Abstract
Zombi pea (Vigna vexillata (L.) A. Rich) is an underutilized crop belonging to the genus Vigna. Two domesticated forms of zombi pea are cultivated as crop plants; seed and tuber forms. The cultivated seed form is present in Africa, while the cultivated tuber form is present in a very limited part of Asia. Genetics of domestication have been investigated in most of cultivated Vigna crops by means of quantitative trait locus (QTL) mapping. In this study, we investigated genetics of domestication in zombi pea by QTL analysis using an F2 population of 139 plants derived from a cross between cultivated tuber form of V. vexillata (JP235863) and wild V. vexillata (AusTRCF66514). A linkage map with 11 linkage groups (LGs) was constructed from this F2 population using 145 SSR, 117 RAD-seq and 2 morphological markers. Many highly segregation distorted markers were found on LGs 5, 6, 7, 8, 10 and 11. Most of the distorted markers were clustered together and all the markers on LG8 were highly distorted markers. Comparing this V. vexillata linkage map with linkage maps of other four Vigna species demonstrated several genome rearrangements in V. vexillata. QTL analysis for 22 domestication-related traits was investigated by inclusive composite interval mapping in which 37 QTLs were identified for 18 traits; no QTL was detected for 4 traits. Number of QTLs detected in each trait ranged from 1 to 5 with an average of only 2.3. Five QTLs for tuber width and three QTLs for tuber weight. Interestingly, 2 QTLs each for tuber width and tuber weight detected on LG2 and LG4 were located at similar position and wild allele increased tuber width and weight. This indicated wild germplasm having small tuber have potential to increase yield of large tuber cultivated type. Large-effect QTLs (PVE > 20%) were on LG4 (pod length), LG5 (leaf size and seed thickness), and LG7 (for seed-related traits). Comparison of domestication-related QTLs of the zombi pea with those of cowpea (Vigna unguiculata), azuki bean (Vigna angularis), mungbean (Vigna radiata) and rice bean (Vigna umbellata) revealed that there was conservation of some QTLs for seed size, pod size and leaf size between zombi pea and cowpea and that QTLs associated with seed size (weight, length, width and thickness) in each species were clustered on same linkage.
Introduction
The genus Vigna is an important taxon of leguminous plants. This genus comprises more than 100 species that distribute in all major continents including Africa, America, Asia, Australia and Europe. Among those species, as high as nine Vigna species are domesticated/cultivated. These domesticated Vigna species include azuki bean (Vigna angularis (Wild.) Ohwi and Ohashi), black gram (Vigna mungo (L.) Hepper), créole bean (Vigna reflexo-pilosa Hayata), mungbean (Vigna radiata (L.) Wilczek), moth bean (Vigna aconitifolia (Jacq.) Maréchal), rice bean (Vigna umbellata (Thunb.) Ohwi and Ohashi), cowpea (Vigna unguiculata (L.) Walps.), Bambara groundnut (Vigna subterranea Verdc.) and zombi pea (Vigna vexillata (L.) A. Rich) [1,2]. The former six species are of Asian origin and belong to the same subgenus Ceratotropis, while the latter three species are of African origin. Cowpea and Bambara groundnut belong to the subgenus Vigna, while zombi pea belongs to the subgenus Plectrotropis. These crops are generally cultivated by resource-poor farmers as a sole crop or a component in various cropping systems.
Among the domesticated Vigna species, zombi pea is the least known crop. Two forms of zombi pea exists; seed type and tuber (storage root) types. The seed type is believed to be domesticated in Sudan (Africa) [3,4], while the tuber type is believed to be domesticated in Bali and Timor, Indonesia (Asia) [5] and India [6,7]. Genetic diversity analysis in a large set of V. vexillata germplasm using simple sequence repeat (SSR; also known as microsatellite) markers revealed clear difference between the two types and suggested that these two types were domesticated from different wild gene pools of V. vexillata [4]. The seed type zombi pea is classified as variety macrosperma. Possibly, this type is domesticated from wild zombi pea of East Africa [4]. It has erect growth habit and large seed size, and is insensitive to day length and no seed dormancy with some degree of pod dehiscence. The variety macrosperma is highly cross-compatible with wild V. vexillata [8]. The tuber type zombi pea has not yet been classified as a variety of V. vexillata. It has viny growth habit and large seed size, and is sensitive to day length and no seed dormancy with some degree of pod shattering. The tuber type is grown principally for fresh edible tuber roots which contain as high as 15% of proteins [5,6]. This cultivated type has been reported to be cross-incompatible with wild V. vexillata and the variety macrosperma [8]. The phenotypic difference between the two cultivated types of V. vexillata is a result of divergent selection during the evolution of the crop. Morphological and physiological differences that occur during the change from wild plants to cultivated crops is known as “domestication syndrome” [9]. Crop domestication is an accelerated evolutionary process that is the result from human intentional and unintentional selection and natural selections [10]. Domestication-related traits in legume crops include plant architecture (determinate growth habit), gigantism in the consumed plant organs (seed size and/or pod size), reduced seed dispersal (indehiscent pod), loss of seed dormancy and no response to photoperiod [11–13].
Knowledge on genetic basis of crop domestication is useful for identification of beneficial gene(s) in the wild relatives of the cultivated crops that can be used in plant breeding. Moreover, such knowledge can also provide insight into crop evolution and agriculture, such that the case of genetic analysis of indehiscent pod in soybean by Funastuki [14] which showed crucial role of the trait in the global expansion of soybean cultivation. Generally, genes controlling crop domestication are identified by quantitative trait loci (QTL) analysis. Among crops of the genus Vigna, QTL mappings of domestication syndrome traits have been carried out in azuki bean [13,15], mungbean [16], rice bean [17], cowpea/yardlong bean [18–21]. The genetics of domestication of zombi pea is of particular interest because two cultivated forms of this legume have experienced different domestications from wild zombi pea. Seed type zombi pea was domesticated from wild zombi pea of Africa, while tuber type zombi pea was domesticated from wild zombi pea of Asia [4,22]. Information on the genetics of domestication-related traits would be useful for zombi pea improvement programs, and for comparative genome studies among members of the genus Vigna. The objectives of this study were to identify QTLs for domestication-related traits in tuber type zombi pea and to compare them with previously reported QTLs for domestication in other Vigna species.
Materials and methods
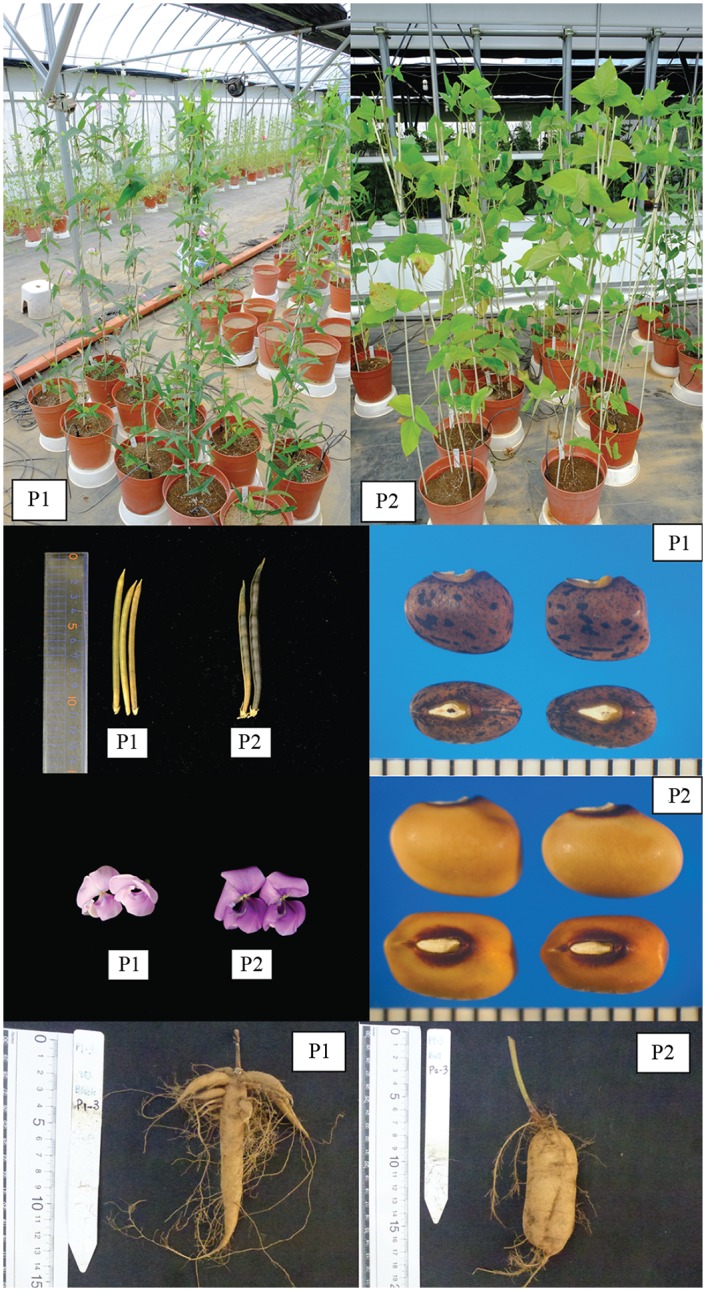
An F2 population of 139 plants was used in this study. The population was developed from a cross between accession JP235863 and accession AusTRCF66514 (Fig 1). JP235863 is a cultivated zombi pea collected from Bali Island, Indonesia which it is cultivated principally for edible tuber root, while AusTRCF66514 is a wild zombi pea from India. AusTRCF66514 was crossed with JP235863 as female and male parents, respectively, to produce F1 hybrid. Only one F1 seed was obtained and was self-pollinated to generate F2 population. One hundred and eighty seven F2 seeds together with parental seeds were sown in pots (1 seed per 1 pot) during June to November 2015 under a greenhouse condition at National Agriculture and Food Research Organization, Tsukuba, Japan. One hundred and fifty eight seeds germinated and developed into plants, but 139 of them grew vigorously and set flowers, and were used for DNA analysis and phenotypic data collection.
Fig 1. Comparison of morphological characteristics between wild V. vexillata accession “AusTRCF66514” and cultivated V. vexillata accession “JP235863” used in this study.
DNA extraction
Total genomic DNA was extracted from young leaves of the parental and the F2 plants using a CTAB method described by Lodhi et al. [23]. DNA quality and quantity was checked by comparing with a known concentration ʎ-DNA (50 and 100 ng/μl) on 1% agarose gel. DNA concentration of each plant was adjusted to 5 ng/μl for PCR analysis. The adjusted DNA was also checked for quality using NanoDrop 8000 (Thermo Fisher Scientific, Wilmington, U.S.A.).
Trait measurements
Twenty-two traits related to domestication in Vigna crops [13,15–17,20] were measured/evaluated (Table 1). Among those traits, seed color (SDC) and presence of black mottles (SDCBM) on seeds were considered as qualitative trait, while the others were considered as quantitative trait. Seedling traits, primary leaf width (LFPW) and primary leaf length (LFPL) were recorded when the first trifoliate was appeared. Stem thickness (STT), 10th node length (STL10), maximum leaf length (LFML) and maximum leaf width (LFMW) were collected when eleventh trifoliate leaf was fully developed. Seed-related traits including number of seeds per pod (SDNPPD) was an average from ten pods, seed width (SDW), seed length (SDL) and seed thickness (SDT) was the average from ten seeds while 100-seed weight (SD100WT) was recorded from 100 seeds of each plant. The number of days from planting to first flowering (FLD) was recorded. Number of days to first mature pod (PDDM) chosen from first flower that had been developed to mature pod. After harvesting all pods, ten pods from each plant were used to measure/recorded for pod width (PDW), pod length (PDL), pod dehiscence (PDT) or number of twist along the length of pod (NTWP). PDT was recorded after the pods were kept in hot air oven at 40°C for 24 hours. Tuber traits (NTB, TBWT, TBW and TBL) were collected at 260 days after planting. Broad-sense heritability of each traits was calculated by using the following equation; , where h2 is broad-sense heritability, is the variance of F2 population, is the variance of JP235863 and is the variance of AusTRCF66514.
Table 1. Domestication-related traits determined in F2 population derived from cross between cultivated (Bali) and wild (India) accessions.
| General attribute | Organ | Trait | Trait abbreviation | QTL/gene | Evaluation |
|---|---|---|---|---|---|
| Pod dehiscence | Pod | Number of twists (count) | PDT | Pdt | Number of twists along the length of shattered pod |
| Gigantism | Seed | 100-seed weight | SD100WT | Sd100wt | Weight of 100 seeds |
| Length (mm) | SDL | Sdl | Maximum distance from top to bottom of the seed | ||
| Width (mm) | SDW | Sdw | Maximum distance from hilum to its opposite side | ||
| Thickness (mm) | SDT | Sdt | Maximum distance between both side of the hilum | ||
| Pod | Length (mm) | PDL | Pdl | Length of straight pod | |
| Width (mm) | PDW | Pdw | Maximum width | ||
| Stem | Thickness (mm) | STT | Stt | Stem diameter under the primary leaf (measured at flowering stage) | |
| Leaf | Primary leaf length (mm) | LFPL | Lfpl | Distance from pulvinus to leaf tip | |
| Primary leaf width (mm) | LFPW | Lfpw | Maximum width | ||
| Maximum leaflet length (mm) | LFML | Lfml | Length of the largest terminal leaflet on leaves between node on first trifoliate leaf and node on ten trifoliate leaf | ||
| Maximum leaflet width (mm) | LFMW | Lfmw | Width of the largest terminal leaflet on leaves between node on first trifoliate leaf and node on tenth trifoliate leaf | ||
| Tuber | Number of tuber | NTB | Ntb | Number of storage root | |
| Tuber width | TBW | Tbw | Maximum width of main tuber | ||
| Tuber length | TBL | Tbl | Distance from top of tuber to bottom of main tuber | ||
| Tuber weight | TBWT | Tbwt | Weight of tubers | ||
| Plant type | Stem | Length (up to 10th node) (cm) | STL10 | Stl10 | Length from node on primary leaf to node 10 of trifoliate leaf |
| Earliness | Flower | Days to first flower (day) | FLD | Fld | Number of days from planting to 1st flowering |
| Pod | Days to maturity of 1st pod (day) | PDDM | Pddm | Number of days from 1stflowering to harvesting of 1st pod | |
| Yield potential | Seed | Number of seeds per pod (seeds/pod) | SDNPPD | Sdnppd | Number of seed per pod |
| Pigmentation | Seed | Seed coat color | SDC | Sdc | Brown (wild) or yellow (cultivated) |
| Black mottle on seed coat | SDCBM | Sdcbm | Present (wild) or absent (cultivated) |
SSR marker analysis
A total of 1,876 SSR primer pairs (827 pairs from azuki bean [24,25], 562 pairs from cowpea [19,26], 318 pairs from mungbean [27–29], and 169 pairs from common bean [30–32]) were screened for polymorphism between the wild and cultivated parents. PCR amplification and fragment analysis for the primers from azuki bean, cowpea, and common bean were performed following the method described by Marubodee et al. [33], while for the primers from mungbean were the same as described by Somta et al. [34]
RAD-seq analysis
RAD-seq analysis was conducted following a modified protocol described by Peterson et al. [35]. Genomic DNA of parents and each F2 plant was digested by double-digest RAD library method. In brief, 10 ng of genomic DNA was digested with EcoRI and BgIII (New England Biolabs, Ipswichs, MA, USA), and purified. Each adaptor with unique 4–8 bp index was ligated to digested DNA samples. The adaptor sequence was as follows: TruSeq_EcoRI_adaptor 1 = A*A*TTGAGATCGGAAGAGCACACGTCTGAACTCCAGTC*A*C TruSeq_EcoRI_adaptor 2 = G*T*CAAGTTTCACAGCTCTTCCGATC*T*C (* = variable index sequences to identify the individual DNA sample). BglII adaptor = A*A*TGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTT*C*CTruSeq_BglII_adaptor 2 = G*A*TCGGAAGAGCTGTGCAGA*C*T. The adaptors were ligated at 37°C overnight in 10 μl volume which contained 1 μl of 10x NEB buffer 2, 0.1 μl of 100x BSA (New England Biolabs), 0.4 μl of 5 uM EcoRI adapter and BgIII adapter, 0.1 μl of 100 mM ATP and 0.5 μl of T4 DNA ligase (Enzymatic, Beverly, MA, USA). The reaction solution was purified with AMPure XP (Beckham Coulter, CA, USA). Three microliter of the purified DNA was used in PCR amplification in 10 μl volume, containing 1μl of each 10 μM index and TruSeq universal primer, 0.3 μl of KOD-Plus-Neo enzyme and 1 μl of 10x PCR buffer (TOYOBO, Osaka, Japan), 0.6 μl of 25 mM MgSO4 and 1 μl of 10 mM dNTP. Thermal cycling was initiated with 94°C step for 2 mins, followed by 20 cycles of 98°C for 10 s, 65°C for 30 s and 68°C for 30 s. The PCR products were pooled and purified again with AMPure XP. The purified DNA was then loaded to a 2.0% agarose gel and fragments with size of about 320 bp were retrieved using E-Gel SizeSelect (Life Technologies, Carlsbad, CA, USA), the library was sequenced with 51 bp single-end read in one lane of HiSeq 2000 (Illumine, San Diego, CA, USA).
Extraction of RAD-tag and bi-allelic RAD-marker detection
RAD-tag sequences were extracted and bi-allelic RAD-markers were detected using Stacks ver. 1.30 [36] following the procedures described by Marubodee et al. [33]. With the Stacks pipeline, the low quality sequences were filtered to obtain 51 bp RAD-tag sequence reads, and classified the RAD-tags sequences into the parental accessions and F2 individuals. Then ran denovo_map.pl program where RAD-tags with less than two sequence mismatches were grouped as a stack, parental stacks with less than three mismatches were estimated as those derived from homologous loci and lists of RAD-tag sequences and their count were constructed for each sample. We called genotypes of F2 individuals only when stacks have more than five RAD-tags (minimum depth of 5).
Linkage map construction using SSR ad RAD-seq markers
A genetic linkage map of the F2 population was constructed using software Join Map 4.0 [37]. Polymorphic markers showing more than 10% missing genotypic data were excluded from linkage analysis. Chi-square analysis was used to test segregation of the marker loci (1:2:1 for co-dominant markers and 3:1 for dominant markers). The markers were grouped using minimum logarithm of the odds (LOD) of 3 and recombination frequency of 0.40. Genetic distance between markers in centimorgan (cM) unit was calculated using Kosambi’s mapping function [38].
QTL analysis
QTLs controlling domestication-related traits were located onto the linkage map by inclusive composite interval mapping (ICIM) method [39] using the software QTL IciMapping 4.1 [40]. Probability value for entering variables in stepwise regression of phenotype on marker variables (PIN) was 0.001. ICIM was performed at every 1 cM. Significant log of odds (LOD) threshold for QTL analysis of each trait was determined by 3,000 permutation test at P = 0.05.
Results
SSR polymorphism
Out of 1,876 SSR primer pairs screened for polymorphism between the parents, 687 pairs (36.6%) were able to amplify DNA of the parents. Primers from azuki bean showed highest amplification rate (51.4%), while those from cowpea showed lowest amplification rate (19.2%). Among the 687 amplifiable primers, only 201 (29.3%) primers showed polymorphism between the parents. Percentage of polymorphic primers ranged from 20.4 (mungbean primers) to 32.7 (azuki bean primers) (S1 Table).
RAD-seq markers
Illumina sequencing with HiSeq 2000 yielded a total of 29,568,103,697 RAD-tag 51-base reads from 578,901,731 raw reads. The number of RAD-tags of parents (AusTRCF66514 and JP235863) and F2 individuals was 3,124,320, 2,700,159 and 573,077,252, respectively. The average number of RAD-tags per F2 individual was 4,122,857. RAD-tags were aligned and clustered into 6,576 stacks. Average fill gaps rated for this 139 F2 individuals was 0.747 (max = 0.949, min = 0). Among these stacks, 479 RAD markers showed homozygote polymorphic genotype between parents. However, among these 479 RAD markers, 362 RAD markers were discarded because they had percentage of missing data more than 10%.
Linkage map analysis
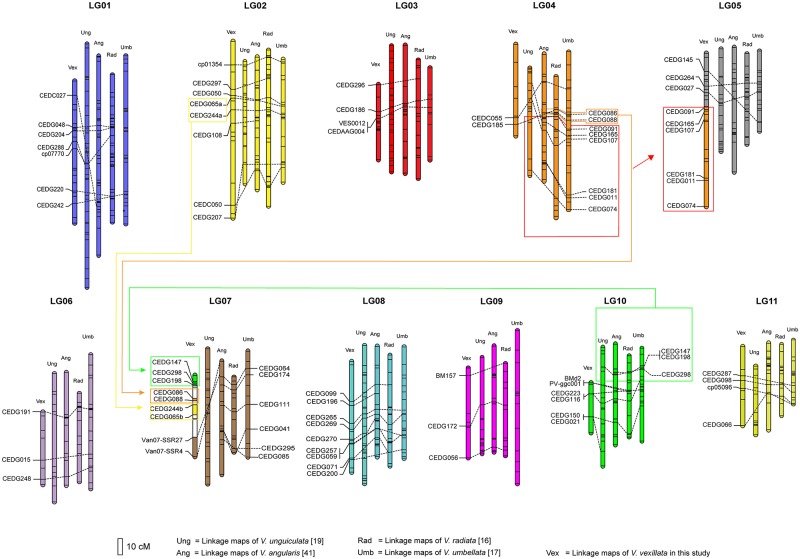
Finally, 264 markers (145 SSR, 117 RAD and 2 morphological markers) were used for the linkage map construction after discarding markers showing identical F2 genotypes, missing data more than 10%. Among these markers, as high as 128 (48.5%) showed segregation distortion. The 264 markers were clustered into 11 linkage groups (LGs 1–11) (Table 2 and Fig 2). The map spanned 704.8 cM in total, with a mean distance between markers of 2.87 cM. The lengths of linkage groups ranged from 30.9 cM (LG10) to 112.7 cM (LG2). A gap between markers more than 15 cM presented on only LG11 (between markers DMBSSR192 and CEDG066).
Table 2. Number of markers and average distance between markers in each linkage group of zombi pea F2 population (AusTRCF66514 x JP235863).
| Linkage group (LG) | No. of SSR markers | No. of RAD-Seq markers | Total number of markers | Total length (cM) | Average distance between markers | |||
|---|---|---|---|---|---|---|---|---|
| mungbean | azuki bean | cowpea | common bean | |||||
| 1 | 2 | 18 | 1 | 3 | 4 | 28 | 87.8 | 3.14 |
| 2 | 2 | 11 | 1 | 2 | 12 | 28 | 112.7 | 4.03 |
| 3 | 3 | 12 | 1 | 0 | 10 | 26 | 48.0 | 1.85 |
| 4 | 1 | 3 | 1 | 0 | 5 | 14 | 56.7 | 4.05 |
| 5 | 1 | 21 | 4 | 1 | 15 | 42 | 95.9 | 2.28 |
| 6 | 0 | 6 | 0 | 0 | 9 | 15 | 45.2 | 3.01 |
| 7 | 2 | 11 | 1 | 1 | 17 | 32 | 51.4 | 1.61 |
| 8 | 1 | 12 | 2 | 0 | 23 | 38 | 68.7 | 1.81 |
| 9 | 1 | 3 | 2 | 1 | 10 | 17 | 59.1 | 3.48 |
| 10 | 0 | 5 | 0 | 2 | 2 | 9 | 30.9 | 3.43 |
| 11 | 1 | 5 | 1 | 0 | 10 | 17 | 48.4 | 2.85 |
| Average | 1.27 | 9.73 | 1.27 | 0.91 | 10.64 | 23.82 | 64.07 | 2.87 |
| Total | 14 | 107 | 14 | 10 | 117 | 262 | 704.8 | 31.54 |
Note SDC and SDCBM on LG4 are morphological markers
Fig 2. A genetic linkage map of V. vexillata developed from F2 population of 139 individuals from crossed between AusTRCF66514 and JP235863.
Map distance and marker names are shown on the left and right side of the linkage groups, respectively. Markers showing significant deviation from the expected segregation ratio at 0.05, 0.01 and 0.001 probability levels are indicated with *, ** and ***, respectively. Marker names with prefixes C_ are RAD-Seq marker. SSR marker names with prefixes CED, Van07 and VES are from azuki bean. Marker names with prefixes VR and DMB-SSR are from mungbean. Marker names with prefixes cp and VM are from cowpea. Marker names with prefixes BM, BMd, SSR-IAC and PV are from common bean. SDC and SDCBM are morphological markers.
LGs 5, 6, 7, 8, 10 and 11 possessed the several/many distorted markers. The distorted markers clustered together. All the markers on LG8 were distorted markers. LGs 5 and 8 possessed many markers showing severe segregation distortion (P< 0.001). All the distorted markers on LG5 showed excessive cultivated parent genotype. Majority of the distorted markers on LGs 6 and 11 and about half of the distorted markers on LG10 showed excessive heterozygous genotype. All the distorted markers on LGs 7 and 8 and about half of the distorted markers on LG10 showed excessive homozygous wild genotype.
Primers CEDG065 and CEDG244 each amplified two different loci (markers). One of the loci from CEDG065 (named as CEDG065a) and one of the loci from CEDG244 (named as CEDG244a) were mapped near each other on LG2. The other loci from CEDG065 (named as CEDG065b) and the other loci from CEDG244 (named as CEDG244b) were mapped adjacent to each other on LG7.This indicated partial genome duplication and orthologous block of LG2 to LG7 or vice versa.
Comparative linkage analysis
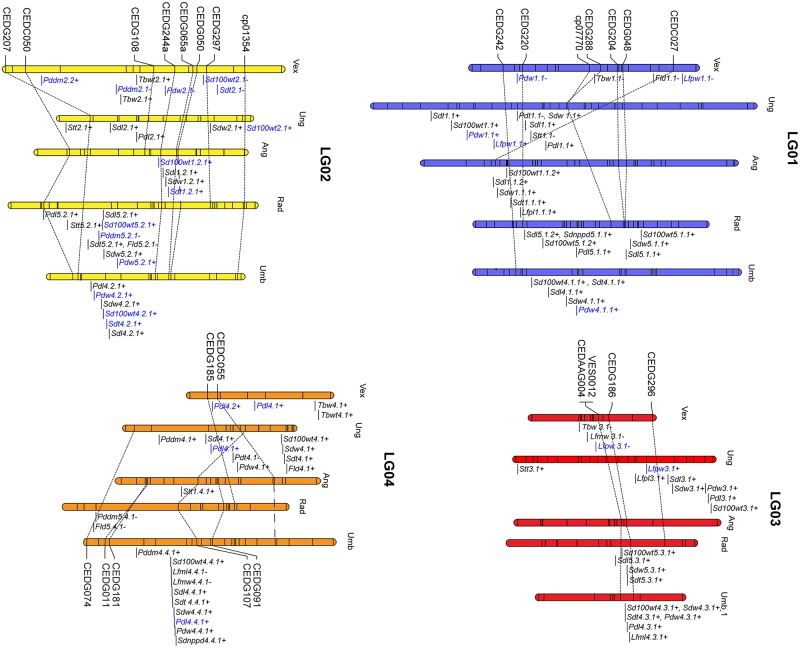
The linkage map of zombi pea developed in this study were compared with previous linkage maps developed for Vigna crops including yardlong bean [19], azuki bean [41], mungbean [16] and rice bean [17] by using common SSR markers (Fig 3). The comparison revealed that in general the linkage groups and orders of the markers among the Vigna species were the same or highly similar. Nonetheless, remarkable macro genome rearrangements were found on LG5 and LG7 of our linkage map for V. vexillata. About a half of LG5 (middle to bottom) appeared to be an orthologous blocks from LG4, while a large portion of LG7 appeared to be orthologous blocks from LG4 and LG10. In addition, a small portion of the LG7 appeared to be duplication from LG2, and vice versa. The length of the zombi pea linkage map developed in this study was shorter that the linkage maps of others Vigna species.
Fig 3. Comparative linkage map among V. vexillata (Vex), V. unguiculata (Ung), V. angularis (Ang), V. radiata (Rad) and V. umbellata (Umb) based on common SSR markers.
Dotted lines connected common SSR markers between linkage maps.
Variation of domestication-related traits
The parents were clearly different in all the traits recorded. The cultivated zombi pea showed higher value of trait value than the wild zombi pea in all the traits except pod shattering, seeds per pod, tubers per plant, and tuber length. For qualitative traits, the cultivated parent had yellow seed, while the wild zombi pea had brown seed with black mottle (Fig 1). The mean, range and standard deviation, and broad-sense heritability of the quantitative traits are shown in Table 3. Mean of the traits of the F2 population was between mean of parents for all traits except pod length, tuber length, 10th node length and number of seeds per pod.
Table 3. Means, standard deviation (S.D.), minimum, maximum and heritability values for the parents and the F2 population that derived from a cross between wild (AusTRCF66514) and cultivated (JP235863).
| Trait | Units | AusTRCF66514 (wild) | JP235863 (cultivated) | F2 population | Heritability (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | Min | Max | Mean | S.D. | Min | Max | Mean | S.D. | Min | Max | |||
| PDT | - | 2.0 | 0.2 | 1.8 | 2.4 | 1.2 | 0.4 | 0.5 | 1.5 | 1.6 | 0.7 | 0.5 | 3.8 | 83.0 |
| SD100WT | g | 2.1 | 0.3 | 1.8 | 2.5 | 7.3 | 0.3 | 6.8 | 7.7 | 4.9 | 1.4 | 2.0 | 8.5 | 95.2 |
| SDL | mm | 3.9 | 0.3 | 3.6 | 4.4 | 6.4 | 0.2 | 6.2 | 6.7 | 5.7 | 0.5 | 4.7 | 6.8 | 79.6 |
| SDW | mm | 3.2 | 0.1 | 3.1 | 3.4 | 4.2 | 0.2 | 4.1 | 4.4 | 4.0 | 0.4 | 3.1 | 5.0 | 87.8 |
| SDT | mm | 2.3 | 0.2 | 1.9 | 2.7 | 3.9 | 0.2 | 3.7 | 4.2 | 3.1 | 0.4 | 2.2 | 4.2 | 63.3 |
| PDL | mm | 85.6 | 1.7 | 83.1 | 89.1 | 94.3 | 16.3 | 61.0 | 112.9 | 75.8 | 12.7 | 41.7 | 108.1 | 16.3 |
| PDW | mm | 3.6 | 0.1 | 3.5 | 3.7 | 5.7 | 0.2 | 5.2 | 5.9 | 5.1 | 0.4 | 4.2 | 6.0 | 80.3 |
| STT | cm | 2.6 | 0.3 | 2.3 | 3.0 | 3.9 | 0.4 | 3.1 | 4.6 | 3.8 | 0.6 | 2.4 | 5.8 | 69.2 |
| LFPL | mm | 29.1 | 2.6 | 24.9 | 34.1 | 54.8 | 6.7 | 43.1 | 63.0 | 49.4 | 7.8 | 28.2 | 71.2 | 58.0 |
| LFPW | mm | 10.0 | 0.8 | 8.6 | 11.8 | 27.9 | 2.7 | 21.2 | 30.3 | 20.9 | 4.7 | 12.8 | 34.7 | 81.8 |
| LFML | mm | 61.4 | 4.9 | 54.7 | 69.3 | 75.4 | 5.5 | 67.3 | 85.3 | 69.3 | 9.9 | 42.3 | 99.0 | 65.2 |
| LFMW | mm | 17.9 | 2.3 | 14.3 | 21.7 | 58.1 | 4.6 | 50.7 | 63.7 | 32.4 | 8.8 | 14.7 | 57.0 | 86.6 |
| NTB | count | 7.5 | 3.3 | 1.0 | 10.0 | 2.1 | 1.4 | 1.0 | 4.0 | 5.0 | 3.0 | 1.0 | 13.0 | 27.7 |
| TBW | mm | 15.8 | 1.7 | 13.2 | 18.5 | 39.4 | 6.6 | 29.0 | 47.3 | 22.6 | 7.3 | 9.3 | 40.0 | 56.3 |
| TBL | mm | 95.7 | 11.4 | 77.5 | 108.0 | 81.5 | 12.4 | 60.2 | 101.0 | 100.0 | 17.0 | 56.9 | 165.6 | 50.7 |
| TBWT | g | 17.8 | 6.5 | 9.1 | 27.8 | 60.7 | 11.9 | 46.9 | 84.1 | 46.0 | 30.2 | 5.8 | 143.0 | 89.9 |
| STL10 | cm | 99.8 | 4.3 | 95.0 | 105.0 | 113.6 | 3.1 | 106.0 | 117.0 | 115.9 | 9.1 | 92.0 | 140.0 | 83.4 |
| FLD | day | 72.4 | 1.6 | 70.0 | 75.0 | 102.6 | 8.4 | 92.0 | 116.0 | 80.7 | 9.1 | 65.0 | 115.0 | 55.3 |
| PDDM | day | 27.8 | 1.4 | 26.0 | 30.0 | 34.0 | 1.4 | 32.0 | 36.0 | 32.8 | 3.2 | 16.0 | 39.0 | 80.5 |
| SDNPPD | count | 12.7 | 0.8 | 10.7 | 13.6 | 7.6 | 2.4 | 3.0 | 11.0 | 4.1 | 1.8 | 1.0 | 9.3 | 5.7 |
| SDC | - | Brown | Yellow | Brown: Yellow | - | |||||||||
| SDCBM | - | Present | Absent | Absent: Present | - | |||||||||
The measured traits in the F2 population showed nearly a normal distribution (Table 3). PDT, SD100WT, SDL, SDW, SDT, PDW, STT, LFPW, LFML, LFMW, TBWT, STL10 and PDDM showed moderate to high heritability (>60%), whereas LFPL, TBW, TBL and FLD showed medium to low heritability (<60%). PDL, NTB and SDNPPD showed heritability lower than 30%.
There was significant and positive correlation (P<0.05) between related traits in F2 population, such as between seed-related traits (SD100WT, SDL, SDW and SDT), between pod width and seed-related traits (SD100WT, SDL, SDW and SDT), between SDNPPD and PDL, between LFPW and LFMW, between TBW and TBWT (S2 Table). Negative correlation was found between TBW, TBWT and SDNPPD, STL10 and LFML and NTB, TBL and FLD (S2 Table). In general, correlation between related traits was moderate to high (>0.50), while correlation between non-related traits was low or none.
QTLs for domestication-related traits
The results of the QTL analysis for each trait in F2 population are shown in Table 4. Out of 22 traits, ICIM did not find any QTL for four traits. Among those 18 traits which QTLs were identified, six traits each had only one QTL.
Table 4. Domestication-related QTLs detected in the F2 population from the cross between wild (AusTRCF66514) and cultivated (JP235863) V. vexillata.
| Traita | QTL name | LGb | Positionc | Flanking markers | LOD | PVEd(%) | Adde | Domf |
|---|---|---|---|---|---|---|---|---|
| PDT | Not detected | |||||||
| SD100WT | Sd100wt2.1- | 2 | 27 | CEDG297 -CEDG050 | 5.38 | 12.78 | -0.72 | 0.19 |
| Sd100wt7.1- | 7 | 4 | SSR-IAC17 -CEDG298 | 10.46 | 28.19 | -1.04 | 0.17 | |
| SDL | Sdl7.1- | 7 | 8 | VR293 -C_14188_198333 | 7.95 | 32.60 | -0.37 | -0.03 |
| SDW | Sdw7.1- | 7 | 3 | C_14239_255581 -CEDG147a | 6.48 | 24.02 | -0.24 | 0.02 |
| SDT | Sdt2.1- | 2 | 24 | VR079 -C_14168_1088157 | 4.10 | 11.90 | -0.18 | 0.01 |
| Sdt5.1- | 5 | 25 | C_20759_179330 -CEDG027 | 9.87 | 29.52 | -0.29 | 0.16 | |
| Sdt8.1- | 8 | 44 | CEDG270 -C_17996_352817 | 3.62 | 9.84 | -0.26 | -0.16 | |
| PDL | Pdl4.1+ | 4 | 27 | SDC -SDCBM | 3.84 | 7.66 | 2.66 | 8.53 |
| Pdl4.2+ | 4 | 48 | CEDC055 -CEDG185 | 9.89 | 20.65 | 2.65 | -14.36 | |
| Pdl5.1+ | 5 | 24 | VES0074 -C_20759_179330 | 4.97 | 9.32 | 1.86 | -10.80 | |
| PDW | Pdw1.1- | 1 | 73 | VES0028-VR276 | 4.04 | 12.71 | -0.19 | -0.01 |
| Pdw2.1- | 2 | 44 | C_14245_70156 -C_13971_317598 | 5.70 | 18.11 | -0.22 | -0.09 | |
| Pdw7.1- | 7 | 3 | C_14239_255581—CEDG147a | 5.13 | 16.28 | -0.21 | 0.03 | |
| STT | Stt7.1- | 7 | 16 | C_21800_94845—C_14963_135079 | 3.44 | 11.25 | -0.30 | -0.08 |
| LFPL | Lfpl8.1- | 8 | 47 | VES0502—CEDG257 | 3.84 | 17.70 | -1.73 | 3.69 |
| LFPW | Lfpw1.1- | 1 | 13 | C_22838_2078041—CEDC027 | 3.65 | 5.90 | -1.55 | -0.52 |
| Lfpw3.1- | 3 | 17 | CEDAAG004—VES1221 | 11.01 | 19.58 | -2.91 | -0.56 | |
| Lfpw5.1- | 5 | 63 | VES0151—CLM808 | 17.37 | 34.03 | -3.47 | -1.11 | |
| LFML | Lfml5.1+ | 5 | 62 | VES0151—CLM808 | 5.49 | 17.19 | 0.53 | 0.14 |
| LFMW | Lfmw3.1- | 3 | 22 | VES0287—C_15258_77506 | 9.88 | 12.72 | -0.45 | 0.18 |
| Lfmw5.1- | 5 | 61 | VES0151—CLM808 | 29.37 | 52.16 | -0.84 | -0.37 | |
| NTB | Not detected | |||||||
| TBW | Tbw1.1- | 1 | 43 | CEDG288—cp07770 | 4.89 | 11.14 | -2.59 | -1.55 |
| Tbw2.1+ | 2 | 67 | C_19917_55403—C_17900_41805 | 3.97 | 9.02 | 2.64 | -0.45 | |
| Tbw3.1- | 3 | 28 | VES0070—C_13980_72787 | 3.61 | 8.30 | -2.64 | -0.58 | |
| Tbw4.1+ | 4 | 3 | cp00800b—C_19884_262390 | 4.00 | 8.88 | 1.00 | 3.60 | |
| Tbw8.1- | 8 | 67 | cp09551—C_13865_516367 | 6.84 | 16.23 | -3.53 | 1.00 | |
| TBL | Not detected | |||||||
| TBWT | Tbwt2.1+ | 2 | 59 | SSR-IAC55—C_20782_361679 | 4.91 | 9.78 | 11.90 | 5.83 |
| Tbwt4.1+ | 4 | 4 | C_19884_262390—C_13828_166928 | 5.72 | 11.22 | 7.37 | 17.38 | |
| Tbwt8.1- | 8 | 57 | C_27072_91617—CEDG071 | 6.98 | 14.66 | -16.65 | 4.27 | |
| STL10 | Not detected | |||||||
| FLD | Fld1.1- | 1 | 21 | CEDG159b—C_14897_42266 | 5.72 | 17.37 | -4.25 | -4.17 |
| PDDM | Pddm2.1- | 2 | 69 | C_17900_41805—C_13837_228097 | 4.72 | 18.92 | -1.71 | 0.70 |
| Pddm2.2+ | 2 | 105 | DMBSSR143—CEDC050 | 3.86 | 15.79 | 0.22 | -2.36 | |
| SDNPPD | Sdnppd 5.1- | 5 | 29 | C_15565_2997—C_23484_701277 | 4.93 | 8.63 | -0.10 | -1.18 |
| Sdnppd7.1+ | 7 | 3 | C_14239_255581—CEDG147a | 11.59 | 25.14 | 0.04 | -2.00 | |
| Sdnppd7.2- | 7 | 47 | C_14010_83045—Van07-4 | 6.52 | 11.89 | -0.25 | 1.33 | |
| Sdnppd8.1+ | 8 | 46 | C_17996_352817—VES0206 | 4.91 | 8.25 | 0.29 | -0.94 | |
| Sdnppd9.1+ | 9 | 25 | C_21498_175788—C_14534_147594 | 3.89 | 6.37 | 0.26 | 0.92 |
aTrait abbreviation is described in Table 1,
b Linkage group,
c Position of QTL on LG,
d Percentage of variance explained by the QTL,
eAdditive effect and
f Dominant effect
Pod dehiscence (PDT)
Both cultivated and wild parents showed small difference in number of twists along the pod (Table 3). In the F2 population, there was little variation of the trait. As a result, no QTL was detected for this trait.
Increase in organ size
Seed size, pod size, stem thickness, leaf size and tuber-related traits were considered.
Seed size (SD100WT, SDL, SDW and SDT)
One to three QTLs for traits related to seed size were located on LGs 2, 5, 7 and 8. At all QTLs detected, alleles from the cultivated parent increase the seed size. The QTLs with the largest phenotypic effects for all four traits were located on LG7 (28.19%, 32.60% and 24.02% for seed weight, seed length and seed width, respectively).
Pod size (PDL and PDW)
The cultivated zombi pea had larger pod size than the wild zombi pea. Three QTLs for each PDL and PDW traits were detected on LGs 1, 2, 4, 5 and 7. The QTLs with the highest contributions to these traits (PVE = 20.65% and 18.11%) were found on LGs 4 and 2. QTLs for both PDL and PDW on LGs 5 and 7 were located close to QTLs for seed size traits (Fig 4).
Fig 4. QTLs detected for domestication-related traits in V. vexillata F2 population of the cross between wild (AusTRCF66514) and cultivated (JP235863) accessions.
The effect of the cultivated parent is indicated after each name. Explanation of trait abbreviation is shown in Table 1. QTL names in blue color are QTLs that are also identified on same linkage group(s) of other Vigna species. Bold QTLs names are common QTLs between/among species.
Stem thickness (STT)
Only one minor QTL (PVE = 11.25%) was detected on LG7 for stem thickness which was influenced by alleles from the cultivated parent that has thicker stem than the wild zombi pea.
Leaf size (LFPL, LFPW, LFML and LFMW)
Only one QTL was detected on LG8 for LFPL. Three QTLs for LFPW were detected on LGs 1, 3 and 5. One QTL was detected on LG5 for LFML. Two QTLs for LFMW were detected on LGs 3 and 5. The QTLs on LG5 for LFPW, LFML and LFMW showed highest PVE and were located very close to each other and likely to be the same locus. Similarly, the QTLs located on LG3 for LFPW and LFMW were located not far from each other. Alleles from the cultivated zombi pea increased leaf width, while alleles from the wild zombi pea increased leaf length.
Tuber traits (NTB, TBW, TBL and TBWT)
No QTL was detected for NTB and TBL. These two traits showed small trait variation and/or low heritability (27.7% for NTB) (Table 3). For TBW, five QTLs were identified on LGs 1, 2, 3, 4 and 8 in which the QTL on the LG8 (Tbw8.1-) had the largest effect (PVE = 16.23%, Table 4). Cultivated alleles on LGs 1, 3, 8 increased TBW. On the contrary, wild alleles on LG 2 (Tbw2.1+) and LG 4 (Tbw4.1+) increased TBW. In case of TBWT, three QTLs were detected on LGs 2, 4 and 8. Again, QTL on the LG8 (Tbwt8.1-) showed the highest PVE (14.66%). Cultivated allele on LG8 increased TBWT. On the contrary, wild alleles on LG 2 (Tbwt2.1+) and LG 4 (Tbwt4.1+) increased TBWT. QTLs for tuber width and tuber weight having phenotypically reverse effect (Tbw2.1+ vs. Tbwt2.1+, and Tbw4.1+ vs. Tbwt4.1+) were co-localized (Fig 4).
Plant type
Stem length was considered.
Stem length (STL10)
No QTL identified for this trait, although the trait showed high heritability (83.4%).
Earliness
Days to first flowering and days to first maturity were considered.
Days to first flowering (FLD)
The cultivated and the wild parents were very different in FLD (102.6 and 72.4 days, respectively). The F2 population showed a high variation for the trait (Table 3). However, only one QTL was detected for this trait. The QTL was on LG1 and accounted for only 17.37% of the trait variation. The alleles from the cultivated parent prolong FLD.
Days to first maturity (PDDM)
Similar to FLD, the cultivated and the wild parents were different in PDDM (34.0 and 27.8 days, respectively). Two QTLs on LG2, Pddm2.1- and Pddm2.2+, were identified for the trait. These QTLs showed similar PVE, 18.92% and 15.79%, respectively. The alleles from the cultivated parent at Pddm2.1- prolong PDDM, while that at Pddm2.2+ hasten PDDM.
Yield potential
Or Number of seed per pod (SDDNPPD). The cultivated parent produced lower SDDNPPD than the wild parent. Five QTLs for SDDNPPD were detected LGs 5, 7, 8 and 9. Two QTLs on LG7 had high effects (PVE = 25.14% and 11.89%). At QTLs Sddnppd5.1- and Sddnppd7.2-, the allele from the cultivated parents increased SDDNPPD, while at the other three QTLs the alleles from the wild parent increased SDDNPPD.
Pigmentation
Seed coat color (SDC) and black mottle on seed coat (SDCBM)
Seed coat color (SDC)
SDC was characterized as a qualitative trait. The cultivated parent had yellow seed coat but wild parent had brown seed coat color. F2 plants segregated for this trait at ratio of 81 (brown) to 24 (yellow), fitting a 3:1 ratio (χ2 = 0.26, P = 0.61). This indicated that SDC is controlled by a single locus, named as Sdc. This locus was mapped as a morphological marker onto LG4 (Fig 2).
Black mottle on seed coat (SDCBM)
SDCBM was also characterized as a qualitative trait. The cultivated Bali had no black mottle, while the wild parent had black mottle on seed coat. F2 progenies segregated for seed coat color at ratio of 76 (black mottle presence) to 29 (black mottle absence), fitting a 3:1 ratio (χ2 = 0.38, P = 0.54). This indicated that SDCBM is controlled by a single locus. The resulted indicated that the presence of SDCBM is controlled by a single dominant locus, named as Sdcbm. Sdcbm was mapped next to the locus Sdc at marker interval VR045 and C_13986_405837 (Fig 2).
Distribution of domestication-related traits
QTLs with large effect (PVE > 20%) were found on four linkage groups, viz. LGs 4, 5 and 7 (Table 4).
LG4
Large-effect QTLs for pod length (Pdl4.2+) was on this linkage group. In addition, seed coat color (SDC) and black mottle (SDCBM) were mapped as morphological makers onto this linkage group.
LG5
Three large-effect QTLs were located to this linkage group; Sdt5.1- for seed thickness and Lfpw5.1- and Lfmw5.1- for leaf width. Lfpw5.1- and Lfmw5.1- with PVE = 34.03% and PVE = 52.16%, respectively were located closed together between markers VES0151 and CLM808. These two QTLs were likely the same locus. In addition, QTLs Sdt5.1-, Pdl5.1+, and Sdnppd5.1- were clustered together on this linkage group (Fig 4).
LG7
Most of large-effect QTLs for seed-related traits (Sd100wt7.1-, Sdl7.1- and Sdw7.1-) were located on this linkage group. These QTLs were clustered with QTLs for pod width (Pdw7.1-), number of seeds per pod (Sdnppd7.1+) and stem thickness (Stt7.1-). In addition, another QTL for number of seeds per pod (Sdnppd7.2-) was also located in this linkage group.
Comparison of QTLs for domestication in V. vexillata with those of other Vigna species
The QTLs for domestication-related traits for zombi pea identified in this study were compared with those for yardlong bean [20], azuki bean [13], mungbean [16], and rice bean [17] based on common SSR markers (Fig 4). The comparison showed that most of the QTLs detected for zombi pea were also found in other Vigna species; they were mapped to the same linkage groups. For example, most of the QTLs for seed-related traits with high PVE in zombi pea were mapped onto LG7 in which several QTLs for seed-related traits were detected for yardlong bean, azuki bean, mungbean and rice bean. Three QTLs, Pddm2.1-, Sdt8.1+ and Sdnppd9.1- were mapped to similar regions with other Vigna species. Nonetheless, none or a few of QTLs for seed-related traits were detected on LG6 and LG10 of zombi pea which is the same case as in other Vigna species except yardlong bean.
Discussion
Fertility of progenies of cultivated zombi pea from Bali x wild zombi pea
A previous study on reproductive compatibility among cultivated zombi pea from Bali (tuber type zombi pea),cultivated zombi pea from Africa (seed type zombi pea), and wild zombi pea from Africa and Australia revealed high compatibility between the African cultivated zombi pea (seed type zombi pea) and the wild form from both Africa and Australia, but revealed various pre- and post-zygotic incompatibility between the Bali cultivated zombi pea (tuber type zombi pea) and the wild form from Africa and Australia [8]. In our present study, we successfully obtained a fertile F2 population for the first time from a cross between the cultivated zombi pea from Bali and Indian wild zombi pea using the former as female parent. This suggested that the Bali cultivated zombi pea constitutes a primary gene pool with the Indian wild zombi pea. The difference between result in our study and that in Damayanti et al. [8] is possibly due to environmental factors. Damayanti et al. [8] noted that even self-pollination of the cultivated zombi pea from Bali results in low pod setting. In our study, when we conducted the hybridization during November 2014 to February 2015, the Bali cultivated zombi pea showed very low pod setting (<3%). When it was grown again during November 2015 to February 2016 and during November 2016 to February 2017, it set more flowers and pods (~15% and ~70%, respectively; Somta and Dachapak, personal observation). In addition to environmental factors, genetic relatedness of the parents appeared to affect the success of hybridization. The wild zombi pea (JP235863 from India) and the Bali cultivated zombi pea were genetically closely related [4], while the wild zombi pea used in the study of Damayanti et al. [8] were from Africa and Australia and they were genetically highly differentiated from the Bali cultivated zombi pea [4].
Transferability and polymorphism of SSR markers
Although as high as 1,876 SSR primers pairs from various Vigna species were screened for polymorphism between the cultivated and wild parents, only 36.6% of them were amplifiable and only 10.7% of them were polymorphic. The low polymorphism of SSR markers was also observed in the mapping parents of V. vexillata used by Marubodee et al. [33] in which only 6.2% of 1,336 SSR markers were polymorphic. In addition, Dachapak et al. [4] reported that only 21.2% of 1,024 SSR markers screened in 6 accessions of V. vexillata from Asia, Africa, Australia and America were polymorphic. However, the percentage of amplifiable SSR markers in Marubodee et al. [33] and Dachapak et al. [4] (65.4% and 58.1%, respectively) was higher than in our study (36.6%). It is worth noting that almost all of the markers used by Marubodee et al. [33] and Dachapak et al. [4] were screened in our study. Nonetheless, these results indicate moderate transferability but low polymorphism of SSR markers from other Vigna species in V. vexillata. High transferability rate of SSRs (>80%) among several other Vigna species have been reported earlier [19,25,29,34,42]. The low transferability rate of SSRs from other Vigna species to V. vexillata is likely due to the fact that those Vigna species and V. vexillata are genetically highly differentiated [43]. Due to the low transferability and polymorphism of SSR markers in V. vexillata, other types of markers (RAD-seq in this study) should be used for genome analysis of V. vexillata.
Linkage map construction and orthologous blocks in zombi pea
Previously, there was only two genetic linkage maps developed for zombi pea [33,44]. The first map contained 14 linkage groups with all were dominant markers (70 RAPD and 47 AFLP) except one co-dominant SSR marker. The map reported by Marubodee et al. [33] comprised 11 linkage groups from 84 SSR and 475 RAD-seq markers. The number of linkage map of zombi pea constructed in our study comprised 11 linkage groups of 145 SSR markers and 117 RAD-seq markers. The number of linkage groups of the maps developed by Marubodee et al. [33] and by this present study corresponded with the chromosome number of V. vexillata. (n = 11).
Previous comparative genome studies in the genus Vigna by means of comparison of linkage maps revealed high genome conservation among the species [17,19,25,42,45]. We compared our V. vexillata linkage map with other Vigna linkage maps using common SSR markers (Fig 3). We found that middle to lower part LG5 of our V. vexillata linkage map was very likely an orthologous block from LG4 and that LG7 of our V. vexillata linkage map was orthologous blocks from LG4 and LG10. In addition, a part of the LG7 appeared to be duplication of LG2 or vice versa (Fig 3). Intraspecific macro-orthologous block rearrangement has been demonstrated in azuki bean by means of comparative linkage mapping and fluorescence in situ hybridization [46,47]. The rearrangement of orthologous block was reciprocal where LG4 and LG6 were intermingled and was found in many accessions of wild azuki bean and it possibly affected fitness of those wild accessions to some environments and seed size [47].
Segregation distortion
Segregation distortion is a normal phenomenon in interspecific hybridization and in even intraspecific hybridization between two highly genetically different genotypes such as between wild and cultivated form of the same species. Segregation distortion of DNA markers has been reported for intraspecific hybridization of several Vigna species including V. radiata [16], V. angularis [41,46], V. umbellata [17], V. mungo [42] and V. unguiculata [19,20]. The percentage of marker distortion in these reports ranged from 3.9% in V. angularis to 48.7% in V. unguiculata. In this study, as high as 48.5% of the markers showed segregation distortion, the value is considered very high as compared to that other reports in Vigna species. The distorted markers were clustered on LGs 5, 6, 7, 8, 10 and 11 (Fig 2). The presence of a gene(s) controlling sterility and/or compatibility may cause segregation distortion of nearby loci and clustering of distorted markers [48]. The highly distorted markers (P< 0.001) were on LGs 5, 7 and 8. The distortions found on LG5 and LG7 possibly stem from the chromosomal rearrangement in these two linkage groups (Fig 2; see also above discussion on orthologous block). The cause of the distortion on LG8 is not known, however, it is possible that this linkage group represents the most genome divergence between the wild and cultivated parents used in this study. It is worth noting that major QTLs for tuber-related traits (Tbw8.1- and Tbwt8.1-) were located on the LG8 (Fig 4 and Table 4). Tuber root trait in the cultivated V. vexillata from Bali used in this study is the most strikingly different adaptive/domestication trait distinguishing the cultivated V. vexillata Bali from other forms of V. vexillata. In yardlong bean (V. unguiculata), major QTLs for pod length and other pod-related traits (the most important domestication trait for this crop) and seed traits were found on LG7 where highly segregation distortions existed [19]. Similarly, in mungbean, major QTLs for seed dormancy, seed productivity and day length sensitivity were located on LG4 that showed high level of segregation distortion [16]. Most of markers mapped on LG11 of intra- and inter-specific crosses of Vigna species always showed segregation distortion [17,19,45]. This is also the case in our study where 58.8% of the markers mapped to LG11 showed segregation distortion (Fig 2). Therefore, LG11 appeared to play an important role in genetic differentiation within and among Vigna species.
Although pod length (PDL) and number of seed per pod (SDNPPD) were highly correlated (r = 0.83) (S2 Table), only one of the QTLs for these two traits were co-located (Table 4 and Fig 4). Since three of the five QTLs, including the largest effect QTL, detected for SDNPPD were mapped to genome regions that showed high segregation distortion, those QTLs may be associated with genetic compatibility and fertility, and thus seed production. In Vigna species, early generation (F1 and F2) hybrid progenies derived from hybridization of distantly-related genotypes always possesses pods with incomplete filled (empty seeds in some locules) [49,50]
QTLs for tuber traits in zombi pea
Most of tubers crops are domesticated from non-legume species such as potato, sweet potato, yam and cassava. In Vigna species, V. vexillata, V. lobatifolia and V. marina produced tuber roots [51]. Tubers produced by legume crops have much more nutrition than non-legume crops especially nitrogen or protein due to nitrogen fixation ability. The protein content in tuber roots of V. vexillata is about 15% which is five-fold higher than that in sweet potato. In potato, tuber weight was controlled by a few QTLs with PVE of 20% or less [52]. Similarly, only three QTLs were identified for tuber weight (TBWT) in V. vexillata with largest PVE of about 15%. Tuber weight of potato has been shown to be negatively correlated with leaf area (leaf size) [53]. On the contrary, cultivated alleles that increase tuber width (Tbw3.1-), primary leaf width (Lfpw3.1-) and maximum leaflet width (Lfmw3.1-) were detected at similar position on LG3, which suggested preiotropic effect of the same gene (Table 4 and Fig 4). It should be noted that alleles from wild V. vexillata increased the tuber weight (Tbwt2.1+, Tbwt4.1+) and tuber width (Tbw2.1+, Tbw4.1+), in spite of the fact that wild V. vexillata has smaller tubers (Table 4 and Fig 1). Map positions of these QTLs are close and therefore tuber weight and tuber width increase by wild allele could be preiotropic effect of the same gene. These reverse effect alleles of wild V. vexillata may account for transgressive segregation found in the F2 population (S1 Fig). In fact, there is a F2 plant (F2-18) with 141–160 g range tuber weight, while tuber weight of cultivated parent is 61–80 g range (S1 Fig). Although QTL could not be detected, tuber number increase by wild Indian parent also contribute tuber weight increase. Therefore, these alleles from the wild V. vexillata will be useful for improvement of tuber yield of the Bali cultivated V. vexillata.
Genomic region and distribution of QTLs for other domestication traits
Many domestication-related traits were studied in Vigna crops including mungbean [16], yardlong bean [19,20], azuki bean [13] and rice bean [17]. In general, these studies found that (i) domestication-related traits were controlled by one or two major QTLs together with some minor QTLs in a narrow genome region, (ii) major QTLs for different traits were clustered and (iii) QTL clusters were not randomly distributed along linkage groups. For examples, major QTLs for seed size, pod size, and seed dormancy were clustered on LG4 of mungbean, and major QTLs for pod length, seed size, pod shattering, leaf size, and stem thickness were clustered on LG7 of yardlong bean. In this study, we did not find clusters of major QTLs for different traits in zombi pea (Fig 4 and Table 4).
Comparison of domestication QTLs between V. vexillata and yardlong bean
Since V. vexillata and is genetically closely related with cowpea/yardlong bean (V. unguiculata) and both of them originate from African, we compared domestication-related QTLs for V. vexillata found in this study with those for yardlong bean reported by [19,20]. Only QTLs having phenotypic effect more than 20% were considered (Table 5).
Table 5. Comparison of major QTLs (PVE > 20%) detected for domestication in F2 population (AusTRCF66514 x JP235863) of Vigna vexillata with the major QTLs (PVE > 20%) detected for domestication in other Vigna species.
| Trait | Zombi pea | Yardlong bean | Azuki bean | Rice bean | Mungbean | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| QTL | PVE | QTL | PVE | QTL | PVE | QTL | PVE | QTL | PVE | |
| SD100WT | Sd100wt7.1 - | 28.2 | Sd100wt7.1+ | 24.6 | Sd100wt2.1.1 + | 27.9 | Sd100wt4.4.1 + | 33.4 | Sd100wt5.8.1+ | 22.2 |
| Sd100wt1.2.1 + | 26.8 | |||||||||
| Sd100wt2.2.1 + | 31.3 | |||||||||
| Sd100wt2.9.1 + | 25.2 | |||||||||
| SDL | Sdl7.1 - | 32.6 | Sdl1.1 + | 20.8 | Sdl2.1.1+ | 31.1 | Sdl4.4.1 + | 28.9 | Sdl5.8.1+ | 20.4 |
| Sdl7.1 + | 26.5 | Sdl1.2.1+ | 24.5 | |||||||
| Sdl2.3.1+ | 20.2 | |||||||||
| SDW | Sdw7.1 - | 24.0 | Sdw7.1+ | 20.9 | Sdw1.2.1+ | 21.2 | Sdw4.4.1 + | 26.3 | dl | dl |
| Sdw2.9.1+ | 21.2 | |||||||||
| SDT | Sdt5.1 - | 29.5 | Sdt3.1 + | 26.4 | Sdt1.2.1 + | 26.0 | Sdt4.4.1 + | 25.2 | Sdt5.8.1+ | 22.7 |
| Sdt2.9.1 + | 23.1 | |||||||||
| PDL | Pdl4.2 + | 20.7 | Pdl7.1 + | 31.0 | Pdl1.7.1+ | 39.1 | Pdl4.4.1 + | 23.1 | Pdl5.7.1+ | 20.5 |
| Pdl1.8.1+ | 22.1 | |||||||||
| LFPW | Lfpw5.1 - | 34.0 | dl | dl | dl | dl | ni | ni | dl | dl |
| LFMW | Lfmw5.1- | 52.2 | ni | ni | dl | dl | Lfmw4.4.1+ | 25.6 | ni | ni |
| SDNPPD | Sdnppd7.1 + | 25.1 | Sdnppd11.1 + | 70.1 | ni | ni | dl | dl | dl | dl |
Note dl = detected but PVE lower than 20% and ni = not investigated
The cultivated zombi pea used in this study and yardlong bean are believed to have been domesticated in Asia, although their origin of species is believed to be in Africa [4,19]. Major QTLs of yardlong bean were detected on four linkage groups (LGs 1, 3, 7 and 11), while large-effect QTLs of zombi pea were found on three linkage groups (LGs 4, 5 and 7). Although some major QTLs were located on LG7 of both species, distributions of QTLs on this linkage group were different.
Seed weight
The 100-seed weight of the cultivated zombi pea was 7.3 g, while the wild zombi pea was only 2.1 g. Two QTLs for this trait were located on LG2 and LG7 (the largest effect (PVE = 28.19%) was on LG7). In yardlong bean, ten QTLs of 100-seed weight were detected on nine linkage groups with the highest QTL effect on LG7 (24.6%). Although the majors QTL for seed weight of zombi pea and yardlong bean were both located on the LG7, they appeared to be different because the QTL region in zombi pea was an orthologous block from LG10 as compared to yardlong bean (Fig 3).
Pod size
Pod size of the cultivated zombi pea is not much longer and thicker than that of the wild zombi pea (94.3 vs. 85.6 mm. for length and 5.7 vs. 3.6 mm. for width, respectively). Six QTLs for pod size in zombi pea was found on LGs 1, 2, 4, 5 and 7 with the largest effect QTL (PVE = 20.7%) on LG4. Seventeen QTLs for pod size in yardlong bean was found on every LGs except LG10 with the largest effect QTL on LG7 for both PDL and PDW (PVE = 31.0% and 31.2%, respectively). One of the QTL for pod size on LG4 might be shared between zombi pea and yardlong bean (Fig 4) was found between markers CEDC055 and CEDG185.
Leaf size
Both primary and mature leaves of the cultivated zombi pea were larger than the wild zombi pea. Seven QTLs were detected for leaf size with highest effect on LG5 (Lfpw5.1- with PVE = 34.03% and Lfmw5.1- with PVE = 52.16%). In yardlong bean, thirteen QTLs for primary leaf were detected on LG1, LG2, LG3, LG6, LG7, LG8, LG9 and LG11, but only one of them (Lfpl7.1+) with highest PVE (20.9%) was found on LG7.
Yield potential
Number of seeds per pod (SDNPPD) of the cultivated zombi pea was lower than that of the wild zombi pea (7.6 and 12.7, respectively). Five QTLs on LG5, LG7, LG8 and LG9 were detected with QTL showing highest PVE on LG7 (25.14%). In yardlong bean, two QTLs were detected on LGs 7 and 11. The QTL on LG11 showed highest PVE (70.1%).
Comparison with azuki bean, mungbean and rice bean
Eight major QTLs (>20% PVE) of SD100WT, SDL, SDW, SDT, PDL, LFPW, LFMW and SDNPPD in zombi pea were compared with domestication-related QTLs from azuki bean [13], mungbean [16] and rice bean [17].
Seed size and weight
Seven QTLs of seed size and weight were detected for zombi pea in which QTLs with high effect were on LG7 (Sd100wt7.1-, Sdl7.1- and Sdw7.1-). In azuki bean, rice bean and mungbean, large-effect QTLs for seed size and weight were co-located on the same region; LGs 1, 2, 3 and 9 for azuki bean, LG4 for rice bean, and LG8 for mungbean. This suggested that major genes controlling seed size with different biological mechanisms exist among domesticated Vigna species.
Pod size
Among the QTLs for pod length and pod width in zombi pea, the QTL Pdl4.2+ on LG4 showed the highest PVE (20.65%). QTL with the highest effect (PVE = 23.1%) for pod length in rice bean was also detected on LG4, while the QTL with the highest effect for pod length in azuki bean and mungbean were on LG7.
Leaf size
Among seven QTLs for leaf size in zombi pea, two QTLs (Lfpw5.1- and Lfmw5.1-) showed PVE higher than 20%; both of which were on LG5. In rice bean, a major QTL for leaf size was located on LG4. No QTL was detected for leaf size in azuki bean and mungbean due to small difference in the mapping parents.
Yield potential
Five QTLs for SDNPPD in zombi pea were located on LGs 5, 7, 8 and 9 with the QTL on LG7 showed highest PVE (25.14%), while three QTLs for SDNPPD in rice bean were detected on LG4 and LG9, and two QTLs for SDNPPD in mungbean were identified on LG1 and LG9. The QTLs on LG7 of zombi pea, rice bean, and mungbean were identified in a similar location and may be the same locus. However, all the QTLs identified in rice bean and mungbean possessed PVE lower than 20%.
Pigmentation
Seed coat color and presence of black mottle on seed coat were treated as morphological markers which were mapped next to each other on the LG4. In azuki bean and mungbean, gene controlling the presence of black mottle on seed coat was also mapped on LG4. This indicated that the gene controlling this trait is highly conserved among species in the genus Vigna.
Conclusions
An F2 of V. vexillata population derived from a partially fertile hybrid between tuber-root-type cultivated form and wild type form were successfully developed. A genetic linkage map was constructed for this population utilizing 145 SSR, 117 RAD-seq and 2 morphological markers and to locate QTL for domestication syndrome traits. In total, 37 QTLs were detected for 18 domestication traits. Eight QTLs with large effect (>20%) were located on 4 out of 11 linkage groups. Domestication traits including seed size, pod size, leaf size, yield potential and seed pigmentation were controlled by one or two major QTLs and a few minor QTLs. QTLs for tubers were found on five different linkage group. Zombi pea had highest number of shared common QTLs with azuki bean, followed by yardlong bean, rice bean and mungbean. QTLs for seed size (SD100WT, SDL, SDW and SDT) were clustered together on different linkage groups of each species, suggesting specific and different seed size increase genes were used independently for each species. This study provides a genetic map and information for marker-assisted selection in zombi pea breeding. QTL analysis revealed potential of wild genetic resources to improve tuber size and yield. The comparative genomic analysis provides more understanding on genome evolution of Vigna species.
Supporting information
(TIFF)
(XLSX)
(XLSX)
Acknowledgments
This research was financially supported by Royal Golden Jubilee PhD programme (RGJ) co-funded by the Thailand Research Fund (TRF) and Kasetsart University.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was financially supported by Royal Golden Jubilee PhD programme (RGJ) co-funded by the Thailand Research Fund (TRF) and Kasetsart University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tomooka N, Kaga A, Isemura T, Vaughan DA, Srinives P, Somta P, et al. Vigna genetic resources. Proceeding of the 14th NIAS international workshop on Genetic Resources, Genetic and Comparative Genomics of Legumes (Glycine and Vigna); 2010. pp. 11–21. [Google Scholar]
- 2.Tomooka N, Naito K, Kaga A, Saksai H, Isemura T, Ogiso-Tanaka E, et al. Evolution, domestication and neo-domestication of the genus Vigna. Plant Genet Resour 2014; 12(1): 168–171. [Google Scholar]
- 3.Ferguson H. The food crops of the Sudan and their relation to environment. Proceeding of Food and Society in the Sudan, Philosophical Society of the Sudan Khartoum, McCorquodale and Co. (Sudan) Ltd; 1954.
- 4.Dachapak S, Somta P, Poonchaivilaisak S, Yimram T, Srinives P. Genetic diversity and structure of the zombi pea (Vigna vexillata (L.) A. Rich) gene pool based on SSR marker analysis. Genetica 2017; 145: 189–200. 10.1007/s10709-017-9957-y [DOI] [PubMed] [Google Scholar]
- 5.Karuniawan A, Iswandi A, Kale PR, Heinzemann J, Grüneberg WJ. Vigna vexillata (L.) A. Rich. cultivated as a root crop in Bali and Timor. Genet Resour Crop Evol 2006; 53(1): 213–217. [Google Scholar]
- 6.Bhattacharyya PK, Ghosh AK, Sanyal B, Ray GD. Grow Vigna vexillata for protein rich tuber cum pulse crop in the northern-eastern hill region. Seed Farms 1984; 10: 33–36. [Google Scholar]
- 7.Asati BS, Yadav DS. Diversity of horticultural crop in north eastern region. ENVIS Bull Himal Ecol 2004; 12: 1–11. [Google Scholar]
- 8.Damayanti F, Lawn RJ, Bielig LM. Genetic compatibility among domesticated and wild accessions of the tropical tuberous legume Vigna vexillata (L.) A. Rich. Crop Pasture Sci 2010; 61: 785–797. [Google Scholar]
- 9.Hammer K. Das Domestikations syndrom. Genet Res Crop Evol 1984; 32:11–34. [Google Scholar]
- 10.Gepts P. Domestication as a long-term selection experiment. Plant Breed Rev 2004; 24(2): 1–44. [Google Scholar]
- 11.Lush WM, Evans LT. The domestication and improvement of cowpea (Vigna unguiculata (L.) Walp.). Euphytica 1981; 30:579–587. [Google Scholar]
- 12.Koinange EMK, Singh SP,Gepts P. Genetic control of the domestication syndrome in common bean. Crop Sci 1996; 36: 1037–1045. [Google Scholar]
- 13.Isemura T, Kaga A, Konishi S, Ando T, Tomooka N, Han OK, et al. Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and their comparison with other warm season legumes. Ann Bot 2007; 100: 1053–1071. 10.1093/aob/mcm155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funatsuki H, Suzuki M, Hirose A, Inaba H, Yamada T, Hajika M, et al. Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc Natl Acad Sci U.S.A. 2014; 111(50): 17797–17802. 10.1073/pnas.1417282111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaga A, Isemura T, Tomooka N, Vaughan DA. The genetics of domestication of the azuki bean (Vigna angularis). Genetics 2008; 178: 1013–1036. 10.1534/genetics.107.078451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isemura T, Kaga A, Tabata S, Somta P, Srinives P, Shimizu T, et al. Construct of genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS One 2012; 7(8): e41304 10.1371/journal.pone.0041304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isemura T, Kaga A, Tomooka N, Shimizu T, Vaughan DA. The genetics of domestication of rice bean, Vigna umbellata. Ann Bot 2010; 106: 927–944. 10.1093/aob/mcq188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andargie M, Pasquet RS, Muluvi GM, Timko MP. Quantitative trait loci analysis of flowering time related traits identified in recombinant inbred lines of cowpea (Vigna unguiculata). Genome 2013; 56(5): 289–294. 10.1139/gen-2013-0028 [DOI] [PubMed] [Google Scholar]
- 19.Kongjaimun A, Kaga A, Tomooka N, Somta P, Shimizu T, Shu Y, et al. An SSR-based linkage map of yardlong bean (Vigna unguiculata (L.) Walp. subsp. unguiculata Sesquipedalis group) and QTL analysis of pod length. Genome 2012a; 55(2): 81–92. [DOI] [PubMed] [Google Scholar]
- 20.Kongjaimun A, Kaga A, Tomooka N, Somta P, Vaughan DA, Srinives P. The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis. Ann Bot 2012b; 109(6): 1185–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo S, Munoz-Amatriain M, Boukar O, Herniter I, Cisse N, Guo YN, et al. Identification of genetic factors controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). bioRxiv 2017; 1–32. 10.1101/202044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garba M, Pasquet RS. The Vigna vexillata (L.) A. Rich. genepool. In: Sorensen M, Estrella JE, Hamann EOJ, Ruiz SAR, editors. Proceedings of the 2nd International Symposium on Tuberous Legumes. 1996 Aug 5–8, Celaya, Guanajuato, Mexico; 1998. pp.61-71.
- 23.Lodhi MA, Ye GN, Weeden NF, Reisch BI. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol Biol Rep 1994; 12:6–13. [Google Scholar]
- 24.Wang XW, Kaga A, Tomooka N, Vaughan DA. The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean [Vigna angularis (Wild.) Ohwi and Ohashi]. Theor Appl Genet 2004; 109: 352–360. 10.1007/s00122-004-1634-8 [DOI] [PubMed] [Google Scholar]
- 25.Chankaew S, Isemura T, Isobe S, Kaga A, Tomooka N, Somta P, et al. Detection of genome donor species of neglected tetraploid crop Vigna reflexo-pilosa (créole bean) and genetic structure of diploid species based on newly developed EST-SSR markers from azuki bean (Vigna angularis). PLoS One 2014; 9(8): e104990.. 10.1371/journal.pone.0104990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li CD, Fatokun CA, Ubi B, Singh BB, Scoles GJ. Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Sci 2001; 41: 189–197. [Google Scholar]
- 27.Somta P, Musch W, Kongsamai B, Chanprame S, Nakasathien S, Toojinda T, et al. New microsatellite markers isolated from mungbean (Vigna radiata (L.) Wilczek). Mol Ecol Resour 2008; 8: 1155–1157. 10.1111/j.1755-0998.2008.02219.x [DOI] [PubMed] [Google Scholar]
- 28.Seehalak W, Somta P, Sommanas W, Srinives P. Microsatellite markers for mungbean developed from sequence database. Mol Ecol Res 2009; 9: 862–864. 10.1111/j.1755-0998.2009.02655.x [DOI] [PubMed] [Google Scholar]
- 29.Tangphatsornruang S, Somta P, Uthaipaisanwong P, Chanprasert J, Sangsrakru D, Seehalak W, et al. Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean (Vigna radiata (L.) Wilczek). BMC Plant Biol 2009; 9: 137 10.1186/1471-2229-9-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu K, Park SJ, Poysa V, Gepts P. Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). J Hered 2000; 91(6): 429–34. [DOI] [PubMed] [Google Scholar]
- 31.Blair MW, Pedraza F, Buendia HF, Gaitan-Solis E, Beebe SE, Gepts P, et al. Development of genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor Appl Genet 2003; 107(8): 1362–1374. 10.1007/s00122-003-1398-6 [DOI] [PubMed] [Google Scholar]
- 32.Hanai LR, de Campos T, Camargo LE, Benchimol LI, de Souza AP, Melotto M, et al. Development, characterization, and comparative analysis of polymorphism at common bean SSR loci isolated from genic and genomic sources. Genome 2007; 50(3): 266–277. 10.1139/g07-007 [DOI] [PubMed] [Google Scholar]
- 33.Marubodee R, Ogiso-Tanaka E, Isemura T, Chankaew S, Kaga A, Nalito K, et al. Construction of an SSR and RAD-marker based molecular linkage map of Vigna vexillata (L.) A. Rich. PLoS One 2015; 10(9): e0138942 10.1371/journal.pone.0138942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somta P, Seehalak W, Srinives P. Development, characterization and cross-species amplification of mungbean (Vigna radiata) genic microsatellite markers. Conserv Genet 2009; 10: 1939–1943. [Google Scholar]
- 35.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo discovery and genotyping in model and non-model species. PLoS One 2012; 7(5): e37135 10.1371/journal.pone.0037135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catchen JM, Aemores A, Hohenlohe P, Cresko W, Postlethwait JH. Stack: building and genotyping loci de novo from short-read sequences. G3 (Bethesda) 2011; 1(3): 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Ooijien JW. 2006. Join Map 4.0, Software for the calculation of genetic linkage maps in experimental populations. Wageningen, Netherlands: Kyazma B. V.
- 38.Kosambi DD. The estimation of map distances from recombination values. Ann Eugenic 1944; 12: 172–175. [Google Scholar]
- 39.Li H, Ye G, Wang J. A modified algorithm for improvement of composite interval mapping. Genetics 2007; 175: 361–374. 10.1534/genetics.106.066811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng L, Li H, Zhang L, Wang J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative locus mapping in biparental populations. Crop J 2015; 3(3): 269–283. [Google Scholar]
- 41.Han OK, Kaga A, Isemura T, Tomooka N, Vaughan DA. A genetic linkage map for azuki bean (Vigna angularis). Theor App Genet 2005; 111: 1278–1287. [DOI] [PubMed] [Google Scholar]
- 42.Chaitieng B, Kaga A, Tomooka N, Isemura T, Huroda Y, Vaughan DA. Development of black gram [Vigna mungo (L.) Hepper] linkage map and its comparison with an azuki bean [Vigna angularis (Wild.) Ohwi and Ohashi] linkage map. Theor Appl Genet 2006; 13(7): 1261–1269. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi Y, Somta P, Muto C, Iseki K, Naito K, Pandiyan M, et al. Novel genetic resources in the genus Vigna unveiled from gene bank accessions. PLoS One 2016; 11(1): e0147568 10.1371/journal.pone.0147568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogundiwin EA, Thottappilly G, Aken’Ova ME, Ekpo EJA, Fatokun CA. Resistance to cowpea mottle carmovirus in Vigna vexillata. Plant Breeding 2002; 121: 517–520. [Google Scholar]
- 45.Somta P, Kaga A, Tomooka N, Kashiwaba K, Isemura T, Chaitieng B, et al. Development of an interspecific Vigna linkage map between Vigna umbellata (Thumb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi and Ohashi and its use in analysis of bruchid resistance and comparative genomics. Plant Breeding 2006; 125(2): 77–84. [Google Scholar]
- 46.Kaga A, Isemura T, Tomooka N, Vaughan DA. The genetics of domestication of the azuki bean (Vigna angularis). Genetics 2008; 178: 1013–1036. 10.1534/genetics.107.078451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Kikuchi S, Muto C, Naito K, Isemura I, Ishimoto M, et al. Reciprocal translocation identified in Vigna angularis dominates the wild population in East Japan. J Plant Res 2015; 128: 653–663. 10.1007/s10265-015-0720-0 [DOI] [PubMed] [Google Scholar]
- 48.Zamir D,Tadmor Y. Unequal segregation of nuclear gene in plants. Bot Gaz 1986; 147:355–358. [Google Scholar]
- 49.Gosal SS, Bajaj YPS. Interspecific hybridization between Vigna mungo and Vigna radiata through embryo culture. Euphytica 1983; 32: 129–l37. [Google Scholar]
- 50.Gupta VP, Plaha P, Rathore PK. Partly fertile interspecific hybrid between a black gram x green gram derivative and an adzuki bean. Plant Breeding 2002; 121: 182–183. [Google Scholar]
- 51.National Research Council. Tropical legumes: resources for the future. The national Academies Press; 1979. [Google Scholar]
- 52.Rak K, Bethke PC, Palta JP. QTL mapping of potato chip color and tuber traits within an autotetraploid family. Mol Breed 2017; 37:15. [Google Scholar]
- 53.Lemaga B, Caesar K. Relationship between number of main stems and yield components of potato (Solanum tuberosum L. cv. Erntestolz) as influenced by different daylengths. Potato Res 1989; 33: 257–267. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.