Abstract
Objective:
Timely initiation of combination antiretroviral therapy (ART) limits latent HIV reservoir size and should also limit reservoir genetic complexity. However, the relationship between these two factors remains unclear, particularly among HIV-infected youth.
Design:
Retrospective analysis of replication-competent latent HIV clones serially isolated by limiting-dilution culture from resting CD4+ T-cell reservoirs from ART-suppressed, young adult participants of a historic phase I therapeutic vaccine trial (PACTG/IMPAACT-P1059).
Methods:
Replication-competent latent HIV clones isolated from resting CD4+ T-cells of 4 perinatally- and 10 non-perinatally-infected young adults (average 22 versus 6 years uncontrolled infection, respectively) were sequenced in Pol and Nef. Within-host HIV sequence datasets were characterized with respect to their genetic diversity and inferred immune escape mutation burden.
Results:
While participants were comparable in terms of sociodemographic and HIV sampling characteristics (e.g. on average, a mean 17 Pol sequences were recovered at 5 timepoints over up to 70 weeks) and the length of ART suppression at study entry (average 3 years), replication-competent HIV reservoir size, genetic diversity, immune escape mutation burden and variant complexity were significantly higher among the perinatally-infected participants who experienced longer durations of uncontrolled viremia. Nevertheless, viral sequences inferred to retain susceptibility to host cellular immune responses were detected in all participants, irrespective of uncontrolled viremia duration.
Conclusions:
HIV elimination in late-suppressed youth may be doubly challenged by larger and more genetically complex reservoirs. Strategies that integrate host and viral genetic complexity to achieve HIV remission or cure may merit consideration in such cases.
Keywords: HIV, latent reservoir, replication-competent, genetic diversity, immune escape, young adults, youth
Introduction
Genetic diversity [1–9] and immune escape [10] within the latent HIV reservoir form barriers to cure. Given that reservoir establishment begins shortly after infection and continues as long as viral replication remains uncontrolled [11–13], timely viremia suppression with combination antiretroviral therapy (ART) should, in theory, limit reservoir complexity. The observation that early ART limits HIV reservoir size in both adult [14–16] and pediatric [17–20] infection supports this; as do the observations that proviral landscapes in elite controllers and early-treated individuals tend to be more homogeneous than viremic controllers and individuals who initiated ART in chronic infection, respectively [5, 21, 22]. However, the effect of uncontrolled viremia duration on reservoir diversity in individuals who did not initiate early suppressive ART remains unclear. This is particularly relevant to persons infected in the decade prior to the availability of combination ART, including perinatally-infected individuals who survived to young adulthood.
Immune escape within the HIV reservoir also remains incompletely characterized in this population. While the majority of latent HIV genomes in adults treated in chronic infection harbored at least one major Human Leukocyte Antigen (HLA) class I-restricted Cytotoxic T-lymphocyte (CTL) escape mutation in Gag, unmutated epitopes - that were subsequently used as targets for reservoir elimination - were also present in all individuals [10]. If the latent reservoir recapitulates within-host HIV evolution [23–25] then CTL epitopes that underwent escape in vivo should be “preserved” in various states of adaptation within it; indeed, a scenario where susceptible and adapted forms of the same epitope co-exist in the replication-competent HIV reservoir could create both challenges and opportunities for cure immunotherapeutics.
Latent HIV reservoir sampling also remains a challenge. Given the high (>90%) burden of defective proviruses [1, 26], direct HIV DNA sequencing may not fully represent genetic diversity within the replication-competent minority that is critical to eradicate [27]. Furthermore, given the propensity of latently HIV-infected cells to undergo clonal expansions [4, 7, 28–30] that can sometimes be short-lived [31], cross-sectional studies may underestimate overall reservoir diversity if such an expansion has recently occurred.
To address these gaps, we genetically characterized replication-competent latent HIV clones isolated from resting CD4+ T-cell reservoirs serially sampled over up to 70 weeks during suppressive ART, from young adult participants of a historic phase I therapeutic vaccine trial (PACTG/IMPAACT-P1059) who differed markedly in terms of their uncontrolled HIV infection duration (due to perinatal acquisition of HIV in the decade before combination ART was available, versus risk behavior later in life) [32, 33]. Although the vaccine was well-tolerated [32] and induced a modest transient reduction in the reservoir [33], reservoir size at trial completion did not significantly differ from baseline. This rare dataset thus offers a unique opportunity to assess replication-competent latent HIV genetic complexity, and investigate its relationship with uncontrolled infection duration, in this key population.
Methods
This study was approved by the Johns Hopkins University School of Medicine and Simon Fraser University Institutional Review Boards. All participants provided written informed consent. This study included 14 of the 20 participants of PACTG/IMPAACT-P1059 for whom replication-competent latent HIV isolates were serially obtained; all participants had plasma HIV RNA <50 copies/ml on ART at trial initiation and maintained viremia suppression throughout follow-up. As previously reported [33], infectious HIV frequencies in resting CD4+ T-cells were quantified in real time at trial screen and entry (week 0), and up to seven visits thereafter (weeks 2, 4, 6, 24, 26, 40 and 72) by end point dilution culture [21, 33]. Resting CD4+ T-cells were enriched from fresh blood, activated to promote virus expression, after which released virus was expanded in CD4+ T-lymphoblasts from HIV-negative donors to quantify original infected cell frequencies in infectious units per million (IUPM) [34, 35]. Nef and partial Pol (HXB2 genomic nucleotides 2253–3254) were amplified from p24-positive culture supernatants by nested RT-PCR using HIV-specific primers, and Sanger sequenced [21]. Sequences were aligned using HIValign [36] (options: MAFFT [37], codon alignment) and edited in AliView v1.18 [38]. Maximum likelihood phylogenies were reconstructed using RAxML v8.2.10 [39] with 100 bootstraps under a generalized time reversible model and visualized using Figtree (http://tree.bio.ed.ac.uk/software/figtree/). Patristic (tip-to-tip phylogenetic) distances were extracted from newick treefiles using Patristic [40]. Pairwise genetic distances were additionally calculated using the dist.dna function in the APE package in R [41]. HLA-associated polymorphisms defined at allele-level resolution in HIV subtype B were published in [42]. HLA-restricted optimal CTL epitopes were defined using the Los Alamos HIV Molecular Immunology Database with recent updates ([43] and C. Brander, personal communication).
Results
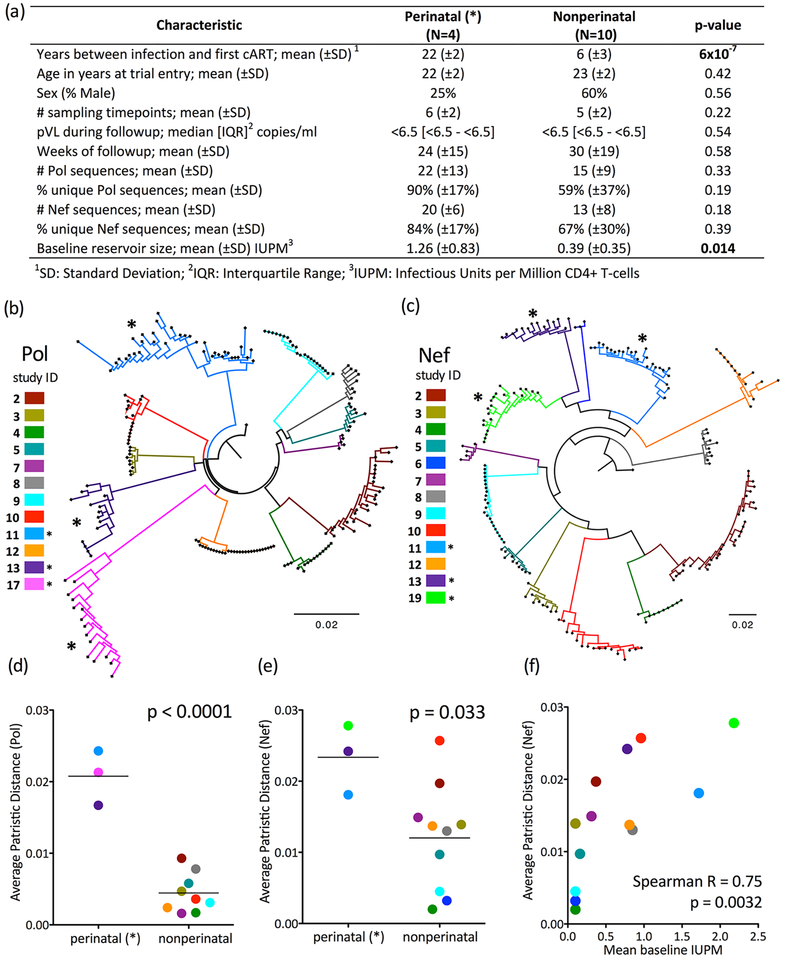
Replication-competent latent HIV sequences were serially sampled from resting CD4+T-cells collected from 14 PACTG/IMPAACT-P1059 participants during suppressive ART (Figure 1A). Participants were stratified into those who acquired HIV perinatally (N=4, in whom mean time between infection and combination ART initiation, henceforth referred to as estimated uncontrolled infection duration, was an estimated 22 years) and those who acquired HIV in adolescence through risk behavior (N=10; mean uncontrolled infection duration 6 years). Consistent with earlier ART limiting reservoir size [14–20], the former had significantly larger reservoirs than the latter (mean baseline IUPM 1.26 vs 0.39, p=0.014), however the groups did not otherwise differ in terms of age, gender or duration of viremia control on ART at study entry (overall median 3.3; range 0.6–6.4 years) [33].
Figure 1: Genetic diversity within the replication-competent latent HIV reservoir increases with untreated infection duration.
Panel A: Participant clinical, immunogenetic, and HIV reservoir dataset characteristics. Throughout all figures, perinatally-infected participants (those with longer uncontrolled infection duration) are denoted by asterisks (*). Panel B: Maximum-likelihood phylogeny relating within-host HIV Pol sequences, colored by participant. All within-host sequence datasets formed monophyletic clades with 100% bootstrap support. Scale in expected nucleotide substitutions per site. Panel C: same as B, but for Nef. All within-host datasets formed monophyletic clades with 100% bootstrap support except those of participants 11 (99%) and 19 (89%). Differences in overall topologies between Pol and Nef trees are attributable to low bootstrap support for the deeper branches (i.e. those that define evolutionary relationships between participants); all were <70% except the subclade comprising participants 2 and 4. No downstream analyses however relied on intra-participant genetic distances. Panel D: Average within-host patristic (tip-to-tip phylogenetic) distances in reservoir Pol sequences; p-value calculated using Student’s T-test. Panel E: Same as panel D, but for Nef. Panel F: Relationship between size and diversity (Nef) of the within-host replication-competent HIV reservoir, assessed by Spearman’s correlation.
To maximize the likelihood that recovered HIV isolates originated from the latent reservoir, analysis was limited to participants who maintained pVL < 50 copies/ml (a single viremia “blip” to 436 copies/ml in participant 2 was excepted). Median pVL during follow-up, assessed using an ultrasensitive assay, was <6.5 copies/ml for both groups (Figure 1A). Given that the vaccine did not ultimately reduce reservoir size [33], and that the latent HIV reservoir is highly stable [5, 11], all HIV sequences recovered from a given participant were pooled together regardless of sampling date to estimate within-host replication-competent reservoir diversity. In total, 204 Pol and 188 Nef sequences were isolated at an average of 5 time points over an average 27 weeks (range 4–70), yielding an overall average 17 Pol and 15 Nef sequences per participant. Groups did not differ in terms of sampling, followup duration or % unique sequences (Figure 1A), though note only Nef sequences were obtained for participants 6 and 19, and only Pol for participant 17 (Figures 1B–C). Overall, 184 (90.1%) Pol and 179 (95.2%) Nef sequences contained no nucleotide mixtures, consistent with clonal HIV outgrowth from endpoint-diluted cell cultures in the majority of wells. Each participant’s HIV sequences formed monophyletic clades with a median 100% bootstrap support (Figures 1B–C).
Identical Pol and/or Nef sequences were recovered in 10 of 14 participants (3 of 4 perinatally-infected and 7 of 10 nonperinatally-infected, p=1.0), consistent with clonal CD4+T-cell expansion as a mechanism of latent HIV reservoir maintenance in youth, regardless of infection mode. Notably, in 9 of these 10 participants, identical sequences were recovered at multiple timepoints up to 70 weeks apart (including participant 4 where the same sequence was recovered at weeks 2, 4, 6 and 72; the sole exception was participant 5, in whom identical sequences were recovered at a single timepoint only). This indicates that clonal descendants of CD4+ T cells harboring replication competent latent HIV tend to persist long-term in infected youth [28].
Replication-competent HIV reservoir diversity, measured in terms of average within-host patristic (tip-to-tip) phylogenetic distances, was significantly higher in perinatally- compared to nonperinatally-infected participants for both Pol (mean 0.21 vs. 0.004 nucleotide substitutions/site, p<0.0001) (Figure 1D) and Nef (mean 0.023 vs. 0.012 nucleotide substitutions/site, p=0.033, Figure 1E). Replication-competent HIV reservoir diversity was also significantly higher in perinatally- compared to nonperinatally-infected participants for both Pol and Nef when measured in terms of mean pairwise genetic distance (p=0.0003 for Pol, p=0.014 for Nef; not shown). Reservoir size correlated strongly with Nef (Spearman’s R=0.75; p=0.0032; Figure 1F) and to a lesser extent Pol (R=0.47; p=0.1) within-host average patristic distances, indicating that larger reservoirs tend to be more genetically diverse (rather than more clonally expanded).
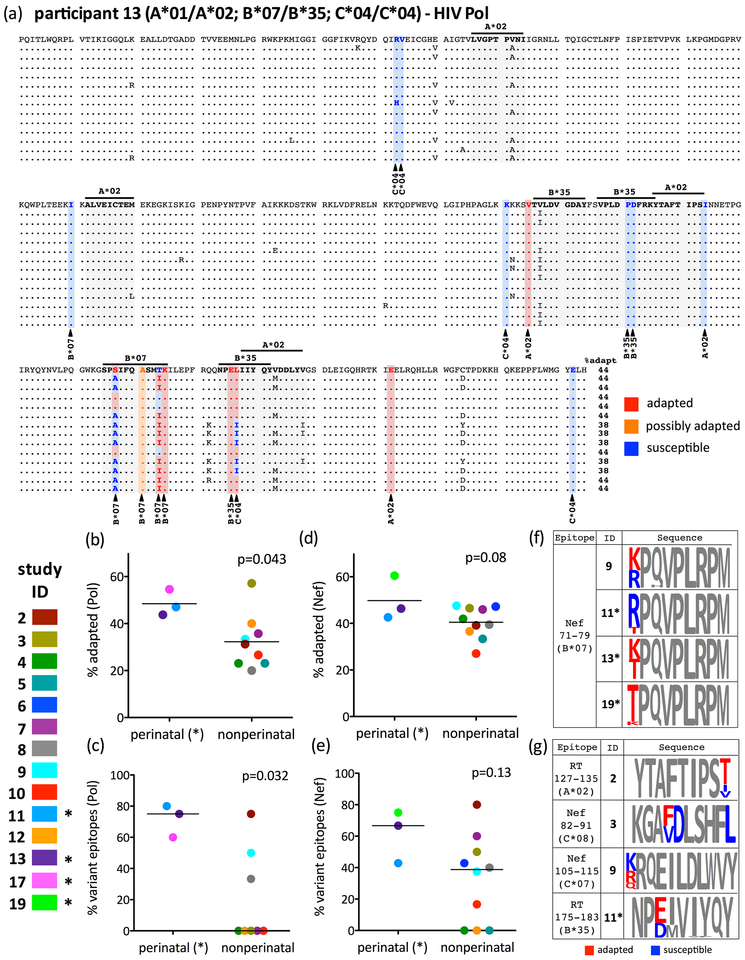
We investigated immune escape two ways. First, we estimated total escape burden by identifying all HIV codons under selection by one or more host HLA alleles and classifying each autologous HIV residue as adapted (inferred escaped) or susceptible, based on published definitions [42] (example in Figure 2A). For each sequence, we calculated the % HLA-associated sites exhibiting an adapted (or possibly adapted) form, and computed the median for each participant’s datasets (e.g. the Pol dataset in Figure 2A is 44% adapted to host HLA). Second, we estimated within-host escape complexity by quantifying the proportion of optimally-described HLA-restricted CTL epitopes exhibiting within-host amino acid variation (e.g. 6/8 [75%] for the Pol dataset in Figure 2A). In Pol, the percentage of HLA-adapted sites was overall higher in perinatally- compared to nonperinatally-infected participants (mean 48% vs. 32%, p=0.043; Figure 2B), as was the percentage of optimally-described Pol CTL epitopes exhibiting within-host amino acid variation (median 75% vs. 0%, p=0.032; Figure 2C). Similar trends were observed for Nef (Figures 2D–E).
Figure 2: Immune escape burden within the replication-competent latent HIV reservoir is complex, and generally increases with untreated infection duration.
Panel A: Pol sequence alignment for participant 13 (non-perinatally infected). The reference sequence (top) was arbitrarily chosen from among those recovered from CD4+T-cells sampled at the earliest timepoint. Sites of HLA-driven adaptation in Pol (defined in [42]) are highlighted, with red, orange and blue denoting adapted (inferred escaped), possibly adapted and susceptible forms, respectively. Optimally-described CTL epitopes restricted by host HLA alleles are shaded in grey. The proportion of HLA-associated sites exhibiting adapted or possibly adapted forms is reported after each sequence. Note the three codons (257, 264 and 277 in this alignment, denoting RT codons 158, 165 and 178), all within HLA-restricted CTL epitopes, where adapted and susceptible forms co-exist within the reservoir. Panel B: Average inferred immune escape burden in Pol (calculated as the median “% adapted” value of all sequences for each participant), stratified by group. P-value calculated using Student’s T-test. Panel C: Percent of HLA-matched optimal CTL epitopes in Pol exhibiting within-host sequence variation, stratified by group. As the data for non-perinatally infected participants are non-normally distributed, the p-value is calculated using the Mann-Whitney U-Test. Panels D, E: Same as panels B and C, but for Nef. Panel F: The B*07-restricted RM9 epitope (Nef codons 71–79) as an example of reservoir immune escape complexity within and between hosts. Letter size is proportional to within-host amino acid prevalence, with red and blue denoting adapted and susceptible forms [42], respectively (all other residues are grey). Note that participant 11 was perinatally-infected, illustrating that HLA-adapted and susceptible forms of a given CTL epitope can co-exist in the reservoir, even in persons who initiated cART two decades afer infection. Panel G: Additional examples of HLA-restricted optimal epitopes in Pol and Nef where adapted and susceptible forms co-exist within an individual’s reservoir.
Notably, however, despite uncontrolled infection durations of >20 years and overall high adaptation levels in some participants, no reservoir was completely adapted to host HLA (Figures 2B, D). Furthermore, on an invidual CTL epitope level, reservoir immune escape complexity differed widely both within and between hosts, an observation that can be illustrated by the HLA-B*07-restricted immunodominant [44] Nef-RM9 epitope (Figure 2F). Four participants (9, 11, 13, 19) expressed HLA-B*07, all of whom exhibited high Nef adaptation. However, while participants 13 and 19’s reservoirs were fully escaped in Nef-RM9, ~45% and ~80% of participants’ 9 and 11’s reservoirs, respectively, harbored sequences that were predicted to retain susceptibility to HLA-B*07-restricted CTL. This indicates that key susceptible epitopes can still be identified even in otherwise highly escaped reservoirs. Indeed, co-existence of HLA-susceptible and adapted forms within the same CTL epitope in an individual’s reservoir occurred commonly: >60% and >30% of participants harbored at least one Pol or Nef epitope respectively where this occurred (examples in Figure 2G). This further supports the reservoir as an archive of within-host HIV evolution [23–25] and suggests that autologous T-cell responses to these epitopes, if effectively re-stimulated, might still be capable of clearing a portion of the reservoir.
Discussion
Serial sampling of the replication-competent HIV reservoir in our young adult cohort supports the notion that reservoir diversity and escape burden continue to increase with uncontrolled infection duration, even in individuals who initiate ART relatively late. Caveats include the study’s modest size, differences in infection route (such that we cannot rule out that higher reservoir complexity is attributable to perinatal transmission rather than uncontrolled infection duration), and that reservoir sampling occurred during administration of an experimental therapeutic HIV vaccine [33]. While the vaccine did not durably reduce reservoir size [33], and we observed no evidence that the vaccine consistently altered overall within-host reservoir diversity (comparisons of the average within-host patristic HIV distances pre- and post-vaccine yielded p=0.8 and p=0.6 for Pol and Nef respectively; not shown), we cannot rule out the possibility that the vaccine may have induced very low-level HIV replication [45] or otherwise perturbed reservoir sequence composition in some participants. Confirmation of our observations in additional cohorts is therefore merited. Nevertheless, our findings may have implications for immmunotherapeutic HIV cure strategies. While, on one hand, HIV elimination in late-treated persons may be doubly challenged by larger and more genetically complex reservoirs, our observation that predicted HLA-susceptible sites were present in all reservoirs, even those of persons who did not achieve sustained virologic suppression until two decades after infection, supports strategies that integrate host and viral genetic data to inform HIV cure immunogen selection.
Acknowledgements:
ZLB and DP designed the study, analyzed and interpreted data, and drafted the manuscript. HS, CZ, KL, BJ, JJ, CKC and TM collected, contributed, analyzed and/or interpreted data. All authors provided critical feedback on the manuscript.
Research reported in this publication was funded by R01HD080474 to D.P and K.L. Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Network was provided by the National Institute of Allergy and Infectious Diseases (awards UM1AI068632, UM1AI068616, and UM1AI106716), the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and the National Institute of Mental Health. Research was also funded in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number UM1AI126617, with co-funding support from the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke (to DP and ZLB), by the Canadian HIV Cure Enterprise Team Grant from the CIHR in partnership with CANFAR and the International AIDS Society (IAS) (HIG-133050 to ZLB), by a project grant from the Canadian Institutes for Health Research (PJT-148621 to ZLB), and a Simon Fraser University Next Big Question fund award (to ZLB). ZLB is supported by a Scholar Award from the Michael Smith Foundation for Health Research. CKC is supported in part by the Duke University Center for AIDS Research (NIH 5P30 AI064518). Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Network (IMPAACT) was provided by the National Institute of Allergy and Infectious Diseases (NIAID) with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH), all components of the National Institutes of Health (NIH), under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC) and UM1AI106716 (IMPAACT LC), and by NICHD contract number HHSN275201800001I. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
We thank Petronella Muresan for assistance with data access and Christian Brander for providing updates to the Los Alamos optimal CTL epitope list. We thank the participants of PACTG/IMPAACT-P1059 without whom this research would not have been possible.
References
- 1.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016,22:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee GQ, Lichterfeld M. Diversity of HIV-1 reservoirs in CD4+ T-cell subpopulations. Curr Opin HIV AIDS 2016,11:383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 2014,20:139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, et al. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4+ T Cells from Effectively Treated Participants. Cell Rep 2017,21:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 2013,110:E4987–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evering TH, Mehandru S, Racz P, Tenner-Racz K, Poles MA, Figueroa A, et al. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog 2012,8:e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009,15:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, et al. Longitudinal Genetic Characterization Reveals That Cell Proliferation Maintains a Persistent HIV Type 1 DNA Pool During Effective HIV Therapy. J Infect Dis 2015,212:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A 2015,112:E1126–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 2015,517:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruff CT, Ray SC, Kwon P, Zinn R, Pendleton A, Hutton N, et al. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J Virol 2002,76:9481–9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 2014,512:74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ledford H HIV rebound dashes hope of ‘Mississippi baby’ cure. In: Nature News; 2014. [Google Scholar]
- 14.Novelli S, Lecuroux C, Avettand-Fenoel V, Seng R, Essat A, Morlat P, et al. Long-term therapeutic impact of the timing of ART in patients diagnosed with primary HIV-1 infection. Clin Infect Dis 2017. [DOI] [PubMed] [Google Scholar]
- 15.Jain V, Hartogensis W, Bacchetti P, Hunt PW, Hatano H, Sinclair E, et al. Antiretroviral therapy initiated within 6 months of HIV infection is associated with lower T-cell activation and smaller HIV reservoir size. J Infect Dis 2013,208:1202–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buzon MJ, Martin-Gayo E, Pereyra F, Ouyang Z, Sun H, Li JZ, et al. Long-term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J Virol 2014,88:10056–10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bitnun A, Samson L, Chun TW, Kakkar F, Brophy J, Murray D, et al. Early initiation of combination antiretroviral therapy in HIV-1-infected newborns can achieve sustained virologic suppression with low frequency of CD4+ T cells carrying HIV in peripheral blood. Clin Infect Dis 2014,59:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Persaud D, Patel K, Karalius B, Rainwater-Lovett K, Ziemniak C, Ellis A, et al. Influence of age at virologic control on peripheral blood human immunodeficiency virus reservoir size and serostatus in perinatally infected adolescents. JAMA Pediatr 2014,168:1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Bonet M, Puertas MC, Fortuny C, Ouchi D, Mellado MJ, Rojo P, et al. Establishment and Replenishment of the Viral Reservoir in Perinatally HIV-1-infected Children Initiating Very Early Antiretroviral Therapy. Clin Infect Dis 2015,61:1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster C, Pace M, Kaye S, Hopkins E, Jones M, Robinson N, et al. Early antiretroviral therapy reduces HIV DNA following perinatal HIV infection. AIDS 2017,31:1847–1851. [DOI] [PubMed] [Google Scholar]
- 21.Persaud D, Ray SC, Kajdas J, Ahonkhai A, Siberry GK, Ferguson K, et al. Slow human immunodeficiency virus type 1 evolution in viral reservoirs in infants treated with effective antiretroviral therapy. AIDS Res Hum Retroviruses 2007,23:381–390. [DOI] [PubMed] [Google Scholar]
- 22.de Azevedo SSD, Caetano DG, Cortes FH, Teixeira SLM, Dos Santos Silva K, Hoagland B, et al. Highly divergent patterns of genetic diversity and evolution in proviral quasispecies from HIV controllers. Retrovirology 2017,14:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambotte O, Chaix ML, Gubler B, Nasreddine N, Wallon C, Goujard C, et al. The lymphocyte HIV reservoir in patients on long-term HAART is a memory of virus evolution. AIDS 2004,18:1147–1158. [DOI] [PubMed] [Google Scholar]
- 24.Kearney MF, Spindler J, Shao W, Yu S, Anderson EM, O’Shea A, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog 2014,10:e1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones BR, Kinloch NN, Horacsek J, Ganase B, Harris M, Harrigan PR, et al. A phylogenetic approach to recover integration dates of latent HIV sequences within-host. PNAS in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013,155:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siliciano JD, Siliciano RF. Assays to Measure Latency, Reservoirs, and Reactivation. Curr Top Microbiol Immunol 2017. [DOI] [PubMed] [Google Scholar]
- 28.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 2016,113:1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, et al. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog 2017,13:e1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest 2017,127:2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, et al. Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc Natl Acad Sci U S A 2018,115:E2575–E2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenough TC, Cunningham CK, Muresan P, McManus M, Persaud D, Fenton T, et al. Safety and immunogenicity of recombinant poxvirus HIV-1 vaccines in young adults on highly active antiretroviral therapy. Vaccine 2008,26:6883–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persaud D, Luzuriaga K, Ziemniak C, Muresan P, Greenough T, Fenton T, et al. Effect of therapeutic HIV recombinant poxvirus vaccines on the size of the resting CD4+ T-cell latent HIV reservoir. AIDS 2011,25:2227–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 2005,304:3–15. [DOI] [PubMed] [Google Scholar]
- 35.Taswell C Limiting dilution assays for the determination of immunocompetent cell frequencies. III. Validity tests for the single-hit Poisson model. J Immunol Methods 1984,72:29–40. [DOI] [PubMed] [Google Scholar]
- 36.Gaschen B, Kuiken C, Korber B, Foley B. Retrieval and on-the-fly alignment of sequence fragments from the HIV database. Bioinformatics 2001,17:415–418. [DOI] [PubMed] [Google Scholar]
- 37.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002,30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsson A AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014,30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014,30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fourment M, Gibbs MJ. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol Biol 2006,6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004,20:289–290. [DOI] [PubMed] [Google Scholar]
- 42.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, et al. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS ONE 2009,4:e6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frahm N, Baker B, Brander C. Identification and Optimal Definition of HIV-Derived Cytotoxic T Lymphocyte (CTL) Epitopes for the Study of CTL Escape, Functional Avidity and Viral Evolution In: HIV Molecular Immunology 2008. Edited by Korber BTM, Brander C, Haynes BF, et al. Los Alamos, New Mexico: Los Alamos National Laboratory, Theoretical Biology and Biophysics; 2008. pp. 3–24. [Google Scholar]
- 44.Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, et al. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med 2006,3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiu C, Cunningham CK, Greenough T, Muresan P, Sanchez-Merino V, Carey V, et al. Identification of ongoing human immunodeficiency virus type 1 (HIV-1) replication in residual viremia during recombinant HIV-1 poxvirus immunizations in patients with clinically undetectable viral loads on durable suppressive highly active antiretroviral therapy. J Virol 2009,83:9731–9742. [DOI] [PMC free article] [PubMed] [Google Scholar]