SUMMARY
Osteoblasts are matrix-depositing cells that can divide and heal bone injuries. Their deep tissue location and the slow progression of bone regeneration challenge attempts to capture osteoblast behaviors in live tissue at high spatiotemporal resolution. Here, we have developed an imaging platform to monitor and quantify individual and collective behaviors of osteoblasts in adult zebrafish scales, skeletal body armor discs that regenerate rapidly after loss. Using a panel of transgenic lines that visualize and manipulate osteoblasts, we find that a founder pool of osteoblasts emerges through de novo differentiation within one day of scale plucking. These osteoblasts undergo division events that are largely uniform in frequency and orientation to establish a primordium. Osteoblast proliferation dynamics diversify across the primordium by two days after injury, with cell divisions focused near, and with orientations parallel to, the scale periphery, occurring coincident with dynamic localization of fgf20a gene expression. In posterior scale regions, cell elongation events initiate in areas soon occupied by mineralized grooves called radii, beginning approximately 2 days postinjury, with patterned osteoblast death events accompanying maturation of these radii. By imaging at single-cell resolution, we detail acquisition of spatiotemporally distinct cell division, motility, and death dynamics within a founder osteoblast pool as bone regenerates.
eTOC
Cox et al. present a model for real time imaging of bone regeneration. When an adult zebrafish loses a scale, de novo differentiation and vigorous Fgf-regulated division establish a pool of mineral-depositing osteoblasts. Osteoblasts then display patterned behaviors in division, shape changes, and death events to sculpt the regenerating structure.
INTRODUCTION
Mammalian bone has the capacity throughout life to regenerate in response to fracture injury. This regeneration is largely mediated by osteoblasts, which appear at injuries, divide, and secrete new bone matrix components. However, there are hurdles to regeneration after a trauma like limb amputation, such as inflammation and scarring, the paucity of mitogens and patterning factors once present in the embryo, and lowered competence of adult tissue to participate in complex morphogenesis [1-3].
By contrast, urodele salamanders like newts and teleosts like zebrafish display robust regeneration of skeletal bone upon amputation [4]. Multiple recent studies have employed retrospective fate-mapping to define lineage relationships during appendage regeneration and infer cellular behaviors like proliferation, motility, and differentiation that might give rise to regenerated structures [5-8]. However, this visualization method has a limited ability to identify dynamic and quickly resolved cell behaviors such as migratory bursts, cell shape changes, orientations of cell divisions, and apoptosis. Long-term, live imaging can capture these dynamic events and has improved understanding of development, homeostasis, and intrinsic responses to insults by stem cell populations [9, 10]. Challenges to real-time studies of skeletal regeneration include tissue depth, complexity, and relatively slow rates of regeneration that may require continuous live imaging over days. Initial live-imaging studies in bony appendages have captured dynamics of cells in multiple tissue types of the limb or fin blastema, albeit with infrequent sampling of a fraction of the structure [8, 11-13].
The teleost body surface is covered by scales, bony plates of integumentary skeleton that contain osteoblast monolayers. Here, to track behaviors of scale osteoblasts in vivo, we adapted protocols for long-term anesthesia of adult zebrafish to enable up to 24 hours of continuous live imaging [14, 15]. With new computational tools and a panel of transgenic lines that express fluorescent and photo-switchable reporter genes in osteoblasts, we captured and quantified behaviors of osteoblasts as they form the initial scale pattern. Our imaging platform tracks an entire osteoblast population and quantifies its dynamic cell behaviors at single-cell resolution in a regenerating appendage. Additionally, the imaging and computational methods presented here should be applicable to a broad range of model organisms and tissues.
RESULTS
Progressive Renewal of the Osteoblast Population during Scale Regeneration
Fish scale development begins in the late larval stage andstage and parallels mammalian dermal bone development, with mature scales bearing structural similarities to bones such as the tibia in their arrangements of collagen fibers [16-18]. In many species including zebrafish, scales undergo significant regenerative growth within days of scale loss due to mating or fighting, or after experimental plucking. Osteoblasts repopulate the scale pocket and begin mineralization within two days in zebrafish, and more complex morphological features mature over the following days to weeks (Figure 1A) [19]. Features of adult scales include organization into an outer, or episquamal, layer of osteoblasts that associate with patterned structures of mineralized bone called circuli and radii, as well as an inner, hyposquamal layer of osteoblasts (Figure 1B) [20, 21]. As flat, disc-shaped surface appendages, scales are conducive to live imaging.
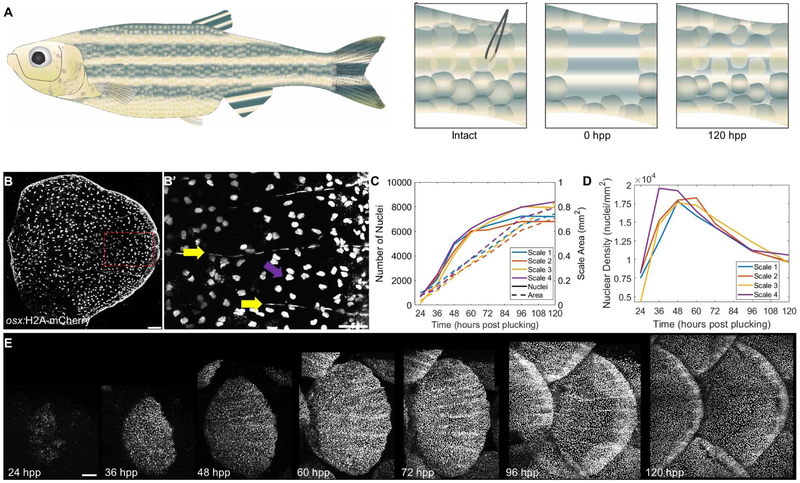
Figure 1. Zebrafish Scale Regeneration.
(A) Model for scale injury. 16-20 scales are plucked with forceps from the caudal peduncle, as its relatively flat surface allows easier z imaging. By 120 hpp, the scales have replenished osteoblast number and much of the mature scale area. (B) Mature scale of an adult osx:H2A-mCherry. fish Scale bar = 100 μm. (B’) Zoom of boxed portion of (B) indicating elongated osteoblast nuclei aligned along the mineralized scale radii (yellow arrows) and rounder nuclei on the bottom, incompletely mineralized layer (magenta arrows). Scale bar = 50 μm. (C) Quantification of osteoblast nuclear number (left axis) and scale area in mm2 (right axis) in four regenerating scales from two fish. (D) Osteoblast nuclear density over time in the four scales quantified in (C). (E) Time course of the first 5 days of scale regeneration in osx:H2A-mCherry fish. Scale bar = 100 μm. See also Figure S1, Videos S1 and S2.
To monitor osteoblast dynamics during scale regeneration, we generated a transgenic line to label nuclei of osterix-expressing cells with mCherry-tagged histones (osx:H2A-mCherry) (Figure 1B). We then plucked scales (4-5 each from four rows, ~16-20 scales total) from the caudal trunk of adult osx:H2A-mCherry transgenic fish and imaged once every 12 hours until 72 hpp, and once every 24 hours after this (Figure 1C). Osteoblasts were first detectable in the scale primordium by about 24 hours post-plucking (hpp). They rapidly increased in number in the 24 h after their first appearance, with 48 hpp scales having on average 740% ± 200% more nuclei than 24 hpp scales (from an average of 550 vs. 4621 nuclei). During this time, increases in osteoblast number outpaced overall scale growth, leading to elevated osteoblast density (Figure 1D). This expansion of the osteoblast population slows over the period from 48 to 72 hpp, with a 42% ± 8% increase to an average of 6562 nuclei. Subsequently, the numbers of osteoblasts plateau, undergoing just a 14% ± 2% increase from 72 to 96 hpp and virtually no change (1% ± 1%) from 96 to 120 hpp (Figure 1C). However, scales continue to increase area, leading to a decrease in density of osteoblast nuclei (Figure 1D-E). Thus, zebrafish scale regeneration involves a rapid expansion of the number of osteoblasts that can conceivably be tracked in toto.
The Osteoblast Founder Pool Arises by De Novo Differentiation
During regeneration of partially amputated zebrafish fins, new ray osteoblasts arise from multiple sources. Genetic fate-mapping of osx-expressing cells first identified cell cycle re-entry by spared osteoblasts near the injury site as a primary source [8, 22]. Ancillary sources can boost their contributions in extreme situations; for instance, the regeneration of amputated fin rays occurs at a normal rate even after virtually all osteoblasts are ablated using nitroreductase-based genetic tools [23]. A recent study found that non-osteoblast precursors located in the joints between ray segments are one of these secondary sources [5].
To investigate the source of the initial pool of osteoblasts in scales regenerating after their complete extirpation, we performed several experiments. First, we genetically ablated osx-expressing cells before plucking scales. In these experiments, metronidazole treatment caused rapid loss of all detectable scale fluorescence from the osx:mCherry-NTR transgene, indicating ablation of virtually all osteoblasts (Figures 2SA’ and S2E) [23]. Interestingly, fish lacking pre-existing osteoblasts regenerated scales at the same rate as controls (Figure S2A-D; n = 7 each for vehicle- and MTZ-treated fish, p = 0.1528 at 48 hpp, p = 0.1740 at 72 hpp). These results indicated that, as for fin regeneration, pre-existing osx-expressing cells are not required for scale regeneration.
Next, to quantify potential de novo osteoblast contributions during the process of regeneration, we generated a transgenic line expressing the green-to-red photoconvertible protein Kaede in osteoblasts (osx:kaede). We photo-converted Kaede to its red fluorescent conformation in adult scales before plucking (Figure 2A-C). At 24 hpp, all new osteoblasts expressed only unconverted green Kaede protein, indicating that these osteoblasts were generated from cells not activating osx regulatory sequences at the time of illumination (Figure 2D-E). We also used osx:kaede animals to investigate the extent to which new osteoblast differentiation contributes to the total osteoblast pool at different stages of regeneration. For these experiments, we imaged different regenerating scales at several time points between 24 and 36 hpp, before photoconverting and imaging the same scales 12 h later (Figure 2F-H). We then calculated the area of the scale containing red protein, representing osteoblasts present at the time of photo-conversion. Our results indicated that de novo osteoblast formation contributes ~50% of the osteoblasts generated from 24-36 hpp or 30-42 hpp. Yet, from 36-48 hpp, only ~10% of osteoblasts arise through de novo differentiation, with the remaining ~90% likely manifest by osteoblast proliferation (n = 6 scales per time point).
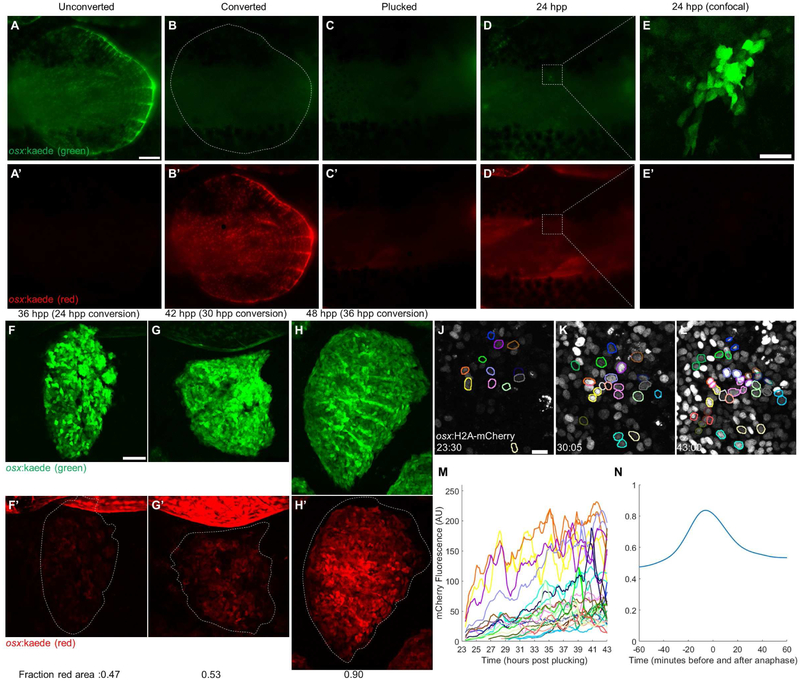
Figure 2. The Initial Scale Osteoblast Pool Regenerates by de novo Differentiation.
(A-E) A single scale was cleared of overlapping scales (A-A’), osteoblasts were photoconverted to red (B-B’), and the scale was removed (C-C’). One day later, all osteoblasts present had no detectable photoconverted protein (D-E’). Panels A-D acquired with a fluorescence dissecting scope (scale bar = 250 μm), panel E acquired with a confocal microscope (scale bar = 50 μm). (F-H) Scales were allowed to regenerate to 24, 30, and 36 hpp, then photoconverted and imaged again at 36 (F-F’), 42 (G-G’), and 48 hpp (H-H’), respectively. The average fraction of red-expressing area for each time point is indicated in the bottom left corner of each image (n = 6 scales per time point). Scale bar = 100 μm. (J-L) Representative pictures from a video of scale regeneration in osx:H2A-mCherry fish, acquired from 23-43 hpp. Colored nuclear outlines indicate tracked nuclei. Outlines present in K and L but not in J represent nuclei first detected later in the video; outlines of the same color indicate daughter nuclei of a dividing nucleus. Scale bar = 10 μm. (M) Fluorescence levels of tracked nuclei. Line colors correspond to outlines in J-L. Splitting of one line into two denotes mitosis. (N) Average fluorescence level of 91 tracked nuclei relative to their peak from 60 minutes before to 60 minutes after anaphase. By 60 minutes postanaphase, nuclear fluorescence decreased 36.1% from its peak but increased 12.5% from the start of tracking. See also Figure S2.
To visualize de novo differentiation in vivo, we developed an imaging platform that maintains adult zebrafish anaesthetized and immobile for up to 24 h (described in Methods and Figure S1). We initiated imaging of osx:H2A-mCherry zebrafish at approximately 24 hpp, when osteoblasts begin to appear beneath the epidermis (Figure 2J). After imaging for 20 h, we reverse-tracked cells from the end of the video and quantified changes in reporter fluorescence levels for hundreds of cells using custom MATLAB software (Figure 2J-L). Our analyses revealed accumulation of osx-driven fluorescence starting from fluorescence levels indistinguishable from fluorescence background, as expected for cells that had only recently activated osx regulatory sequences (Figure 2M). Imaging across multiple z planes showed little to no change in tissue depth of new osteoblasts, indicating that changes in fluorescence are not due to cells moving into the plane of focus and arguing against emergence of a once-cloaked population of differentiated osteoblasts. In several nuclei we observed a rapid increase and decrease in fluorescence associated with chromosome condensation and segregation, indicative of cell division (Figure 2N). In total, our observations indicate that scale regeneration begins with de novo osteoblast differentiation to create a founder pool by 24 hpp. Within the next day, direct duplication of osteoblasts rapidly supersedes additional de novo differentiation.
Spatiotemporally Distinct Osteoblast Proliferation Dynamics during Scale Regeneration
To identify spatiotemporal patterns of osteoblast behaviors during regeneration, we quantified the regional distribution of nuclei over the course of regeneration (see Experimental Procedures). We found higher density of osteoblast nuclei at the center relative to the entire scale until about 48 hpp. From that point, the distribution begins to change, and, by 72 hpp, becomes dense at the peripheries (Figure 3A, Table S1). This peripheral packaging of osteoblasts was unlikely to result from regionally biased differentiation events, given that de novo osteoblast production was greatly reduced by 36 hpp (see Figure 2F-H above).
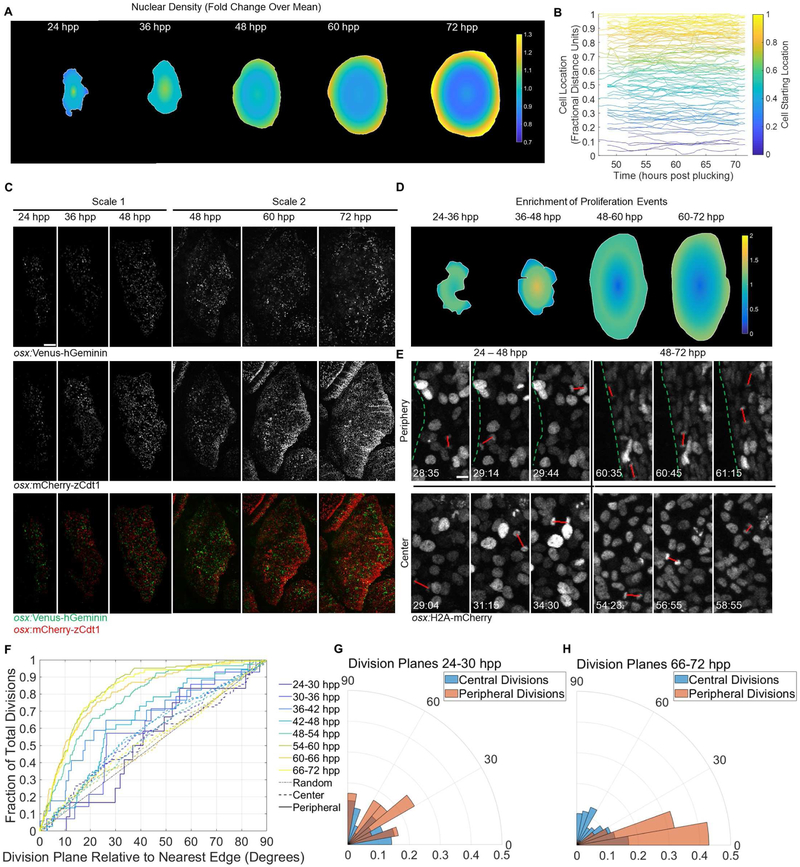
Figure 3. Osteoblast Proliferation Dynamics Acquire Peripheral Bias during Scale Regeneration.
(A) Heat map showing nuclear density across regenerating scales over time. n = 6 scales per time point. Color bar indicates relative change in nuclear density over entire scale at that time point. (B) Relative radial distance of 113 individually tracked nuclei in videos of a 48-72 hpp regenerating scale. Each line represents an individual nucleus, color-coded based on starting point. (C). osx:Venus-hGeminin; osx:mCherry-zCdt1 fish imaged from 24-48 (left three columns) and 48-72 (right three columns) hpp. Top row: osx:Venus-hGeminin (S/G2/M cells). Middle row: osx:mCherry-zCdt1 (G1/G0 cells). Bottom row: overlay. Distribution of cell division shifts outward to the scale periphery over time. Scale bar = 100 μm. (D) Heat map showing changes in proliferation relative to osteoblast density in scales over time. The distribution of osteoblast divisions from 24-72 hpp was divided by the density distribution calculated in (A). (E) Panels from two osx:H2A-mCherry videos indicating orientation planes of divisions (red lines) near the edge (top row) and in the center (bottom row) of the scale from 24-48 hpp (left column) and 48-72 hpp (right column). Dotted green lines indicate scale edge. Scale bar = 10 μm. (F) Cumulative distribution function of division orientations (degrees) shown in (E). Divisions binned in 6-h windows. Solid lines indicate divisions in the outer 10% of the scale area; dashed lines indicate divisions in the inner 90%. Gray dotted line indicates randomly oriented divisions. Fraction of divisions on the y axis is cumulative, i.e. at x = 10 degrees, the y value indicates the number of divisions in that time bin within 0-10 degrees of parallel to the edge, divided by all divisions in the bin; at x = 20, y value indicates all divisions within 0-20 degrees of parallel, etc. until the fraction reaches 1.0. (G-H) Probability polar histograms indicating division plane relative to plane of scale edge for the first (G) and last (H) time bins from (F). See also Figure S3 and Table S1.
To test whether changes in cell migration and/or proliferation account for changes in osteoblast density, we performed long-term (20-h), continuous live imaging on six scales, three each in the 24 to 48 hpp and 48 to 72 hpp windows (Videos S1 and S2). First, we assigned a distance value between 0 and 1.0 for each nucleus based on its location between the centroid (0) and nearest scale edge (1.0) and tracked this fractional distance for the duration of the video (n = 592 tracks, at least 75 tracks per video, selected tracks shown in Figure 3B). With this system, nuclei could change physical location without changing fractional distance if they moved outward at the same rate of scale growth. We found that the average nucleus changed its location less than 0.01 fractional distance units. Based on the relatively stable position of nuclei in the scale at 48 hpp, we concluded that cell migration was unlikely to account for the peripherally biased nuclear density.
To detect bias in osteoblast proliferation events, we generated osx:Venus-hGeminin and osx:mCherry-zCdt1 transgenic lines that express components of the fluorescent ubiquitination-based cell cycle indicator (FUCCI) system, in which fluorophores are tagged with degradation sequences of cell cycle proteins [24]. Live imaging of these reporters in regenerating scales indicated reciprocal oscillating of Venus (S/G2/M phases) and mCherry (G1/G0) fluorescence, enabling discrimination of cell cycle phases. Mimicking the maps of nuclear density, early regenerating scales appeared to have an even or slightly center-biased distribution of Venus-hGeminin+ osteoblasts, whereas scales at 72 hpp appeared to contain more Venus-hGeminin+ osteoblasts near the periphery (Figure 3C). Quantification of these images revealed a statistically significant increase in the ratio of Venus to mCherry expression at the center of the scales at 36 hpp and at the edges of the scales at 72 hpp (p < 10−10 and p < 10−17 chi-square test, n = 4 scales imaged at 24-48 hpp and 4 scales imaged at 48-72 hpp). At other time points, the difference between peripheral and center fluorescence was not significant. These FUCCI data suggested an emerging bias in proliferation dynamics during scale regeneration.
As relative levels of fluorescent reporters alone are not sufficient to describe proliferation rates, we analyzed videos of regenerating osx:H2A-mCherry scales from 24 to 72 hpp and placed all divisions into 6-h bins. Three of the four bins prior to 48 hpp showed a slight bias toward centrally located divisions; only the earliest (24-30 hpp) was not significantly oriented to central divisions (Figure S3A). Across these four time bins, between 5% and 12% of divisions occurred within 0.1 fractional units of the scale edge (0.9-1.0). By contrast, from 48-72 hpp, divisions occurred more frequently farther from the center of the scale than random, with the cumulative distribution function plot of each bin moving successively farther from expected random values: 19%, 26%, 32%, and finally 36% of divisions occurred near the scale periphery (Figure S3A). We used these data to generate heat maps projected onto a “virtual scale” indicating the regions of increased proliferation. Importantly, these maps resembled our FUCCI images and revealed increased proliferation rates near scale edges (Figure 3C-D). Thus, as scale regeneration progresses, the spatial dynamics of proliferation events shift from uniformity to a peripheral bias.
To identify additional regional division biases, we analyzed the angles of division for all mitotic events (Figure 3E), calculating in degrees the absolute value of each angle of division minus the angle of the closest scale edge. We found that 24-48 hpp osteoblasts displayed a slight overall bias to divide in parallel with the edge beginning with the 30-36 hpp bin, but interior and peripheral populations did not show a significant difference in directionality of division planes (Figure 3F). By contrast, from 48-72 hpp, peripheral osteoblasts in all time bins were biased toward dividing in parallel with the closest edge compared to their interior counterparts. This change was especially apparent when comparing division planes between the first and last time bins (Figure 3G-H). Thus, as scale regeneration progresses, osteoblasts at the extreme boundaries of the scale develop a bias for division that maintains their positions in a circular, leading edge of osteoblasts, as opposed to changing position through perpendicular divisions.
During our videos from 48 to 72 hpp, we began to notice apparent differences in hyposquamal and episquamal osteoblast behaviors. In earlier phases of scale regeneration, it is difficult to assign cells to the episquamal or hyposquamal layer. Thus, we analyzed tracked osteoblasts from 48 to 72 hpp for differences in behaviors, focusing specifically on an 18-h window between 52 and 70 hpp that the three videos shared. Episquamal cells moved at a slightly higher average velocity and underwent larger overall displacement from their initial to final position in two of the three videos. The third video showed a similar trend but did not reach significance (Figure S3B-C; displacement: p < 10−7 for scales 1 and 2, p = 0.25 for scale 3; velocity: p <10−15 for scale 1, p = 10−4 for scale 2, p = 0.11 for scale 3). We also found that episquamal cells proliferate at a lower rate (6%) than hyposquamal cells (26%; Figure S3D; p = 0.022, n = 3 scales). Overall, osteoblasts display heterogeneous dynamics across three dimensions of the scale during regeneration.
Osteoblast Proliferation Dynamics Track Fgf Ligand Expression
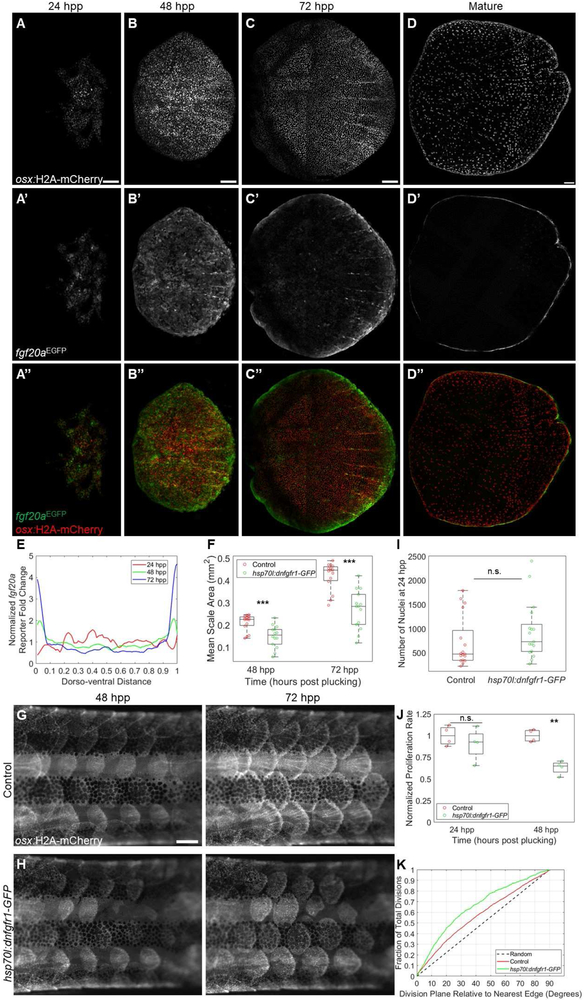
We hypothesized that spatiotemporal changes in osteoblast division were likely responding to signaling dynamics. Previously, Rohner and colleagues reported that mutations in the zebrafish fgfr1a gene perturb scale development, leading to fewer but enlarged scales in adult animals [25]. Aman et al. recently found that pharmacological pan-Fgf receptor inhibition arrested initiation and growth of scales in juvenile fish [26]. To identify dynamism of potential Fgf signals during scale regeneration, we examined scale fluorescence in an EGFP enhancer trap line that reports expression of the fgf20a ligand gene [27]. fgf20a is expressed in scale peripheries in growing juvenile zebrafish, and compound mutant zebrafish heterozygous for both fgfr1a and fgf20a mutations show scale growth defects [28]. Confocal imaging indicated that the fgf20aEGFP reporter is expressed during scale regeneration (Figure 4A-D). Upon scale plucking, expression was first detectable at ~24 hpp and relatively evenly distributed across the dorsoventral axis (Figure 4A). By 48 hpp, reporter fluorescence levels at the dorsal and ventral peripheries (within 10% of the edge) were 1.5 and 1.6-fold the levels of the mean reporter expression, respectively, and exceeded two-fold at points within the peripheries (Figure 4B). The distinction was more conspicuous at 72 hpp, when dorsal and ventral extremes expressed EGFP at 2.1 and 2.7-fold the levels of the mean reporter expression, reaching 4.6 fold at the most extreme periphery of the scale (Figure 4C). Sections through scales of fgf20aEGFP fish revealed reporter expression in basal p63+ cells and in osteoblasts (Figure S4). We occasionally found EGFP expression in basal cells at 24 hpp without detectable expression in presumptive osteoblast regions, indicating fgf20a may be induced in basal cells before osteoblasts (Supplemental Figure 4D). Conversely, at 72 hpp, EGFP was widespread in osteoblasts but comparatively lower in basal cells (Figures S4C and SF).
Figure 4. Fibroblast Growth Factor Signaling Modulates Scale Regeneration.
(A-C”) osx:H2A-mCherry; fgf20aEGFP regenerating scales at 24 (A-A”), 48 (B-B”), and 72 (C-C”) hpp. fgf20a enhancer trap fluorescence (middle row) is initially distributed evenly but later localizes to the scale periphery. Scale bar = 100 μm. (D-D”) Mature adult osx:H2A-mCherry; fgf20aEGFP zebrafish scale with fluorescence restricted to the scale periphery. Scale bar = 100 μm. (E) Quantification of fold change relative to mean EGFP expression in regenerating scales from 24-72 hpp. (F) Box and whisker plot of average scale area for wild-type (red circles) and hsp70l:dnfgfr1-GFP (green circles) fish at 48 and 72 hpp. hsp70l:dnfgfr1-GFP fish had smaller scales at both time points (n=15 control, 14 hsp70l:dnfgfr1-GFP fish). (G-H) Regenerating scales at 48 (left column) and 72 (right column) hpp in heat-shocked osx:H2A-mCherry; hsp70l:dnfgfr1-GFP fish (H) and osx:H2A-mCherry siblings lacking the heat shock transgene (G). Scale bar = 500 μm. (I) Box and whisker plot comparing number of nuclei counted in individual control and hsp70l:dnfgfr1-GFP fish at 24 hpp. No significant difference was detected (n = 17 control scales, 17 hsp70l:dnfgfr1-GFP scales). (J) Box and whisker plot showing proliferation rate of control and hsp70l:dnfgfr1-GFP scales imaged in the 24 hpp+ range and 48 hpp+ range, normalized to the mean proliferation rate of time-matched controls. Proliferation is significantly reduced in hsp70l:dnfgfr1-GFP fish at 48 hpp+ (n = 4 control and 4 hsp70l:dnfgfr1-GFP scales, 2 each imaged from 48-58 hpp and 53-63 hpp) but at 24 hpp+ (n = 4 control and 4 hsp70l:dnfgfr1-GFP fish, 2 each imaged from 24-34 hpp and 27-37 hpp). (K) Cumulative distribution function of division plane relative to nearest scale edge for control and hsp70l:dnfgfr1-GFP scales imaged from 48 hpp onward. hsp70l:dnfgfr1-GFP fish show a greater bias toward dividing in line with the plane of the nearest edge. See also Figure S4.
To examine requirements for Fgf signaling in scale regeneration, we used a heat-shock inducible hsp70l:dnfgfr1-GFP line to inhibit Fgf receptors [29]. Mature scales did not show any significant size difference between hsp70l:dnfgfr1-GFP fish and control siblings (n = 9 control, 9 transgenic fish, p = 0.95). However, daily heat shocks inhibited regeneration of plucked scales, reducing average scale area by 33% at 48 hpp and 37% at 72 hpp (Figure 4F-H; n = 15 wild-type, 14 hsp70l:dnfgfr1-GFP, p < 0.001 at 48 hpp, p < 10−4 at 72 hpp). To address the mechanism of inhibition, we quantified the number of osteoblasts present in scales at 24 hpp, at which point de novo differentiation has generated the starting pool. No significant difference was observed, suggesting no prominent role for Fgf signaling in osteoblast differentiation (Figure 4I; p = 0.25, n = 17 wild-type scales from 5 fish, 17 hsp70l:dnfgfr1-GFP scales from 5 fish). We assessed potential differences in proliferation by taking multiple 10-h videos during the first and second days post-plucking and marking each division event (a total of 4,955 divisions from scales across 80 total h of imaging). While we did not find a significant decrease in proliferation at 24 hpp, hsp70l:dnfgfr1-GFP scale osteoblasts imaged in the 48 hpp range showed a 37% decrease in proliferation rate relative to controls (Figure 4J; p < 0.01, n = 4 control scales from 2 fish, 4 hsp70l:dnfgfr1-GFP scales from 2 fish). The cumulative distribution functions of division locations were similar between control and hsp70l:dnfgfr1-GFP scales (data not shown), but we detected a significant bias toward divisions in parallel with scale edges in transgenic tissue (Figure 4K; p < 10−7, n = 2,163 divisions from control and 613 divisions from hsp70l:dnfgfr1-GFP fish). Thus, our evidence indicates that the proliferation dynamics of osteoblasts associate spatiotemporally with expression of a ligand with the potential to trigger a key signaling pathway during scale regeneration.
Osteoblast Behaviors Sculpt Regenerating Radii
As scales mature, their outer layers form mineralized canals called radii that harbor blood vessels[20]. A recent study by Rasmussen and colleagues found that radii also contain nerves, and scale growth and radial tracts of osteoblasts are essential for proper vascularization and innervation of juvenile zebrafish skin [30]. We noticed that queues of osteoblasts representing nascent radii first appear during regeneration at approximately 48 hpp and spread anteriorly across the scale surface (Supplemental Figure 5A-G). To determine how osteoblasts form radii, we tracked the location and shape of cells that initiate radius formation using the osx:EGFP-CAAX line to mark osteoblast membranes. We imaged three different scales beginning shortly before 48 hpp and then tracked cells for 10-12 h (Figure 5A-B; Video S3) [31]. We measured cell shape at 2-h intervals in 199 cells (132 non-radius, 67 radius cells) and calculated the aspect ratio of each cell. The major axes of cells within the growing radius aligned with the direction of radius growth, arguing that changes in aspect ratio might contribute to growth of the radius (Figure 5C). Indeed, over the course of imaged radius growth, radial osteoblasts increased their aspect ratio from an average of 1.72 ± 0.08 vs. 2.2 ± 0.1, a 27% increase. Conversely, non-radius osteoblasts showed a 9% aspect ratio decrease, from 1.81 ± 0.06 vs. 1.64 ± 0.05 on average (Figure 5D). Chi-square tests confirmed a significant difference in the average aspect ratios of the two populations and a significant increase in radius aspect ratio, but the decrease in non-radius aspect ratio was on the edge of significance (p < 10−10 for comparison of populations, p < 0.001 for radius cells, and p = 0.0956 for non-radius cells). Notably, the two populations were not statistically different at the start of imaging but became so by 50 hpp (unpaired t-test, p < 10−5 at 50 hpp). Increases in the aspect ratio of radius cells were due to elongation of the major axis (14.6 ± 0.5 μm vs. 17.0 ± 0.7 μm, p = 0.003), accompanied by a small decrease in the minor axis that was not statistically significant (8.8 ± 0.3 μm vs. 8.1 ± 0.2 μm, p = 0.1471) (Figure 5E). On the contrary, the small decrease in the aspect ratio of non-radius cells was due to a small decrease in the major axis (16.6 ± 0.4 μm vs. 15.4 ± 0.3 μm, p = 0.0101), but no significant change in the minor axis (9.5 ± 0.2 μm vs. 9.7 ± 0.2 μm, p = 0.8083) (Figure 5E). Changes in cell area did not achieve statistical significance by chi-square tests (Figure 5F; radius cells, 97 ± 5 μm2 vs. 100 ± 4 μm2, p = 0.853; non-radius cells, 115 ± 4 μm2 vs. 109 ± 3 μm2, p = 0.117). However, the difference in area between the two populations displayed the opposite trend from aspect ratio, starting as statistically significant but losing that distinction by 50 hpp (unpaired t-test, p = 0.323 at 50 hpp). These data reveal cell shape changes along the radial axis as a major morphogenetic event associated with radius growth.
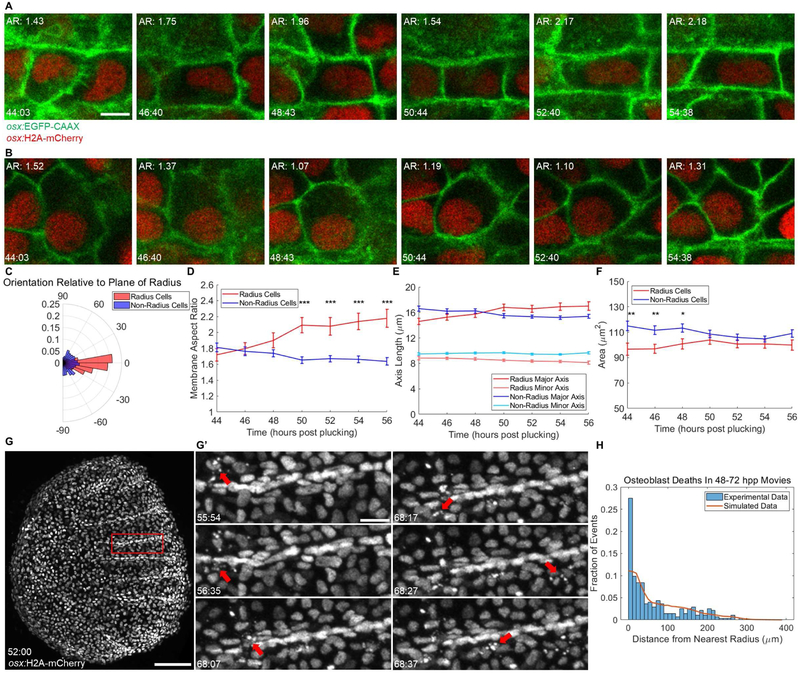
Figure 5. Scale Radii Regenerate by Patterned Osteoblast Shape Changes and Cell Death Events.
(A-B) Individual tracked radius- (A) and non-radius-forming (B) osteoblasts over during a 10-h video, shown at approximately 2-h intervals. Top left of each panel: membrane aspect ratio. Bottom left: time post-plucking (hours:minutes). Scale bar = 5 μm. (C) Polar histogram indicating orientation of the major axis of tracked osteoblasts to the plane of radius growth (n = 132 non-radial osteoblasts, 67 radial osteoblasts, imaged 10-12 h). (D) Average membrane aspect ratio for radius- (red line) and non-radius-forming cells (blue line) over time. By 50 hpp. the aspect ratio of radius osteoblasts is significantly greater than non-radius osteoblasts. (E) Data from (D) decomposed into changes in length of major (darker shading) and minor (lighter shading) axes of radius (red) and non-radius cells (blue). (F) Changes in mean areas of radius and non-radius osteoblasts. Increases in radius cell area and decreases in nonradius cell area are not individually significant but result in loss of a significant difference between areas of radius and non-radius cells. (G) A regenerating scale expressing osx:H2A-mCherry at 52 hpp, the first time point of a 20-h video. Scale bar = 100 μm. (G’) Panels depicting nuclear fragmentation (red arrows) at various times from the region outlined in (G). Scale bar = 20 μm. (H) Histogram of cell death data from three >20-h videos spanning 48-72 hpp total. 273 hand-counted cell deaths were plotted by distance from the nearest radius (blue bars, bins of 10 microns). Observed data were compared to the results of 1000 simulations of randomly placed cell deaths (273,000 random cell deaths, orange line). See also Figure S5 and Video S3.
In a previous study, apoptotic bodies were associated with osteoblasts at the site of developing and healing mammalian bone [32, 33]. Strikingly, we observed in our videos many instances of active fragmentation by osteoblast nuclei within and near forming radii (Figure 5G-G’). In 24-48 hpp scales prior to radius formation, apoptosis was rare and appeared unbiased. However, as regeneration progressed, osteoblast apoptosis was evident in and around osteoblasts forming radii at the scale posterior. In each of 3 videos acquired from 48-72 hpp, osteoblast nuclei fragmented at a significantly elevated rate within 10 microns of a nearby growing radius (Figure 5H). Nearly a third of all cell deaths occurred in the area neighboring the radius (Figure 5D), despite the radii comprising only 1.3% of the scale area on average. Radius-associated deaths tended to occur near the anterior of the growing radius, and there did not appear to be a difference in proliferation rate near radii (Figure S5H-J). Thus, live imaging and cell tracking enables quantification of regulated osteoblast behaviors – shape changes in certain cells and death in others - that accompany the formation of scale radii.
DISCUSSION
Recently developed platforms for large-scale, single-cell sequencing and transcriptome analysis can infer relationships of cell populations without the need for visualization. However, these methods alone do not track cell dynamics that influence tissue structure, such as timing and location of cell divisions [34-36]. Furthermore, as we show in this study, cell death is common in growth and regeneration, and retrospective cell sequencing excludes cells that do not survive to the point of cell harvest. Real-time imaging over long timescales, by contrast, can identify these and other dynamic events that underlie tissue regeneration. Here, we combine a panel of new transgenic lines with an imaging and analysis platform to capture osteoblast behaviors during bone regeneration. Our tools access a cell population that has not been imaged in vivo during regeneration, and our findings provide quantitative information to explain the cellular basis of integumentary bone regeneration.
Previous work reported that original scale osteoblasts in juvenile zebrafish arise from recruitment and differentiation of fibroblasts below the basal epidermis [37]. A recent study addressing the mechanisms of scale regeneration concluded that de novo differentiation was the primary source of new osteoblasts, which then proliferate at very low rates [38]. By contrast, we found that scale regeneration involves a high degree of osteoblast division, especially in the second and third day after injury, and that this direct duplication accounts for most of scale growth. Iwasaki et al. did not track single cell dynamics across long timescales, and they assayed proliferation at time points subsequent to the burst of osteoblast division we observed. Our work highlights the importance of live imaging to reveal cellular mechanisms of regeneration that can escape other methods of analysis.
Our findings suggest that de novo differentiation alone is insufficient to reconstruct plucked scales; instead, a succession of differentiation, proliferation, and parallel routes of shape change or cell death are employed. These phases are not temporally isolated: osteoblasts initiate division before neighboring cells have activated osx expression, and morphogenesis of scale radii begins while osteoblasts continue to proliferate. The mitotic phase is dampened long before mature scale size is recovered, indicating a regenerative program in which cells are replenished rapidly and then later sculpted for detail. Our data suggest that Fgf20a ligand, or more generally Fgf signaling, may guide aspects of this osteoblast proliferation. A recent report indicated that that pharmacological inhibition of Fgf signaling halted both initiation and growth of juvenile scales, whereas our transgenic methods indicate a requirement for Fgf signaling in osteoblast proliferation from 48 hpp onward, but not de novo osteoblast differentiation or earlier proliferation [26]. This possible discrepancy may be due to differences in research tools or mechanisms between the two contexts. The significance of the effects of Fgf receptor inhibition on osteoblast division planes we observed is unclear but may contribute to the reduced size of regenerated scales.
The prevalence of nuclear fragmentation near growing radii indicates that cell death may have functional relevance during radius morphogenesis. It is possible that excess osteoblasts may be necessary to initiate mineralization early in regeneration, which begins around 48 hpp [19]. Then, the release of factors from dying cells or changes in mechanical tension could potentially have instructive or permissive roles in elongation of radii.
The imaging methods presented here are applicable to skin, eyes, and other shallow-depth tissues in zebrafish. Key refinements would increase the length of imaging sessions, for example, by automation of water delivery and oxygen maintenance. New transgenic lines that report real-time signaling pathway readouts in osteoblasts can be integrated to generate molecular models to explain cell behaviors. Or, the osteoblast reporters we describe here can be combined with reporters representing other cell types like skin epithelium [12], to enable imaging of in vivo interactions between key cell types. We expect that cataloged datasets of long-term in vivo imaging of cellular and subcellular events will deconstruct tissue regeneration to a resolution that can inform regenerative medicine.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kenneth Poss (kenneth.poss@duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish
Zebrafish of Ekkwill and Ekkwill/AB strains were maintained between 26-28.5°C with a 14:10 hour light:dark cycle. Fish between 3 and 12 months old were used for experiments. For heat shocks, fish were placed on an independent system that maintained 26-28.5°C water for all but the 1.75 hou rs of the day at the beginning of the light cycle, during which the water was heated to a maximum of 38°C. Zebrafish were placed in the heat shock system the evening before injury. Heat shock occurred between 8:00 am and 9:45 am the following morning, and scales were plucked after ubiquitous expression of EGFP was detected, at approximately 12:00 pm, or 4 hours after onset of heat shock. For osteoblast ablation, 2 osx:mCherry-NTR fish each were placed in a VWR 90 × 50 mm glass dish containing 60 mL fish facility system water (control) or 60 mL system water with 10 μM metronidazole (ablation). Fish were kept in the dark and treated overnight (16 h), then rinsed and returned to system water. For Kaede photoconversion of intact mature scales, fish was anesthetized in tricaine as described in the Imaging section below and placed in a petri dish under the 10X objective of a Zeiss Axio Examiner Z1 microscope. A single scale was identified and focused as quickly as possible using light from an HXP 120V lamp passed through the GFP fluorescence filter, then the filter was switched to blue fluorescence for 1 minute to photoconvert the scale. Transgenic lines used in this study were Tg(osx:H2A-mCherry)pd310 (described below), Tg(osx:mCherry-zCdt1)pd270 (described below), Tg(osx:Venus-hGeminin)pd271 (described below), Tg(osx:kaede)pd64 (described below), Tg(osx:mCherry-NTR)pd46 [23], Tg(osx:EGFP-CAAX)pd51 [31], Tg(hsp70l:dnfgfr1-GFP)pd1 [29], and Tg(fgf20aEGFP)/HGN21A [27].
METHOD DETAILS
Scale injury
Fish were placed in a dish containing 0.09% phenoxyethanol in system water. One fish was anesthetized at a time in phenoxyethanol solution until swimming ceased and operculum movement slowed, then removed from solution and placed on a flat surface such as a petri dish lid. Fish were viewed under a fluorescent dissecting scope so that scales could be distinguished by their fluorescent reporters. For non-transgenic fish or those without fluorescence in the scales, fish were viewed under a normal stereomicroscope. Approximately 16-20 scales, 4-5 each from 4 rows on the trunk of the fish, were removed with forceps from the caudal peduncle of the fish. For experiments in which mature osx:kaede scales were photoconverted, a single scale was left in the center of the otherwise denuded region. All other scales were removed to ensure overlapping tissue would not prevent full photoconversion of a scale. After scale removal, fish were returned to system water to recover from anesthesia, before a return to circulating water.
Construction of transgenic zebrafish
The H2A-mCherry-SV40 polyA sequence was obtained from a plasmid constructed by Dr. Jingli Cao, amplified by PCR using the primers 5’-TTTTTGGATCCGCCGCCACCATGGCAGGTGGAAAAGCAGG-3’ and 5’-TTTTTCTCGAGTTACTTGTACAGCTCGTCCATGC-3’, digested using XhoI, then partially digested using BamHI due to the presence of a BamHI site in the ORF [40]. The fragment was then ligated into a pBluescript SK plasmid that had been modified to contain ISceI sites in the MCS and into which osx:mCherry had already been subcloned. Venus-hGeminin and mCherry-zCdt1 fragments were generated by restriction digest with XhoI and a partial digest with BamHI from a TOPO vector containing each sequence. The mCherry sequence of the aforementioned osx:mCherry vector with ISceI sites was removed by digestion with BamHI and XhoI, and the FUCCI sequences were each ligated into the osx-containing vector. osx:kaede plasmid was constructed by PCR amplification of Kaede sequence with BamHI and XhoI overhangs, digestion with BamHI and XhoI, then subcloning into the osx-containing vector. Plasmids were cut using I-SceI enzyme for 30 minutes at 37°C before injection into AB or EK embryos at the one-cell stage.
Microscopy
All confocal images were acquired using a Leica SP8 confocal microscope and LAS X software, with a 25x FLUOTAR water immersion lens (Leica 15506374). Full images of scales were acquired at 0.75x zoom, resulting in 0.606 μm pixel size. Videos of radii were captured at 3x zoom, resulting in 0.152 μm pixel size. Whenever possible, a z step size equivalent to pixel length and width was used to ensure cubic pixels and allow three-dimensional image permutation and rotation. When time constraints required, z step sizes up to the optical section size (1.7 μm at 1 AU pinhole width) were used. A Zeiss AxioZoom V16 and Zen Pro 2012 software was used to acquire certain low-magnification images. Zeiss Axio Examiner Z1 microscope was used for Kaede photoconversion (described above in Zebrafish section).
Histology
Zebrafish trunks were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. Scales were fixed in PFA for 1 h at room temperature (RT). Tissues were rinsed 3x in phosphate buffered saline (PBS), suspended in 1.5% agarose/5% sucrose in water and placed in 30% sucrose at 4°C overnight or until sin king to the bottom of a 1.5 mL tube. Then, they were frozen in Tissue Freezing Medium (General Data TFM-5) and sectioned by cryostat at 14 μm/section. Slides were rehydrated in PBS with 0.1% Tween (PBST), then blocked in PBST with 10% heat-inactivated newborn calf serum and 1% DMSO (NCS-BPT) with 2% horse serum for 1 h at 37°C. Primary antibody was applied for 3 h at 37°C. Slides were washed in PBST, then secondary antibody was applied for 1 h at 37°C. Slides were washed in PBST, then treated with mounting medium/DAPI.
Imaging
For long-term live imaging (Figure S1A-B), zebrafish were anesthetized in 0.01% tricaine in system water. Fish were allowed to swim in tricaine solution for approximately 15-30 min, or until all swimming ceased but gill movement continued. While awaiting anesthesia, the imaging dish was prepared as follows: plexiglass plate(s) (Figure S1C; two plates were stacked in our imaging setup, but one thicker plate could be used) with a hole 6 cm in diameter were held in place in the center of a plexiglass dish (Supplemental Figure 1D) with nontoxic modeling clay, then a bed of 1% agarose in system water was poured in the center of the plate (Supplemental Figure 1E). A glass microscope slide was then placed at one end of the bed to allow elevation of the caudal fin and attached with modeling clay. For each experiment, one fish was placed on its side on the agarose bed, and its caudal fin was set on a glass slide to bring the caudal peduncle of the trunk parallel with the platform. Several drops of diluted tricaine were placed near the head prior to mounting. Cooling 1% agarose was applied in a circle around the regenerating scale(s) of interest, covering the caudal fin, the trunk anterior to the scale, and the areas of platform dorsal and ventral to the scale. Agarose did not cover or contact the regenerating tissue. Once the agarose solidified, 120 mL of diluted tricaine, or just enough to cover the fish and keep a consistent water level over the area to be imaged, was added to the dish. A peristaltic pump (Cole Parmer Peristaltic Pump #EW-73160-32) with silicone tubing was loaded with system water. The outflow tubing was attached via a cut 200 mL plastic pipette tip to Tygon B-44-4X (0.7 mm inner diameter/2.4 mm outer diameter) tubing to fit inside the mouth. Gill movement was monitored; when movement slowed, system water was applied for approximately 30 seconds at a rate of 3.5 mL/min or until regular rhythm was restored. Approximately once every 4 h, water in the dish was completely replaced with freshly prepared diluted tricaine using a serological pipette. Fish were imaged using a Leica SP8 confocal microscope. After imaging was completed, diluted tricaine water was completely replaced by system water. The peristaltic pump was turned on until gill movement quickened, and the fish was released from the agarose and returned to system water. Based on comparison of nuclear counts from longer, coarser-resolution time courses and manually counted divisions from long-term videos, it is possible that regeneration kinetics are slowed somewhat by long-term anesthesia. However, inspection of fish in the days after long-term imaging did not reveal any noticeable structural defects in scale regeneration.
Image analysis
To measure contributions of converted osx:kaede cells to regenerating scales, the red area of the scale was calculated and divided by the total (red + green) area of the scale. Red area was detected using a MATLAB script to find pixels above threshold background levels that were set for each image based on pixel intensity values outside the scale.
Nuclear counting was performed manually in FIJI (ImageJ) using three-dimensional stacks of each scale [39]. To plot cell density, a MATLAB script was used to determine the location of each individual nucleus and plot its relative location between the centroid of the scale and the edge, whose coordinates were determined by hand-drawing or thresholding osx:EGFP-CAAX signal when available.
For nuclear tracking during live imaging, we wrote a custom MATLAB GUI (available at https://github.com/bdcox/cell_tracking_gui) to display the scale in three spatial dimensions and over time. To achieve unbiased tracking, we randomly generated coordinates and used them to select which nuclei to track. All nuclei were manually tracked, and osx-directed expression data was extracted from a small circular area around the centroid of each nucleus.
For location and division plane of nuclear division analyses, each video was played through 100 square pixels (approximately 36 μm2) at a time, and lines were drawn manually in ImageJ to connect sister nuclei as soon as they were resolvable after telophase. Image stacks were read into MATLAB, and custom scripts calculated plane of division based on the direction of each line. Outlines of each scale over time were extracted using thresholding of the membrane (EGFP-CAAX) channel in MATLAB. For early videos in which an outline could not be generated computationally due to the patchy nature of osteoblast populations, an outline was hand-drawn in ImageJ. Each mitosis was binned based on its relative location between the centroid and nearest point on the scale outline. To test for statistically significant non-random distribution of mitoses, we performed 1000 simulations for each scale video in which a number of mitotic locations and planes of division equal to and time-matched with the real events were randomly generated. Real data was compared to randomly generated data using the Kolmogorov-Smirnov test. Statistical calculations were performed in MATLAB.
For proliferation analyses in control and hsp70l:dnfgfr1-GFP fish, videos were analyzed 100 square pixels at a time as described above, and proliferation rates of each scale were normalized to the mean rate of proliferation for control scales at that time point. Two scales each from a single fish for control and hsp70l:dnfgfr1-GFP were imaged at 24, 27, 48, and 53 hpp.
For quantifying osteoblast density, we assigned a distance value to every nucleus based on its relative position between the centroid (distance = 0.0) and nearest edge (distance = 1.0). Because the scale is not a perfect circle, centroid-edge distance is not a constant radius. This variation, along with the growth in scale size over time, necessitated measuring distance in relative rather than raw metric units. We then calculated the raw and normalized densities of nuclei in intervals of one tenth (0.1) of centroid-edge distance in 12-h intervals between 24 and 72 hpp.
To quantify regional changes in fluorescence in osx:Venus-hGeminin;osx:mCherry-zCdt1 and fgf20aEGFP;osx:H2A-mCherry fish, we cropped a representative section of each scale 100-μm wide that included the entire dorsal-ventral length of the scale and divided it into dorsal-ventral sections such that the dorsal tip was assigned 0 and the ventral tip assigned 1.0. The ratio of Venus to mCherry expression was then measured. The edge region was defined as 0-0.1 or 0.9-1.0. For fgf20aEGFP analysis, we normalized EGFP fluorescence to mCherry fluorescence to account for dimming of signal in extremes of the scale that is likely due to tissue depth at those locations, and we applied a Savitzky-Golay filter to the mean of 6 scales per time point. To quantify aspect ratios, membranes were manually traced in a MATLAB GUI. The regionprops function was applied to each membrane to measure area, major and minor axis length of an ellipse fitted to the membrane outline, and orientation. Aspect ratio was calculated by dividing the major by the minor axis. To assess the significance of changes between radius and non-radius cells, an unpaired t-test was used. To assess internal changes in radius and non-radius cells, a paired t-test was used and time points from 46 to 54 hpp were analyzed, as this was the range which was common to each video of radius growth.
For analyses of cell death and proliferation with respect to their proximity to radii, each video was similarly played through as above, with dots marking every observed nuclear fragmentation or division. Radii were colored manually in ImageJ based on high intensity of osteoblast reporters, then the modified images were read into MATLAB to obtain coordinates of radii. We then calculated distance from each event to the nearest growing radius and again performed random simulations and tested for statistical significance using the K-S test. To test whether average location of cell death along each radius changed over time, we applied a chi-square test to 2 hour bins of cell death events and compared it to the overall mean location of cell deaths across all time points.
QUANTIFICATION AND STATISTICAL ANALYSIS
All average values in figures are depicted as mean ± SEM. For box plots, the top and bottom of each box represent the 25th and 75th percentile, respectively. The line in the middle of each box is the median. Whiskers extend to all data points not deemed outliers (>1.5 interquartile ranges from the top or bottom of box). Sample size, p values, and specific tests used (t test, chi-square test, Kolmogorov-Smirnov test) are indicated in the text in the Results section and Supplemental Figure Legends. Chi-square tests were performed with n-1 degrees of freedom. Statistical calculations were performed using MATLAB or Microscoft Excel. *, p <0.05. ns, p > 0.05
DATA AND SOFTWARE AVAILABILITY
MATLAB Cell Tracking GUI is available at https://github.com/bdcox/cell_tracking_gui.
Supplementary Material
Video S1. Long-Term Live Imaging of an osx:H2A-mCherry Scale on the Second Day of Regeneration. Related to Figure 1. A single scale regenerating between 23:30 hpp and 43:00 hpp. De novo activation of osx-driven fluorescence can be visualized throughout the area of the scale. The edges of intact scales are present at the top of the frame. Left to right: anterior to posterior. Scale bar = 100 μm.
Video S2. Long-Term Live Imaging of an osx:H2A-mCherry Scale on the Third Day of Regeneration. Related to Figure 1. A single scale regenerating between 52:02 hpp and 71:32 hpp. Little to no de novo activation of osx-driven fluorescence is visible. Proliferation is biased toward the scale edges. Nuclei in nascent radii at the posterior half of the scale align and elongate, and nuclear fragmentation near radii can be observed. Left to right: anterior to posterior. Scale bar = 100 μm.
Video S3. Dual Color Imaging in osx:H2A-mCherry; osx:GFP-CAAX Fish during Scale Radii Formation. Related to Figure 5. Zoom on a section of a single scale regenerating between 44:03 hpp and 54:23 hpp. Osteoblasts can be observed aligning along the anterior-posterior axis of the scale and changing axis ratio (cells from this and other videos quantified in Figure 5C). Magenta: osx:H2A-mCherry, yellow: osx:GFP-CAAX. Left to right: anterior to posterior. Scale bar = 50 μm.
Highlights.
Zebrafish scales are a model for real-time in vivo imaging of bone regeneration
De novo osteoblasts differentiation creates the initial osteoblast pool
Fgf signaling regulates osteoblast proliferation during scale regeneration
Regeneration involves patterned osteoblast division, shape changes, and death
ACKNOWLEDGMENTS
We thank J. Burris, S. Miller, L. Frauen, and T. Curtis for zebrafish care, A. Dickson for artwork, S. Johnson for fabricating the imaging dish, and A. Puliafito for assistance with MATLAB programming. B.D.C. and V.A.T. were supported by Graduate Research Fellowships (1106401) from the National Science Foundation. A.D. was supported by Early (P2ELP3_172293) and Advanced (P300PA_177838) Postdoc.Mobility fellowships from the Swiss National Science Foundation. This work was supported by a Whitehead Faculty Scholar Award to S.D.T., and a grant (R01 GM074057) from NIH to K.D.P.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes 5 figures, one table, and 3 videos.
REFERENCES
- 1.D'Ippolito G, Schiller PC, Ricordi C, Roos BA, and Howard GA (1999). Age-related osteogenic potential of mesenchymal stromal stem cells from human vertebral bone marrow. J Bone Min Res 14, 1115–1122. [DOI] [PubMed] [Google Scholar]
- 2.Han M, Yang X, Farrington JE, and Muneoka K (2003). Digit regeneration is regulated by Msx1 and BMP4 in fetal mice. Development 130, 5123–5132. [DOI] [PubMed] [Google Scholar]
- 3.Dolan CP, Dawson LA, and Muneoka K (2018). Digit Tip Regeneration: Merging Regeneration Biology with Regenerative Medicine. Stem Cells Translat Med 7, 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tornini VA, and Poss KD (2014). Keeping at arm's length during regeneration. Dev Cell 29, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ando K, Shibata E, Hans S, Brand M, and Kawakami A (2017). Osteoblast Production by Reserved Progenitor Cells in Zebrafish Bone Regeneration and Maintenance. Dev Cell 43, 643–650 e643. [DOI] [PubMed] [Google Scholar]
- 6.Shibata E, Ando K, Murase E, and Kawakami A (2018). Heterogeneous fates and dynamic rearrangement of regenerative epidermis-derived cells during zebrafish fin regeneration. Development 145. [DOI] [PubMed] [Google Scholar]
- 7.Sandoval-Guzman T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, Tazaki A, Joven A, Tanaka EM, and Simon A (2014). Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell 14, 174–187. [DOI] [PubMed] [Google Scholar]
- 8.Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell 20, 713–724. [DOI] [PubMed] [Google Scholar]
- 9.Mesa KR, Rompolas P, Zito G, Myung P, Sun TY, Brown S, Gonzalez DG, Blagoev KB, Haberman AM, and Greco V (2015). Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature 522, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown S, Pineda CM, Xin T, Boucher J, Suozzi KC, Park S, Matte-Martone C, Gonzalez DG, Rytlewski J, Beronja S, et al. (2017). Correction of aberrant growth preserves tissue homeostasis. Nature 548, 334–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tornini VA, Puliafito A, Slota LA, Thompson JD, Nachtrab G, Kaushik AL, Kapsimali M, Primo L, Di Talia S, and Poss KD (2016). Live Monitoring of Blastemal Cell Contributions during Appendage Regeneration. Curr Biol 26, 2981–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CH, Puliafito A, Cox BD, Primo L, Fang Y, Di Talia S, and Poss KD (2016). Multicolor Cell Barcoding Technology for Long-Term Surveillance of Epithelial Regeneration in Zebrafish. Dev Cell 36, 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Currie JD, Kawaguchi A, Traspas RM, Schuez M, Chara O, and Tanaka EM (2016). Live Imaging of Axolotl Digit Regeneration Reveals Spatiotemporal Choreography of Diverse Connective Tissue Progenitor Pools. Dev Cell 39, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wynd BM, Watson CJ, Patil K, Sanders GE, and Kwon RY (2017). A Dynamic Anesthesia System for Long-Term Imaging in Adult Zebrafish. Zebrafish 14, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C, Volkery S, and Siekmann AF (2015). Intubation-based anesthesia for long-term time-lapse imaging of adult zebrafish. Nat Protoc 10, 2064–2073. [DOI] [PubMed] [Google Scholar]
- 16.Metz JR, de Vrieze E, Lock EJ, Schulten IE, and Flik G (2012). Elasmoid scales of fishes as model in biomedical bone research. J App Ichthyol 28, 382–387. [Google Scholar]
- 17.Sire JY, and Akimenko MA (2004). Scale development in fish: a review, with description of sonic hedgehog (shh) expression in the zebrafish (Danio rerio). Intl J Dev Biol 48, 233–247. [DOI] [PubMed] [Google Scholar]
- 18.Parichy DM, Elizondo MR, Mills MG, Gordon TN, and Engeszer RE (2009). Normal Table of Post-Embryonic Zebrafish Development: Staging by Externally Visible Anatomy of the Living Fish. Dev Dyn 238, 2975–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Vrieze E, Sharif F, Metz JR, Flik G, and Richardson MK (2011). Matrix metalloproteinases in osteoclasts of ontogenetic and regenerating zebrafish scales. Bone 48, 704–712. [DOI] [PubMed] [Google Scholar]
- 20.Sire JY, Quilhac A, Bourguignon J, and Allizard F (1997). Evidence for participation of the epidermis in the deposition of superficial layer of scales in zebrafish (Danio rerio): A SEM and TEM study. J Morphol 231, 161–174. [DOI] [PubMed] [Google Scholar]
- 21.Waterman RE (1970). Fine structure of scale development in the teleost, Brachydanio rerio. Anat Rec 168, 361–379. [DOI] [PubMed] [Google Scholar]
- 22.Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simoes M, Leon J, Roehl H, Cancela ML, and Jacinto A (2011). Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development 138, 3897–3905. [DOI] [PubMed] [Google Scholar]
- 23.Singh SP, Holdway JE, and Poss KD (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell 22, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S, and Miyawaki A (2009). Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A 106, 20812–20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohner N, Bercsenyi M, Orban L, Kolanczyk ME, Linke D, Brand M, Nusslein-Volhard C, and Harris MP (2009). Duplication of fgfr1 permits Fgf signaling to serve as a target for selection during domestication. Curr Biol 19, 1642–1647. [DOI] [PubMed] [Google Scholar]
- 26.Aman AJ, Fulbright AN, and Parichy DM (2018). Wnt/beta-catenin regulates an ancient signaling network during zebrafish scale development. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata E, Yokota Y, Horita N, Kudo A, Abe G, Kawakami K, and Kawakami A (2016). Fgf signalling controls diverse aspects of fin regeneration. Development 143, 2920–2929. [DOI] [PubMed] [Google Scholar]
- 28.Daane JM, Rohner N, Konstantinidis P, Djuranovic S, and Harris MP (2016). Parallelism and Epistasis in Skeletal Evolution Identified through Use of Phylogenomic Mapping Strategies. Mol Biol Evol 33, 162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y, Grill S, Sanchez A, Murphy-Ryan M, and Poss KD (2005). Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132, 5173–5183. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen JP, Vo NT, and Sagasti A (2018). Fish Scales Dictate the Pattern of Adult Skin Innervation and Vascularization. Dev Cell 46, 344–359.e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nachtrab G, Kikuchi K, Tornini VA, and Poss KD (2013). Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development 140, 3754–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landry P, Sadasivan K, Marino A, and Albright J (1997). Apoptosis is coordinately regulated with osteoblast formation during bone healing. Tissue & Cell 29, 413–419. [DOI] [PubMed] [Google Scholar]
- 33.Jilka RL, Weinstein RS, Bellido T, Parfitt AM, and Manolagas SC (1998). Osteoblast programmed cell death (apoptosis): modulation by growth factors and cytokines. J Bone and Min Res 13, 793–802. [DOI] [PubMed] [Google Scholar]
- 34.Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, and Junker JP (2018). Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotech 36, 469–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, and Klein AM (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, and Schier AF (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, and Nusslein-Volhard C (2008). Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet 4, e1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasaki M, Kuroda J, Kawakami K, and Wada H (2018). Epidermal regulation of bone morphogenesis through the development and regeneration of osteoblasts in the zebrafish scale. Dev Biol 437, 105–119. [DOI] [PubMed] [Google Scholar]
- 39.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Meth 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao J, Wang J, Jackman CP, Cox AH, Trembley MA, Balowski JJ, Cox BD, De Simone A, Dickson AL, Di Talia S, et al. (2017). Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue. Dev Cell 42, 600–615.e604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Long-Term Live Imaging of an osx:H2A-mCherry Scale on the Second Day of Regeneration. Related to Figure 1. A single scale regenerating between 23:30 hpp and 43:00 hpp. De novo activation of osx-driven fluorescence can be visualized throughout the area of the scale. The edges of intact scales are present at the top of the frame. Left to right: anterior to posterior. Scale bar = 100 μm.
Video S2. Long-Term Live Imaging of an osx:H2A-mCherry Scale on the Third Day of Regeneration. Related to Figure 1. A single scale regenerating between 52:02 hpp and 71:32 hpp. Little to no de novo activation of osx-driven fluorescence is visible. Proliferation is biased toward the scale edges. Nuclei in nascent radii at the posterior half of the scale align and elongate, and nuclear fragmentation near radii can be observed. Left to right: anterior to posterior. Scale bar = 100 μm.
Video S3. Dual Color Imaging in osx:H2A-mCherry; osx:GFP-CAAX Fish during Scale Radii Formation. Related to Figure 5. Zoom on a section of a single scale regenerating between 44:03 hpp and 54:23 hpp. Osteoblasts can be observed aligning along the anterior-posterior axis of the scale and changing axis ratio (cells from this and other videos quantified in Figure 5C). Magenta: osx:H2A-mCherry, yellow: osx:GFP-CAAX. Left to right: anterior to posterior. Scale bar = 50 μm.