Abstract
Background
Sentinel lymph node (SLN) status is an important prognostic factor for patients with breast cancer, which is currently determined by invasive SLN biopsy in clinical practice.
Purpose
To non-invasively predict SLN metastasis in breast cancer using DCE-MRI intra- and peritumoral radiomics features combined with or without clinicopathologic characteristics of the primary tumor.
Study type
Retrospective.
Population
A total of 163 breast cancer patients (55 positive SLN and 108 negative SLN).
Field strength/sequence
1.5T, T1-weighted DCE-MRI.
Assessment
A total of 590 radiomic features were extracted for each patient from both intratumoral and peritumoral regions-of-interest (ROIs). To avoid overfitting, the dataset was randomly separated into a training set (~67%) and a validation set (~33%). The prediction models were built with the training set using logistic regression on the most significant radiomic features in the training set combined with or without clinicopathologic characteristics. The prediction performance was further evaluated in the independent validation set.
Statistical tests
Mann-Whitney U-test, Spearman correlation, lasso regression, logistic regression and receiver operating characteristic (ROC) analysis were performed.
Results
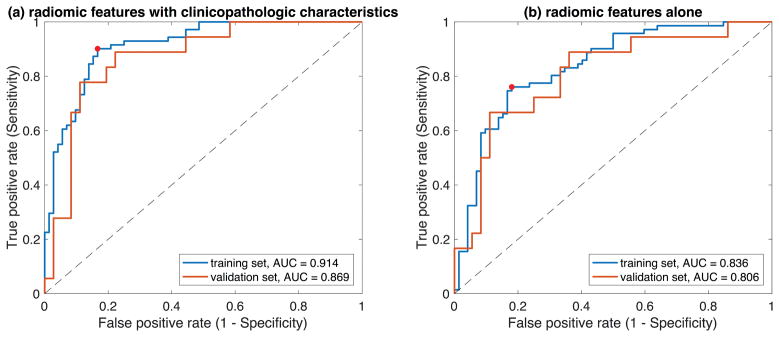
Combining radiomic features with clinicopathologic characteristics, 6 features were automatically selected in the training set to establish the prediction model of SLN metastasis. In the independent validation set, the area under ROC curve (AUC) was 0.869 (NPV=0.886). Using radiomic features alone in the same procedure, 4 features were selected and the validation set AUC was 0.806 (NPV=0.824).
Data Conclusion
This is the first attempt to demonstrate the feasibility of using DCE-MRI radiomics to predict SLN metastasis in breast cancer. Clinicopathologic characteristics improved the prediction performance. This study provides non-invasive methods to evaluate SLN status for guiding further treatment of breast cancer patients, and can potentially benefit those with negative SLN, by eliminating unnecessary invasive lymph node removal and the associated complications, which is a step further towards precision medicine.
Keywords: Breast Cancer, Sentinel Lymph Node Metastasis, Radiomics, DCE-MRI, Precision Medicine
Introduction
Breast cancer is the most commonly diagnosed cancer in females and accounts for 29% of all new cancer diagnoses in women(1). The 5-year relative survival rate for women diagnosed with localized breast cancer is up to 98.6%. However, for breast cancer patients with regional lymph node metastasis, the survival rate declines to 84.4%(2). The sentinel lymph node (SLN) is the first draining site to be affected during tumor spreading, thus SLN status offers a valuable prognostic factor to guide treatment decisions. There is a general consensus that complete axillary lymph node (ALN) dissection (ALND) is not necessary for patients without SLN metastasis(3) due to serious associated complications(4). Recently, ALND is being debated even for patients with positive SLN due to its lack of overall survival improvement compared to SLN dissection alone(5) for women with early-stage breast cancer, no palpable axillary adenopathy, and 1 or 2 positive SLNs. Therefore, non-invasive assessment of SLN status is extremely important: for patients with negative SLN, no lymph nodes need to be removed. Moreover, for patients with positive SLN, treatments including SLN dissection, ALND or radiation therapy should be further evaluated by clinicians depending on the tumor’s characteristics.
However, SLN status is clinically determined by SLN dissection, also called SLN biopsy (SLNB), which is the widely accepted diagnostic staging procedure of care for primary treatment of early breast cancer. SLNB has several drawbacks. First, it is an invasive procedure linked to potentially significant complications, such as shoulder dysfunction, nerve damage, lymphedema and upper arm numbness(6–8). Second, SLNB has been considered a controversial procedure, owing to the unstandardized radiopharmaceuticals, injection sites, and the level of the experience of the operator, etc(9). Third, SLNB is associated with a potentially high false-negative rate ranging from 5.5% to 16.7% (10). Therefore, non-invasive methods are highly desirable to preoperatively evaluate SLN metastasis.
The Memorial Sloan-Kettering Cancer Center has developed a model (MSKCC nomogram) to predict the likelihood of SLN metastasis based on clinical and histopathological information such as patient age, tumor size and histologic type(11). However, the area under the receiver operating characteristic (ROC) curve (AUC) was relatively low at 0.754, which hinders its clinical adoption. Although several similar nomograms(12–14) and validation studies(15–18) were published later, the reported AUC values were all lower than 0.8, which is insufficient to reliably guide clinical practice.
Recently, with the advancement in pattern recognition tools, radiomics has drawn increased interest. Radiomics is a process of converting medical images into mineable data by extracting high-throughput quantitative features. The subsequent analysis of these features can provide potential non-invasive biomarkers for clinical-decision support(19–22). Radiomics has been used in breast cancer to predict malignancy(23), pathological complete response to neoadjuvant chemotherapy (NAC) (24), etc. Radiomics combined with clinicopathologic risk factors can increase prediction power(20) and has been applied to predict regional lymph node metastasis in colorectal cancer(25) and bladder cancer(26).
In the current literature, only one paper(27) evaluated SLN in breast cancer using radiomic analysis of the primary tumor. In the work by Dong et al(27), T2-weighted fat-suppression (T2FS) and diffusion-weighted MRI (DWI) was used to predict SLN metastasis, and the texture features extracted from the intratumoral regions yielded an AUC of 0.805 in the validation set. However, DWI has not been incorporated into routine clinical breast MRI examinations and is not available for every patient. In addition, patient and tumor characteristics were regarded as statistically insignificant predictors in their method, which is in disagreement with the well-established associations between SLN metastasis and tumor size, tumor location, histological grade (28,29). Furthermore, this method along with most radiomic studies focused on intratumoral features, while peritumoral regions surrounding tumors have been shown to contain valuable information in cancer studies of the breast(24), liver(30,31) and brain(32). The usefulness of clinicopathologic characteristics and peritumoral features in predicting SLN metastasis for breast cancer patients remains under-investigated.
The purpose of this study is to develop a non-invasive radiomic signature from preoperative dynamic contrast-enhanced MRI (DCE-MRI) of the primary tumor combined with or without clinicopathologic factors to predict SLN metastasis in patients with invasive breast cancer. We hypothesize that the intratumoral and peritumoral features extracted from the routine DCE-MRI plus clinicopathologic information can provide more accurate prediction performance.
Methods and materials
Study Population
This retrospective study was approved by our institutional review board. The inclusion criteria were: (1) Preoperative DCE-MRI performed between June 2013 and June 2017; (2) Patients with invasive breast carcinoma confirmed by histopathology from biopsy or resection specimen; (3) Patients received SLNB/ALND (SLN was considered to be positive when cancer cells were present in the removed SLN). (4) Patients did not receive NAC prior to baseline biopsy or MRI examination. Exclusion criteria included the following: (1) Patients without SLNB or dissection and without PET/CT examination; (2) Patients with very small region-of-interest (ROI) (less than 64 voxels). (3) MRI examination after NAC.
The clinical and histopathological data were collected from patient medical records and included age, tumor location, histological type and grade of invasive carcinoma, molecular subtype [according to the status of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor-2 (HER2) and proliferation marker Ki-67], lymph-vascular invasion (LVI) and multifocality. Note that the tumor size was also included in this study but used as a radiomic feature (volume) introduced in the later section.
Finally, a total of 163 patients (55 positive SLN and 108 negative SLN) who met the criteria and with DCE-MRI of the same spatial resolution and completed clinicopathologic characteristics were included in this study. The details of the 7 clinical and histopathological characteristics were shown in Table 1.
Table 1.
Clinical and histopathological characteristics.
| Group | Patients with positive SLN (n=55) | Patients with negative SLN (n=108) | p valuea |
|---|---|---|---|
| Age (mean±SD) | 55.1±12.9 | 55.5±12.6 | 0.844 |
| Tumor location (UIQ or not) | 0.343 | ||
| Yes | 2 (3.6%) | 8 (7.4%) | |
| No | 53 (96.4%) | 100 (92.6%) | |
| Histological type | 0.259 | ||
| Invasive ductal carcinoma | 49 (89.1%) | 88 (81.5%) | |
| Invasive lobular carcinoma | 6 (10.9%) | 16 (14.8%) | |
| Others | 0 (0.0%) | 4 (3.7%) | |
| Histological grade | <0.001 | ||
| I | 1 (1.8%) | 27 (25.0%) | |
| II | 34 (61.8%) | 56 (51.9%) | |
| III | 20 (36.4%) | 25 (23.1%) | |
| Molecular subtype | 0.392 | ||
| Luminal A | 16 (29.1%) | 45 (41.7%) | |
| Luminal B | 29 (52.7%) | 49 (45.4%) | |
| HER2 positive | 4 (7.3%) | 4 (3.7%) | |
| Triple negative | 6 (10.9%) | 10 (9.3%) | |
| LVI status | <0.001 | ||
| Positive | 35 (63.6%) | 9 (8.3%) | |
| Negative | 20 (36.4%) | 99 (91.7%) | |
| Number of other lesions in the ipsilateral breast | 0.862 | ||
| 0 | 51 (92.7%) | 100 (92.6%) | |
| 1 | 3 (5.5%) | 6 (5.6%) | |
| 2 | 1 (1.8%) | 1 (0.9%) | |
| 3 | 0 | 1 (0.9%) |
Two-tailed two-sample t-test with unequal variances was used to compare the age. Other characteristics were tested by chi-square cross-tabulation.
Abbreviations: SLN – sentinel lymph node, UIQ – upper inner quadrant, SD – standard deviation, HER2 – human epidermal growth factor receptor-2, LVI – lymph-vascular invasion.
MRI Examination
MRI scans were performed with 8-channel breast coils on 1.5T GE Signa HDxt scanners (GE healthcare, Wauwatosa, Wisconsin). The whole MRI protocol included high resolution sagittal T1-weighted, axial fast spin-echo T2-weighted, axial STIR T2-weighted and sagittal T1-weighted DCE-MRI sequences. Only DCE-MRI images were collected in this study. The gadolinium contrast agent, Magnevist (Schering, Berlin, Germany) was intravenously injected at a dose of 0.2ml/kg at a rate of 2 ml/s with a power injector, followed by a 20-ml saline flush at a rate of 2 ml/s. One pre-contrast and four post-contrast phases were acquired using a sagittal VIBRANT multiphase sequence with the following parameters: repetition time (TR)= 4.46~7.80ms; echo time (TE) = 1.54~4.20ms; flip angle=10°; matrix =256X256; pixel size = 0.7×0.7 mm2; slice thickness = 2mm.
Analysis Workflow
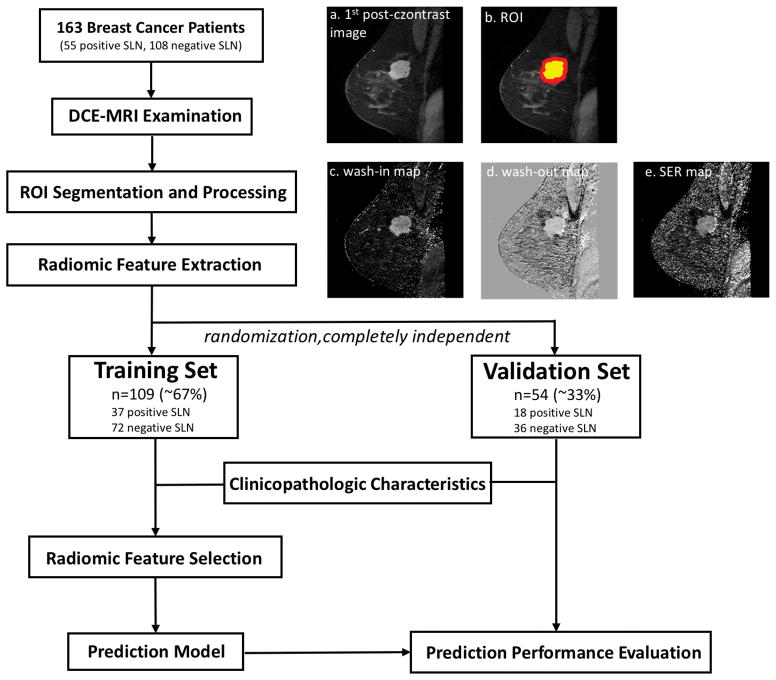
As shown in Figure 1, the prediction workflow includes ROI segmentation and processing, radiomic feature extraction, feature selection and prediction model generation (using a training set) and prediction performance evaluation (using an independent validation set).
Figure 1.
The workflow of this study. 163 breast cancer patients with DCE-MRI scans were included in this study. The ROIs were identified on (a) the 1st post-contrast image. (b) shows the corresponding intratumoral and peritumoral ROIs: the yellow region is the original intratumoral ROI covering the enhancing tumor drawn by the radiologist, while the red region indicates the peritumoral ROI after dilation; Radiomic features were extracted from (c) wash-in map, (d) wash-out map, and (e) SER map. The dataset was then randomly separated into a training set (~67%) and a validation set (~33%). The prediction model was built in the training set after combining clinical and histopathological information and was further tested in the completely independent validation set.
ROI Segmentation and Processing
ROIs covering the whole tumor were manually drawn slice by slice on the first post-contrast images using MRIcron (open source software; version 4AUGUST2014 Build) by a radiologist who had 11 years of experience in breast MRI. Because most of the MR images were from diagnostic MRI and MR images were obtained after biopsy (within one week), we tried to include the whole lesions on each slice, avoiding the biopsy tract, the hemorrhagic areas caused by biopsy and the biopsy ring artifacts.
The peritumoral regions were obtained by dilating the original ROI by approximately 4mm in 3D using MATLAB 2017b. Radiomic features were extracted from both the original intratumoral ROIs and the peritumoral ROIs. A representative DCE-MRI image and its corresponding ROIs were shown in Figure 1(a) and (b).
Although the main analysis was conducted using validation set ROIs from the same radiologist, a first-year radiology resident also independently delineated the ROIs for patients in the validation set in order to evaluate the effect of ROI segmentation and the inter-reader reliability of the prediction model.
Radiomic Feature Extraction
As shown in the workflow, three maps independent of original MR signal intensity were first generated to allow direct comparison across patients: wash-in maps ((S1-S0)/S0)×100%, wash-out maps ((S1-S4)/S1))×100% and signal enhancement ratio (SER) maps ((S1-S0)/(S4-S0))×100%, where S0, S1, and S4 are the pre-contrast, first post-contrast and fourth (the last) post-contrast images, respectively.
Four categories of features were extracted using either LIFEx v3.42(33) or MATLAB 2017b: (1) shape features, (2) histogram features, (3) texture features, and (4) Laws features(34). Table 2 provides a list of the extracted features along with the detailed descriptions. A total of 590 radiomic features were extracted for each patient. Because of the large amount of radiomic features available, overfitting is a concern. As a result, we divided out data set into separate training set and validation set to address overfitting concerns.
Table 2.
List of the extracted features with descriptions.
| Category (Quantity) | Featurese | Description |
|---|---|---|
|
Shape features N=8a |
Volume, Solidity, Eccentricity, Equivalent diameter, Extent, Surface area, Sphericity, and Compacity. | Describe the morphological and geometric characters of a breast tumor. |
|
Histogram features N=54b |
Minvalue, Maxvalue, Meanvalue, Stdvalue, Skewness, Kurtosis, Entropy (log10), Entropy (log2), and Energy (gray level discretizationf) | Reflect the distribution of intensities of individual voxels in the intratumoral and peritumoral ROIs. |
|
Texture features N=192c |
||
| Gray-level co-occurrence matrix (GLCM)(43) | Homogeneity, Energy, Contrast, Correlation, Entropy (log10), Entropy (log2), and Dissimilarity. | Measure the spatial complexity of the voxel values in the intratumoral and peritumoral ROIs, describing the degree of heterogeneity. |
| Neighborhood gray-level different matrix (NGLDM)(41) | Coarseness, Contrast, and Busyness. | |
| Gray-level run length matrix (GLRLM)(44) | SRE, LRE, LGRE, HGRE, SRLGE, SRHGE, LRLGE, LRHGE, GLNU, RLNU, and RP | |
| Gray-level zone length matrix (GLZLM)(45) | SZE, LZE, LGZE, HGZE, SZLGE, SZHGE, LZLGE, LZHGE, GLNU, ZLNU, and ZP | |
|
Laws features (46) N=336d |
Predefined 14 filters were applied to identify the structure patterns corresponding to level (L), edge (E), spot (S), ripple (R) and wave (W), deriving from the combinations of the 5 kernel vectors: L5= [1 4 6 4 1], E5= [−1 −2 0 2 1], S5= [−1 0 2 0 −1], R5= [1 −4 6 −4 1], and W5= [−1 2 0 −2 −1]. The 14 filters were L5E5, L5S5, L5R5, L5W5, E5E5, E5S5, E5R5, E5W5, S5S5, S5R5, S5W5, R5R5, R5W5, W5W5. Skewness, Kurtosis, Entropy (log10) and Energy were calculated. Relative resampling with 128 gray levels was used. | Detect the level, edge, spot, ripple, and wave textural patterns of the images, which are able to capture patterns of inconsistent enhancement and abnormal structure, such as speckling and rippling within the intratumoral and peritumoral regions (24,32). |
extracted from the original intratumoral ROIs.
9 per map per ROI (3 maps, both intratumoral and peritumoral ROIs).
32 per map per ROI (3 maps, both intratumoral and peritumoral ROIs).
4 per filter per map per ROI (14 filters, 3 maps, both intratumoral and peritumoral ROIs).
The first 6 shape features and Laws features were calculated in Matlab 2017b, other features were calculated in LIFEx 3.42.
The same gray level discretization procedure was performed for histogram features and texture featrues: 128 gray levels and absolute resampling (for intratumoral ROIs, wash-in map: 0~640; wash-out map: −156~100; SER map: −1280~1280; for peritumoral ROIs, wash-in map: 0~640, wash-out map: −540~100, SER map: −1280~1280).
Abbreviations: ROI – region of interest, SER – signal enhancement ratio, SRE – Short-Run Emphasis, LRE – Long-Run Emphasis, LGRE – Low Gray-level Run Emphasis, HGRE – High Gray-level Run Emphasis, SRLGE – Short-Run Low Gray-level Emphasis, SRHGE – Short-Run High Gray-level Emphasis, LRLGE – Long-Run Low Gray-level Emphasis, LRHGE – Long-Run High Gray-level Emphasis, GLNU – Gray-Level Non-Uniformity for run, RLNU – Run Length Non-Uniformity, RP – Run Percentage, SZE – Short-Zone Emphasis, LZE – Long-Zone Emphasis, LGZE – Low Gray-level Zone Emphasis, HGZE – High Gray-level Zone Emphasis, SZLGE – Short-Zone Low Gray-level Emphasis, SZHGE – Short-Zone High Gray-level Emphasis, LZLGE – Long-Zone Low Gray-level Emphasis, LZHGE – Long-Zone High Gray-level Emphasis, GLNU – Gray-Level Non-Uniformity for zone, ZLNU – Zone Length Non-Uniformity, ZP – Zone Percentage.
Feature Selection and Prediction Model Construction (training set)
The dataset was randomly divided into two independent subsets: a training set (~67%, 109 patients with 37 positive SLN) and a validation set (~33%, 55 patients with 19 positive SLN). The training set was used for feature selection and prediction model generation.
The first step is data rebalance in the training set, as the class imbalance can lead to a significant bias of the prediction model (samples tend to be classified into the majority group). We adopted the adaptive synthetic sampling approach for imbalanced learning algorithm(35) to improve the class balance by synthesizing new samples for the minority group. The radiomic feature selection was conducted on the rebalanced training set (71 positive SLN and 72 negative SLN).
For feature selection, a backward selection method was first performed using Mann-Whitney U-test to identify those significant radiomic features with p-value<0.05 between patients with positive and negative SLN. Next, in order to avoid overfitting from redundant features, groups of highly correlated radiomic features (Spearman |ρ|≥0.95) were identified; and each group was represented by the one with the highest discrimination power, that is, AUC furthest from 0.5 on predicting SLN metastasis. Then, the remaining radiomic features combined with and without the 7 clinicopathologic characteristics (Table 1) were given to the least absolute shrinkage selection operator (LASSO) regression. A logistic LASSO model with 3-fold cross-validation was performed to select the most important predictors, i.e. those features with nonzero coefficients when the minimum cross-validation error plus one standard deviation was reached.
The selected most important features were used to establish logistic regression models to predict SLN metastasis in breast cancer. The ROC curves were plotted with the optimal thresholds determined by maximizing the Youden index (sensitivity+specificity–1). The AUC, sensitivity, specificity and negative predictive value (NPV) were then calculated in the training set. All of the procedures in this section were conducted in MATLAB R2017b.
Prediction Performance Evaluation (validation set)
The generated prediction models were further tested in the independent validation set using the same thresholds determined in the training set. The corresponding ROC curve, AUC, sensitivity, specificity and NPV were also calculated in MATLAB R2017b.
Results
In the training set, 311 out of 590 radiomic features had a Mann-Whitney U-test p-value<0.05 between patients with positive and negative SLN. After selecting the representative features from each group of highly correlated features, 114 radiomic features were left. Combined the 114 radiomic features with the 7 clinicopathologic characteristics, the 3-fold cross-validation LASSO model selected 6 final features for the logistic regression prediction model, as shown in Table 3. Table 4 reports the prediction performance measures in the training and validation sets, and Figure 2(a) demonstrates the corresponding ROC curves. The same procedure was performed but only using the 114 radiomic features (no clinicopathologic characteristics) in the 3-fold cross-validation LASSO model. Four features were selected as shown in Table 3, and the prediction performance is illustrated in Table 4 and Figure 2(b).
Table 3.
Selected features in the prediction models.
| Selected features | |
|---|---|
| Radiomic features plus clinicopathologic characteristics | GLZLM_LZLGE (wash-in map, intratumoral ROI); GLCM_Entropy (log10) (wash-out map, intratumoral ROI); NGLDM_Contrast (wash-out map, intratumoral ROI); NGLDM_Coarseness (wash-in map, peritumoral ROI); Laws_Skewness after L5S5 (SER map, peritumoral ROI); LVI. |
| Radiomic features alone | GLCM_Entropy (log10) (wash-out map, intratumoral ROI); NGLDM_Contrast (wash-out map, intratumoral ROI); NGLDM_Coarseness (wash-in map, peritumoral ROI); Laws_Skewness after L5S5 (SER map, peritumoral ROI); |
Table 4.
Prediction performance in training and validation sets.
| Radiomic features plus clinicopathologic characteristics | radiomic features alone | |||
|---|---|---|---|---|
| Training set | Validation set | Training set | Validation set | |
| AUC | 0.914 | 0.869 | 0.836 | 0.806 |
| Sensitivity | 0.901 | 0.778 | 0.761 | 0.667 |
| Specificity | 0.833 | 0.861 | 0.819 | 0.778 |
| NPV | 0.896 | 0.886 | 0.776 | 0.824 |
Figure 2.
ROC curves of (a) adding clinicopathologic characteristics and (b) using radiomic features alone for the training and the independent validation sets. The red points on the ROC curves of the training set is the optimal cut-off point (threshold) determined using the Youden index.
As shown in Table 4, the combination of DCE-MRI radiomic features with clinicopathologic characteristics achieved a high AUC of 0.869 in the independent validation set, which outperformed the prediction model using DCE-MRI radiomics alone (AUC=0.806). More interestingly, for those patients predicted to have negative SLN, 31 out of 35 had a negative histopathology, achieving a high NPV of 0.886. Although the prediction performance decreased when only using DCE-MRI radiomic features, the result is still acceptable and very comparable to the previous study using radiomic features from DWI and T2FS (AUC=0.805)(27). Note that DWI is currently not integrated into the routine clinical breast MRI protocol in many hospitals.
As a post hoc analysis to demonstrate the usefulness of peritumoral features extracted from DCE-MRI in SLN metastasis prediction, we performed the same analysis as above with only 299 intratumoral features for selection. The AUC of the independent validation set decreased to 0.819 when clinicopathologic characteristics were used and 0.752 when only intratumoral features were used.
When the radiomic features in the validation set were re-calculated using ROIs drawn by the first-year resident (a second reader), the prediction performance for combined radiomic features and clinicopathologic characteristics decreased to AUC=0.836, Sensitivity=0.722, Specificity=0.806, and NPV=0.853. Such decrease is expected, and the prediction result remains superior to previous work. In particular, the NPV is very comparable to the one derived from the ROIs drawn by the first reader.
Discussion
This work is the first attempt to predict SLN metastasis in breast cancer non-invasively using DCE-MRI radiomics and demonstrates promising prediction performance in an independent validation set. Particularly, with the help of clinicopathologic information, the prediction model achieved a high AUC of 0.869 and NPV of 0.886. Our study focuses on predicting SLN metastasis by radiomic analysis, providing a potential non-invasive biomarker of SLN status, which can be used to guide the further treatment planning in breast cancer. A high NPV could potentially offer greater benefits for patients with negative SLN, which accounts for about 65% of all breast cancer patients(36), helping them eliminate the unnecessary invasive lymph node removal and avoid the overtreatment of axillary fossa along with the associated serious complications.
Radiomics is a relatively new technique that provides potential biomarkers for clinical outcomes by image feature extraction and analysis(21). Most radiomic studies focused only on intratumoral regions, while in this study, we also extracted features from the peritumoral regions. Many breast cancer studies have found that crucial information can be indicated by changes in the area surrounding tumors, such as peritumoral lymphatic vessel invasion(37), peritumoral lymphocytic infiltration(38) and peritumoral edema(39). A 2017 study(24) extracted both intratumoral and peritumoral radiomic features from pretreatment breast DCE-MRI, and successfully predicted pathological complete response to NAC. Peritumoral features have also been shown to be associated with intrahepatic metastasis of hepatocellular carcinoma(30,31). For predicting SLN metastasis in breast cancer, our results showed that some peritumoral features were selected into the final prediction models and in turn improved the prediction performance as demonstrated in the post hoc analysis. These data suggest that peritumoral features from DCE-MRI contain valuable information on tumor metastasis and should be included in further related radiomic studies.
Radiomic features are usually difficult to intuitively interpret, but they are able to capture the heterogeneity and complexity of the tumor microenvironment. Taking some selected features in Results as examples, NGLDM_Contrast actually calculates the intensity difference between neighboring regions(40); NGLDM_Coarseness represents the level of spatial rate of change in intensity(41); GLCM_Entropy describes the randomness of gray-level voxel pairs(40); and Laws features are able to detect patterns of inconsistent enhancement and abnormal structure(24,32). Whereas these data might be impossible for humans to perceive, they are predictive of tumor status and easily captured by radiomic analysis.
Our prediction performance outperforms previous prediction models for SLN metastasis in breast cancer which only used clinicopathologic characteristics of the primary tumor. Previous studies have shown that clinicopathologic characteristics are associated with SLN metastasis, and can even be independent predictors(28,29). Several nomograms, such as MSKCC nomogram, have been developed(11–14) to predict the likelihood of SLN metastasis only based on the clinicopathologic information including patient age, tumor size, tumor location, LVI, multifocality, histologic tumor type, etc. However, these nomograms and their following validation studies all failed to reach an AUC > 0.8(11–18). In this study, we combined DCE-MRI radiomic features with clinicopathologic characteristics to effectively improve the prediction performance, achieving a higher AUC of 0.869 on the independent validation set.
The prediction performance slightly decreased (AUC=0.836) when validation set features were generated using ROIs drawn by a second reader. This is likely due to the fact that the prediction model was trained using the features generated by ROIs drawn by the first reader. Also, the second reader is a first-year resident while the first reader is a radiologist with 11-years of experience in breast imaging. Although there was variability in the manually drawn ROIs, the prediction performance is still good and achieved a high NPV of 0.853. Our next step is to develop an automated and reliable breast lesion segmentation based on machine learning, and to further establish a fully automated pipeline of non-invasive SLN status prediction for breast cancer patients.
The routine T2 weighted imaging was not included in the present analysis as this study focuses on the feasibility of DCE-MRI radiomics for SLN status prediction. In fact, the tumor boundaries were not clear on T2 weighted images, making it difficult to delineate the ROIs. Moreover, a previous study (27) investigated T2FS radiomics, and the prediction performance for SLN metastasis was not satisfying (AUC=0.77). We believe DCE-MRI is able to provide more information about the tumor’s characteristics compared to T2 weighted imaging. Further work is expected to combine different MRI contrasts in radiomic analysis for SLN status prediction.
One might argue that some clinicopathologic characteristics such as LVI are available only after surgical biopsy. In fact, many patients receive excisional biopsies before SLN biopsy, offering all necessary clinicopathologic information available (11). However, we agree that this might not be the case across different institutes. Therefore, for patients without LVI, we also established a prediction model based on DCE-MRI radiomic features alone. The prediction performance (AUC=0.806) is very comparable to a 2017 study(27) utilizing radiomic features from T2FS and DWI to predict SLN metastasis in breast cancer (AUC=0.805). It is noted that DCE-MRI has become a critical part of a routine clinical breast MRI protocol, while DWI is not available in every hospital.
The current study has some limitations. As a retrospective study, inherent variations and biases may influence the results. One such variation is the change of TR and TE among DCE-MRI acquisitions; however, difference in the original MR signal intensity was not a major concern in this study as the radiomic features were extracted from three ratio maps which reflect contrast agent concentration changes. Although the ratios are functions of TR, these functions are relatively slow varying(42). A carefully designed prospective study is warranted to investigate the findings of this study. Another limitation is that most of the DCE-MRI images were collected after biopsy of the primary tumor. Although we tried to avoid the hemorrhagic or edema areas caused by the biopsy during ROI drawing, this problem may still potentially affect feature calculation. Furthermore, although our method exhibits superior or comparable performance in predicting SLN metastasis compared to the previous studies, different populations may lead to different prediction outcomes. Further studies are needed to validate the prediction performance on a larger dataset from different centers and scanners; we also expect the prediction performance to further improve by incorporating more advanced radiomic features and clinicopathologic characteristics.
In conclusion, this is the first study to demonstrate the feasibility of using radiomic analysis of DCE-MRI to predict SLN metastasis in breast cancer. This method provides potential non-invasive biomarkers of SLN status, which is a very important prognostic factor for guiding treatment decisions and eliminating unnecessary invasive lymph node removal for patients with negative SLN. This study is a step towards precision medicine and personalized treatment for breast cancer patients. Further studies are expected to improve the prediction model and validate the findings using multicenter data.
Acknowledgments
Grant Support: This work was supported by the National Key Research and Development Program of China (2017YFC1309100), National Institutes of Health (R03CA223052), Carol M. Baldwin Foundation for Breast Cancer Research (2017-Huang). YG would like to thank the support from National Natural Science Foundation of China No. 61601302 and Shenzhen peacock plan (NO.KQTD2016053112051497)
Abbreviation key
- ALN
axillary lymph node
- ALND
axillary lymph node dissection
- AUC
area under the receiver operating characteristic curve
- DCE-MRI
dynamic contrast-enhanced MRI
- DWI
diffusion-weighted MRI
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor-2
- LASSO
least absolute shrinkage selection operator
- LVI
lymph-vascular invasion
- NAC
neoadjuvant chemotherapy
- NPV
negative predictive value
- PR
progesterone receptor
- ROC
receiver operating characteristic
- ROI
regions-of-interest
- SER
signal enhancement ratio
- SLN
sentinel lymph node
- SLNB
sentinel lymph node biopsy
- T2FS
T2-weighted fat-suppression
- TE
echo time
- TR
repetition time
- UIQ
upper inner quadrant
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA: a cancer journal for clinicians. 2014;64(4):252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 3.Lyman GH, Temin S, Edge SB, et al. Sentinel lymph node biopsy for patients with early-stage breast cancer: American Society of Clinical Oncology clinical practice guideline update. Journal of Clinical Oncology. 2014;32(13):1365–1383. doi: 10.1200/JCO.2013.54.1177. [DOI] [PubMed] [Google Scholar]
- 4.Recht A, Houlihan MJ. Axillary lymph nodes and breast cancer. A review. Cancer. 1995;76(9):1491–1512. doi: 10.1002/1097-0142(19951101)76:9<1491::aid-cncr2820760902>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Giuliano AE, Ballman KV, McCall L, et al. Effect of axillary dissection vs no axillary dissection on 10-year overall survival among women with invasive breast cancer and sentinel node metastasis: the ACOSOG Z0011 (Alliance) randomized clinical trial. Jama. 2017;318(10):918–926. doi: 10.1001/jama.2017.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kootstra J, Hoekstra-Weebers JE, Rietman H, et al. Quality of life after sentinel lymph node biopsy or axillary lymph node dissection in stage I/II breast cancer patients: a prospective longitudinal study. Annals of surgical oncology. 2008;15(9):2533–2541. doi: 10.1245/s10434-008-9996-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilke LG, McCall LM, Posther KE, et al. Surgical complications associated with sentinel lymph node biopsy: results from a prospective international cooperative group trial. Annals of surgical Oncology. 2006;13(4):491–500. doi: 10.1245/ASO.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Mansel R, Goyal A, Newcombe R. Objective assessment of lymphedema, shoulder function and sensory deficit after sentinel node biopsy for invasive breast cancer: ALMANAC trial. Breast Cancer Research and Treatment. 2004;88:S12. [Google Scholar]
- 9.Manca G, Rubello D, Tardelli E, et al. Sentinel lymph node biopsy in breast cancer: indications, contraindications, and controversies. Clinical nuclear medicine. 2016;41(2):126–133. doi: 10.1097/RLU.0000000000000985. [DOI] [PubMed] [Google Scholar]
- 10.Hindié E, Groheux D, Brenot-Rossi I, Rubello D, Moretti J-L, Espié M. The sentinel node procedure in breast cancer: nuclear medicine as the starting point. Journal of Nuclear Medicine. 2011;52(3):405–414. doi: 10.2967/jnumed.110.081711. [DOI] [PubMed] [Google Scholar]
- 11.Bevilacqua JLB, Kattan MW, Fey JV, Cody HS, III, Borgen PI, Van Zee KJ. Doctor, what are my chances of having a positive sentinel node? A validated nomogram for risk estimation. Journal of Clinical Oncology. 2007;25(24):3670–3679. doi: 10.1200/JCO.2006.08.8013. [DOI] [PubMed] [Google Scholar]
- 12.Veerapong JJB, Mittendorf E, Harrell R, Bassett R, Yi M, Ross M, Meric-Bernstam F, Babiera G, Kuerer H, Lucci A, Bedrosian I, Brodt J, Jakub J, Hunt K, Hwang R. A validated risk assessment of sentinel lymph node involvement in breast cancer patients. Society of Surgical Oncology Annual Cancer Symposium; 2011. [Google Scholar]
- 13.Reyal F, Rouzier R, Depont-Hazelzet B, et al. The molecular subtype classification is a determinant of sentinel node positivity in early breast carcinoma. PLoS One. 2011;6(5):e20297. doi: 10.1371/journal.pone.0020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J-y, Chen J-j, Yang B-l, et al. Predicting sentinel lymph node metastasis in a Chinese breast cancer population: assessment of an existing nomogram and a new predictive nomogram. Breast cancer research and treatment. 2012;135(3):839–848. doi: 10.1007/s10549-012-2219-x. [DOI] [PubMed] [Google Scholar]
- 15.Klar M, Foeldi M, Markert S, Gitsch G, Stickeler E, Watermann D. Good prediction of the likelihood for sentinel lymph node metastasis by using the MSKCC nomogram in a German breast cancer population. Annals of surgical oncology. 2009;16(5):1136–1142. doi: 10.1245/s10434-009-0399-3. [DOI] [PubMed] [Google Scholar]
- 16.Qiu P-f, Liu J-j, Wang Y-s, et al. Risk factors for sentinel lymph node metastasis and validation study of the MSKCC nomogram in breast cancer patients. Japanese journal of clinical oncology. 2012;42(11):1002–1007. doi: 10.1093/jjco/hys150. [DOI] [PubMed] [Google Scholar]
- 17.Van la Parra R, Francissen C, Peer P, et al. Assessment of the Memorial Sloan-Kettering Cancer Center nomogram to predict sentinel lymph node metastases in a Dutch breast cancer population. European journal of cancer. 2013;49(3):564–571. doi: 10.1016/j.ejca.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Ngo C, Mouttet D, De Rycke Y, et al. Validation over time of a nomogram including HER2 status to predict the sentinel node positivity in early breast carcinoma. European Journal of Surgical Oncology. 2012;38(12):1211–1217. doi: 10.1016/j.ejso.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. European journal of cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambin P, Leijenaar RT, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nature reviews Clinical oncology. 2017 doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 22.Mazurowski MA. Radiogenomics: what it is and why it is important. Journal of the American College of Radiology. 2015;12(8):862–866. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 23.Bickelhaupt S, Paech D, Kickingereder P, et al. Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. Journal of Magnetic Resonance Imaging. 2017;46(2):604–616. doi: 10.1002/jmri.25606. [DOI] [PubMed] [Google Scholar]
- 24.Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Research. 2017;19(1):57. doi: 10.1186/s13058-017-0846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y-q, Liang C-h, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. Journal of Clinical Oncology. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 26.Wu S, Zheng J, Li Y, et al. A Radiomics Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. Clinical Cancer Research. 2017;23(22):6904–6911. doi: 10.1158/1078-0432.CCR-17-1510. [DOI] [PubMed] [Google Scholar]
- 27.Dong Y, Feng Q, Yang W, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. European Radiology. 2017:1–10. doi: 10.1007/s00330-017-5005-7. [DOI] [PubMed] [Google Scholar]
- 28.Fujii T, Yajima R, Tatsuki H, et al. Significance of lymphatic invasion combined with size of primary tumor for predicting sentinel lymph node metastasis in patients with breast cancer. Anticancer research. 2015;35(6):3581–3584. [PubMed] [Google Scholar]
- 29.La Verde N, Biagioli E, Gerardi C, et al. Role of patient and tumor characteristics in sentinel lymph node metastasis in patients with luminal early breast cancer: an observational study. SpringerPlus. 2016;5(1):114. doi: 10.1186/s40064-016-1720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budhu A, Forgues M, Ye Q-H, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer cell. 2006;10(2):99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X-D, Zhang J-B, Zhuang P-Y, et al. High expression of macrophage colony-stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. Journal of clinical oncology. 2008;26(16):2707–2716. doi: 10.1200/JCO.2007.15.6521. [DOI] [PubMed] [Google Scholar]
- 32.Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings. European radiology. 2017;27(10):4188–4197. doi: 10.1007/s00330-016-4637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nioche C, Orlhac F, Boughdad S, et al. A freeware for tumor heterogeneity characterization in PET, SPECT, CT, MRI and US to accelerate advances in radiomics. Journal of Nuclear Medicine. 2017;58(supplement 1):1316–1316. [Google Scholar]
- 34.Laws KI. Image Processing INST. University of Southern California; Los Angeles: 1980. Textured image segmentation. [Google Scholar]
- 35.He H, Bai Y, Garcia EA, Li S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. Neural Networks, 2008 IJCNN 2008(IEEE World Congress on Computational Intelligence) IEEE International Joint Conference on: IEEE; 2008. pp. 1322–1328. [Google Scholar]
- 36.Kuijs V, Moossdorff M, Schipper R, et al. The role of MRI in axillary lymph node imaging in breast cancer patients: a systematic review. Insights into imaging. 2015;6(2):203–215. doi: 10.1007/s13244-015-0404-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasparini G, Weidner N, Bevilacqua P, et al. Tumor microvessel density, p53 expression, tumor size, and peritumoral lymphatic vessel invasion are relevant prognostic markers in node-negative breast carcinoma. Journal of Clinical Oncology. 1994;12(3):454–466. doi: 10.1200/JCO.1994.12.3.454. [DOI] [PubMed] [Google Scholar]
- 38.Ocaña A, Diez-Gónzález L, Adrover E, Fernández-Aramburo A, Pandiella A, Amir E. Tumor-infiltrating lymphocytes in breast cancer: ready for prime time? Journal of Clinical Oncology. 2015;33(11):1298–1299. doi: 10.1200/JCO.2014.59.7286. [DOI] [PubMed] [Google Scholar]
- 39.Uematsu T. Focal breast edema associated with malignancy on T2-weighted images of breast MRI: peritumoral edema, prepectoral edema, and subcutaneous edema. Breast Cancer. 2015;22(1):66–70. doi: 10.1007/s12282-014-0572-9. [DOI] [PubMed] [Google Scholar]
- 40.Orlhac F, Nioche C, Buvat I. Technical appendix—local image features extraction——LIFEx—. Paris: 2016. [Google Scholar]
- 41.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Transactions on systems, man, and Cybernetics. 1989;19(5):1264–1274. [Google Scholar]
- 42.Chen P-C, Lin D-J, Jao J-C, Hsiao C-C, Lin L-M, Pan H-B. The Impact of Flip Angle and TR on the Enhancement Ratio of Dynamic Gadobutrol-enhanced MR Imaging: In Vivo VX2 Tumor Model and Computer Simulation. Magnetic Resonance in Medical Sciences. 2015;14(3):193–202. doi: 10.2463/mrms.2014-0048. [DOI] [PubMed] [Google Scholar]
- 43.Haralick RM, Shanmugam K. Textural features for image classification. IEEE Transactions on systems, man, and cybernetics. 1973;(6):610–621. [Google Scholar]
- 44.Xu D-H, Kurani AS, Furst JD, Raicu DS. Run-length encoding for volumetric texture. Heart. 2004;27:25. [Google Scholar]
- 45.Thibault G, Fertil B, Navarro C, et al. Shape and texture indexes application to cell nuclei classification. International Journal of Pattern Recognition and Artificial Intelligence. 2013;27(01):1357002. [Google Scholar]
- 46.Laws KI. Rapid texture identification. IMAGE PROC FOR MISSILE GUID. 1980;238:376–381. [Google Scholar]