, Berberov EM, Allan B, Potter AA, Köster W. 2010. Salmonella enterica serovar Enteritidis tatB and tatC mutants are impaired in Caco-2 cell invasion in vitro and show reduced systemic spread in chickens. Infect Immun 78:3493–3505. doi: 10.1128/IAI.00090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weatherspoon-Griffin N, Zhao G, Kong W, Kong Y, Morigen Andrews-Polymenis H, McClelland M, Shi Y. 2011. The CpxR/CpxA two-component system up-regulates two Tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J Biol Chem 286:5529–5539. doi: 10.1074/jbc.M110.200352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Craig M, Sadik AY, Golubeva YA, Tidhar A, Slauch JM. 2013. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol Microbiol 89:887–902. doi: 10.1111/mmi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ercoli G, Tani C, Pezzicoli A, Vacca I, Martinelli M, Pecetta S, Petracca R, Rappuoli R, Pizza M, Norais N, Soriani M, Aricò B. 2015. LytM proteins play a crucial role in cell separation, outer membrane composition, and pathogenesis in nontypeable Haemophilus influenzae. mBio 6:e02575-14. doi: 10.1128/mBio.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frirdich E, Biboy J, Adams C, Lee J, Ellermeier J, Gielda LD, Dirita VJ, Girardin SE, Vollmer W, Gaynor EC. 2012. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog 8:e1002602. doi: 10.1371/journal.ppat.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frirdich E, Vermeulen J, Biboy J, Soares F, Taveirne ME, Johnson JG, DiRita VJ, Girardin SE, Vollmer W, Gaynor EC. 2014. Peptidoglycan ld-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the DL-carboxypeptidase Pgp1. J Biol Chem 289:8007–8018. doi: 10.1074/jbc.M113.491829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ha R, Frirdich E, Sychantha D, Biboy J, Taveirne ME, Johnson JG, DiRita VJ, Vollmer W, Clarke AJ, Gaynor EC. 2016. Accumulation of peptidoglycan O-acetylation leads to altered cell wall biochemistry and negatively impacts pathogenesis factors of Campylobacter jejuni. J Biol Chem 291:22686–22702. doi: 10.1074/jbc.M116.746404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. 2010. Relaxation of peptidoglycan cross-linking promotes Helicobacter pylori's helical shape and stomach colonization. Cell 141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang G, Lo LF, Forsberg LS, Maier RJ. 2012. Helicobacter pylori peptidoglycan modifications confer lysozyme resistance and contribute to survival in the host. mBio 3:e00409-12. doi: 10.1128/mBio.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ragland SA, Schaub RE, Hackett KT, Dillard JP, Criss AK. 2017. Two lytic transglycosylases in Neisseria gonorrhoeae impart resistance to killing by lysozyme and human neutrophils. Cell Microbiol 19:12662. doi: 10.1111/cmi.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subashchandrabose S, Smith S, DeOrnellas V, Crepin S, Kole M, Zahdeh C, Mobley HL. 2015. Acinetobacter baumannii genes required for bacterial survival during bloodstream infection. mSphere 1:e00013-15. doi: 10.1128/mSphere.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun YH, Bakshi S, Chalmers R, Tang CM. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat Med 6:1269–1273. doi: 10.1038/81380. [DOI] [PubMed] [Google Scholar]
- 85.Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. 2006. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol 4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 86.Canavessi AM, Harms J, de Leon Gatti N, Splitter GA. 2004. The role of integrase/recombinase xerD and monofunctional biosynthesis peptidoglycan transglycosylase genes in the pathogenicity of Brucella abortus infection in vitro and in vivo. Microb Pathog 37:241–251. doi: 10.1016/j.micpath.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 87.Zhao Y, Blumer SE, Sundin GW. 2005. Identification of Erwinia amylovora genes induced during infection of immature pear tissue. J Bacteriol 187:8088–8103. doi: 10.1128/JB.187.23.8088-8103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen K, Antalis P, Gladitz J, Sayeed S, Ahmed A, Yu S, Hayes J, Johnson S, Dice B, Dopico R, Keefe R, Janto B, Chong W, Goodwin J, Wadowsky RM, Erdos G, Post JC, Ehrlich GD, Hu FZ. 2005. Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae. Infect Immun 73:3479–3491. doi: 10.1128/IAI.73.6.3479-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol Microbiol 44:73–88. doi: 10.1046/j.1365-2958.2002.02877.x. [DOI] [PubMed] [Google Scholar]
- 90.Dörr T, Cava F, Lam H, Davis BM, Waldor MK. 2013. Substrate specificity of an elongation-specific peptidoglycan endopeptidase and its implications for cell wall architecture and growth of Vibrio cholerae. Mol Microbiol 89:949–962. doi: 10.1111/mmi.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vollmer W, Bertsche U. 2008. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta 1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 92.Scheffers DJ, Pinho MG. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell-wall recycling. Ann N Y Acad Sci 1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim JK, Lee HJ, Kikuchi Y, Kitagawa W, Nikoh N, Fukatsu T, Lee BL. 2013. Bacterial cell wall synthesis gene uppP is required for Burkholderia colonization of the stinkbug gut. Appl Environ Microbiol 79:4879–4886. doi: 10.1128/AEM.01269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Templin MF, Ursinus A, Höltje JV. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J 18:4108–4117. doi: 10.1093/emboj/18.15.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hervé M, Boniface A, Gobec S, Blanot D, Mengin-Lecreulx D. 2007. Biochemical characterization and physiological properties of Escherichia coli UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase. J Bacteriol 189:3987–3995. doi: 10.1128/JB.00087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tans-Kersten J, Gay J, Allen C. 2000. Ralstonia solanacearum AmpD is required for wild-type bacterial wilt virulence. Mol Plant Pathol 1:179–185. doi: 10.1046/j.1364-3703.2000.00023.x. [DOI] [PubMed] [Google Scholar]
- 99.Folkesson A, Eriksson S, Andersson M, Park JT, Normark S. 2005. Components of the peptidoglycan-recycling pathway modulate invasion and intracellular survival of Salmonella enterica serovar Typhimurium. Cell Microbiol 7:147–155. doi: 10.1111/j.1462-5822.2004.00443.x. [DOI] [PubMed] [Google Scholar]
- 100.Soo PC, Wei JR, Horng YT, Hsieh SC, Ho SW, Lai HC. 2005. Cell envelope architecture, hemolysin production, and cell attachment ability in Serratia marcescens. Infect Immun 73:6075–6084. doi: 10.1128/IAI.73.9.6075-6084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Elamin AA, Steinicke S, Oehlmann W, Braun Y, Wanas H, Shuralev EA, Huck C, Maringer M, Rohde M, Singh M. 2017. Novel drug targets in cell wall biosynthesis exploited by gene disruption in Pseudomonas aeruginosa. PLoS One 12:e0186801. doi: 10.1371/journal.pone.0186801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Das D, Hervé M, Feuerhelm J, Farr CL, Chiu HJ, Elsliger MA, Knuth MW, Klock HE, Miller MD, Godzik A, Lesley SA, Deacon AM, Mengin-Lecreulx D, Wilson IA. 2011. Structure and function of the first full-length murein peptide ligase (Mpl) cell wall recycling protein. PLoS One 6:e17624. doi: 10.1371/journal.pone.0017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scheurwater E, Reid CW, Clarke AJ. 2008. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol 40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 104.Koraimann G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci 60:2371–2388. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Herlihey FA, Clarke AJ. 2017. Controlling autolysis during flagella insertion in Gram-negative bacteria. Adv Exp Med Biol 925:41–56. doi: 10.1007/5584_2016_52. [DOI] [PubMed] [Google Scholar]
- 106.Korotkov KV, Sandkvist M, Hol WGJ. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Howard SP, Gebhart C, Langen GR, Li G, Strozen TG. 2006. Interactions between peptidoglycan and the ExeAB complex during assembly of the type II secretin of Aeromonas hydrophila. Mol Microbiol 59:1062–1072. doi: 10.1111/j.1365-2958.2005.05003.x. [DOI] [PubMed] [Google Scholar]
- 108.Howard SP. 2013. Assembly of the type II secretion system. Res Microbiol 164:535–544. doi: 10.1016/j.resmic.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 109.Li G, Miller A, Bull H, Howard SP. 2011. Assembly of the type II secretion system: identification of ExeA residues critical for peptidoglycan binding and secretin multimerization. J Bacteriol 193:197–204. doi: 10.1128/JB.00882-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vanderlinde EM, Strozen TG, Hernández SB, Cava F, Howard SP. 2017. Alterations in peptidoglycan cross-linking suppress the secretin assembly defect caused by mutation of GspA in the type II secretion system. J Bacteriol 199:e00617-16. doi: 10.1128/JB.00617-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zahrl D, Wagner M, Bischof K, Bayer M, Zavecz B, Beranek A, Ruckenstuhl C, Zarfel GE, Koraimann G. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455–3467. doi: 10.1099/mic.0.28141-0. [DOI] [PubMed] [Google Scholar]
- 112.García-Gómez E, Espinosa N, de la Mora J, Dreyfus G, González-Pedrajo B. 2011. The muramidase EtgA from enteropathogenic Escherichia coli is required for efficient type III secretion. Microbiology 157:1145–1160. doi: 10.1099/mic.0.045617-0. [DOI] [PubMed] [Google Scholar]
- 113.Burkinshaw BJ, Deng W, Lameignère E, Wasney GA, Zhu H, Worrall LJ, Finlay BB, Strynadka NC. 2015. Structural analysis of a specialized type III secretion system peptidoglycan-cleaving enzyme. J Biol Chem 290:10406–10417. doi: 10.1074/jbc.M115.639013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu YC, Lin CN, Wang SH, Ng SC, Hu WS, Syu WJ. 2010. A putative lytic transglycosylase tightly regulated and critical for the EHEC type three secretion. J Biomed Sci 17:52. doi: 10.1186/1423-0127-17-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oh HS, Kvitko BH, Morello JE, Collmer A. 2007. Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effector proteins into plant cells. J Bacteriol 189:8277–8289. doi: 10.1128/JB.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hausner J, Hartmann N, Jordan M, Büttner D. 2017. The predicted lytic transglycosylase HpaH from Xanthomonas campestris pv. vesicatoria associates with the type III secretion system and promotes effector protein translocation. Infect Immun 85:e00788-16. doi: 10.1128/IAI.00788-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zupan J, Hackworth CA, Aguilar J, Ward D, Zambryski P. 2007. VirB1* promotes T-pilus formation in the vir-type IV secretion system of Agrobacterium tumefaciens. J Bacteriol 189:6551–6563. doi: 10.1128/JB.00480-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kohler PL, Hamilton HL, Cloud-Hansen K, Dillard JP. 2007. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J Bacteriol 189:5421–5428. doi: 10.1128/JB.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Santin YG, Cascales E. 2017. Domestication of a housekeeping transglycosylase for assembly of a type VI secretion system. EMBO Rep 18:138–149. doi: 10.15252/embr.201643206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weber BS, Hennon SW, Wright MS, Scott NE, de Berardinis V, Foster LJ, Ayala JA, Adams MD, Feldman MF. 2016. Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. mBio 7:e01253-16. doi: 10.1128/mBio.01253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rambow-Larsen AA, Weiss AA. 2002. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J Bacteriol 184:2863–2869. doi: 10.1128/JB.184.11.2863-2869.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Herlihey FA, Moynihan PJ, Clarke AJ. 2014. The essential protein for bacterial flagella formation FlgJ functions as a β-N-acetylglucosaminidase. J Biol Chem 289:31029–31042. doi: 10.1074/jbc.M114.603944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Herlihey FA, Osorio-Valeriano M, Dreyfus G, Clarke AJ. 2016. Modulation of the lytic activity of the dedicated autolysin for flagellum formation SltF by flagellar rod proteins FlgB and FlgF. J Bacteriol 198:1847–1856. doi: 10.1128/JB.00203-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.De la Mora J, Ballado T, González-Pedrajo B, Camarena L, Dreyfus G. 2007. The flagellar muramidase from the photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol 189:7998–8004. doi: 10.1128/JB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roure S, Bonis M, Chaput C, Ecobichon C, Mattox A, Barrière C, Geldmacher N, Guadagnini S, Schmitt C, Prévost MC, Labigne A, Backert S, Ferrero RL, Boneca IG. 2012. Peptidoglycan maturation enzymes affect flagellar functionality in bacteria. Mol Microbiol 86:845–856. doi: 10.1111/mmi.12019. [DOI] [PubMed] [Google Scholar]
- 126.Chaban B, Hughes HV, Beeby M. 2015. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol 46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 127.Lovewell RR, Collins RM, Acker JL, O'Toole GA, Wargo MJ, Berwin B. 2011. Step-wise loss of bacterial flagellar torsion confers progressive phagocytic evasion. PLoS Pathog 7:e1002253. doi: 10.1371/journal.ppat.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rosenthal RS, Nogami W, Cookson BT, Goldman WE, Folkening WJ. 1987. Major fragment of soluble peptidoglycan released from growing Bordetella pertussis is tracheal cytotoxin. Infect Immun 55:2117–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heiss LN, Moser SA, Unanue ER, Goldman WE. 1993. Interleukin-1 is linked to the respiratory epithelial cytopathology of pertussis. Infect Immun 61:3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Luker KE, Collier JL, Kolodziej EW, Marshall GR, Goldman WE. 1993. Bordetella pertussis tracheal cytotoxin and other muramyl peptides: distinct structure-activity relationships for respiratory epithelial cytopathology. Proc Natl Acad Sci U S A 90:2365–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Woodhams KL, Chan JM, Lenz JD, Hackett KT, Dillard JP. 2013. Peptidoglycan fragment release from Neisseria meningitidis. Infect Immun 81:3490–3498. doi: 10.1128/IAI.00279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chan JM, Dillard JP. 2016. Neisseria gonorrhoeae crippled its peptidoglycan fragment permease to facilitate toxic peptidoglycan monomer release. J Bacteriol 198:3029–3040. doi: 10.1128/JB.00437-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cloud KA, Dillard JP. 2002. A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production. Infect Immun 70:2752–2757. doi: 10.1128/IAI.70.6.2752-2757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cloud-Hansen KA, Hackett KT, Garcia DL, Dillard JP. 2008. Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J Bacteriol 190:5989–5994. doi: 10.1128/JB.00506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jacobs C, Huang LJ, Bartowsky E, Normark S, Park JT. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J 13:4684–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Garcia DL, Dillard JP. 2008. Mutations in ampG or ampD affect peptidoglycan fragment release from Neisseria gonorrhoeae. J Bacteriol 190:3799–3807. doi: 10.1128/JB.01194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. 2009. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol 191:2012–2022. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lenz JD, Hackett KT, Dillard JP. 2017. A single dual-function enzyme controls the production of inflammatory NOD agonist peptidoglycan fragments by Neisseria gonorrhoeae. mBio 8:e01464-17. doi: 10.1128/mBio.01464-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Burroughs MH, Cabellos C, Prasad S, Tuomanen E. 1992. Bacterial components and the pathophysiology of injury to the blood-brain barrier: does cell wall add to the effects of endotoxin in gram-negative meningitis? J Infect Dis 165:S82–S85. doi: 10.1093/infdis/165-Supplement_1-S82. [DOI] [PubMed] [Google Scholar]
- 140.Burroughs MH, Prasad S, Cabellos C, Mendelman P, Tuomanen E. 1993. The biological activities of peptidoglycan in experimental Haemophilus influenzae meningitis. J Infect Dis 167:464–468. doi: 10.1093/infdis/167.2.464. [DOI] [PubMed] [Google Scholar]
- 141.Burroughs M, Rozdzinski E, Geelen S, Tuomanen E. 1993. A structure-activity relationship for induction of meningeal inflammation by muramyl peptides. J Clin Invest 92:297–302. doi: 10.1172/JCI116565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Mémet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 143.Noto JM, Peek RM. 2012. The Helicobacter pylori cag pathogenicity island. Methods Mol Biol 921:41–50. doi: 10.1007/978-1-62703-005-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Boneca IG. 2005. The role of peptidoglycan in pathogenesis. Curr Opin Microbiol 8:46–53. doi: 10.1016/j.mib.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 145.Graham JE, Peek RM, Krishna U, Cover TL. 2002. Global analysis of Helicobacter pylori gene expression in human gastric mucosa. Gastroenterology 123:1637–1648. doi: 10.1053/gast.2002.36589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bartoleschi C, Pardini MC, Scaringi C, Martino MC, Pazzani C, Bernardini ML. 2002. Selection of Shigella flexneri candidate virulence genes specifically induced in bacteria resident in host cell cytoplasm. Cell Microbiol 4:613–626. doi: 10.1046/j.1462-5822.2002.00216.x. [DOI] [PubMed] [Google Scholar]
- 147.Girardin SE, Tournebize R, Mavris M, Page AL, Li X, Stark GR, Bertin J, DiStefano PS, Yaniv M, Sansonetti PJ, Philpott DJ. 2001. CARD4/Nod1 mediates NF-κB and JNK activation by invasive Shigella flexneri. EMBO Rep 2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nigro G, Fazio LL, Martino MC, Rossi G, Tattoli I, Liparoti V, De Castro C, Molinaro A, Philpott DJ, Bernardini ML. 2008. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cell Microbiol 10:682–695. doi: 10.1111/j.1462-5822.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- 149.Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, Le Bourhis L, Karrar A, Viala J, Mak J, Hutton ML, Davies JK, Crack PJ, Hertzog PJ, Philpott DJ, Girardin SE, Whitchurch CB, Ferrero RL. 2010. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol 12:372–385. doi: 10.1111/j.1462-5822.2009.01404.x. [DOI] [PubMed] [Google Scholar]
- 150.Pradipta AR, Fujimoto Y, Hasegawa M, Inohara N, Fukase K. 2010. Characterization of natural human nucleotide-binding oligomerization domain protein 1 (Nod1) ligands from bacterial culture supernatant for elucidation of immune modulators in the environment. J Biol Chem 285:23607–23613. doi: 10.1074/jbc.M110.137893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schwechheimer C, Sullivan CJ, Kuehn MJ. 2013. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry 52:3031–3040. doi: 10.1021/bi400164t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Thay B, Damm A, Kufer TA, Wai SN, Oscarsson J. 2014. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-κB activation. Infect Immun 82:4034–4046. doi: 10.1128/IAI.01980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cecil JD, O'Brien-Simpson NM, Lenzo JC, Holden JA, Chen YY, Singleton W, Gause KT, Yan Y, Caruso F, Reynolds EC. 2016. Differential responses of pattern recognition receptors to outer membrane vesicles of three periodontal pathogens. PLoS One 11:e0151967. doi: 10.1371/journal.pone.0151967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cañas MA, Fábrega MJ, Giménez R, Badia J, Baldomà L. 2018. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front Microbiol 9:498. doi: 10.3389/fmicb.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jan AT. 2017. Outer membrane vesicles (OMVs) of Gram-negative bacteria: a perspective update. Front Microbiol 8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yu YJ, Wang XH, Fan GC. 2018. Versatile effects of bacterium-released membrane vesicles on mammalian cells and infectious/inflammatory diseases. Acta Pharmacol Sin 39:514–533. doi: 10.1038/aps.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schwechheimer C, Kulp A, Kuehn MJ. 2014. Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol 14:324. doi: 10.1186/s12866-014-0324-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schwechheimer C, Rodriguez DL, Kuehn MJ. 2015. NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli. Microbiologyopen 4:375–389. doi: 10.1002/mbo3.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Alhazmi A. 2018. NOD-like receptor(s) and host immune responses with Pseudomonas aeruginosa infection. Inflamm Res 67:479–493. doi: 10.1007/s00011-018-1132-0. [DOI] [PubMed] [Google Scholar]
- 161.Moreira LO, Zamboni DS. 2012. NOD1 and NOD2 signaling in infection and inflammation. Front Immunol 3:328. doi: 10.3389/fimmu.2012.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, Thomas BJ, Malosse C, Gantier MP, Casillas LN, Votta BJ, Bertin J, Boneca IG, Sasakawa C, Philpott DJ, Ferrero RL, Kaparakis-Liaskos M. 2014. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 15:623–635. doi: 10.1016/j.chom.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 163.Dagil YA, Arbatsky NP, Alkhazova BI, L'vov VL, Mazurov DV, Pashenkov MV. 2016. The dual NOD1/NOD2 agonism of muropeptides containing a meso-diaminopimelic acid residue. PLoS One 11:e0160784. doi: 10.1371/journal.pone.0160784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Girardin SE, Travassos LH, Hervé M, Blanot D, Boneca IG, Philpott DJ, Sansonetti PJ, Mengin-Lecreulx D. 2003. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem 278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 165.Caruso R, Warner N, Inohara N, Núñez G. 2014. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cigana C, Curcurù L, Leone MR, Ieranò T, Lorè NI, Bianconi I, Silipo A, Cozzolino F, Lanzetta R, Molinaro A, Bernardini ML, Bragonzi A. 2009. Pseudomonas aeruginosa exploits lipid A and muropeptides modification as a strategy to lower innate immunity during cystic fibrosis lung infection. PLoS One 4:e8439. doi: 10.1371/journal.pone.0008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vijayrajratnam S, Pushkaran AC, Balakrishnan A, Vasudevan AK, Biswas R, Mohan CG. 2016. Bacterial peptidoglycan with amidated meso-diaminopimelic acid evades NOD1 recognition: an insight into NOD1 structure-recognition. Biochem J 473:4573–4592. doi: 10.1042/BCJ20160817. [DOI] [PubMed] [Google Scholar]
- 168.Rosenzweig HL, Martin TM, Jann MM, Planck SR, Davey MP, Kobayashi K, Flavell RA, Rosenbaum JT. 2008. NOD2, the gene responsible for familial granulomatous uveitis, in a mouse model of uveitis. Invest Ophthalmol Vis Sci 49:1518–1524. doi: 10.1167/iovs.07-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rosenzweig HL, Jann MM, Vance EE, Planck SR, Rosenbaum JT, Davey MP. 2010. Nucleotide-binding oligomerization domain 2 and Toll-like receptor 2 function independently in a murine model of arthritis triggered by intra-articular peptidoglycan. Arthritis Rheum 62:1051–1059. doi: 10.1002/art.27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Saha S, Qi J, Wang S, Wang M, Li X, Kim YG, Núñez G, Gupta D, Dziarski R. 2009. PGLYRP-2 and Nod2 are both required for peptidoglycan-induced arthritis and local inflammation. Cell Host Microbe 5:137–150. doi: 10.1016/j.chom.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Correa RG, Milutinovic S, Reed JC. 2012. Roles of NOD1 (NLRC1) and NOD2 (NLRC2) in innate immunity and inflammatory diseases. Biosci Rep 32:597–608. doi: 10.1042/BSR20120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jeong YJ, Kim CH, Song EJ, Kang MJ, Kim JC, Oh SM, Lee KB, Park JH. 2012. Nucleotide-binding oligomerization domain 2 (Nod2) is dispensable for the innate immune responses of macrophages against Yersinia enterocolitica. J Microbiol 50:489–495. doi: 10.1007/s12275-012-1534-6. [DOI] [PubMed] [Google Scholar]
- 173.Benoit M, Bechah Y, Capo C, Murray PJ, Mege JL, Desnues B. 2009. Role of the cytoplasmic pattern recognition receptor Nod2 in Coxiella burnetii infection. Clin Microbiol Infect 15:154–155. doi: 10.1111/j.1469-0691.2008.02270.x. [DOI] [PubMed] [Google Scholar]
- 174.Oliveira FS, Carvalho NB, Zamboni DS, Oliveira SC. 2012. Nucleotide-binding oligomerization domain-1 and -2 play no role in controlling Brucella abortus infection in mice. Clin Dev Immunol 2012:861426. doi: 10.1155/2012/861426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sônego F, Castanheira FV, Czaikoski PG, Kanashiro A, Souto FO, França RO, Nascimento DC, Freitas A, Spiller F, Cunha LD, Zamboni DS, Alves-Filho JC, Cunha FQ. 2014. MyD88-, but not Nod1- and/or Nod2-deficient mice, show increased susceptibility to polymicrobial sepsis due to impaired local inflammatory response. PLoS One 9:e103734. doi: 10.1371/journal.pone.0103734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Travassos LH, Carneiro LA, Girardin SE, Boneca IG, Lemos R, Bozza MT, Domingues RC, Coyle AJ, Bertin J, Philpott DJ, Plotkowski MC. 2005. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J Biol Chem 280:36714–36718. doi: 10.1074/jbc.M501649200. [DOI] [PubMed] [Google Scholar]
- 177.Watanabe T, Asano N, Fichtner-Feigl S, Gorelick PL, Tsuji Y, Matsumoto Y, Chiba T, Fuss IJ, Kitani A, Strober W. 2010. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest 120:1645–1662. doi: 10.1172/JCI39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zilbauer M, Dorrell N, Elmi A, Lindley KJ, Schüller S, Jones HE, Klein NJ, Núnez G, Wren BW, Bajaj-Elliott M. 2007. A major role for intestinal epithelial nucleotide oligomerization domain 1 (NOD1) in eliciting host bactericidal immune responses to Campylobacter jejuni. Cell Microbiol 9:2404–2416. doi: 10.1111/j.1462-5822.2007.00969.x. [DOI] [PubMed] [Google Scholar]
- 179.Berrington WR, Iyer R, Wells RD, Smith KD, Skerrett SJ, Hawn TR. 2010. NOD1 and NOD2 regulation of pulmonary innate immunity to Legionella pneumophila. Eur J Immunol 40:3519–3527. doi: 10.1002/eji.201040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mavrogiorgos N, Mekasha S, Yang Y, Kelliher MA, Ingalls RR. 2014. Activation of NOD receptors by Neisseria gonorrhoeae modulates the innate immune response. Innate Immun 20:377–389. doi: 10.1177/1753425913493453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Geddes K, Rubino S, Streutker C, Cho JH, Magalhaes JG, Le Bourhis L, Selvanantham T, Girardin SE, Philpott DJ. 2010. Nod1 and Nod2 regulation of inflammation in the Salmonella colitis model. Infect Immun 78:5107–5115. doi: 10.1128/IAI.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Claes AK, Steck N, Schultz D, Zähringer U, Lipinski S, Rosenstiel P, Geddes K, Philpott DJ, Heine H, Grassl GA. 2014. Salmonella enterica serovar Typhimurium ΔmsbB triggers exacerbated inflammation in Nod2 deficient mice. PLoS One 9:e113645. doi: 10.1371/journal.pone.0113645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shimada K, Chen S, Dempsey PW, Sorrentino R, Alsabeh R, Slepenkin AV, Peterson E, Doherty TM, Underhill D, Crother TR, Arditi M. 2009. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog 5:e1000379. doi: 10.1371/journal.ppat.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jeong YJ, Kang MJ, Lee SJ, Kim CH, Kim JC, Kim TH, Kim DJ, Kim D, Núñez G, Park JH. 2014. Nod2 and Rip2 contribute to innate immune responses in mouse neutrophils. Immunology 143:269–276. doi: 10.1111/imm.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kovarova M, Hesker PR, Jania L, Nguyen M, Snouwaert JN, Xiang Z, Lommatzsch SE, Huang MT, Ting JP, Koller BH. 2012. NLRP1 dependent pyroptosis leads to acute lung injury and morbidity in mice. J Immunol 189:2006–2016. doi: 10.4049/jimmunol.1201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cohen TS, Prince AS. 2013. Activation of inflammasome signaling mediates pathology of acute Pseudomonas aeruginosa pneumonia. J Clin Invest 123:1630–1637. doi: 10.1172/JCI66142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hsu LC, Ali SR, McGillivray S, Tseng PH, Mariathasan S, Humke EW, Eckmann L, Powell JJ, Nizet V, Dixit VM, Karin M. 2008. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci U S A 105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Reubold TF, Hahne G, Wohlgemuth S, Eschenburg S. 2014. Crystal structure of the leucine-rich repeat domain of the NOD-like receptor NLRP1: implications for binding of muramyl dipeptide. FEBS Lett 588:3327–3332. doi: 10.1016/j.febslet.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 189.Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M, Underhill DM. 2016. Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell 166:624–636. doi: 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kang MJ, Jo SG, Kim DJ, Park JH. 2017. NLRP3 inflammasome mediates interleukin-1β production in immune cells in response to Acinetobacter baumannii and contributes to pulmonary inflammation in mice. Immunology 150:495–505. doi: 10.1111/imm.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Iannitti RG, Napolioni V, Oikonomou V, De Luca A, Galosi C, Pariano M, Massi-Benedetti C, Borghi M, Puccetti M, Lucidi V, Colombo C, Fiscarelli E, Lass-Flörl C, Majo F, Cariani L, Russo M, Porcaro L, Ricciotti G, Ellemunter H, Ratclif L, De Benedictis FM, Talesa VN, Dinarello CA, van de Veerdonk FL, Romani L. 2016. IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis. Nat Commun 7:10791. doi: 10.1038/ncomms10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bergsbaken T, Fink SL, Cookson BT. 2009. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Latz E, Xiao TS, Stutz A. 2013. Activation and regulation of the inflammasomes. Nat Rev Immunol 13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Man SM, Kanneganti TD. 2015. Regulation of inflammasome activation. Immunol Rev 265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Grailer JJ, Canning BA, Kalbitz M, Haggadone MD, Dhond RM, Andjelkovic AV, Zetoune FS, Ward PA. 2014. Critical role for the NLRP3 inflammasome during acute lung injury. J Immunol 192:5974–5983. doi: 10.4049/jimmunol.1400368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lin CK, Kazmierczak BI. 2017. Inflammation: a double-edged sword in the response to Pseudomonas aeruginosa Infection. J Innate Immun 9:250–261. doi: 10.1159/000455857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nau R, Eiffert H. 2005. Minimizing the release of proinflammatory and toxic bacterial products within the host: a promising approach to improve outcome in life-threatening infections. FEMS Immunol Med Microbiol 44:1–16. doi: 10.1016/j.femsim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 198.Ragland SA, Criss AK. 2017. From bacterial killing to immune modulation: recent insights into the functions of lysozyme. PLoS Pathog 13:e1006512. doi: 10.1371/journal.ppat.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Callewaert L, Van Herreweghe JM, Vanderkelen L, Leysen S, Voet A, Michiels CW. 2012. Guards of the great wall: bacterial lysozyme inhibitors. Trends Microbiol 20:501–510. doi: 10.1016/j.tim.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 200.Ibrahim HR, Thomas U, Pellegrini A. 2001. A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J Biol Chem 276:43767–43774. doi: 10.1074/jbc.M106317200. [DOI] [PubMed] [Google Scholar]
- 201.Ibrahim HR, Matsuzaki T, Aoki T. 2001. Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett 506:27–32. doi: 10.1016/S0014-5793(01)02872-1. [DOI] [PubMed] [Google Scholar]
- 202.Nash JA, Ballard TN, Weaver TE, Akinbi HT. 2006. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J Immunol 177:519–526. doi: 10.4049/jimmunol.177.1.519. [DOI] [PubMed] [Google Scholar]
- 203.Torrens G, Pérez-Gallego M, Moya B, Munar-Bestard M, Zamorano L, Cabot G, Blázquez J, Ayala JA, Oliver A, Juan C. 2017. Targeting the permeability barrier and peptidoglycan recycling pathways to disarm Pseudomonas aeruginosa against the innate immune system. PLoS One 12:e0181932. doi: 10.1371/journal.pone.0181932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Callewaert L, Michiels CW. 2010. Lysozymes in the animal kingdom. J Biosci 35:127–160. doi: 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- 205.Callewaert L, Aertsen A, Deckers D, Vanoirbeek KGA, Vanderkelen L, Van Herreweghe JM, Masschalck B, Nakimbugwe D, Robben J, Michiels CW. 2008. A new family of lysozyme inhibitors contributing to lysozyme tolerance in Gram-negative bacteria. PLoS Pathog 4:e1000019. doi: 10.1371/journal.ppat.1000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Monchois V, Abergel C, Sturgis J, Jeudy S, Claverie J. 2001. Escherichia coli ykfE ORFan gene encodes a potent inhibitor of C-type lysozyme. J Biol Chem 276:18437–18441. doi: 10.1074/jbc.M010297200. [DOI] [PubMed] [Google Scholar]
- 207.Deckers D, Masschalck B, Aertsen A, Callewaert L, Van Tiggelen CG, Atanassova M, Michiels CW. 2004. Periplasmic lysozyme inhibitor contributes to lysozyme resistance in Escherichia coli. Cell Mol Life Sci 61:1229–1237. doi: 10.1007/s00018-004-4066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Deckers D, Vanlint D, Callewaert L, Aertsen A, Michiels CW. 2008. Role of the lysozyme inhibitor Ivy in growth or survival of Escherichia coli and Pseudomonas aeruginosa bacteria in hen egg white and in human saliva and breast milk. Appl Environ Microbiol 74:4434–4439. doi: 10.1128/AEM.00589-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Vanderkelen L, Ons E, Van Herreweghe JM, Callewaert L, Goddeeris BM, Michiels CW. 2012. Role of lysozyme inhibitors in the virulence of avian pathogenic Escherichia coli. PLoS One 7:e45954. doi: 10.1371/journal.pone.0045954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Derbise A, Pierre F, Merchez M, Pradel E, Laouami S, Ricard I, Sirard JC, Fritz J, Lemaître N, Akinbi H, Boneca IG, Sebbane F. 2013. Inheritance of the lysozyme inhibitor Ivy was an important evolutionary step by Yersinia pestis to avoid the host innate immune response. J Infect Dis 207:1535–1543. doi: 10.1093/infdis/jit057. [DOI] [PubMed] [Google Scholar]
- 211.Wang C, Hu Y, Sun B, Li J, Sun L. 2013. Edwardsiella tarda Ivy, a lysozyme inhibitor that blocks the lytic effect of lysozyme and facilitates host infection in a manner that is dependent on the conserved cysteine residue. Infect Immun 81:3527–3533. doi: 10.1128/IAI.00503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li MF, Wang C, Sun L. 2015. Edwardsiella tarda MliC, a lysozyme inhibitor that participates in pathogenesis in a manner that parallels Ivy. Infect Immun 83:583–590. doi: 10.1128/IAI.02473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Leysen S, Vanderkelen L, Van Asten K, Vanheuverzwijn S, Theuwis V, Michiels CW, Strelkov SV. 2012. Structural characterization of the PliG lysozyme inhibitor family. J Struct Biol 180:235–242. doi: 10.1016/j.jsb.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 214.Leysen S, Van Herreweghe JM, Callewaert L, Heirbaut M, Buntinx P, Michiels CW, Strelkov SV. 2011. Molecular basis of bacterial defense against host lysozymes: X-ray structures of periplasmic lysozyme inhibitors PliI and PliC. J Mol Biol 405:1233–1245. doi: 10.1016/j.jmb.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 215.Um SH, Kim JS, Kim K, Kim N, Cho HS, Ha NC. 2013. Structural basis for the inhibition of human lysozyme by PliC from Brucella abortus. Biochemistry 52:9385–9393. doi: 10.1021/bi401241c. [DOI] [PubMed] [Google Scholar]
- 216.Clarke CA, Scheurwater EM, Clarke AJ. 2010. The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J Biol Chem 285:14843–14847. doi: 10.1074/jbc.C110.120931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Callewaert L, Masschalck B, Deckers D, Nakimbugwe D, Atanassova M, Aertsen A, Michiels CW. 2005. Purification of Ivy, a lysozyme inhibitor from Escherichia coli, and characterisation of its specificity for various lysozymes. Enzyme Microb Technol 37:205–211. doi: 10.1016/j.enzmictec.2005.03.001. [DOI] [Google Scholar]
- 218.Van Herreweghe JM, Vanderkelen L, Callewaert L, Aertsen A, Compernolle G, Declerck PJ, Michiels CW. 2010. Lysozyme inhibitor conferring bacterial tolerance to invertebrate type lysozyme. Cell Mol Life Sci 67:1177–1188. doi: 10.1007/s00018-009-0241-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vanderkelen L, Van Herreweghe JM, Vanoirbeek KG, Baggerman G, Myrnes B, Declerck PJ, Nilsen IW, Michiels CW, Callewaert L. 2011. Identification of a bacterial inhibitor against g-type lysozyme. Cell Mol Life Sci 68:1053–1064. doi: 10.1007/s00018-010-0507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Vanderkelen L, Van Herreweghe JM, Callewaert L, Michiels CW. 2011. Goose-type lysozyme inhibitor (PliG) enhances survival of Escherichia coli in goose egg albumen. Appl Environ Microbiol 77:4697–4699. doi: 10.1128/AEM.00427-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Callewaert L, Vanoirbeek KGA, Lurquin I, Michiels CW, Aertsen A. 2009. The Rcs two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme. J Bacteriol 191:1979–1981. doi: 10.1128/JB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Joslin SN, Pybus C, Labandeira-Rey M, Evans AS, Attia AS, Brautigam CA, Hansen EJ. 2015. A Moraxella catarrhalis two-component signal transduction system necessary for growth in liquid media affects production of two lysozyme inhibitors. Infect Immun 83:146–160. doi: 10.1128/IAI.02486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pérez-Gallego M, Torrens G, Castillo-Vera J, Moya B, Zamorano L, Cabot G, Hultenby K, Albertí S, Mellroth P, Henriques-Normark B, Normark S, Oliver A, Juan C. 2016. Impact of AmpC derepression on fitness and virulence: the mechanism or the pathway? mBio 7:e01783-16. doi: 10.1128/mBio.01783-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Fumeaux C, Bernhardt TG. 2017. Identification of MupP as a new peptidoglycan recycling factor and antibiotic resistance determinant in Pseudomonas aeruginosa. mBio 8:e00102-17. doi: 10.1128/mBio.00102-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vadlamani G, Thomas MD, Patel TR, Donald LJ, Reeve TM, Stetefeld J, Standing KG, Vocadlo DJ, Mark BL. 2015. The β-lactamase gene regulator AmpR is a tetramer that recognizes and binds the d-Ala-d-Ala motif of its repressor UDP-N-acetylmuramic acid (MurNAc)-pentapeptide. J Biol Chem 290:2630–2643. doi: 10.1074/jbc.M114.618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dik DA, Domínguez-Gil T, Lee M, Hesek D, Byun B, Fishovitz J, Boggess B, Hellman LM, Fisher JF, Hermoso JA, Mobashery S. 2017. Muropeptide binding and the X-ray structure of the effector domain of the transcriptional regulator AmpR of Pseudomonas aeruginosa. J Am Chem Soc 139:1448–1451. doi: 10.1021/jacs.6b12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Langaee TY, Dargis M, Huletsky A. 1998. An ampD gene in Pseudomonas aeruginosa encodes a negative regulator of AmpC β-lactamase expression. Antimicrob Agents Chemother 42:3296–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Juan C, Maciá MD, Gutiérrez O, Vidal C, Pérez JL, Oliver A. 2005. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 49:4733–4738. doi: 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zamorano L, Reeve TM, Deng L, Juan C, Moyá B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. 2010. NagZ inactivation prevents and reverts β-lactam resistance, driven by AmpD and PBP 4 mutations, in Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:3557–3563. doi: 10.1128/AAC.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Moya B, Dötsch A, Juan C, Blázquez J, Zamorano L, Haussler S, Oliver A. 2009. β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zamorano L, Reeve TM, Juan C, Moyá B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. 2011. AmpG inactivation restores susceptibility of pan-β-lactam-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 55:1990–1996. doi: 10.1128/AAC.01688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yang TC, Chen TF, Tsai JJ, Hu RM. 2014. NagZ is required for beta-lactamase expression and full pathogenicity in Xanthomonas campestris pv. campestris str. 17. Res Microbiol 165:612–619. doi: 10.1016/j.resmic.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 233.Zhang W, Lee M, Hesek D, Lastochkin E, Boggess B, Mobashery S. 2013. Reactions of the three AmpD enzymes of Pseudomonas aeruginosa. J Am Chem Soc 135:4950–4953. doi: 10.1021/ja400970n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rivera I, Molina R, Lee M, Mobashery S, Hermoso JA. 2016. Orthologous and paralogous AmpD peptidoglycan amidases from Gram-negative bacteria. Microb Drug Resist 22:470–476. doi: 10.1089/mdr.2016.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liu C, Wang X, Chen Y, Hao H, Li X, Liang J, Duan R, Li C, Zhang J, Shao S, Jing H. 2016. Three Yersinia enterocolitica AmpD homologs participate in the multi-step regulation of chromosomal cephalosporinase, AmpC. Front Microbiol 7:1282. doi: 10.3389/fmicb.2016.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Juan C, Moyá B, Pérez JL, Oliver A. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob Agents Chemother 50:1780–1787. doi: 10.1128/AAC.50.5.1780-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Moya B, Juan C, Albertí S, Pérez JL, Oliver A. 2008. Benefit of having multiple ampD genes for acquiring β-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock REW, Martínez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to β-lactam antibiotics. Antimicrob Agents Chemother 54:4159–4167. doi: 10.1128/AAC.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Tsutsumi Y, Tomita H, Tanimoto K. 2013. Identification of novel genes responsible for overexpression of ampC in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 57:5987–5993. doi: 10.1128/AAC.01291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wang K, Chen YQ, Salido MM, Kohli GS, Kong JL, Liang HJ, Yao ZT, Xie YT, Wu HY, Cai SQ, Drautz-Moses DI, Darling AE, Schuster SC, Yang L, Ding Y. 2017. The rapid in vivo evolution of Pseudomonas aeruginosa in ventilator-associated pneumonia patients leads to attenuated virulence. Open Biol 7:170029. doi: 10.1098/rsob.170029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Diaz Caballero J, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YC, Waters VJ, Hwang DM, Guttman DS. 2015. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. mBio 6:e00981-15. doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Moradali MF, Ghods S, Rehm BHA. 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Dziarski R, Gupta D. 2010. Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun 16:168–174. doi: 10.1177/1753425910366059. [DOI] [PubMed] [Google Scholar]
- 244.Kashyap DR, Rompca A, Gaballa A, Helmann JD, Chan J, Chang CJ, Hozo I, Gupta D, Dziarski R. 2014. Peptidoglycan recognition proteins kill bacteria by inducing oxidative, thiol, and metal stress. PLoS Pathog 10:e1004280. doi: 10.1371/journal.ppat.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Gowda RN, Redfern R, Frikeche J, Pinglay S, Foster JW, Lema C, Cope L, Chakravarti S. 2015. Functions of peptidoglycan recognition proteins (Pglyrps) at the ocular surface: bacterial keratitis in gene-targeted mice deficient in Pglyrp-2, -3 and -4. PLoS One 10:e0137129. doi: 10.1371/journal.pone.0137129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lee J, Geddes K, Streutker C, Philpott DJ, Girardin SE. 2012. Role of mouse peptidoglycan recognition protein PGLYRP2 in the innate immune response to Salmonella enterica serovar Typhimurium infection in vivo. Infect Immun 80:2645–2654. doi: 10.1128/IAI.00168-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bobrovsky P, Manuvera V, Polina N, Podgorny O, Prusakov K, Govorun V, Lazarev V. 2016. Recombinant human peptidoglycan recognition proteins reveal antichlamydial activity. Infect Immun 84:2124–2130. doi: 10.1128/IAI.01495-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Boneca IG. 2009. Mammalian PGRPs in the spotlight. Cell Host Microbe 5:109–111. doi: 10.1016/j.chom.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 249.Goldman SM, Kamel F, Ross GW, Jewell SA, Marras C, Hoppin JA, Umbach DM, Bhudhikanok GS, Meng C, Korell M, Comyns K, Hauser RA, Jankovic J, Factor SA, Bressman S, Lyons KE, Sandler DP, Langston JW, Tanner CM. 2014. Peptidoglycan recognition protein genes and risk of Parkinson's disease. Mov Disord 29:1171–1180. doi: 10.1002/mds.25895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zulfiqar F, Hozo I, Rangarajan S, Mariuzza RA, Dziarski R, Gupta D. 2013. Genetic association of peptidoglycan recognition protein variants with inflammatory bowel disease. PLoS One 8:e67393. doi: 10.1371/journal.pone.0067393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bishop RE, Weiner JH. 1992. Coordinate regulation of murein peptidase activity and AmpC beta-lactamase synthesis in Escherichia coli. FEBS Lett 304:103–108. doi: 10.1016/0014-5793(92)80598-B. [DOI] [PubMed] [Google Scholar]
- 252.Rhazi N, Galleni M, Page MI, Frère JM. 1999. Peptidase activity of beta-lactamases. Biochem J 341(Pt 2):409–413. [PMC free article] [PubMed] [Google Scholar]
- 253.Morosini MI, Ayala JA, Baquero F, Martínez JL, Blázquez J. 2000. Biological cost of AmpC production for Salmonella enterica serotype Typhimurium. Antimicrob Agents Chemother 44:3137–3143. doi: 10.1128/AAC.44.11.3137-3143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Damblon C, Zhao GH, Jamin M, Ledent P, Dubus A, Vanhove M, Raquet X, Christiaens L, Frère JM. 1995. Breakdown of the stereospecificity of dd-peptidases and beta-lactamases with thiolester substrates. Biochem J 309:431–436. doi: 10.1042/bj3090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Henderson TA, Young KD, Denome SA, Elf PK. 1997. AmpC and AmpH, proteins related to the class C beta-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol 179:6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.González-Leiza SM, de Pedro MA, Ayala JA. 2011. AmpH, a bifunctional dd-endopeptidase and dd-carboxypeptidase of Escherichia coli. J Bacteriol 193:6887–6894. doi: 10.1128/JB.05764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Trépanier S, Prince A, Huletsky A. 1997. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob Agents Chemother 41:2399–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Okazaki A, Avison MB. 2008. Induction of L1 and L2 β-lactamase production in Stenotrophomonas maltophilia is dependent on an AmpR-type regulator. Antimicrob Agents Chemother 52:1525–1528. doi: 10.1128/AAC.01485-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lindquist S, Lindberg F, Normark S. 1989. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC beta-lactamase gene. J Bacteriol 171:3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lodge J, Busby S, Piddock L. 1993. Investigation of the Pseudomonas aeruginosa ampR gene and its role at the chromosomal ampC beta-lactamase promoter. FEMS Microbiol Lett 111:315–320. [DOI] [PubMed] [Google Scholar]
- 261.Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Høiby N, Mathee K. 2005. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB β-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob Agents Chemother 49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. 2012. The regulatory repertoire of Pseudomonas aeruginosa AmpC β-lactamase regulator AmpR includes virulence genes. PLoS One 7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Balasubramanian D, Kumari H, Jaric M, Fernandez M, Turner KH, Dove SL, Narasimhan G, Lory S, Mathee K. 2014. Deep sequencing analyses expands the Pseudomonas aeruginosa AmpR regulon to include small RNA-mediated regulation of iron acquisition, heat shock and oxidative stress response. Nucleic Acids Res 42:979–998. doi: 10.1093/nar/gkt942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Balasubramanian D, Kumari H, Mathee K. 2015. Pseudomonas aeruginosa AmpR: an acute-chronic switch regulator. Pathog Dis 73:1–14. doi: 10.1111/2049-632X.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A. 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Cabot G, López-Causapé C, Ocampo-Sosa AA, Sommer LM, Domínguez MÁ, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Plesiat P, Oliver A. 2016. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother 60:7415–7423. doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sánchez-Diener I, Zamorano L, López-Causapé C, Cabot G, Mulet X, Peña C, Del Campo R, Cantón R, Doménech-Sánchez A, Martínez-Martínez L, Arcos SC, Navas A, Oliver A. 2017. Interplay among resistance profiles, high-risk clones, and virulence in the Caenorhabditis elegans Pseudomonas aeruginosa infection model. Antimicrob Agents Chemother 61:e01586-17. doi: 10.1128/AAC.01586-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Doty SL, Chang M, Nester EW. 1993. The chromosomal virulence gene, chvE, of Agrobacterium tumefaciens is regulated by a LysR family member. J Bacteriol 175:7880–7886. doi: 10.1128/jb.175.24.7880-7886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Harris SJ, Shih YL, Bentley SD, Salmond GP. 1998. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol Microbiol 28:705–717. doi: 10.1046/j.1365-2958.1998.00825.x. [DOI] [PubMed] [Google Scholar]
- 270.Kovacikova G, Lin W, Skorupski K. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol Microbiol 53:129–142. doi: 10.1111/j.1365-2958.2004.04121.x. [DOI] [PubMed] [Google Scholar]
- 271.Russell DA, Byrne GA, O'Connell EP, Boland CA, Meijer WG. 2004. The LysR-type transcriptional regulator VirR is required for expression of the virulence gene vapA of Rhodococcus equi ATCC 33701. J Bacteriol 186:5576–5584. doi: 10.1128/JB.186.17.5576-5584.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Hernández-Lucas I, -Hernández AL, Encarnación S, Fernández-Mora M, Martínez-Batallar AG, Salgado H, Oropeza R, Calva E. 2008. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J Bacteriol 190:1658–1670. doi: 10.1128/JB.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Stock JB, Park P, Surette MG, Levit M. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p 25–51. In Hoch JA, Silhavy TJ (ed), Two-component signal transduction. ASM Press, Washington, DC. [Google Scholar]
- 274.Alksne LE, Rasmussen BA. 1997. Expression of the AsbA1, OXA-12, and AsbM1 beta-lactamases in Aeromonas jandaei AER 14 is coordinated by a two-component regulon. J Bacteriol 179:2006–2013. doi: 10.1128/jb.179.6.2006-2013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Avison MB, Horton RE, Walsh TR, Bennett PM. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J Biol Chem 276:26955–26961. doi: 10.1074/jbc.M011186200. [DOI] [PubMed] [Google Scholar]
- 276.Zamorano L, Moyà B, Juan C, Mulet X, Blázquez J, Oliver A. 2014. The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to β-lactams, fitness, biofilm growth, and global regulation. Antimicrob Agents Chemother 58:5084–5095. doi: 10.1128/AAC.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Huang HH, Chen WC, Lin CW, Lin YT, Ning HC, Chang YC, Yang TC. 2017. Relationship of the CreBC two-component regulatory system and inner membrane protein CreD with swimming motility in Stenotrophomonas maltophilia. PLoS One 12:e0174704. doi: 10.1371/journal.pone.0174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Tiwari S, Jamal SB, Hassan SS, Carvalho PVSD, Almeida S, Barh D, Ghosh P, Silva A, Castro TLP, Azevedo V. 2017. Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: an overview. Front Microbiol 8:1878. doi: 10.3389/fmicb.2017.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Haag AF, Bagnoli F. 2017. The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr Top Microbiol Immunol 409:145–198. doi: 10.1007/82_2015_5019. [DOI] [PubMed] [Google Scholar]
- 280.González LJ, Vila AJ. 2012. Carbapenem resistance in Elizabethkingia meningoseptica is mediated by metallo-β-lactamase BlaB. Antimicrob Agents Chemother 56:1686–1692. doi: 10.1128/AAC.05835-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Girlich D, Kolb A, Naas T, Nordmann P. 2009. Characterization of regulatory element Rp3 of regulation of beta-lactamases from Ralstonia pickettii. FEMS Microbiol Lett 301:50–56. doi: 10.1111/j.1574-6968.2009.01796.x. [DOI] [PubMed] [Google Scholar]
- 282.Docquier J, Pantanella F, Giuliani F, Thaller MC, Amicosante G, Galleni M, Frère JM, Bush K, Rossolini GM. 2002. CAU-1, a subclass B3 metallo-β-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome. Antimicrob Agents Chemother 46:1823–1830. doi: 10.1128/AAC.46.6.1823-1830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Gao CH, Yang M, He ZG. 2011. An ArsR-like transcriptional factor recognizes a conserved sequence motif and positively regulates the expression of phoP in mycobacteria. Biochem Biophys Res Commun 411:726–731. doi: 10.1016/j.bbrc.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 284.Saha RP, Chakrabarti P. 2006. Molecular modeling and characterization of Vibrio cholerae transcription regulator HlyU. BMC Struct Biol 6:24. doi: 10.1186/1472-6807-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Sahly H, Navon-Venezia S, Roesler L, Hay A, Carmeli Y, Podschun R, Hennequin C, Forestier C, Ofek I. 2008. Extended-spectrum β-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesins in Klebsiella pneumoniae. Antimicrob Agents Chemother 52:3029–3034. doi: 10.1128/AAC.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Schaufler K, Semmler T, Pickard DJ, de Toro M, de la Cruz F, Wieler LH, Ewers C, Guenther S. 2016. Carriage of extended-spectrum beta-lactamase-plasmids does not reduce fitness but enhances virulence in some strains of pandemic Escherichia coli lineages. Front Microbiol 7:336. doi: 10.3389/fmicb.2016.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hennequin C, Robin F, Cabrolier N, Bonnet R, Forestier C. 2012. Characterization of a DHA-1-producing Klebsiella pneumoniae strain involved in an outbreak and role of the AmpR regulator in virulence. Antimicrob Agents Chemother 56:288–294. doi: 10.1128/AAC.00164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Ramirez MS, Traglia GM, Lin DL, Tran T, Tolmasky ME. 2014. Plasmid-mediated antibiotic resistance and virulence in Gram-negatives: the Klebsiella pneumoniae paradigm. Microbiol Spectr 2(5):PLAS-0016-2013. doi: 10.1128/microbiolspec.PLAS-0016-2013. [DOI] [PubMed] [Google Scholar]
- 289.Gharrah MM, Mostafa El-Mahdy A, Barwa RF. 2017. Association between virulence factors and extended spectrum beta-lactamase producing Klebsiella pneumoniae compared to nonproducing isolates. Interdiscip Perspect Infect Dis 2017:7279830. doi: 10.1155/2017/7279830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Aoki S, Hirakata Y, Kondoh A, Gotoh N, Yanagihara K, Miyazaki Y, Tomono K, Yamada Y, Kohno S, Kamihira S. 2004. Virulence of metallo-β-lactamase-producing Pseudomonas aeruginosa in vitro and in vivo. Antimicrob Agents Chemother 48:1876–1878. doi: 10.1128/AAC.48.5.1876-1878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Dubois D, Prasadarao NV, Mittal R, Bret L, Roujou-Gris M, Bonnet R. 2009. CTX-M β-lactamase production and virulence of Escherichia coli K1. Emerg Infect Dis 15:1988–1990. doi: 10.3201/eid1512.090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 293.Cortés P, Blanc V, Mora A, Dahbi G, Blanco JE, Blanco M, López C, Andreu A, Navarro F, Alonso MP, Bou G, Blanco J, Llagostera M. 2010. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl Environ Microbiol 76:2799–2805. doi: 10.1128/AEM.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Valat C, Auvray F, Forest K, Métayer V, Gay E, Peytavin de Garam C, Madec JY, Haenni M. 2012. Phylogenetic grouping and virulence potential of extended-spectrum-β-lactamase-producing Escherichia coli strains in cattle. Appl Environ Microbiol 78:4677–4682. doi: 10.1128/AEM.00351-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Peirano G, Mulvey GL, Armstrong GD, Pitout JD. 2013. Virulence potential and adherence properties of Escherichia coli that produce CTX-M and NDM β-lactamases. J Med Microbiol 62:525–530. doi: 10.1099/jmm.0.048983-0. [DOI] [PubMed] [Google Scholar]
- 296.Lavigne JP, Cuzon G, Combescure C, Bourg G, Sotto A, Nordmann P. 2013. Virulence of Klebsiella pneumoniae isolates harboring blaKPC-2 carbapenemase gene in a Caenorhabditis elegans model. PLoS One 8:e67847. doi: 10.1371/journal.pone.0067847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Göttig S, Riedel-Christ S, Saleh A, Kempf VA, Hamprecht A. 2016. Impact of blaNDM-1 on fitness and pathogenicity of Escherichia coli and Klebsiella pneumoniae. Int J Antimicrob Agents 47:430–435. doi: 10.1016/j.ijantimicag.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 298.Marciano DC, Karkouti OY, Palzkill T. 2007. A fitness cost associated with the antibiotic resistance enzyme SME-1 β-lactamase. Genetics 176:2381–2392. doi: 10.1534/genetics.106.069443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Fernández A, Pérez A, Ayala JA, Mallo S, Rumbo-Feal S, Tomás M, Poza M, Bou G. 2012. Expression of OXA-type and SFO-1 β-lactamases induces changes in peptidoglycan composition and affects bacterial fitness. Antimicrob Agents Chemother 56:1877–1884. doi: 10.1128/AAC.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Cordeiro NF, Chabalgoity JA, Yim L, Vignoli R. 2014. Synthesis of metallo-β-lactamase VIM-2 is associated with a fitness reduction in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 58:6528–6535. doi: 10.1128/AAC.02847-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Gaillot O, Clément C, Simonet M, Philippon A. 1997. Novel transferable β-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J Antimicrob Chemother 39:85–87. doi: 10.1093/jac/39.1.85. [DOI] [PubMed] [Google Scholar]
- 302.Koeck JL, Arlet G, Philippon A, Basmaciogullari S, Thien HV, Buisson Y, Cavallo JD. 1997. A plasmid-mediated CMY-2 β-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol Lett 152:255–260. doi: 10.1111/j.1574-6968.1997.tb10436.x. [DOI] [PubMed] [Google Scholar]
- 303.Hossain A, Reisbig MD, Hanson ND. 2004. Plasmid-encoded functions compensate for the biological cost of AmpC overexpression in a clinical isolate of Salmonella typhimurium. J Antimicrob Chemother 53:964–970. doi: 10.1093/jac/dkh240. [DOI] [PubMed] [Google Scholar]
- 304.Derouaux A, Sauvage E, Terrak M. 2013. Peptidoglycan glycosyltransferase substrate mimics as templates for the design of new antibacterial drugs. Front Immunol 4:78. doi: 10.3389/fimmu.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Sauvage E, Terrak M. 2016. Glycosyltransferases and transpeptidases/penicillin-binding proteins: valuable targets for new antibacterials. Antibiotics 5:12. doi: 10.3390/antibiotics5010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Livermore DM, Mushtaq S, Warner M, Vickers A, Woodford N. 2017. In vitro activity of cefepime/zidebactam (WCK 5222) against Gram-negative bacteria. J Antimicrob Chemother 72:1373–1385. doi: 10.1093/jac/dkw593. [DOI] [PubMed] [Google Scholar]
- 307.Galley NF, O'Reilly AM, Roper DI. 2014. Prospects for novel inhibitors of peptidoglycan transglycosylases. Bioorg Chem 55:16–26. doi: 10.1016/j.bioorg.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Moya B, Barcelo IM, Bhagwat S, Patel M, Bou G, Papp-Wallace KM, Bonomo RA, Oliver A. 2017. WCK 5107 (zidebactam) and WCK 5153 are novel inhibitors of PBP2 showing potent “β-lactam enhancer” activity against Pseudomonas aeruginosa, including multidrug-resistant metallo-β-lactamase-producing high-risk clones. Antimicrob Agents Chemother 61:e02529-16. doi: 10.1128/AAC.02529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Stubbs KA, Balcewich M, Mark BL, Vocadlo DJ. 2007. Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated beta-lactam resistance. J Biol Chem 282:21382–21391. doi: 10.1074/jbc.M700084200. [DOI] [PubMed] [Google Scholar]
- 310.Stubbs KA, Bacik JP, Perley-Robertson GE, Whitworth GE, Gloster TM, Vocadlo DJ, Mark BL. 2013. The development of selective inhibitors of NagZ: increased susceptibility of Gram-negative bacteria to β-lactams. Chembiochem 14:1973–1981. doi: 10.1002/cbic.201300395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Lamers RP, Nguyen UT, Nguyen Y, Buensuceso RNC, Burrows LL. 2015. Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. Microbiologyopen 4:879–895. doi: 10.1002/mbo3.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Paharik AE, Schreiber HL, Spaulding CN, Dodson KW, Hultgren SJ. 2017. Narrowing the spectrum: the new frontier of precision antimicrobials. Genome Med 9:110. doi: 10.1186/s13073-017-0504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]