Abstract
SNARE proteins constitute the core of the exocytotic membrane fusion machinery. Fusion occurs when vesicle-associated and target membrane-associated SNAREs zipper into trans-SNARE complexes (“SNAREpins”), but the number required is controversial and the mechanism of cooperative fusion poorly understood. We developed a highly coarse-grained molecular dynamics simulation to access the long fusion timescales, which revealed a two-stage process. First, zippering energy was dissipated and cooperative entropic forces assembled the SNAREpins into a ring; second, entropic forces expanded the ring, pressing membranes together and catalyzing fusion. We predict that any number of SNAREs fuses membranes, but fusion is faster with more SNAREs.
Introduction
Membrane fusion during exocytosis, trafficking and other fundamental cellular processes is mediated by a cellular fusion machinery whose core consists of the SNARE proteins [1–3]. During exocytosis, this machinery fuses the vesicle and target phospholipid membranes, creating a fusion pore that allows release of vesicle contents [4, 5]. To facilitate precisely timed release of neurotransmitters (NTs) at neuronal synapses, the demands on the machinery at pre-synaptic axon terminals are particularly severe. The machinery must respond to action potential-evoked influx of Ca2+ through voltage-gated Ca channels and fuse the synaptic vesicle and presynaptic plasma membranes, all on sub millisecond timescales [6, 7]. Synaptotagmin is the Ca2+sensor [8, 9], while other components include complexin, which clamps spontaneous and asynchronous release but activates synchronous release [10, 11], and Munc13 and Munc18 which play roles in vesicle docking and priming and SNARE complex assembly [12].
The actual membrane fusion step is thought to be executed by the core SNARE machinery, when the SNARE domain of the vesicle-associated v-SNARE synaptobrevin forms a coiled-coil complex (“SNAREpin”) with the plasma-membrane associated t-SNAREs SNAP25 and syntaxin [1]. Zippering of the v-SNARE into the binary SNAP-25/syntaxin t-SNARE acceptor complex [13] pulls the membranes into close proximity and catalyzes their fusion, but the mechanism remains poorly understood. Several SNAREpins are required for fusion, but how the SNAREpins cooperate is unknown and the reported number required varies widely depending on the experimental study [14–22].
A common view is that some or all of the ~ 65 kT of zippering energy [23] is transduced into membrane energy sufficient to fuse the membranes. In this picture, the number of SNAREs required for fusion would depend on a comparison of the energy needed to fuse the membranes, with the amount of zippering energy released. However, no such transduction mechanism is established. One model is that the linker domains (LDs) connecting the SNARE motifs to the transmembrane domains (TMDs) are stiff, storing zippering energy as LD bending energy that could deform and fuse membranes [24, 25]. However, as this mechanism would presumably be abolished by any flexibility in the LD, it appears inconsistent with experiments showing, by contrast, a progressive reduction in fusion when flexible residues of increasing length are introduced into the LDs [26–28].
A second possibility is that the zippering energy is simply dissipated [29]. Indeed, the expected SNARE complexation time of s [23, 30, 31] is much less than fusion timescales, suggesting zippering completes well before fusion. How then do the SNAREs accomplish fusion? In a recent simulation study, we showed that the nanoscale confinement of the membranes imposed by full SNARE zippering is sufficient in itself to catalyze fusion, driven by entropic forces among SNAREpins and membranes [29]. Experiment and mathematical modeling also suggest a post-fusion role for such entropic forces, whereby SNAREs regulate the size of the fusion pore and its release kinetics [32].
Here we develop a molecular dynamics (MD) simulation of SNARE-mediated fusion at the neuronal synapse, extending our previous study of equilibrium properties [29]. Implementing a real dynamics, we could directly measure fusion times from the waiting times for a spontaneous fluctuation in membrane energy to reach an assumed fusion threshold. Our highly coarse-grained (CG) representation preserves principal biophysical properties of the SNARES while enabling simulations to access the long timescale collective effects of fusion, a major obstacle for all-atom or moderately CG approaches [33–35]. Our simulations, representing the final stages of evoked fusion when unclamped SNAREs are left unfettered to execute fusion, revealed a 2-stage process: (1) SNAREs became fully zippered and were spontaneously organized into a ring at the fusion site by entropic forces; (2) entropic forces expanded the ring and pulled the membranes into nanoscale proximity so that fusion was rapidly triggered by a thermal membrane energy fluctuation that reached the fusion threshold, recently measured experimentally [36]. With more SNAREs at the fusion site entropic forces were greater, so that fusion was faster; 16 SNAREpins were required to attain of order the millisecond timescales of neurotransmitter release.
Results
Model
We developed a real time MD simulation of neuronal SNAREpins bridging a 40 nm diameter vesicle and a planar target membrane (Fig. 1A, B). To capture the collective behavior of many SNARE complexes over long fusion timescales required radical coarse-graining [29]. For model details, see Supplementary Material.
Fig. 1.
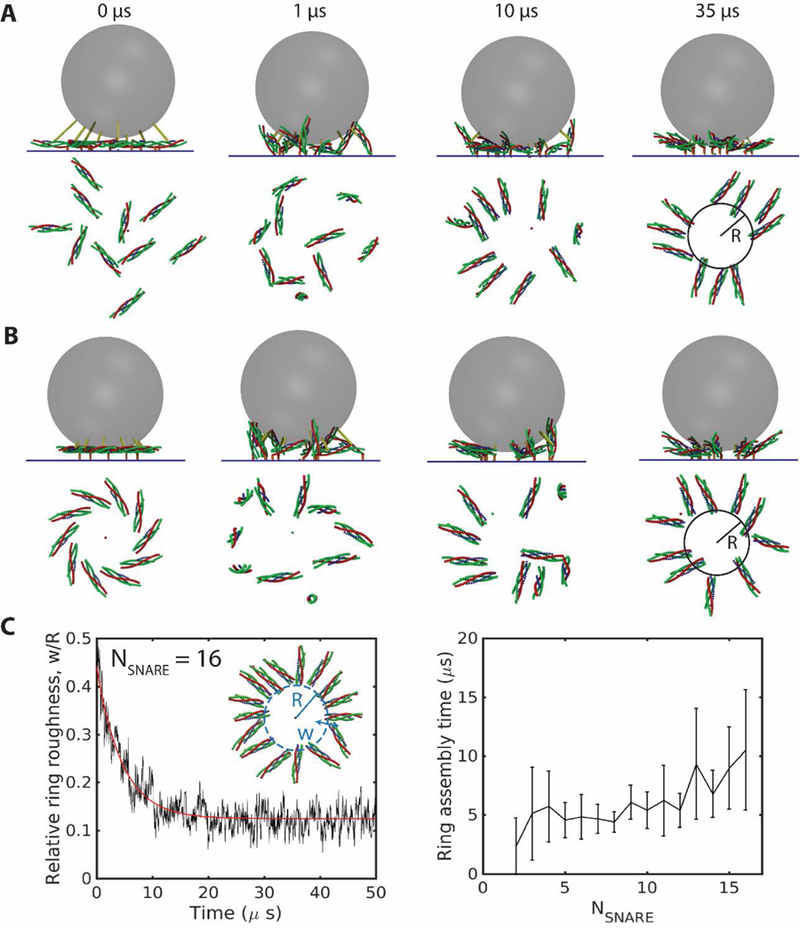
SNAREpins spontaneously assemble into a ring. Simulations of the model described in text, parameters as in Tables S1, S2. (A), (B) Snapshots of simulations with SNARE complexes bridging a synaptic vesicle membrane and the plasma membrane. Side views (top) and top views of the SNAREs (bottom) at the indicated times. From either a random, half-zippered initial condition (A), or a plausible ordered pre-fusion initial configuration, (B), the SNAREpins become fully zippered and self-assemble into a ring. (C) Relative inner ring roughness versus time for 16 SNAREpins starting from a random initial condition (left). The relative roughness decreases (red curve is best fit exponential, with time constant 4.5 µs) to a minimum of ~ 0.12 in the fully assembled ring. Ring assembly times from exponential fits versus number of SNAREs (right). More SNAREpins need longer to assemble. Error bars are standard deviation, n=20.
Coarse-grained representation of SNAREs.
The SNARE complex is a 16-layer coiled coil of two α-helices from SNAP25, one from VAMP, and one from syntaxin [37, 38]. Our model represents each α-helix by 16 beads, each bead representing four structured residues belonging to one layer. A bead carries the total charge of its residues (table S2), uniformly distributed over its surface. The t-SNARE motifs are structured [3, 13] while the VAMP motif can zipper/unzip freely. The uncomplexed segments of VAMP and syntaxin, connecting the complexed segments to the vesicle and target membrane, respectively, are assumed unstructured and represented as wormlike chains [23].
Interactions.
On a bead-by-bead basis, SNAREpins interact with other SNAREpins and the membranes electrostatically (Table S2), and sterically via repulsive Lennard-Jones potentials. Unstructured segments exert tensile force on the last structured bead (closest to the C-terminus). The membranes are continuous, uniformly charged surfaces. The membrane energy is the total interaction energy between vesicle and target membranes. Here is the van der Waals energy determined using experimentally measured lipid bilayer Hamaker constants [39], the electrostatic energy due to the negatively charged lipids, and is the work done by short-ranged hydration forces when the membrane separation at the point of closest approach is [40]. Here and are determined using composition weighted averages of measurements in pure lipid species [39, 40]. All forces are determined by taking the gradient of the corresponding energy.
Molecular dynamics.
SNAREpins translate with velocity and rotate with angular velocity about their center of mass at R, determined by the total force and torque on the constituent beads,
where is the scalar translational drag coefficient of the SNAREpin, the rotational drag matrix of the SNAREpin, the total force due to interactions on bead and is the random thermal force on bead obeying the fluctuation dissipation theorem, The drag coefficient of a bead obeys the Stokes law, where is the viscosity of water and nm the bead radius (table S1).
The vesicle translates with velocity following the force balance
where is the vesicle drag coefficient, the total force on the vesicle due to interactions, and the thermal noise force. In simulations, the translational and rotational velocities are determined at each step, and the positions and orientations of all components are then updated by Euler’s method.
Zippering dynamics of SNARE complexes.
VAMP zippers (or unzips) stochastically in a sequence of layer-by-layer events, each converting four unstructured VAMP residues into one VAMP bead in the SNARE complex (or the reverse). The rates are and [41], where is a representative rate for assembly or disassembly of α-helices [23, 30]. The change in energy when one additional bead zippers, equals the change in energy due to the binding of those VAMP residues into the SNARE complex, plus the change in the worm-like chain energy of the uncomplexed segment of VAMP. The binding energies obey the measured zippering energy landscape [23].
Calculation of fusion times.
Fusion occurs when a fluctuation of the total membrane energy first reaches the fusion energy threshold, which we set to consistent with experiment [36]. Since the waiting time for a fluctuation of this magnitude was beyond our computer resources, we instead used a range of lower and computationally accessible hypothetical values of We found that the waiting times depended exponentially on , as expected since large energy fluctuations are rare. We then extrapolated to the experimental value of
Initial configuration of SNARE complexes.
In simulations, the SNARE complexes were initially half-zippered (i.e., VAMP assembled into the SNARE complex from the N terminus to the ionic layer [42, 43]). We tested two initial configurations: (1) A disordered configuration, with randomly oriented SNAREpins (Fig. 1A), and (2) an ordered ring-like configuration, representing a plausible arrangement immediately following unclamping of the SNAREs after injection of Ca2+ (Fig. 1B). We refer to the latter as the “unclamped” configuration, motivated by a hypothesis that a ring-like Synaptotagmin oligomer clamps fusion by separating the vesicle and plasma membranes, and is disassembled when [Ca2+] is elevated [44]. Accordingly, we arranged the SNAREpins in a ring with positions and orientations reproducing those in a recent synaptotagmin-SNAREpin chimera crystal structure [45].
Fig. 2.
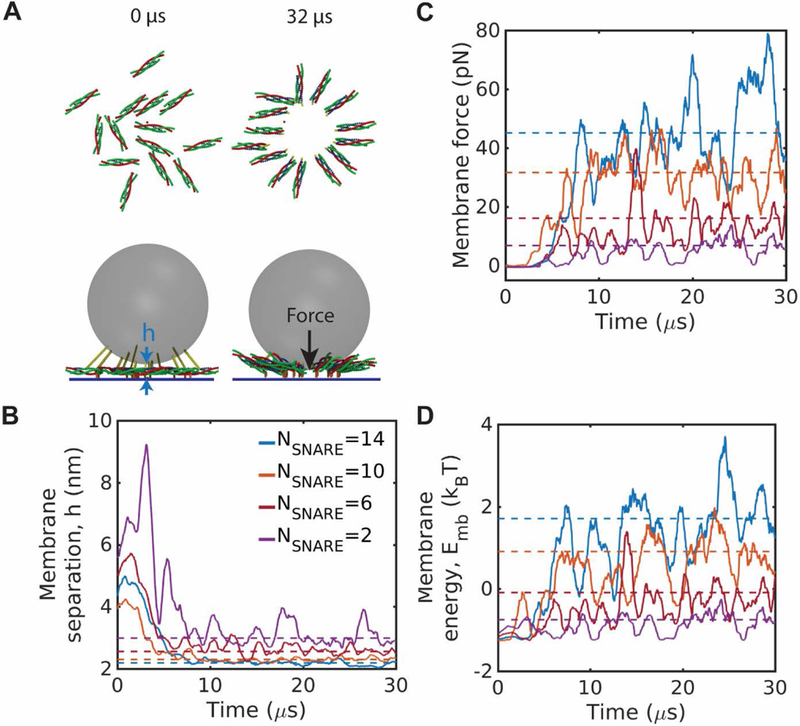
Spontaneous assembly and expansion of SNAREpin rings presses membranes together and increases membrane energy. (A) Snapshots from a simulation with 14 SNAREpins. Starting from a random initial condition as in Fig. 1A, the SNAREpins assemble into a ring that is expanded, pulling the membranes into close proximity due to vesicle curvature. (B)-(D) Time-dependence of membrane separation at the point of closest approach, (B), force on the membranes, (C), and membrane energy, (D), as rings assemble from random initial conditions. Ring assembly presses the membranes together and increases their energy. Dotted lines show mean values during the final 15 µs. Time plots are coarse-grained over 100 ns.
SNAREpins spontaneously zipper and assemble into a ring
We performed simulations with varying numbers of SNAREpins . For any initial configuration and number of SNAREpins, after a few μs the SNAREpins self-assembled into a ring centered on the point of closest approach of the vesicle and target membranes. Typical assembly sequences of 10 SNAREpins from a disordered initial configuration or an ordered unclamped initial configuration are shown in Figs. 1A and 1B, respectively. Since the SNAREpins were initially half-zippered, they first pitched upward, but as zippering proceeded and the uncomplexed VAMP segment shortened, the C-terminal ends of the SNAREs were pulled inwards and in the final state formed an inner ring of radius , with the N-terminal ends pointing radially outwards.
To quantify the assembly process, we monitored the time evolution of the inner ring roughness, the root mean square deviation of the radius from its mean value , averaged over the SNAREpins of the ring. The relative roughness, , decreased to a minimum in the final steady state ring, Fig. 1C. Thus, the organization became increasingly regular over time. Defining the ring assembly time as the time constant for the decay of roughness, from best fit exponentials the assembly time increased moderately with number of SNAREpins, from for two SNAREpins to ~11 µs for 16 SNAREpins.
Assembly of the ring from these initial conditions, in which all SNAREpins were half-zippered, was accompanied by complete zippering of the SNARE motifs up to layer +8. Zippering occurred in a few μs. For example, for SNAREpins, the last SNAREpin zippered after 19±6 µs (n=20 runs). The process was irreversible: once zippered, small zippering/unzipping fluctuations occurred of mean magnitude less than one bead. Thus, the zippering energy is rapidly dissipated.
Assembly of SNAREpin rings presses membranes together and increases membrane energy
As the SNAREpin ring assembled and the SNARE motifs became fully zippered, the vesicle and target membranes were pulled into close proximity over ~5–10 µs (Fig. 2A). Membrane separation at the point of closest approach, , decreased to a steady state value of ~2–3 nm depending on the number of SNAREpins (Fig. 2B), and the membrane energy increased substantially as the membranes were drawn closer (Fig. 2D). The force pressing the membranes together increased to significant values, e.g. ~45 pN for 14 SNAREpins (Fig. 2C).
Entropic forces expand the SNAREpin ring and pull membranes together
The mechanism that pressed the membranes together and increased their energy was ring expansion, driven by entropic forces among the SNAREpins and between SNAREpins and membranes. The SNAREpins and membranes interact via short range steric-electrostatic forces that prohibit overlap. Thus, more configurations are available in a ring of larger radius, where the SNAREpins are more spaced and can orient in more polar directions. In other words, the ring tends to expand because the entropy is then greater. Due to the vesicle curvature, expansion of the ring pulls the membranes closer together with a certain force and increases their energy.
To explicitly illustrate the entropic force effect, we performed simulations with an artificial initial condition with fully zippered SNAREpins in a small ring of inner radius R = 4 nm. Due to repeated collisions of the SNAREpins with each other and with the membranes, the SNAREpins moved radially outward and after a time of order a microsecond the ring had expanded to a greater steady state radius (Fig. 3A,B). Simultaneous with this entropically driven ring expansion, the membrane separation decreased and the membrane force and energy increased (Fig. 3B). The final equilibrium radius is determined by a force balance: the mean force that presses the membranes together due to SNAREpin ring expansion is then equal to the mean opposing force, dominated by the hydration force. (Equivalently, the entropic free energy decrease due to ring expansion plus the membrane-membrane energy increase is a minimum.)
Fig. 3.
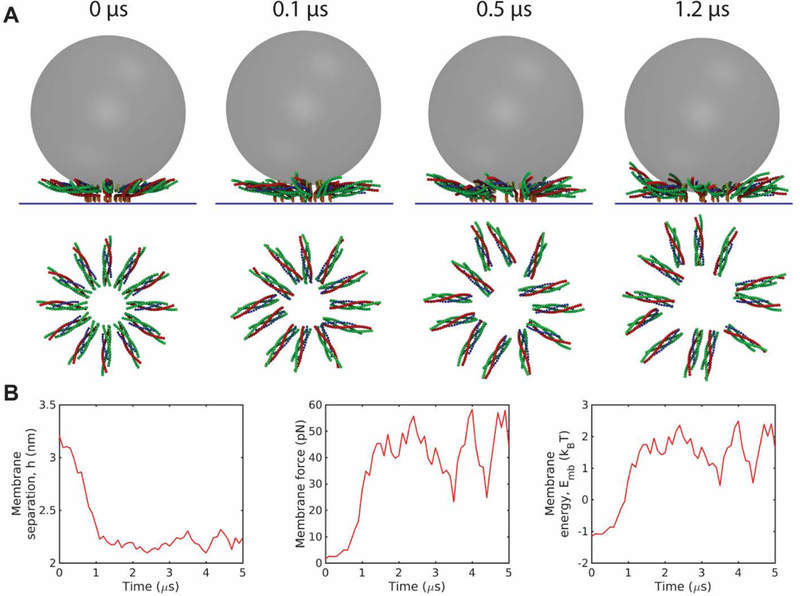
Entropic forces expand the SNAREpin ring and pull the membranes together. (A) Snapshots of a simulation starting from an initial condition with 12 fully zippered SNAREpins in a ring of atypically small inner radius, 4 nm. Entropic forces among the SNAREpins and membranes rapidly expand the ring to a radius of ~ 10 nm. (B) Time-dependence of membrane separation, force and energy during the simulation of (A). The rapid ring expansion forces the membranes together, boosting the force on the membranes and their energy in 1–2 μs. Time plots are coarse-grained over 100 ns.
With more SNAREpins, assembled rings are larger, more ordered and press membranes closer together
Once the SNAREpins were fully assembled into a ring, all SNARE motifs were fully zippered, and the relative ring roughness had a steady-state value of ~12–15% depending on the number of SNAREs (Fig. 4A,B), compared to ~30–50% in the initial condition (Fig. 1C). With more SNAREpins at the fusion site, entropic forces are expected to be greater, so that assembled rings should be larger. This expectation was confirmed by typical snapshots of the ring after assembly, shown for = 4, 10 and 16 in Fig. 4A. Assembled rings with more SNAREpins were indeed larger: the inner ring radius increased from nm for one SNAREpin to nm for 16 SNAREpins (Fig. 4B). Once a SNAREpin ring was assembled, it fluctuated about a steady state. Even with very few SNAREpins fluctuations about a perfect ring were small, as measured by the relative roughness of the inner ring (Fig. 4B).
Fig. 4.
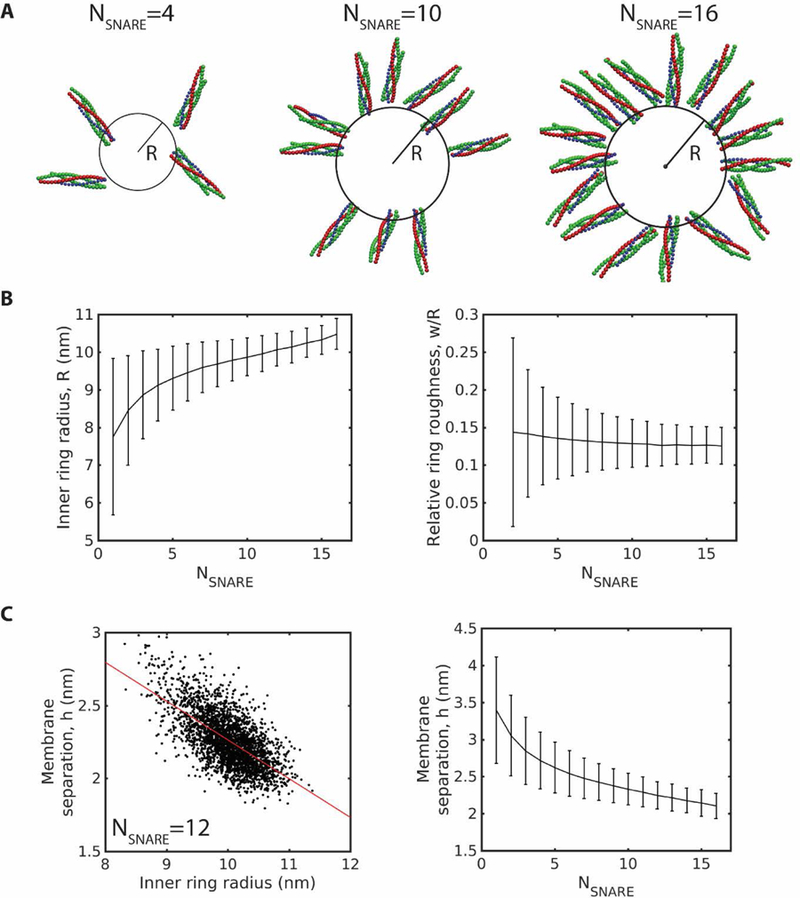
With more SNAREpins, assembled rings are larger, more ordered, and press membranes closer together with higher energies. Mean values are obtained by time averaging over 60 µs. Error bars: SD over the same time frame. (A) Snapshots of typical simulated rings with 4, 10, and 16 SNAREpins after reaching steady state. (B) Steady state ring radius and relative roughness versus number of SNAREpins. (C) Scatter plot of instantaneous membrane separation and inner ring radius for a steady state ring of 12 SNAREpins reveals a negative correlation left. Red line: linear fit, . Membrane separation versus number of SNAREpins in steady state rings, right.
With more SNAREpins, the membranes were pulled closer together (Fig. 4C), as expected from the larger rings. The geometric relationship between ring size and membrane separation was also apparent within a single ring, where fluctuations in membrane separation were strongly negatively correlated with fluctuations in ring radius (Fig. 4C).
With more SNAREpins, fluctuations in membrane energy are larger
Once the SNAREpin ring was assembled and equilibrated, the membrane energy exhibited large fluctuations. With more SNAREpins, the fluctuations increased dramatically while the mean energy increased only moderately. The sensitivity of fluctuations to the number of SNAREs was reflected by the maximum energy reached during a given time interval. For example, in one set of simulations, during a 50 µs time interval the peak membrane energy for assembled rings of and 16 SNAREpins was and , respectively (Fig. 5A). The respective mean energies were and 2.6 .
Fig. 5.
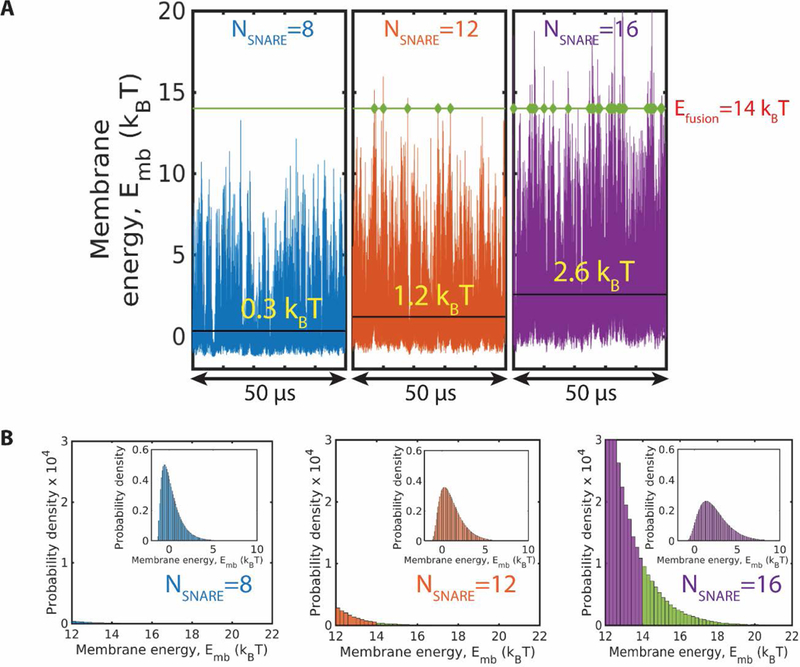
With more SNAREpins, membrane energy fluctuations are greater and the fusion energy threshold is reached sooner. (A) Membrane energy versus time in three simulations with 8, 12 and 16 SNAREpins, respectively. For a hypothetical fusion energy of (green line), the instants when the fluctuating energy attains the fusion energy value are indicated. With more SNAREpins, energy fluctuations are greater and the fusion threshold is reached more frequently. Mean membrane energy values are indicated (black lines). (B) Tails of the membrane energy probability distributions for the 3 simulations of (A). Insets show the full distributions. The weighting for energies above the fusion threshold (shown green) increases with the number of SNAREpins.
An important consequence of the larger energy fluctuations with more SNAREs present is that waiting times to attain a given energy for the first time are smaller. Consider, for example, a hypothetical value of the fusion energy threshold, . With 16 SNAREpins, this energy is reached frequently, once every ~2 µs; with 12 SNAREpins once every ~17 µs; and with 8 SNAREpins, the energy was never attained during a simulation of 50 µs (Fig. 5A).
The reduced waiting times for more SNAREpins was reflected in the probability distributions of membrane energies. With more SNAREpins, energy distributions were broader, having more weighting for high energies (Fig. 5B). The total probability for energies exceeding the threshold of was , and for 16, 12 and 8 SNAREpins, respectively.
High energies are achieved rarely and require long wait times
The time for membrane fusion to occur is the waiting time for an energy fluctuation to reach the fusion threshold. We first tested a range of hypothetical values of the fusion threshold . Fig. 6A shows the fluctuations in the membrane energy for a ring of 16 SNAREpins over 100 µs. For fusion thresholds of and the waiting times to reach the threshold for the first time were 2.1 µs, 6.8 µs and 20.1 µs, respectively. Thus, the waiting time increases rapidly with increasing energy threshold.
Fig. 6.
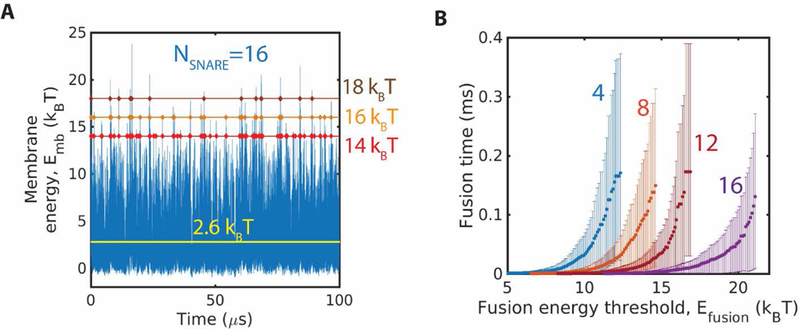
Waiting times for membrane energy fluctuations to attain a threshold value increase with the energy value and decrease with number of SNAREs. (A) Membrane energy versus time in a simulation with 16 SNAREpins. Three hypothetical fusion energy thresholds are indicated. Energy fluctuations reach 14 once every ~2 , 16 once every ~7 , and 18 once every ~20 , The mean energy is indicated (black line). (B) Fusion time versus hypothetical value of the fusion energy threshold, , for four values of the number of SNAREs, . The fusion time is the mean waiting time for the first instant that an energy fluctuation reaches . Fusion time increases with and . Error bars are 95% confidence intervals.
Next, we used our simulations to measure hypothetical fusion times (i.e. waiting times) versus the hypothetical fusion threshold energy and the number of SNAREpins in the ring. The fusion time increased dramatically with fusion energy threshold and decreased dramatically with number of SNAREpins (Fig. 6B). Provided was high enough that the waiting time for fusion was long the mean wait time grew exponentially with . Least squares exponential fits were good for every number of SNAREpins (Fig. 7A).
Fig. 7.
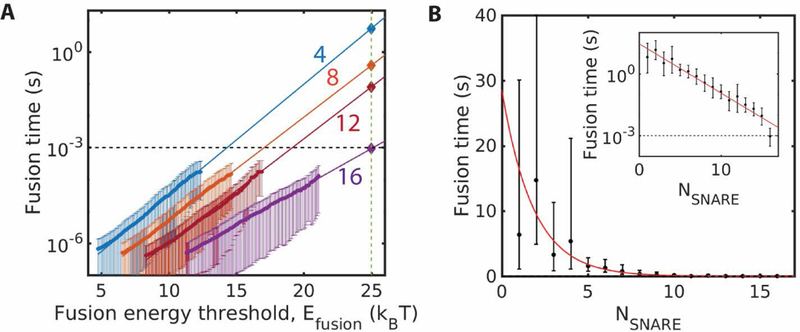
Any number of SNAREs can catalyze fusion, but fusion is faster with more SNAREs. Error bars are 95% confidence intervals from least-squares fits. (A) Simulated fusion times versus hypothetical fusion energy threshold for four values of the number of SNAREs. Exponential fits to the measured data are extrapolated to the experimental fusion energy, to obtain the predicted fusion times. (B) Predicted fusion times versus number of SNAREs, using the procedure of (A). Red curve: best fit exponential
Calculation of fusion times
To calculate the actual fusion time requires using the actual experimental value of the fusion threshold, which we take to be However, since our simulations could not be run for long enough to realize such high membrane energies, we used our exponential fits to extrapolate the measured waiting times for lower values of (Fig. 6B) to the experimental value.
The extrapolation procedure is shown in Fig. 7A for four values of the number of SNAREpins. The predicted fusion times depend strongly on the number of SNAREs. For 4 SNAREpins, the fusion time exceeds 1s, while 16 SNAREpins are predicted to catalyze fusion after just ms (95% confidence interval: 0.5–2.3 ms).
Repeating this procedure for many values of , the predicted mean fusion times are plotted in Fig. 7B for . The data is approximately fit by an exponential relation,
where and .
Discussion
Computational modeling of the cellular membrane fusion apparatus is a daunting challenge, because many proteins act collectively on extremely long timescales in computer simulation terms. Simulating even the core SNARE machinery alone is extremely difficult, as many SNARE complexes likely act in concert over timescales 1 ms or greater. All-atom and somewhat coarse-grained computational approaches continue to make profound contributions to the understanding of these phenomena [33, 35, 46]. However, at present to capture the collective behavior of many SNAREs over ~ 1 ms timescales requires even more coarse-grained representations.
Here we addressed this challenge by developing a molecular dynamics description with SNAREs systematically but radically coarse-grained, to such a degree that we could simulate the collective behavior of up to 20 SNAREpins on timescales up to ~ 100 μs. The results are consistent with our earlier study of the long time equilibrated behavior of the SNAREpins using a Monte Carlo approach which can only describe equilibrium [29]. There we estimated fusion times using a scaling argument for the timescales and by fitting to an in vitro measurement of fusion time [20]. The present model follows events in time, without fitting parameters. Thus we could track the spontaneous assembly of the SNAREpin ring in time, and measure waiting times for fluctuations in membrane energies to reach a fusion threshold whose value is taken from experiment [36].
SNARE-mediated fusion is a two-stage process: ring assembly followed by membrane confinement
Simulations revealed two consecutive episodes leading to membrane fusion. (1) Regardless of the initial arrangement, over ~ 10 µs the SNAREpins became fully zippered and spontaneously organized themselves into a ring centered on the fusion site with radially-oriented SNAREpins (Fig. 1). Ring assembly occurred spontaneously, with no energetic barrier. The assembled ring is expanded, clearing a central zone where the membranes will meet and fuse (Figs. 1, 3). (2) In the second phase, the tendency of the assembled ring to remain expanded presses the membranes into nanoscale proximity (Fig. 2). In microscopic terms this is a lengthy phase: the membranes remain confined, and after a waiting time ranging from ~10s (for one SNAREpin) to ~ 1 ms (for 16 SNAREpins) a thermal fluctuation provides sufficient energy to fuse the vesicle and target membranes (Figs. 5–7).
Entropic forces drive SNARE-mediated fusion
Entropic forces, intrinsically cooperative in nature, drive the spontaneous processes (1) and (2) above. SNAREpins are bulky, ~10 nm-long complexes that have trouble fitting together into a small space. Entropically, several SNAREpins are much happier outwardly oriented in a ring configuration, similarly to the behavior of rod-like molecules in disclination defects in nematic liquid crystals [47]. Entropic forces also drive ring expansion since an expanded ring affords the SNAREpins more orientational and translational entropy (Fig. 3), which in turn presses the membranes together with forces of up to ~ 50 pN for larger numbers of SNAREpins (Fig. 2). The proximity of the membranes, and the forces pressing them together, elevate the frequency of high membrane energy fluctuations and thereby provoke their fusion. Thus, our model suggests that the cooperativity among SNAREs that fuses membranes is entropic in nature.
Any number of SNAREs can catalyze fusion, but fusion is faster with more SNAREs
The fusion mechanism emerging from our model differs fundamentally from a common view, that the SNARE complexation energy is somehow pumped into the membranes to drive fusion. This view leads to an expectation of a requirement on the number of SNAREs for fusion, given a fusion energy threshold . In our simulations, the complexation energy is completely dissipated long before fusion; this is hardly surprising, given a collection of partially complexed trans SNAREpins, as there is nothing to prevent irreversible energetically downhill zippering on μs timescales.
We find the energy for fusion is instead provided by thermal fluctuation, assisted by entropically generated force that presses the membranes together. The waiting time for a sufficient fluctuation is lower for more SNAREpins, as the entropic forces are then greater. Hence, more SNAREs fuse membranes faster (Fig. 7), consistent with the large spread in reported SNARE requirements for fusion [14–22]. Given that any number can catalyze fusion, different apparent requirements may emerge from different experimental methods depending on the timescale probed [29]. Further, these conclusions suggest that, in the interests of efficiency, the number of SNAREpins used at synaptic terminals to achieve membrane fusion may equal the minimum number required to achieve the submillisecond timescales necessary for proper neurotransmission.
Is it plausible that a spontaneous thermal fluctuation generates the energy for fusion? Indeed, our analysis demonstrates this. (1) Without any fitting parameters, for 16 SNAREpins MD simulations generated fusion times of order 1 ms, in the physiological range (Fig. 7). That is, waiting times for a sufficient energy fluctuation are consistent with physiological fusion times. (2) For larger numbers of SNAREpins, forces of ~ 50 pN press the membranes together (Fig. 2C). Acting over a scale of 1 nm (of order the membrane separation) such a force performs ~ 12 of work, sufficient to substantially lower the fusion barrier, [36].
We stress that in the fusion mechanism emerging from this study the zippering energy, though dissipated, is far from wasted. Zippering serves the vital purpose of bringing and maintaining the membranes into very close proximity; helped by SNAREpin expansion, it is this proximity which is the mechanism of fusion. A closely related function of zippering is to create the ~ 10 nm long rigid SNARE complexes that interact entropically. Thus, the upstream origin of the entropic forces that drive fusion is zippering, because zippering produces the bulky cylindrically-shaped SNARE complexes, and crowds them into a very small space by pulling the membranes close together.
Our objective here is to access long timescales to study the essentials of collective SNARE-mediated fusion, and accordingly the model adopts a number of simplifications. As membrane phospholipids are not represented, it cannot represent the fusion process itself, which may involve hemifused or other intermediates [48–52]. Instead, fusion is indicated when the membrane energy attains the fusion threshold. Since TMDs may perturb bilayer packing [35, 53, 54] and help nucleate fusion, we view the predicted fusion rates as lower bounds, and our prediction of a 16 SNAREpin requirement for ~ 1 ms fusion can be viewed as an upper bound.
Ca-evoked membrane fusion in cells is a three-stage process
Our model can be thought of in two contexts. (1) It describes in vitro measurements of fusion mediated by SNAREs alone, for example assays measuring fusion between small unilamellar vesicles and supported bilayers [20, 29, 55]. Since no machinery is present that might hold back full zippering of the SNAREpins before fusion, we expect the basic assumptions of our model are valid. (2) It describes the final stage of evoked release in cells, if one assumes that, following Ca-evoked unclamping, the SNAREs are ultimately left unhindered to execute fusion. Whether or not this is true is unknown. If so, full zippering presumably occurs within microseconds, leaving fully zippered SNAREpins to execute fusion (possibly assisted by other factors affecting the membranes, such as Syt-mediated remodeling [56] or Ca-mediated adhesion and increase of tension and interleaflet tension [50, 51, 57]). Half-zippered pre-fusion states have been proposed, for example [43], but as yet no mechanism is established that could transduce energy from the final zippering episode (half- to fully-zippered) into membrane fusion energy. Whether a machinery exists that couples zippering energy into membrane disruption and fusion remains an exciting question. Until that question is settled, the simplest assumption is that no such mechanism exists, the assumption implicit in our model. Indeed, our model suggests that no such mechanism is needed, since it predicts ~ 1ms fusion is achieved with sufficiently many SNAREs acting on their own.
In conclusion, we suggest that Ca-evoked membrane fusion proceeds in three stages at neuronal synapses. First, as is well accepted, the fusion machinery senses Ca and unclamps the SNARE complexes, mediated by Syt and other components [8, 9, 12, 56, 58]. We propose that the last two stages are then driven by unhindered SNAREs, as described above: full zippering and entropically-driven SNAREpin ring assembly, followed by entropically-driven membrane confinement and fusion.
Supplementary Material
Acknowledgements
This work was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM117046. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We acknowledge computing resources from Columbia University’s Shared Research Computing Facility project. We thank Sathish Thiyagarajan for helpful discussions.
References
- 1.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P & Rothman JE (1993) SNAP receptors implicated in vesicle targeting and fusion, Nature 362, 318–24. [DOI] [PubMed] [Google Scholar]
- 2.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH & Rothman JE (1998) SNAREpins: minimal machinery for membrane fusion, Cell 92, 759–72. [DOI] [PubMed] [Google Scholar]
- 3.Jahn R & Scheller RH (2006) SNAREs - engines for membrane fusion, Nature Reviews Molecular Cell Biology 7, 631–643. [DOI] [PubMed] [Google Scholar]
- 4.Chandler DE & Heuser JE (1980) Arrest of membrane fusion events in mast cells by quick-freezing, J Cell Biol 86, 666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindau M & Alvarez de Toledo G (2003) The fusion pore, Biochim Biophys Acta 1641, 167–73. [DOI] [PubMed] [Google Scholar]
- 6.Sabatini BL & Regehr WG (1996) Timing of neurotransmission at fast synapses in the mammalian brain, Nature 384, 170–2. [DOI] [PubMed] [Google Scholar]
- 7.Sabatini BL & Regehr WG (1999) Timing of synaptic transmission, Annu Rev Physiol 61, 521–42. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Chacon R, Konigstorfer A, Gerber SH, Garcia J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C & Sudhof TC (2001) Synaptotagmin I functions as a calcium regulator of release probability, Nature 410, 41–9. [DOI] [PubMed] [Google Scholar]
- 9.Sorensen JB, Fernandez-Chacon R, Sudhof TC & Neher E (2003) Examining synaptotagmin 1 function in dense core vesicle exocytosis under direct control of Ca2+, J Gen Physiol 122, 265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraudo CG, Eng WS, Melia TJ & Rothman JE (2006) A clamping mechanism involved in SNARE-dependent exocytosis, Science 313, 676–80. [DOI] [PubMed] [Google Scholar]
- 11.Tang J, Maximov A, Shin OH, Dai H, Rizo J & Sudhof TC (2006) A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis, Cell 126, 1175–87. [DOI] [PubMed] [Google Scholar]
- 12.Sudhof TC (2013) Neurotransmitter release: the last millisecond in the life of a synaptic vesicle, Neuron 80, 675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasshauer D & Margittai M (2004) A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly, J Biol Chem 279, 7613–21. [DOI] [PubMed] [Google Scholar]
- 14.Hua Y & Scheller RH (2001) Three SNARE complexes cooperate to mediate membrane fusion, Proc Natl Acad Sci U S A 98, 8065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha R, Ahmed S, Jahn R & Klingauf J (2011) Two synaptobrevin molecules are sufficient for vesicle fusion in central nervous system synapses, Proc Natl Acad Sci U S A 108, 14318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohrmann R, de Wit H, Verhage M, Neher E & Sorensen JB (2010) Fast Vesicle Fusion in Living Cells Requires at Least Three SNARE Complexes, Science 330, 502–505. [DOI] [PubMed] [Google Scholar]
- 17.Acuna C, Guo Q, Burré J, Sharma M, Sun J & Südhof TC (2014) Microsecond Dissection of Neurotransmitter Release: SNARE-Complex Assembly Dictates Speed and Ca2+ Sensitivity, Neuron 82, 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Bogaart G, Holt MG, Bunt G, Riedel D, Wouters FS & Jahn R (2010) One SNARE complex is sufficient for membrane fusion, Nature structural & molecular biology 17, 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi L, Shen QT, Kiel A, Wang J, Wang HW, Melia TJ, Rothman JE & Pincet F (2012) SNARE proteins: one to fuse and three to keep the nascent fusion pore open, Science 335, 1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karatekin E, Di Giovanni J, Iborra C, Coleman J, O’Shaughnessy B, Seagar M & Rothman JE (2010) A fast, single-vesicle fusion assay mimics physiological SNARE requirements, Proc Natl Acad Sci U S A 107, 3517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domanska MK, Kiessling V, Stein A, Fasshauer D & Tamm LK (2009) Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion, J Biol Chem 284, 32158–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez JM, Kreutzberger AJ, Kiessling V, Tamm LK & Jahn R (2014) Variable cooperativity in SNARE-mediated membrane fusion, Proc Natl Acad Sci U S A 111, 12037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, Rothman JE & Zhang Y (2012) Single reconstituted neuronal SNARE complexes zipper in three distinct stages, Science 337, 1340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lentz BR & Lee J (1999) Poly (ethylene glycol)(PEG)-mediated fusion between pure lipid bilayers: a mechanism in common with viral fusion and secretory vesicle release?(Review), Molecular membrane biology 16, 279–296. [DOI] [PubMed] [Google Scholar]
- 25.Knecht V & Grubmuller H (2003) Mechanical coupling via the membrane fusion SNARE protein syntaxin 1A: a molecular dynamics study, Biophys J 84, 1527–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNew JA, Weber T, Engelman DM, Sollner TH & Rothman JE (1999) The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion, Molecular Cell 4, 415–421. [DOI] [PubMed] [Google Scholar]
- 27.Kesavan J, Borisovska M & Bruns D (2007) v-SNARE actions during Ca2+-triggered exocytosis, Cell 131, 351–363. [DOI] [PubMed] [Google Scholar]
- 28.Zhou P, Bacaj T, Yang X, Pang ZP & Sudhof TC (2013) Lipid-anchored SNAREs lacking transmembrane regions fully support membrane fusion during neurotransmitter release, Neuron 80, 470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mostafavi H, Thiyagarajan S, Stratton BS, Karatekin E, Warner JM, Rothman JE & O’Shaughnessy B (2017) Entropic forces drive self-organization and membrane fusion by SNARE proteins, Proc Natl Acad Sci U S A 114, 5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubelka J, Hofrichter J & Eaton WA (2004) The protein folding ‘speed limit’, Curr Opin Struct Biol 14, 76–88. [DOI] [PubMed] [Google Scholar]
- 31.Xi Z, Gao Y, Sirinakis G, Guo H & Zhang Y (2012) Single-molecule observation of helix staggering, sliding, and coiled coil misfolding, Proc Natl Acad Sci U S A 109, 5711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z, Bello OD, Thiyagarajan S, Auclair SM, Vennekate W, Krishnakumar SS, O’Shaughnessy B & Karatekin E (2017) Dilation of fusion pores by crowding of SNARE proteins, Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP & Marrink SJ (2008) The MARTINI Coarse-Grained Force Field: Extension to Proteins, J Chem Theory Comput 4, 819–34. [DOI] [PubMed] [Google Scholar]
- 34.Fortoul N, Singh P, Hui CY, Bykhovskaia M & Jagota A (2015) Coarse-Grained Model of SNARE-Mediated Docking, Biophys J 108, 2258–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risselada HJ, Kutzner C & Grubmuller H (2011) Caught in the act: visualization of SNARE-mediated fusion events in molecular detail, Chembiochem 12, 1049–55. [DOI] [PubMed] [Google Scholar]
- 36.Francois-Martin C, Rothman JE & Pincet F (2017) Low energy cost for optimal speed and control of membrane fusion, Proc Natl Acad Sci U S A 114, 1238–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutton RB, Fasshauer D, Jahn R & Brunger AT (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution, Nature 395, 347–353. [DOI] [PubMed] [Google Scholar]
- 38.Stein A, Weber G, Wahl MC & Jahn R (2009) Helical extension of the neuronal SNARE complex into the membrane, Nature 460, 525–U105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leckband D & Israelachvili J (2001) Intermolecular forces in biology, Q Rev Biophys 34, 105–267. [DOI] [PubMed] [Google Scholar]
- 40.Rand RP & Parsegian VA (1989) Hydration Forces between Phospholipid-Bilayers, Biochimica Et Biophysica Acta 988, 351–376. [Google Scholar]
- 41.Thompson PA, Eaton WA & Hofrichter J (1997) Laser temperature jump study of the helix<==>coil kinetics of an alanine peptide interpreted with a ‘kinetic zipper’ model, Biochemistry 36, 9200–10. [DOI] [PubMed] [Google Scholar]
- 42.Li F, Pincet F, Perez E, Giraudo CG, Tareste D & Rothman JE (2011) Complexin activates and clamps SNAREpins by a common mechanism involving an intermediate energetic state, Nat Struct Mol Biol 18, 941–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F, Kummel D, Coleman J, Reinisch KM, Rothman JE & Pincet F (2014) A half-zippered SNARE complex represents a functional intermediate in membrane fusion, J Am Chem Soc 136, 3456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Bello O, Auclair SM, Wang J, Coleman J, Pincet F, Krishnakumar SS, Sindelar CV & Rothman JE (2014) Calcium sensitive ring-like oligomers formed by synaptotagmin, Proc Natl Acad Sci U S A 111, 13966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q, Lai Y, Bacaj T, Zhao M, Lyubimov AY, Uervirojnangkoorn M, Zeldin OB, Brewster AS, Sauter NK, Cohen AE, Soltis SM, Alonso-Mori R, Chollet M, Lemke HT, Pfuetzner RA, Choi UB, Weis WI, Diao J, Sudhof TC & Brunger AT (2015) Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis, Nature 525, 62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma S & Lindau M (2016) The mystery of the fusion pore, Nat Struct Mol Biol 23, 5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandrasekhar S (1992) Liquid Crystals, 2nd edn, Cambridge University Press. [Google Scholar]
- 48.Hernandez JM, Stein A, Behrmann E, Riedel D, Cypionka A, Farsi Z, Walla PJ, Raunser S & Jahn R (2012) Membrane fusion intermediates via directional and full assembly of the SNARE complex, Science 336, 1581–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diao J, Grob P, Cipriano DJ, Kyoung M, Zhang Y, Shah S, Nguyen A, Padolina M, Srivastava A, Vrljic M, Shah A, Nogales E, Chu S & Brunger AT (2012) Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion, eLife 1, e00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warner JM & O’Shaughnessy B (2012) The hemifused state on the pathway to membrane fusion, Phys Rev Lett 108, 178101. [DOI] [PubMed] [Google Scholar]
- 51.Warner JM & O’Shaughnessy B (2012) Evolution of the hemifused intermediate on the pathway to membrane fusion, Biophys J 103, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozlovsky Y & Kozlov MM (2002) Stalk model of membrane fusion: solution of energy crisis, Biophys J 82, 882–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNew JA, Weber T, Parlati F, Johnston RJ, Melia TJ, Söllner TH & Rothman JE (2000) Close Is Not Enough Snare-Dependent Membrane Fusion Requires an Active Mechanism That Transduces Force to Membrane Anchors, Journal of cell biology 150, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Z, Thiyagarajan S, O’Shaughnessy B & Karatekin E (2017) Regulation of Exocytotic Fusion Pores by SNARE Protein Transmembrane Domains, Front Mol Neurosci 10, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stratton BS, Warner JM, Wu Z, Nikolaus J, Wei G, Wagnon E, Baddeley D, Karatekin E & O’Shaughnessy B (2016) Cholesterol Increases the Openness of SNARE-Mediated Flickering Fusion Pores, Biophys J 110, 1538–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunger AT, Leitz J, Zhou QJ, Choi UB & Lai Y (2018) Ca2+-Triggered Synaptic Vesicle Fusion Initiated by Release of Inhibition, Trends in Cell Biology 28, 631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nikolaus J, Stockl M, Langosch D, Volkmer R & Herrmann A (2010) Direct visualization of large and protein-free hemifusion diaphragms, Biophys J 98, 1192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rizo J & Rosenmund C (2008) Synaptic vesicle fusion, Nat Struct Mol Biol 15, 665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


