Abstract
Background
The progression towards low-cost and rapid next-generation sequencing has uncovered a multitude of variants of unknown significance (VUS) in both patients and asymptomatic “healthy” individuals. A VUS is a rare or novel variant for which disease pathogenicity has not been conclusively demonstrated or excluded, and thus cannot be definitively annotated. VUS therefore pose critical clinical interpretation and risk-assessment challenges, and new methods are urgently needed to better characterize their pathogenicity.
Methods
To address this challenge and showcase the uncertainty surrounding genomic variant interpretation, we recruited a “healthy” asymptomatic individual, lacking cardiac-disease clinical history, carrying a hypertrophic cardiomyopathy (HCM)-associated genetic variant (NM_000258.2:c.170C>A, NP_000249.1:p.Ala57Asp) in the sarcomeric gene MYL3, reported by the ClinVar database to be “likely pathogenic”. Human induced pluripotent stem cells (iPSCs) were derived from the heterozygous VUSMYL3(170C>A) carrier, and their genome was edited using CRISPR/Cas9 to generate four isogenic iPSC lines: (1) corrected “healthy” control, (2) homozygous VUSMYL3(170C>A), (3) heterozygous frameshift mutation (fs) MYL3(170C>A/fs), and (4) known heterozygous MYL3 pathogenic mutation (NM_000258.2:c.170C>G), at the same nucleotide position as VUSMYL3(170C>A)), lines. Extensive assays including measurements of gene expression, sarcomere structure, cell size, contractility, action potentials, and calcium handling were performed on the isogenic iPSC-derived cardiomyocytes (iPSC-CMs).
Results
The heterozygous VUSMYL3(170C>A)-iPSC-CMs did not show an HCM phenotype at the gene expression, morphology, or functional levels. Furthermore, genome-edited homozygous VUSMYL3(170C>A)- and frameshift mutation MYL3(170C>A/fs)-iPSC-CMs lines were also asymptomatic, supporting a benign assessment for this particular MYL3 variant. Further assessment of the pathogenic nature of a genome-edited isogenic line carrying a known pathogenic MYL3 mutation, MYL3(170C>G), and a carrier-specific iPSC-CMs line, carrying a, MYBPC3(961G>A) HCM variant demonstrated the ability of this combined platform to provide both pathogenic and benign assessments.
Conclusions
Our study illustrates the ability of CRISPR/Cas9 genome-editing of carrier-specific iPSCs to elucidate both benign and pathogenic HCM functional phenotypes in a carrier-specific manner in a dish. As such, this platform represents a promising VUS risk-assessment tool that can be used for assessing HCM-associated VUS specifically, and VUS in general, and thus significantly contribute to the arsenal of precision medicine tools available in this emerging field.
Keywords: VUS, HCM, CRISPR/Cas9, iPSCs
INTRODUCTION
Continued advancement in the field of medical genetics towards low-cost, and rapid next-generation sequencing has enabled increased genetic screening for disease-causing genetic mutations, not only in patients but also in “healthy” individuals. Because of this, genetic testing has uncovered a multitude of novel unclassified genetic variants, also known as variant(s) of unknown significance (VUS).1, 2 A VUS is a genetic change for which pathogenicity has not been confirmed.3 Consequently, clinical genetic testing often reports VUS with ambiguous and uncertain pathogenicity. This can cause anxiety and stress in otherwise asymptomatic individuals and families who are found to carry a VUS, especially when a poorly characterized variant is erroneously reported as “likely pathogenic” or even “pathogenic”.4 VUS therefore poses serious clinical interpretation and risk-assessment challenges that limit the ability of healthcare providers to effectively counsel and treat individuals with unclear genetic predisposition to disease.
Hypertrophic cardiomyopathy (HCM), a common cause of sudden cardiac death (SCD), is a heterogeneous disease with vast genetic variability in the human genome, including a high degree of novel (“private”) mutations.5 The findings of numerous VUS associated with HCM in “healthy” populations have created ambiguity in genetic test reports, posing a growing and unresolved challenge for cardiovascular precision medicine.6 In silico tools such as PolyPhen2,7 AlignGVGD,8 and SIFT9 have been developed to enable the prediction of the pathogenic likelihood of gene mutations by attempting to calculate the possible impact of an amino acid substitution on protein damage. However, these are only in silico predictions, and a variant classified as “likely pathogenic” or even “pathogenic” may actually be benign.10 Hence, it is imperative to develop new experimental systems, ideally of human origin, which can help assess the functional pathogenic potential of a VUS in a patient-specific manner.
In the case of heart disease, recent advances in induced pluripotent stem cell (iPSC) biology are generating groundbreaking opportunities for the study of genetic cardiomyopathy and familial arrhythmogenic syndromes such as Long QT,11 catecholaminergic polymorphic ventricular tachycardia (CPVT),12 Brugada syndrome13 and HCM.14, 15 Cardiomyocytes derived from iPSCs (iPSC-CMs) enable recapitulation of abnormal pathogenic cardiac phenotypes in a disease- and patient-specific manner.16 Further strengthening this “disease in a dish” platform, the field of genome editing now allows interrogation of genetic mutations via genotype-phenotype correlation in a patient-specific manner using iPSCs.17–20 With the aid of clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9) and single-stranded oligodeoxynucleotide (ssODN) mediated genome editing technologies, a genetic mutation can be readily introduced or corrected in iPSCs to create isogenic patient-specific iPSC lines in a dish.21, 22 The generated isogenic iPSC lines share the same genetic background and differ exclusively at the mutation site. This advantage eliminates confounding factors due to different genetic backgrounds, allowing the genotype-phenotype relationship to be deciphered in a more precise and measurable manner. Taken together, the powerful combination of iPSC-based disease modeling and tools such as CRISPR/Cas9-mediated genome editing present an unprecedented opportunity to study VUS functional phenotypes in a patient-specific manner.
To highlight the potential application of CRISPR/Cas9 and iPSC disease models to clinical genetic testing, we have chosen to study a VUS with contradictory genotype-phenotype reports. The VUS is a missense variant (NM_000258.2:c.170C>A, NP_000249.1:p.Ala57Asp) in MYL3, a protein-coding gene that is a prominent component of the sarcomere complex.23 The VUS was discovered in an asymptomatic carrier without clinical nor family history (3 generations) associated with suspicion of cardiac disease and sudden cardiac death (SCD). Despite the asymptomatic nature of the individual, this VUS is surprisingly predicted with confidence using in silico tools to be pathogenically damaging to protein function. Moreover, conflicting interpretations of pathogenicity are reported in the ClinVar database.24 Some genetic testing resources classify this variant as a VUS, while others classify it as a likely pathogenic mutation having the potential to cause HCM. In this study, we use a combined CRISPR/Cas9-iPSC approach to evaluate the pathogenicity of this HCM-associated VUS. These approaches will be critical in the future to mitigate patient and family anxiety when they are confronted with unclear genetic predisposition to a disease, allowing more targeted therapies to reduce unnecessary harmful medical interventions and testing while cutting cost.
METHODS
Detailed methods are available in the Data Supplement. The data, analytic methods, and study materials for the purposes of reproducing the results or replicating procedures can be made upon request to the corresponding author who manages the information.
Genomic variant detection in heart disease-related genes
The study was carried out under Stanford Institutional Review Board (IRB) and that the subjects gave informed consent. 54 healthy subjects without cardiac-disease clinical history and presenting with normal ECG and echocardiography findings were recruited by the Stanford Cardiovascular Institute Biobank. Genetic testing of these subjects for this study was approved by Stanford University Institutional Review Board. Genomic DNA was isolated using DNeasy Blood and Tissue kits (Qiagen). Ion Torrent libraries from the 54 individual DNA samples were prepared with the Ion AmpliSeq™ Kit for Chef DL8 (Thermo Fisher Scientific) using a custom AmpliSeq panel (Thermo Fisher Scientific), which covers 135 cardiomyopathy and congenital heart disease DNA panel genes associated with sudden cardiac death.25 Next, the libraries were sequenced on an Ion PI™ Chip (Thermo Fisher Scientific) with the Ion Proton system using the Ion PI™ Hi-Q™ Sequencing 200 Kit (Thermo Fisher Scientific). Sequence reads were aligned to human genome hg19 using Ion Torrent Suite Software (v5.0) and genomic variants were detected by Torrent Variant Caller plugin. VCF files of all samples were then annotated by Ion Reporter (ionreporter.thermofisher.com). Annotations of genetic variants were from ClinVar.24
iPSC Generation
The study was carried out under Stanford Institutional Review Board (IRB) and Stem Cell Research Oversight (SCRO) Committee guidelines and that the subjects gave informed consent. Patients’ peripheral blood mononuclear cells (PBMCs) were collected and reprogrammed from a MYL3(170C>A) VUS carrier, a MYBPB3(961G>A) variant carrier, and two healthy individuals26 using the CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific).
Genome Editing
iPSCs were transfected with CRISPR/Cas9 and ssODN using the Lipofectamine 3000 Reagent (Thermo Fisher Scientific). For each well of a 6-well plate, 1 μg CRISPR/Cas9 vectors and 4 μg ssODN were used for transfection. For each genome edited line (isogenic corrected, isogenic homozygous VUSMYL3(170C>A), frameshift mutation MYL3(170C>A/fs), and isogenic heterozygous MYL3(170C>G)), several clones were generated, three of which were continuously propagated and used for cardiomyocytes differentiation and characterization.
Statistical Analysis
For statistical analysis, Student’s t-test was used to compare the difference between two data sets. For comparison among multiple groups, one-way or two-way ANOVA was used, where appropriate, and Holm-Sidak or Tukey after-tests were used for all pairwise comparisons, depending on the properties of the data sets. A P value <0.05 was considered to be statistically significant. The presented data was derived from three replicates, acquired from three differentiations performed on three separate occasions. All data in bar graphs were presented as mean ± standard error mean.
RESULTS
Uncovering a likely pathogenic HCM variant in an asymptomatic individual
We recruited 54 healthy individuals without clinical suspicion for heart disease from the Stanford Cardiovascular Institute iPSC Biobank, then sequenced their DNA using a custom 135 cardiomyopathy and congenital heart disease DNA panel genes associated with sudden cardiac death to determine whether they carried “pathogenic” or “likely pathogenic” mutations.25 Based on the ClinVar annotations24 of the resulting 592 unique non-synonymous genetic variants across these 54 individuals, the majority (78%) were categorized as benign, likely benign or VUS (Supplemental Figure S1A). Interestingly, 17 individuals each carried one variant annotated as “likely pathogenic”, among them 4 individuals carried a “likely pathogenic” HCM-related VUS (Supplemental Table S1).
Further complicating these conflicting variant findings is the fact that most individuals lack detailed family clinical histories. Out of the 4 identified “likely pathogenic” HCM VUS individuals, only one individual had multigenerational family members that were available and willing to undergo further echocardiography, authorize open access to their clinical and family history, and agree to a direct interview, in addition to genetic screening. We therefore chose to study this individual’s missense variant, (NM_000258.2:c.170C>A, NP_000249.1:p.Ala57Asp) in the sarcomeric gene MYL3 (Figure 1A, 1B). Importantly, this variant has not been previously reported in the literature and presents conflicting ClinVar reports, with some classifying the variant as a VUS and others as likely pathogenic (Table 1).24
Figure 1. Confirmation of the likely pathogenic VUSMYL3(170C>A) uncovered in an asymptomatic carrier.
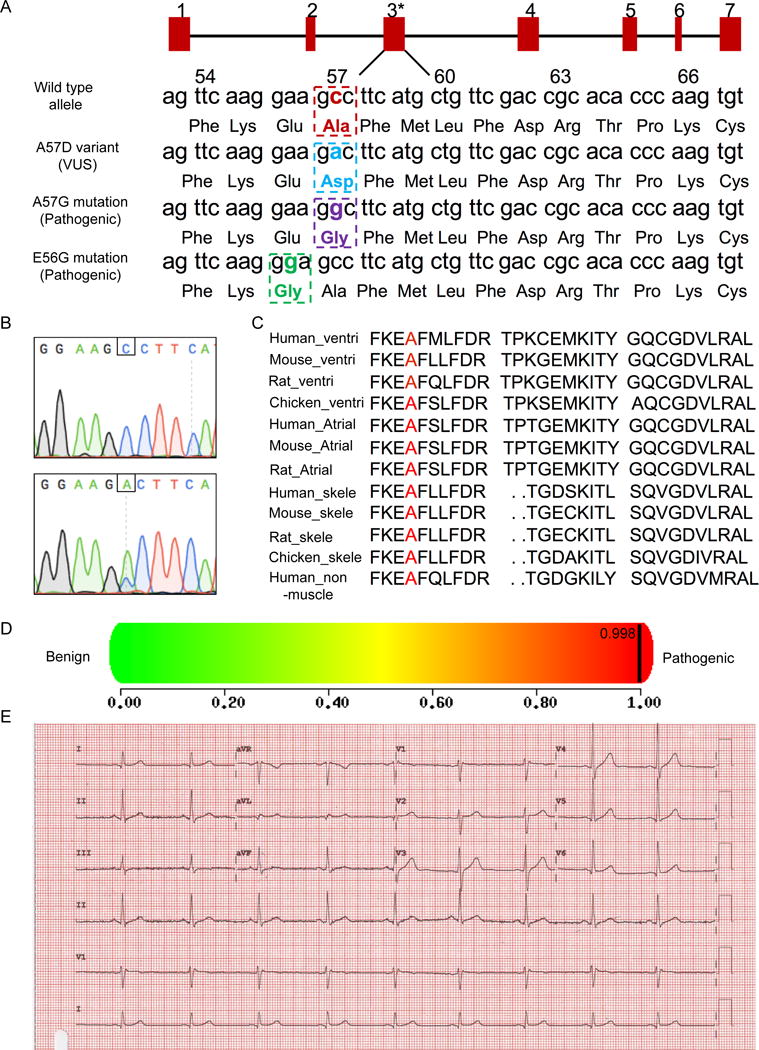
(A) Schematic illustration of the nucleotide and amino acid position of the VUS (NM_000258.2:c.170C>A, NP_000249.1:p.Ala57Asp) and two known pathogenic mutations (A57G, E56G) in the MYL3 gene. The red rectangles represent exons 1-7. Asterisks displays VUSMYL3(170C>A) position. Numbers 54-66 annotate amino acids positions. (B) Sanger sequencing confirmation of the VUSMYL3(170C>A). Upper panel: Sequencing result of a healthy control individual, lower panel: sequencing result of the heterozygous VUSMYL3(170C>A) carrier. Nucleotide position of the VUSMYL3(170C>A) is displayed by a framing square. (C) Alignment of regions flanking the VUSMYL3(170C>A) (red font) in MYL3 protein showing evolutionary conservation of the mutated residue across species and isoforms. (D) A prediction score of the VUSMYL3(170C>A) predicted by the in silico tool PolyPhen-2. (E) The VUSMYL3(170C>A) carrier’s ECG recording.
Table 1.
ClinVar report for VUSMYL3(170C>A)
| Clinical significance (Last evaluated) | Collection method | Condition(s) (Mode of inheritance) | Submitter – Study name | Description |
|---|---|---|---|---|
| Uncertain significance (Aug 29, 2013) | clinical testing | not specified [MedGen] | Laboratory for Molecular Medicine, Partners HealthCare Personalized Medicine | None |
| Uncertain significance (May 18, 2017) | clinical testing | not specified [MedGen] | GeneDx | Based on the currently available information, it is unclear whether this variant is a pathogenic variant or a rare benign variant |
| Uncertain significance (Nov 30, 2015) | clinical testing | Cardiomyopathy [MedGen | Orphane] | Division of Genomic Diagnostics, The Children’s Hospital of Philadelphia | None |
| Likely pathogenic (Oct 31, 2016) | clinical testing | not provided [MedGen] | Praxis fuer Humangenetik Tuebingen | Ala57Asp variant may be pathogenic, additional studies are needed to fully assess its clinical significance |
| Uncertain significance (Jul 7, 2014) | clinical testing | Primary familial hypertrophic cardiomyopathy [MedGen | Orphanet|OMIM] | Blueprint Genetics | Found together with likely pathogenic MYBPC3 (NM_000256.3:c.3800G>A) |
MYL3 gene encodes MYL3 protein, which is an important modulator of sarcomeric myosin cross-bridge kinetics essential for the modulation of cardiac muscle contraction.23 The A57D variant is a non-conservative amino acid substitution (Figure 1C), which is likely to affect secondary protein structure as these residues differ in polarity, charge, size, and other properties.27 Missense variants in nearby residue (E56G) and at the same residue (A57G) have been reported in the Human Gene Mutation Database in association with HCM (Figure 1A), further supporting the functional importance of this region of the protein and a strong association of this particular locus with the HCM disease.28 The VUSMYL3(170C>A) was then evaluated using an in silico analysis tool, PolyPhen2, on a damaging score scale of 0-1, 0 being benign and 1 being pathogenic (damaging to the protein function).7 Surprisingly, despite the asymptomatic cardiac history of the 46 years old VUSMYL3(170C>A) carrier, the in silico analysis tool displayed a high score of 0.998 (Figure 1D), confidently predicting the VUSMYL3(170C>A) to be pathogenic and very damaging to the function of the MYL3 protein. This was further validated using additional in silico tools (SIFT and AlignGVGD). Moreover, interrogation of this VUSMYL3(170C>A) using the Exome Aggregation Consortium (ExAC) database showed that this mutation is observed in only 14 out of 121,338 individuals (Supplemental Table S2), indicating again this variant is infrequent and likely pathogenic.
At the carrier level, an electrocardiogram (ECG) assessment was conducted and did not detect an abnormal phenotype (Figure 1E). Similarly, echocardiogram did not show any evidence of hypertrophic cardiomyopathy (Table 2). To further assess the asymptomatic nature of this HCM-associated VUSMYL3(170C>A), seven family members representing three generations underwent Sanger sequencing. Four of the seven family members, including the studied proband, (70, 48, 46, 11 years of age) were found to carry the “likely pathogenic” VUSMYL3(170C>A) in their genomes (Supplemental Figure S1B, 1C). Similar to the proband, none of them displayed any phenotypic evidence of HCM, as is depicted by their echocardiograph results in Table 2, nor proarrhythmic clinical history (e.g., syncope or resuscitation).
Table 2.
Echocardiography results for individuals with MYL3 c.170C>A or MYBPC3 c. 961G>A.
| MYL3 c.170C>A Individual I.2 | MYL3 c.170C>A Individual II.2 | MYL3 c.170C>A Individual II.1 | MYL3 c.170C>A Individual III.2 | MYBPC3 c.961G>A Patient | |
|---|---|---|---|---|---|
| LVID (cm) | 4.5 | 4.7 | 5.4 | 4.4 | 4.5 |
| Septal WT (cm) | 0.85 | 0.82 | 0.92 | 0.67 | 1.8 |
| Posterior WT (cm) | 0.87 | 0.88 | 0.85 | 0.74 | 0.92 |
| Relative WT | 0.38 | 0.36 | 0.33 | 0.32 | 0.41 |
| LV mass index (g/m2) | 75 | 66 | 84 | 65 | 151 |
| LVEF (%) | 70 | 68 | 61 | 67 | 48.7 |
| E (cm/s) | 69 | 62 | 51 | 90 | 58.7 |
| E/A | 1.3 | 0.9 | 1 | 2 | 1.4 |
| e’ average (cm/s) | 6.5 | 7.0 | 10 | 13 | 7.1 |
| E/e’ | 11 | 8.9 | 5.1 | 6.9 | 8.5 |
| LAV index (mL/m2) | 28 | 23 | 21 | 24 | 41.2 |
| Estimated RAP (mmHg) | 3 | 3 | 3 | 3 | 10 |
| Septal curvature | Normal | Normal | Normal | Normal | Reverse curvature |
LVID: left ventricular diastolic diameter, WT: wall thickness, LV: left ventricular, LVEF: left ventricular ejection fraction, E: peak early diastolic transmitral flow velocities, E/A: peak early and late diastolic transmitral flow velocities ratio, E/e’: peak early diastolic transmitral flow velocities and peak early diastolic mitral annular velocities ratio, LAV: left atrial volume, RAP: right atrial pressure. MYL3 c.170C>A individuals: I.2 70 yo mother; II.1 48 yo brother; II.2 46 proband; III.2 11 yo daughter.
Generation of an isogenic corrected “healthy” control iPSC line using CRISPR/Cas9
One crucial limitation of patient-specific iPSCs as a unique tool for the study of human disease has been the inability to perform experiments under genetically defined conditions. This is particularly relevant for the study of this specific VUSMYL3(170C>A) case, as heterogeneous genetic background among individuals may disguise the subtle phenotype in vitro due to the asymptomatic nature of the VUSMYL3(170C>A) carrier. Accordingly, in addition to the control iPSC lines derived from two non-consanguineous healthy individuals,26 an isogenic genetically corrected control line was generated from iPSCs of the heterozygous VUSMYL3(170C>A) carrier to help evaluate disease-relevant changes. The VUSMYL3(170C>A) carrier’s peripheral blood mononuclear cells (PBMCs) were collected and reprogrammed to iPSCs (Supplemental Figure S2). CRISPR/Cas9 were then used to generate the isogenic corrected control cell line (Figures 2A, 2B). Three independent isogenic corrected clones were generated and confirmed by Sanger sequencing (Figure 2C, Supplemental Figure S3A). To exclude potential off-target cuttings by CRISPR/Cas9, ten genomic loci with the highest sequence homology to the guide RNA’s binding site were checked by Sanger sequencing, separately for each one of the three clones. Sequencing results showed that the generated isogenic control cell line did not harbor any insertions or deletions at these loci (Supplemental Table S3).
Figure 2. Genome-editing of an isogenic corrected iPSC line using CRISPR.
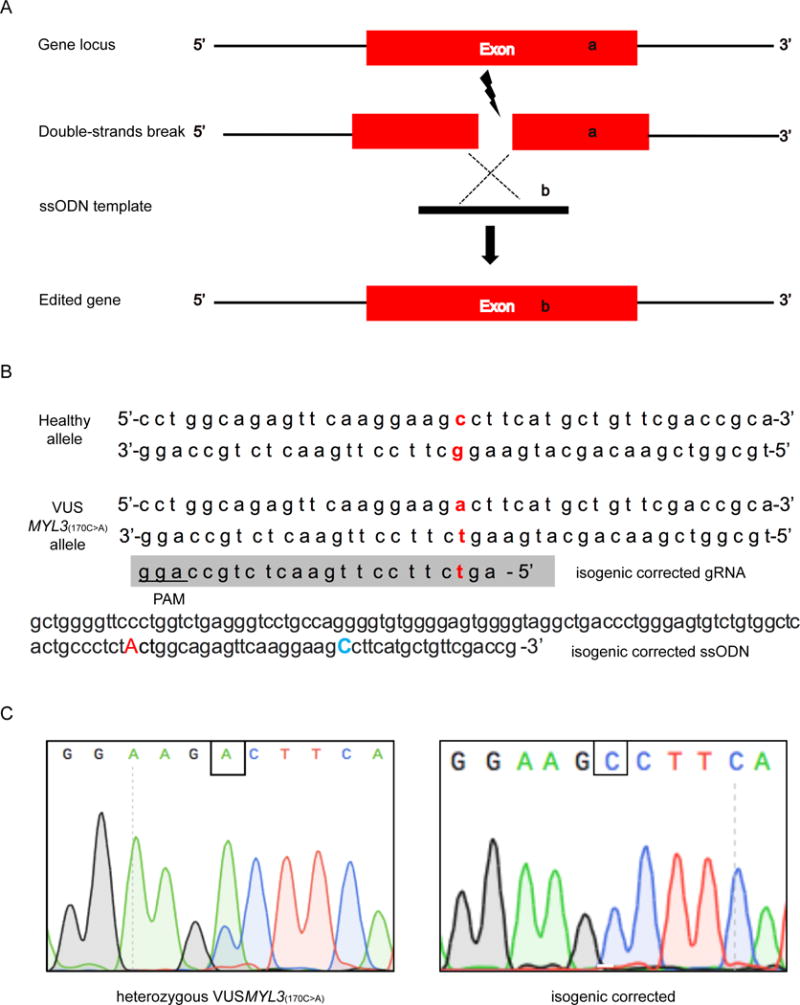
(A) Schematic view of CRISPR/Cas9 and ssODN mediated genome editing. (B) Sequence of gRNA and ssODN for generating the isogenic corrected iPSC line. The blue capitalized ‘C’ annotates the desired Cytidine to be introduced to the VUSMYL3(170C>A) allele. The red lower-case letters display nucleotides positions of the VUSMYL3(170C>A), while the red capitalized ‘A’ represents a silent mutation. (C) Sanger sequencing confirmation of the isogenic corrected iPSC line (clone 1). Additional clones are presented in Supplemental Figure S3.
Interrogation of cell morphology and gene expression of the heterozygous VUSMYL3(170C>A) iPSC-CMs
Multiple studies have reported myocyte sarcomere disarray and increased cell size to be features of HCM.14, 15 Therefore, to test whether the heterozygous VUSMYL3(170C>A)-iPSC-CMs display similar sarcomeric disarray and increased cell size, we conducted immunostaining of sarcomeric proteins and cell size measurements on the VUSMYL3(170C>A)-iPSC-CMs and compared it to the two healthy controls and isogenic corrected control (consisting of 3 clones) iPSC-CMs (Supplemental Figure S3B, 3C). Interestingly, immunostaining using sarcomere protein antibody anti-cardiac troponin T and anti sarcomeric-α-Actinin showed that the heterozygous VUSMYL3(170C>A)-iPSC-CMs display normal sarcomere distribution, similar to the two healthy controls and isogenic control iPSC-CM counterparts (Figure 3A, Supplemental Figure S3D). Furthermore, cell size analysis revealed no significant differences in cell size distribution and average cell area among the tested iPSC-CM lines (healthy controls, isogenic corrected control, and heterozygous VUSMYL3(170C>A)) (Figure 3B, 3C, 3D, Supplemental Figure S3E). Overall, iPSC-CMs carrying the HCM-associated VUSMYL3(170C>A) do not express HCM-relevant abnormal morphological features.
Figure 3. Cell morphology and gene expression analysis of the heterozygous VUSMYL3(170C>A) iPSC-CMs.
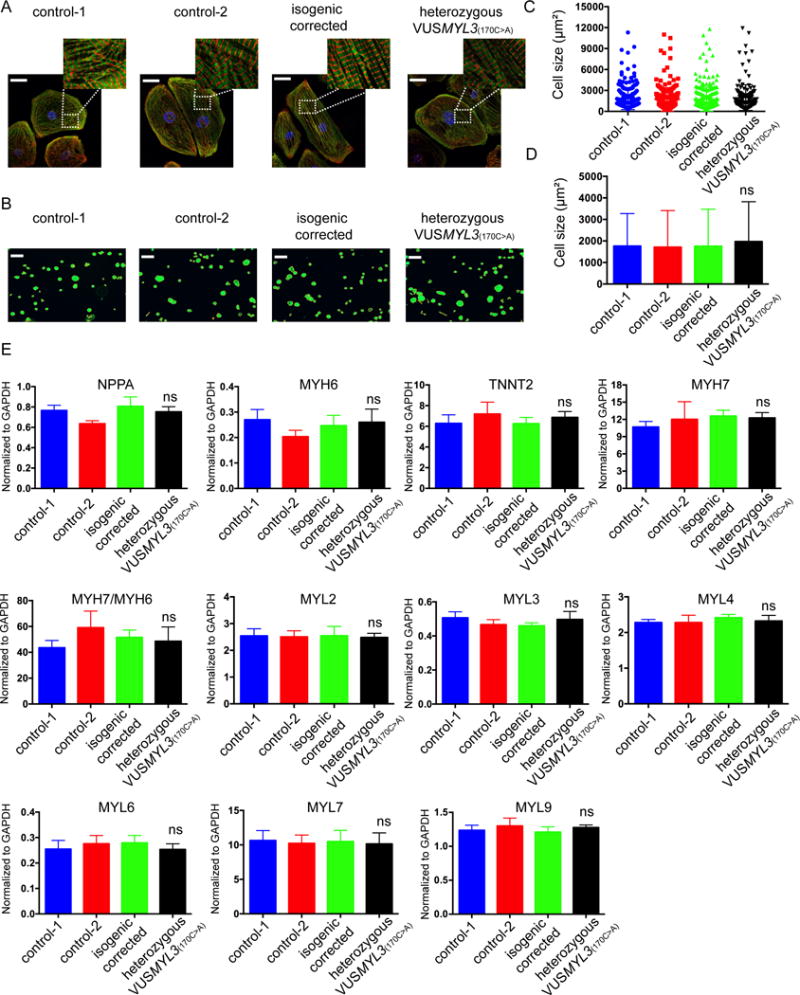
(A) Sarcomere immunostaining analysis of iPSC-CMs differentiated from the two healthy control, isogenic corrected, and heterozygous VUSMYL3(170C>A) lines, displaying sacromeric-α-actinin (red) and troponin T (green) stainings. Scale bars represents 10 μm. (B) A schematic representation of cell size measurements using Phalloidin. Scale bars represents 150 μm. (C) Distribution of iPSC-CMs size. (D) iPSC-CMs size statistical analysis (n≥200 cells per line). The heterozygous VUSMYL3(170C>A)-iPSC-CMs revealed no significant difference (ns) in average cell area compared with the tested iPSC-CM lines (healthy controls-1, healthy control-2 and isogenic corrected control), (E) Gene expression analysis of differentiated iPSC-CMs at day 45-50 dpd. The heterozygous VUSMYL3(170C>A)-iPSC-CMs revealed no significant difference in gene expression compared with the aforementioned tested iPSC-CM lines.
Cardiomyocytes derived from HCM patients upregulate natriuretic peptide precursor A (NPPA) expression and have high β-myosin/α-myosin ratios.14, 29 In addition, HCM patient-specific iPSC-CMs were previously reported to display upregulated expression of a group of genes that include NPPA, TNNT2, MYL2, MYL4, and MYH7.14, 15 Accordingly, gene expression analysis was performed on the VUSMYL3(170C>A)-iPSC-CMs at 45-50 days post-differentiation (dpd). The heterozygous VUSMYL3(170C>A)-iPSC-CMs displayed normal gene expression similar to their control counterparts, exhibiting no increased expression of the aforementioned genes, nor an elevation of the β-myosin/α-myosin ratio (Figure 3E, Supplemental Figure S4). Next, we further interrogated the potential effect of the VUSMYL3(170C>A) on the expression level of additional myosin light chain (MYL) genes that are highly expressed in cardiomyocytes (MYL3, MYL6, MYL7 and MYL9).30 Interestingly, the heterozygous VUSMYL3(170C>A)-iPSC-CMs expressed normal gene expression for all the tested myosin light chain genes (Figure 3E, Supplemental Figure S4). Altogether, these data show that the VUSMYL3(170C>A)-iPSC-CMs do not display the HCM-associated phenotype at the gene expression level.
Functional assessments of the heterozygous VUSMYL3(170C>A) iPSC-CMs
HCM is caused by mutations in sarcomere genes. As such, HCM-related changes in the molecular sarcomeric motor apparatus impair cardiac function, in many cases leading to increased contractility.31, 32 To test for potential MYL3 mutation-related functional alterations in the contractile apparatus of VUSMYL3(170C>A)-iPSC-CMs, contractility assays were performed. Similar to the healthy control and isogenic corrected control lines, the heterozygous VUSMYL3(170C>A)-iPSC-CMs displayed comparable contractility phenotype (Figure 4A), without any abnormal changes in beating rate, contraction and relaxation velocities (Figure 4B, 4C, 4D, Supplemental Figure S5A), contraction-relaxation distance, or contraction-relaxation duration (Supplemental Figure S5B).
Figure 4. Comprehensive functional analysis of the heterozygous VUSMYL3(170C>A) iPSC-CMs.
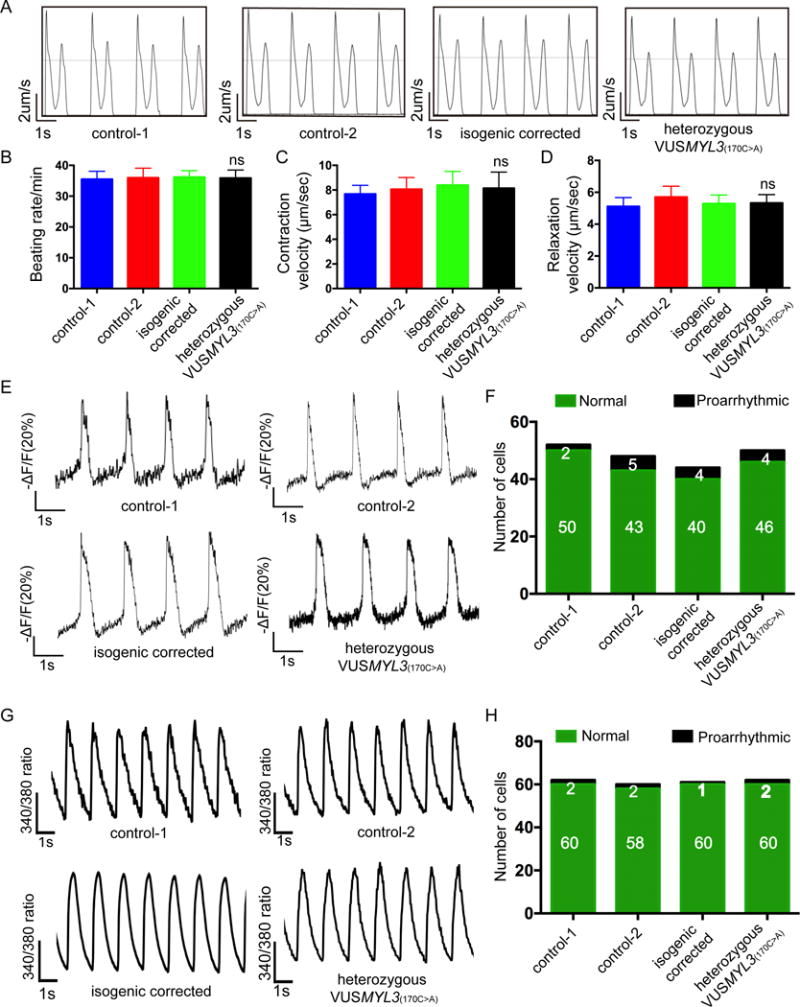
(A) Representative contractility traces of iPSC-CMs from the two healthy control, isogenic corrected, and the heterozygous VUSMYL3(170C>A) lines. (B-D) Beating rate, contraction velocity, and relaxation velocity, respectively. The heterozygous VUSMYL3(170C>A)-iPSC-CMs revealed no significant difference (ns) in beating rate, contraction velocity, and relaxation velocity, respectively, compared with the aforementioned tested iPSC-CM lines. (E) Representative action potential traces. (F) Number of iPSC-CMs displaying proarrhythmic activity. (G) Representative Ca2+ transients. (H) Number of iPSC-CMs showing proarrhythmic calcium transients.
Arrhythmia is a clinical hallmark of HCM.33, 34 Recent studies demonstrated the ability of patient- and disease-specific iPSC-CMs to recapitulate this abnormal electrophysiological phenotype using the iPSC technology.14, 15 Accordingly, VUSMYL3(170C>A)-iPSC-CMs were electrophysiologically assessed using a fluorescent voltage sensor with fast kinetics for imaging high-frequency electrical activity, applied as a membrane voltage change indicator, known as accelerated sensor of action potentials (ASAP).35 The VUSMYL3(170C>A)-iPSC-CMs did not display a clear proarrhythmic potential (Figure 4E), with only 8% of the heterozygous VUSMYL3(170C>A)-iPSC-CMs displaying delayed afterdepolarizations (DADs), a percentage similar to that detected in two healthy controls and isogenic corrected control lines (4%, 10%, and 9%, respectively) (Figure 4F, Supplemental Figure S5C). In summary, in contrast to the in silico pathogenic prediction, the lack of proarrhythmic activity presented by the VUSMYL3(170C>A)-iPSC-CMs clearly reflects the asymptomatic nature of the VUSMYL(170C>A) carrier.
Previous studies interrogating the various mechanisms underlying arrhythmia in patients with HCM have implicated abnormal Ca2+ homeostasis as one of the proarrhythmic mediators.36, 37 A recent study conducted on patient-specific HCM iPSC-CMs also revealed high levels of diastolic Ca2+, contributing to the disease proarrhythmic nature.14 To test for a proarrhythmic Ca2+-handling phenotype, we next recorded Ca2+ transients. Compared to the two healthy controls and the isogenic corrected control iPSC-CMs lines, the heterozygous VUSMYL3(170C>A)-iPSC-CMs did not exhibit abnormal proarrhythmic Ca2+ transients (Figure 4G, 4H, Supplemental Figure S5D), in accordance with the previous electrophysiological findings. In conclusion, the genetic, morphological, and functional assessments of the heterozygous VUSMYL3(170C>A)-iPSC-CMs all display an asymptomatic phenotype, similarly to that exhibited clinically by the VUSMYL3(170C>A) carrier.
Comprehensive analysis of a generated isogenic homozygous VUSMYL3(170C>A) iPSC line
The heterozygous VUSMYL3(170C>A)-iPSC-CMs did not show a pathogenic phenotype, despite a high in silico assessment score confidently predicting it to be pathogenically damaging. This predicament may be the result of the VUSMYL3(170C>A) being a disease-causing variant in the recessive state, and therefore explains the lack of pathogenic phenotype observed at the iPSC-CMs level. Unfortunately, since this variant is a VUS by definition, there is no support for recessive inheritance of this VUSMYL3(170C>A). To overcome this possible predicament, we next created a homozygous VUSMYL3(170C>A) line using a CRISPR/Cas9 (Supplemental Figure S6A), generating three independent clones for characterization. Sanger sequencing confirmed that the heterozygous VUSMYL3(170C>A) line was turned into a homozygous VUSMYL3(170C>A) line (Figure 5A, Supplemental Figure S6B), and no off-target mutation was detected in the three homozygous VUSMYL3(170C>A) iPSC clones (Supplemental Table S4). Interestingly, genome-edited homozygous VUSMYL3(170C>A)-iPSC-CMs displayed similar genetic, morphological, and functional assessment results, compared to those displayed by the heterozygous VUSMYL3(170C>A) and isogenic corrected control lines. The genome-edited homozygous VUSMYL3(170C>A)-iPSC-CMs also did not exhibit the following: (1) elevation in HCM-related genes (Figure 5C, Supplemental Figure S7A); (2) alterations in the gene expression level of the aforementioned myosin light chain genes (Figure 5C, Supplemental Figure S7A) (3) increased cell size (Figure 5D, Supplemental Figure S7B); (4) disarray of sarcomere organization (Figure 5E, Supplemental Figure S7C); (5) alterations in contractility (Figure 5F-H, Supplemental Figure S7D-F); (6) electrophysiological proarrhythmic phenotype (Figure 5I, 5J, Supplemental Figure S7G); or (7) irregular Ca2+-handling (Figure 5K, 5L, Supplemental Figure S7H). In conclusion, the lack of a pathogenic phenotype in all three individually generated genome-edited homozygous VUSMYL3(170C>A)-iPSC-CMs clones, further supports the interpretation that the VUSMYL3(170C>A) variant is benign.
Figure 5. Comprehensive assays for assessing the homozygous VUSMYL3(170C>A) and the heterozygous frameshift mutation MYL3(170C>A/fs) iPSC-CMs.
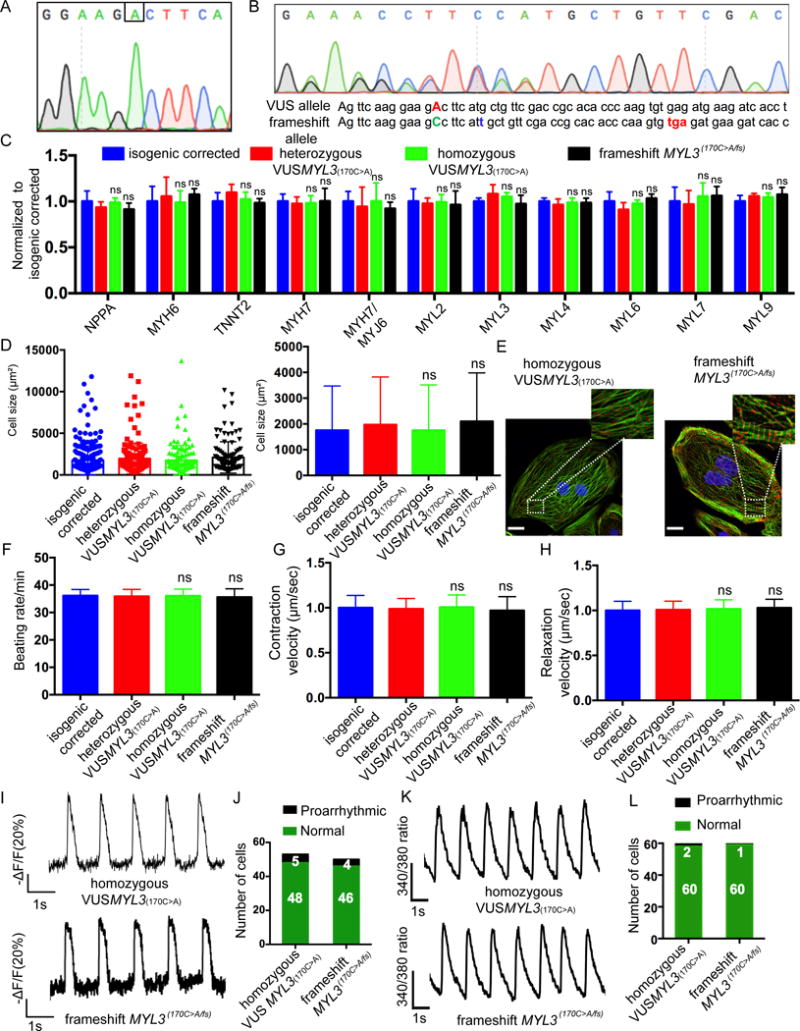
(A) Sanger sequencing confirmation of the isogenic homozygous VUSMYL3(170C>A) iPSC (clone1). Additional clones are presented in Supplemental Figure S6. (B) Sanger sequencing confirmation of the isogenic heterozygous frameshift mutation MYL3(170C>A/fs)-iPSC (clone1). Additional clones are presented in Supplemental Figure S6. The red capitalized ‘A’ represents the VUSMYL3(170C>A). The blue lower-case ‘t’ represents non-homologous end joining (NHEJ)-mediated 1bp insertion. The red lower-case letters (tga) display the premature stop codon. (C) Gene expression analysis. The homozygous VUSMYL3(170C>A)-iPSC-CMs and the frameshift mutation MYL3(170C>A/fs)-iPSC-CMs revealed no significant difference in gene expression compared with the isogenic corrected and the heterozygous VUSMYL3(170C>A) iPSC-CMs. (D) Cell size assessment (n≥200 cells per line). The homozygous VUSMYL3(170C>A)-iPSC-CMs and frameshift mutation MYL3(170C>A/fs)-iPSC-CMs revealed no significant difference (ns) in cell size distribution (left panel) and average cell area (right panel) compared with isogenic corrected and heterozygous VUSMYL3(170C>A). (E) Sarcomere immunostaining analysis displaying sacromeric-α-Actinin (red) and troponin T (green) stainings. Scale bars represents 10 μm. (F-H) Beating rate, contraction velocity, and relaxation velocity analysis, respectively. The homozygous VUSMYL3(170C>A) and frameshift mutation MYL3(170C>A/fs)-iPSC-CMs revealed no significant difference in beating rate, contraction velocity, and relaxation velocity, respectively, compared with isogenic corrected and heterozygous VUSMYL3(170C>A). (I) Representative action potential traces. (J) Number of iPSC-CMs displaying proarrhythmic activity. (K) Representative calcium transients. (L) Number of iPSC-CMs showing proarrhythmic calcium transients.
To further confirm the benign phenotype interpretation, we generated a premature stop codon (frameshift mutation) in the healthy MYL3 allele of the heterozygous VUSMYL3(170C>A) cell line, thus generating a cell line that lacks a healthy MYL3 allele (loss of heterozygosity) and thus only expresses the variant allele. Three individual frameshift mutation clones were generated using the same gRNA as the generation of the homozygous VUSMYL3(170C>A) line. To test the integrity of the frameshift mutation line, the presence of a premature stop codon in the MYL3 gene was validated by Sanger sequencing of the genomic DNA (Figure 5B, Supplementary Figure S6C). Sanger sequencing of the RT-PCR product validated the degradation of the frameshift allele mRNA. Thus only the mutated allele mRNA was identified (Supplementary Figure S6D). Next, three frameshift mutation clones were assessed at the genetic, morphological and functional levels, and compared to the heterozygous VUSMYL3(170C>A) and isogenic corrected control phenotypes. Similar to the homozygous VUSMYL3(170C>A) line, we found no evidence of a HCM phenotype in the frameshift mutation cell line (Figure 5C-L, Supplementary Figure S7A-H), further strengthening our conclusion that VUSMYL3(170C>A) is a benign genomic variant.
Comprehensive analysis of symptomatic HCM iPSC-CMs lines
To rule out the possibility that the benign prediction for the VUSMYL3(170C>A) was simply due to a lack of sensitivity on part of the iPSC-CMs platform, we next assayed two different HCM disease-associated mutations, displaying severe clinical HCM-related symptoms at the carrier level, in iPSC-CMs as positive controls. To validate that the benign assessment is not the result of the inability of the iPSC-CM platform to recapitulate a pathogenic MYL3 mutation phenotype in vitro, a known autosomal dominant pathogenic mutation, MYL3 c.170C>G, at the same nucleotide position as the VUSMYL3(170C>A), was studied. This mutation was reported to develop a severe HCM phenotype, including SCD, with disease penetrance in 78% of the genotyped MYL3 c.170C>G mutation carriers, and an onset as early as 28 years of age.27 For this purpose, the MYL3 c.170C>G mutation was introduced into the heterozygous VUSMYL3(170C>A) iPSCs using CRISPR/Cas9 (Supplementary Figure S8A). Accordingly, three independent iPSC clones (named MYL3(170C>G)) were generated and were tested by Sanger sequencing to confirm that the MYL3 c.170C>A variant allele was replaced with the MYL3 c.170C>G mutation (Figure 6A, Supplemental Figure S8B) without off-target cutting (Supplemental Table S5). Next, to further elucidate the capability of the iPSC-CMs platform to recapitulate a pathogenic phenotype of a symptomatic carrier, a heterozygous HCM missense genetic variant in the sarcomeric gene MYBPC3 (MYBPC3 c.961G>A, p.Val321Met) (Figure 6A) was studied using a carrier-specific iPSC-CMs line. Note that the carrier presented an early HCM onset at the young age of 23 (Table 2), is implanted with a cardioverter defibrillator, and has a strong family history of SCD, as early as 34 years of age.
Figure 6. Comprehensive assays for assessing the MYL3(170C>G) and MYBPC3(961G>A) iPSC-CMs.
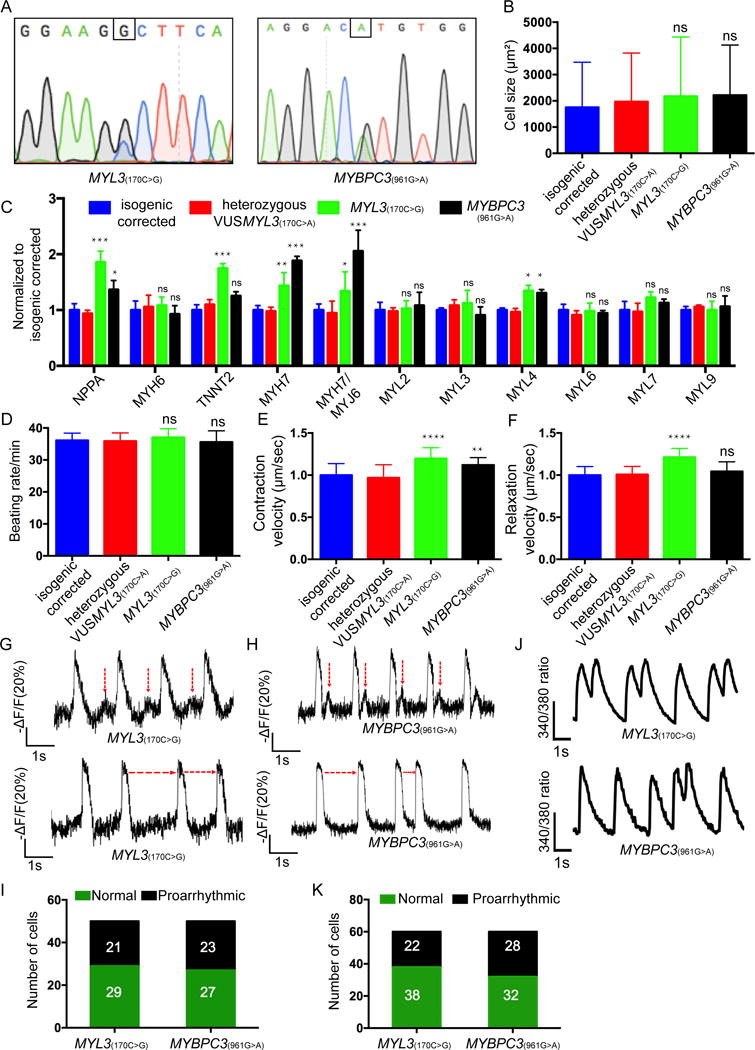
(A) Sanger sequencing confirmation of the isogenic MYL3(170C>G) clone1,(Additional clones are presented in Supplemental Figure S8) and MYBPC3(961G>A) iPSC line. (B) Cell size assessment (n≥200 cells per line) displayed as mean ± SD. The MYL3(170C>G) and MYBPC3(961G>A) iPSC-CMs revealed no significant difference (ns) in average cell area compared with isogenic corrected and heterozygous VUSMYL3(170C>A). (C) HCM-related gene expression analysis. The MYL3(170C>G) and MYBPC3(961G>A) iPSC-CMs revealed no significant difference in gene expression compared with the isogenic corrected and the heterozygous VUSMYL3(170C>A) iPSC-CM lines. (D-F) Beating rate, contraction velocity, and relaxation velocity, respectively. The MYL3(170C>G) and MYBPC3(961G>A) iPSC-CMs revealed no significant difference in beating rate, contraction velocity, and relaxation velocity, respectively, compared with isogenic corrected and heterozygous VUSMYL3(170C>A). (G-H) Representative action potentials traces displaying DADs (highlighted by red vertical arrows) and varying inter-beats intervals (indicated by red horizontal arrows) recorded from MYL3(170C>G) and MYBPC3(961G>A) iPSC-CMs, respectively. (I) Number of MYL3(170C>G) and MYBPC3(961G>A) iPSC-CMs displaying proarrythmic activity (J) Representative Ca2+ transients displaying DADs. (K) Number of MYL3(170C>G) and MYBPC3(961G>A) iPSC-CMs depicting proarrhythmic calcium transients. *p < 0.1, ** p < 0.01, *** p < 0.001, **** p < 0.0001, compared with isogenic corrected and heterozygous VUSMYL3(170C>A) lines.
The iPSC lines were then evaluated for pathogenic morphological changes and functional proarrhythmic potential. Similar to the heterozygous VUSMYL3(170C>A)-iPSC-CMs, both MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs exhibited no significant difference in cell size (Figure 6B, Supplementary Figure S8C) and had normal sarcomere distribution (Supplementary Figure S8D). However, while heterozygous VUSMYL3(170C>A)-iPSC-CMs displayed normal expression of HCM-related genes (NPPA, TNNT2, MYL2, and MYH7), these HCM-related genes, including the β-myosin/α-myosin ratio, were elevated in the MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs (Figure 6C, Supplementary Figure S8E). While both MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs did not exhibit a change in expression level of the myosin light chain genes (MYL2, MYL3, MYL6, MYL7 and MYL9) in comparison to the isogenic corrected and heterozygous VUSMYL3(170C>A)-iPSC-CMs, a significant increase in the MYL4 expression was noted (Figure 6C), as has been previously reported by multiple HCM-related studies.38–40 Contractility measurements did not display a statistically significant difference in beating rate among the tested lines (Figure 6D, Supplementary Figure S8F). Both MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs displayed faster contraction velocity in comparison to the corrected isogenic control line and heterozygous VUSMYL3(170C>A)-iPSC-CMs (Figure 6E, Supplementary Figure S8F). MYL3(170C>G)-iPSC-CMs displayed faster relaxation velocity (Figure 6F, Supplemental Figure S8F), as previously documented for this specific MYL3(170C>G) gene mutation,41, 42 and as validated in this study by three independent MYL3(170C>G)-iPSC-CMs clones. The MYBPC3(961G>A)-iPSC-CMs did not exhibit a significant difference in relaxation velocity compared with the isogenic corrected control and heterozygous VUSMYL3(170C>A)-iPSC-CMs (Figure 6F). Notably, while only 8% of heterozygous VUSMYL3(170C>A)-iPSC-CMs displayed a proarrhythmic electrophysiological phenotype in the form of DADs (Figure 4F), 42% and 46% of the MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs, respectively, exhibited DADs, displaying higher amplitude and shorter coupling intervals (Figure 6G, 6I, Supplemental Figure S8G). In addition, both presented significant variability in beat-to-beat intervals of action potentials (Figure 6H), another notable proarrhythmic characteristic. In accordance with the electrophysiological findings, 37% and 47% of the MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs, respectively, exhibited an abnormal Ca2+ phenotype, presenting double-peaked Ca2+ transients (Figure 6J, K, Supplemental Figure S8H) that was previously shown to be a proarrhythmic manifestation in HCM iPSC-CM14 as well as in another familial arrhythmogenic syndrome iPSC-CM study.12 These events were virtually absent in the VUSMYL3(170C>A)-iPSC-CMs. Importantly, Fura2 Ca2+ imaging further demonstrated a significant increase in diastolic Ca2+ in both MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs, when compared to the corrected and heterozygous VUSMYL3(170C>A)-iPSC-CMs lines (Supplementary Figure S8I).
Taken together, similarly to the symptomatic nature of both MYL3(170C>G) and MYBPC3(961G>A) carriers, an evident symptomatic functional phenotype was recapitulated by the MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs, respectively, highlighting the potential of the iPSC-CM platform to successfully assess both pathogenic and benign phenotypes.
DISCUSSION
In this study, we combine the powerful strengths of iPSC-based disease modeling and CRISPR/Cas9-mediated genome editing technology to demonstrate their unique potential as a personalized VUS risk-assessment platform for determining the pathogenicity of VUS. To highlight the dilemma and uncertainty that VUS presents for patients and clinicians around the world, we screened dozens of “healthy” individuals in search of a VUS that can clearly showcase the uncertainty surrounding genomic variant interpretation: a variant in a healthy individual that is annotated as “likely pathogenic”. By combining both aforementioned technologies for the study of such a VUS (MYL3 c.170C>G associated with HCM), we were able to: 1) recapitulate the asymptomatic phenotype of the VUSMYL3(170C>A) carrier by showing a lack of HCM-related gene expression profile, morphological phenotype, and proarrhythmic activity in the heterozygous VUSMYL3(C>A)-iPSC-CMs; 2) confirm the asymptomatic clinical phenotype as a benign assessment in a genome-edited isogenic homozygous VUSMYL3(170C>A) line, as well as in a heterozygous frameshift mutation MYL3(170C>A/fs) line; 3) validate the ability of the combined iPSC-CMs and genome-editing platform to recapitulate a pathogenic phenotype of a symptomatic carrier.
HCM is a prevalent hereditary cardiac disorder linked by both animal models and patient-specific iPSC-CMs studies to arrhythmia and SCD.14, 15, 43 In contrast to these models, the carrier-specific VUSMYL3(170C>A)-iPSC-CMs did not display a proarrhythmic phenotype. To validate whether the observed asymptomatic nature of the VUSMYL3(170C>A)-iPSC-CMs is the result of a recessive missense mutation, we also generated a homozygous VUSMYL3(170C>A) line. Interestingly, similar to the heterozygous cells, the phenotype displayed by all three homozygous iPSC-CMs clones was an asymptomatic one as well. The asymptomatic assessment of the homozygous line was further strengthened by an asymptomatic phenotype detected in the genome-edited heterozygous frameshift mutation MYL3(170C>A/fs) line (expressing solely the c.170C>A variant allele). Collectively, these data confirm that the lack of pathogenic phenotype in heterozygous VUSMYL3(170C>A)-iPSC-CMs was not the result of a recessive gene.
HCM is a complex and heterogeneous disease encompassing a wide variety of mutations and variants.29, 32 To validate whether the benign asymptomatic assessments of both the heterozygous VUSMYL3(170C>A) and the genome-edited homozygous VUSMYL3(170C>A)-iPSC-CMs are not the result of a potential inability on part of the iPSC-CMs platform to recapitulate a pathogenic MYL3 mutation phenotype, we next genome-edited iPSCs with a known disease-causing mutation in the MYL3 gene (MYL3 c.170C>G), at the same nucleotide position as the VUSMYL3(170C>A)27. Next, to further test the sensitivity of this cellular platform to recapitulate a HCM VUS, iPSCs from an HCM patient with a MYBPC3 c.961G>A variant were generated from a carrier presenting classical clinical HCM symptoms. We found that both the MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs did show a HCM pathogenic phenotype, for example upregulation of HCM-related genes expression, elevation in β-myosin/α-myosin ratio, irregular contractility, proarrhythmic electrophysiological phenotype (DADs), and an abnormal Ca2+ phenotype, similar to the results from previous HCM iPSC-CMs and HCM animal studies.14, 15, 41, 44, 45 The HCM-related phenotypes observed in both the MYL3(170C>G)- and MYBPC3(961G>A)-iPSC-CMs, effectively demonstrate that the lack of phenotype exhibited by both the heterozygous and homozygous VUSMYL3(170C>A)-iPSC-CMs was not due to a technical artifact or faulty detection sensitivity on part of the iPSC-CMs platform, but rather a reflection of a benign phenotype.
Over the past decade, a large range of studies, using similar assays to those presented in the current study, have demonstrated the intriguing ability of the iPSC-CMs platform to recapitulate disease phenotype presented by the patient at bedside at the patient-derived cellular level in the dish, as been previously reported for genetic cardiomyopathy and arrhythmogenic syndromes such as: LQT, CPVT, Brugada syndrome, familial dilated cardiomyopathy (DCM), and HCM.11, 12, 14, 15, 46 Similarly, iPSC-CMs have been also shown to recapitulate a normal “healthy” phenotype in cells derived from “healthy” subjects, as documented by countless studies. This striking recapitulation capability of the iPSC-CMs platform is also vivid in this study. The chosen carrier is a healthy 46 years old active, athletic, healthy individual, that although has been phenotyped with a “likely pathogenic” HCM MYL3 c.170C>A VUS, does not have a clinical cardiac history, presents normal ECG, and normal echocardiography results. Further, supporting the asymptomatic nature of the carrier is the carrier’s impressive family history, consisting of 3 generations of asymptomatic carriers as old as 70 years of age. Impressively, the proband derived-cells reflect the proband’s asymptomatic nature. In a similar manner, in the case of the MYL3(170C>G) mutation, the cells also strikingly recapitulated the carriers’ clinical phenotype. MYL3(170C>G) mutation carriers have been shown to experience HCM-related symptoms including SCD, with disease penetrance of 78% and an onset as early as 28 and as late as 38 years of age. The genome-edited MYL3(170C>G)-iPSC-CMS nicely recapitulate the symptomatic nature of the carriers. Reinforcing these findings, an additional carrier displaying a HCM variant demonstrating a clinical HCM onset as early as 23 years of age and a family history of SCD at an age as young as 34, was studied. In this case, the carrier-derived iPSC-CMs were also able to recapitulate a pathogenic phenotype. Altogether, the presented evidence here demonstrates the capability of the carrier-specific iPSC-CMs platform to reveal both a benign and pathogenic phenotype of prospective variants in a manner that recapitulates the carrier’s clinical presentation.
Collectively, our data demonstrate the unique potential of combining iPSC-based disease modeling and CRISPR/Cas9-mediated genome editing technology as a personalized VUS risk-assessment platform that can facilitate the study of the growing VUS phenomenon. Linking together iPSC and genome editing technologies presents an unprecedented opportunity to elucidate the pathogenicity of VUS in a dish in a carrier-specific manner. Importantly, these efforts will help improve the study and diagnostics of VUS specifically and the promotion of personalized and precision medicine in general, better guiding clinicians in their choice of therapy and providing a clearer result for the VUS carriers.
Supplementary Material
CLINICAL PERSPECTIVE.
What is new?
Our study demonstrates for the first time the unique potential of combining iPSC-based disease modeling and CRISPR/Cas9-mediated genome editing technology as a personalized risk-assessment platform for determining the pathogenicity of a variant of uncertain significance (VUS) in a patient-specific manner.
What are the clinical implications?
This personalized risk-assessment platform will help improve the study and diagnostics of VUS specifically and the promotion of personalized and precision medicine in general, better guiding clinicians in their choice of therapy and providing a clearer result for VUS carriers.
Acknowledgments
We would like to thank Blake Wu for editing the manuscript. We thank Andrew Olson from Stanford Neuroscience Microscopy Service (NMS), Jon Mulholland and Cedric Espenel from Stanford Cell Sciences Imaging Facility (CSIF) for their help with confocal imaging.
FUNDING SOURCES
This study was funded by Stanford Frankenstein@200 grant (HC), the National Institutes of Health (NIH) grants K08 HL119251 (KDW), R01 HL126527, R01 HL130020, and R01 HL113006 (JCW).
Footnotes
DISCLOSURES
None
References
- 1.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Comm ALQA Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eccles BK, Copson E, Maishman T, Abraham JE, Eccles DM. Understanding of BRCA VUS genetic results by breast cancer specialists. BMC Cancer. 2015;15:936. doi: 10.1186/s12885-015-1934-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi Y, Yang S, Nykamp K, Garcia J, Lincoln SE, Topper SE. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9:13. doi: 10.1186/s13073-017-0403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrucelli N, Lazebnik N, Huelsman KM, Lazebnik RS. Clinical interpretation and recommendations for patients with a variant of uncertain significance in BRCA1 or BRCA2: A survey of genetic counseling practice. Genet Test. 2002;6:107–113. doi: 10.1089/10906570260199357. [DOI] [PubMed] [Google Scholar]
- 5.Maron BJ, Maron MS, Semsarian C. Genetics of Hypertrophic cardiomyopathy after 20 years clinical perspectives. J Am Coll Cardiol. 2012;60:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 6.Roma-Rodrigues C, Fernandes AR. Genetics of hypertrophic cardiomyopathy: advances and pitfalls in molecular diagnosis and therapy. The Application of Clinical Genetics. 2014;7:195–208. doi: 10.2147/TACG.S49126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juang JM, Lu TP, Lai LC, Hsueh CH, Liu YB, Tsai CT, Lin LY, Yu CC, Hwang JJ, Chiang FT, Yeh SS, Chen WP, Chuang EY, Lai LP, Lin JL. Utilizing multiple in silico analyses to identify putative causal SCN5A variants in Brugada syndrome. Sci Rep. 2014;4:3850. doi: 10.1038/srep03850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora S, Huwe PJ, Sikder R, Shah M, Browne AJ, Lesh R, Nicolas E, Deshpande S, Hall MJ, Dunbrack RL, Jr, Golemis EA. Functional analysis of rare variants in mismatch repair proteins augments results from computation-based predictive methods. Cancer Biol Ther. 2017;18:519–533. doi: 10.1080/15384047.2017.1326439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7:Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 12.Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, Belhassen B, Nof E, Glikson M, Gepstein L. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol. 2012;60:990–1000. doi: 10.1016/j.jacc.2012.02.066. [DOI] [PubMed] [Google Scholar]
- 13.Liang P, Sallam K, Wu H, Li Y, Itzhaki I, Garg P, Zhang Y, Vermglinchan V, Lan F, Gu M, Gong T, Zhuge Y, He C, Ebert AD, Sanchez-Freire V, Churko J, Hu S, Sharma A, Lam CK, Scheinman MM, Bers DM, Wu JC. Patient-specific and genome-edited induced pluripotent stem cell-derived cardiomyocytes elucidate single-cell phenotype of Brugada syndrome. J Am Coll Cardiol. 2016;68:2086–2096. doi: 10.1016/j.jacc.2016.07.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT, Wu JC. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han L, Li Y, Tchao J, Kaplan AD, Lin B, Li Y, Mich-Basso J, Lis A, Hassan N, London B, Bett GC, Tobita K, Rasmusson RL, Yang L. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc Res. 2014;104:258–269. doi: 10.1093/cvr/cvu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson KD, Wu JC. Induced pluripotent stem cells. Jama-J Am Med Assoc. 2015;313:1613–1614. doi: 10.1001/jama.2015.1846. [DOI] [PubMed] [Google Scholar]
- 17.Hendriks WT, Warren CR, Cowan CA. Genome editing in human pluripotent stem cells: approaches, pitfalls, and solutions. Cell Stem Cell. 2016;18:53–65. doi: 10.1016/j.stem.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma N, Liao BJ, Zhang H, Wang LL, Shan YL, Xue YT, Huang K, Chen SB, Zhou XX, Chen Y, Pei DQ, Pan GJ. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free beta-thalassemia induced pluripotent stem cells. Journal of Biological Chemistry. 2013;288:34671–34679. doi: 10.1074/jbc.M113.496174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma N, Shan YL, Liao BJ, Kong GY, Wang C, Huang K, Zhang H, Cai XJ, Chen SB, Pei DQ, Chen NS, Pan GJ. Factor-induced reprogramming and zinc finger nuclease-aided gene targeting cause different genome instability in β-thalassemia induced pluripotent stem cells (iPSCs) Journal of Biological Chemistry. 2015;290:12079–12089. doi: 10.1074/jbc.M114.624999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 22.Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez OM, Jones M, Guzman G, Szczesna-Cordary D. Myosin essential light chain in health and disease. Am J Physiol Heart Circ Physiol. 2007;292:H1643–1654. doi: 10.1152/ajpheart.00931.2006. [DOI] [PubMed] [Google Scholar]
- 24.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, Karapetyan K, Katz K, Liu C, Maddipatla Z, Malheiro A, McDaniel K, Ovetsky M, Riley G, Zhou G, Holmes JB, Kattman BL, Maglott DR. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson KD, Shen P, Fung E, Karakikes I, Zhang A, InanlooRahatloo K, Odegaard J, Sallam K, Davis RW, Lui GK, Ashley EA, Scharfe C, Wu JC. A rapid, high-quality, cost-effective, comprehensive and expandable targeted next-generation sequencing assay for inherited heart diseases. Circ Res. 2015;117:603–611. doi: 10.1161/CIRCRESAHA.115.306723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmstrom A, Matsa E, Zhang Y, Kumar A, Fan AC, Del Alamo JC, Wu SM, Moslehi JJ, Mercola M, Wu JC. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci Transl Med. 2017;9:eaaaf2584. doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee W, Hwang TH, Kimura A, Park SW, Satoh M, Nishi H, Harada H, Toyama J, Park JE. Different expressivity of a ventricular essential myosin light chain gene Ala57Gly mutation in familial hypertrophic cardiomyopathy. Am Heart J. 2001;141:184–189. doi: 10.1067/mhj.2001.112487. [DOI] [PubMed] [Google Scholar]
- 28.Andersen PS, Hedley PL, Page SP, Syrris P, Moolman-Smook JC, McKenna WJ, Elliott PM, Christiansen M. A novel myosin essential light chain mutation causes hypertrophic cardiomyopathy with late onset and low expressivity. Biochem Res Int. 2012;2012:685108. doi: 10.1155/2012/685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 30.England J, Loughna S. Heavy and light roles: myosin in the morphogenesis of the heart. Cell Mol Life Sci. 2013;70:1221–1239. doi: 10.1007/s00018-012-1131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belus A, Piroddi N, Scellini B, Tesi C, Amati GD, Girolami F, Yacoub M, Cecchi F, Olivotto I, Poggesi C. The familial hypertrophic cardiomyopathy-associated myosin mutation R403Q accelerates tension generation and relaxation of human cardiac myofibrils. J Physiol-London. 2008;586:3639–3644. doi: 10.1113/jphysiol.2008.155952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsiglia JD, Pereira AC. Hypertrophic cardiomyopathy: how do mutations lead to disease? Arq Bras Cardiol. 2014;102:295–304. doi: 10.5935/abc.20140022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 34.Maron BJ, Shirani J, Poliac LC, Mathenge R, Roberts WC, Mueller FO. Sudden death in young competitive athletes. Clinical, demographic, and pathological profiles. JAMA. 1996;276:199–204. [PubMed] [Google Scholar]
- 35.Chamberland S, Yang HH, Pan MM, Evans SW, Guan S, Chavarha M, Yang Y, Salesse C, Wu H, Wu JC, Clandinin TR, Toth K, Lin MZ, St-Pierre F. Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. Elife. 2017;6:e25690. doi: 10.7554/eLife.25690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adabag AS, Maron BJ, Appelbaum E, Harrigan CJ, Buros JL, Gibson CM, Lesser JR, Hanna CA, Udelson JE, Manning WJ, Maron MS. Occurrence and frequency of arrhythmias in hypertrophic cardiomyopathy in relation to delayed enhancement on cardiovascular magnetic resonance. J Am Coll Cardiol. 2008;51:1369–1374. doi: 10.1016/j.jacc.2007.11.071. [DOI] [PubMed] [Google Scholar]
- 37.Wolf CM, Moskowitz IP, Arno S, Branco DM, Semsarian C, Bernstein SA, Peterson M, Maida M, Morley GE, Fishman G, Berul CI, Seidman CE, Seidman JG. Somatic events modify hypertrophic cardiomyopathy pathology and link hypertrophy to arrhythmia. Proc Natl Acad Sci U S A. 2005;102:18123–18128. doi: 10.1073/pnas.0509145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaub MC, Hefti MA, Zuellig RA, Morano I. Modulation of contractility in human cardiac hypertrophy by myosin essential light chain isoforms. Cardiovasc Res. 1998;37:381–404. doi: 10.1016/s0008-6363(97)00258-7. [DOI] [PubMed] [Google Scholar]
- 39.Morano M, Zacharzowski U, Maier M, Lange PE, Alexi-Meskishvili V, Haase H, Morano I. Regulation of human heart contractility by essential myosin light chain isoforms. J Clin Invest. 1996;98:467–473. doi: 10.1172/JCI118813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petzhold D, Lossie J, Keller S, Werner S, Haase H, Morano I. Human essential myosin light chain isoforms revealed distinct myosin binding, sarcomeric sorting, and inotropic activity. Cardiovasc Res. 2011;90:513–520. doi: 10.1093/cvr/cvr026. [DOI] [PubMed] [Google Scholar]
- 41.Kazmierczak K, Paulino EC, Huang W, Muthu P, Liang J, Yuan CC, Rojas AI, Hare JM, Szczesna-Cordary D. Discrete effects of A57G-myosin essential light chain mutation associated with familial hypertrophic cardiomyopathy. Am J Physiol Heart Circ Physiol. 2013;305:H575–589. doi: 10.1152/ajpheart.00107.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kazmierczak K, Yuan CC, Liang J, Huang W, Rojas AI, Szczesna-Cordary D. Remodeling of the heart in hypertrophy in animal models with myosin essential light chain mutations. Front Physiol. 2014;5:353. doi: 10.3389/fphys.2014.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geisterfer-Lowrance AA, Christe M, Conner DA, Ingwall JS, Schoen FJ, Seidman CE, Seidman JG. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–4. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 44.Marsiglia JDC, Pereira AC. Hypertrophic Cardiomyopathy: how do mutations lead to disease? Arq Bras Cardiol. 2014;102:295–303. doi: 10.5935/abc.20140022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witjas-Paalberends ER, Ferrara C, Scellini B, Piroddi N, Montag J, Tesi C, Stienen GJ, Michels M, Ho CY, Kraft T, Poggesi C, van der Velden J. Faster cross-bridge detachment and increased tension cost in human hypertrophic cardiomyopathy with the R403Q MYH7 mutation. J Physiol. 2014;592:3257–3272. doi: 10.1113/jphysiol.2014.274571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


