SUMMARY
T cell differentiation in the thymus proceeds in an ordered sequence of developmental events characterized by variable expression of CD4 and CD8 coreceptors. Here, we report that immature single-positive (ISP) thymocytes are molecularly distinct from all other T cell populations in the thymus in their expression of a gene profile that is dependent on the transcription factor BRD4. Conditional deletion of BRD4 at various stages of thymic differentiation reveals that BRD4 selectively regulates the further differentiation of ISPs by targeting cell cycle and metabolic pathways, but it does not affect the extensive proliferation that results in the generation of ISPs. These studies lead to the conclusion that the ISP subpopulation is not a hybrid transitional state but a molecularly distinct subpopulation that is selectively dependent on BRD4.
In Brief
Thymocytes differentiate from immature DN to ISP, DP, and single-positive thymocytes. Gegonne et al. report the finding that BRD4 is required at the transition from immature ISP to DP thymocytes but not for the differentiation of DN thymocytes or the maturation of conventional single-positive thymocytes from the DP stage.
Graphical Abstract
INTRODUCTION
The generation of T cells in the thymus results from the sequential differentiation of a series of thymocyte precursors (Mingueneau et al., 2013; Rothenberg et al., 2016; Vacchio et al., 2016). The earliest thymic immigrants from the bone marrow do not express any of the markers associated with mature T cells, namely the T cell receptor (TCR) and CD4/CD8 coreceptor molecules, and are called double negatives (DNs). Within the thymus, the DN cells undergo a series of maturation steps punctuated by cycles of proliferation, rearrangement of the TCRβ gene, and intracellular expression of the TCRβ protein. Further differentiation to the immature single-positive (ISP) stage is accompanied by cell surface expression of CD8. The transition of the ISP to the double-positive (DP), CD4+CD8+ thymocytes, requires a single round of cell cycle, leading to surface expression of both CD4 and TCRαβ (Yu et al., 2004). DP thymocytes differentiate into either mature CD4+ or CD8+ single-positive thymocytes that emigrate from the thymus to seed peripheral organs. DP thymocytes also give rise to invariant natural killer T (iNKT) cells; CD4+ single-positive (SP) thymocytes generate Foxp3+ regulatory T cells (Tregs).
An analysis of the transcriptional landscape of T cell differentiation concluded that it was accompanied by gradual changes in gene expression, punctuated by a sharp decrease in transcription in the transition to the DP stage (Mingueneau et al., 2013). Although they constitute only a small fraction (~0.5%) of the total thymocyte population, the ISPs represent a critical step in thymic differentiation. It has been established that expression of the transcription factors TCF-1, LEF-1, and RORγt is necessary for the transition from ISP to DP (You et al., 2009), but a detailed molecular characterization of the ISP has not been done.
Thymic development is accompanied by large changes in cellular proliferation as thymocytes transition from DN to ISP to DP stages of differentiation. DN thymocytes undergo multiple rounds of proliferation, dependent on the expression of c-Myc (Dose et al., 2006). In contrast, DP thymocytes do not proliferate and do not express c-Myc (Mingueneau et al., 2013). A requirement for c-MYC for the single round of cell cycle that ISPs undergo remains to be established. c-Myc gene expression is known to be regulated by the bromodomain protein 4 (BRD4) (Zuber et al., 2011). BRD4 is a transcriptional and epigenetic regulator that plays a pivotal role in cancer and inflammatory diseases. In many cell types, proliferation depends on BRD4, which functions throughout cell cycle: as a mitotic bookmark, at the G1/S transition (Dey et al., 2000, 2009; Mochizuki et al., 2008), and at the G2/M transition through its interactions with various cell cycle factors (Farina et al., 2004). Deletion of BRD4 arrests cells at G1-S transition and is growth inhibitory to NIH 3T3 and mouse embryonic fibroblasts (MEFs) (Devaiah et al., 2016a; Dey et al., 2009; Maruyama et al., 2002; Mochizuki et al., 2008).
BRD4 regulates lineage-specific gene expression of both innate and adaptive immune cells (Bolden et al., 2014; Cheung et al., 2017; Dey et al., 2000; Mele et al., 2013; Schmidt et al., 2015) and is required for macrophage secretion of inflammatory cytokines and myogenic differentiation and in vitro differentiation of Th17 T cells, (Cheung et al., 2017; Roberts et al., 2017).
The dependence on both robust proliferation and expression of lineage-specific genes during thymic differentiation suggested that BRD4 may play a critical role in this process. The present study was undertaken to establish a detailed gene expression profile of DN, ISP, and DP pre-selection thymocytes and to determine whether BRD4 affects their patterns of gene expression, proliferation, and differentiation. We identify the ISP as a cell type that is molecularly distinct from either the DN or DP subpopulations. Furthermore, by conditionally deleting BRD4 at various stages of thymic differentiation, we have established that BRD4 selectively targets gene expression in the ISP cells: deletion of BRD4 in ISPs downregulates cell cycle and metabolic pathways, leading to a block in the transition to the DP stage. BRD4 is not necessary either for proliferation at the DN stage or for the subsequent maturation of conventional CD4 and CD8 single-positive thymocytes from the DP stage. These studies lead to two conclusions: (1) that the ISP subpopulation is not a hybrid transitional state but a molecularly distinct subpopulation, and (2) that ISP differentiation is selectively dependent on BRD4.
RESULTS
BRD4 Expression Is Required Early in Thymocyte Development
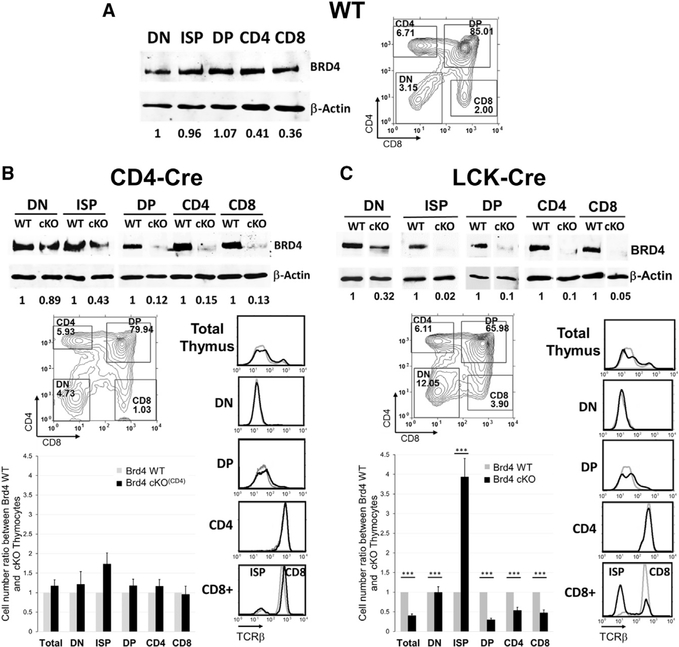
BRD4 protein is expressed in the thymus and at similar levels across the different stages of thymocyte development. The levels of BRD4 in the thymus are comparable to those in peripheral CD4 and CD8 T cells and B cells (Figures 1A, S1A, and S1B). To assess its role in thymocyte development, BRD4 was conditionally deleted at various stages of thymocyte development (Figures S1C and S1D). In thymi deleted of BRD4 by CD4-Cre, BRD4 protein levels were already reduced by 90% at the DP stage, allowing us to determine whether BRD4 affects the development of DP or CD4 and CD8 single-positive thymocytes (Figure 1B). Despite the depletion of BRD4 protein, the total numbers and distribution of DN, ISP, DP, CD4+, and CD8+ thymocytes, as well as their surface expression of TCRβ and co-receptor molecules, were not significantly different from those of the wild-type (WT) (Figure 1A). Thus, deletion of BRD4 by CD4-Cre did not significantly affect the differentiation of DP or their progression to single-positive thymocytes, even though it is expressed in those populations.
Figure 1. Brd4 Deletion by LCK-Cre, but Not CD4-Cre, Affects Thymocyte Development.
The pattern of BRD4 expression and effect on thymocyte numbers and differentiation was determined for WT (A), BRD4 f/- CD4-Cre+ (cKO(CD4)) (B), and Brd4 f/- LCK-Cre+ (cKO) (C) thymocytes.
(A, left, and B and C, top) Immunoblot analysis of BRD4 and β-Actin in different populations of thymocytes. Level of BRD4 expression in thymocytes is normalized to β-Actin and quantitated relative to DN.
(A, right, and B and C, bottom) Thymocytes from either Brd4 wild-type (WT) (A, right) or BRD4 f/- CD4-Cre+ (cKO(CD4)) (B, bottom ) and Brd4 f/- LCK-Cre+ (cKO) (C, bottom) mice were analyzed by flow cytometry (FACS) based on their CD4 and CD8 cell surface expression to determine the distribution of DN, DP, and CD4 and CD8 singlepositive cells. Surface TCRβ for each cell population was determined in parallel (right). (WT, gray line, cKO, black line). Cell number of each population was determined from the percentage and normalized relative to the WT cell numbers (bar graph).
(B) The results represent mean ± SEM of five independent analyses with eight Brd4 f/- CD4-Cre+ mice and seven WT mice. None of the differences observed between WT and knockout populations is statistically significant.
(C) The results represent mean ± SEM of 13 independent analyses with 19 Brd4 f/- LCK-Cre+ mice and 19 WT mice. ***p < 10−4; t test.
See also Figure S1.
To further define the stage(s) of thymocyte development during which BRD4 expression is required, we deleted BRD4 with LCK-Cre (BRD4_cKO), which is expressed at an earlier stage than CD4-Cre (Figure S1D). Accordingly, BRD4 protein levels were markedly reduced in DN thymocytes in the BRD4_cKO thymus, relative to the WT littermates; deletion was complete by the ISP stage (Figures 1C, S1E, and S2A).
The early loss of BRD4 at the DN stage by LCK-Cre-mediated deletion was accompanied by an aberrant distribution of thymocytes among the four subpopulations: whereas the percentage of DP was decreased, the percentages of DN and CD8+ thymocytes were markedly increased. (Figure 1C). The BRD4-deleted thymi were also significantly smaller than those of the WT, resulting largely from a decrease of almost 65% in the DP population (Figure 1C, bottom). The numbers of CD4+ and CD8+ thymocytes were also proportionally reduced.
In contrast to the aberrant patterns of co-receptor expression, TCRβ expression in the LCK-Cre BRD4-depleted thymus paralleled that of the WT in all subpopulations, with one notable exception (Figure 1C). Within the CD8+ population, a large subpopulation of the BRD4-deleted cells did not express surface TCRβ. The increase in the proportion of CD8+ thymocytes that lack surface TCRβ expression indicated an accumulation of ISP thymocytes, which are characterized by the expression of CD8, but not TCR, on their cell surface (Figure 1C, bottom) (Takahama et al., 1992). Taken together, these findings identify a block in differentiation at the ISP stage in the absence of BRD4, which resulted in an increased proportion of ISPs and a failure of normal developmental processes.
Deletion of BRD4 by VAV-Cre, which is first expressed in hematopoietic stem cells (HSCs) in the bone marrow (Figure S1D) (Georgiades et al., 2002), was embryonic lethal, with only two live births of more than 160 total (A.D., unpublished data). The two pups that survived had few thymocytes, among multiple other defects in hematopoiesis (A.D., unpublished data), consistent with the previous observation that adult bone marrow depletion of BRD4 by short hairpin RNA (shRNA) suppresses hematopoiesis (Bolden et al., 2014) and with the finding that germline deletion of BRD4 results in embryonic lethality while heterozygosity results reduced proliferation rates and anatomical defects (Houzelstein et al., 2002).
Taken together, these results lead to the conclusion that BRD4 is required during the early stage(s) of thymic development, but is not required for the maturation of phenotypically normal thymocytes at the DP or conventional single-positive stages. Thus, BRD4 is critical during a narrow window of thymic differentiation prior to the DP stage and focused on the ISP stage.
BRD4 Deletion Blocks Thymocyte Development at the ISP Stage
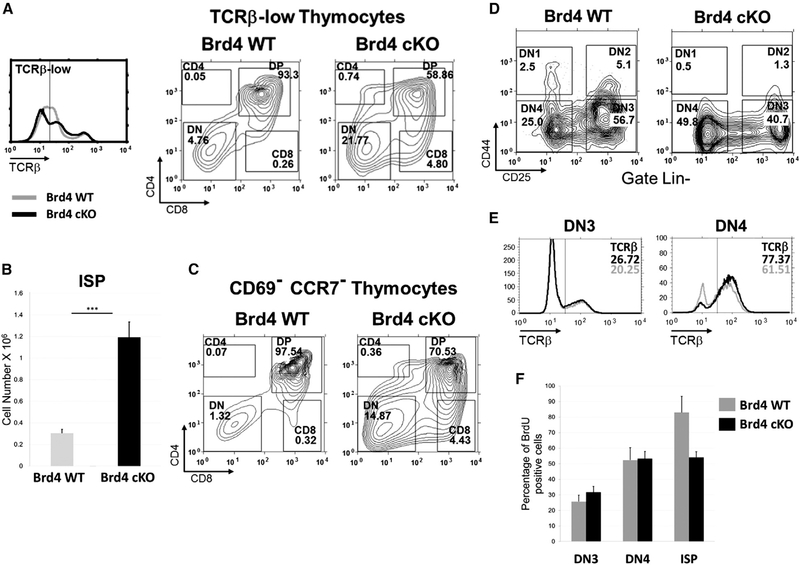
The increased fraction of TCRβ-low/negative CD8+ thymocytes in the thymus deleted of BRD4 by LCK-Cre (Figure 1C) indicated an accumulation of ISPs. To directly examine the ISP populations in WT and BRD4-deficient thymus, we determined the fraction of CD8+ ISP cells within the total surface TCRβ-low/negative thymocyte population (Figure 2A). The deletion of BRD4 (BRD4 cKO) was accompanied by a large increase in TCRβ-low/negative cells in the thymus, relative to the WT (Figure 2A, left). Among the total TCR β-low/negative cells in the thymus (gated to the left of the vertical line in Figure 2A, left), the percentage of CD8+ ISP cells increased nearly 20-fold to 4.8% in the BRD4-deleted thymus, compared with 0.26% in the WT (Figure 2A, middle and right panels). Similarly, within the total thymocyte population, the WT consisted of 0.16% ISP, whereas the ISP population in the BRD4-deleted thymus increased by 10-fold to 1.6%, consistent with a block in differentiation (data not shown). This increased percentage of ISP cells led to a correspondingly higher absolute number of ISP in the BRD4-deficient thymus (Figure 2B).
Figure 2. BRD4 Regulates the Transition from ISP to DP.
(A) Thymocytes from BRD4 WT (gray line) and BRD4 Brd4 f/- LCK-Cre+ (cKO) (black line) were analyzed by FACS for surface TCRβ expression (left panel). Cells with low surface TCRβ (left of vertical line) were analyzed by FACS for CD4 and CD8 cell surface expression. ISPs were identified as CD8-positive, surface TCRβ-low cells. BRD4 WT, middle panel; BRD4 cKO, right panel.
(B) Quantification of ISPs in thymi from BRD4 WT and BRD4 cKO. The results represent mean ± SEM of 13 independent analyses with 19 Brd4 cKO mice and 19 WT mice. ***p < 10−4; t test.
(C) CD69low, CCR7low thymocytes from Brd4 WT and Brd4 cKO mice were analyzed for the expression of CD4 and CD8. The data are representative of three independent experiments.
(D) DN thymocytes from BRD4 WT or BRD4 cKO, defined as negative for lineage commitment markers (Lin-) were categorized into the four stages of differentiation based on cell surface CD25 and CD44 markers as described in the Experimental Procedures. The data are representative of four independent experiments.
(E) Intracellular TCRβ expression in DN3 and DN4 thymocytes from WT (gray line) and Brd4 cKO (black line) mice. DN3 (left) and DN4 (right) thymocytes were stained as described in the Experimental Procedures and fixed, and intracellular TCRβ expression was analyzed by FACS. The data are representative of two independent experiments.
(F) BrdU incorporation by DN3, DN4, and ISP thymocytes from WT and Brd4 cKO. The three cell types were analyzed for BrdU incorporation 4 hr after injection.Data shown summarize three independent experiments for DN and two for ISP, and represent mean ± SEM. No significant differences were observed in the DN3 and DN4 populations between WT and cKO.
See also Figure S2.
The block in the transition from ISP to DP in the BRD4deficient thymus was further documented by analyzing the distribution of the immature pre-selection thymocyte subsets. Pre-selection thymocytes are characterized by their CD69low and CCR7low phenotype. In WT mice, these pre-selection CD69lowCCR7low thymocytes consisted primarily of DP thymocytes (97.5%), with a small fraction of DN (1.3%) and ISP (0.32%) cells (Figure 2C). In striking contrast, in the BRD4-deficient thymus, the percentage of CD8+ (ISP) cells increased sharply, representing 4.4% of the CD69lowCCR7low population, about a 14-fold increase relative to the WT. The identity of the CD8+ cells as ISPs was confirmed by the demonstration that they express CD80αβ, CD24, but no surface TCR0γδ (Table S2) (data not shown). Thus, loss of BRD4 leads to an accumulation of ISP cells, reflecting a block in the transition from ISP to DP thymocytes.
We next examined the possibility that BRD4 also regulates DN maturation. The four DN subpopulations, DN1–4, are distinguished by their expression of the cell surface markers CD25 and CD44. Deletion of BRD4 by LCK-Cre resulted in a shift in the distribution of DN cells, with an increase in the percentage of the more differentiated DN4 (Figure 2D). A decrease in the size of DN4 thymocytes also accompanied the loss of BRD4 (Figure S2B). Although modest, these changes in DN cells in the absence of BRD4 led us to examine whether their maturation was defective.
Maturation from DN cells to ISP cells requires both the rearrangement of the TCRβ genes and extensive proliferation (Rothenberg et al., 2016; Takahama et al., 1992). A defect in either TCRβ rearrangement or proliferation might result in a failure of BRD4-deleted DN cells to differentiate. We first examined the extent of TCRβ rearrangement, as assessed by internal staining of DN3 and DN4 cells. No differences were detected in the level of intracellular TCRβ between the BRD4-deficient and WT thymocytes, indicating that BRD4 is not required for the rearrangement or expression of TCRβ (Figure 2E). Therefore, the block in differentiation resulting from BRD4 deficiency occurs subsequent to, and is independent of, TCRβ rearrangement.
To determine whether BRD4 deletion affects the proliferation of DN thymocytes, we examined their ability to incorporate bromodeoxyuridine (BrdU). Mice were injected peritoneally with BrdU and their DN thymocytes analyzed by flow cytometry for BrdU incorporation 4 hr post-injection. The extent of BrdU incorporation was indistinguishable between the BRD4-deleted and WT thymocytes at DN1, DN2, DN3, or DN4 stages, despite the fact that BRD4 levels were already reduced by over two-thirds in total BRD4 deleted (BRD4f/-LCK-Cre+) DN thymocytes (Figures 2F, S2A, and S2C). Thus, BRD4 is not required for proliferation of DN thymocytes. In contrast, the ISP cells incorporated significantly less BrdU, suggesting a defect in proliferation at the ISP stage (Figure 2F).
The failure to detect defects in either TCRβ rearrangement or proliferation in BRD4-deficient DN3 or DN4 cells indicates that deletion of BRD4 at the DN2/3 stage does not significantly impair maturation of DN thymocytes through to the DN4 stage. Thus, BRD4 is not required for the differentiation of DN thymocytes. Rather, a major role of BRD4 in thymocyte development is in regulating the transition from ISP to DP.
ISP Thymocytes Are a Molecularly Distinct Thymocyte Subpopulation
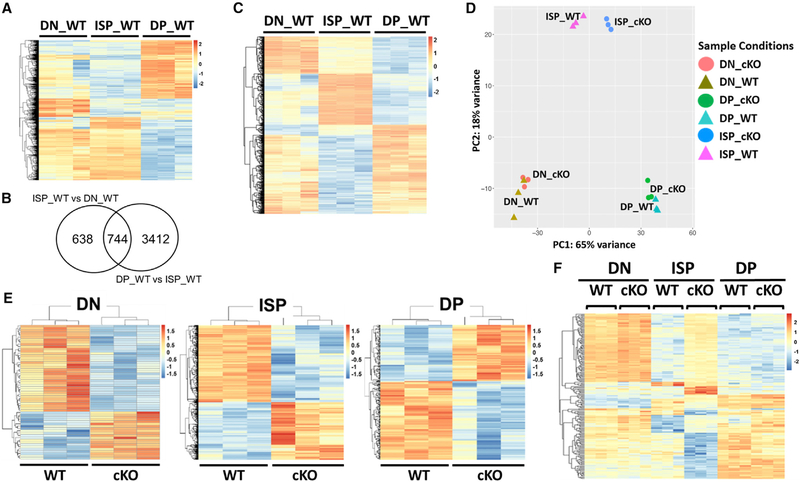
To begin to understand the molecular basis of the block in differentiation in thymocytes deleted of BRD4 by LCK-Cre, we first examined the gene expression profiles of DN, ISP, and DP thymocytes from WT thymus by RNA sequencing (RNA-seq) to determine whether the ISP is simply a transitional cell or a molecularly distinct subpopulation. Each of the transitions from one developmental stage to the next was accompanied by large changes in gene expression (Figure 3A). The greatest differences in gene expression in the WT were associated with the ISP subpopulation in its transition to the DP stage (Figure 3A; Table S1). Furthermore, 744 genes (7.5% of the total) were differentially expressed in ISPs relative to both the DN and the DP subpopulations (Figures 3B and3C).
Figure 3. Brd4 Deletion Preferentially Targets ISP Thymocytes.
All of the heatmaps represent genes with a signal intensity fold change > 2 and FDR <0.01.
(A) Heatmap comparing RNA expression profiles of DN, ISP, and DP thymocytes from WT thymus.
(B) Venn diagram summarizing the differences in gene expression between WT ISP thymocytes and WT DN and DP, respectively.
(C) Heatmap of the 744 genes differentially expressed in WT ISP relative to DN and DP.
(D) Principal-component analysis (PCA) of gene expression profiles of DN, ISP, and DP thymocytes from BRD4 WT and BRD4 cKO.
(E) Heatmaps illustrating the effects of BRD4 deletion by LCK-Cre (cKO) relative to WT on gene expression in DN, ISP, and DP thymocytes.
(F) Heatmap comparing the expression of the 194 BRD4-dependent genes of the 744 that are differentially expressed genes in WT ISP relative to DN and DP.
See also Figure S3.
Pathway analyses revealed that the gene sets that are differentially expressed in the WT ISP population compared to both WT DN and DP are primarily associated with cell cycle and metabolic pathways (Figure S3A). Most of the changes in gene expression between DN and ISP were clustered in immune and T cell development pathways, as well as cell cycle pathways (Figure S3B). Consistent with the known quiescence of DP cells, the transition from ISP to DP cells was accompanied by a downregulation of cell cycle pathways and an upregulation of cell death and T cell development pathways (Figure S3C).
Principal-component analysis (PCA) highlighted the fact that the ISP population is molecularly distinct and no more related to either DN or DP than they are to each other (Figure 3D). These findings document the ISP as a cell population whose gene expression profile is not simply a hybrid intermediate state between DN and DP and may be required to support the transition to DP.
BRD4 Targets a Set of ISP-Specific Transcripts
We next examined by RNA-seq the effect on gene expression of BRD4 deletion in DN, ISP, and DP thymocyte subpopulations (Figure 3E). Deletion of BRD4 did not reduce gene expression globally: All the subpopulations, in both WT and BRD4 cKO, expressed approximately the same total number of genes. However, BRD4 deletion selectively affected levels of gene expression (Table S1). Pathway analysis revealed that many genes associated with immune cell function are differentially expressed in all thymocyte subpopulations as a result of BRD4 deletion (Table S2).
The largest effect of BRD4 deletion on gene expression was observed in the ISP cells (Figure 3E; Table S1). Among the 1,168 genes that were differentially expressed between BRD4deficient and WT ISP cells, 194 were among those differentially expressed in WT ISP relative to both DN or DP (Figure 3F). Although BRD4 deletion affects a number of gene sets (Figure S4A), the pathways most affected were those related to metabolism and cell growth, as well as immune signaling pathways (Figure S4C), which were all downregulated in the absence of BRD4. Thus, BRD4 functions as a regulator of the major ISP pathways. Importantly, these results indicate that the ISPs are a molecularly distinct thymocyte subpopulation that is selectively regulated by BRD4.
Although we detected no significant phenotypic defects in either the DN or DP subpopulations in the absence of BRD4, we did observe modest differential gene expression in both subpopulations. Deletion of BRD4 resulted in the differential expression of only 107 genes in the DN population, despite the two-thirds loss of BRD4 (Table S1). These changes may account for the small differences observed in the size of DN and the DN4/ 3 ratio. Similarly, a relatively modest number of genes were differentially expressed in BRD4_cKO DP cells compared to WT (Table S1). The overall effect of BRD4 is as repressor of the major DN and DP immune and signaling pathways (Table S1; Figures S4B and S4D).
Interestingly, PCA and t-distributed stochastic neighbor embedding (t-SNE) analysis highlighted the fact that the differences among the DN, ISP, and DP subpopulations are notably greater than the differences between the WT and BRD4_cKO within a subpopulation (Figure 3D).
BRD4 Regulates Glycolytic and MYC Pathways in ISP Thymocytes
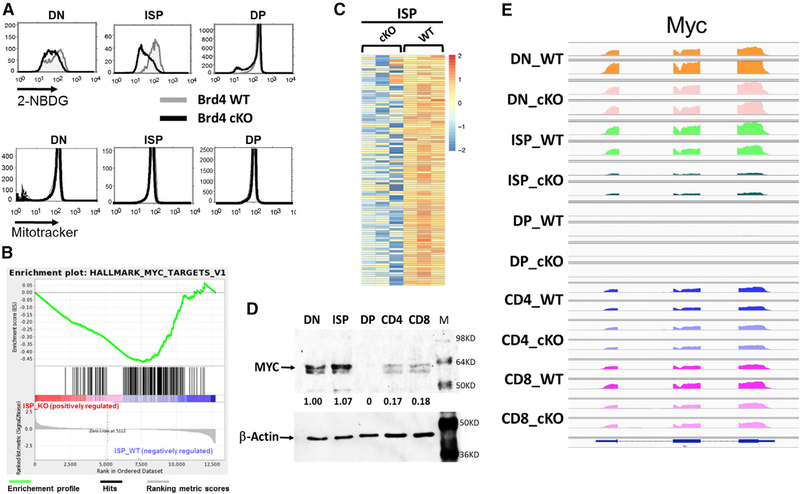
Deletion of BRD4 affected expression not only of immune pathways but also profoundly affected glycolytic pathways in ISP thymocytes (Figures 4 and S4C), leading to the prediction that BRD4_cKO ISP cells should be defective in glycolysis. To test this prediction, we assessed the ability of BRD4_cKO ISP thymocytes to incorporate the glucose analog, 2-NBDG (Figure 4A, top). As predicted, BRD4 cKO ISP thymocytes were impaired in glucose uptake. In contrast, BRD4 deletion did not affect glucose uptake in DP. Glucose uptake in DN thymocytes was slightly reduced in the absence of BRD4. Mitochondrial pathways were not identified in the analyses; accordingly, mitochondrial mass was not detectably affected by the absence of BRD4 in any of the thymocyte subpopulations (Figure 4A, bottom).
Figure 4. BRD4 Regulates Glycolytic and c-Myc Pathways in ISP Thymocytes.
(A) Top: glucose uptake in WT and BRD4 cKO DN, ISP, and DP thymocytes, as measured by uptake of the glucose analog 2-NBDG and FACS. WT, gray; BRD4 cKO, black. Bottom: mitochondrial mass of Brd4 WT and cKO DN, ISP, and DP thymocytes, as assessed by MitoTracker uptake and FACS.
(B) Enrichment plot of the c-Myc target gene set in ISPs. The c-Myc_targets_v1 gene set is significantly downregulated in the Brd4 cKO (p < 0.0001). The green curve shows the running sum of enrichment score (ES) for ranked genes. The hash marks under the plot represent the genes that are the leading-edge subset, which accounts for the gene set’s enrichment signal.
(C) Heatmap showing the normalized expression of the top 105 downregulated myc_target_v1 genes in BRD4 cKO ISP thymocytes.
(D) Immunoblot analysis of c-MYC and β-Actin in different populations of thymocytes.
(E) IGV visualization of Myc gene expression in WT DN, ISP, DP, CD4, and CD8 thymocytes. IGV data range: 0–1,000.
See also Figure S4.
BRD4 deletion in ISP cells also significantly repressed many MYC target pathways but increased expression of other genes, notably inhibitors of cell cycle progression such as Cdkn1b and 2d (Figures 4B and4C). To characterize the role of BRD4 in regulating MYC target pathways, we examined Myc RNA and proteins levels in the individual WT thymocyte subpopulations (Figures 4D and4E). Myc RNA was robustly expressed in all subpopulations except for the DPs, which express no detectable MYC consistent with their quiescent phenotype. Accordingly, MYC protein levels are highest in the ISPs and DNs, somewhat lower in CD4+ and CD8+ thymocytes, and undetectable in DPs (Figure 4D).
BRD4 deletion only eliminated Myc RNA expression in the ISP thymocytes (Figures 4E and S4C). Myc RNA levels in the DNs were unaffected, consistent with our in vivo observations that deletion of BRD4 did not abrogate the proliferation of DN thymocytes (Figure 2F). Myc expression in CD4+ or CD8+ SPs was similarly unaffected (Figure 4E). Thus, Myc expression is only BRD4 dependent at the ISP stage.
These findings validate the predictions of the RNA-seq data, indicating that the RNA-seq data provide an accurate assessment of the molecular phenotypes of the thymocyte subpopulations. Importantly, the RNA-seq and functional data establish ISP thymocytes as a molecularly distinct subpopulation, not a hybrid transitional state, that is selectively regulated by BRD4.
BRD4-DeficientISPsFailtoMaturetoDPsduringInVitro Culture
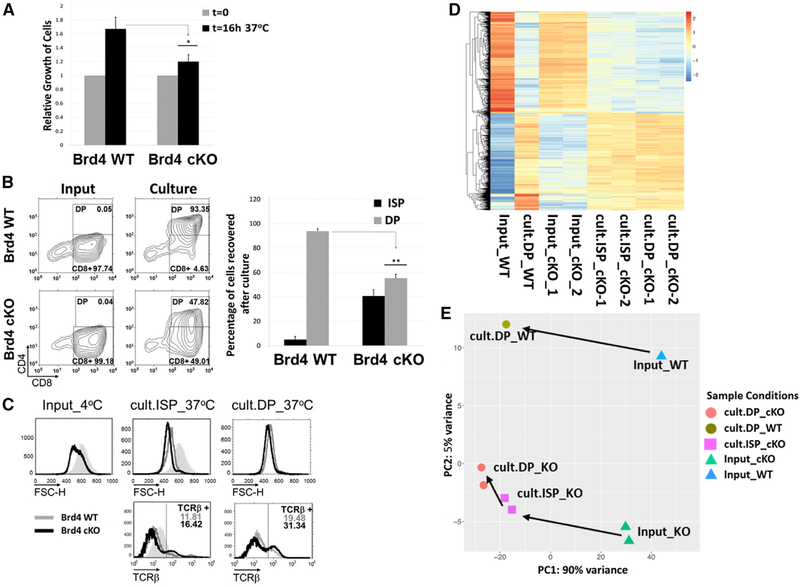
The RNA-seq data provided a static molecular description of each of the thymocyte subpopulations. However, they did not define the dynamic changes in gene expression that accompany the transition from ISP to DP. Maturation of ISP to DP requires a single round of cell cycle (Yu et al., 2004) and can be reproduced in an in vitro culture, allowing us to identify those genes dynamically associated with differentiation. Following an overnight culture, WT ISPs doubled in number and all matured to DP thymocytes (Figures 5A and5B). In contrast, BRD4-deficient ISPs did not divide and were severely impaired in their ability to differentiate into DP cells. Less than one-half of the BRD4_cKO ISPs expressed CD4 after the overnight culture and at levels lower than the WT CD4+ cells (Figures 5A and5B). However, those BRD4deficient ISPs that expressed CD4+ following the in vitro culture expressed surface TCRs at levels comparable to the WT, indicating that BRD4 is not necessary for surface TCR expression (Figure 5C, bottom). Furthermore, our results lead to the conclusion that, although BRD4 is not required for DN proliferation, it is necessary for the single round of cell cycle that leads to the transition from ISP to DP. This raises the intriguing possibility that the requirements for cell division in these populations are distinct.
Figure 5. BRD4 cKO ISPs Are Blocked in Their Maturation into DP Thymocytes In Vitro.
(A) ISPs from BRD4 cKO and WT thymi were cultured overnight, and the extent of their proliferation was quantified as cell recovery (percent of input) after theovernight culture; data are the mean ± SEM of four independent experiments. *p < 0.05; t test.
(B) Maturation of ISP into DP (culture panel), relative to the input (input panels) of ISPs from WT (top) and cKO (bottom), was assessed by flow-cytometricmeasurement of CD4 and CD8 surface marker expression. The histogram (right) represents the mean ± SEM of four independent cultures of Brd4 cKO ISPs and four WT ISPs. **p < 0.005; t test.
(C) FACS of the input ISPs remaining after culture (cult.ISP_37˚C) and DP appearing after culture (cult.DP_37˚C) (from B) assessing their size by forward scatter (top) and TCRβ surface expression (bottom). WT, gray; Brd4 cKO, black; shaded peak, WT ISP input. The data are representative of four independent experiments.
(D) The heatmap was plotted based on 3,194 genes that are differentially expressed between DP_WT after overnight culture (cult.DP_WT) and input ISP_WT(Input_WT). Input ISPs from Brd4 cKO mice (Input_cKO), remaining cKO ISPs (cult.ISP_cKO), and cKO DPs (cult.DP_cKO) after overnight culture.
(E) PCA representation of the WT and cKO input ISP and thymocyte populations after overnight culture.
See also Figure S5.
During normal development in the WT thymus, the transition to the DP stage is accompanied by a reduction in the size of the thymocytes. The BRD4-deficient ISPs harvested from the thymus were smaller than WT ISPs, presumably reflecting the loss of MYC (Figure 5C, top). Nevertheless, the ISPs that progressed to the CD4+ stage in culture, as well as those that did not, underwent the further reduction in size associated with the normal transition to DP (Figure 5C, top). We conclude that although BRD4-deficient ISP undergo some of the phenotypic changes associated with differentiation—indicative of signaling—they are severely restricted in their ability to mature to the DP stage, as manifested by an impaired ability to undergo cell division and reflected in the accumulation of ISPs in the BRD4-deficient thymus.
To characterize the molecular events associated with the transition from ISP to DP thymocytes during the in vitro culture, RNAseq analysis was performed on each of the four cell populations (Figure 5D). Reflecting what was observed during in vivo maturation, the in vitro maturation of WT ISP (Input_WT) to DP (cult.DP_WT) was associated with large changes in gene expression (Figure 5D; Input_WT versus cult.DP_WT). Among the 3,194 genes that were differentially expressed, cell cycle-related and metabolic pathway genes predominated; very few immune pathways were differentially expressed (Figure S5). Thus, maturation of WT ISP thymocytes to the DP stage is primarily dependent on metabolic and cell cycle genes.
In the absence of BRD4, ISP cells only partially progressed to mature DP thymocytes, as evidenced by the lack of proliferation and low levels of CD4 expression (Figures 5A and5B). About one-half of the genes differentially expressed between ISP and DP in the WT (1548 genes) were also differentially expressed in the BRD4-deficient cells following the in vitro culture. These common genes are mainly cell cycle-related genes (Figure S5A). The genes that were differentially expressed in the transition from ISP to DP in the WT cells, but not in the BRD4-deficient cells, were mainly in metabolic pathways (Figure S5B). These results suggest that the primary defect in the maturation of ISP to DP in the absence of BRD4 may be metabolic, leading to a failure of cell cycle (Figure S5C).
The nature of the defect in maturation in the absence of BRD4 is best visualized in a PCA (Figure 5E). During in vitro culture, as observed in vivo (Figure 3E), the maturation of WT ISP (Input_WT) to DP thymocytes (cult.DP_WT) was accompanied by large changes in the expression profile. The BRD4_cKO ISP (Input_cKO) also underwent large changes in expression profile but were unable to fully differentiate to the DP stage (cult.DP_cKO). The CD4+ cells (cult.DP_cKO) that emerged from the overnight culture of BRD4_cKO ISP (Input_cKO) were markedly different from WT DP (cult.DP_WT) cells in their gene expression profile. Interestingly, the gene expression profile of the BRD4_cKO ISPs (cult.ISP_cKO) remaining after overnight culture was more closely related to the cells that became CD4+ (cult.DP_cKO) than to the original, input ISPs (Input-cKO). Thus, although the BRD4_cKO ISPs can undergo some reprogramming in the DP-developmental pathway, they are unable to fully reprogram their expression within the time frame of the in vitro culture to become DP thymocytes, likely due to a failure in metabolic pathways.
Taken together, our results indicate that BRD4 selectively regulates the transition from ISP to DP through its targeting of genes in metabolic pathways, as well as of MYC and its targets.
BRD4 Is Required for the Generation of Tregs and iNKT Cells, but Not Conventional CD4+ or CD8+ Cells
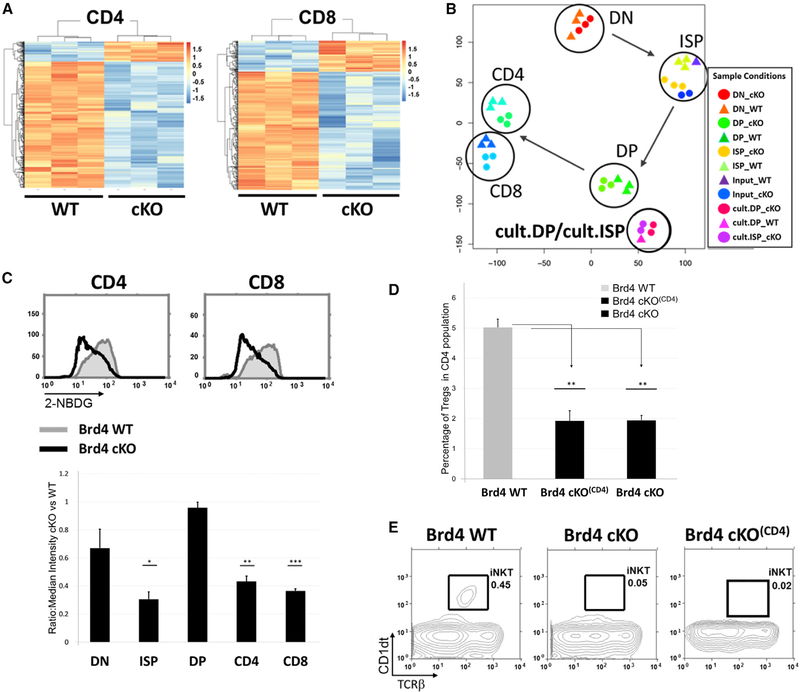
Although BRD4 is required for the efficient progression of ISP to the DP stage, the DPs that develop in the absence of BRD4 further differentiate to conventional CD4+ and CD8+ single-positive thymocytes (Figure 1C). This leads to the question of the effect of BRD4 loss on their gene expression profiles. Only a relatively small number of genes were differentially expressed in the absence of BRD4 in either CD4+ or CD8+ cells (Figures 6A and6B; Table S1). Of the differentially expressed genes, the majority were positively regulated by BRD4 in both CD4+ (63%) and CD8+ (79%). This is in contrast to either the ISP or DP thymocytes where one-half of the genes were negatively regulated by BRD4 (Figure 3E; Table S1). Among the major pathways activated by BRD4 in both CD4+ and CD8+ are immune response pathways (Figures S6A and S6B). Although TCRβ-high single-positive thymocytes can escape the BRD4 restriction and emigrate to the periphery, they are not functional and do not efficiently proliferate in response to stimulation (Figure S6D). In addition, BRD4-deficient CD4+ and CD8+ thymocytes and splenic T cells display impaired glucose uptake, although the major metabolic pathways are not affected (Figures 6C and S6C).
Figure 6. Conventional CD4+ and CD8+ Single-Positive Thymocytes Develop in the Absence of BRD4, but the Generation of Treg and iNKT Cells Is Affected.
(A) Heatmapscomparing gene expression levelsin BRD4 cKO andWTCD4andCD8single-positivethymocyteswithsignalintensityfold change > 2 andFDR <0.01.
(B) t-SNE representation summarizing the relationships of both in situ and in vitro thymocyte populations based on their gene expression profiles.
(C) Left: glucose uptake in WT and BRD4 cKO CD4+ and CD8+ thymocytes, as assessed by uptake of the glucose analog 2-NBDG and FACS. WT, gray; BRD4 cKO, black. Right: 2-NBDG uptake median intensity comparison between Brd4 cKO and WT thymocytes. The data are representative of three independent experiments. ***p <10−4, **p <0.005, *p <0.01; t test.
(D) The presence of CD4+ Treg cells in BRD4-deleted and WT thymi was assessed by FACS based on intracellular staining for Foxp3. BRD4 deletion was mediated by either the LCK-Cre transgene (Brd4 cKO) or the CD4-Cre transgene (Brd4 cKO(CD4)). The graph is the summary of three Brd4 WT, four Brd4 cKO(CD4), and two Brd4 cKO mice. **p < 0.005.
(E) The presence of iNKT cells in Brd4 WT, Brd4 cKO, and BRD4 cKO(CD4) thymi was assessed by FACS based on CD1 dt and TCRb surface expression. Theresults are representative of two independent experiments.
See also Figure S6.
Although conventional CD4+ and CD8+ single-positive thymocytes develop in the absence of BRD4, we asked whether other thymocyte subpopulations were affected. Among the genes that were downregulated in CD4+ thymocytes in the absence of BRD4 was Foxp3, which is required for the differentiation of Tregs (Hori et al., 2003). This led us to ask whether the development of Tregs depended on BRD4 expression. Remarkably, deletion of BRD4 by either LCK-Cre or CD4-Cre led to a significant reduction in both percentage and number of Tregs (Figure 6D), suggesting that there is a distinct requirement for BRD4 expression after the development of CD4+ and CD8+ single-positive thymocytes. While BRD4 is necessary for the generation of Tregs, it is not required for maintenance of Tregs: deletion of BRD4 by Foxp3-Cre did not affect Treg development or the number of Tregs in thymus or peripheral tissues (Figure S6E). However, BRD4-deficient Tregs are markedly impaired in their response to stimulation (Figure S6F).
iNKT cell development is dependent on CD1d selection on DP thymocytes (Bendelac et al., 1995; Zajonc and Kronenberg, 2009). BRD4 deletion did not affect CD1d1 levels in DP thymocytes (Table S2). Thus, it was to find that deletion of BRD4 by either LCK-Cre or CD4-Cre resulted in a loss of iNKT cells (Figure 6E). Interestingly, BRD4 deletion by LCK-Cre resulted in a decrease in CD1d1 expression in CD4+ single-positive thymocytes. This suggests that development of both iNKT and Treg subpopulations requires BRD4 expression beyond the DP stage.
Given the profound effect that BRD4 has on the differentiation of these thymocyte subpopulations, it is remarkable that its deletion targets relatively few genes. This is best visualized in a PCA of all of the thymocyte subpopulations (Figure 6B), which reveals that BRD4-deleted cells still cluster closely with their WT counterpart. Notably, both the BRD4-deleted and WT ISP subpopulations are more related to one another than to either their DN precursors or DP successors, further documenting the ISP as a distinct subpopulation.
Taken together, our results demonstrate that BRD4 selectively regulates the transition from ISP to DP: it is not required either for DN differentiation or the further differentiation of conventional CD4+ and CD8+ single-positive thymocytes. However, it is required for the generation of Tregs and iNKT cells.
DISCUSSION
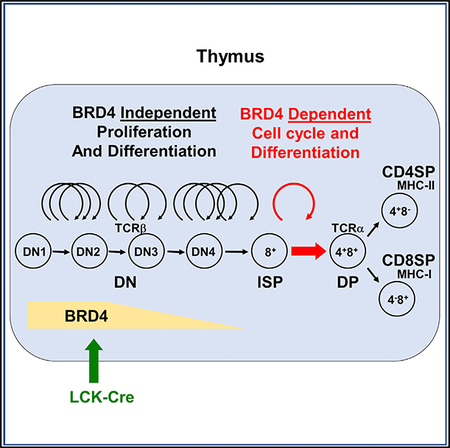
BRD4 is being widely investigated as a therapeutic target in inflammatory diseases, as well as a variety of solid and hematological cancers (for review, see Andrieu et al., 2016a, and Devaiah et al., 2016b). It is a chromatin-binding protein with both kinase and acetyl transferase activities that plays an active role in regulating transcription by linking chromatin structure and transcription (Devaiah et al., 2012, 2016a; Dey et al., 2009). In the present study, we have examined the in vivo requirement for BRD4 during thymocyte development. The stages of thymocyte differentiation—DN, ISP, DP, and singlepositive —are characterized by their cell surface expression of the markers CD4 and CD8. Among these, the ISP subpopulation is the least well characterized and has been considered a transitional cell between the DN and DP stages of differentiation (Mingueneau et al., 2013). Our findings now identify ISP thymocytes as a discrete subpopulation with a gene expression profile distinct from either the DN precursors or the DP successors. Although BRD4 is expressed throughout all stages of thymocyte differentiation, the requirement for BRD4 is restricted to limited stages of the thymic differentiation program. Among pre-selection thymocytes, only the transition from ISP to DP is BRD4 dependent; among post-selection thymocytes, the generation of both Tregs and iNKT is abrogated in the absence of BRD4 expression. In ISP cells, BRD4 primarily regulates genes in metabolic pathways; deletion of BRD4 results in a failure to support the single round of cell cycle associated with differentiation to the DP stage. It is not required for the proliferation associated with DN stage amplification or the maturation from DP to conventional single-positive thymocytes where no proliferation is involved. We propose a model in which the transition from the highly proliferative DN stage to the quiescent DP stage requires a reprogramming through the ISP stage that is regulated by BRD4.
BRD4 is expressed throughout thymocyte differentiation and at equivalent levels among the subpopulations. However, among the pre-selection thymocytes, BRD4 depletion selectively affects the ISP subpopulation. The expression of over 1,100 genes—most in immune, cell cycle, and metabolic pathways—is affected. The loss of BRD4 resulted in increases in expression of about half of the genes and decreases in the other half, indicating that BRD4 acts as both a transcriptional activator and silencer. The cell cycle and metabolic pathways that are most affected by BRD4 deletion are largely downregulated by BRD4 loss. This results in BRD4-deleted ISPs that are deficient in metabolic activity.
The maturation of WT thymocytes is accompanied by large changes in gene expression. The largest changes occur in the transition from ISP to DP. Of particular note, the gene expression profile of the ISP subpopulation is distinct from both the DN and the DP; nearly 10% of its genes are uniquely differentially expressed. Thus, the ISP is not simply a transitional population. Rather, the ISPs are a molecularly and functionally distinct thymocyte subpopulation. (Whether the ISPs are a homogeneous population or consist of further subpopulations cannot be distinguished by the present data and will require single-cell analyses beyond the scope of this study.) Although the maturation from ISP to DP is accomplished during a single round of cell cycle, it is accompanied by changes in expression of approximately 40% of the genes. These differentially expressed genes map to immune, cell cycle, notch signaling, and metabolic pathways. Among those genes are the transcription factors, TCF-1, LEF-1, and the orphan nuclear receptor, RORγt, which are necessary for the transition from ISP to DP. Significantly, their expression is not affected by BRD4 deletion and does not account for the block in differentiation of BRD4-deleted ISP. Rather, depletion of BRD4 results in the downregulation of metabolic pathways necessary for the transition from ISPs to quiescent DP cells. The present findings are consistent with previous reports that BRD4, as well as BRD2, regulate metabolic gene pathways in pancreatic β-cells (Deeney et al., 2016).
ISPs have been reported to represent a transitional state between Notch dependence in DN thymocytes and Notch independence in DP thymocytes (Xiong et al., 2011). BRD4 contributes to this transition, since depletion of BRD4 in ISP thymocytes leads to downregulation of Notch signaling pathways. This finding is consistent with a previous report that BRD4 regulates the Jagged1/Notch1 pathway in breast cancer cells (Andrieu et al., 2016b). Interestingly, in contrast to its effect in ISPs, BRD4 depletion leads to an upregulation of Notch signaling pathways in DP and DN, consistent with the interpretation that the ISPs are a molecularly distinct population.
Among the ISP genes that are regulated by BRD4 is Myc. Although Myc is expressed in all thymocyte subpopulations (except for DP), BRD4 only regulates Myc expression and that of its downstream targets in ISP. While DN and ISP thymocytes express Myc at approximately equal levels, DN thymocytes are highly proliferative, whereas ISPs undergo only a single round of proliferation. This leads to the conclusion that proliferation of ISP thymocytes is BRD4 dependent but proliferation of DN thymocytes is not. While Myc is known to be regulated by BRD4 in many, but not all, cancer cell lines, the mechanisms underlying this selective regulation of Myc by BRD4 in ISP thymocytes and other cell types remain to be determined.
Deletion of BRD4 prevents the cell cycle required for the transition from ISP to DP, indicating that BRD4 plays a critical role in the coordination of cell cycle exit and differentiation. The role of chromatin modifiers in this transition has been well documented in a number of systems, including myogenesis and neurogenesis (Ruijtenberg and van den Heuvel, 2016; Villarreal, 1991). In previous studies, we have demonstrated that BRD4 is a histone acetyl transferase that remodels chromatin. Thus, the requirement for BRD4 in regulating the switch from proliferation to differentiation, which has not been previously reported, may reflect its chromatin-modifying activity. It is worth noting that, among the BET proteins, only BRD4 has histone acetyltransferase (HAT) activity.
In sharp contrast to the dramatic effects of BRD4 deletion on ISP thymocytes, the DN, DP, and single-positive thymocyte subpopulations are only modestly affected. In the absence of BRD4, DN thymocytes differentiate normally, as evidenced by their ability to proliferate and undergo TCRβ rearrangement, consistent with the limited effect on gene expression. Similarly, BRD4 plays a relatively small role in DP or single-positive thymocytes, which are phenotypically normal in the absence of BRD4. Among the genes that are downregulated in BRD4-deficient CD4+ thymocytes is Foxp3, accounting for the failure of Tregs to develop in the absence of BRD4. iNKT cells development is also abrogated in the absence of BRD4. Taken together, these findings lead to the conclusion that BRD4 is a major determining factor in only a subset of thymocytes, namely ISP, Treg, and iNKT cells.
In conclusion, the present studies have identified the ISP as a molecularly distinct cell type that is selectively regulated by BRD4. Further maturation to the DP thymocyte stage requires a reprogramming of gene expression that depends on the BRD4-regulated pathways of MYC, cell cycle, and metabolism. BRD4-independent TCR signaling and DP transcription factors are necessary, but not sufficient.
EXPERIMENTAL PROCEDURES
Generation and Fluorescence-Activated Cell-Sorting Analysis of the BRD4 KO Mice
The design of the Brd4 knockout (Brd4-) and Brd4-floxed (Brd4f) alleles, described previously (Devaiah et al., 2016a), as well as the strategy for generating the different BRD4 KO mice are summarized in Figures S1C and S1D and Supplemental Experimental Procedures. The experiments were approved by the National Cancer Institute Animal Care and Use Committee (ASP 18–417, 18–430) and mice were cared for in accordance with National Institutes of Health guidelines. Thymocytes were prepared, counted, and assessed for CD4, CD8, and TCRβ surface protein expression by flow cytometry (fluorescence-activated cell sorting [FACS]) using an LSRII or LSR Fortessa (Becton Dickinson). Dead cells were excluded from the analysis by propidium iodide gating. 105 events in the live-cell gate were acquired; data were analyzed using EIB-Flow Control software developed at the National Institutes of Health. For internal staining (TCRβ, BrdU) and small populations (ISPs, Tregs), 106 events were acquired. Gates were tightly defined based on control thymocyte staining. t test (two-tailed, unequal variance) was used.
Antibodies
All antibodies are described in Supplemental Experimental Procedures.
Immunoblotting
Extracts of 5 × 105 thymocytes or lymph node lymphocytes were isolated using the TER1 buffer (Thermo Fisher; FNN0071); soluble lysate fractions were analyzed by immunoblotting using antibodies as indicated. Tubulin and β-actin antibodies were used as loading controls.
Purification of Thymocytes and Lymph Node T Cells
DP thymocytes were purified by flow cytometry based on the expression of both CD4 and CD8 surface markers and the lack of CD69 at the surface. CD4 thymocytes were pre-purified using the Life Technology Dynabeads Untouched Mouse CD4 Cells Kit and sorted by flow cytometry based on the expression of CD4 and TCRβ at the surface. DN, ISP, and CD8 thymocytes were pre-purified using the Life Technology Dynabeads Untouched Mouse CD8 Cells Kit and then sorted by FACS, respectively, on their lack of CD4, CD8, and TCRβ surface expression for DNs, the expression of CD8 and low TCRβ expression for the ISPs, and finally the expression of CD8 and TCRβ at the surface for CD8 single-positive cells. Lymph node CD4 and CD8 T cells were purified using the EasySep Mouse CD4+ or CD8+ T Cell Isolation Kit (StemCell Technologies), respectively, followed by FACS based on CD4, CD8, and TCRβ expression.
BrdU Incorporation
Mice were injected in the peritoneal cavity with 1 mg of BrdU 2 and 4 hr before harvesting the thymi. The BrdU analysis used either 5 × 106 total thymocytes or 2 × 106 enriched non-committed thymocytes. The non-committed thymocytes were enriched by depletion of CD4+ and/or CD8+ cells with a mix of anti-CD4 and anti-CD8 microbeads on MACS columns (Miltenyi Biotec). The DN thymocytes were stained using a cocktail of antibodies recognizing surface proteins expressed on lineage committed cells (TCRβ, CD4, CD8, B220, NK1–1, MACI, GR-1, GL3) and CD44 and CD25 antibodies; ISP thymocytes were stained with the same cocktail of antibodies minus CD8, and then fixed and treated according to the procedure described in the BrdU Flow Kit (BD Pharmingen; 559619). DN and ISP thymocytes were identified by the absence of staining with their respective lineage cocktail of antibodies. The subpopulations of DN and the ISP were defined based on the expression of CD44 and CD25 or CD8, respectively, and their ability to proliferate assessed by flow cytometry of BrdU incorporation.
TCRβ Intracellular Staining
5 × 106 DN thymocytes were identified as above, fixed using the Cytofix/Cytoperm solution and the procedure recommended by BD Pharmingen, internally stained with an anti-TCRβ antibody, and analyzed by FACS.
Foxp3 Intracellular Staining
3 × 106 thymocytes were first labeled for CD4, CD8, and TCRβ surface expression, and then fixed using the Foxp3/Transcription Factor Staining Buffer Kit from eBioscience (00–5523-00). Treg thymocytes were internally stained with an anti-Foxp3 antibody and analyzed by FACS.
RNA-Seq Analysis
DN, ISP, DP, CD4, and CD8 thymocytes from WT and Brd4 cKO (Brd4f/- LCK-Cre+/−) males (6–8 weeks old) were purified as described above and evaluated post-purification by FACS. The purity of each thymocyte population ranged from 95% to 100%. RNA-seq analysis was done as described before (Devaiah et al., 2016a). RNA was isolated from WT and BRD4-deleted cells using the MicroRNeasy-Plus Kit (QIAGEN). Libraries were made using the Clontech SMARTer Universal Low Input RNA Kit for sequencing and sequenced as pair-end reads on an HiSeq platform according to established procedures. RNA-seq reads were aligned to mouse reference genome (mm10) with STAR aligner (Dobin et al., 2013). Raw read counts were obtained using htseq-count (Anders et al., 2015) and normalized for further analysis using the built-in normalization algorithms of DESeq2 (Love et al., 2014). All differential expression analysis was performed with DESeq2, and a gene was considered as differentially expressed between two groups when its fold change > 2 and false-discovery rate (FDR)-adjusted p value <0.01.
Statistical testing, PCA, hierarchical clustering, and t-SNE (van der Maaten and Hinton, 2008) were performed in R. Gene set enrichment analysis (GSEA) for the differentially expressed genes was performed using Hallmark gene set collection from MsigDB (https://www.broadinstitute.org/gsea/). GSEA uses the list rank information without using a threshold, allowing the ability to focus more on the biological processes that are distributed across an entire network of genes and subtle at the level of individual genes (Subramanian et al., 2005). Read distribution was visualized using the IGV platform (http://software.broadinstitute.org/software/igv). The percentages of reads aligned to the coding regions were similar across the different samples. GO and Pathway analysis were performed using Metascape.org.
Glucose Uptake
2 × 106 cells were washed once with serum-free RPMI then resuspended in 1 mL of warmed serum-free RPMI. Mitotracker Green (Thermo Fisher; M7514) was diluted to a 1 μM working solution, and then added to the cells at the final concentration of 100 nM. Cells were incubated for 20 min at 37˚C, and then washed once prior to staining with surface markers.
Mitochondrial Mass Evaluation
2 × 106 cells were washed once with serum-free/glucose-free RPMI and resuspended in 1 mL of warmed serum-free/glucose-free RPMI. 2-NBDG (Thermo Fisher; N 13195) was diluted to a 1 μM in DMSO, and then added to a final concentration of 100 μM. Cells were incubated for 20 min at 37˚C, and then washed once prior to staining with surface markers.
ISP Culture
WT or Brd4-deleted ISP thymocytes were purified as described above using the Life Technology Dynabeads Untouched Mouse CD8 Cells Kit, and then sorted by FACS based on their CD8 surface expression and low TCRβ surface expression. Equal numbers of WT and Brd4-deleted cells were incubated for 16 hr in RPMI medium supplemented with 10% fetal calf serum, 1 mM Napyruvate, MEM non-essential amino acids (1×), and 2-mercaptoethanol; proliferation was assayed by counting the cells after culture. The cells were also analyzed by FACS for CD4 surface expression.
Supplementary Material
Highlights.
ISP thymocyte differentiation is selectively dependent on BRD4
Deletion of BRD4 in ISPs downregulates cell cycle and metabolic pathways
BRD4 is not necessary for proliferation of the DN thymocytes
Treg and iNKT cell differentiation is BRD4 dependent
ACKNOWLEDGMENTS
The authors gratefully acknowledge Drs. Remy Bosselut, Richard Hodes, Hyun Park, and Ranjan Sen for their critical reading of the manuscript and helpful discussions, and Dr. Jie Mu and Dr. Thomas Ciucci for their technical assistance and data analysis support, respectively. We thank Mike Kruhlak for his help in microscopy; the members of the flow cytometry facility, Dr. Susan Sharrow, Tony Adams, Larry Granger, and Assiatu Crossman for their advice; and the members of the D.S.S. lab for discussions. This research was supported by the Intramural Research Program of the NIH National Cancer Institute Center for Cancer Research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.06.007.
DATA AND SOFTWARE AVAILABILITY
The accession number for all of the RNA-seq data reported in this paper is GEO: GSE109255.
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrieu G, Belkina AC, and Denis GV (2016a). Clinical trials for BET inhibitors run ahead of the science. Drug Discov. Today. Technol 19, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrieu G, Tran AH, Strissel KJ, and Denis GV (2016b). BRD4 regulates breast cancer dissemination through Jagged1/Notch1 signaling. Cancer Res. 76, 6555–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, and Brut-kiewicz RR (1995). CD1 recognition by mouse NK1+ T lymphocytes. Science 268, 863–865. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, Clevers H, and Lowe SW (2014). Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 8, 1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KL, Zhang F, Jaganathan A, Sharma R, Zhang Q, Konuma T, Shen T, Lee JY, Ren C, Chen CH, et al. (2017). Distinct roles of Brd2 and Brd4 in potentiating the transcriptional program for Th17 cell differentiation. Mol. Cell 65, 1068–1080.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeney JT, Belkina AC, Shirihai OS, Corkey BE, and Denis GV (2016). BET bromodomain proteins Brd2, Brd3 and Brd4 selectively regulate metabolic pathways in the pancreatic β-cell. PLoS One 11, e0151329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ 3rd, and Singer DS (2012). BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc. Natl. Acad. Sci. USA 109, 6927–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, Dey A, Ozato K, and Singer DS (2016a). BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol 23, 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Gegonne A, and Singer DS (2016b). Bromodomain 4: a cellular Swiss army knife. J. Leukoc. Biol 100, 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, and Ozato K (2000). A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol 20, 6537–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, McNally J, and Ozato K (2009). Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell 20, 4899–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose M, Khan I, Guo Z, Kovalovsky D, Krueger A, von Boehmer H, Khazaie K, and Gounari F (2006). c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood 108, 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Hattori M, Qin J, Nakatani Y, Minato N, and Ozato K (2004). Bromodomain protein Brd4 binds to GTPase-activating SPA-1, modulating its activity and subcellular localization. Mol. Cell. Biol 24, 9059–9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, and Print CG (2002). VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 34, 251–256. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, and Sakaguchi S (2003). Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061. [DOI] [PubMed] [Google Scholar]
- Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, and Beddington RS (2002). Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22, 3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, Sciortino S, Shuman J, Hurwitz J, and Ozato K (2002). A mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol 22, 6509–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele DA, Salmeron A, Ghosh S, Huang HR, Bryant BM, and Lora JM (2013). BET bromodomain inhibition suppresses TH17-mediated pathology. J. Exp. Med 210, 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, et al. ; Immunological Genome Consortium (2013). The transcriptional landscape of ab T cell differentiation. Nat. Immunol 14, 619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, and Ozato K (2008). The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem 283, 9040–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TC, Etxaniz U, Dall’Agnese A, Wu SY, Chiang CM, Brennan PE, Wood MJA, and Puri PL (2017). BRD3 and BRD4 BET bromodomain proteins differentially regulate skeletal myogenesis. Sci. Rep 7, 6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Ungerbäck J, and Champhekar A (2016). Forging T-lymphocyte identity: intersecting networks of transcriptional control. Adv. Immunol 129, 109–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg S, and van den Heuvel S (2016). Coordinating cell proliferation and differentiation: antagonism between cell cycle regulators and cell typespecific gene expression. Cell Cycle 15, 196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SF, Larsen BD, Loft A, Nielsen R, Madsen JG, and Mandrup S (2015). Acute TNF-induced repression of cell identity genes is mediated by NFĸB-directed redistribution of cofactors from super-enhancers. Genome Res. 25, 1281–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama Y, Shores EW, and Singer A (1992). Negative selection of precursor thymocytes before their differentiation into CD4+CD8+ cells. Science 258, 653–656. [DOI] [PubMed] [Google Scholar]
- Vacchio MS, Ciucci T, and Bosselut R (2016). 200 million thymocytes and I: a beginner’s survival guide to T cell development. Methods Mol. Biol 1323, 3–21. [DOI] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Villarreal LP (1991). Relationship of eukaryotic DNA replication to committed gene expression: general theory for gene control. Microbiol. Rev 55, 512–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Armato MA, and Yankee TM (2011). Immature single-positive CD8+ thymocytes represent the transition from Notch-dependent to Notch-independent T-cell development. Int. Immunol 23, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Li Q, Wu C, Kim J, Ottinger M, and Howley PM (2009). Regulation of aurora B expression by the bromodomain protein Brd4. Mol. Cell. Biol 29, 5094–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Erman B, Park JH, Feigenbaum L, and Singer A (2004). IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORgammat: impact on thymocyte development. J. Exp. Med 200, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, and Kronenberg M (2009). Carbohydrate specificity of the recognition of diverse glycolipids by natural killer T cells. Immunol. Rev 230, 188–200. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. (2011). RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.