Abstract
Purine metabolism is depending on a large amount of enzymes to ensure cellular homeostasis. Among these enzymes, we have been interested in the 5′-nucleotidase cN-II and its role in cancer biology and in response of cancer cells to treatments. This protein has been cited and studied in a large number of papers published during the last decade for its involvement in non-cancerous pathologies such as hereditary spastic paraplegia, schizophrenia, and blood pressure regulation. Here, we review these articles in order to give an overview of the recently discovered clinical relevance of cN-II.
Keywords: GWAS, Diseases, Genetic variants, Mutations, NT5C2, Purine metabolism
Introduction
Cytosolic 5′-nucleotidase II (cN-II) is a ubiquitously expressed and highly conserved enzyme that dephosphorylates purine nucleoside monophosphates (preferentially IMP and GMP) into their corresponding nucleosides and inorganic phosphate. Its enzymatic activity and biochemical features have been fairly well characterized on purified recombinant protein [1], and the crystal structure is known since 2007 [2]. Since our first observation on its prognostic value in nucleoside analogue-treated patients with acute myeloid leukemia showing that patients with high expression of cN-II in leukemic blasts have a worse outcome than those with a lower expression [3], the implication of cN-II in cancer cells and in the response to anticancer treatment has been extensively demonstrated [4]. Indeed, shRNA-based cell models with decreased cN-II expression are more sensitive to purine nucleoside analogues and nucleobases as compared to control cells [5]. Additional work with cell models and animals suggest important biological roles of this enzyme both in cancer cells and in physiological conditions. The role of the enzymatic activity of cN-II and the modifications in purine nucleotide pools in these observations is not always demonstrated. We showed that cN-II expression in human neuroblastoma cells and in lung cancer cells correlated to cell proliferation [6, 7], whereas it inhibition in human breast cancer cells was associated with a better defense towards reactive oxygen species and a better adaptability to glucose deprivation in culture media [8]. Further, the siRNA-mediated inhibition of cN-II expression in murine skeletal muscles induced an increase in the AMP/ATP ratio and a subsequent increase in the activation of AMPK [9], even though this was not confirmed in cN-II deficient mice [10].
In parallel to such biological studies, a number of genetic studies as well as genome wide association studies (GWAS) have identified the cN-II encoding gene NT5C2 or some genetic variants therein as being associated to various pathological conditions. Here, we give an overview of these recently published data that show or suggest clinical importance of this enzyme in hereditary spastic paraplegia, psychiatric disorders, blood pressure, and body mass index.
Hereditary spastic paraplegia
Hereditary spastic paraplegia (HSP) is a group of neurodegenerative disorders with various genetic origins and clinical presentations [11, 12]. The locus harboring the NT5C2 gene (10q24.3-q25.1) was identified as being associated with HSP in a consanguineous family (Table 1) [13]. This locus, named SPG45, contains 87 genes, and the authors suggested MRPL43 to be the best candidate for the functionality of the observed disease, due to its role in mitochondria regulation and protein-folding. The association between HSP and NT5C2 was further confirmed in another study on 55 families using whole-exome sequencing, where 5 families showed mutations in this gene [14]. Out of the 5 mutations observed, 3 induced a modification on the protein with two premature stop codons (R29X and R149X) and a frameshift (S409Vfs436X) and two were splice site mutations (c.175 and c.988). Of interest, two other proteins involved in nucleotide metabolism, the nucleotidase CD39/ENTPD1 and AMP-deaminase 2 (AMPD2), were also found to be mutated in HSP families. Both these enzymes, as well as cN-II, regulate the purine nucleotide pools through their enzymatic activity. This, together with the fact that purine nucleotides have protective roles in the brain [18], reinforces the possibility of the implication of cN-II activity in the development of HSP. NT5C2 mutations in SPG45 and the autosomal recessive transmission mode were later validated with the observation of another splice site mutation (c.1159) resulting in a shortened cN-II protein in two brothers [15], a deletion of amino-acids 258–271 in three siblings [16] and a missense mutation (L460P) in three patients from another family [17]. NT5C2 is since recognized as being the functional gene in the SPG45 locus.
Table 1.
Deleterious mutations and genetic modifications observed in HSP SPG45 patients
| Patients | Mutations | Reference |
|---|---|---|
| 5 members of consanguineous family | 10q24.3-q25.1 | [13] |
| 2 siblings of a consanguineous family | c.175+1 = splice variant | [14] |
| 2 sisters of a consanguineous family | c.988-1 = splice variant | |
| 2 sisters of a consanguineous family | c1225Gdel = premature stop at 436 | |
| 3 siblings of a consanguineous family | R29stop | |
| 2 brothers of a consanguineous family | R149stop | |
| 2 brothers Qatari consanguineous family | c.1159+1 = deletion of exon 14 | [15] |
| 3 siblings Iranian consanguineous family | p.258-271del | [16] |
| 3 members of consanguineous family | L460P | [17] |
Schizophrenia
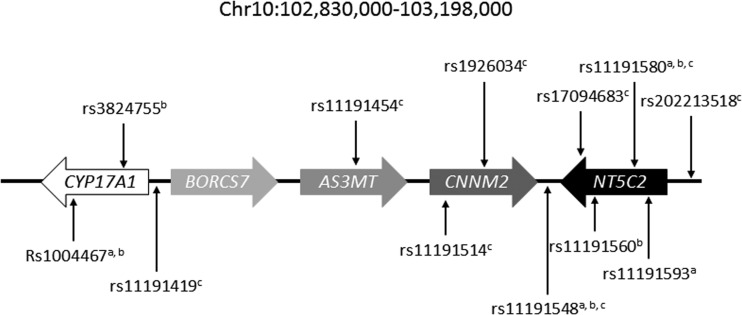
A large number of GWAS studies on different ethnic populations has been performed in order to identify genetic causes for schizophrenia and other psychiatric diseases, and the NT5C2-containing locus was reported in several cases (Fig. 1). rs11191580 was first identified in a meta-analysis from 17 independent studies and confirmed within a validation set [19], and later in a South Chinese Han population [20] as well as for bipolar disorders in a Latino cohort [21]. rs11191580 was also confirmed in a meta-analysis, but the role of AS3MT through rs7085104 situated on the same locus was rather suggested as an important genetic variant in this study [22]. rs17094683 was found in another GWAS study [23] and rs1926034 in a Swedish cohort [24]. Later, rs11191454 in AS3MT situated close to NT5C2 within 10q24 and in strong linkage disequilibrium with many surrounding SNP was associated with five psychiatric disorders [25]. The NT5C2-containing genomic region was further studied in a Chinese population, and the association with schizophrenia was again confirmed for rs11191419 and rs11191514 [26]. However, based on bioinformatics prediction of the involvement of genetic variants in gene regulation, the authors suggested that NT5C2 was not functional in this correlation, but that rather other genes of the same cluster (AS3MT, CNNM2, and CALHM1) would be responsible for the clinical onset of schizophrenia. This was at least partially contested by a study on the cis-regulatory effect of SNP in this region [27]. Indeed, rs11191419 and rs202213518 influenced NT5C2 gene expression as shown by differential allelic expression studies. This is in line with our previous study showing the presence of genetic regulation of NT5C2 [28]. In addition to this, rs11191548 was shown to alter the binding site of miR-1/206/613 as the sequence with the minor allele (G) did not response to miR-1/206613 [29]. Even though these results do not show a clear functional role of cN-II in schizophrenia, at least one SNP (rs11191548) identified in GWAS studies regulates NT5C2 gene expression.
Fig. 1.
Genetic variants within the studied genomic region with possible association between discussed pathologies and NT5C2. a blood pressure; b BMI/body fat; c schizophrenia
Blood pressure
Genetic variants within the locus 10q24 were associated with high blood pressure in a large study including more than 84,000 individuals from European and Indian Asian origin [30]. A T at the position of rs11191548 in the intergenic region between NT5C2 and CNNM2 was associated with a higher systolic blood pressure as compared to patients with a C at the same position (p = 3.10−7). This was later confirmed in studies on Europeans [31] and Asians [32], as well as on Asians in a study where rs11191580 also was associated with blood pressure [33]. The same region was also identified in another large-scale study, but with a better score for a genetic variant further away from NT5C2 (rs1004467) [34]. Also, rs11191593 was associated with blood pressure [35]. Finally, a gene expression study showed that military pilots with hypertension had a lower cN-II expression than control pilots [36]. Interestingly, there was also a decreased expression of CD39/ENTPD1, confirming an additional link between these two proteins after the one in HSP. The direct involvement of cN-II in the regulation of blood pressure is not clearly demonstrated and needs further studies to be confirmed. Indeed, rs1004467 and rs11191548 have for example been reported to modulate CYP17A1 and therefor potentially be functional through this gene even though the latter SNP is quite far from this gene.
Body mass index and body fat
Increased body mass and fat are risk factors for a number of diseases, such as high blood pressure. One SNP (rs11191548) already identified as a risk factor for increased blood pressure was also associated with subcutaneous fat area, in particular in women in a Japanese cohort [37]. However, the T-allele which is associated with increased blood pressure was here found to correlate with decreased fat. Another SNP (rs1004467), situated in the nearby CYP17A1 gene and in linkage disequilibrium with rs11191548, was also associated with modified body fat. Another SNP (rs11191580) that is situated in a NT5C2 intron and that is in complete linkage disequilibrium with rs11191548 was later found associated with body mass index in East Asian people [38], whereas rs11191560 also situated in an NT5C2 intron and between rs11191548 and rs11191580 was associated with higher body mass index in a European meta-analysis [39]. Finally, rs3824755 was also associated to BMI in a series of more than 17,000 individuals [40] and further shown to increase within the population together with BMI [41]. However, this SNP is situated within the CYP17A1 gene and not in NT5C2 as reported in some of these papers, making the role for cN-II less evident. As for blood pressure, these studies are solely showing correlations, and no functional involvement of cN-II in the regulation of BMI and body fat exists today.
Relapsed leukemia and resistance to chemotherapy
In addition to the cited work on associations between genetic variants or germ-line mutations and pathologies, tumor-specific NT5C2-mutations have been reported to play a major role in leukemia relapse and their resistance to chemotherapy. This was first seen with hyperactivating mutations in relapsed pediatric patients with acute lymphoblastic leukemia (ALL) [42, 43]. Additional mutations were found in a subsequent study that also showed the relapse-specificity of the mutations [44]. Such mutations were then shown to be very rare in new leukemia and solid tumors, with only one case in adult acute myeloid leukemia (E240Q) and one in colorectal carcinoma (R363Q) from a total of 2496 tumors [45]. One ALL patient with testicular relapses showed also NT5C2 mutations (R367Q and D407V) [46], whereas they were quite frequent in a series of 67 relapse leukemia patients [47] as well as in relapsed T-ALL patients (5/13 patients) [48] and acute promyelocytic leukemia patients [49]. The most frequent mutation (R367Q) was recently shown to be a key driver in the evolution of relapsed ALL [50], and additional mutations were found in more patients by the same group [51, 52]. Overall, these studies show that NT5C2 mutations are frequent in relapsed leukemia (Table 2) and that this has a direct consequence on the response to treatments. However, this is known to be due to the enzymatic activity of cN-II, at least for the mutants for which the mutation has been shown to be associated with an enzymatic hyperactivity.
Table 2.
NT5C2 mutations reported in various cancer patients at diagnosis or relapse. A total of 129 mutations are reported. ALL acute lymphoblastic leukemia, AML acute myeloid leukemia, APL acute promyelocytic leukemia, CC colorectal cancer
| Reference | [43] | [42] | [44] | [48] | [45] | [46] | [47] | [50] | [52] | [51] | [49] | Total |
| Cancer | ALL | ALL | ALL | ALL | AML and CC | ALL | ALL | ALL | ALL | ALL | APL | |
| R34Q | 1 | 1 | ||||||||||
| R39Q | 2 | 1 | 2 | 3 | 8 | |||||||
| R195Q | 1 | 1 | ||||||||||
| R238G/Q/W/L | 5 | 3 | 5 | 4 | 2 | 5 | 1 | 25 | ||||
| E240Q | 1 | 1 | ||||||||||
| R291W | 1 | 1 | ||||||||||
| K359Q | 1 | 1 | 2 | |||||||||
| S360P | 1 | 1 | ||||||||||
| R363Q | 1 | 1 | ||||||||||
| R367Q | 13 | 1 | 5 | 4 | 1 | 7 | 7 | 1 | 14 | 1 | 54 | |
| L375F | 2 | 1 | 3 | |||||||||
| p396-400del | 1 | 1 | ||||||||||
| D384N | 1 | 1 | ||||||||||
| K404N/ins | 1 | 1 | 1 | 3 | ||||||||
| D407A/V/H/Y/E | 1 | 1 | 1 | 2 | 1 | 3 | 9 | |||||
| S408R | 1 | 1 | 2 | |||||||||
| p408-415del | 1 | 1 | ||||||||||
| P414S/A | 1 | 1 | 1 | 2 | 5 | |||||||
| D415G | 1 | 1 | ||||||||||
| S445F | 1 | 1 | 2 | |||||||||
| R446Q | 1 | 1 | ||||||||||
| V454M | 1 | 1 | ||||||||||
| R478S | 1 | 1 | ||||||||||
| T489M | 1 | 1 | ||||||||||
| Q523stop | 1 | 1 | ||||||||||
| P533L | 1 | 1 |
Conclusive remarks
The studies reviewed here are mostly based on the association between genetic variants or mutations and the risk to develop given diseases, but do rarely give any information about the potential molecular functionality of cN-II in the observed pathologies. Therefore, additional studies are warranted and dependent on various cell and animal models. cN-II deficient mice were recently published in a study on the role of cN-II in contracting muscles [10]. No particular phenotype or behavior was described in this paper, but these mice could constitute a model of choice for studying the development of diseases discussed in this review.
cN-II is an extremely well-conserved protein with more than 90% homology as far as to fish [53]. Such sequence conservation not only limited to the active site is strongly suggestive of additional roles independent from the enzymatic activity and rather through protein-protein interactions. We already identified and confirmed physical interaction with the inflammation protein IPAF, even though we still have not deciphered the role of this interaction [53]. Additional interactions are highly possible and could explain many of the observed phenotypes, but request a large amount of additional research, especially due to the fact that such interactions could be specific over time during development as well as in particular cells expressing the two interacting proteins at the same time and in the same subcellular location. One possibility within this hypothesis is the presumed interaction between cN-II and the multimodal protein Kidins220 [53]. This physical interaction has not yet been confirmed in mammalian cells, but Kidins220 has been suggested to be implicated in Alzheimer’s disease [54] and a recent study also showed a potential role of cN-II in this disease [55].
Another explanation is of course that all roles of cN-II are due to its enzymatic activity and regulation of the purine metabolism. This could be supported by the involvement of other genes in this metabolism as they could also have important effects on purine homeostasis. As already mentioned, AMPD2 and CD39/ENTPD1 were also involved in HSP [14]. AMPD2 converts AMP into IMP by deamination, whereas CD39 degrades extracellular ATP into AMP, and both enzymes thus regulate purine nucleotide pools together with cN-II. Other studies have shown that these are involved in cognitive disorders for CD39/ENTPD1 [56], in the maturation of neurons together with CD73 [57], and in the neurodegenerative brainstem disorder pontocerebellar hypoplasia for AMPD2 [58–61]. Finally, purine metabolism is shown to be a major regulator both in Parkinson’s disease and in schizophrenia [62, 63]. As both several enzymes of purine metabolism are associated with neurological diseases and purine metabolism and pools are regulating such pathologies, it is highly possible that the enzymatic activity of cN-II and its role in the maintenance of balanced purine pools is involved in the development of HSP-patients in particular.
In conclusion, the implication of the genomic region surrounding NT5C2 on chromosome 10 is undoubtedly associated to and involved in discussed pathologies. Further studies are warranted to conclude on the functional role of cN-II in the corresponding pathogenesis.
Funding
LPJ received funding for research on cN-II from Olav Raagholt og Gerd Meidel Raagholts stiftelse for forskning.
Conflict of interest
Lars Petter Jordheim declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Allegrini S, Scaloni A, Careddu MG, Cuccu G, D'Ambrosio C, Pesi R, Camici M, Ferrara L, Tozzi MG. Mechanistic studies on bovine cytosolic 5′-nucleotidase II, an enzyme belonging to the HAD superfamily. Eur J Biochem. 2004;271:4881–4891. doi: 10.1111/j.1432-1033.2004.04457.x. [DOI] [PubMed] [Google Scholar]
- 2.Wallden K, Stenmark P, Nyman T, Flodin S, Graslund S, Loppnau P, Bianchi V, Nordlund P. Crystal structure of human cytosolic 5′-nucleotidase II: insights into allosteric regulation and substrate recognition. J Biol Chem. 2007;282:17828–17836. doi: 10.1074/jbc.M700917200. [DOI] [PubMed] [Google Scholar]
- 3.Galmarini CM, Graham K, Thomas X, Calvo F, Rousselot P, El Jafaari A, Cros E, Mackey JR, Dumontet C (2001) Expression of high Km 5′-nucleotidase in leukemic blasts is an independent prognostic factor in adults with acute myeloid leukemia. Blood 98:1922–1926 [DOI] [PubMed]
- 4.Jordheim LP, Chaloin L. Therapeutic perspectives for cN-II in cancer. Curr Med Chem. 2013;20:4292–4303. doi: 10.2174/0929867311320340008. [DOI] [PubMed] [Google Scholar]
- 5.Jordheim LP, Puy JY, Cros-Perrial E, Peyrottes S, Lefebvre I, Perigaud C, Dumontet C. Determination of the enzymatic activity of cytosolic 5′-nucleotidase cN-II in cancer cells: development of a simple analytical method and related cell line models. Anal Bioanal Chem. 2015;407:5747–5758. doi: 10.1007/s00216-015-8757-4. [DOI] [PubMed] [Google Scholar]
- 6.Cividini F, Cros-Perrial E, Pesi R, Machon C, Allegrini S, Camici M, Dumontet C, Jordheim LP, Tozzi MG. Cell proliferation and drug sensitivity of human glioblastoma cells are altered by the stable modulation of cytosolic 5′-nucleotidase II. Int J Biochem Cell Biol. 2015;65:222–229. doi: 10.1016/j.biocel.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Pesi R, Petrotto E, Colombaioni L, Allegrini S, Garcia-Gil M, Camici M, Jordheim LP, Tozzi MG (2018) Cytosolic 5′-nucleotidase II silencing in a human lung carcinoma cell line opposes cancer phenotype with a concomitant increase in p53 phosphorylation. Int J Mol Sci 19. 10.3390/ijms19072115 [DOI] [PMC free article] [PubMed]
- 8.Bricard G, Cadassou O, Cassagnes LE, Cros-Perrial E, Payen-Gay L, Puy JY, Lefebvre-Tournier I, Tozzi MG, Dumontet C, Jordheim LP. The cytosolic 5′-nucleotidase cN-II lowers the adaptability to glucose deprivation in human breast cancer cells. Oncotarget. 2017;8:67380–67393. doi: 10.18632/oncotarget.18653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkarni SS, Karlsson HK, Szekeres F, Chibalin AV, Krook A, Zierath JR. Suppression of 5′-nucleotidase enzymes promotes AMP-activated protein kinase (AMPK) phosphorylation and metabolism in human and mouse skeletal muscle. J Biol Chem. 2011;286:34567–34574. doi: 10.1074/jbc.M111.268292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kviklyte S, Vertommen D, Yerna X, Andersen H, Xu X, Gailly P, Bohlooly YM, Oscarsson J, Rider MH. Effects of genetic deletion of soluble 5′-nucleotidases NT5C1A and NT5C2 on AMPK activation and nucleotide levels in contracting mouse skeletal muscles. Am J Physiol Endocrinol Metab. 2017;313:E48–E62. doi: 10.1152/ajpendo.00304.2016. [DOI] [PubMed] [Google Scholar]
- 11.Fink JK. Hereditary spastic paraplegia: clinical principles and genetic advances. Semin Neurol. 2014;34:293–305. doi: 10.1055/s-0034-1386767. [DOI] [PubMed] [Google Scholar]
- 12.Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol. 2014;261:518–539. doi: 10.1016/j.expneurol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Dursun U, Koroglu C, Kocasoy Orhan E, Ugur SA, Tolun A. Autosomal recessive spastic paraplegia (SPG45) with mental retardation maps to 10q24.3-q25.1. Neurogenetics. 2009;10:325–331. doi: 10.1007/s10048-009-0191-3. [DOI] [PubMed] [Google Scholar]
- 14.Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, Masri A, Kayserili H, Al-Aama JY, Abdel-Salam GMH, Karminejad A, Kara M, Kara B, Bozorgmehri B, Ben-Omran T, Mojahedi F, El Din Mahmoud IG, Bouslam N, Bouhouche A, Benomar A, Hanein S, Raymond L, Forlani S, Mascaro M, Selim L, Shehata N, Al-Allawi N, Bindu PS, Azam M, Gunel M, Caglayan A, Bilguvar K, Tolun A, Issa MY, Schroth J, Spencer EG, Rosti RO, Akizu N, Vaux KK, Johansen A, Koh AA, Megahed H, Durr A, Brice A, Stevanin G, Gabriel SB, Ideker T, Gleeson JG. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsaid MF, Ibrahim K, Chalhoub N, Elsotouhy A, El Mudehki N, Abdel Aleem A. NT5C2 novel splicing variant expands the phenotypic spectrum of Spastic Paraplegia (SPG45): case report of a new member of thin corpus callosum SPG-Subgroup. BMC Med Genet. 2017;18(33):33. doi: 10.1186/s12881-017-0395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darvish H, Azcona LJ, Tafakhori A, Ahmadi M, Ahmadifard A, Paisan-Ruiz C (2017) Whole genome sequencing identifies a novel homozygous exon deletion in the NT5C2 gene in a family with intellectual disability and spastic paraplegia. NPJ Genom Med 2. 10.1038/s41525-017-0022-7 [DOI] [PMC free article] [PubMed]
- 17.Straussberg R, Onoufriadis A, Konen O, Zouabi Y, Cohen L, Lee JYW, Hsu CK, Simpson MA, McGrath JA (2017) Novel homozygous missense mutation in NT5C2 underlying hereditary spastic paraplegia SPG45. Am J Med Genet A 173:3109–3113. 10.1002/ajmg.a.38414 [DOI] [PubMed]
- 18.Matute C, Cavaliere F. Neuroglial interactions mediated by purinergic signalling in the pathophysiology of CNS disorders. Semin Cell Dev Biol. 2011;22:252–259. doi: 10.1016/j.semcdb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Schizophrenia Psychiatric Genome-Wide Association Study C Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Jiang J, Long J, Ling W, Huang G, Guo X, Su L. The rs11191580 variant of the NT5C2 gene is associated with schizophrenia and symptom severity in a South Chinese Han population: evidence from GWAS. Rev Bras Psiquiatr. 2017;39:104–109. doi: 10.1590/1516-4446-2016-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez S, Gupta J, Villa E, Mallawaarachchi I, Rodriguez M, Ramirez M, Zavala J, Armas R, Dassori A, Contreras J, Flores D, Jerez A, Ontiveros A, Nicolini H, Escamilla M. Replication of genome-wide association study (GWAS) susceptibility loci in a Latino bipolar disorder cohort. Bipolar Disord. 2016;18:520–527. doi: 10.1111/bdi.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Chang H, Peng T, Li M, Xiao X. Evidence of AS3MT(d2d3)-associated variants within 10q24.32-33 in the genetic risk of major affective disorders. Mol Neuropsychiatry. 2017;2:213–218. doi: 10.1159/000452998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aberg KA, Liu Y, Bukszar J, McClay JL, Khachane AN, Andreassen OA, Blackwood D, Corvin A, Djurovic S, Gurling H, Ophoff R, Pato CN, Pato MT, Riley B, Webb T, Kendler K, O'Donovan M, Craddock N, Kirov G, Owen M, Rujescu D, St Clair D, Werge T, Hultman CM, Delisi LE, Sullivan P, van den Oord EJ. A comprehensive family-based replication study of schizophrenia genes. JAMA Psychiatry. 2013;70:573–581. doi: 10.1001/jamapsychiatry.2013.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergen SE, O'Dushlaine CT, Ripke S, Lee PH, Ruderfer DM, Akterin S, Moran JL, Chambert KD, Handsaker RE, Backlund L, Osby U, McCarroll S, Landen M, Scolnick EM, Magnusson PK, Lichtenstein P, Hultman CM, Purcell SM, Sklar P, Sullivan PF. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 2012;17:880–886. doi: 10.1038/mp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cross-Disorder Group of the Psychiatric Genomics C Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan F, Zhang T, Li L, Fu D, Lin H, Chen G, Chen T. Two-stage replication of previous genome-wide association studies of AS3MT-CNNM2-NT5C2 gene cluster region in a large schizophrenia case-control sample from Han Chinese population. Schizophr Res. 2016;176:125–130. doi: 10.1016/j.schres.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Duarte RRR, Troakes C, Nolan M, Srivastava DP, Murray RM, Bray NJ. Genome-wide significant schizophrenia risk variation on chromosome 10q24 is associated with altered cis-regulation of BORCS7, AS3MT, and NT5C2 in the human brain. Am J Med Genet B Neuropsychiatr Genet. 2016;171:806–814. doi: 10.1002/ajmg.b.32445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordheim LP, Nguyen-Dumont T, Thomas X, Dumontet C, Tavtigian SV. Differential allelic expression in leukoblast from patients with acute myeloid leukemia suggests genetic regulation of CDA, DCK, NT5C2, NT5C3, and TP53. Drug Metab Dispos. 2008;36:2419–2423. doi: 10.1124/dmd.108.023184. [DOI] [PubMed] [Google Scholar]
- 29.Hauberg ME, Holm-Nielsen MH, Mattheisen M, Askou AL, Grove J, Borglum AD, Corydon TJ. Schizophrenia risk variants affecting microRNA function and site-specific regulation of NT5C2 by miR-206. Eur Neuropsychopharmacol. 2016;26:1522–1526. doi: 10.1016/j.euroneuro.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Wellcome Trust Case Control C, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O'Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O'Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB (2009) Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 41:666–676. 10.1038/ng.361
- 31.International Consortium for Blood Pressure Genome-Wide Association S. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, consortium CA, Consortium CK, KidneyGen C, EchoGen c, consortium C-H. Aspelund T, Garcia M, Chang YP, O'Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, WL MA, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, CA MK, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O'Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, Katsuya T, Yokota M, Kim YJ, Ong RT, Nabika T, Gu D, Chang LC, Kokubo Y, Huang W, Ohnaka K, Yamori Y, Nakashima E, Jaquish CE, Lee JY, Seielstad M, Isono M, Hixson JE, Chen YT, Miki T, Zhou X, Sugiyama T, Jeon JP, Liu JJ, Takayanagi R, Kim SS, Aung T, Sung YJ, Zhang X, Wong TY, Han BG, Kobayashi S, Ogihara T, Zhu D, Iwai N, Wu JY, Teo YY, Tai ES, Cho YS, He J. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Kim YK, Dorajoo R, Li H, Lee IT, Cheng CY, He M, Sheu WH, Guo X, Ganesh SK, He J, Lee J, Liu J, Hu Y, Rao DC, Tsai FJ, Koh JY, Hu H, Liang KW, Palmas W, Hixson JE, Han S, Teo YY, Wang Y, Chen J, Lu CH, Zheng Y, Gui L, Lee WJ, Yao J, Gu D, Han BG, Sim X, Sun L, Zhao J, Chen CH, Kumari N, He Y, Taylor KD, Raffel LJ, Moon S, Rotter JI, Ida Chen YD, Wu T, Wong TY, Wu JY, Lin X, Tai ES, Kim BJ, Kelly TN. Genome-wide association study meta-analysis of long-term average blood pressure in East Asians. Circ Cardiovasc Genet. 2017;10:e001527. doi: 10.1161/CIRCGENETICS.116.001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly TN, Takeuchi F, Tabara Y, Edwards TL, Kim YJ, Chen P, Li H, Wu Y, Yang CF, Zhang Y, Gu D, Katsuya T, Ohkubo T, Gao YT, Go MJ, Teo YY, Lu L, Lee NR, Chang LC, Peng H, Zhao Q, Nakashima E, Kita Y, Shu XO, Kim NH, Tai ES, Wang Y, Adair LS, Chen CH, Zhang S, Li C, Nabika T, Umemura S, Cai Q, Cho YS, Wong TY, Zhu J, Wu JY, Gao X, Hixson JE, Cai H, Lee J, Cheng CY, Rao DC, Xiang YB, Cho MC, Han BG, Wang A, Tsai FJ, Mohlke K, Lin X, Ikram MK, Lee JY, Zheng W, Tetsuro M, Kato N, He J. Genome-wide association study meta-analysis reveals transethnic replication of mean arterial and pulse pressure loci. Hypertension. 2013;62:853–859. doi: 10.1161/HYPERTENSIONAHA.113.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao XC, Yang SH, Yan YQ, Zhang X, Zhang L, Jiao B, Jiang S, Yu ZB. Identification of differential gene expression profile from peripheral blood cells of military pilots with hypertension by RNA sequencing analysis. BMC Med Genet. 2018;11(59):59. doi: 10.1186/s12920-018-0378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hotta K, Kitamoto A, Kitamoto T, Mizusawa S, Teranishi H, Matsuo T, Nakata Y, Hyogo H, Ochi H, Nakamura T, Kamohara S, Miyatake N, Kotani K, Komatsu R, Itoh N, Mineo I, Wada J, Yoneda M, Nakajima A, Funahashi T, Miyazaki S, Tokunaga K, Masuzaki H, Ueno T, Chayama K, Hamaguchi K, Yamada K, Hanafusa T, Oikawa S, Yoshimatsu H, Sakata T, Tanaka K, Matsuzawa Y, Nakao K, Sekine A. Genetic variations in the CYP17A1 and NT5C2 genes are associated with a reduction in visceral and subcutaneous fat areas in Japanese women. J Hum Genet. 2012;57:46–51. doi: 10.1038/jhg.2011.127. [DOI] [PubMed] [Google Scholar]
- 38.Wen W, Zheng W, Okada Y, Takeuchi F, Tabara Y, Hwang JY, Dorajoo R, Li H, Tsai FJ, Yang X, He J, Wu Y, He M, Zhang Y, Liang J, Guo X, Sheu WH, Delahanty R, Guo X, Kubo M, Yamamoto K, Ohkubo T, Go MJ, Liu JJ, Gan W, Chen CC, Gao Y, Li S, Lee NR, Wu C, Zhou X, Song H, Yao J, Lee IT, Long J, Tsunoda T, Akiyama K, Takashima N, Cho YS, Ong RT, Lu L, Chen CH, Tan A, Rice TK, Adair LS, Gui L, Allison M, Lee WJ, Cai Q, Isomura M, Umemura S, Kim YJ, Seielstad M, Hixson J, Xiang YB, Isono M, Kim BJ, Sim X, Lu W, Nabika T, Lee J, Lim WY, Gao YT, Takayanagi R, Kang DH, Wong TY, Hsiung CA, Wu IC, Juang JM, Shi J, Choi BY, Aung T, Hu F, Kim MK, Lim WY, Wang TD, Shin MH, Lee J, Ji BT, Lee YH, Young TL, Shin DH, Chun BY, Cho MC, Han BG, Hwu CM, Assimes TL, Absher D, Yan X, Kim E, Kuo JZ, Kwon S, Taylor KD, Chen YD, Rotter JI, Qi L, Zhu D, Wu T, Mohlke KL, Gu D, Mo Z, Wu JY, Lin X, Miki T, Tai ES, Lee JY, Kato N, Shu XO, Tanaka T. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23:5492–5504. doi: 10.1093/hmg/ddu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Magi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Zhao JH, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Leach IM, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stancakova A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Arnlov J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Bluher M, Bohringer S, Bonnycastle LL, Bottcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Chen YI, Clarke R, Daw EW, de Craen AJM, Delgado G, Dimitriou M, Doney ASF, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe HJ, Grallert H, Grammer TB, Grassler J, Gronberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga JJ, James AL, Jeff JM, Johansson A, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindstrom J, Lo KS, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PKE, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Muller G, Muller-Nurasyid M, Musk AW, Nagaraja R, Nothen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Smith AV, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundstrom J, Swertz MA, Swift AJ, Syvanen AC, Tan ST, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh HW, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, LifeLines Cohort S, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gadin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee JY, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JRB, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, van’t Hooft FM, Vinkhuyzen AAE, Westra HJ, Zheng W, Zondervan KT, Consortium AD, Group A-BW, Consortium CAD, Consortium CK, Glgc, Icbp, Investigators M, Mu TC, Consortium MI, Consortium P, ReproGen C, Consortium G, International Endogene C. Heath AC, Arveiler D, Bakker SJL, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrieres J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gejman PV, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hypponen E, Illig T, Jacobs KB, Jarvelin MR, Jockel KH, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, JJP K, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Marchand LL, Lehtimaki T, Lyssenko V, Mannisto S, Marette A, Matise TC, Mckenzie CA, Mcknight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, PAF M, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schunkert H, Schwarz PEH, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tonjes A, Tregouet DA, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Volker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PIW, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui J, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, Marz W, Melbye M, Metspalu A, Moebus S, Munroe PB, Njolstad I, Oostra BA, CNA P, Pedersen NL, Perola M, Perusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann HE, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O'Connell JR, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, Franke L, Frayling TM, MI MC, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, RJF L, Speliotes EK. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samaan Z, Lee YK, Gerstein HC, Engert JC, Bosch J, Mohan V, Diaz R, Yusuf S, Anand SS, Meyre D, Epi DGI. Obesity genes and risk of major depressive disorder in a multiethnic population: a cross-sectional study. J Clin Psychiatry. 2015;76:e1611–e1618. doi: 10.4088/JCP.14m09720. [DOI] [PubMed] [Google Scholar]
- 41.Abadi A, Alyass A, Robiou du Pont S, Bolker B, Singh P, Mohan V, Diaz R, Engert JC, Yusuf S, Gerstein HC, Anand SS, Meyre D. Penetrance of polygenic obesity susceptibility loci across the body mass index distribution. Am J Hum Genet. 2017;101:925–938. doi: 10.1016/j.ajhg.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, Tang Z, Zumbo P, Li S, Zavadil J, Levine RL, Cardozo T, Hunger SP, Raetz EA, Evans WE, Morrison DJ, Mason CE, Carroll WL. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45:290–294. doi: 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, Paietta E, Racevskis J, Rowe JM, Tallman MS, Paganin M, Basso G, Hof J, Kirschner-Schwabe R, Palomero T, Rabadan R, Ferrando A. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19:368–371. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, Song G, Easton J, Harvey RC, Wheeler DA, Ma J, Doddapaneni H, Vadodaria B, Wu G, Nagahawatte P, Carroll WL, Chen IM, Gastier-Foster JM, Relling MV, Smith MA, Devidas M, Guidry Auvil JM, Downing JR, Loh ML, Willman CL, Gerhard DS, Mullighan CG, Hunger SP, Zhang J (2015) Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 6:6604. 10.1038/ncomms7604 [DOI] [PMC free article] [PubMed]
- 45.Oh HR, Choi YJ, Yoo NJ, Lee SH. Leukemia relapse-associated mutation of NT5C2 gene is rare in de novo acute leukemias and solid tumors. Pathol Oncol Res. 2016;22:223–224. doi: 10.1007/s12253-015-9965-0. [DOI] [PubMed] [Google Scholar]
- 46.Ding LW, Sun QY, Mayakonda A, Tan KT, Chien W, Lin DC, Jiang YY, Xu L, Garg M, Lao ZT, Lill M, Yang H, Yeoh AE, Koeffler HP. Mutational profiling of acute lymphoblastic leukemia with testicular relapse. J Hematol Oncol. 2017;10(65):65. doi: 10.1186/s13045-017-0434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter-Pechanska P, Kunz JB, Hof J, Zimmermann M, Rausch T, Bandapalli OR, Orlova E, Scapinello G, Sagi JC, Stanulla M, Schrappe M, Cario G, Kirschner-Schwabe R, Eckert C, Benes V, Korbel JO, Muckenthaler MU, Kulozik AE. Identification of a genetically defined ultra-high-risk group in relapsed pediatric T-lymphoblastic leukemia. Blood Cancer J. 2017;7:e523. doi: 10.1038/bcj.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kunz JB, Rausch T, Bandapalli OR, Eilers J, Pechanska P, Schuessele S, Assenov Y, Stutz AM, Kirschner-Schwabe R, Hof J, Eckert C, von Stackelberg A, Schrappe M, Stanulla M, Koehler R, Avigad S, Elitzur S, Handgretinger R, Benes V, Weischenfeldt J, Korbel JO, Muckenthaler MU, Kulozik AE. Pediatric T-cell lymphoblastic leukemia evolves into relapse by clonal selection, acquisition of mutations and promoter hypomethylation. Haematologica. 2015;100:1442–1450. doi: 10.3324/haematol.2015.129692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehmann-Che J, Bally C, Letouze E, Berthier C, Yuan H, Jollivet F, Ades L, Cassinat B, Hirsch P, Pigneux A, Mozziconacci MJ, Kogan S, Fenaux P, de The H (2018) Dual origin of relapses in retinoic-acid resistant acute promyelocytic leukemia. Nat Commun 9:2047. 10.1038/s41467-018-04384-5 [DOI] [PMC free article] [PubMed]
- 50.Tzoneva G, Dieck CL, Oshima K, Ambesi-Impiombato A, Sanchez-Martin M, Madubata CJ, Khiabanian H, Yu J, Waanders E, Iacobucci I, Sulis ML, Kato M, Koh K, Paganin M, Basso G, Gastier-Foster JM, Loh ML, Kirschner-Schwabe R, Mullighan CG, Rabadan R, Ferrando AA. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature. 2018;553:511–514. doi: 10.1038/nature25186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dieck CL, Tzoneva G, Forouhar F, Carpenter Z, Ambesi-Impiombato A, Sanchez-Martin M, Kirschner-Schwabe R, Lew S, Seetharaman J, Tong L, Ferrando AA (2018) Structure and mechanisms of NT5C2 mutations driving thiopurine resistance in relapsed lymphoblastic leukemia. Cancer Cell 34:136–147 e136. 10.1016/j.ccell.2018.06.003 [DOI] [PMC free article] [PubMed]
- 52.Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, Sanchez-Martin M, Carpenter Z, Penson A, Perez-Garcia A, Eckert C, Nicolas C, Balbin M, Sulis ML, Kato M, Koh K, Paganin M, Basso G, Gastier-Foster JM, Devidas M, Loh ML, Kirschner-Schwabe R, Palomero T, Rabadan R, Ferrando AA. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2016;113:11306–11311. doi: 10.1073/pnas.1608420113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cividini F, Tozzi MG, Galli A, Pesi R, Camici M, Dumontet C, Jordheim LP, Allegrini S. Cytosolic 5′-nucleotidase II interacts with the leucin rich repeat of NLR family member Ipaf. PLoS One. 2015;10:e0121525. doi: 10.1371/journal.pone.0121525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez-Menendez C, Gamir-Morralla A, Jurado-Arjona J, Higuero AM, Campanero MR, Ferrer I, Hernandez F, Avila J, Diaz-Guerra M, Iglesias T. Kidins220 accumulates with tau in human Alzheimer’s disease and related models: modulation of its calpain-processing by GSK3beta/PP1 imbalance. Hum Mol Genet. 2013;22:466–482. doi: 10.1093/hmg/dds446. [DOI] [PubMed] [Google Scholar]
- 55.Alonso-Andres P, Albasanz JL, Ferrer I, Martin M (2018) Purine-related metabolites and their converting enzymes are altered in frontal, parietal and temporal cortex at early stages of Alzheimer’s disease pathology. Brain Pathol. 10.1111/bpa.12592 [DOI] [PMC free article] [PubMed]
- 56.Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, Zecha A, Mohseni M, Puttmann L, Vahid LN, Jensen C, Moheb LA, Bienek M, Larti F, Mueller I, Weissmann R, Darvish H, Wrogemann K, Hadavi V, Lipkowitz B, Esmaeeli-Nieh S, Wieczorek D, Kariminejad R, Firouzabadi SG, Cohen M, Fattahi Z, Rost I, Mojahedi F, Hertzberg C, Dehghan A, Rajab A, Banavandi MJ, Hoffer J, Falah M, Musante L, Kalscheuer V, Ullmann R, Kuss AW, Tzschach A, Kahrizi K, Ropers HH. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 57.Matyash M, Zabiegalov O, Wendt S, Matyash V, Kettenmann H. The adenosine generating enzymes CD39/CD73 control microglial processes ramification in the mouse brain. PLoS One. 2017;12:e0175012. doi: 10.1371/journal.pone.0175012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akizu N, Cantagrel V, Schroth J, Cai N, Vaux K, McCloskey D, Naviaux RK, Van Vleet J, Fenstermaker AG, Silhavy JL, Scheliga JS, Toyama K, Morisaki H, Sonmez FM, Celep F, Oraby A, Zaki MS, Al-Baradie R, Faqeih EA, Saleh MA, Spencer E, Rosti RO, Scott E, Nickerson E, Gabriel S, Morisaki T, Holmes EW, Gleeson JG. AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell. 2013;154:505–517. doi: 10.1016/j.cell.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marsh AP, Lukic V, Pope K, Bromhead C, Tankard R, Ryan MM, Yiu EM, Sim JC, Delatycki MB, Amor DJ, McGillivray G, Sherr EH, Bahlo M, Leventer RJ, Lockhart PJ. Complete callosal agenesis, pontocerebellar hypoplasia, and axonal neuropathy due to AMPD2 loss. Neurol Genet. 2015;1:e16. doi: 10.1212/NXG.0000000000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marsh AP, Yap P, Tan T, Pope K, White SM, Chong B, McGillivray G, Boys A, Stephenson SE, Leventer RJ, Stark Z, Lockhart PJ. A novel AMPD2 mutation outside the AMP deaminase domain causes pontocerebellar hypoplasia type 9. Am J Med Genet A. 2017;173:820–823. doi: 10.1002/ajmg.a.38076. [DOI] [PubMed] [Google Scholar]
- 61.Kortum F, Jamra RA, Alawi M, Berry SA, Borck G, Helbig KL, Tang S, Huhle D, Korenke GC, Hebbar M, Shukla A, Girisha KM, Steinlin M, Waldmeier-Wilhelm S, Montomoli M, Guerrini R, Lemke JR, Kutsche K. Clinical and genetic spectrum of AMPD2-related pontocerebellar hypoplasia type 9. Eur J Hum Genet. 2018;26:695–708. doi: 10.1038/s41431-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia-Esparcia P, Hernandez-Ortega K, Ansoleaga B, Carmona M, Ferrer I. Purine metabolism gene deregulation in Parkinson’s disease. Neuropathol Appl Neurobiol. 2015;41:926–940. doi: 10.1111/nan.12221. [DOI] [PubMed] [Google Scholar]
- 63.O’Donovan SM, Sullivan C, Koene R, Devine E, Hasselfeld K, Moody CL, McCullumsmith RE. Cell-subtype-specific changes in adenosine pathways in schizophrenia. Neuropsychopharmacology. 2018;43:1667–1674. doi: 10.1038/s41386-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]