Abstract
Subseafloor sedimentary environments harbor a remarkable number of microorganisms that constitute anaerobic and aerobic microbial ecosystems beneath the ocean margins and open-ocean gyres, respectively. Microbial biomass and diversity richness generally decrease with increasing sediment depth and burial time. However, there has been a long-standing debate over the contribution and distribution of Archaea in the subseafloor sedimentary biosphere. Here we show the global quantification of archaeal and bacterial 16S rRNA genes in 221 sediment core samples obtained from diverse oceanographic settings through scientific ocean drilling using microfluidic digital PCR. We estimated that archaeal cells constitute 37.3% of the total microbial cells (40.0% and 12.8% in the ocean margin and open-ocean sites, respectively), corresponding to 1.1 × 1029 cells on Earth. In addition, the relative abundance of archaeal 16S rRNA genes generally decreased with the depth of water in the overlying sedimentary habitat, suggesting that Archaea may be more sensitive to nutrient quality and quantity supplied from the overlying ocean.
Subject terms: Microbial ecology, Environmental microbiology
Marine sediment covers approximately 70% of the Earth’s surface, representing one of the largest microbial habitats on Earth. Previous studies, through scientific ocean drilling, revealed that microorganisms are globally distributed in strictly aerobic sediment columns of the oligotrophic open-ocean gyre [1–4] and in strictly anaerobic organic-rich sediments along the continental margins [5–10], even down to 2–2.5 km below the ocean floor [11–13]. Consequently, the current estimate of the global subseafloor cell number in sediments is 2.9 × 1029 cells, corresponding to 4 Pg of biomass carbon (i.e., 0.18–3.6% of the total living biomass on Earth [2]), which is two orders of magnitude lower than the previous estimates [5, 9, 14].
Despite these extensive explorations of microbial biomass in various subseafloor sedimentary habitats over the decades, abundance and distribution of Archaea remain debatable [15]. Considering differences in physiology between Bacteria and Archaea, they could respond differently to environmental settings and thus result in population shift. For example, the more rigid and less permeable cell membrane of Archaea is believed to be favorable for surviving energetically challenging conditions in the deep subseafloor biosphere [16]. Using quantitative real-time PCR with domain-specific primer (and probe) sets, it was reported that the copy numbers of archaeal 16S rRNA genes were several orders of magnitude lower than the total number or even below the quantification limit [7, 8, 17]. However, lack of advanced technology at the time of these investigations could have compromised the accuracy and thus the reliable quantification of archaeal 16S rRNA genes: (1) condensed humic substances were always co-extracted from organic-rich sediment samples, which might inhibit PCR reactions and spectrophotometric quantification [18]; (2) sequence mismatches of the domain-specific primers may not amplify some major archaeal linages [9, 19–21]; (3) DNA extractability between archaeal and bacterial cells may be significantly different [9, 22]; and (4) experimental contaminations easily occur under the standard laboratory condition for molecular microbial ecology [23]. Consequently, compilation of the existing molecular quantification data obtained using multiple methods and different quality controls resulted in highly scattered values [24]. Comparing with the conventional quantitative PCR, digital PCR (dPCR) can relieve the problems (1) listed above because quantity obtained by dPCR is independent from amplification efficiency, and thus that allows direct comparison between different samples from various sedimentary settings.
As an alternative way to assess archaeal abundance in subseafloor sediments, Lipp et al. [9] studied archaeal and bacterial intact polar lipids (IPLs) as live biomarker proxies, showing that at least 87% of IPLs were attributable to archaeal cell membranes, whereas analysis of the relative abundances of archaeal 16S rRNA genes, assessed by slot-blot hybridization and quantitative real-time PCR combined with a physical cell destruction method for DNA extraction, yielded a value of approximately 35–40%. This study pointed out that Archaea contribute to subseafloor sedimentary biomass more than previously expected. However, follow-up radiotracer incubation experiments demonstrated that the degradation rates of archaeal IPLs in sediments are one to two orders of magnitude lower than those of bacterial IPLs, and 50–96% of archaeal IPLs are considered to represent fossil signals [25].
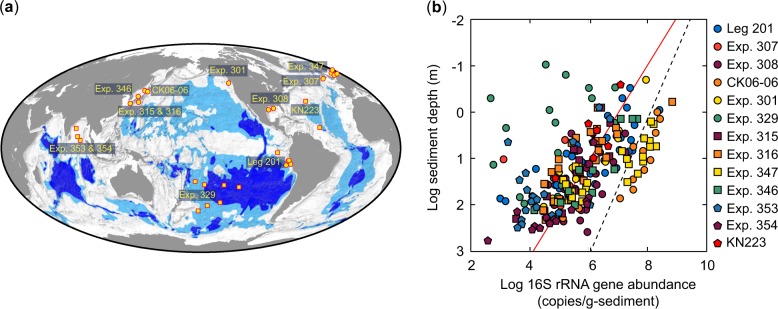
These previous reports suggest that the true nature of archaeal abundance and distribution in the subseafloor sedimentary biosphere remain unclear and should be reconsidered. In the present study, we analyzed 221 sediment cores collected from 0.2–392.2 m below seafloor at 38 drilling sites during 13 scientific expeditions since the first microbiology-dedicated Ocean Drilling Program (ODP) Leg 201 in 2002 (Fig. 1a, Supplementary Table S1). To maintain high levels of quality control and quality assessment for the molecular quantification analysis, we performed sediment sub-sampling, DNA extraction and purification, and quantification of 16S rRNA genes at the same place, at the same time, under consistent experimental conditions (see Supplementary Text).
Fig. 1.
Site locations and 16S rRNA gene abundance. a Site locations are plotted on the map showing regions where dissolved oxygen and aerobic activity may occur throughout the sediment [3]. Circles and squares indicate marginal and open-ocean sites, respectively. Sediment samples were collected at different depths from the surface to 392 m below seafloor during 13 scientific drilling cruises from 38 drilling sites. In total, we analyzed 221 sediment samples. Leg 201, Ocean Drilling Program (ODP) Leg 201 Peru Deep Biosphere; Exp. 301, Integrated Ocean Drilling Program (IODP) Expedition 301 Juan de Fuca Hydrogeology; Exp. 307, IODP Expedition 307 Modern Carbonate Mounds: Porcupine Drilling; Exp. 308, IODP Expedition 308 Gulf of Mexico Hydrogeology; Exp. 315 and 316, IODP Expedition 315 and 316 NanTroSEIZE; Exp. 353, IODP Expedition 346 Asian Monsoon; Exp. 347, IODP Expedition 347 Baltic Sea Paleoenvironment; Exp. 353, IODP Expedition 353 Indian Monsoon Rainfall; Exp. 354, IODP Expedition 354 Bengal Fan; CK06-06, the Chikyu shakedown cruise offshore Shimokita; KN223, R/V Knorr cruise 223 in North Atlantic. b Depth distribution of prokaryotic 16S rRNA abundance quantified by microfluidic digital PCR (Supplementary Table S1). The red line indicates the regression line generated using least squares analysis with the abundance of 16S rRNA gene [log (16S rRNA gene abundance) = 7.03 − 0.97 log (depth), r2 = 0.38]. The dashed black line shows the regression line [log (cell count) = 8.05 − 0.68 log (depth), r2 = 0.70] of total direct cell count [10]. That regression line is based on cells per mL of sediment instead of copies per gram for dPCR
The abundance of archaeal and bacterial 16S rRNA genes was measured by microfluidic dPCR [18], showing a logarithmically decreasing trend with increasing sediment depth (Fig. 1b). The equation for the regression line relating gene abundance to depth is:
which is relatively in good agreement with that of the previous data of acridine orange direct cell count [5, 10]:
Those regression lines indicate that the quantity of 16S rRNA gene obtained by dPCR is generally lower than cell counts due to that fact that not full recovery of DNA from sedimentary cells cannot be achieved. By comparing the published data of direct counts of cells and dPCR data in this study, we obtain the following formula between those two data:
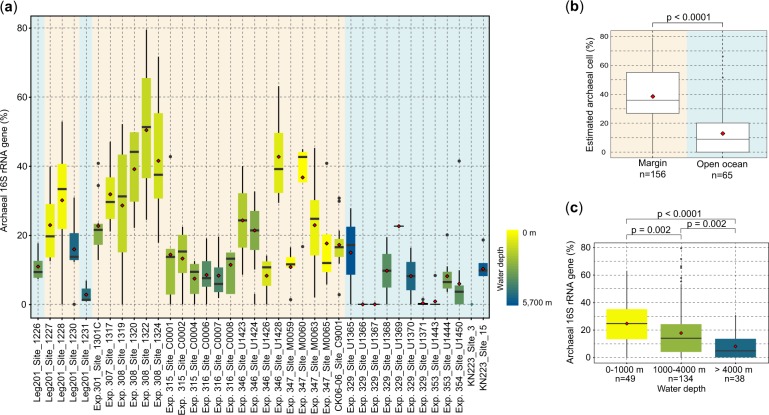
The mean relative abundance of archaeal 16S rRNA genes at each drilling site notably varied (Fig. 2a, Supplementary Fig. S1). The sediment samples used in this study were obtained from 24 marginal ocean sites and 14 open-ocean sites, representing the anaerobic and aerobic microbial ecosystems in the subseafloor sedimentary biosphere, respectively. Our microfluidic dPCR analysis shows that the relative abundance of archaeal 16S rRNA genes to the total 16S rRNA genes (i.e., archaeal and bacterial 16S rRNA genes) is 22.6% and 5.9% in average for the marginal ocean and open-ocean sites, respectively (Fig. 2b). This difference indicates that archaeal contribution to the anaerobic microbial ecosystem is more prominent than to the aerobic ecosystem. The highest archaeal abundance of up to 50.4% was observed at the Integrated Ocean Drilling Program Site 1322 in the Mars-Ursa salt-withdrawal basin on the north-eastern Gulf of Mexico continental slope (Fig. 2a, Supplementary Table S1), where nutrients and energy substrates were additionally supplied from the continent via the Mississippi River to the slope deposit [20, 26].
Fig. 2.
Boxplot of archaeal proportion in microbial 16S rRNA gene (%) at each drilling site determined by microfluidic digital PCR. a The edges of the box are the first and third quartile, red diamonds are average proportions, and gray dots are outliers (see also Supplementary Fig. S1, Supplementary Table S1). The color of the box indicates water depth; light blue, marginal ocean sites, and light brown, open-ocean sites. b The summary of estimated archaeal cell proportion in marginal ocean sites (n = 156) and open-ocean sites (n = 65). The relative abundance of archaeal cells to the total microbial cells in marginal ocean sedimentary habitats was significantly greater (Mann–Whitney U test, p < 0.05) than that in open-ocean sedimentary habitats. c The summary of archaeal 16S rRNA gene proportions in the three different water depth classes, 0–1000, 1000–4000, and >4000 m (see also Supplementary Fig. S2). The relative abundance of archaeal 16S rRNA gene in these classes are significantly different (Mann–Whitney U test, p < 0.05). A high relative abundance of archaeal 16S rRNA genes is observed for the shallow class
To evaluate the population size of archaeal cells based on our dPCR dataset, we applied the average copy number of the 16S rRNA gene on genomes of Archaea and Bacteria, 1.7 and 4.7 copies/genome, respectively (rrnDB version 5.2) [27]. As a result, we estimated that 40.0% and 12.8% of the total microbial cells are Archaea in marginal anaerobic and open-ocean aerobic communities, respectively (Fig. 2b). If we can assume that open-ocean sediments generally contain 10% of all microbial cells in the global subseafloor sediments as suggested by the biogeographic biomass distribution model [2], archaeal cells account for 37.3% of all microbial cells in the global subseafloor sedimentary biosphere. Importantly, despite the variance in archaeal population depending on the oceanographic setting, the relative abundance of Archaea in the subseafloor sedimentary biosphere is relatively similar to the estimate in the global oceans at 41.9% [28]. This suggests that Archaea comprise a biomass comparable to Bacteria throughout the surface and subsurface microbial ecosystem in the ocean.
Upon comparing dPCR data between near-seafloor sediment samples and sediment samples from deeper parts, we identified several sites where the relative abundances of archaeal 16S rRNA genes were higher in deeper horizons than in near-seafloor sediment samples (for example, sites 1226, 1301, 1320, 1322, 1324, M0060, and U1428). This trend was previously observed in IPL-based analyses [9] and other studies of shallow sediments; however, we observed decreasing trends for other sites (Supplementary Fig. S2). Several factors are conceivable for these trends in archaeal fractions: (1) archaeal cells may be more eco-physiologically resistant and/or adaptable to low-energy flux habitats than most bacterial cells [16, 29, 30]; (2) biological and geophysical migration of surface sedimentary conditions may stimulate bacterial growth than archaeal growth [31]; (3) some of Archaea may specifically utilize deeply buried recalcitrant substrates, such as mineralized detrital proteins and/or humic derivatives [32, 33], and therefore, some archaeal fractions may be retained as essential ecosystem functions (for example, methanogenesis and acetogenesis) against the selective environmental pressure of geophysical and energetic constrains during burial [4, 30, 34]; and/or (4) geophysical, sedimentological, and hydrogeological characteristics and formation stability may constrain the supply of water, nutrients, and energy substrates, and subsequently, have an impact on the fraction of archaeal community in subseafloor sedimentary microbial ecosystems [7, 8, 12, 35].
Interestingly, our data also reveal that the relative abundance of archaeal 16S rRNA genes generally decreases with water depth at the drilling site (Fig. 2c, Supplementary Fig. S2). Here we categorized the water depths of study sites into three groups: epi-mesopelagic (0−1000 m); bathy-pelagic (1000−4000 m); and abyss-pelagic depths (>4000 m). Consequently, the relative abundance of archaeal 16S rRNA gene in epi-mesopelagic water depths was found to be the highest at 24.7% among the three depth categories. The relative abundance of archaeal 16S rRNA genes was significantly low (p < 0.05, t-test) in the deeper bathy- (17.8%) and abyss-pelagic (8.2%) water depths (Fig. 2c, Supplementary Fig. S2). These results suggest that water depth is one of the key environmental factors constraining the relative abundance of Archaea in the total microbial community. One of the possible explanations for why the water depth constrains archaeal population in the subseafloor biosphere may be the quality and quantity of consumable organic matter deposited from the photosynthetic zone in the water column down to the seafloor [36]. Alternatively, the differences in organic matter may cause different consumption rate of oxygen, and therefore different oxygen concentrations. In addition, any other factor correlated with water depth could also affect archaeal population in the sedimentary habitat. To answer this question, further integrated investigations for evaluating the interaction between organic matter and archaeal metabolic characteristics in the subseafloor sedimentary biosphere are needed [15].
In conclusion, this study provides the best estimates to date of the global distribution and abundance of Archaea in the marginal ocean (anaerobic) and open-ocean (aerobic) sedimentary habitats. Our findings in this global dPCR survey confirm that archaeal biomass significantly contributes to the subseafloor sedimentary biosphere (37.3% of the total microbial cells). Nevertheless, metabolic, physiological, and evolutionary functions of subseafloor sedimentary microbes remain largely unknown. These are the foci of our ongoing research using the global subseafloor sediment core samples.
Electronic supplementary material
Acknowledgements
We thank all the crew, drilling team members, and shipboard scientists for core sampling during ODP Leg 201; IODP Expeditions 301, 307, 308, 315, 316, 329, 346, 347, 353, and 354; the Chikyu shakedown cruise CK06-06; and the R/V Knorr cruise KN223. This research used samples and/or data provided by the Ocean Drilling Program (ODP), Integrated Ocean Drilling Program, and International Ocean Discovery Program (IODP). The authors specially thank R. R. Adhikari, A. H. Kaksonen, Y. Morono, N. Xiao, T. Nunoura, and G.-I. Uraomoto for their assistance in sediment core sampling as shipboard scientists during IODP Expeditions, and L. P. Wömer, K.-U. Hinrichs, and S. D’Hondt for their useful inputs and discussion. We are also grateful for the technical assistance of S. Hashimoto, T. Terada, and M. Tsutsumi at the Kochi Institute for Core Sample Research, JAMSTEC. This work was partially supported by the Japan Society for the Promotion of Science Grant-in-Aid for Science Research (no. 26251041 to FI and 17H03956 to TH) and the Deep Carbon Observatory (DCO) Deep Life Community pilot project to TH. This is a contribution to the DCO.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0253-3) contains supplementary material, which is available to authorized users.
References
- 1.D’Hondt S, Spivak AJ, Pockalny R, Ferdelman TG, Fischer JP, Kallmeyer J, et al. Subseafloor sedimentary life in the South Pacific Gyre. Proc Natl Acad Sci USA. 2009;106:11651–56. doi: 10.1073/pnas.0811793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA. 2012;109:16213–6. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Hondt S, Inagaki F, Zarikian CA, Abrams LJ, Dubois N, Engelhardt T, et al. Presence of oxygen and aerobic communities from sea floor to basement in deep-sea sediments. Nat Geosci. 2015;8:299–304. doi: 10.1038/ngeo2387. [DOI] [Google Scholar]
- 4.Walsh EA, Kirkpatrick JB, Rutherford SD, Smith DC, Sogin M, D’Hondt S. Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 2015;10:979–89. doi: 10.1038/ismej.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkes RJ, Cragg BA, Bale SJ, Getliff JM, Goodman K, Rochelle PA, et al. Deep bacterial biosphere in Pacific Ocean sediments. Nature. 1994;371:410–3. doi: 10.1038/371410a0. [DOI] [Google Scholar]
- 6.D’Hondt S, Jørgensen BB, Miller DJ, Batzke A, Blake R, Cragg BA, et al. Distributions of microbial activities in deep subseafloor sediments. Science. 2004;306:2216–21. doi: 10.1126/science.1101155. [DOI] [PubMed] [Google Scholar]
- 7.Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K, et al. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl Environ Microbiol. 2003;69:7224–35. doi: 10.1128/AEM.69.12.7224-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci USA. 2006;103:2815–20. doi: 10.1073/pnas.0511033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipp JS, Morono Y, Inagaki F, Hinrichs KU. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature. 2008;454:991–4. doi: 10.1038/nature07174. [DOI] [PubMed] [Google Scholar]
- 10.Parkes RJ, Cragg B, Roussel E, Webster G, Weightman A, Sass H. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar Geol. 2014;352:409–25. doi: 10.1016/j.margeo.2014.02.009. [DOI] [Google Scholar]
- 11.Ciobanu MC, Burgaud G, Dufresne A, Breuker A, Rédou V, Maamar SB, et al. Microorganisms persist at record depths in the subseafloor of the Canterbury Basin. ISME J. 2014;8:1370–80. doi: 10.1038/ismej.2013.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inagaki F, Hinrichs KU, Kubo Y, Bowles MW, Heuer VB, Hong WL, et al. Exploring deep microbial life in coal-bearing sediment down to ~2.5 km below the ocean floor. Science. 2015;349:420–4. doi: 10.1126/science.aaa6882. [DOI] [PubMed] [Google Scholar]
- 13.Trembath-Reichert E, Morono Y, Ijiri A, Hoshino T, Dawson KS, Inagaki F, et al. Methyl-compound use and slow growth characterize microbial life in 2-km-deep subseafloor coal and shale beds. Proc Natl Acad Sci USA. 2017;114:E9206–15. doi: 10.1073/pnas.1707525114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinrichs KU, Inagaki F. Downsizing the deep biosphere. Science. 2012;338:204–5. doi: 10.1126/science.1229296. [DOI] [PubMed] [Google Scholar]
- 16.Valentine DL. Opinion: adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 2007;5:316–23. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- 17.Schippers A, Neretin LN. Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR. Environ Microbiol. 2006;8:1251–60. doi: 10.1111/j.1462-2920.2006.01019.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino T, Inagaki F. Molecular quantification of environmental DNA using microfluidics and digital PCR. Syst Appl Microbiol. 2012;35:390–5. doi: 10.1016/j.syapm.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J. 2008;2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 20.Nunoura T, Soffientino B, Blazejak A, Kakuta J, Oida H, Schippers A, et al. Subseafloor microbial communities associated with rapid turbidite deposition in the Gulf of Mexico continental slope (IODP Expedition 308) FEMS Microbiol Ecol. 2009;69:410–24. doi: 10.1111/j.1574-6941.2009.00718.x. [DOI] [PubMed] [Google Scholar]
- 21.Hoshino T, Inagaki F. A comparative study of microbial diversity and community structure in marine sediments using poly (A) tailing and reverse transcription PCR. Front Microbiol. 2013;4:160. doi: 10.3389/fmicb.2013.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morono Y, Terada T, Hoshino T, Inagaki F. Hot-alkaline DNA extraction method for deep-subseafloor archaeal communities. Appl Environ Microbiol. 2014;80:1985–94. doi: 10.1128/AEM.04150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morono Y, Inagaki F. Analysis of low-biomass microbial communities in the deep biosphere. Adv Appl Microbiol. 2016;95:149–78. doi: 10.1016/bs.aambs.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd KG, May MK, Kevorkian RT, Steen AD. Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl Environ Microbiol. 2013;79:7790–9. doi: 10.1128/AEM.02090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie S, Lipp JS, Wegener G, Ferdelman TG, Hinrichs KU. Turnover of microbial lipids in the deep biosphere and growth of benthic archaeal populations. Proc Natl Acad Sci USA. 2013;110:6010–4. doi: 10.1073/pnas.1218569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flemings PB, Behrman JH, John CM, the Expedition 308 Scientists. Proceedings of the integrated ocean drilling program, Vol. 308. College Station, TX: Integrated Ocean Drilling Program Management International Inc.; 2006. 10.2204/iodp.proc.308.2006.
- 27.Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2014;43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–10. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 29.Schlegel K, Leone V, Faraldo-Gómez JD, Müller V. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc Natl Acad Sci USA. 2012;109:947–52. doi: 10.1073/pnas.1115796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoehler TM, Jørgensen BB. Microbial life under extreme energy limitation. Nat Rev Microbiol. 2013;11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Andersen TJ, Morono Y, Inagaki F, Jørgensen BB, Lever MA. Bioturbation as a key driver behind the dominance of Bacteria over Archaea in near-surface sediment. Sci Rep. 2017;7:2400. doi: 10.1038/s41598-017-02295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, et al. Predominant archaea in marine sediments degrade detrital proteins. Nature. 2013;496:215–8. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- 33.He Y, Li M, Perumal V, Feng X, Fang J, Xie J, et al. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol. 2016;1:16035. doi: 10.1038/nmicrobiol.2016.35. [DOI] [PubMed] [Google Scholar]
- 34.Starnawski P, Batailon T, Ettema TJG, Jochum LM, Schreiber L, Chen X, et al. Microbial community assembly and evolution in subseafloor sediment. Proc Natl Acad Sci USA. 2017;114:2940–5. doi: 10.1073/pnas.1614190114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ijiri A, Inagaki F, Kubo Y, Adhikari RR, Hattori S, Hoshino T, et al. Deep-biosphere methane production stimulated by geofluids in the Nankai accretionary complex. Sci Adv. 2018;4:eaao4631. doi: 10.1126/sciadv.aao4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lutz MJ, Caldeira K, Dunbar RB, Behrenfeld MJ. Seasonal rhythms of net primary production and particulate organic carbon flux to depth describe the efficiency of biological pump in the global ocean. J Geophys Res Oceans. 2007;112:C10. doi: 10.1029/2006JC003706. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.