Abstract
Cardiovascular disease (CVD), the leading cause of death and morbidity in the Western world, begins with lipid accumulation in the arterial wall, which is the initial step in atherogenesis. Alterations in lipid metabolism result in increased risk of cardiometabolic disorders, and treatment of lipid disorders remains the most common strategy aimed at reducing the incidence of CVD. Work done over the past decade has identified numerous classes of non-coding RNA molecules including microRNAs (miRNAs) and long-non-coding RNAs (lncRNAs) as critical regulators of gene expression involved in lipid metabolism and CVD, mostly acting at post-transcriptional level. A number of miRNAs, including miR-33, miR-122 and miR-148a, have been demonstrated to play important role in controlling the risk of CVD through regulation of cholesterol homeostasis and lipoprotein metabolism. lncRNAs are recently emerging as important regulators of lipid and lipoprotein metabolism. However, much additional work will be required to fully understand the impact of lncRNAs on CVD and lipid metabolism, due to the high abundance of lncRNAs and the poor-genetic conservation between species. This article reviews the role of miRNAs and lncRNAs in lipid and lipoprotein metabolism and their potential implications for the treatment of CVD.
Keywords: miRNAs, lncRNAs, cholesterol metabolism, cardiovascular disease, atherosclerosis
Graphical abstract
INTRODUCTION
Cardiovascular disease (CVD) is the leading cause of death and morbidity in the Western world. Alterations in lipid metabolism result in increased risk of cardiometabolic disorders, including type-2 diabetes and atherosclerosis. Pharmaceutical treatment of lipid disorders remains the most commonly used strategy aimed at reducing the incidence of CVD. Work done over the past decade has identified numerous classes of non-coding RNA molecules including microRNAs (miRNAs) and long-non-coding RNAs (lncRNAs) as critical regulators of gene expression involved in lipid metabolism and CVD mostly acting at a post-transcriptional level [1–3]. miRNAs are small non-coding RNAs containing about 22 nucleotides in length found in plants, animals and some viruses. miRNAs directly bind to the 3′ untranslated region of target mRNAs and control numerous biological processes through RNA silencing and post-transcriptional regulation of gene expression [1, 2]. A single miRNA can regulate many (hundreds of) putative targets and a single transcript may be regulated by multiple microRNAs [1, 4]. miRNAs have been identified as key regulators of multiple cardiometabolic pathologies, including obesity, insulin resistance, atherosclerosis and heart failure [5–9]. Many miRNAs have been shown to be dysregulated in different disease states and alterations in miRNA expression or genetic variants associated with miRNAs or miRNAs binding site loci have been associated with the progression of cardiometabolic diseases. This implies a unique therapeutic potential of miRNAs in these diseases. In particular, emerging evidence demonstrates that miRNAs are critical regulators of lipid and lipoprotein metabolism and promising therapeutic targets for the treatment of CVD [8, 10–13].
While miRNAs are well studied and have an established role in the regulation of lipid metabolism and CVD, the contribution of lncRNAs in lipid homeostasis has just started to emerge [3, 8, 14, 15]. lncRNAs are a heterogeneous group of transcribed RNA molecules ranging from 200 to 100,000 nucleotides in length. It is estimated that there are about 15,900 lncRNAs in human genome annotated by Genecode (Version 22) [14, 16], however, the lncRNA annotations are rapidly increasing with over 170,000 human lncRNA transcripts by NONCODE database (http://www.bioinfo.org/noncode/) [17]. lncRNAs are categorized based on genome location into long intergenic ncRNAs (lincRNAs), natural antisense transcripts (NATs), enhancer-like ncRNAs (eRNAs), and transcribed ultra-conserved regions (T-UCRs) [18, 19]. Multiple studies have shown that numerous lncRNAs are regulated during development, exhibit cell type-specific expression patterns, localize to specific subcellular compartments, and are associated with physiological and pathological functions involved in cholesterol metabolism and CVD [3, 8, 16]. However, the mechanisms by which many of the lncRNAs exert their effects are poorly understood and the poor conservation of lncRNAs across species poses a problem for the development of lncRNA based therapies. This article reviews the role of miRNAs and lncRNAs in lipid and lipoprotein metabolism and their potential implication for the treatment of CVD.
miRNAs REGULATION OF HIGH DENSITY LIPOPROTEIN (HDL) METABOLISM AND REVERSE CHOLESTEROL TRANSPORT
Cholesterol is an essential structural component of cell membranes and myelin sheaths. In addition to its structural requirement, cholesterol is also a biosynthetic precursor of bile acids, steroid hormones and vitamin D. Abnormal levels of cholesterol are highly associated with the progression of cardiometabolic diseases including type-II diabetes and atherosclerosis [20, 21]. Mammalian cells cannot degrade cholesterol, thus must be removed through a physiological process known as reverse cholesterol transport (RCT). During RCT, excess cholesterol is transported from the peripheral tissues to the liver, where cholesterol can be reutilized or excreted into the feces [22, 23]. Plasma HDL and its major protein component, apolipoprotein AI (ApoAI), are the primary sterol transporters capable of facilitating cholesterol mobilization from the peripheral cells to the liver following cholesterol efflux by the ATP-binding cassette transporters ABCG1 and ABCA1 [22, 23]. The HDL receptor scavenger receptor class B type 1 (SR-B1) mediates RCT by facilitating cholesterol efflux from macrophages and the uptake of HDL cholesterol by hepatocytes for routing to the bile [24]. Several transporters localized to the bile canaliculus have been shown to play essential roles in the biliary secretion process including adenosine triphosphate (ATP)-binding cassette transporter B11 (ABCB11), ABCB4, ABCG5/G8 and ATPase class I type 8B member 1 (ATP8B1) [25]. RCT-mediated cholesterol efflux from foam cells can prevent atherosclerotic lesion progression and induce regression of pre-existing plaques. By mediating the excretion of cholesterol and bile acids, RCT lowers the level of cholesterol in circulation and has relevant implications for CVD [26, 27]. Over the past decade, miRNAs have been identified as critical regulators of HDL-C metabolism involved in HDL biogenesis, cellular cholesterol efflux, HDL-C uptake and bile acid transport [10].
Regulation of HDL-C metabolism and RCT by miR-33
Of all the miRNAs involved in regulation of HDL-C metabolism and RCT, the miR-33 family are the most studied and best understood. miR-33a and miR-33b are intronic miRNAs encoded within the genes for the SREBP-2 and SREBP-1 transcription factors. SREBP-1 and SREBP-2 are two of the primary transcription factors responsible for regulation of lipid metabolism. In response to low sterol levels SREBP-2 is activated, leading to the induction of genes involved in cholesterol synthesis and uptake [28]. SREBP-2 also induces the expression of its own transcript, which leads to a concurrent induction of miR-33a. Under these conditions, miR-33a assists in the SREBP-2 mediated regulation of cholesterol levels by targeting ABCA1 and ABCG1 to prevent further removal of cholesterol from the cell [29–31]. More recently, experiments done with macrophages from miR-33 deficient mice demonstrated that miR-33 plays an important role in regulating cholesterol efflux in vivo as well as in vitro [32]. Similar to the coordinated effects of SREBP-2 and miR-33a, miR-33b is induced along with its host gene, SREPB-1, and has been shown to limit oxidation of fatty acids by targeting numerous transcripts including those encoding CPT-1, CROT, and HADHβ [33]. This supports the primary function of SREBP-1, which is responsible for induction of genes involved in fatty acid synthesis. In addition to their ability to regulate intracellular cholesterol levels and fatty acid oxidation, miR-33a and miR-33b have been shown to be key regulators of other steps in the reverse cholesterol transport pathway, including HDL-C biogenesis and bile acid synthesis/secretion. While ABCA1 is one of the key transport molecules involved in cholesterol efflux from cells, it is also required for the synthesis of the HDL-C molecules that are primarily responsible for transport of cholesterol back to the liver. As such, miR-33 is also an important regulator of HDL-C biogenesis by targeting ABCA1 in the liver [34, 35]. Finally, miR-33a and miR-33b are also involved in the last stage of RCT by regulating factors involved in the synthesis (CYP7A1) and secretion (ATP8B1 and ABCB11) of bile acids, which allow removal of cholesterol from the body through the feces [36, 37]. Through its ability to regulate many different aspects of RCT, including the efflux of cholesterol from peripheral tissues, transport to the liver, and removal in the form of bile acids, miR-33 has been shown to be a very potent regulator of RCT.
Targeting miR-33 for the treatment of atherosclerosis
Because of its important role in regulation of RCT, multiple studies have sought to determine whether inhibition of miR-33 may provide a reliable therapeutic approach for the treatment of atherosclerosis. Current treatments for cardiovascular disease focus primarily on strategies to reduce levels of circulating LDL-C and thereby limit the accumulation of cholesterol within atherosclerotic plaques. More recently, considerable effort has been made to develop strategies for promoting the removal of lipids from macrophages and other cells that make up the plaque by raising circulating HDL-C levels. Unfortunately, current therapeutics, while successful at raising circulating HDL-C levels, have not had a substantial impact on the risk of cardiovascular events [38, 39]. However, the ability of miR-33 to impact many different functions related to RCT suggests that anti-miR-33 therapies may be able to increase both the amount and function of circulating HDL-C, thereby providing a more promising approach for treatment of patients with CVD. Since its identification, many different studies have shown that inhibition or genetic deletion of miR-33 leads to elevated expression of ABCA1 in the liver and increased circulating HDL-C in mice and non-human primates [30, 31, 40–42]. Moreover inhibition of miR-33 in mice was found to promote the regression of atherosclerotic plaques [43], and further work demonstrated that inhibition or genetic deletion of miR-33 could reduce plaque progression [44–46]. However, not all studies have found significant changes in circulating HDL-C levels or atherosclerotic plaque size in response to anti-miR-33 therapeutics [47]. While the reason behind these discrepancies is not well understood, numerous differences in the experimental design of these studies, including the type of inhibitory molecules used, length and timing of the treatment, and type of diet could contribute to the different outcomes of these studies. Consistent with the idea that miR-33 may impact atherogenesis by directly promoting cholesterol efflux from plaque macrophages, our group recently demonstrated that genetic ablation of miR-33 specifically in macrophages and other hematopoietic cells protects against the progression of atherosclerosis without altering plasma HDL-C levels [32].
Other regulatory functions of miR-33
In addition to its direct regulation of RCT, miR-33 has been shown to control a myriad of other biologic functions that could also play an important role in its effects on atherosclerosis, additional forms of vascular disease, and other disease states. As well as being a key regulator of HDL-C metabolism and fatty acid oxidation, miR-33 has also been reported to target factors involved in cholesterol transport. NPC1 is an important regulator of intracellular cholesterol trafficking and has been shown to be directly targeted by miR-33 [30]. Similarly, oxysterol binding protein-like 6 (OSBPL6) has also been identified as a miR-33 target involved in the transport of intracellular cholesterol [48]. Additional work will be needed to clarify how these targets effect the ability of miR-33 to regulate the efflux of HDL-C and determine whether they may also impact the transport of LDL/VLDL cholesterol. Indeed, the effect of miR-33 on VLDL metabolism remains controversial, with different studies yielding contradictory results. Long term treatment with miR-33 inhibitors was found to increase levels of circulating triglycerides (TAGs) and promote accumulation of lipids in the liver [49]. Similarly, another group demonstrated that inhibition of miR-33 could increase VLDL-C secretion and circulating TAGs by targeting N-ethylmaleimide-sensitive factor (NSF) [50]. These findings are in direct contradiction to an earlier report in which VLDL-C was found to be lower in non-human primates treated with miR-33 inhibitors, as well as a number of studies in which inhibition of miR-33 increased circulating HDL-C without affecting plasma VLDL-C levels [29–31, 51]. Similarly, recent work by our group has demonstrated that genetic deficiency in miR-33 results in increased total cholesterol and TAGs in Ldlr−/− mice fed a Western diet [32], while loss of miR-33 in Apoe−/− animals was not found to impact levels of either total cholesterol or TAGs [45]. It is not currently clear why different studies have found such disparate effects on VLDL-C and TAGs or what impact this may have on the development of CVD.
While the direct regulation of ABCA1 and ABCG1 is likely the primary means by which miR-33 regulated cholesterol efflux, miR-33 has also been shown to regulate important factors involved in nutrient sensing and energy regulation. This may also be important for its ability to mediate cholesterol efflux, as ABCA1 and ABCG1 require ATP utilization in order to remove cholesterol from the cell. The miR-33 target AMPK is a nutrient sensitive kinase that regulates numerous metabolic functions including activation of peroxisome proliferator–activated receptor γ coactivator 1-α (PGC1-α). PGC1-α is the primary transcription factor involved in regulation of mitochondrial biogenesis, and its expression has also been shown to be regulated by miR-33 [33]. More recently, Karunakaran and colleagues have demonstrated that the ability of miR-33 to target PGC1-α as well as other factors involved in mitochondrial function, such as pyruvate dehydrogenase kinase isozyme 4 (PDK4), and solute carrier family 25 (SLC25A25) can alter mitochondrial respiration and thereby impact ATP availability and cholesterol efflux by ABCA1 [52]. Similarly, miR-33 was found to regulate factors involved in autophagy, which was also shown to be involved in the ability of miR-33 to promote cholesterol efflux [46]. Regulation of autophagy was also suggested to be involved in the effects of miR-33 on efferocytosis, although other work in miR-33 deficient macrophages showed increased uptake of apoptotic cells without any changes in genes related to autophagy [32]. The ability of miR-33 to regulate energy balance through regulation of fatty acid metabolism, mitochondrial function and autophagy has been shown to impact many different aspects of immune response including T-cell priming [53], bacterial killing by macrophages [54], and macrophage polarization [55]. The reduced inflammatory response of macrophages treated with miR-33 inhibitors is believed to contribute to its reduction of atherosclerotic plaque size, and a recent report shows that miR-33 deficient animals are protected from abdominal aortic aneurysm due to reduced inflammation [56].
miR-33 has also been demonstrated to regulate the proliferation of numerous cell types by targeting a number of different factors, including key mediators of the cell cycle (CDK6, CyclinD1). This has been shown to impact a number of different biologic functions including adipocyte differentiation [57], liver regeneration [58], and tumor growth/metastasis [59–61]. More recently the effect of miR-33 on cellular proliferation has also been shown to impact late stages of CVD. Work by Nishiga et al. demonstrated that mice deficient in miR-33 have reduced cardiac fibrosis following transverse aortic constriction (TAC). This work further demonstrates that miR-33 deficient animals had reduced cardiac function following TAC due to reduced fibroblast proliferation and impaired cardiac remodeling [62]. Moreover, miR-33 was found to reduce intimal hyperplasia following vein grafts due to reduced smooth muscle cell proliferation [63]. This work suggests that inhibition of miR-33 for the treatment of atherosclerosis could actually have a detrimental effect on cardiac function following heart failure or vein graft bypass surgery. Additionally, whole body deletion of miR-33 was found to promote the development of obesity and metabolic dysfunction [64], and this was found to offset the beneficial effects that occurred within plaque macrophages resulting in no changes in total plaque size [32]. Even more recently work by our group has demonstrated that the lack of miR-33 results in impaired function and reduced responsiveness to insulin in a number of key metabolic tissues, including the liver and adipose tissue, on both chow and high fat diets (HFD). This work further demonstrates that the primary driving force behind both the obesity and metabolic dysfunction is an increase in food consumption [65]. Overall, the ability of miR-33 to regulate many different factors involved in the development of CVD makes it an exciting therapeutic target, but the vast array of different physiologic functions it controls also raises important concerns about the potential for unintended consequences. The most common methods for targeting miRNAs relies on antisense oligonucleotide (ASO)-based approaches and miRNA mimetics, however, the requirement for chemical modifications, hybridization-associated off-target effects, toxicity and drug delivery problems make the leap from pre-clinical to clinical models a challenge [66].
miRNA regulation of ABCA1 expression
In addition to the well-known role of miR-33 in controlling ABCA1 expression and cholesterol efflux, other miRNAs including miR-144, miR-148a, miR-758, miR-26, miR-106b, miR-27, miR-145, miR-20a/b and miR-10b, have been shown to regulate ABCA1 expression at the post-transcriptional level in a variety of different cell types, such as macrophages, pancreatic β-cells, neurons, enterocytes and hepatocytes [67–76]. Two independent groups identified ABCA1 as a direct target of miR-144, and inhibition of miR-144 increased the expression of ABCA1 in hepatocytes and macrophages, as well as raising plasma HDL levels in mice [67, 77]. Conversely, overexpression of miR-144 using miR-144-3p mimics (agomir) accelerates pathological progression of atherosclerosis in Apoe−/− mice through enhanced expression of inflammatory factors, decreased cholesterol efflux, reduced circulating HDL-C and impaired RCT in vivo [78]. Two independent studies identified miR-128-1 and miR-148a as key regulators of the expression of ABCA1 [79]. Overexpression of both miR-128-1 or miR-148a resulted in reduced hepatic ABCA1 expression and plasma HDL-C levels in hyperlipidemic mice, however, only silencing of miR-148a significantly increased hepatic ABCA1 expression and circulating HDL-C levels. Both miRNAs also regulate the expression of ABCA1 in macrophages and their overexpression and inhibition attenuated and enhanced ABCA1 expression and cholesterol efflux respectively [75, 79].
Recent data demonstrate that intestinal microorganisms could influence lipid metabolism, triggering development of metabolic and cardiovascular diseases including atherosclerosis [80]. Intestinal microbiota can regulate RCT by modulating the expression of miR-10b [74]. Protocatechuic acid (PCA), a gut microbiota metabolite of cyanidin-3 to 0-β-glucoside (Cy-3-G), promotes cholesterol efflux from macrophages and increases cellular ABCA1 and ABCG1 expression [74]. Importantly, Cy-3-G consumption promotes macrophage RCT and regresses atherosclerotic lesions by the regulation of the miRNA-10b-ABCA1/ABCG1 cascade in Apoe−/− mice [74]. In neuronal cells, miR-106b represses ABCA1 expression and impairs cellular cholesterol efflux [71]. miR-106b increases levels of secreted amyloid β (Aβ) by increasing Aβ production and preventing Aβ clearance, which can be rescued by overexpression of miR-106b-resistant ABCA1 [71]. Additionally, miR-145 post-transcriptionally regulates ABCA1 expression in HepG2 cells and in murine pancreatic islets [73], while miR-758, miR-26, miR-27 and miR-20a/b inhibit ABCA1 expression and cholesterol efflux in mouse and human macrophage cell lines [69, 70, 72, 76]. Notably, miR-20a/b treatment promotes atherosclerotic development in Apoe−/− mice accompanied by decreased ABCA1 expression in the liver, attenuated RCT, and impaired formation of nascent HDL. Alternatively, miR-20a/b silencing attenuates the progression of atherosclerosis [76].
ABCA1 expression is transcriptionally regulated by liver X receptors (LXRs), oxysterol-activated transcription factors that control cellular cholesterol and fatty acid homeostasis in mammals [81, 82]. Aside from repressing ABCA1 expression through directly binding to its 3′UTR, miRNAs have also been shown to regulate transcriptional activation of ABCA1 by controlling LXR expression. LXR is directly targeted and transcriptionally regulated by miR-1, miR-206, miR-613, and miR-155 [70, 83, 84]. miR-1, miR-206 and miR-613 suppress lipogenesis by targeting LXRα and attenuate the expression of LXRα target genes including SREBP-1c, fatty acid synthase (FAS), carbohydrate responsive element-binding protein (chREBP) and acetyl-CoA carboxylase 1 (ACC1) [84, 85]. In addition, miR-155 has been shown to be essential in the development of non-alcoholic hepatosteatosis in mice by inhibiting LXRα and its target genes involved in glucose regulation, fatty acid uptake, and lipid metabolism [83]. LXR and retinoic-X-receptor (RXR) form a heterodimer to mediate the transcriptional activation of ABCA1. Repression of RXRα by miR-128-2 inhibits LXR-induced ABCA1 expression, which is demonstrated as a regulator of cholesterol homeostasis [86]. Together, these results demonstrate that the expression of ABCA1 is targeted by numerous miRNAs in different cell types indicating the importance of ABCA1 for regulation of different biologic functions and the critical role of microRNAs in the control of ABCA1 levels.
miRNA regulation of SR-B1 expression and function
In addition to ABCA1/G1-mediated cholesterol efflux, scavenger receptor class B member 1 (SR-B1) mediates bidirectional flux of cholesterol between cells and HDL-C, thus modulating the composition and structure of HDL-C particles and facilitating the delivery of cholesterol to steroidogenic tissues and the liver [87, 88]. The absence of SR-B1 results in early onset of occlusive atherosclerotic coronary artery disease and cardiac dysfunction in Apoe−/− mice [89]. A number of reports have identified miRNAs, including miR-185, miR-96, miR-233, miR-125a and miR-455, which bind directly to the 3′ UTR of SR-B1 and inhibit its expression. This leads to impaired HDL-C uptake in hepatic cells, whereas antagonism of these miRNAs displays opposite effects [90, 91]. Interestingly, decreased expression of miR-96 and miR-185 coincided with increased levels of hepatic SR-B1 in Apoe−/− mice fed a high-fat diet [90]. Furthermore, miR-185 serves as a relevant regulator of lipid metabolism under normal physiological or pathological conditions by targeting SR-B1, not only in the liver, but also in macrophages. miR-185 suppresses SR-B1 expression and selective HDL-C uptake in human THP-1 monocytic cell line [90]. While these studies demonstrate that the expression of these miRNAs influences SR-B1 mediated cholesterol efflux and delivery in vitro, further studies will be important for defining their relevance in regulating cholesterol metabolism in vivo.
miRNA REGULATION OF LDL-C METABOLISM
High levels of plasma LDL-C is the major risk factor for developing atherosclerosis. Plasma LDL-C levels are regulated by the rate of hepatic very low-density lipoprotein (VLDL) production and LDL-C clearance via the hepatic LDLR. Recent studies have identified miRNAs that control circulating LDL-C by regulating both processes [92].
miRNA regulation of VLDL secretion
miR-122 and miR-30c were the first miRNAs identified as critical regulators of plasma LDL-C in vivo by controlling VLDL secretion [93–96]. Genetic ablation or antisense oligonucleotide (ASO) inhibition of miR-122 results in significantly reduced plasma cholesterol and TAG levels in WT mice fed a chow or HFD [93, 97, 98]. The decreases in circulating cholesterol and TAGs observed after miR-122 inhibition or genetic deletion reflects reductions in both the HDL-C and VLDL fractions accompanied by lower levels of serum ApoB-100 and ApoE [93, 99]. Genetic deficiency of miR-122 in mice promotes hepatic steatosis accompanied by accumulation of both cholesterol and TAGs, as well as infiltration of inflammatory cells in the liver [97, 99]. Mechanistically, miR-122−/− mice show a significant reduction in hepatic microsomal TAG transfer protein (MTP), which is required for hepatic VLDL assembly and secretion [99]. Despite, these promising findings, germline and liver-specific deletion of miR-122 results in greater tumor incidence and higher tumor burden in the liver [97, 99]. Levels of circulating miR-122 was identified as a novel biomarker associated with liver injury and risk of developing metabolic syndrome and type 2 diabetes (T2D) [100].
Similar to miR-122, miR-30c also regulates the expression of MTP. miR-30c directly targets the 3′UTR region of MTP and significantly decrease hepatic VLDL secretion. Additionally, miR-30c inhibits hepatic lipid synthesis by targeting lysophosphatidyl glycerol acyltransferase 1 (LPGAT1), an enzyme involved in phospholipid synthesis [96]. Of note, hepatic overexpression of miR-30c using lentivirus or miRNA mimics lowered plasma cholesterol levels and de novo hepatic lipogenesis in Western diet-fed Apoe−/− mice, attenuating the progression of atherosclerosis [94, 96]. Importantly, post-transcriptional inhibition of MTP expression and VLDL secretion by miR-30c did not result in steatosis in the liver, suggesting that miR-30c might have therapeutic potential as an anti-hyperlipidemic and anti-atherosclerotic agent [96].
miRNA regulation of LDLR expression and activity
The clearance of LDL by the LDLR and the role of LDLR in cardiovascular disease have been extensively studied [101, 102]. Defects in the LDLR are the most common cause of familial hypercholesterolemia (FH) characterized by very high circulating LDL-C levels and premature coronary artery disease [101]. Recently, several studies have identified miRNAs that control the expression of LDLR in hepatic cell lines, including miR-27a/b, miR-128-1, miR-130b, miR-148a, miR-185, miR-199a, and miR-301 [75, 79]. Among these, only three miRNAs, miR-128-1, miR-148a and miR-185 were confirmed to control plasma LDL-C in vivo [75, 79, 103, 104]. miR-148a inhibits the expression of LDLR and antagonism of miR-148a in vivo markedly increases hepatic LDLR levels and enhances LDL-C clearance [75, 79]. Interestingly, miR-148a inhibition also increases hepatic ABCA1 expression and circulating HDL-C levels, demonstrating the therapeutic potential of inhibiting miR-148a to ameliorate an elevated LDL-C/HDL-C ratio, a prominent risk factor for CVD [75]. The relevance of miR-148a in regulating circulating lipoproteins was recently corroborated in humans by the recent identification of SNPs (rs4722551 and rs4719841) present in the promoter region of miR-148a, which were associated with significant alterations in plasma cholesterol, LDL-C and TAG levels [79]. Besides LDLR and ABCA1, miR-148a has been shown to directly target the 3′UTR of other genes involved in lipid metabolism, including PGC1-α, AMPK, and insulin induced gene (INSIG)-1 [79]. Näär and colleagues also identified miR-128-1 using a GWAS meta-analysis as a relevant regulator of plasma lipids in humans. Similar to miR-148a, miR-128-1 also targets the 3′UTR of the LDLR and ABCA1 [79] and its inhibition reduced circulating LDL-C. Notably, long-term antisense antagonism of miR-128-1 in mice significantly improved glucose homeostasis and insulin sensitivity by enhancing the expression of the insulin receptor (INSR) and insulin receptor substrate 1 (IRS-1) [79]. The effects of anti-miR-128-1 treatment on circulating cholesterol and TAGs, as well as on insulin sensitivity and glucose homeostasis, highlight its potential contribution to numerous aspects of the metabolic syndrome [79]. In addition to miR-148a and miR-128-1, another study has demonstrated that miR-185 also regulates cholesterol uptake by directly targeting the LDLR and the LDLR-destabilizing RNA binding protein, RNA-binding KH-type splicing regulatory protein (KSRP) [103, 104]. Consistently, antagonism of miR-185 in vivo shows both, cholesterol-lowering and anti-atherosclerosis effects in Apoe−/− mice [103].
Other miRNAs regulating LDL-C metabolism
Hepatic LDLR expression is transcriptionally regulated by the SREBP transcription factors, which also control the transcription of most of the genes associated with cholesterol and fatty acid biosynthesis. SREBP2 expression and activation is promoted under conditions of cholesterol-depletion and mediates the transcriptional upregulation of LDLR expression in patients treated with statins, a competitive inhibitor of the rate-controlling enzyme of the cholesterol synthesis pathway, HMG-CoA reductase [105]. Interestingly, the Osborne group identified a polycistronic miRNA locus, miR-96/182/183, that is upregulated in the liver of mice treated with statins and ezetimibe, a drug that inhibits cholesterol absorption in the small intestine [106]. Additional studies demonstrated that miR-96/182/183 expression is regulated directly by SREBP2, and two members of the cluster, miR-96 and miR-182, inhibit INSIG-2 and FBXW7 [106]. Since INSIG-2 and FBXW7 are negative regulators of SREBP activation by preventing SREBP proteolytic processing and enhancing nuclear degradation respectively, this regulatory mechanism suggests that induction of miR-96/182/183 works in a feed-forward manner to further increase SREBP transcription and activation during low cholesterol conditions. Indeed, overexpression of all three pre-miRNAs is sufficient to increase SREBP activity and enhance lipid synthesis in Hela cells. However, antagomirs of miR-96 and miR-182 cannot alter hepatic or circulating cholesterol levels in mice [106]. Additional experiments in other animal models will be important to elucidate the relevance of this miRNA cluster in regulating lipoprotein metabolism. miR-185 was also found to be regulated by the SREBP-1 transcription factor by binding to the SRE region in its promoter [104]. HFD feeding in mice induces the expression of miR-185, which correlated with increased circulating cholesterol levels and decreased SREBP-2 expression by directly targeting the 3′UTR of SREBP-2 [104]. Importantly, overexpression of miR-185 results in decreased LDLR protein expression and LDL uptake, as well as impaired HMGCR activity in vitro, which mirrors the decreased SREBP-2 activity [104].
LncRNAs AND CHOLESTEROL METABOLISM
In addition to microRNAs, it has been recently demonstrated that a number of lncRNAs control lipid metabolism in different tissues and cells. This section summarizes the recent findings in this area highlighting the specific contribution of those lncRNAs that influence circulating lipids.
MeXis
MeXis (macrophage-expressed LXR-induced sequence) has recently been identified as a sequence induced by LXR in macrophages [107]. MeXis is encoded in the proximity of the Abca1 locus and is required for full LXR-mediated ABCA1 transcription in response to LXR activators. Interestingly, MeXis is located in the nucleus and controls chromosome architecture in the Abca1 locus. MeXis interacts directly with DDX17, an established nuclear receptor coactivator, that is enriched at the LXR-binding sites in the Abca1 enhancer region in macrophages. To study the contribution of MeXis in regulation of macrophage ABCA1 expression and atherogenesis in mice, Tontonoz’s group generated MeXis deficient mice. The authors found that absence of MeXis significantly reduced ABCA1 expression in macrophages and enhanced the progression of atherosclerosis. Of note, a similar transcript (TCONS00016111) is encoded in the proximity of the ABCA1 locus in humans and its overexpression or inhibition influences ABCA1 levels and cholesterol efflux in a human macrophage cell line (THP-1). These findings correlate with the significant association between a SNP overlapping the TCONS00016111 transcript and human coronary disease and suggest that the LXR-MeXis-ABCA1 axis might control macrophage sterol metabolism in humans. Since LXR activators have shown deleterious effects in vivo, due to promotion of hepatic fatty acid synthesis and steatosis, the identification of MeXis as a specific regulator of LXR-induced ABCA1 expression in macrophages open a novel avenue for promoting RCT in vivo without causing adverse effects.
LeXis
Similar to MeXis, the same group identified another LXR-regulated transcript [LeXis (Liver-expressed LXR-induced sequence)], encoded in the proximity of the Abca1 locus in mouse hepatocytes [108]. Hepatic LeXis levels influence the expression of genes associated with cholesterol biosynthesis and hepatic cholesterol content. Mice lacking LeXis have elevated expression of cholesterol biosynthetic enzymes and increased hepatic cholesterol content. Similar effects were observed in mice transduced with adenoviral vectors expressing shRNA targeting LeXis. Conversely, overexpression of LeXis significantly attenuates the expression of cholesterogenic genes in the liver. To determine the molecular mechanism by which LeXis control the expression of cholesterol biosynthetic genes, the authors preformed mass spectrometry analysis of LeXis interacting proteins. Interestingly, they found Raly, a heterogeneous ribonucleoprotein previously linked to SREBP-2, as a novel LeXis binding partner. Raly is associated with the promoter regions of cholesterol biosynthetic genes and its occupancy is reduced in presence of LeXis. Indeed, the effect of LeXis in regulating the expression of cholesterol biosynthetic genes appears to be mediated by Raly, as its ability to control hepatic cholesterol metabolism is lost when Raly is knocked down. Similar to MeXis, LeXis has a putative lncRNA (TCONS16452) in the human genome region adjacent to ABCA1 gene. However, its relevance in regulation of cholesterol metabolism in human or human hepatic cell lines remains to be elucidated.
ApoA4-AS
ApoA4-AS is a lncRNA identified in the liver of obese mice [109]. Similarly, this non-coding RNA is upregulated in human samples from subjects with fatty liver compared with healthy controls. ApoA4-AS has a similar pattern of expression that ApoA4 gene. Interestingly, circulating ApoA4 levels correlate with HDL plasma levels. ApoA4-AS interacts directly with HuR, a RNA binding protein that controls mRNA stability and translation, promoting ApoA4 mRNA stability [109]. Silencing ApoA4-AS in vivo did not influence hepatic TAG content but significantly reduced plasma TAG and cholesterol levels. However, how ApoA4-AS regulates circulating lipids is not known. The authors suggest that ApoA4-AS might influence VLDL secretion by reducing hepatic ApoA4 levels, which have been associated with hepatic VLDL production. Further studies will be important to define the mechanism by which ApoA4-As controls lipoprotein metabolism and hepatic lipid homeostasis.
LncHR1
Hepatitis C virus (HCV) infection induces SREBP expression and hepatic lipid accumulation. Using this premise, Li and colleagues identified LncHR1 in human hepatoma cells (Huh7) inoculated HCV. LncHR1 is located in the nucleus and the cytoplasm and controls the expression SREBP-1-responsive genes. LncHR1 overexpression attenuated the expression of SREBP-1 and FAS and reduced lipid droplet (LD) formation in Huh7 cells. Conversely, silencing of Huh7 enhanced SREBP-1 and FAS expression and promoted LD accumulation [110]. These results were further confirmed in vivo using a transgenic mouse model that overexpressed LncHR1 (LncHR1TG). Similar to the results obtained in vitro, hepatic SREBP-1 and FAS expression was attenuated in LncHR1TG mice compared to WT mice. Remarkably, plasma TAG levels were also reduced in the transgenic mice compared to WT mice. However, the mechanism by which LncHR1 controls circulating lipids is still poorly understood. Further experiments will be important to determine whether circulating TAGs are affected by reduced VLDL production, catabolism, or clearance.
ApoA1-AS
ApoA1-AS is a lncRNA located in the APO gene cluster that encodes several APO genes including ApoA1, ApoA4, ApoA5 and ApoC3. ApoA1-AS is an antisense transcript that regulates the expression of ApoA1 by influencing the binding of LSD1 and SUZ12 to the APO cluster [111]. Both, LSD1 and SUZ12 epigenetically modified this genetic locus and inhibited ApoA1, ApoA4 and ApoC3 expression. Most importantly antagonism ApoA1-AS in monkeys increased hepatic expression and circulating levels of ApoA1. It will be very interesting for future studies to analyze the impact of ApoA1-AS silencing on regulation of lipoprotein metabolism and HDL function.
LncLSTR
LncLSTR is a liver enriched LncRNA, whose expression is regulated by metabolic state [112]. LncLSTR expression is reduced during fasting state and significantly upregulated upon refeeding. However, the exact molecular mechanism that controls it expression during metabolic adaptation is not known. To define the specific contribution of LncLSTR in regulating lipid metabolism, Li et al inhibited the expression of LncLSTR using shRNA adenovirus. The results showed a significant reduction in circulating TAGs in WT and Apoe−/− mice. Mechanistically, the authors found that LncLSTR significantly increases the expression of ApoC2, a positive regulator of lipoprotein lipase (LPL), the enzyme that controls the triglyceride-rich lipoprotein (TRL) catabolism and fatty acid uptake and storage in oxidative tissues.
Lnc-HC
Lnc-HC is a liver and adipocyte expressed LncRNA that regulates the expression of CYP7a1, an important enzyme in the cholesterol biosynthesis pathway, and ABCA1 [113]. Inhibition of Lnc-HC significantly increased the expression of CYP7a1 and ABCA1 in hepatocytes. Mechanistically the authors found that Lnc-HC interacts with hnRNPA2B1, a RNA binding protein. This RNA-protein complex interacts with CYP7a1 and ABCA1 and regulates the expression of both transcripts. Further experiments will be important to define whether these mechanisms operate in vivo.
DYNLRB2
DYNLRB2 is a LincRNA expressed in human macrophages treated with oxidized-LDL (ox-LDL) [114]. Upregulation of DYNLRB2 in macrophage foam cells enhances ABCA1 expression and the G protein-coupled receptor 119 (GPR119), promoting cholesterol efflux and decreasing neutral lipid accumulation. While the role of DYNLRB2 in regulating lipoprotein metabolism and atherogenesis is not known, absence of DYNLRB2 reduced circulating lipids and attenuated atherosclerosis, suggesting that DYNLRB2 might influence lipoprotein metabolism and the progression of atherosclerosis.
HULC
HULC is a LincRNA highly upregulated in hepatocellular carcinoma (HCC) and contributes malignant development by supporting abnormal lipid metabolism in hepatoma cells [115, 116]. HULC expression is positively correlated with the acyl-CoA synthetase subunit ACSL1 in clinical HCC tissues. Mechanistically, HULC elicits the methylation of CpG islands in the miR-9 promoter, resulting in the suppression of miR-9 expression and activation of the transcription factor PPARA and its subsequent transactivation of ACSL1, which promotes lipogenesis and accumulation of triglycerides and cholesterol in hepatoma cells [115]. The upregulation of HULC by ACSL1 through transcription factor RXRA forms a positive feedback loop with HULC/miR-9/PPARA/ACSL1/cholesterol/RXRA/HULC in hepatoma cells, providing new insights into the mechanism of abnormal lipid metabolism mediated by HULC in the development of HCC.
CONCLUDING REMARKS
Both miRNAs and lncRNAs have been demonstrated to be important regulators of lipid metabolism capable of substantially impacting the development of atherosclerosis and other cardiometabolic diseases. While the ability of miRNAs regulating cholesterol metabolism has been well established, major hurdles still remain in the development and utilization of miRNA based therapeutic approaches for the treatment of CVD in humans. While the ability of miRNAs to bind many different targets allows for control of very robust and complex regulatory networks as evidenced by the ability of miR-148 to both elevate HDL-C and reduce LDL-C levels, this promiscuity also raises concerns over possible unintended consequences of miRNA-based therapies. The legitimacy of these concerns is demonstrated by the metabolic dysfunction of mice lacking miR-33 and the increased incidence of hepatocellular carcinoma following miR-122 inhibition. However, with the advent of new tools for identifying physiologically relevant miRNA targets and exploring the impact of individual miRNA-target interactions, the field of miRNA research is entering a new phase that will facilitate the development of novel approaches to modulate miRNA biology in a more targeted and specific manner.
Conversely, research on lncRNAs is still in its infancy and a great deal of additional work is still needed to characterize the multitudes of lncRNAs that have been discovered. These efforts are hindered by the poor conservation of lncRNAs across species and the varied mechanisms by which lncRNAs have been demonstrated to impact cellular functions. Indeed, a number of potential lncRNAs have been found to actually code for proteins of short peptides demonstrating that these are not in fact lncRNAs at all. While the challenged in this burgeoning new field are daunting, the dramatic impact of lncRNAs on lipid metabolism and other biologic functions necessitates a continued effort to understand this important new system of biologic regulation and progress in this field is moving forward at a rapid rate. The dysregulation of both lncRNAs and miRNAs in different diseases states makes these attractive candidates for therapeutic interventions, as approaches to restore levels of these molecules could provide an effective mechanism to curb disease progression without disrupting normal biologic functions. While many challenges remain in understanding these new classes of non-coding RNAs and utilizing this for the treatment of human disease, these novel classes of regulatory molecules provide great promise for the development of novel therapies.
Figure.
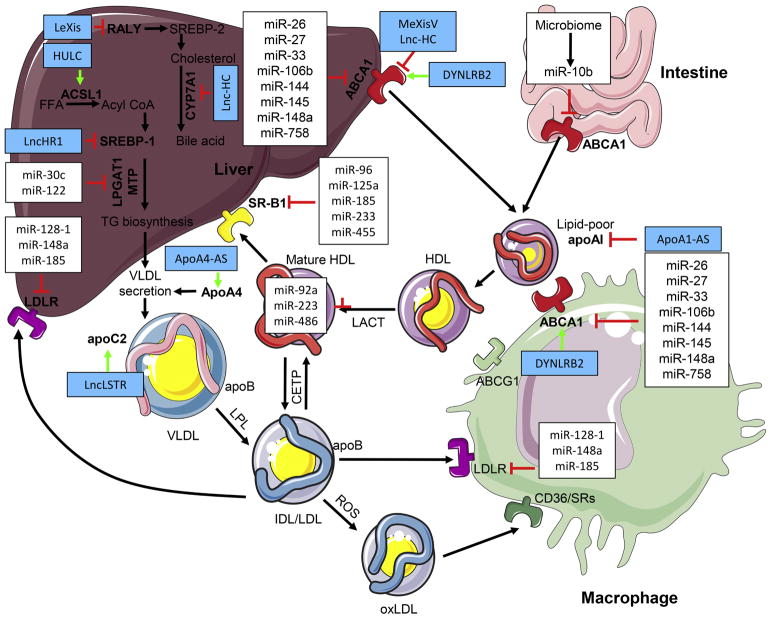
Non-coding RNAs regulation of high-density lipoprotein (HDL) and low-density lipoprotein (LDL) metabolism. miRNAs have been shown to repress genes involved in HDL and LDL homeostasis in different tissues as indicated. In the liver and intestine, ATP-binding cassette transporter A1 (ABCA1) is directly targeted by multiple miRNAs, by which reduces cholesterol efflux to lipid-poor apolipoprotein AI (apoAI) that generate nascent HDL. The repression of ABCA1 by miRNAs in peripheral cells such as macrophages inhibits the cholesterol efflux to lipid-poor apoAI. Through the action of lecithin-cholesterol acyltransferase (LCAT), free cholesterol on nascent HDL is esterified forming cholesterol esters and converting nascent HDL to mature HDL. Circulating miRNAs, such as miR-92a, miR-223 and miR-486, are association with HDL particles. These miRNAs are transported by HDL to target tissues or cells (such as liver and endothelial cells) to repress the expression of their target genes. The cholesterol hepatic clearance and excretion from HDL is reduced by targeting scavenger receptor B-I (SR-B1) by miRNAs. Moreover, miRNAs repress the expression of microsomal triglyceride transfer protein (MTP) and lysophosphatidylglycerol acyltransferase 1 (LPGAT1) involved in very low-density lipoprotein (VLDL) secretion. VLDL is converted in the circulation to LDL and intermediate-density lipoprotein (IDL) by lipoprotein lipase (LPL). Cholesterol ester-rich IDL and LDL can return to the liver and be taken up through interaction with the LDL receptor (LDLR), which can be regulated by miRNAs to decrease LDL clearance. Blue boxes highlight lncRNAs, which regulate genes that control cholesterol and fatty acid metabolism. CETP, cholesteryl ester transfer protein; SRs, scavenger receptors; LeXis, liver-expressed LXR-induced sequence; MeXis, macrophage-expressed LXR-induced sequence; LncLSTR, liver specific triglyceride regulator; ApoA1-AS, ApoA1 antisense; ApoA4-AS, ApoA4 antisense; HUR, human antigen R; ACSL1, acyl-CoA synthetase subunit; FFA, free fatty acid; SREBP, sterol regulatory element-binding protein. Figure was created using the Servier Medical Art illustration resources (http://www.servier.com).
Acknowledgments
C.F.-H. is supported in part by NIH Grant R35HL135820 and Foundation Leducq Transatlantic Network of Excellence in Cardiovascular Research and AHA Established Investigator Award (16EIA27550004).
Footnotes
CONFLICT OF INTEREST STATEMENT
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Hernando C, Moore KJ. MicroRNA modulation of cholesterol homeostasis. Arterioscler Thromb Vasc Biol. 2011;31(11):2378–82. doi: 10.1161/ATVBAHA.111.226688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price NL, Fernandez-Hernando C. miRNA regulation of white and brown adipose tissue differentiation and function. Biochim Biophys Acta. 2016;1861(12 Pt B):2104–2110. doi: 10.1016/j.bbalip.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239–50. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh AK, Aryal B, Zhang X, Fan Y, Price NL, Suarez Y, Fernandez-Hernando C. Posttranscriptional regulation of lipid metabolism by non-coding RNAs and RNA binding proteins. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail. 2016;18(5):457–68. doi: 10.1002/ejhf.495. [DOI] [PubMed] [Google Scholar]
- 10.Canfran-Duque A, Lin CS, Goedeke L, Suarez Y, Fernandez-Hernando C. Micro-RNAs and High-Density Lipoprotein Metabolism. Arterioscler Thromb Vasc Biol. 2016;36(6):1076–84. doi: 10.1161/ATVBAHA.116.307028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Hernando C. Antiatherogenic properties of high-density lipoprotein-enriched microRNAs. Arterioscler Thromb Vasc Biol. 2014;34(6):e13–4. doi: 10.1161/ATVBAHA.114.303542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Hernando C, Ramirez CM, Goedeke L, Suarez Y. MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol. 2013;33(2):178–85. doi: 10.1161/ATVBAHA.112.300144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goedeke L, Aranda JF, Fernandez-Hernando C. microRNA regulation of lipoprotein metabolism. Curr Opin Lipidol. 2014;25(4):282–8. doi: 10.1097/MOL.0000000000000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–69. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Zheng L, Wang Q, Hu YW. Emerging roles and mechanisms of long noncoding RNAs in atherosclerosis. Int J Cardiol. 2017;228:570–582. doi: 10.1016/j.ijcard.2016.11.182. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z. Progress and prospects of long noncoding RNAs in lipid homeostasis. Mol Metab. 2016;5(3):164–70. doi: 10.1016/j.molmet.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, Li Z, Bu D, Sun N, Zhang MQ, Chen R. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44(D1):D203–8. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–33. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aryal B, Rotllan N, Fernandez-Hernando C. Noncoding RNAs and atherosclerosis. Curr Atheroscler Rep. 2014;16(5):407. doi: 10.1007/s11883-014-0407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104(4):503–16. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 21.Lusis AJ. Atherosclerosis. Nature. 2000;407(6801):233–41. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rader DJ, Hovingh GK. HDL and cardiovascular disease. Lancet. 2014;384(9943):618–25. doi: 10.1016/S0140-6736(14)61217-4. [DOI] [PubMed] [Google Scholar]
- 23.Rosenson RS, Brewer HB, Jr, Davidson WS, Fayad ZA, Fuster V, Goldstein J, Hellerstein M, Jiang XC, Phillips MC, Rader DJ, Remaley AT, Rothblat GH, Tall AR, Yvan-Charvet L. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 2012;125(15):1905–19. doi: 10.1161/CIRCULATIONAHA.111.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linton MF, Tao H, Linton EF, Yancey PG. SR-BI: A Multifunctional Receptor in Cholesterol Homeostasis and Atherosclerosis. Trends Endocrinol Metab. 2017;28(6):461–472. doi: 10.1016/j.tem.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dikkers A, Tietge UJ. Biliary cholesterol secretion: more than a simple ABC. World J Gastroenterol. 2010;16(47):5936–45. doi: 10.3748/wjg.v16.i47.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linsel-Nitschke P, Tall AR. HDL as a target in the treatment of atherosclerotic cardiovascular disease. Nat Rev Drug Discov. 2005;4(3):193–205. doi: 10.1038/nrd1658. [DOI] [PubMed] [Google Scholar]
- 27.Nijstad N, Gautier T, Briand F, Rader DJ, Tietge UJ. Biliary sterol secretion is required for functional in vivo reverse cholesterol transport in mice. Gastroenterology. 2011;140(3):1043–51. doi: 10.1053/j.gastro.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 28.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328(5985):1566–9. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–3. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107(27):12228–32. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price NL, Rotllan N, Canfran-Duque A, Zhang X, Pati P, Arias N, Moen J, Mayr M, Ford DA, Baldan A, Suarez Y, Fernandez-Hernando C. Genetic Dissection of the Impact of miR-33a and miR-33b during the Progression of Atherosclerosis. Cell Rep. 2017;21(5):1317–1330. doi: 10.1016/j.celrep.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108(22):9232–7. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, Kaminski WE, Hahmann HW, Oette K, Rothe G, Aslanidis C, Lackner KJ, Schmitz G. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22(4):347–51. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 35.Oram JF, Vaughan AM. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Curr Opin Lipidol. 2000;11(3):253–60. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 36.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012;4(9):882–95. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T, Francl JM, Boehme S, Chiang JY. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology. 2013;58(3):1111–21. doi: 10.1002/hep.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vergeer M, Bots ML, van Leuven SI, Basart DC, Sijbrands EJ, Evans GW, Grobbee DE, Visseren FL, Stalenhoef AF, Stroes ES, Kastelein JJ. Cholesteryl ester transfer protein inhibitor torcetrapib and off-target toxicity: a pooled analysis of the rating atherosclerotic disease change by imaging with a new CETP inhibitor (RADIANCE) trials. Circulation. 2008;118(24):2515–22. doi: 10.1161/CIRCULATIONAHA.108.772665. [DOI] [PubMed] [Google Scholar]
- 39.Kaur N, Pandey A, Negi H, Shafiq N, Reddy S, Kaur H, Chadha N, Malhotra S. Effect of HDL-raising drugs on cardiovascular outcomes: a systematic review and meta-regression. PLoS One. 2014;9(4):e94585. doi: 10.1371/journal.pone.0094585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107(40):17321–6. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–7. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, Hansen HF, Whittaker S, Henry S, Brookes R, Najafi-Shoushtari SH, Chung RT, Whetstine JR, Gerszten RE, Kauppinen S, Naar AM. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5(212):212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121(7):2921–31. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice--brief report. Arterioscler Thromb Vasc Biol. 2013;33(8):1973–7. doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− mice. J Am Heart Assoc. 2012;1(6):e003376. doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouimet M, Ediriweera H, Afonso MS, Ramkhelawon B, Singaravelu R, Liao X, Bandler RC, Rahman K, Fisher EA, Rayner KJ, Pezacki JP, Tabas I, Moore KJ. microRNA-33 Regulates Macrophage Autophagy in Atherosclerosis. Arterioscler Thromb Vasc Biol. 2017;37(6):1058–1067. doi: 10.1161/ATVBAHA.116.308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marquart TJ, Wu J, Lusis AJ, Baldan A. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2013;33(3):455–8. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouimet M, Hennessy EJ, van Solingen C, Koelwyn GJ, Hussein MA, Ramkhelawon B, Rayner KJ, Temel RE, Perisic L, Hedin U, Maegdefessel L, Garabedian MJ, Holdt LM, Teupser D, Moore KJ. miRNA Targeting of Oxysterol-Binding Protein-Like 6 Regulates Cholesterol Trafficking and Efflux. Arterioscler Thromb Vasc Biol. 2016;36(5):942–51. doi: 10.1161/ATVBAHA.116.307282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goedeke L, Salerno A, Ramirez CM, Guo L, Allen RM, Yin X, Langley SR, Esau C, Wanschel A, Fisher EA, Suarez Y, Baldan A, Mayr M, Fernandez-Hernando C. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol Med. 2014;6(9):1133–41. doi: 10.15252/emmm.201404046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen RM, Marquart TJ, Jesse JJ, Baldan A. Control of very low-density lipoprotein secretion by N-ethylmaleimide-sensitive factor and miR-33. Circ Res. 2014;115(1):10–22. doi: 10.1161/CIRCRESAHA.115.303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karunakaran D, Richards L, Geoffrion M, Barrette D, Gotfrit RJ, Harper ME, Rayner KJ. Therapeutic Inhibition of miR-33 Promotes Fatty Acid Oxidation but Does Not Ameliorate Metabolic Dysfunction in Diet-Induced Obesity. Arterioscler Thromb Vasc Biol. 2015;35(12):2536–43. doi: 10.1161/ATVBAHA.115.306404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karunakaran D, Thrush AB, Nguyen MA, Richards L, Geoffrion M, Singaravelu R, Ramphos E, Shangari P, Ouimet M, Pezacki JP, Moore KJ, Perisic L, Maegdefessel L, Hedin U, Harper ME, Rayner KJ. Macrophage Mitochondrial Energy Status Regulates Cholesterol Efflux and Is Enhanced by Anti-miR33 in Atherosclerosis. Circ Res. 2015;117(3):266–78. doi: 10.1161/CIRCRESAHA.117.305624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein Geltink RI, O’Sullivan D, Corrado M, Bremser A, Buck MD, Buescher JM, Firat E, Zhu X, Niedermann G, Caputa G, Kelly B, Warthorst U, Rensing-Ehl A, Kyle RL, Vandersarren L, Curtis JD, Patterson AE, Lawless S, Grzes K, Qiu J, Sanin DE, Kretz O, Huber TB, Janssens S, Lambrecht BN, Rambold AS, Pearce EJ, Pearce EL. Mitochondrial Priming by CD28. Cell. 2017;171(2):385–397 e11. doi: 10.1016/j.cell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouimet M, Koster S, Sakowski E, Ramkhelawon B, van Solingen C, Oldebeken S, Karunakaran D, Portal-Celhay C, Sheedy FJ, Ray TD, Cecchini K, Zamore PD, Rayner KJ, Marcel YL, Philips JA, Moore KJ. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol. 2016;17(6):677–86. doi: 10.1038/ni.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ouimet M, Ediriweera HN, Gundra UM, Sheedy FJ, Ramkhelawon B, Hutchison SB, Rinehold K, van Solingen C, Fullerton MD, Cecchini K, Rayner KJ, Steinberg GR, Zamore PD, Fisher EA, Loke P, Moore KJ. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest. 2015;125(12):4334–48. doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakao T, Horie T, Baba O, Nishiga M, Nishino T, Izuhara M, Kuwabara Y, Nishi H, Usami S, Nakazeki F, Ide Y, Koyama S, Kimura M, Sowa N, Ohno S, Aoki H, Hasegawa K, Sakamoto K, Minatoya K, Kimura T, Ono K. Genetic Ablation of MicroRNA-33 Attenuates Inflammation and Abdominal Aortic Aneurysm Formation via Several Anti-Inflammatory Pathways. Arterioscler Thromb Vasc Biol. 2017;37(11):2161–2170. doi: 10.1161/ATVBAHA.117.309768. [DOI] [PubMed] [Google Scholar]
- 57.Price NL, Holtrup B, Kwei SL, Wabitsch M, Rodeheffer M, Bianchini L, Suarez Y, Fernandez-Hernando C. SREBP-1c/MicroRNA 33b Genomic Loci Control Adipocyte Differentiation. Mol Cell Biol. 2016;36(7):1180–93. doi: 10.1128/MCB.00745-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramirez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, Baldan A, Esplugues E, Fernandez-Hernando C. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle. 2012;11(5):922–33. doi: 10.4161/cc.11.5.19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian F, Wei H, Tian H, Qiu Y, Xu J. miR-33a is downregulated in melanoma cells and modulates cell proliferation by targeting PCTAIRE1. Oncol Lett. 2016;11(4):2741–2746. doi: 10.3892/ol.2016.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu N, Li Z, Yu Z, Yan F, Liu Y, Lu X, Yang W. MicroRNA-33b suppresses migration and invasion by targeting c-Myc in osteosarcoma cells. PLoS One. 2014;9(12):e115300. doi: 10.1371/journal.pone.0115300. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Zhang C, Zhang Y, Ding W, Lin Y, Huang Z, Luo Q. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein Cell. 2015;6(12):881–9. doi: 10.1007/s13238-015-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishiga M, Horie T, Kuwabara Y, Nagao K, Baba O, Nakao T, Nishino T, Hakuno D, Nakashima Y, Nishi H, Nakazeki F, Ide Y, Koyama S, Kimura M, Hanada R, Nakamura T, Inada T, Hasegawa K, Conway SJ, Kita T, Kimura T, Ono K. MicroRNA-33 Controls Adaptive Fibrotic Response in the Remodeling Heart by Preserving Lipid Raft Cholesterol. Circ Res. 2017;120(5):835–847. doi: 10.1161/CIRCRESAHA.116.309528. [DOI] [PubMed] [Google Scholar]
- 63.Huang K, Bao H, Yan ZQ, Wang L, Zhang P, Yao QP, Shi Q, Chen XH, Wang KX, Shen BR, Qi YX, Jiang ZL. MicroRNA-33 protects against neointimal hyperplasia induced by arterial mechanical stretch in the grafted vein. Cardiovasc Res. 2017;113(5):488–497. doi: 10.1093/cvr/cvw257. [DOI] [PubMed] [Google Scholar]
- 64.Horie T, Nishino T, Baba O, Kuwabara Y, Nakao T, Nishiga M, Usami S, Izuhara M, Sowa N, Yahagi N, Shimano H, Matsumura S, Inoue K, Marusawa H, Nakamura T, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat Commun. 2013;4:2883. doi: 10.1038/ncomms3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price NL, Singh AK, Rotllan N, Goedeke L, Wing A, Canfran-Duque A, Diaz-Ruiz A, Araldi E, Baldan A, Camporez JP, Suarez Y, Rodeheffer MS, Shulman GI, de Cabo R, Fernandez-Hernando C. Genetic Ablation of miR-33 Increases Food Intake, Enhances Adipose Tissue Expansion, and Promotes Obesity and Insulin Resistance. Cell Rep. 2018;22(8):2133–2145. doi: 10.1016/j.celrep.2018.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–38. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 67.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel A, Zavadil J, Castrillo A, Kim J, Suarez Y, Fernandez-Hernando C. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res. 2013;112(12):1592–601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Aguiar Vallim TQ, Tarling EJ, Kim T, Civelek M, Baldan A, Esau C, Edwards PA. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ Res. 2013;112(12):1602–12. doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011;31(11):2707–14. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 2012;586(10):1472–9. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 71.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C, Kim J. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol. 2012;235(2):476–83. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goedeke L, Rotllan N, Ramirez CM, Aranda JF, Canfran-Duque A, Araldi E, Fernandez-Hernando A, Langhi C, de Cabo R, Baldan A, Suarez Y, Fernandez-Hernando C. miR-27b inhibits LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels in mice. Atherosclerosis. 2015;243(2):499–509. doi: 10.1016/j.atherosclerosis.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang MH, Zhang LH, Wijesekara N, de Haan W, Butland S, Bhattacharjee A, Hayden MR. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arterioscler Thromb Vasc Biol. 2013;33(12):2724–32. doi: 10.1161/ATVBAHA.113.302004. [DOI] [PubMed] [Google Scholar]
- 74.Wang D, Xia M, Yan X, Li D, Wang L, Xu Y, Jin T, Ling W. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ Res. 2012;111(8):967–81. doi: 10.1161/CIRCRESAHA.112.266502. [DOI] [PubMed] [Google Scholar]
- 75.Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, Horton JD, Lasuncion MA, Naar AM, Suarez Y, Fernandez-Hernando C. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med. 2015;21(11):1280–9. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang B, Wang X, Song X, Bai R, Yang H, Yang Z, Xiao C, Bian Y. MicroRNA-20a/b regulates cholesterol efflux through post-transcriptional repression of ATP-binding cassette transporter A1. Biochim Biophys Acta. 2017;1862(9):929–938. doi: 10.1016/j.bbalip.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 77.de Aguiar Vallim T, Tarling E, Kim T, Civelek M, Baldan A, Esau C, Edwards P. MicroRNA-144 Regulates Hepatic ABCA1 and Plasma HDL Following Activation of the Nuclear Receptor FXR. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu YW, Hu YR, Zhao JY, Li SF, Ma X, Wu SG, Lu JB, Qiu YR, Sha YH, Wang YC, Gao JJ, Zheng L, Wang Q. An agomir of miR-144-3p accelerates plaque formation through impairing reverse cholesterol transport and promoting pro-inflammatory cytokine production. PLoS One. 2014;9(4):e94997. doi: 10.1371/journal.pone.0094997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, Hatoum I, Shah N, Lu Y, Kristo F, Psychogios N, Vrbanac V, Lu YC, Hla T, de Cabo R, Tsang JS, Schadt E, Sabeti PC, Kathiresan S, Cohen DE, Whetstine J, Chung RT, Fernandez-Hernando C, Kaplan LM, Bernards A, Gerszten RE, Naar AM. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med. 2015;21(11):1290–7. doi: 10.1038/nm.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chistiakov DA, Bobryshev YV, Kozarov E, Sobenin IA, Orekhov AN. Role of gut microbiota in the modulation of atherosclerosis-associated immune response. Front Microbiol. 2015;6:671. doi: 10.3389/fmicb.2015.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bensinger SJ, Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454(7203):470–7. doi: 10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- 82.Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov. 2014;13(6):433–44. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 83.Miller AM, Gilchrist DS, Nijjar J, Araldi E, Ramirez CM, Lavery CA, Fernandez-Hernando C, McInnes IB, Kurowska-Stolarska M. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One. 2013;8(8):e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ou Z, Wada T, Gramignoli R, Li S, Strom SC, Huang M, Xie W. MicroRNA hsa-miR-613 targets the human LXRalpha gene and mediates a feedback loop of LXRalpha autoregulation. Mol Endocrinol. 2011;25(4):584–96. doi: 10.1210/me.2010-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhong D, Huang G, Zhang Y, Zeng Y, Xu Z, Zhao Y, He X, He F. MicroRNA-1 and microRNA-206 suppress LXRalpha-induced lipogenesis in hepatocytes. Cell Signal. 2013;25(6):1429–37. doi: 10.1016/j.cellsig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 86.Adlakha YK, Khanna S, Singh R, Singh VP, Agrawal A, Saini N. Pro-apoptotic miRNA-128-2 modulates ABCA1, ABCG1 and RXRalpha expression and cholesterol homeostasis. Cell Death Dis. 2013;4:e780. doi: 10.1038/cddis.2013.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ji Y, Jian B, Wang N, Sun Y, Moya ML, Phillips MC, Rothblat GH, Swaney JB, Tall AR. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J Biol Chem. 1997;272(34):20982–5. doi: 10.1074/jbc.272.34.20982. [DOI] [PubMed] [Google Scholar]
- 88.Jian B, de la Llera-Moya M, Ji Y, Wang N, Phillips MC, Swaney JB, Tall AR, Rothblat GH. Scavenger receptor class B type I as a mediator of cellular cholesterol efflux to lipoproteins and phospholipid acceptors. J Biol Chem. 1998;273(10):5599–606. doi: 10.1074/jbc.273.10.5599. [DOI] [PubMed] [Google Scholar]
- 89.Braun A, Trigatti BL, Post MJ, Sato K, Simons M, Edelberg JM, Rosenberg RD, Schrenzel M, Krieger M. Loss of SR-BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E-deficient mice. Circ Res. 2002;90(3):270–6. doi: 10.1161/hh0302.104462. [DOI] [PubMed] [Google Scholar]
- 90.Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, Hong B. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33(10):1956–64. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu Z, Shen WJ, Kraemer FB, Azhar S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol Cell Biol. 2012;32(24):5035–45. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goedeke L, Wagschal A, Fernandez-Hernando C, Naar AM. miRNA regulation of LDL-cholesterol metabolism. Biochim Biophys Acta. 2016;1861(12 Pt B):2047–2052. doi: 10.1016/j.bbalip.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 94.Irani S, Pan X, Peck BC, Iqbal J, Sethupathy P, Hussain MM. MicroRNA-30c Mimic Mitigates Hypercholesterolemia and Atherosclerosis in Mice. The Journal of biological chemistry. 2016;291(35):18397–409. doi: 10.1074/jbc.M116.728451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 96.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19(7):892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–83. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122(8):2884–97. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramirez CM, Goedeke L, Rotllan N, Bonora E, Hughes AD, Santer P, Fernandez-Hernando C, Tilg H, Willeit J, Kiechl S, Mayr M. Circulating MicroRNA-122 Is Associated With the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes. 2017;66(2):347–357. doi: 10.2337/db16-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brown MS, Goldstein JL. Expression of the familial hypercholesterolemia gene in heterozygotes: mechanism for a dominant disorder in man. Science. 1974;185(4145):61–3. doi: 10.1126/science.185.4145.61. [DOI] [PubMed] [Google Scholar]
- 102.Goldstein JL, Brown MS, Stone NJ. Genetics of the LDL receptor: evidence that the mutations affecting binding and internalization are allelic. Cell. 1977;12(3):629–41. doi: 10.1016/0092-8674(77)90263-x. [DOI] [PubMed] [Google Scholar]
- 103.Jiang H, Zhang J, Du Y, Jia X, Yang F, Si S, Wang L, Hong B. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis. 2015;243(2):523–32. doi: 10.1016/j.atherosclerosis.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 104.Yang M, Liu W, Pellicane C, Sahyoun C, Joseph BK, Gallo-Ebert C, Donigan M, Pandya D, Giordano C, Bata A, Nickels JT., Jr Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J Lipid Res. 2014;55(2):226–38. doi: 10.1194/jlr.M041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, Hammer RE, Moon YA, Horton JD. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci U S A. 2005;102(15):5374–9. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jeon TI, Esquejo RM, Roqueta-Rivera M, Phelan PE, Moon YA, Govindarajan SS, Esau CC, Osborne TF. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013;18(1):51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sallam T, Jones M, Thomas BJ, Wu X, Gilliland T, Qian K, Eskin A, Casero D, Zhang Z, Sandhu J, Salisbury D, Rajbhandari P, Civelek M, Hong C, Ito A, Liu X, Daniel B, Lusis AJ, Whitelegge J, Nagy L, Castrillo A, Smale S, Tontonoz P. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. 2018;24(3):304–312. doi: 10.1038/nm.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sallam T, Jones MC, Gilliland T, Zhang L, Wu X, Eskin A, Sandhu J, Casero D, Vallim TQ, Hong C, Katz M, Lee R, Whitelegge J, Tontonoz P. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534(7605):124–8. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qin W, Li X, Xie L, Li S, Liu J, Jia L, Dong X, Ren X, Xiao J, Yang C, Zhou Y, Chen Z. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res. 2016;44(13):6423–33. doi: 10.1093/nar/gkw341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li D, Cheng M, Niu Y, Chi X, Liu X, Fan J, Fan H, Chang Y, Yang W. Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. International journal of biological sciences. 2017;13(3):349–357. doi: 10.7150/ijbs.16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Halley P, Kadakkuzha BM, Faghihi MA, Magistri M, Zeier Z, Khorkova O, Coito C, Hsiao J, Lawrence M, Wahlestedt C. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6(1):222–30. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li P, Ruan X, Yang L, Kiesewetter K, Zhao Y, Luo H, Chen Y, Gucek M, Zhu J, Cao H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21(3):455–67. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lan X, Yan J, Ren J, Zhong B, Li J, Li Y, Liu L, Yi J, Sun Q, Yang X, Sun J, Meng L, Zhu W, Holmdahl R, Li D, Lu S. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology. 2016;64(1):58–72. doi: 10.1002/hep.28391. [DOI] [PubMed] [Google Scholar]
- 114.Hu YW, Yang JY, Ma X, Chen ZP, Hu YR, Zhao JY, Li SF, Qiu YR, Lu JB, Wang YC, Gao JJ, Sha YH, Zheng L, Wang Q. A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. Journal of lipid research. 2014;55(4):681–97. doi: 10.1194/jlr.M044669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75(5):846–57. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 116.Wang Y, Chen F, Zhao M, Yang Z, Li J, Zhang S, Zhang W, Ye L, Zhang X. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J Biol Chem. 2017;292(37):15395–15407. doi: 10.1074/jbc.M117.783738. [DOI] [PMC free article] [PubMed] [Google Scholar]